Significance
We created a dataset of dinoflagellate transcriptomes to resolve internal phylogenetic relationships of the group. We show that the dinoflagellate theca originated once, through a process that likely involved changes in the metabolism of cellulose, and suggest that a late origin of dinosterol in the group is at odds with dinoflagellates being the source of this important biomarker before the Mesozoic. We also show that nonphotosynthetic dinoflagellates have retained nonphotosynthetic plastids with vital metabolic functions, and propose that one of these may be the evolutionary source of dinoflagellate bioluminescence. Finally, we reconstruct major molecular and morphological transitions in dinoflagellates and highlight the role of horizontal gene transfer in the origin of their unique nuclear architecture.
Keywords: dinoflagellates, phylogeny, theca, plastids, dinosterol
Abstract
Dinoflagellates are key species in marine environments, but they remain poorly understood in part because of their large, complex genomes, unique molecular biology, and unresolved in-group relationships. We created a taxonomically representative dataset of dinoflagellate transcriptomes and used this to infer a strongly supported phylogeny to map major morphological and molecular transitions in dinoflagellate evolution. Our results show an early-branching position of Noctiluca, monophyly of thecate (plate-bearing) dinoflagellates, and paraphyly of athecate ones. This represents unambiguous phylogenetic evidence for a single origin of the group’s cellulosic theca, which we show coincided with a radiation of cellulases implicated in cell division. By integrating dinoflagellate molecular, fossil, and biogeochemical evidence, we propose a revised model for the evolution of thecal tabulations and suggest that the late acquisition of dinosterol in the group is inconsistent with dinoflagellates being the source of this biomarker in pre-Mesozoic strata. Three distantly related, fundamentally nonphotosynthetic dinoflagellates, Noctiluca, Oxyrrhis, and Dinophysis, contain cryptic plastidial metabolisms and lack alternative cytosolic pathways, suggesting that all free-living dinoflagellates are metabolically dependent on plastids. This finding led us to propose general mechanisms of dependency on plastid organelles in eukaryotes that have lost photosynthesis; it also suggests that the evolutionary origin of bioluminescence in nonphotosynthetic dinoflagellates may be linked to plastidic tetrapyrrole biosynthesis. Finally, we use our phylogenetic framework to show that dinoflagellate nuclei have recruited DNA-binding proteins in three distinct evolutionary waves, which included two independent acquisitions of bacterial histone-like proteins.
Dinoflagellates comprise approximately 2,400 named extant species, of which approximately half are photosynthetic (1). However, this represents a fraction of their estimated diversity: in surface marine waters, dinoflagellates are some of the most abundant and diverse eukaryotes known (2). Dinoflagellates’ ecological significance befits their abundance: photosynthetic species are dominant marine primary producers, and phagotrophic species play an important role in the microbial loop through predation and nutrient recycling. Approximately 75–80% of the toxic eukaryotic phytoplankton species are dinoflagellates, and they cause shellfish poisoning and harmful algal blooms of global importance. Symbiotic genera like Symbiodinium participate in interactions with metazoans and are essential for the formation of reef ecosystems, and parasitic forms play a central role in the collapse of harmful algal blooms, including those caused by dinoflagellates themselves (3). Dinoflagellates synthesize important secondary metabolites including sterols, polyketides, toxins, and dimethylsulfide, and several of them have evolved bioluminescence. They have a nonnucleosomal system of nuclear DNA packaging, widespread trans-splicing in mRNAs, and highly unusual plastid and mitochondrial genomes with complex transcript modifications (4–8). Their photosynthesis relies on unique light-harvesting complexes, and its frequent loss in the group makes dinoflagellates a model for understanding the basis of evolutionary reliance on nonphotosynthetic plastid organelles.
Detailed understanding of dinoflagellate biology has been limited by a paucity of sequence data, especially unusual features such as the organization of their very large and complex nuclear genomes (9, 10). Poorly resolved dinoflagellate trees have further complicated predictions of how specific metabolic pathways evolved and how they are distributed in uncultured members of the group. To date, molecular phylogenies have established the deep-branching positions of Oxyrrhis marina (here included in the dinoflagellates) and the parasitic Syndiniales [possibly several lineages (11)], but the internal relationships in the so-called core dinoflagellates, that is, all other orders and most species in the group, have remained unresolved except at low taxonomic levels (12–14). Traditionally, dinoflagellate taxonomy has been based on their tabulation, the arrangement of vesicles in the cell cortex that may or may not contain cellulosic thecal plates (collectively the theca). Whether the dinoflagellate theca originated once or multiple times has been controversial. Dinoflagellates have left a fossil record that is one of the richest among protists, and many preserve a detailed record of tabulation through reflection of thecal plates that provide insights into the history of some modern taxa, as well as extinct groups. They have also left an extensive biogeochemical record (i.e., sterols), but reconciling this evidence with poorly resolved gene phylogenies has been difficult (15, 16).
We circumvented the difficulties inherent to the sequencing of large dinoflagellate genomes by compiling a phylogenetically representative transcriptomic dataset to illuminate dinoflagellate biology and evolution. We infer a strongly resolved phylogeny for dinoflagellates and provide phylogenetic evidence for a single origin of the theca, which coincides with major predicted changes in cellulose metabolism. We propose a model for the evolution of tabulation, and show that pre-Mesozoic biomarkers that have often been associated with the group are unlikely to come from dinoflagellate sources. Three distantly related, nonphotosynthetic dinoflagellates were found to be dependent on plastid metabolism, and we propose that this dependency is likely to apply to all free-living (i.e., nonparasitic) dinoflagellates and that plastidial metabolites are likely to represent the evolutionary origin of dinoflagellate bioluminescence. Finally, we reconstruct character evolution in dinoflagellates and show that their modern-day biology was shaped by stepwise molecular, metabolic, and morphological innovations, including nuclear DNA-binding proteins of a bacterial origin.
Results and Discussion
Dinoflagellate Phylogeny.
Representative, strongly resolved phylogeny for dinoflagellates.
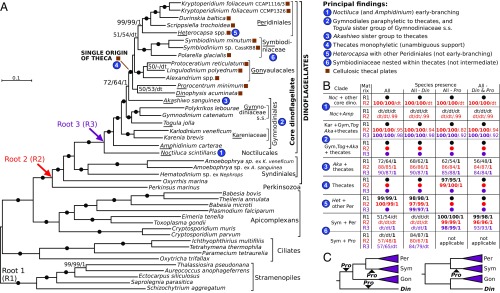
An inability to resolve dinoflagellate relationships has hindered evolution-driven predictions of their biology and a full integration of the group’s rich fossil record with molecular-based schemes of evolution. Our aim was to overcome these limitations by erecting a framework for character mapping rooted in a representative phylogeny of all major dinoflagellate lineages. We generated transcriptomes from key species lacking deep-coverage sequence data—Noctiluca scintillans, Togula jolla, Protoceratium reticulatum, Polarella glacialis, Hematodinium spp., Amphidinium carterae, and two isolates of Amoebophrya sp. parasites together with their hosts, Karlodinium veneficum and Akashiwo sanguinea—and complemented these with data from recent sequencing projects (9, 17–19) (SI Appendix, Table S1). Sequences were added into alignments of conserved proteins previously used in eukaryotic phylogenies (20), and their orthology was verified in individual protein trees (Materials and Methods); 101 orthologous alignments with the fewest missing data were selected and concatenated into three phylogenetic matrices that differ by the root (Fig. 1A and SI Appendix, Table S1). The matrices include six dinoflagellate lineages previously absent in multiprotein phylogenies: Noctilucales, Gymnodiniaceae s.s., Togula, Akashiwo, Prorocentrales, and Dinophysiales, representing a broadly sampled large dinoflagellate datasets. Maximum-likelihood and Bayesian inferences on all three matrices gave consistent and well-supported topologies (Fig. 1 A and B). Relationships between the outgroups and the early-branching Oxyrrhis, Hematodinium, and Amoebophyra spp. are fully resolved and congruent with earlier studies (11). Core dinoflagellates are monophyletic, and several longstanding issues about their relationships can be resolved (Fig. 1).
Fig. 1.
Multiprotein phylogeny of dinoflagellates. (A) Best maximum-likelihood tree (IQ-Tree) of dinoflagellates and relatives based on 101-protein dataset (root 1 matrix, 43 species, 29,400 sites). Branches show ultrafast bootstraps (IQ-Tree)/nonparametric bootstraps (RAxML)/posterior probabilities (PhyloBayes) (dash indicates <50/50/0.5 support; filled circles indicate 100/100/1 support; dt indicates a different topology). Roots of alternative matrices (Perkinsus, root 2, 30,780 sites; and Noctiluca, root 3, 30,988 sites) are shown by arrows. (B) Overview of branch supports for principal findings (taxon and matrix abbreviations as underlined in A) in phylogenies of 12 matrices that differ by their root (R1–R3) and species presence (All, All - Din, All - Pro, All - Din & Pro; SI Appendix, Table S1). (C) Two placements of Dinophysis (Din) relative to Gon, Per, and Sym thecates and a variable position of Prorocentrum (Pro) as identified in phylogenies of the 12 matrices (SI Appendix, Table S3, provides tree topology tests).
Early position of Noctilucales and athecate paraphyly.
Athecate dinoflagellates have long confounded dinoflagellate molecular phylogenies as a result of their intermixing with thecate taxa, for example within the so-called Gymnodiniales–Peridiniales–Prorocentrales (GPP) complex (21), or as a result of the unstable position of certain outliers like the Noctilucales, which have at times been placed as basal or nested deeply inside the group (12, 16, 22). Our analyses resolve these issues and help reconcile dinoflagellate morphological and molecular data in several important ways. First, we find that athecate dinoflagellates represent a paraphyletic assemblage with respect to the thecates (Fig. 1A), suggesting that earlier mixed groupings like the GPP complex are artifacts caused by limited phylogenetic resolution. Second, N. scintillans and A. carterae are the earliest-branching core dinoflagellates, with Noctiluca positioned at the base in most analyses, except for Bayesian inferences on Root 2 matrix, in which it is also basal but together with Amphidinium (Fig. 1B). Statistical evaluation of alternative tree topologies by approximately unbiased test and expected-likelihood weights test rejects topologies other than Noctiluca representing the earliest branch of core dinoflagellates (P = 0.01; see SI Appendix, Table S2 and SI Materials and Methods). This position is reinforced by the absence of a cox3 split in Noctiluca (as detailed later) and resolves the long-problematic position of the Noctilucales (12–14, 16, 22), making them central to understanding the biology of the core dinoflagellate ancestor. Third, the previously mysterious Togula (23) is related to the Gymnodiniaceae sensu stricto (a clade represented here by Gymnodinium s.s. and Polykrikos). Finally, Akashiwo is placed as the sister taxon to thecate dinoflagellates in all analyses, although an alternative topology as a sister to Gymnodiaceae s.s. and Togula cannot be rejected (SI Appendix, Table S2). Statistical support for the monophyly of Akashiwo and the thecates increases when the divergent outgroup sequences are excluded in both phylogenies and tree topology tests (Fig. 1B and SI Appendix, Table S2). This suggests that the relationship is likely genuine, making Akashiwo the closest investigated athecate relative of thecate dinoflagellates. (Fig. 1B). Overall, the order Gymnodiniales represents multiple paraphyletic lineages at the base of the core dinoflagellates, despite their close morphological similarity [Akashiwo, the Kareniaceae, and even one member of the Noctilucales were, until recently, classified in the genus Gymnodinium (13, 24)], suggesting that their conserved morphological characteristics were ancestral to all core dinoflagellates.
Monophyly of thecate dinoflagellates and nested position of Symbiodiniaceae.
In molecular phylogenies, thecate dinoflagellates have been mixed with athecate species and are only exceptionally recovered as monophyletic in specific datasets and with low support (12, 14). Our large-scale phylogenies, which include all five major thecate groups, recover thecate dinoflagellates as monophyletic, always with maximal or near-maximal support (Fig. 1 A and B). This provides unambiguous phylogenetic support for the single origin of the dinoflagellate theca. Peridiniales, Gonyaulacales, and Symbiodiniaceae (represented by Symbiodinium and Polarella) are monophyletic in all our analyses. The long-problematic Heterocapsa, previously placed at the base of dinoflagellates (25), or away from the Peridiniales (14), is strongly resolved as the sister group to other Peridiniales (Fig. 1 A and B), a position consistent with its modified peridinialean tabulation (26). The placement of Prorocentrum and Dinophysis, both representatives of poorly sampled and morphologically divergent orders, remains unresolved within the thecates: Dinophysis is placed at the base of the Gonyaulacales or of all thecates with low support, and the position of Prorocentrum is even more unstable (Fig. 1C). Analyses excluding Dinophysis, Prorocentrum, or both confirm the common origin and monophyly and of the other thecate lineages, that is, the Gonyaulacales, Symbiodiniaceae, and Peridiniales inclusive of Heterocapsa (Fig. 1B). The branching order of these core thecate lineages is also conserved: the Gonyaulacales always branch comparatively early, and the Symbiodiniaceae are always late-branching within the thecates and consistently recovered close to the Peridiniales. This topology is weakly supported, but support increases when the problematic Prorocentrum is excluded (Fig. 1C). An exhaustive testing of alternative tree topologies (SI Appendix, Table S3 and SI Materials and Methods) rejects all topologies in which the Symbiodiniaceae appear as the sister group of other thecates at the significance level of P = 0.05 (and also at P = 0.01 except for a single dataset in which both Dinophysis and Prorocentrum are absent). Symbiodiniaceae (Symbiodinium, Polarella, and related forms) are frequently classified together with the early fossil genus Suessia as the “Suessiales” (26) or even within the “Suessiaceae” (27, 28), but, if this is correct, the Symbiodiniaceae should appear as the sister group of all other living thecates, a topology never recovered in phylogenies. Morphological evidence does not support the combination of the two groups either: although tabulations in symbiodiniaceans and suessiaceans have more series of thecal plates than most thecate dinoflagellates, determining the homologies of individual plates is not possible (26, 29). Thus, we use the family Symbiodiniaceae (26) for the clade uniting Symbiodinium, Polarella, and their modern relatives (27, 28) to separate them from the exclusively fossil Suessiaceae (Suessia and related forms). It remains possible (but not likely) that the Suessiaceae developed their theca independently, but all other fossil and modern thecate lineages seem to have originated from a common ancestor. Four independent lines of evidence support this: monophyly of the modern thecates in multiprotein phylogenies (Fig. 1), rapid emergence of fossils reflecting the possession of the theca during the early Mesozoic (30), similarities in tabulation patterns between different thecate lineages (15, 26), and the presence of theca-associated cellulases of a common evolutionary origin in modern thecates (Fig. 2).
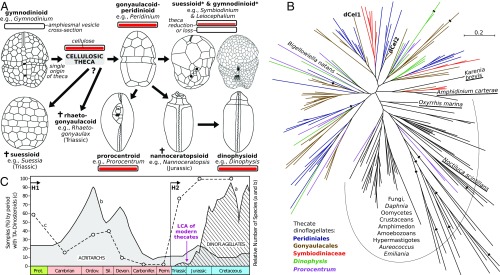
Fig. 2.
Thecal evolution and dinoflagellate paleohistory. (A) Phylogeny-driven model of changes between major modern and fossil (crosses) tabulational types. Gymnodinoid tabulation with numerous small, empty amphiesmal vesicles is ancestral and gave rise to the gonyaulacoid–peridinioid tabulation with a few large, cellulose-rich thecal plates. Suessioid and gymnodinioid tabulations in modern Symbiodiniaceae and Borghiellaceae (asterisk) are derived independently of the standard gymnodiniod and Triassic suessioid tabulations (Suessia), and are characterized by decrease or loss of cellulose content. Prorocentroid and dinophysioid tabulations are derived from the gonyaulacoid–peridiniod tabulation (the latter probably via a nannoceratopsioid intermediate). Triassic suessioid and rhaetogonyaulacoid tabulations may represent evolutionary intermediates or independent experiments in thecal plate reduction. (B) Maximum-likelihood phylogeny (IQ-Tree) of 184 eukaryotic GH7 proteins reveals cellulases in athecate dinoflagellates (underlined) and their radiation in the thecate (color-coded). Black rectangles indicate 50% reduction in branch length. Known GH7 cellulases in P. lunula (dCel1) and Lingulodinium polyedrum (dCel2) are shown. Further details are provided in SI Appendix, Fig. S1 and Table S4. (C) Alternative hypotheses (H1 and H2) on the first emergence of triaromatic dinosteranes attributable to dinoflagellates or their direct ancestors (H2 is preferred by our data). Relative species numbers of dinoflagellates (a) and acritarchs (b) and percentage of dinosterane-positive samples (c; see ref. 35 for sample data) from the Proterozoic (green), Paleozoic (red), and Mesozoic (blue) are shown together with the predicted emergence of the last common ancestor (LCA) of modern thecates. Reprinted with permission from refs. 26, 28 (www.sciencedirect.com/science/journal/14344610), 35 (permission conveyed through Copyright Clearance Center, Inc.), and 74.
Thecal Evolution and Dinoflagellate Paleohistory.
Phylogeny-driven model for theca origin, evolution, and loss.
Most thecate dinoflagellates (both living and fossil) belong to the Gonyaulacales and Peridiniales, two orders with tabulations involving five to six latitudinal series of thecal plates. The details of these tabulations are consistently distinct and longstanding in the fossil record, a pattern consistent with the fact that, in molecular phylogenies, the two orders are not closely related within the thecates (Fig. 1). These patterns suggest that dinoflagellates with gonyaulacoid–peridinoid tabulations originated comparatively early: the extinct rhaetogonyaulacoids (Fig. 2A) in the Middle to Late Triassic (31) and true, modern-looking gonyaulacoids and peridinoids in the later Early Jurassic. Even if the phylogenetic position of the Dinophysiales and Prorocentrales in molecular trees remains unresolved, their tabulation patterns are morphologically divergent and unlikely to represent ancestral or transitional states: the fossil Nannoceratopsis suggests, for example, that the dinophysioid tabulation type is evolutionarily derived (Fig. 2A). As explained earlier, we suggest that the suessioid and gymnodinioid tabulations of the Symbiodiniaceae and their sister group, the Borghiellaceae (27), are also derived secondarily from gonyaulacoid–peridinioid ancestors and originated by a secondary increase in plate number (Fig. 2A); they do not represent early intermediates in theca evolution, as considered by some earlier models (15, 32). In contrast, the Late Triassic suessioid fossils such as Suessia could represent an intermediate stage between gymnodinioid and gonyaulacoid–peridinioid tabulation types or an independent example of decrease in primary plate number from gymnodinioid ancestors (Fig. 2A). All in all, paleontological and molecular phylogenetic data suggest that all living thecate dinoflagellates originated from ancestors with a gonyaulacoid–peridinoid tabulation and argue for the derived position of the Symbiodiniaceae. The model is limited by the incompleteness of the fossil record and will be further developed by understanding the tabulations and phylogenies of little known or morphologically divergent incertae sedis thecates like Heterodinium, Thecadinium, or Cladopyxis (26). No simple scenario [plate decrease, increase, and fragmentation models (32)] can account for the evolution of thecal tabulation from a phylogeny-driven perspective (Fig. 1): secondary increase in plate number is observed not only in symbiodiniaceans but also in Pyrophacus (Gonyaulacales), a genus with a multiplated tabulation derived from ancestors with a gonyaulacoid tabulation, whereas other thecates have gone through a process of plate decrease, e.g., Dinophysiales and Prorocentrales (in the hyposome) and the Late Triassic to Middle Jurassic fossil Valvaeodinium. Our model also strongly suggests that the theca can be lost: some species in the Symbiodiniaceae and Borghiellaceae lack visible cellulose in amphiesmal vesicles altogether (28, 33), and their phylogenetic positions suggest that their thecae were lost more than once (Fig. 2A). Finally, a broad, negative relationship between the number and relative surface area of amphiesmal vesicles and the amount of cellulose contained in them emerges. The Gymnodiniales have numerous, small amphiesmal vesicles that lack cellulose, whereas the Gonyaulacales, Peridiniales, Prorocentrales, and Dinophysiales have few, large amphiesmal vesicles containing thick thecal plates, the ancestral state for all living thecate dinoflagellates (Fig. 2A). Symbiodiniaceans that have moderate plate numbers in 7–10 latitudinal series have only thin cellulosic plates, but those members of the Symbiodiniaceae and Borghiellaceae that reverted to a gymnodinoid tabulation often lack cellulose altogether (Fig. 2A) (e.g refs. 28, 33, but see also ref. 27). Additional data for example from the Borghiellaceae and Pyrophacus will make it possible to test these trends, but, as things stand now, it seems that the acquisition of thick cellulosic plates within amphiesmal vesicles is constrained with their surface area and number. Subsequent reductions and losses of cellulose in the Symbiodiniaceae and Borghelliaceae relaxed this constraint, leading to a partial or complete reversal to numerous small-sized amphiesmal vesicles.
Origin of theca coincides with onset of cellulase radiation.
The origin of the dinoflagellate theca is intimately linked to the biosynthesis of cellulose, its building material, but investigations into the details of cellulose production in dinoflagellates have been limited to rare ultrastructural and labeling studies (34). Recently, production of a highly expressed cellulase [dCel1 from Glycosyl hydrolase family 7 (GH7)] was shown to be coupled to the cell cycle progression in Crypthecodinium cohnii and was immunolocalized to the cell wall in several dinoflagellates, suggesting an important role in cellulose processing during division (31). We identified multiple diversified paralogs of GH7 genes in all thecates and one to three closely related paralogs in four athecate dinoflagellates in our dataset (SI Appendix, Table S4). A eukaryote-wide phylogeny of 184 slow-evolving GH7 protein sequences (Fig. 2B and SI Appendix, Fig. S1 and SI Materials and Methods) suggests that the thecate paralogs are derived by multiple rounds of duplication followed by selective lineage sorting. The branching pattern is poorly resolved, but indicates a common origin for most thecate GH7 proteins together with sequences from the athecate Karenia brevis and A. carterae and algae Bigelowiella natans and Thalassiosira oceanica (the latter two are nested within dinoflagellates and were presumably spread horizontally). Some duplications in the thecate GH7 occurred at the level of genera or orders, but at least eight and possibly twice as many paralogs apparently originated earlier (SI Appendix, Fig. S1)—presumably in the common ancestor of all thecates. These observations suggest that the radiation of GH7 genes in thecate dinoflagellates is linked to the evolutionary origin and subsequent evolution of the theca. The GH7 protein identified in K. brevis (SI Appendix, Table S4) likely corresponds to the dCel1 homolog previously immunolocalized in the cell cortex (31). Interestingly, A. sanguinea, the likely sister group of thecate dinoflagellates, is immunopositive for that same protein (31), although the corresponding GH7 sequence remains unknown (our mixed transcriptome of Akashiwo cells infected by Amoebophrya sp. lacks it). The function of GH7 enzymes in athecate species has not been studied, but they are likely involved in the metabolism of cellulose or related polysaccharides, which may have been an important precondition for the acquisition of the cellulosic thecal plates. Unlike cellulose breakdown, cellulose biosynthesis in dinoflagellates is not understood at the molecular level (34). We identified three types of algal cellulose synthase (CESA-like) homologs in thecate and athecate dinoflagellates, candidates for elucidating their cellulose biosynthesis (SI Appendix, Table S4).
Dinosterol is absent in deep-branching dinoflagellates.
The diversity and abundance of dinoflagellates in Mesozoic and younger sediments correlates with levels of triaromatic dinosterols, derivatives of the fossilizing biomarker 4-methyl sterol, dinosterol (4α, 23, 24R-trimethyl-5α-cholest-22E-en-3β-ol) (15, 35). Dinosteranes also occur in Late Proterozoic and early Paleozoic sediments that are often enriched with acritarchs (microfossils of uncertain origin, some of which have been speculatively attributed to dinoflagellates or their direct ancestors), and this has led to the proposal that dinoflagellates are ancient and acquired dinosterol biosynthesis early in their evolution (35–37). We compared this hypothesis (Fig. 2C, H1) to a Mesozoic origin of the dinoflagellate dinosterol (Fig. 2C, H2) by mapping sterol distribution onto our updated phylogeny of dinoflagellates (Fig. 1). Dinosterol and other 4-methyl sterols are absent from all dinoflagellate relatives with known sterol profiles, including ciliates, perkinsids, apicomplexans, Chromera, and Vitrella, but also Oxyrrhis (38) and Amoebophrya, which likely only acquires 4-methyl sterols from its host (39, 40). In core dinoflagellates, 4-methyl sterols are ubiquitous, but dinosterol itself is absent in three of their earliest branches: Noctiluca, Amphidinium, and the Kareniaceae (e.g., refs. 41–43). Gyrodinium dominans, likely another early core dinoflagellate (14), also lacks dinosterol (38). This suggests that dinosterol appeared first in the last common ancestor of Gymnodiniaceae s.s., Akashiwo, and thecate dinoflagellates (although broader testing for its presence in early-branching dinoflagellates is needed). We suggest that pre-Mesozoic dinosteranes are unlikely to originate from dinoflagellates for four reasons. First, dinosteranes from the Late Proterozoic and early Paleozoic greatly predate unambiguous dinoflagellate fossils, and dinosterol presence in modern species is restricted to close relatives of the thecates (Fig. 1), which originated in the early Mesozoic. Second, Paleozoic acritarch microfossils bear no demonstrable morphological similarity to dinoflagellates (26). Third, dinosteranes prevalence in Paleozoic and Proterozoic samples is highly variable compared with Mesozoic samples (35). They seem to be entirely absent from the Carboniferous and Permian (35), a discontinuity that contrasts with their almost universal preservation in Mesozoic and younger sediments and species. Finally, small amounts of dinosterol are known from a modern species of diatom (44), and traces of dinosteranes are also present in Archean bitumens, where dinoflagellates could not have possibly existed (45). All this suggests that different organisms in different geological eras evolved dinosterol biosynthesis independently of dinoflagellates and that dinosterol production by certain acritarchs ended with their mid-Paleozoic extinction. We also note that the phylogenetic distance between the origin of dinosterol-producing athecates and the origin of modern thecate dinoflagellates (see Fig. 1) is consistent with the time lapse between the Early Triassic dinosterane increase and the appearance of modern thecate orders in the Early Jurassic sediments (Fig. 2C). We therefore suggest that abundant dinosteranes in some Scythian (Early Triassic) sediments predating the earliest thecate fossils (Middle Triassic) (35) are derived from athecate dinoflagellates alone, which gained the ability to produce dinosterols near the Permian/Triassic boundary and became abundant shortly after it (Fig. 2C, H2).
Plastid Metabolism and Dependency.
Plastid metabolism in nonphotosynthetic dinoflagellates.
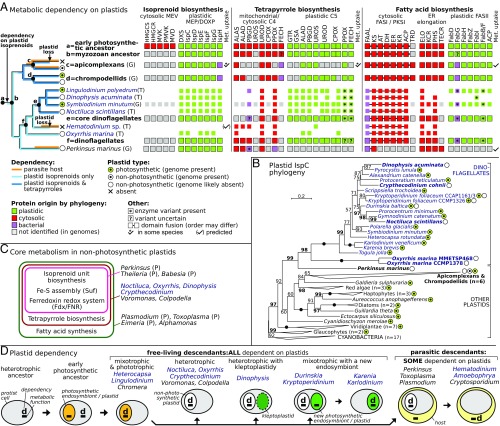
Approximately half of the described dinoflagellate species are nonphotosynthetic and are traditionally considered to lack plastids. The other half contains a photosynthetic peridinin-pigmented plastid that, in some lineages, has been replaced by other types of plastids. The peridinin plastid was inherited from the plastid in the common photosynthetic ancestor of dinoflagellates and apicomplexans (46, 47), but whether cryptic, nonpigmented plastids have been retained in nonphotosynthetic dinoflagellates remains contentious: Crypthecodinium and Oxyrrhis appear to contain plastid-derived genes (48, 49), whereas Hematodinium lacks all traces of the organelle (50). We investigated whether plastid and cytosolic pathways for isoprenoid, tetrapyrrole, and fatty acid biosynthesis were present in two distantly related nonphotosynthetic dinoflagellates, N. scintillans and O. marina, as well as in Dinophysis acuminata, a fundamentally nonphotosynthetic species that nevertheless carries kleptoplastids. For each metabolic enzyme in these pathways, we elaborated a single protein phylogeny and classified its origin as plastidic (in a clade with photosynthetic eukaryotes only), cytosolic (in a clade containing heterotrophic eukaryotes), or bacterial (in a clade with bacteria, putative recent horizontal transfer), a methodology informed by published localizations in model eukaryotes (e.g., ref. 51) and by in silico targeting predictions in selected proteins (Fig. 3 and SI Appendix, SI Materials and Methods).
Fig. 3.
Plastid metabolism and dependency in nonphotosyntetic dinoflagellates. (A) Phylogeny-driven reconstruction of plastid and nonplastid variants of core metabolism (isoprenoid, tetrapyrrole, and fatty acid biosynthesis) in genomes (marked as “G”) or transcriptomes (“T”) of dinoflagellates and relatives. Individual enzymes (SI Appendix, Table S5) were classified by protein phylogenies and color-coded as to their presence/absence and origin. The data suggest that Oxyrrhis, Noctiluca, and Dinophysis are metabolically dependent on plastids. Metabolite (Met.) uptake was summarized from the literature. (B) Maximum-likelihood phylogeny (IQ-Tree) reveals IspCs of cyanobacterial origin in nonphotosynthetic dinoflagellates and relatives (bold); ultrafast bootstraps at branches are shown (>50 shown; ≥95 highlighted; filled circles, 100). (C) Three grades in functional organization of core metabolic pathways in nonphotosynthetic plastids in dinoflagellates (blue) and relatives (“P” represents parasites). (D) Model for evolutionary dependency on plastids in dinoflagellates and relatives, which is applicable to other eukaryotes. Ancestral dependency (marked as “d”) on plastid metabolism (loss of cytosolic isoprenoid biosynthesis; later reinforced by the loss of C4 tetrapyrrole biosynthesis in some taxa) led to retention of plastids in all free-living and many parasitic descendants. The dependency can be transferred onto a new plastidial symbiont (Kareniaceae) or host organism (in parasites dependent solely on host-derived metabolites); only the latter leads to an outright loss of the plastid.
All three investigated dinoflagellates contain an isoprenoid pathway of plastid origin (all seven enzymes are present in Noctiluca and Dinophysis) and lack the cytosolic pathway variant (Fig. 3A), This is exemplified by their retention of cyanobacterial IspC enzymes (Fig. 3B), which branch among orthologs from photosynthetic dinoflagellates and other algae. Similarly, all three nonphotosynthetic dinoflagellates contain multiple components of the plastid tetrapyrrole pathway (an essentially complete enzyme set is present in Noctiluca and Dinophysis), but only two to three components of that in mitochondria and the cytosol. Comparing our data to the Symbiodinium minutum genome, we propose that a single tetrapyrrole pathway of a predominantly plastid origin that initiates from glutamate (Fig. 3A, GTR and GSA) is present in all core dinoflagellates, a feature typical of eukaryotic plastids [mitochondrial aminolevulinic acid synthase (ALA) synthase is present in the early-branching Hematodinium, Oxyrrhis, and Perkinsus (50)]. None of the three nonphotosynthetic dinoflagellates contain proteins for plastid fatty acid biosynthesis, suggesting that this pathway is dispensable in dinoflagellates in the absence of photosynthesis (Fig. 3A; FabI in Dinophysis is unusual; SI Appendix, SI Materials and Methods). Genes for plastid iron–sulfur cluster assembly (SufB, C, D), ferredoxin (Fd) redox system [i.e., Fd NADP+ reductase (FNR)], and triose phosphate membrane translocators (TPTs) are also present in the three species (SI Appendix, Table S5).
Plastid protein targeting and genome loss.
We further investigated 56 protein sequences in Noctiluca, Oxyrrhis, and Dinophysis of a plastidic origin (SI Appendix, Table S5). Most are incomplete, but seven are complete (they contain a partial spliced leader at the 5′ terminus of the corresponding transcript), and another 28 carry an extension of more than 50 aa at their N terminus. Proteins from the latter two categories were tested for the presence of plastid-targeting peptides in silico, and 17 of them carry bipartite targeting signatures comprising signal and transit peptides (SI Appendix, Table S6). Thirteen of these contain a phenylalanine at or near the predicted signal peptide cleavage site, and three Oxyrrhis proteins contain a second transmembrane region, all characteristics of targeting to plastids but not to other subcellular compartments in dinoflagellates (52, 53). In silico predictions have limited accuracy, but the consistent presence of N-terminal extensions and signal peptides in proteins is congruent only with a plastidic origin. For example, cyanobacterial Fds in Noctiluca and Dinophysis with four conserved cysteine residues required for Fe-S formation contain N-terminal extensions with signal and transit peptides for plastid targeting (truncated in Oxyrrhis; SI Appendix, Fig. S2). Noctiluca and Dinophysis also contain a plastid-targeted Fd NADP+ reductase (i.e., FNR; SI Appendix, Tables S5 and S6), suggesting that their Fd–FNR redox system might have a similar function to that in the nonphotosynthetic plastid of Plasmodium (54). SufB and ClpC have essential functions in plastids, but, in apicomplexans, they also constitute key barriers to the loss of the plastid genome (47, 55). SufB carries a bipartite plastid-targeting signature in Oxyrrhis (SI Appendix, Table S6), an apparently incomplete N-terminal extension in Dinophysis, and is encoded on GC-rich contigs in all three species (55–66.7% GC), all typical of a nuclear but not plastidial localization. Similarly to sufB, all three nonphotosynthetic dinoflagellates contain plastid-like clpC fragments on GC-rich, likely nuclear contigs (SI Appendix, SI Materials and Methods). SufB and clpC are also nucleus-encoded in Perkinsus and Symbiodinium (47, 56), and this indicates that both genes were relocated from the plastid genome early in their evolution. Because plastids in photosynthetic dinoflagellates encode only photosystem genes (7, 56) and ancestral reconstruction identifies no additional barriers to genome loss (47), evidence increasingly indicates that plastid genomes in nonphotosynthetic dinoflagellates and Perkinsus were lost with the loss of photosynthesis.
Principles of plastid dependency in dinoflagellates and eukaryotes.
Noctiluca, Oxyrrhis, and Dinophysis are metabolically dependent on cryptic plastids for the biosynthesis of isoprenoid units, and Noctiluca and Dinophysis for tetrapyrroles; evidence for this are multiple proteins in pathways of plastidial origin (as determined by phylogenies), presequences for plastid targeting, the absence of cytosolic pathway variants, and plastid localization of homologs in model species (Fig. 3 and SI Appendix, Tables S5 and S6). A full relocalization of either pathway to the cytosol is unprecedented in any organism, and the dependency on plastid pathways is supported by the fact that we obtain similar results from three distantly related heterotrophs and also from closely related phototrophs, one of which has genome data available (9). Additional plastid pathways—Fd redox system and Fe-S assembly—are present in Noctiluca, Oxyrrhis, and Dinophysis; these are essential for the function of the plastid but not for the host cell. Metabolism of amino acids remains insufficiently known in dinoflagellates and is absent in the plastids of apicomplexans (51). A comparison of nonphotosynthetic plastids in the broader group (Fig. 3C) reveals three functional grades in core metabolism that reflect dispensability of individual pathways: the biosynthesis of isoprenoid units (and required cofactors) is ubiquitous and is the only core plastid pathway in piroplasmid apicomplexans and Perkinsus, whereas plastid fatty acid biosynthesis was retained only in apicomplexans and Alphamonas (47) (Fig. 3 A and C).
The pattern of plastid dependency in dinoflagellates parallels that in apicomplexans and chrompodellids [chromerids and colpodellids (47)] and reinforces conclusions that their common ancestor had a plastid (46) and was reliant on it for isoprenoid units after it lost the capability to synthesize them in the cytosol (47). Despite rare secondary losses of plastids in certain parasites (50, 57) and ongoing uncertainties about plastid presence in some organisms (e.g., gregarines, Psammosa, Eudubosquella), plastids are indispensable in all free-living members of this group yet examined (Fig. 3A) (47, 48), including multiple uncultured forms (58). This pattern suggests that the metabolic dependency on plastids in free-living species cannot be bypassed by obtaining the relevant compounds from the environment or ingested prey (Fig. 3D). Rather, it has only increased with time as redundant cytosolic and mitochondrial pathways continue being lost (Fig. 3A) (47). For example, the loss of mitochondrial delta-ALA in core dinoflagellates has extended their plastid dependency to tetrapyrrole biosynthesis (Fig. 3A), much like in apicomplexans and chrompodellids (47). Most parasites retain plastids (59, 60), but their dependency on the organelle can be reduced or bypassed completely by the uptake of host metabolites (Fig. 3A) (57)—searches in transcriptomic data indicate that Amoebophrya parasites lack the plastid, which was likely lost in their common ancestor with Hematodinium (50). Based on these patterns, we suggest that all free-living (but not all parasitic) dinoflagellates rely on plastid organelles that are derived from the ancestral peridinin plastid (Fig. 3D). These include phagotrophs (Noctiluca), osmotrophs (Crypthecodinium), and species with kleptoplastidy (Dinophysis) and new endosymbionts (Durinskia) except where these endosymbionts have substituted metabolite dependency on the ancestral plastid (likely in the Kareniaceae; Fig. 3D). This provides a broad rationale for why dinoflagellates with diatom endosymbionts contain two types of plastid isoprenoid and tetrapyrrole pathways (61) (Fig. 3 B and D). It also explains why Dinophysis contains a plastid Fd and TPT of a dinoflagellate ancestry (62): both proteins contain bipartite targeting presequences with signal peptides (SI Appendix, Fig. S2 and Table S6; the latter was truncated in ref. 62), suggesting they are targeted into a cryptic three-membrane plastid (Fig. 3D), not the kleptoplastid as previously argued (62). Finally, we emphasize that a complete loss of a plastid organelle has never been confirmed in free-living eukaryotes, and we posit that this would be hard to achieve in established endosymbioses given the dependency patterns that exist in free-living dinoflagellates and related organisms (Fig. 3 A and D) (47).
Plastid tetrapyrroles and the evolution of bioluminescence.
Several species of dinoflagellates are bioluminescent (63). In the photosynthetic species Pyrocystis lunula, the light-emitting compound luciferin has an open tetrapyrrole structure thought to be synthesized from the structurally similar chlorophyll a (64): the organism incorporates radioactively labeled chlorophyll precursors into chlorophyll and luciferin, suggesting that their biosynthesis is linked (65). However, other bioluminescent dinoflagellates like Noctiluca, Protoperidinium, and certain Polykrikos species are nonphotosynthetic (63) and not known to synthesize chlorophyll. The prediction that they acquire chlorophyll from their prey (66) is inconsistent with prey-independent bioluminescence in at least one of them, Protoperidinium crassipes (67). Our finding of the plastid tetrapyrrole pathway in Noctiluca, which also leads to the precursors of chlorophyll, offers an alternative explanation of luciferin presence: it may be obtained by biosynthesis rather than scavenging, at least in some species. The plastid tetrapyrrole pathway is apparently indispensable as a key requirement for heme synthesis in all core dinoflagellates (Fig. 3A), and could therefore account for luciferin production in any bioluminescent dinoflagellate, irrespective of the presence of photosynthesis. This biosynthesis scenario also opens the possibility that luciferin is not derived via chlorophyll per se, but via an earlier intermediate in its biogenesis, perhaps a chlorophyllide or chlorine-like tetrapyrrole. Although this remains to be tested experimentally, our finding of the plastid tetrapyrrole pathway supports the possibility that bioluminescence in nonphotosynthetic dinoflagellates relies on a biosynthetic machinery repurposed from heme and chlorophyll production.
Character Evolution in Dinoflagellates.
Nuclear evolution: Stepwise horizontal gene gain.
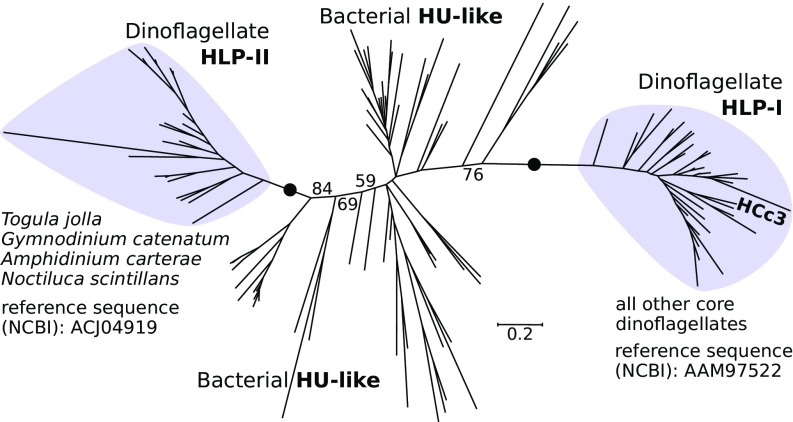
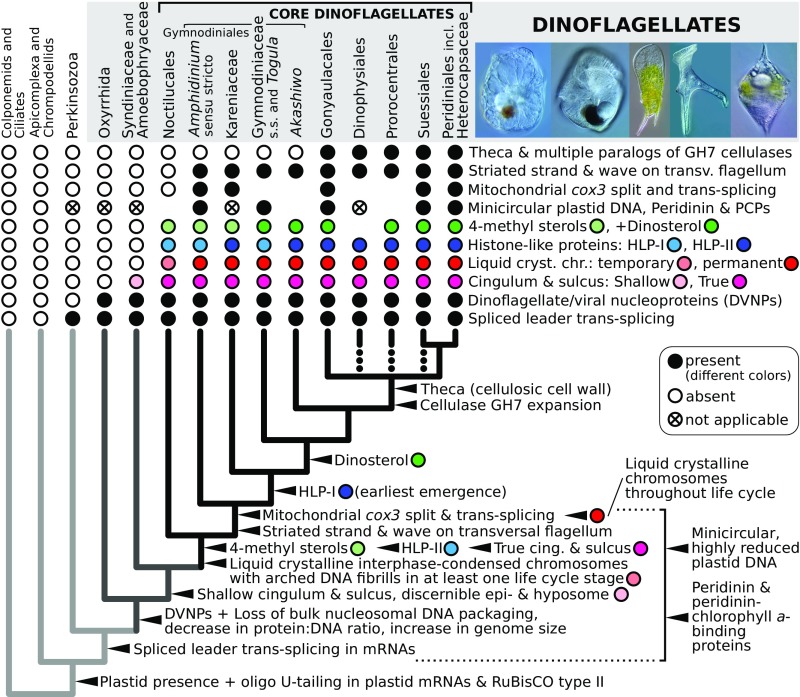
Dinoflagellates have unique nuclei that have lost bulk nucleosomal DNA packaging, and instead condense DNA by using two types of basic proteins that are different from histones. Dinoflagellate/viral nucleoproteins (DVNPs) are similar to uncharacterized proteins from phycodnaviruses, are distributed in all dinoflagellates yet examined, and represent a family of basic proteins with high DNA-binding affinity (4). In contrast, dinoflagellate histone-like proteins (HLPs) are of bacterial origin and have been found only in certain core dinoflagellate species; they are primarily detected at the chromosome periphery, where they are predicted to organize extended DNA loops during transcription (68). We identified DVNPs in all transcriptomes in our dataset, confirming their ubiquitous distribution among dinoflagellates. Our searches also confirm that HLPs are absent in all early-branching taxa (Oxyrrhis, Hematodinium, and Amoebophrya spp.) and are ubiquitous in core dinoflagellates. Unexpectedly, however, we found that HLPs in Noctiluca, Amphidinium, Togula, and Gymnodinium are dissimilar in sequence to HLPs in other dinoflagellates despite their similar length and structure (SI Appendix, Table S4). We reconstructed the phylogeny of dinoflagellate HLPs together with a representative selection of their closest orthologs, the bacterial HU-like proteins (HLPs in other eukaryotes are not closely related to those in dinoflagellates). The outcome confirms a wide separation between the dinoflagellate type known previously, HLP-I [e.g., HCc3 in Crypthecodinium (68)], and the HLP-II (Fig. 4 and SI Appendix, Fig. S3). Interestingly, HLP-I and HLP-II have mutually exclusive distributions (Fig. 4 and SI Appendix, Fig. S3), which suggest that HLP-II rather than HLP-I was ancestral to core dinoflagellates. HLP-I most likely appeared in the ancestor of Kareniaceae and other core dinoflagellates (it temporarily coexisted with HLP-I followed by selective loss or spread horizontally later between the Kareniaceae and thecates and Akashiwo; Fig. 5). Because HLP-I and HLP-II are monophyletic but not closely related to each other, and HU-like proteins are present in a wide range of bacterial phyla, the dinoflagellate HLPs are likely derived from HU-like proteins and not vice versa (this is in contrast to DVNPs, in which the direction of transfer with phycodnaviruses cannot be established). The unique molecular architecture of dinoflagellate nuclei thus resulted from at least three independent waves of protein gain (Fig. 5). The recruitment of DVNPs took place in the group’s ancestor, leading to a decrease in the nuclear protein:DNA ratio and potentially the loss of bulk nucleosomal packaging and increase in the genome size in dinoflagellates. HLPs were acquired later than DVNPs by at least two independent horizontal transfers from different bacterial donors. The initial gain of HLPs in the ancestor core dinoflagellates coincided with the emergence of liquid crystalline chromosomes with arched DNA fibrils, which are condensed permanently in most species.
Fig. 4.
Evolution of histone-like proteins. Phylogeny of bacterial (HU-like) and dinoflagellate HLPs reveals a dinoflagellate-type histone-like protein, HLP-II, in early-branching core dinoflagellates. HLP-II has a mutually exclusive distribution with HLP-I (e.g., the characterized HCc3 in C. cohnii, in bold). Further details are provided in SI Appendix, Fig. S3 and Table S4.
Fig. 5.
Model for character evolution in dinoflagellates. Ancestral character states (filled circles) of conserved traits are reconstructed on the consensus phylogeny of dinoflagellates and their relatives by parsimony (arrowheads). Dotted branches in the thecate lineages indicate uncertain placement. Gaps indicate missing data, and “not applicable” denotes plastid genome absence or the presence of a different plastid genome type (Kareniaceae). The vertical square bracket indicates an evolutionary range in which traits emerged. Photos of dinoflagellates (by G. S. Gavelis), left to right: Kofoidinium sp. (Noctilucales), Nematodinium sp. (Gymnodiniaceae s.s.), Neoceratium praelongum (Gonyaulacales), Dinophysis miles (Dinophysiales), and Heterocapsa sp. (Peridiniales).
Organelle evolution: Plastid reduction and mitochondrial cox3 split.
Evidence of a dependency on plastids in nonphotosynthetic dinoflagellates (Fig. 3) corroborates earlier conclusions that the common ancestor of dinoflagellates and apicomplexans was photosynthetic (46) and dependent on plastid-generated isoprenoids (47). Our phylogeny also supports the prediction that more than a dozen descendant lineages of this dinoflagellate–apicomplexan ancestor have lost photosynthesis (46, 69). At least two parasites, Cryptosporidium and Hematodinium, have lost the plastid outright, but this is not the case in other parasites and in any free-living lineages that have been investigated with sufficient detail (six independent transitions to heterotrophy). We thus posit that plastid loss in dinoflagellates and apicomplexans is less frequent than their retention after the loss of photosynthesis, and is limited to a few parasites (47). After the split with apicomplexans but at least by the time Amphidinium diverged, the dinoflagellate plastid acquired the photosynthetic carotenoid peridinin, peridin–chlorophyll binding proteins, and a reduced, minicircular genome (6). Our results suggest that during this transition the plastid sufB and clpC genes (key barriers to plastid genome loss in apicomplexans) were relocated to the nucleus in dinoflagellates. This made the dinoflagellate plastid genome dispensable in the absence of photosynthesis, likely explaining why all heterotrophic representatives studied to date appear to lack it. In at least four distantly related photosynthetic dinoflagellates, the expression of plastid genes is accompanied by substitutional editing of corresponding mRNAs (Fig. 5) (7). The origin of plastid editing is, however, uncertain: it appeared some time after the divergence of apicomplexans and chrompodellids (70) and possibly became more widespread after the divergence of Amphidinium (71), but pinpointing its origin more precisely will require an analysis on deep-branching photosynthetic dinoflagellates such as Spatulodinium pseudonoctiluca (13).
The mtDNA in at least five lineages of core dinoflagellates including Amphidinium contains a unique feature: cox3 is split in the same region into two fragments that are trans-spliced at the RNA level (8, 72). The split is absent in Hematodinium and earlier diverging species, but, to our knowledge, its presence in the Noctilucales was not known until now (Fig. 5). We identified a cox3 contig in the Noctiluca transcriptome corresponding to a full-length protein (terminated by a canonical stop codon rare in the group; SI Appendix, SI Materials and Methods). Mapping individual RNA read pairs onto the contig demonstrated continuous transcription across the split region and provides no support for the existence of two transcripts and their trans-splicing. PCR amplification by using Noctiluca genomic DNA as a template produced a single product spanning both sides of the cox3 split, the identity of which was confirmed by sequencing (SI Appendix, SI Materials and Methods). Because the phylogenetic distribution and the unique character of the cox3 split are indicative of a single evolutionary origin, the uninterrupted cox3 in Noctiluca corroborates the early position of the Noctilucales among core dinoflagellates (Fig. 1).
Character map: Framework for evolutionary and functional predictions.
By using parsimony, we reconstructed ancestral character states of major conserved morphological and molecular traits at different points of the dinoflagellate phylogeny (Fig. 5 and SI Appendix, SI Materials and Methods). Newly mapped transitions include the gain of 4-methyl sterols, dinosterol, nuclear HLP-I and II, the mitochondrial cox3 split, the theca, and the gain of multiple paralogs of GH7 cellulases. Two additional transitions map at the common ancestor of Amphidinium and later-diverging taxa: the gain of condensed liquid crystalline chromosomes throughout the life cycle and the gain of a proteinaceous striated rod in the transverse flagellum, which produces a strongly pronounced flagellar wave (Fig. 5). The corresponding characteristics in the Noctilucales are little understood as yet—chromosomes in one of their life stages, the trophont, are relaxed and the transversal flagellum in their gametes is trailing, wave-less, and contains only a thin filament in place of the striated rod (73). Detailed analysis is required to determine whether these states represent true evolutionarily intermediates or secondary modifications associated with the unusual morphology of this order. The origin of other dinoflagellate characteristics was established previously and is reinforced within our framework: gain of plastids, RuBisCO form II and oligoU-tailing in plastid mRNAs before the split with apicomplexans (46), and the acquisition of spliced leader trans-splicing of mRNAs in their common ancestor with perkinsids (Fig. 5). DVNPs, ubiquitous in the species in our dataset, are ancestral to dinoflagellates and associated with changes in protein:DNA ratio and genome size. The ancestor of syndinians and core dinoflagellates had a life stage with a shallow sulcus and cingulum (flagellar grooves), the latter dividing the cell into an upper episome and a lower hyposome, a transitional morphology between short flagellar grooves in Oxyrrhis and Psammosa and deeply engraved perpendicular flagellar grooves in core dinoflagellates (Fig. 5). Altogether, most transitions map to the branch corresponding to the ancestor of core dinoflagellates, but other characteristics are scattered widely along the evolutionary backbone (Fig. 5). Thus, the ecological success of dinoflagellates has resulted from a series of independent changes to the morphology, metabolism, and molecular biology of their ancestors.
Conclusions
We used sequence data to illuminate dinoflagellate biology and evolution. Evidence from our multiprotein phylogenies resolves numerous issues relating to dinoflagellate relationships, provides strong support for the single origin of the theca, and helps reconcile several apparent contradictions in dinoflagellate fossil, biogeochemical, and molecular data (Figs. 1 and 2). The origin of the theca coincides with a radiation of cell wall-localized cellulases involved in cell division (Fig. 2B). Plastid biosynthetic pathways exist in the nonphotosynthetic Noctiluca, Oxyrrhis, and Dinophysis, and cytosolic pathway variants do not (Fig. 3). This suggests that all free-living dinoflagellates are metabolically dependent on plastids that have taken over important cellular functions, apparently early in the evolution of the group; plastidial tetrapyrrole biosynthesis may also explain the existence of bioluminescent luciferin in nonpigmented dinoflagellates. The origin of the liquid crystalline nuclei coincides with the acquisition of bacterial histone-like proteins, which occurred in two distinct evolutionary phases (Fig. 4), suggesting that horizontal gene transfers were the ultimate origin of key dinoflagellate features. By producing a map of the major transitions in the evolutionary history of dinoflagellates (Fig. 5), we provide a predictional framework that will facilitate the investigation of many aspects of the group’s cell biology (nuclear organization, plastid evolution), molecular biology, and paleobiology.
Materials and Methods
RNA was extracted by RNAqueous kit or TRIzol Plus RNA kit. Paired-end 50-bp or 100-bp Illumina sequence reads were generated and assembled in Trinity version 2 or as part of the Marine Microbial Eukaryote Transcriptome Sequencing Project pipeline (19). Phylogenetic matrices were prepared from alignments in MAFFT version 7.215 stripped of hypervariable sites in Block Mapping and Gathering with Entropy version 1.1. Phylogenies were computed in IQ-Tree (1,000 ultrafast bootstraps), RAxML version 8 (300 nonparametric bootstraps), and Phylobayes (where applicable). Plastid targeting signals were analyzed in SignalP 4.1 (D-score cutoff 0.45) and ChloroP 1.1 at 0.45 cTP-score cutoff. Species culturing and sequencing, phylogenetic inferences, and analyses of plastid metabolism and protein targeting are detailed in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Bill MacMillan for technical support and Patrick Keeling for facilities and support. This work was supported by a University College London Excellence Fellowship (to J.J.), a CIFAR Global Scholar Fellowship (to J.J.), a University of British Columbia Four-Year PhD Fellowship (to J.J.), Gordon and Betty Moore Foundation Grant 2637 to the National Center for Genome Resources (NCGR), National Science and Engineering Research Council of Canada Grant NSERC 2014-05258 (to B.S.L. and G.S.G.), a Tula Foundation Grant to Patrick Keeling (F.B.), the Centre for Microbial Biodiversity and Evolution (F.B.), Australian Research Council Grant DP1093395 (to S.G.G.), Science Foundation Ireland Grant 13/SIRG/2125 (to S.G.G.), NSF Grant EF-0629625 (to C.F.D. and T.R.B.), and Canadian Institute for Health Research Grant MOP-42517 to Patrick Keeling. MMETSP samples were sequenced, assembled, and annotated at NCGR. This is ESS contribution no. 20160099.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the iMicrobe database (project code CAM_P_0001000) and GenBank Transcriptome Shotgun Assembly (TSA) Sequence Database (accession nos. GELK00000000 and GEMP00000000).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614842114/-/DCSupplemental.
References
- 1.Gómez F. A quantitative review of the lifestyle, habitat and trophic diversity of dinoflagellates (Dinoflagellata, Alveolata) Syst Biodivers. 2012;10(3):267–275. [Google Scholar]
- 2.de Vargas C, et al. Tara Oceans Coordinators Ocean plankton.Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348(6237):1261605. doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- 3.Velo-Suárez L, Brosnahan ML, Anderson DM, McGillicuddy DJ., Jr A quantitative assessment of the role of the parasite Amoebophrya in the termination of Alexandrium fundyense blooms within a small coastal embayment. PLoS One. 2013;8(12):e81150. doi: 10.1371/journal.pone.0081150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gornik SG, et al. Loss of nucleosomal DNA condensation coincides with appearance of a novel nuclear protein in dinoflagellates. Curr Biol. 2012;22(24):2303–2312. doi: 10.1016/j.cub.2012.10.036. [DOI] [PubMed] [Google Scholar]
- 5.Wong JTY, New DC, Wong JCW, Hung VKL. Histone-like proteins of the dinoflagellate Crypthecodinium cohnii have homologies to bacterial DNA-binding proteins. Eukaryot Cell. 2003;2(3):646–650. doi: 10.1128/EC.2.3.646-650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, Green BR, Cavalier-Smith T. Single gene circles in dinoflagellate chloroplast genomes. Nature. 1999;400(6740):155–159. doi: 10.1038/22099. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Morse D. Rampant polyuridylylation of plastid gene transcripts in the dinoflagellate Lingulodinium. Nucleic Acids Res. 2006;34(2):613–619. doi: 10.1093/nar/gkj438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nash EA, Nisbet RER, Barbrook AC, Howe CJ. Dinoflagellates: A mitochondrial genome all at sea. Trends Genet. 2008;24(7):328–335. doi: 10.1016/j.tig.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Shoguchi E, et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol. 2013;23(15):1399–1408. doi: 10.1016/j.cub.2013.05.062. [DOI] [PubMed] [Google Scholar]
- 10.Lin S, et al. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science. 2015;350(6261):691–694. doi: 10.1126/science.aad0408. [DOI] [PubMed] [Google Scholar]
- 11.Bachvaroff TR, et al. Dinoflagellate phylogeny revisited: Using ribosomal proteins to resolve deep branching dinoflagellate clades. Mol Phylogenet Evol. 2014;70:314–322. doi: 10.1016/j.ympev.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoppenrath M, Leander BS. Dinoflagellate phylogeny as inferred from heat shock protein 90 and ribosomal gene sequences. PLoS One. 2010;5(10):e13220. doi: 10.1371/journal.pone.0013220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gómez F, Moreira D, López-García P. Molecular phylogeny of noctilucoid dinoflagellates (Noctilucales, Dinophyceae) Protist. 2010;161(3):466–478. doi: 10.1016/j.protis.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Orr RJS, Murray SA, Stüken A, Rhodes L, Jakobsen KS. When naked became armored: An eight-gene phylogeny reveals monophyletic origin of theca in dinoflagellates. PLoS One. 2012;7(11):e50004. doi: 10.1371/journal.pone.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fensome RA, Saldarriaga JF, Taylor “Max” FJR (1999) Dinoflagellate phylogeny revisited: Reconciling morphological and molecular based phylogenies. Grana 38(2):66–80.
- 16. Saldarriaga JF, Taylor “Max” FJR, Cavalier-Smith T, Menden-Deuer S, Keeling PJ (2004) Molecular data and the evolutionary history of dinoflagellates. Eur J Protistol 40(1):85–111.
- 17.Imanian B, Keeling PJ. Horizontal gene transfer and redundancy of tryptophan biosynthetic enzymes in dinotoms. Genome Biol Evol. 2014;6(2):333–343. doi: 10.1093/gbe/evu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gavelis GS, White RA, Suttle CA, Keeling PJ, Leander BS. Single-cell transcriptomics using spliced leader PCR: Evidence for multiple losses of photosynthesis in polykrikoid dinoflagellates. BMC Genomics. 2015;16(1):528. doi: 10.1186/s12864-015-1636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keeling PJ, et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12(6):e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burki F, Okamoto N, Pombert J-F, Keeling PJ. The evolutionary history of haptophytes and cryptophytes: Phylogenomic evidence for separate origins. Proc Biol Sci. 2012;279(1736):2246–2254. doi: 10.1098/rspb.2011.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saunders GW, Hill DRA, Sexton JP, Andersen RA. Small-subunit ribosomal RNA sequences from selected dinoflagellates: Testing classical evolutionary hypotheses with molecular systematic methods. In: Bhattacharya D, editor. Origins of Algae and Their Plastids. Springer; Vienna: 1997. pp. 237–259. [Google Scholar]
- 22.Fukuda Y, Endoh H. Phylogenetic analyses of the dinoflagellate Noctiluca scintillans based on beta-tubulin and Hsp90 genes. Eur J Protistol. 2008;44(1):27–33. doi: 10.1016/j.ejop.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Jørgensen MF, Murray S, Daugbjerg N. A new genus of athecate interstitial dinoflagellates, Togula gen. nov., previously encompassed within Amphidinium sensu lato: Inferred from light and electron microscopy and phylogenetic analyses of partial large subunit ribosomal DNA sequences. Phycol Res. 2004;52(3):284–299. [Google Scholar]
- 24.Daugbjerg N, Hansen G, Larsen J, Moestrup Ø. Phylogeny of some of the major genera of dinoflagellates based on ultrastructure and partial LSU rDNA sequence data, including the erection of three new genera of unarmoured dinoflagellates. Phycologia. 2000;39(4):302–317. [Google Scholar]
- 25.Zhang H, Bhattacharya D, Lin S. A three-gene dinoflagellate phylogeny suggests monophyly of prorocentrales and a basal position for Amphidinium and Heterocapsa. J Mol Evol. 2007;65(4):463–474. doi: 10.1007/s00239-007-9038-4. [DOI] [PubMed] [Google Scholar]
- 26.Fensome RA, et al. 1993. A Classification of Living and Fossil Dinoflagellates. Micropaleontology Special Publication 7 (American Museum of Natural History, New York)
- 27.Moestrup Ø, Lindberg K, Daugbjerg N. Studies on woloszynskioid dinoflagellates IV: The genus Biecheleria gen. nov. Phycol Res. 2009;57(3):203–220. [Google Scholar]
- 28.Takahashi K, Moestrup Ø, Jordan RW, Iwataki M. Two new freshwater woloszynskioids Asulcocephalium miricentonis gen. et sp. nov. and Leiocephalium pseudosanguineum gen. et sp. nov. (Suessiaceae, Dinophyceae) lacking an apical furrow apparatus. Protist. 2015;166(6):638–658. doi: 10.1016/j.protis.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Medlin LK, Fensome RA. 2013. Dinoflagellate macroevolution: Some considerations based on an integration of molecular, morphological and fossil evidence. Biological and Geological Perspectives of Dinoflagellates, eds Lewis JM, Marret F, Bradley L. The Micropalaeontological Society, Special Publications (Geological Society, London), pp 255–266.
- 30.Fensome RA, MacRae RA, Moldowan JM, Taylor FJR, Williams GL. The early Mesozoic radiation of dinoflagellates. Paleobiology. 1996;22(3):329–338. [Google Scholar]
- 31.Bujak JP, Williams GL. The evolution of dinoflagellates. Can J Bot. 1981;59(11):2077–2087. [Google Scholar]
- 32.Hansen G, Daugbjerg N, Henriksen P. Baldinia anauniensis gen. et sp. nov.: A “new” dinoflagellate from Lake Tovel, N. Italy. Phycologia. 2007;46(1):86–108. [Google Scholar]
- 33.Sekida S, Horiguchi T, Okuda K. Development of thecal plates and pellicle in the dinoflagellate Scrippsiella hexapraecingula (Peridiniales, Dinophyceae) elucidated by changes in stainability of the associated membranes. Eur J Phycol. 2004;39(1):105–114. [Google Scholar]
- 34.Kwok ACM, Wong JTY. The activity of a wall-bound cellulase is required for and is coupled to cell cycle progression in the dinoflagellate Crypthecodinium cohnii. Plant Cell. 2010;22(4):1281–1298. doi: 10.1105/tpc.109.070243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moldowan JM, et al. Chemostratigraphic reconstruction of biofacies: Molecular evidence linking cyst-forming dinoflagellates with pre-Triassic ancestors. Geology. 1996;24(2):159–162. [Google Scholar]
- 36.Moldowan JM, Talyzina NM. Biogeochemical evidence for dinoflagellate ancestors in the early cambrian. Science. 1998;281(5380):1168–1170. doi: 10.1126/science.281.5380.1168. [DOI] [PubMed] [Google Scholar]
- 37.Summons RE, Walter MR. Molecular fossils and microfossils of prokaryotes and protists from Proterozoic sediments. Am J Sci. 1990;290:212–244. [Google Scholar]
- 38.Chu F-LE, et al. Sterol production and phytosterol bioconversion in two species of heterotrophic protists, Oxyrrhis marina and Gyrodinium dominans. Mar Biol. 2008;156(2):155–169. [Google Scholar]
- 39.Place AR, Bai X, Kim S, Sengco MR, Wayne Coats D. Dinoflagellate host-parasite sterol profiles dictate karlotoxin sensitivity(1) J Phycol. 2009;45(2):375–385. doi: 10.1111/j.1529-8817.2009.00649.x. [DOI] [PubMed] [Google Scholar]
- 40.Leblond JD, Sengco MR, Sickman JO, Dahmen JL, Anderson DM. Sterols of the syndinian dinoflagellate Amoebophrya sp., a parasite of the dinoflagellate Alexandrium tamarense (Dinophyceae) J Eukaryot Microbiol. 2006;53(3):211–216. doi: 10.1111/j.1550-7408.2006.00097.x. [DOI] [PubMed] [Google Scholar]
- 41.Teshima SI, Kanazawa A, Tago A. Sterols of the dinoflagellate Noctiluca milialis. Mem Fac Fish Kagoshima Univ. 1980;29:319–326. [Google Scholar]
- 42.Withers NW, Goad LJ, Goodwin TW. A new sterol, 4α-methyl-5α-ergosta-8(14),24(28)-dien-3β-ol, from the marine dinoflagellate Amphidinium carterae. Phytochemistry. 1979;18(5):899–901. [Google Scholar]
- 43.Leblond JD, Chapman PJ. A survey of the sterol composition of the marine dinoflagellates Karenia brevis, Karenia mikimotoi, and Karlodinium micrum: Distribution of sterols within other members of the class Dinophyceae. J Phycol. 2002;38(4):670–682. [Google Scholar]
- 44.Volkman JK, Barrett SM, Dunstan GA, Jeffrey SW. Geochemical significance of the occurrence of dinosterol and other 4-methyl sterols in a marine diatom. Org Geochem. 1993;20(1):7–15. [Google Scholar]
- 45.Brocks JJ, Buick R, Summons RE, Logan GA. A reconstruction of Archean biological diversity based on molecular fossils from the 2.78 to 2.45 billion-year-old Mount Bruce Supergroup, Hamersley Basin, Western Australia. Geochim Cosmochim Acta. 2003;67(22):4321–4335. [Google Scholar]
- 46.Janouškovec J, Horák A, Oborník M, Lukeš J, Keeling PJ. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci USA. 2010;107(24):10949–10954. doi: 10.1073/pnas.1003335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Janouškovec J, et al. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc Natl Acad Sci USA. 2015;112(33):10200–10207. doi: 10.1073/pnas.1423790112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanchez-Puerta MV, Lippmeier JC, Apt KE, Delwiche CF. Plastid genes in a non-photosynthetic dinoflagellate. Protist. 2007;158(1):105–117. doi: 10.1016/j.protis.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 49.Slamovits CH, Keeling PJ. Plastid-derived genes in the nonphotosynthetic alveolate Oxyrrhis marina. Mol Biol Evol. 2008;25(7):1297–1306. doi: 10.1093/molbev/msn075. [DOI] [PubMed] [Google Scholar]
- 50.Gornik SG, et al. Endosymbiosis undone by stepwise elimination of the plastid in a parasitic dinoflagellate. Proc Natl Acad Sci USA. 2015;112(18):5767–5772. doi: 10.1073/pnas.1423400112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seeber F, Soldati-Favre D. Metabolic pathways in the apicoplast of apicomplexa. Int Rev Cell Mol Biol. 2010;281:161–228. doi: 10.1016/S1937-6448(10)81005-6. [DOI] [PubMed] [Google Scholar]
- 52.Nassoury N, Cappadocia M, Morse D. Plastid ultrastructure defines the protein import pathway in dinoflagellates. J Cell Sci. 2003;116(pt 14):2867–2874. doi: 10.1242/jcs.00517. [DOI] [PubMed] [Google Scholar]
- 53.Patron NJ, Waller RF, Archibald JM, Keeling PJ. Complex protein targeting to dinoflagellate plastids. J Mol Biol. 2005;348(4):1015–1024. doi: 10.1016/j.jmb.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 54.Pandini V, et al. Ferredoxin-NADP+ reductase and ferredoxin of the protozoan parasite Toxoplasma gondii interact productively in vitro and in vivo. J Biol Chem. 2002;277(50):48463–48471. doi: 10.1074/jbc.M209388200. [DOI] [PubMed] [Google Scholar]
- 55.Howe CJ, Purton S. The little genome of apicomplexan plastids: Its raison d’etre and a possible explanation for the ‘delayed death’ phenomenon. Protist. 2007;158(2):121–133. doi: 10.1016/j.protis.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 56.Mungpakdee S, et al. Massive gene transfer and extensive RNA editing of a symbiotic dinoflagellate plastid genome. Genome Biol Evol. 2014;6(6):1408–1422. doi: 10.1093/gbe/evu109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abrahamsen MS, et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304(5669):441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 58.Janouškovec J, Horák A, Barott KL, Rohwer FL, Keeling PJ. Global analysis of plastid diversity reveals apicomplexan-related lineages in coral reefs. Curr Biol. 2012;22(13):R518–R519. doi: 10.1016/j.cub.2012.04.047. [DOI] [PubMed] [Google Scholar]
- 59.McFadden GI, Reith ME, Munholland J, Lang-Unnasch N. Plastid in human parasites. Nature. 1996;381(6582):482. doi: 10.1038/381482a0. [DOI] [PubMed] [Google Scholar]
- 60.Matsuzaki M, Kuroiwa H, Kuroiwa T, Kita K, Nozaki H. A cryptic algal group unveiled: A plastid biosynthesis pathway in the oyster parasite Perkinsus marinus. Mol Biol Evol. 2008;25(6):1167–1179. doi: 10.1093/molbev/msn064. [DOI] [PubMed] [Google Scholar]
- 61.Hehenberger E, Imanian B, Burki F, Keeling PJ. Evidence for the retention of two evolutionary distinct plastids in dinoflagellates with diatom endosymbionts. Genome Biol Evol. 2014;6(9):2321–2334. doi: 10.1093/gbe/evu182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wisecaver JH, Hackett JD. Transcriptome analysis reveals nuclear-encoded proteins for the maintenance of temporary plastids in the dinoflagellate Dinophysis acuminata. BMC Genomics. 2010;11:366. doi: 10.1186/1471-2164-11-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marcinko CLJ, Painter SC, Martin AP, Allen JT. A review of the measurement and modelling of dinoflagellate bioluminescence. Prog Oceanogr. 2013;109:117–129. [Google Scholar]
- 64.Topalov G, Kishi Y. Chlorophyll Catabolism leading to the skeleton of dinoflagellate and Krill luciferins: Hypothesis and model studies. Financial support from the National Institutes of Health (NS 12108) is gratefully acknowledged. Angew Chem Int Ed Engl. 2001;40(20):3892–3894. [PubMed] [Google Scholar]
- 65.Wu C, Akimoto H, Ohmiya Y. Tracer studies on dinoflagellate luciferin with [15N]-glycine and [15N]-l-glutamic acid in the dinoflagellate Pyrocystis lunula. Tetrahedron Lett. 2003;44(6):1263–1266. [Google Scholar]
- 66.Liu L, Hastings JW. Two different domains of the luciferase gene in the heterotrophic dinoflagellate Noctiluca scintillans occur as two separate genes in photosynthetic species. Proc Natl Acad Sci USA. 2007;104(3):696–701. doi: 10.1073/pnas.0607816103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamaguchi A, Horiguchi T. Culture of the heterotrophic dinoflagellate Protoperidinium crassipes (Dinophyceae) with noncellular food items(1) J Phycol. 2008;44(4):1090–1092. doi: 10.1111/j.1529-8817.2008.00547.x. [DOI] [PubMed] [Google Scholar]
- 68.Chan Y-H, Wong JTY. Concentration-dependent organization of DNA by the dinoflagellate histone-like protein HCc3. Nucleic Acids Res. 2007;35(8):2573–2583. doi: 10.1093/nar/gkm165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saldarriaga JF, Taylor FJ, Keeling PJ, Cavalier-Smith T. Dinoflagellate nuclear SSU rRNA phylogeny suggests multiple plastid losses and replacements. J Mol Evol. 2001;53(3):204–213. doi: 10.1007/s002390010210. [DOI] [PubMed] [Google Scholar]
- 70.Janouškovec J, et al. Split photosystem protein, linear-mapping topology, and growth of structural complexity in the plastid genome of Chromera velia. Mol Biol Evol. 2013;30(11):2447–2462. doi: 10.1093/molbev/mst144. [DOI] [PubMed] [Google Scholar]
- 71.Barbrook AC, et al. Polyuridylylation and processing of transcripts from multiple gene minicircles in chloroplasts of the dinoflagellate Amphidinium carterae. Plant Mol Biol. 2012;79(4-5):347–357. doi: 10.1007/s11103-012-9916-z. [DOI] [PubMed] [Google Scholar]
- 72.Jackson CJ, Waller RF. A widespread and unusual RNA trans-splicing type in dinoflagellate mitochondria. PLoS One. 2013;8(2):e56777. doi: 10.1371/journal.pone.0056777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Soyer M-O. Etude ultrastructurale de l’endoplasme et des vacuoles chez deux types de Dinoflagellés appartenant aux genres Noctiluca (Suriray) et Blastodinium (Chatton) Z Zellforsch Mikrosk Anat. 1970;105(3):350–388. [PubMed] [Google Scholar]
- 74.Lee SY, et al. Morphological characterization of Symbiodinium minutum and S. psygmophilum belonging to clade B. Algae. 2014;29(4):299–310. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.