Significance
This study explores the structural basis for the difference between the closely related Na+,K+-ATPase and H+,K+-ATPase with respect to ion transport stoichiometry, electrogenicity, and cation selectivity, which has long been an enigma. A cysteine in the membrane domain of Na+,K+-ATPase is substituted by arginine in H+,K+-ATPase and has been found to be replaced by phenylalanine in a patient with alternating hemiplegia of childhood. These substitutions affect the functioning of the Na+-specific site of Na+,K+-ATPase profoundly. The positively charged side chain of the arginine present in H+,K+-ATPase serves as an internal cation that replaces bound Na+, thus explaining how H+,K+-ATPase avoids binding and transporting Na+ at this site, thereby performing electroneutral transport, despite the general structural and functional resemblance to the electrogenic Na+,K+-ATPase.
Keywords: H+,K+-pump; Na+,K+-pump; electrogenicity; internal cation; alternating hemiplegia of childhood
Abstract
Na+,K+-ATPase and H+,K+-ATPase are electrogenic and nonelectrogenic ion pumps, respectively. The underlying structural basis for this difference has not been established, and it has not been revealed how the H+,K+-ATPase avoids binding of Na+ at the site corresponding to the Na+-specific site of the Na+,K+-ATPase (site III). In this study, we addressed these questions by using site-directed mutagenesis in combination with enzymatic, transport, and electrophysiological functional measurements. Replacement of the cysteine C932 in transmembrane helix M8 of Na+,K+-ATPase with arginine, present in the H+,K+-ATPase at the corresponding position, converted the normal 3Na+:2K+:1ATP stoichiometry of the Na+,K+-ATPase to electroneutral 2Na+:2K+:1ATP stoichiometry similar to the electroneutral transport mode of the H+,K+-ATPase. The electroneutral C932R mutant of the Na+,K+-ATPase retained a wild-type–like enzyme turnover rate for ATP hydrolysis and rate of cellular K+ uptake. Only a relatively minor reduction of apparent Na+ affinity for activation of phosphorylation from ATP was observed for C932R, whereas replacement of C932 with leucine or phenylalanine, the latter of a size comparable to arginine, led to spectacular reductions of apparent Na+ affinity without changing the electrogenicity. From these results, in combination with structural considerations, it appears that the guanidine+ group of the M8 arginine replaces Na+ at the third site, thus preventing Na+ binding there, although allowing Na+ to bind at the two other sites and become transported. Hence, in the H+,K+-ATPase, the ability of the M8 arginine to donate an internal cation binding at the third site is decisive for the electroneutral transport mode of this pump.
The Na+,K+-ATPase and H+,K+-ATPase are closely related members of the P-type ATPase family of membrane transporters (1, 2). The Na+,K+-ATPase is an essential ion pump present in the cell membrane of all mammalian cells, where it carries out electrogenic exchange of three internal Na+ ions for two external K+ ions per ATP molecule split. It is widely accepted that the transport of Na+ and K+ occurs by a consecutive mechanism in which Na+ is occluded in the phosphorylated enzyme and released on the external side before K+ binds and is transported into the cell, also involving an occluded intermediate (3) (the reaction cycle is shown in Fig. S1). Crystal structures of the Na+,K+-ATPase show that two Na+ ions bind at sites that overlap substantially with the K+ sites (so-called sites I and II), whereas the third Na+ ion is bound at a distinct Na+-specific site (site III) (1, 4, 5). Two isoforms of H+,K+-ATPase, with different expression profiles, perform electroneutral H+,K+-exchange by a mechanism basically similar to the Na+,K+-ATPase mechanism (6, 7). The nongastric H+,K+-ATPase participates in K+ reabsorption in the kidney and colon, whereas the better characterized gastric H+,K+-ATPase is responsible for acid secretion into the lumen of the stomach and has been proposed to have two different stoichiometries: 1H+:1K+:1ATP at low luminal pH and 2H+:2K+:1ATP at neutral pH (6–8). A low-resolution crystal structure of the H+,K+-ATPase obtained at acidic pH in the presence of the K+ congener Rb+ shows that one bound Rb+ ion occupies a site equivalent to site II of the Na+,K+-ATPase, whereas the occupancy of site I is very low (8).
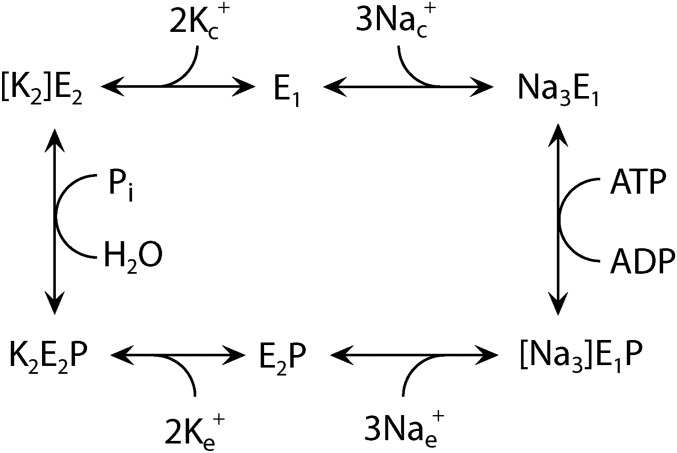
Fig. S1.
Na+,K+-ATPase reaction cycle. E1 and E2 represent the main conformational states of the enzyme that are Na+ selective and K+ selective, respectively. P indicates phosphorylation from ATP at the conserved aspartate of the P-type ATPase signature sequence. During the transport process, the Na+ and K+ ions become occluded [i.e., bound in the protein without access to the medium on either side of the membrane (shown in brackets)]. Free ions are labeled c and e for cytoplasmic and extracellular, respectively. The H+,K+-ATPase functions by a similar, but electroneutral, mechanism, with the 3 Na+ being replaced by 1 or 2 H+ and 1 or 2 K+ being countertransported.
The structural basis for the difference between Na+,K+-ATPase and H+,K+-ATPase with respect to stoichiometry, electrogenicity, and specific cation selectivity (H+ vs. Na+) is not well understood. Most of the residues involved in Na+ and K+ binding in the Na+,K+-ATPase are fully conserved in the H+,K+-ATPase (E329, Y773, T774, T776, E781, D810, Q925, and D928; rat α1 Na+,K+-ATPase numbering). One of the cation binding aspartates of transmembrane helix M6 of Na+,K+-ATPase (D806 in the rat α1 enzyme) is conserved in the nongastric H+,K+-ATPase and substituted by glutamate in gastric H+,K+-ATPase. A serine involved in Na+ and K+ binding in the Na+,K+-ATPase (S777 in rat α1 Na+,K+-ATPase), located in transmembrane helix M5, is nonconservatively replaced by a lysine in the H+,K+-ATPases (alignment is shown in Fig. 1). The occupation of site I by this lysine with its potential positive charge might account, in part, for the difference in transport stoichiometry between H+,K+-ATPase and Na+,K+-ATPase (7, 9), but consistent results for gain of electrogenicity upon mutation of this lysine in the gastric and nongastric H+,K+-ATPases have not been obtained (7, 10). It remains to be explained how the H+,K+-ATPases avoid binding Na+ (or H+) at the site corresponding to site III of Na+,K+-ATPase (4), given the conservation of most coordinating residues of site III (Y773, T776, D810, Q925, and D928).
Fig. 1.
Alignment of transmembrane segments M5-M6 and M8 of Na+,K+-ATPases and H+,K+-ATPases. Arrows pinpoint important residues differing between Na+,K+-ATPases and H+,K+-ATPases. Residue numbering corresponds to the rat α1 Na+,K+-ATPase.
Sequence alignment shows that a cysteine conserved in the α-subunit transmembrane helix M8 of all Na+,K+-ATPases is replaced by arginine in H+,K+-ATPases (Fig. 1). Insertion of arginine in place of the cysteine in the model of the Na+,K+-ATPase structure indicates that the positively charged guanidine+ group of the arginine side chain reaches into site III, where it might substitute for the bound Na+ (Fig. 2). Thus, it may be hypothesized that this arginine functions as an internal cation conferring electroneutrality of transport. In the present study, we have used biochemical assays and electrophysiology to examine this hypothesis by studying the functional consequences of replacing the M8 cysteine of Na+,K+-ATPase with arginine and several other substituents, including phenylalanine, which is present in a patient with alternating hemiplegia of childhood (11). Our results demonstrate that the arginine substitution makes the Na+,K+-ATPase electroneutral like the H+,K+-ATPase, while maintaining high Na+,K+-ATPase activity and K+ transport rate. This finding, in turn, suggests that the ability of the arginine of M8 to work as an internal cation substituent for Na+ and prevent Na+ binding at the third Na+-specific site is decisive for the electroneutral transport mode of H+,K+-ATPase.
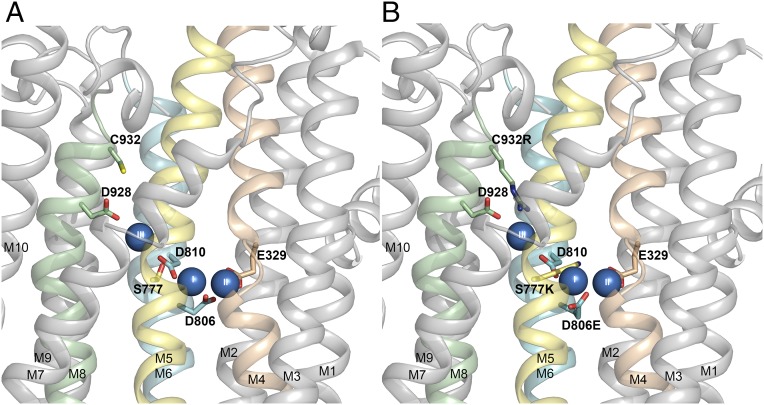
Fig. 2.
Structural features relevant to the present study. Side view of the transmembrane domain of the Na+,K+-ATPase structure with bound Na+ ions (blue spheres numbered I, II, and III), cytoplasmic side upward [Protein Data Bank (PDB) ID code 3WGV, protomer B (4)]. (A) Wild type with C932 and selected ion binding residues in stick representation. (B) Same structure as in A, but with C932, S777, and D806 replaced with the residues found in the gastric H+,K+-ATPase (alignment shown in Fig. 1).
Results
Mutants Studied.
The cysteine in transmembrane helix M8 of rat α1 Na+,K+-ATPase was replaced with the positively charged arginine, the residue present at the equivalent position in H+,K+-ATPase, as well as with the neutral residues alanine, leucine, and phenylalanine. Wild type and mutants were expressed in COS-1 cells for enzymatic measurements on isolated leaky membrane preparations and K+ uptake measurements in intact cells. The corresponding Xenopus α1 wild type and mutants were expressed together with β3 in Xenopus oocytes for electrophysiological measurements.
Functional Studies on Isolated Membranes.
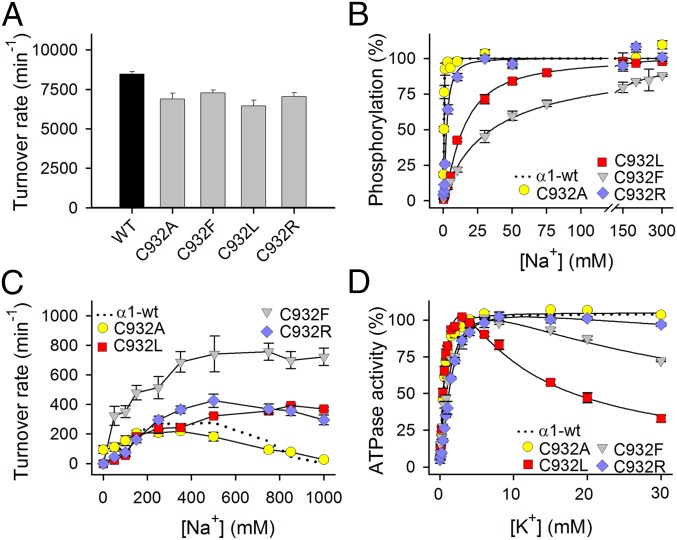
The Na+,K+-ATPase activity per molecule (turnover rate) was determined under conditions optimal for the wild type. For C932R, the turnover rate was 83% of the wild type, and for the other C932 mutants, it was likewise only slightly reduced (Fig. 3A and Table S1).
Fig. 3.
Functional properties of C932 mutants in isolated, permeabilized membranes. (A) Na+,K+-ATPase activity was determined at 37 °C in the presence of 30 mM histidine buffer (pH 7.4), 130 mM NaCl, 20 mM KCl, 3 mM ATP, 3 mM MgCl2, 1 mM EGTA, and 10 μM ouabain. The turnover rate was calculated as the ratio between the Na+,K+-ATPase activity and the active site concentration. (B) Phosphorylation was carried out for 10 s at 0 °C in 20 mM Tris (pH 7.5), 3 mM MgCl2, 1 mM EGTA, 2 μM [γ-32P]ATP, 10 μM ouabain, 20 μg of oligomycin/mL, and the indicated concentration of Na+ with various concentrations of N-methyl-d-glucamine to maintain the ionic strength. (C) Na+-ATPase activity was measured as described for A except for the absence of K+ and the presence of Na+, as indicated on the abscissa. The turnover rate was calculated as described for A. (D) Na+,K+-ATPase activity was measured as described for A except that the Na+ concentration was 40 mM and the K+ concentration was varied. Error bars indicate SE. Nonlinear regression was used to fit data (lines) in B and D (equations are provided in SI Methods). Wild-type data reproduced from the study by Holm et al. (14) are shown as dotted lines. Statistics are shown in Table S1.
Table S1.
Functional analysis of mutants
| Enzyme | Maximal enzyme turnover rate,* min−1 | K0.5 (Na+),† μM | K0.5 (K+),† μM | K0.5 (VO43−),† μM | K0.5 (ATP),† μM | Inhibition of Na+-ATPase at high Na+‡ |
| Wild type α1 | 8,474 ± 165 (n = 11) | 483 ± 18 (n = 8) | 624 ± 13 (n = 14) | 2.5 ± 0.1 (n = 23) | 432 ± 25 (n = 9) | ++ |
| C932A | 6,896 ± 354 (n = 6) | 476 ± 29 (n = 6) | 518 ± 19 (n = 3) | 2.5 ± 0.1 (n = 3) | 494 ± 40 (n = 3) | +++ |
| 1.0× | 1.2× ↓ | 1.0× | 1.1× ↑ | |||
| C932F | 7,281 ± 179 (n = 6) | 35,292 ± 3,498 (n = 7) | 996 ± 35 (n = 3) | 5.1 ± 0.2 (n = 5) | 220 ± 75 (n = 4) | − |
| 73.1× ↑ | 1.6× ↑ | 2.1× ↑ | 2.0× ↓ | |||
| C932L | 6,448 ± 383 (n = 6) | 14,339 ± 794 (n = 5) | 459 ± 7 (n = 3) | 3.7 ± 0.2 (n = 3) | 231 ± 26 (n = 3) | − |
| 29.7× ↑ | 1.4× ↓ | 1.5× ↑ | 1.9× ↓ | |||
| C932R | 7,031 ± 258 (n = 6) | 2,440 ± 212 (n = 6) | 1,310 ± 54 (n = 3) | 3.1 ± 0.1 (n = 5) | 189 ± 10 (n = 4) | + |
| 5.1× ↑ | 2.1× ↑ | 1.2× ↑ | 2.3× ↓ |
SEs and number of independent determinations (n) are indicated. Fold change (×) relative to wild type is indicated by number with arrow.
Data illustrated in Fig. 3A. The Na+,K+-ATPase activity was determined at 37 °C in the presence of 30 mM histidine buffer (pH 7.4), 130 mM NaCl, 3 mM ATP, 3 mM MgCl2, 1 mM EGTA, ouabain to inhibit the endogenous enzyme, and 20 mM KCl. The enzyme turnover rate was calculated as the ratio between the Na+,K+-ATPase activity and the active site concentration (maximum phosphorylation from [γ-32P]ATP measured at 0 °C in the presence of 150 mM NaCl and oligomycin).
Extracted from the data in Fig. 3 B and D and Fig. S2 A and B by nonlinear regression fitting procedures (equations are shown in SI Methods). The best fit shown as a line in the figures.
Semiquantitative interpretation of the data in Fig. 3C (inhibition at high Na+ indicated by +, and lack of inhibition indicated by −).
The interaction with Na+ at the cytoplasmic-facing transport sites was studied by determining the Na+ dependence of phosphorylation from [γ-32P]ATP (compare the reaction cycle in Fig. S1). C932F and C932L were very disruptive, reducing the apparent Na+ affinity (increasing K0.5 for Na+) as much as 73- and 30-fold, respectively, whereas the effect of the C932R substitution was much less pronounced (fivefold), and C932A was wild type-like (Fig. 3B and Table S1).
K+ activates dephosphorylation of the Na+,K+-ATPase by binding at sites I and II in their extracellular-facing configuration in E2P, thereby stimulating the ATPase activity. In the absence of K+, Na+ can substitute for K+ (also known as “Na+-ATPase activity”), although with much less efficiency (12) (∼40-fold lower turnover rate for wild type; compare Fig. 3 A and C). Under these conditions, Na+ binding at site III in its extracellular-facing “leaving” configuration leads to slowing of the pump cycle, which is manifested as inhibition of Na+-ATPase at high Na+ concentrations. At 1 M Na+, 100% inhibition was observed for C932A and wild-type pumps, whereas inhibition was abolished in C932F and C932L. For C932R, the activity was only reduced by 25% at 1 M Na+, indicating a considerable decrease of the affinity for inhibitory Na+ or of its efficacy (Fig. 3C and Table S1). It is also noteworthy that Na+-ATPase activity of C932F was markedly enhanced relative to wild type in the whole range of Na+ concentrations, suggesting that the structural perturbation by the phenylalanine increases the efficacy of activation of dephosphorylation by Na+ bound at site I and/or II, thus revealing interference with the normal K+ selectivity of these sites [compare previous observations of similar effects for certain other mutations perturbing the Na+ binding sites (13, 14)].
The K+ dependence of the Na+,K+-ATPase activity is shown in Fig. 3D. C932F and C932R displayed mild reduction of apparent K+ affinity for activation relative to wild type (only 1.6- and 2.1-fold, respectively; Table S1). In addition to the K+ activation phase, a distinct inhibition phase was observed at high K+ concentrations for C932L and C932F, most pronounced for C932L. Such an inhibition is believed to reflect K+ competition at one or more of the cytoplasmic-facing Na+ sites and indicates overall loss of the effective Na+ selectivity (14), which is in accordance with the reduced Na+ affinity of these mutants. C932A was wild type-like, and only a diminutive K+ inhibition was observed for C932R in accordance with the relatively minor reduction of Na+ affinity in C932R. The mutational effects on the E1-E2 distribution (the main conformational states of the enzyme that are Na+-selective and K+-selective, respectively) were determined in assays for ATP and vanadate binding (Fig. S2 and Table S1), which demonstrated that the low affinity for Na+ detected for some of the mutants is not an indirect effect of an E2 shift of the E1-E2 conformational equilibrium.
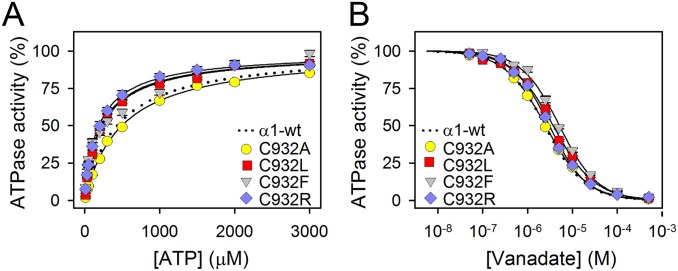
Fig. S2.
Apparent affinities for ATP and vanadate. (A) Na+,K+-ATPase activity was determined at 37 °C in the presence of 30 mM histidine buffer (pH 7.4), 130 mM NaCl, 20 mM KCl, 3 mM MgCl2, 1 mM EGTA, 10 μM ouabain, and the indicated concentrations of ATP. (B) Na+,K+-ATPase activity was determined under conditions as described for A, except that the ATP concentrations was 3 mM and the vanadate concentration was varied as indicated. For mutants, average values are shown, with error bars indicating SE (seen when larger than symbols). Nonlinear regression was used to fit data as shown by lines (equations are shown in SI Methods). Wild-type data were reproduced from the study by Holm et al. (14) and are shown as dotted lines. Statistics are shown in Table S1. The Na+,K+-ATPase inhibitor vanadate binds exclusively to the E2 form, whereas ATP binds with high affinity to the E1 form and only with low affinity to the E2 form. It is thus feasible to analyze the E1-E2 distribution by determining the ATP and vanadate concentration dependences of the Na+,K+-ATPase activity. C932A displayed wild-type–like apparent affinities for ATP and vanadate, indicating that the E1-E2 distribution is undisturbed. For C932R, C932F, and C932L the changes in affinities for ATP and vanadate indicated a slight shift of the E1-E2 distribution in favor of E1. Hence, the low affinity for Na+ detected for some of the mutants is not an indirect effect of an E2 shift of the E1-E2 conformational equilibrium.
Electrophysiological Studies of C932 Mutants in Intact Cells.
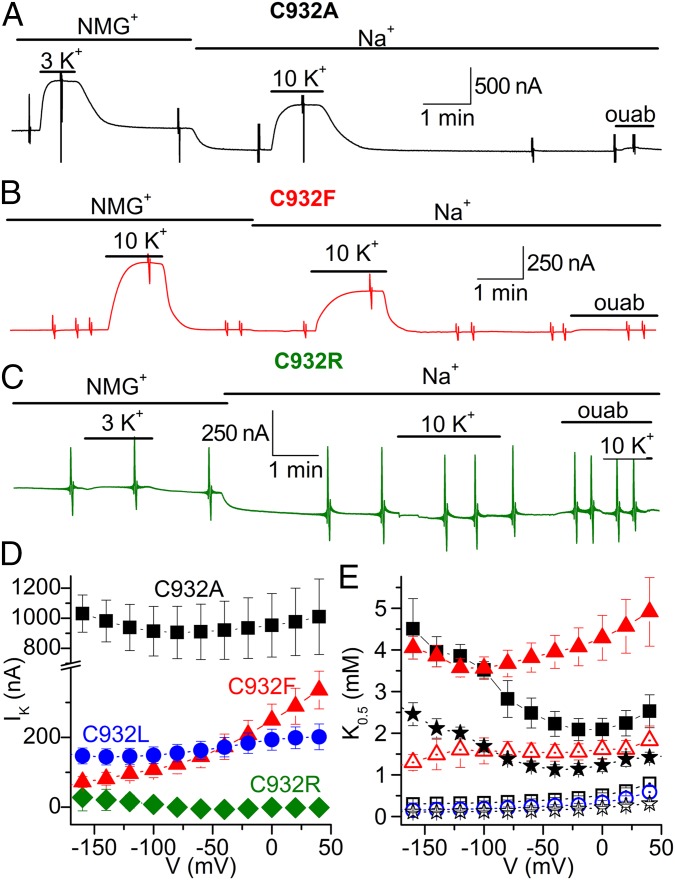
The pump currents were studied in Xenopus oocytes under voltage clamp. Fig. 4 A, B, and D shows that the mutants C932A (for simplicity, we maintain rat α1 numbering throughout the article), C932L, and C932F all displayed K+-induced outward pump current due to electrogenic transport, as is well known for wild-type Na+,K+-ATPase (15). In contrast, C932R lacked such current (Fig. 4 C and D). Taken together, the normal Na+,K+-ATPase activity (Fig. 3A) and the absence of outward current indicate that C932R performs electroneutral transport.
Fig. 4.
K+-induced pump currents in Na+,K+-ATPase C932 mutants. (A–C) Representative slow time-base recordings obtained at −50 mV in oocytes expressing C932 mutants. Application of K+ (3 or 10 mM) in the absence [presence of 125 mM N-methyl-d-glucamine (NMG+)] or presence of 125 mM external Na+ induced robust outward currents in C932A (A) and C932F (B), but failed to induce current in C932R (C). Larger values in A are due to larger protein expression, which is variable from oocyte to oocyte. The vertical deflections in the traces correspond to application of 100-ms-long square voltage pulses to obtain current/voltage relationships. ouab, ouabain. (D) Average maximal K+-induced (IK) current in NMG+ as a function of voltage. (E) Voltage dependence of the K0.5 for activation of current induced by external K+ in the presence of Na+ (solid symbols) or in the absence of Na+ (open symbols). For D and E, the symbols represent C932A (squares), C932F (triangles), C932L (circles), and C932R (diamonds). Data for wild type (stars) were previously reported [average of two independent datasets (16, 17)]. To obtain the maximal current and K0.5 values in D and E, the K+ concentration dependence of the K+-induced current was obtained at each voltage and fitted with a Hill equation (17). Error bars indicate SE (n = 5–12).
Fig. 4D illustrates the maximal pump current in the absence of external Na+. C932R showed lack of current for the whole voltage range studied. C932A and C932L showed current, but with little voltage dependence, as previously described for the wild type in the absence of external Na+ (15–17). However, the maximal pump current of C932F increased throughout the voltage range studied, possibly reflecting the extremely reduced affinity for cytoplasmic Na+, which allows a positive voltage to enhance the pump rate by supporting Na+ entry into the site.
Fig. 4E shows the voltage dependence of the apparent affinity for K+ determined by fitting the Hill equation to dose–response curves obtained for each voltage in the absence (C932A, C932L, C932F, and wild type) and presence (C932A, C932F, and wild type) of external Na+. With external Na+ present, the wild-type Na+,K+-pump shows a distinct U-shaped voltage dependence of the K0.5 for K+, which can be understood in terms of high-field access channels that the Na+ and K+ ions must transit to reach their binding sites from the extracellular side (15, 18, 19). The voltage dependence of C932A was wild type-like, but for C932F, the effect of negative voltage was much less pronounced, resulting in a shift of the position of the minimum toward more negative potentials in good accordance with the markedly reduced affinity of C932F for Na+ deduced from the biochemical data. Also in good agreement with the enzymatic assays, C932F exhibited a reduced K+ affinity relative to wild type.
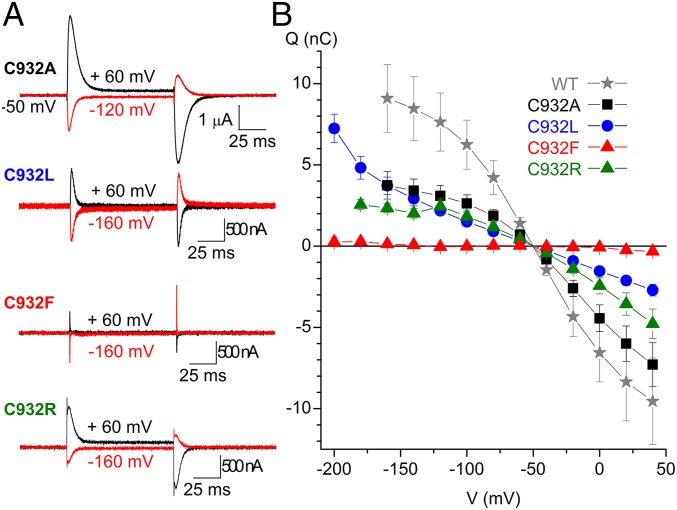
External Na+ binding by the mutants was further evaluated by measuring the transient charge movement in the presence of 125 mM Na+ (Fig. 5). In wild-type pumps, such current transients represent the voltage-dependent redistribution between [Na3]E1P and E2P intermediates (20) (see reaction intermediates in Fig. S1). Fig. 5A shows representative ouabain-sensitive currents elicited by pulses to different voltages from the holding potential (−50 mV). The integral of the transient currents observed when the voltage was returned to holding was plotted as a function of the voltage pulse to obtain the so-called charge-voltage (Q-V) curves shown in Fig. 5B and in the normalized form for the wild type, C932A, and C932R in Fig. 6A. The Q-V curve of C932A exhibited a wild-type–like slope with 29 mV more positive V0.5 than the wild type, corresponding to an approximately twofold increase of Na+ affinity (20, 21), which matches the biochemical data for the affinity for external Na+ (Fig. 3C). C932F lacked significant transients in the voltage range studied (down to −200 mV) in agreement with its very low Na+ affinity. The Q-V curve obtained for C932L was composite, with a shallower component and a second, low-affinity component that appears below −140 mV (impossible to study in greater detail because oocytes do not withstand voltages below −200 mV). This low-affinity component may reflect Na+ interaction with the Na+ exclusive site III exhibiting very low affinity for external Na+, although higher than the affinity of C932F, in accordance with the enzymatic studies. Surprisingly, C932R presented robust current transients, although corresponding to a Q-V curve with a shallower slope and with V0.5 shifted to a more positive voltage compared with the wild type and C932A (Figs. 5 and 6A). Hence, a partial reaction of C932R appears to be electrogenic, even though the overall pump cycle of this mutant is electroneutral. However, in contrast to the wild-type Na+,K+-ATPase and the C932A mutant, C932R showed only a slight difference (at very negative voltages) between the Q-V relations obtained in the presence and absence of external Na+ (Fig. 6B). Altogether, the large current transients observed with C932R provide clear evidence that this mutant is well expressed in the oocytes, thus corroborating that the lack of steady-state outward currents is due to electroneutral transport mode.
Fig. 5.
Transient currents in C932 mutants. (A) Ouabain-sensitive currents were determined in the presence of 125 mM external Na+. The current elicited by 100-ms-long voltage pulses in the presence of 10 mM ouabain was subtracted from the currents elicited by the same pulse protocol in the absence of inhibitor. (B) Q-V curves for WT (n = 10), C932A (n = 10), C932L (n = 6), C932F (n = 5), and C932R (n = 6). The charge plotted in B was obtained by integration of the current at the end of pulses for varying voltages as exemplified in A. Error bars indicate SE.
Fig. 6.
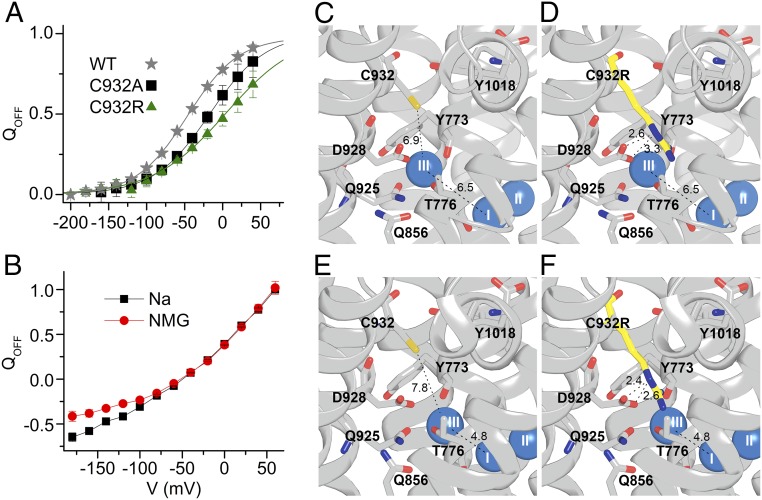
Binding of extracellular Na+ by C932R and structural interpretation. (A) Normalized Q-V relation for WT, C932A, and C932R (data in Fig. 5B). Data were fitted with the Boltzmann distribution (lines), giving parameters V0.5 (center of distribution) and k (slope factor) (17). Parameters from the best fit (in millivolts) were V0.5 = −46.4 ± 0.7, k = 33.8 ± 0.8 for WT; V0.5 = −17.0 ± 6.2, k = 36.6 ± 3.8 for C932A; and V0.5 = 4.3 ± 9.8, k = 48.2 ± 4.2 for C932R. QOFF, holding voltage. (B) Normalized Q-V for C932R measured in the presence and absence (NMG) of 125 mM external Na+. (C–F) Structures based on PDB ID code 3WGV, protomers A and B (chains C and A, respectively, in the PDB file), with C932 being replaced by arginine in D and F. Wild-type protomer B (C), C932R protomer B (D), wild-type protomer A (E), and C932R protomer A (F) are shown. Side views with the cytoplasmic side up are shown. The bound Na+ ions are shown as blue spheres. Broken lines with numbers indicate distances in angstroms.
K+ Transport and ATP Hydrolysis in Intact Cells.
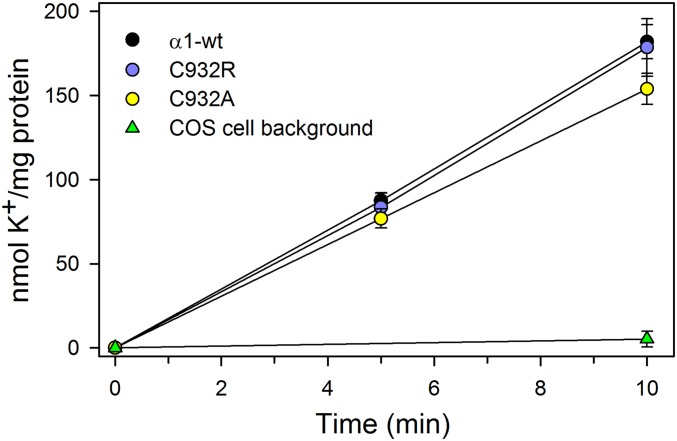
To determine whether the mutations affect the K+ transport steps of the Na+,K+-pump cycle, we measured active accumulation of K+ in COS-1 cells expressing C932R, C932A, or wild-type Na+,K+-ATPase, using 86Rb+ as a K+ congener (14, 22). The results obtained following correction for variance in protein expression show little or no difference in K+ uptake rates between the mutants and wild-type pumps (Fig. 7), further supporting electroneutral transport by the C932R mutant.
Fig. 7.
K+ transport into intact cells. Uptake of the K+ congener 86Rb+ at 37° C in COS-1 cells stably expressing either wild type (α1-wt) or mutants (C932R and C932A) was determined following incubation for the indicated time intervals. K+ uptake was calculated by multiplying the relative 86Rb+ uptake per milligram of protein by the K+ concentration in the uptake medium and is expressed per milligram of total protein normalized to represent identical expression levels of wild type and mutants. The nonspecific uptake obtained at 10 mM ouabain was subtracted from the data obtained at 5 μM ouabain (inhibiting the endogenous Na+,K+-ATPase). The indicated background corresponds to nontransfected COS-1 cells. Error bars indicate SE (n ≥ 3).
The Na+,K+-ATPase activity under the conditions used for measurements of K+ uptake in the intact cell system was estimated using the leaky plasma membrane Na+,K+-ATPase preparation in the presence of 130 mM K+ and 15 mM Na+. Under these cytoplasmic-like conditions, the cytoplasmic-facing Na+ sites are not saturated, whereas the extracellular-facing sites are saturated by K+. The turnover rates determined under these near-physiological conditions were only ∼20% of the maximal turnover rate in Table S1, but very similar among wild type and the two mutants: 1,559 ± 79 min−1 (n = 2) for wild type, 1,774 ± 81 min−1 (n = 3) for C932A, and 1,273 ± 166 min−1 (n = 3) for C932R. Hence, we conclude that C932R does not affect the ratio between K+ uptake and ATP hydrolysis.
Discussion
This study addresses the structural basis for the difference in electrogenicity between Na+,K+-ATPase and H+,K+-ATPase. Our results pinpoint the importance of an arginine in H+,K+-ATPase at the position corresponding to C932 (rat α1 numbering) in transmembrane helix M8 of Na+,K+-ATPase. We demonstrate unequivocally that the C932R mutation makes the Na+,K+-ATPase electroneutral, without affecting the ATPase turnover rate or the stoichiometry of K+ transport in intact cells. Hence, the C932R Na+,K+-ATPase mutant exports two Na+ ions and imports two K+ ions per ATP hydrolyzed instead of the normal three Na+ ions and two K+ ions. The presence of arginine at this position in both H+,K+-ATPase isoforms thereby explains the electroneutrality of these pumps.
According to the crystal structure of the Na+,K+-ATPase with three Na+ ions bound (4), Na+ site III is formed by side-chain oxygen ligands of T776, S777, Q925, and D928, as well as the π-electrons of the aromatic side chain and the backbone oxygen of Y773. C932 is located one α-helical turn closer to the intracellular side than D928 in M8 (Fig. 2A), but the side-chain sulfhydryl group of C932 is not important for Na+ or K+ binding, because replacement of C932 with alanine did not lower the apparent affinities for the ions (Table S1). On the other hand, replacement of C932 with the bulky hydrophobic residues phenylalanine and leucine resulted in a dramatic reduction of the apparent Na+ affinity from both sides of the membrane, and the phenylalanine, but not the leucine, disturbed K+ binding slightly. Hence, in C932L, Na+ sites I and II, which alternately accommodate Na+ and K+, are largely undisturbed, whereas Na+ site III is disturbed, which can be explained by collision of the leucine side chain with Y773 and D928 (Fig. S3A). The phenylalanine side chain is more bulky with its aromatic ring (Fig. S3B), explaining why its impact reaches to site I, thus affecting K+ binding. Recently, the homologous replacement of C927 of the α3-isoform of Na+,K+-ATPase with phenylalanine, tyrosine, or tryptophan was identified in patients with the neurological disease alternating hemiplegia of childhood (11). The very low Na+ affinity likely results in a rise of the intracellular Na+ concentration that contributes to the pathophysiology (22).
Fig. S3.
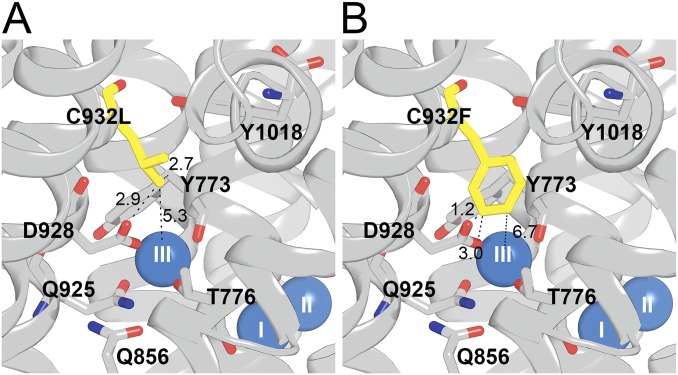
Structural features of mutants C932L (A) and C932F (B). Side views with cytoplasmic side up are shown, based on PDB ID code 3WGV [protomer B (chain A in the PDB file)], with C932 being replaced. Selected residues (rat α1 numbering) are shown as sticks and colored according to the elements (carbon, gray; oxygen, red; nitrogen, blue), except that the substituent is highlighted by the yellow color of carbon atoms. The bound Na+ ions are shown as blue spheres. Broken lines with numbers indicate distances in angstroms.
In contrast to the conspicuous effects of phenylalanine and leucine, we found only a relatively modest, fivefold reduction of Na+ affinity for C932R, relative to the wild type, despite the fact that the side chain of the arginine has a larger volume, and thus would be expected to be at least as perturbing as the side chain of the phenylalanine. It should be noted that the K0.5 determined for Na+ is an apparent affinity for Na+ activation of phosphorylation, which requires binding of Na+ at all three sites in the wild type. Thus, the K0.5 for Na+ depends on the properties of all three Na+ sites and their cooperative interaction. Because the arginine side chain reaches into site III in the C932R mutant (Figs. 2B and 6D), the most reasonable explanation of the relatively small perturbation of the apparent Na+ affinity seen for C932R is that the positively charged arginine guanidine+ group, similar in size and chemistry to partially hydrated Na+ (23), can act like Na+ at site III. If Na+ were absent from site III, the partially charged nitrogen atoms of the arginine side chain in C932R would interact in place of Na+ with the site III liganding residues D928, T776, and Y773. Guanidinium derivatives have, in fact, been shown to be high-affinity antagonists of Na+ ions in the occlusion sites of Na+,K+-ATPase (24), and on the basis of electrophysiological studies, guanidinium+ ions were proposed to interact specifically at Na+ site III (16, 17), which is consistent with biochemical trypsinolysis studies (25). Thus, in accordance with the illustrations in Figs. 2B and 6D, the arginine guanidine+ group seems structurally capable of replacing Na+ at site III as an internal cation without preventing occupation of the other two sites by Na+.
The sequence of cytoplasmic Na+ binding at the three sites of the Na+,K+-ATPase is unsettled (4, 26). The present data are consistent with the crystal structure-based proposal that the Na+ ion at site III is the first ion to bind, followed by occupation of sites I and II, respectively, in a sequential and cooperative binding mechanism (4). According to this hypothesis, the guanidine+ group of the arginine at site III in the C932R mutant would replace the binding of the first Na+ ion in the sequence, preparing the Na+,K+-ATPase for the consecutive binding of Na+ at sites I and II, thereby accomplishing the reaction cycle with only two Na+ ions being translocated for each ATP hydrolyzed. The relatively minor reduction of the apparent affinity of sites I/II for Na+ in C932R could be due to interference with the cooperative interaction between these sites and site III and/or to the tethered cation forcing an order of binding of cytoplasmic Na+ different from the normally preferred order. In any case, our data are fully consistent with phosphorylation being controlled by binding of the last Na+ to a site different from site III, in agreement with the evolutionary attractive hypothesis proposing that site II, the only transport site conserved in all P-type ATPases, is the last to be occupied from the cytoplasm, triggering the phosphorylation reaction in all P-type ATPases (4, 14).
Although the overall pump cycle of C932R was electroneutral, the voltage-dependent transient current exhibited by C932R indicated that at least one partial reaction is electrogenic. Because the Q-V relationship of C932R was only mildly sensitive to the presence of external Na+ (at very negative voltages corresponding to strongly reduced affinity; Fig. 6B), the major voltage-dependent reaction step of this mutant is a conformational change rather than a Na+ binding/dissociation step, probably the E1P-to-E2P transition (10, 27, 28). The low affinity of C932R for external Na+ is consistent with the slight inhibition seen in the Na+-ATPase assay at very high Na+ concentrations (Fig. 3C), thus reflecting the binding of Na+ to site III of the C932R mutant from the extracellular side, albeit with much reduced affinity. The exact structural basis for this Na+ binding cannot be established in the absence of crystal structure(s) of the Na+-bound intermediate(s) following [Na3]E1P in the enzyme cycle, but it is worth noting that the position of the Na+ ion at site III deviates slightly between the two protomers in the available crystal structure (4), as seen by comparing Fig. 6 C and D with Fig. 6 E and F. In protomer A, the Na+ ion at site III is 1.7 Å closer to the Na+ ion in site I (and thus to transmembrane segment M5) compared with protomer B, and the inserted arginine approaches D928 more closely in protomer A than in protomer B. It seems likely that the two protomers represent different stages of the dynamic translocation process, and the presence of the arginine excludes Na+ binding at site III less efficiently in the state represented by protomer A than in the state represented by protomer B. The overlap between the arginine side chain and Na+ might be reduced even more in the intermediates following [Na3]E1P in the process toward extracellular release of Na+. Thus, it is reasonable to expect that these intermediates are able to bind Na+ at site III from the extracellular side, albeit with reduced affinity.
A mechanism where arginine works as an internal cation replacing Na+ has previously been revealed for a carnitine transporter (CaiT), which is closely related to Na+-coupled substrate symporters, but is Na+ independent, because an arginine undergoing conformational changes mimics Na+ binding and unbinding (29). An equivalent observation is the ability of negatively charged amino acid residues to substitute for Cl− in neurotransmitter, Na+,Cl−-cotransporters to make these transporters independent of Cl− (30). Hence, the present finding adds an example of the general use of charged amino acid side chains to substitute for transported ions, when a change in transport stoichiometry is advantageous.
The lysine of transmembrane helix M5 replacing S777 of Na+,K+-ATPase in the H+,K+-ATPases has previously been attributed a role as an internal cation similar to the present proposal for the arginine of M8 (7, 9). In favor of this function of the M5 lysine is the finding that alanine substitution confers electrogenicity to the nongastric H+,K+-ATPase, thus raising the number of cations transported toward the extracellular side per cycle (7). Although a similar gain of electrogenicity was not observed for the corresponding mutant of gastric H+,K+-ATPase (10), the conformational change from E1P to E2P was proposed to result in the formation of a salt bridge between the M5 lysine and the M6 glutamate in the gastric H+,K+-ATPase, thereby expelling the bound H+ toward the extracellular side (9, 10). In combination with the present results, the previous findings with the M5 lysine suggest that both the lysine and the M8 arginine contribute internal cations that, together, ensure the electroneutrality and ability of H+,K+-ATPases to work against a steep concentration gradient.
Methods
For biochemical analysis, mutations were introduced into cDNA encoding the ouabain-resistant rat α1-isoform Na+,K+-ATPase. Mutants and wild type were expressed in COS-1 cells under ouabain selection pressure (13, 22). Studies of enzyme function were performed on isolated plasma membranes made leaky to allow access of incubation media from both sides of the membrane. Ouabain was present to inhibit the endogenous Na+,K+-ATPase. ATPase activity was determined at 37 °C by following the liberation of Pi (14). The Na+ dependence of phosphorylation from [γ-32P]ATP was determined at 0 °C in the presence of oligomycin to stabilize the phosphoenzyme and was quantified by phosphor imaging of bands separated by SDS/PAGE (14). The active site concentration was determined as the maximum phosphorylation from [γ-32P]ATP measured in the presence of 150 mM NaCl and oligomycin. Cellular K+ transport at 37 °C was determined using the K+ congener 86Rb+ (22), and the uptake data were normalized to represent identical expression levels as calculated from the active site concentration.
For electrophysiological studies, mutations were introduced into an ouabain-resistant version (Q120R/N131D mutant) of Xenopus α1 Na+,K+-ATPase. Oocytes were injected with the cRNA and loaded with Na+ 1–2 h before two electrode voltage-clamp recordings in external solution with and without 125 mM Na+ (31). Further details are provided in SI Methods.
SI Methods
Biochemical Studies.
Mutations were introduced by PCR into cDNA encoding the rat α1-isoform, which is ouabain-resistant. Mutants and wild type were expressed in COS-1 cells in the presence of 10 μM ouabain (13, 14). The cDNA was sequenced full-length before transfection as well as following isolation of stable transfectants, where it had become integrated in the genome. Functional analysis was carried out on the plasma membrane fraction of the cells made leaky by use of either sodium deoxycholate or alamethicin to allow access of ions and ATP from both sides of the membrane.
ATPase activity was determined by following the liberation of Pi within the linear time range (up to 20 min) using a colorimetric method (13, 14). The ATPase activity associated with the expressed exogenous enzyme was calculated by subtracting the ATPase activity measured at 10 mM ouabain (inhibiting all ATPase activity) from the ATPase activity measured at 10 μM ouabain (inhibiting the endogenous COS-1–cell Na+,K+-ATPase).
To determine the K+ transport activity, COS-1 cells were grown to confluence in culture medium containing 5 μM ouabain, and were incubated for 5 or 10 min at 37 °C and 5% (vol/vol) CO2 in serum-free medium containing 154 mM Na+ and 5.3 mM K+, to which had been added 86Rb+ (0.5 μCi/mL), and 5 μM ouabain (to inhibit the endogenous Na+,K+-ATPase) or 10 mM ouabain (to inhibit all Na+,K+-ATPase). Uptake of 86Rb+ was terminated by washing the cells with an ice-cold stop solution. Following incubation in 0.2 M NaOH for 2 h at 4 °C, the cells were scraped off, and the amount of 86Rb+ in the suspension was determined by liquid scintillation counting. K+ uptake was calculated by multiplying the relative 86Rb+ uptake per milligram of protein by the K+ concentration in the uptake medium (14, 22).
For determination of phosphorylation, the enzyme was mixed with [γ-32P]ATP in the presence of the indicated Na+ concentration at 0 °C, followed by acid quenching 10 s later. The precipitated 32P-labeled phosphoenzyme was washed twice by centrifugation, and SDS/PAGE was performed under acid conditions (pH 6.0), ensuring stability of the acyl/phosphoryl bond during electrophoresis (13, 14). The radioactivity associated with the separated Na+,K+-ATPase band was quantified by phosphor imaging using a cyclone storage phosphor system (PerkinElmer Life Sciences). To inhibit the endogenous COS-1–cell Na+,K+-ATPase, the membranes were preincubated with 10 μM ouabain, which was present also during the phosphorylation reaction. The level of background phosphorylation was determined in a reaction mix containing a high concentration of K+ (50 mM) instead of Na+, because K+ prevents phosphorylation by reversing the reaction cycle, and the K+ background (only a tiny fraction of the total 32P-incorporation) was subtracted before calculation of the phosphorylation level.
Data points were fitted by nonlinear regression using the SigmaPlot program (SPSS, Inc.), and the best fits are shown as lines in the figures. For fitting of the Na+ dependence of phosphorylation and ATP dependence of ATPase activity, the Hill equation for binding of an activating ligand (L) was used:
Amax is the maximum level of ATPase activity or phosphorylation corresponding to infinite ligand concentration. For K+ dependence of ATPase activity, a second Hill term was added to represent inhibition. For fitting of the vanadate dependence of ATPase activity, the Hill equation modified to describe binding of an inhibiting ligand was used:
Electrophysiological Studies.
For electrophysiological experiments, the Xenopus α1 mutants and β3 subunits subcloned in the pSD5 vector were linearized with Bgl II and transcribed with an SP6 mMessage mMachine kit (Ambion). Mutations were introduced into a template corresponding to wild-type Xenopus α1 made ouabain-resistant by introduction of the mutations Q120R/N131D. At 3 to 6 d after cRNA injection (α/β 1:1 molar ratio), oocytes were incubated for 1–2 h in a Na+-loading solution containing 150 mM Hepes, 20 mM tetraethylammonium Cl, and 0.2 mM EGTA (pH 7.2 with NaOH, osmolarity of 240 mOsm/kg) supplemented with 1 μM ouabain to inhibit endogenous pumps (31). They were kept until recording in Na+ external solution. External solutions were composed of 120 mM NaOH (or 120 mM N-methyl-d-glucamine), 10 mM Hepes, 5 mM Ba(OH)2, 1 mM Mg(OH)2, and 0.5 mM Ca(OH)2 (pH 7.6, with methanesulfonic acid).
Two-electrode voltage-clamp recording was performed with an OC-725C amplifier (Warner Instruments) and with a Digidata 1440 A/D board, a Minidigi 1A, and pClamp 10 software (all from Molecular Devices). Signals were acquired at 10 kHz and filtered at 2 kHz. Slow time-base recording with the Minidigi 1A were performed at 1 KHz, offline-filtered (10 Hz), and decimated for display. Current and voltage electrodes filled with 3 M KCl had resistances between 0.4 and 1 MΩ. Agar bridges connected the AgCl pellet bath electrodes to the Cl-free external solutions. The curves for individual cells were normalized to the total charge from the Boltzmann fit (Fig. 6A) or the charge moved by the pulse at +60 mV in the presence of Na+ (Fig. 6B).
Error Bars in Data Figures.
In all data figures, average values are shown, with error bars indicating SE (seen only when larger than the size of the symbols).
Structural Figures.
Structural figures were made using PyMOL software. For mutants, the most probable rotamer of the introduced substituted side chain is shown based on energy minimization of the side-chain orientation with PyMOL, keeping Na+ III in the structure to illustrate the overlap with the arginine guanidine+ group despite steric clashes of the arginine side chain with Na+ III and M5 residues.
Acknowledgments
We thank Kirsten Lykke Pedersen, Janne Petersen, Randi Scheel, Nina Juste, Adam Bernal, and Sukanyalakshmi Chebrolu for expert technical assistance. This work was supported, in part, by grants from the Danish Medical Research Council (to B.V.), the Novo Nordisk Foundation (to B.V.), the Riisfort Foundation (to B.V.), the Lundbeck Foundation (to B.V. and A.P.E.), the Foundation for the Advancement of Medical Science (to B.V.), and the MEMBRANES Center, Aarhus University (to B.V.), as well as by Grants MCB-1515434 from NSF and R15-NS081570-01 from the NIH (to P.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617951114/-/DCSupplemental.
References
- 1.Morth JP, et al. Crystal structure of the sodium-potassium pump. Nature. 2007;450(7172):1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 2.Abe K, Tani K, Nishizawa T, Fujiyoshi Y. Inter-subunit interaction of gastric H+,K+-ATPase prevents reverse reaction of the transport cycle. EMBO J. 2009;28(11):1637–1643. doi: 10.1038/emboj.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glynn IM. Annual review prize lecture. ‘All hands to the sodium pump’. J Physiol. 1993;462(1):1–30. doi: 10.1113/jphysiol.1993.sp019540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanai R, Ogawa H, Vilsen B, Cornelius F, Toyoshima C. Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state. Nature. 2013;502(7470):201–206. doi: 10.1038/nature12578. [DOI] [PubMed] [Google Scholar]
- 5.Shinoda T, Ogawa H, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 A resolution. Nature. 2009;459(7245):446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- 6.Shin JM, Munson K, Vagin O, Sachs G. The gastric HK-ATPase: Structure, function, and inhibition. Pflugers Arch. 2009;457(3):609–622. doi: 10.1007/s00424-008-0495-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnay M, Crambert G, Kharoubi-Hess S, Geering K, Horisberger JD. Electrogenicity of Na,K- and H,K-ATPase activity and presence of a positively charged amino acid in the fifth transmembrane segment. J Biol Chem. 2003;278(21):19237–19244. doi: 10.1074/jbc.M300946200. [DOI] [PubMed] [Google Scholar]
- 8.Abe K, Tani K, Friedrich T, Fujiyoshi Y. Cryo-EM structure of gastric H+,K+-ATPase with a single occupied cation-binding site. Proc Natl Acad Sci USA. 2012;109(45):18401–18406. doi: 10.1073/pnas.1212294109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koenderink JB, Swarts HG, Willems PH, Krieger E, De Pont JJ. A conformation-specific interhelical salt bridge in the K+ binding site of gastric H,K-ATPase. J Biol Chem. 2004;279(16):16417–16424. doi: 10.1074/jbc.M400020200. [DOI] [PubMed] [Google Scholar]
- 10.Dürr KL, Seuffert I, Friedrich T. Deceleration of the E1P-E2P transition and ion transport by mutation of potentially salt bridge-forming residues Lys-791 and Glu-820 in gastric H+/K+-ATPase. J Biol Chem. 2010;285(50):39366–39379. doi: 10.1074/jbc.M110.133470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panagiotakaki E, et al. Italian IBAHC Consortium French AHC Consortium International AHC Consortium Clinical profile of patients with ATP1A3 mutations in Alternating Hemiplegia of Childhood-a study of 155 patients. Orphanet J Rare Dis. 2015;10:123. doi: 10.1186/s13023-015-0335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan JH, Hollis RJ. External Na dependence of ouabain-sensitive ATP:ADP exchange initiated by photolysis of intracellular caged-ATP in human red cell ghosts. Nature. 1980;288(5791):587–589. doi: 10.1038/288587a0. [DOI] [PubMed] [Google Scholar]
- 13.Toustrup-Jensen M, Vilsen B. Importance of Glu282 in transmembrane segment M3 of the Na+,K+-ATPase for control of cation interaction and conformational changes. J Biol Chem. 2002;277(41):38607–38617. doi: 10.1074/jbc.M203665200. [DOI] [PubMed] [Google Scholar]
- 14.Holm R, Einholm AP, Andersen JP, Vilsen B. Rescue of Na+ affinity in aspartate 928 mutants of Na+,K+-ATPase by secondary mutation of glutamate 314. J Biol Chem. 2015;290(15):9801–9811. doi: 10.1074/jbc.M114.625509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rakowski RF, Vasilets LA, LaTona J, Schwarz W. A negative slope in the current-voltage relationship of the Na+/K+ pump in Xenopus oocytes produced by reduction of external [K+] J Membr Biol. 1991;121(2):177–187. doi: 10.1007/BF01870531. [DOI] [PubMed] [Google Scholar]
- 16.Ratheal IM, et al. Selectivity of externally facing ion-binding sites in the Na/K pump to alkali metals and organic cations. Proc Natl Acad Sci USA. 2010;107(43):18718–18723. doi: 10.1073/pnas.1004214107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yaragatupalli S, Olivera JF, Gatto C, Artigas P. Altered Na+ transport after an intracellular alpha-subunit deletion reveals strict external sequential release of Na+ from the Na/K pump. Proc Natl Acad Sci USA. 2009;106(36):15507–15512. doi: 10.1073/pnas.0903752106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rakowski RF. Charge movement by the Na/K pump in Xenopus oocytes. J Gen Physiol. 1993;101(1):117–144. doi: 10.1085/jgp.101.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sagar A, Rakowski RF. Access channel model for the voltage dependence of the forward-running Na+/K+ pump. J Gen Physiol. 1994;103(5):869–893. doi: 10.1085/jgp.103.5.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmgren M, Rakowski RF. Charge translocation by the Na+/K+ pump under Na+/Na+ exchange conditions: Intracellular Na+ dependence. Biophys J. 2006;90(5):1607–1616. doi: 10.1529/biophysj.105.072942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Capendeguy O, Geering K, Horisberger JD. A third Na+-binding site in the sodium pump. Proc Natl Acad Sci USA. 2005;102(36):12706–12711. doi: 10.1073/pnas.0505980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toustrup-Jensen MS, et al. Relationship between intracellular Na+ concentration and reduced Na+ affinity in Na+,K+-ATPase mutants causing neurological disease. J Biol Chem. 2014;289(6):3186–3197. doi: 10.1074/jbc.M113.543272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hille B. The permeability of the sodium channel to organic cations in myelinated nerve. J Gen Physiol. 1971;58(6):599–619. doi: 10.1085/jgp.58.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.David P, Mayan H, Cohen H, Tal DM, Karlish SJD. Guanidinium derivatives act as high affinity antagonists of Na+ ions in occlusion sites of Na+,K+-ATPase. J Biol Chem. 1992;267(2):1141–1149. [PubMed] [Google Scholar]
- 25.Mahmmoud YA, Shattock M, Cornelius F, Pavlovic D. Inhibition of K+ transport through Na+,K+-ATPase by capsazepine: Role of membrane span 10 of the α-subunit in the modulation of ion gating. PLoS One. 2014;9(5):e96909. doi: 10.1371/journal.pone.0096909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneeberger A, Apell H-J. Ion selectivity of the cytoplasmic binding sites of the Na,K-ATPase: II. Competition of various cations. J Membr Biol. 2001;179(3):263–273. doi: 10.1007/s002320010051. [DOI] [PubMed] [Google Scholar]
- 27.Goldshlegger R, Karlish SJ, Rephaeli A, Stein WD. The effect of membrane potential on the mammalian sodium-potassium pump reconstituted into phospholipid vesicles. J Physiol. 1987;387:331–355. doi: 10.1113/jphysiol.1987.sp016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stengelin M, Fendler K, Bamberg E. Kinetics of transient pump currents generated by the (H,K)-ATPase after an ATP concentration jump. J Membr Biol. 1993;132(3):211–227. doi: 10.1007/BF00235739. [DOI] [PubMed] [Google Scholar]
- 29.Kalayil S, Schulze S, Kühlbrandt W. Arginine oscillation explains Na+ independence in the substrate/product antiporter CaiT. Proc Natl Acad Sci USA. 2013;110(43):17296–17301. doi: 10.1073/pnas.1309071110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zomot E, et al. Mechanism of chloride interaction with neurotransmitter:sodium symporters. Nature. 2007;449(7163):726–730. doi: 10.1038/nature06133. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell TJ, Zugarramurdi C, Olivera JF, Gatto C, Artigas P. Sodium and proton effects on inward proton transport through Na/K pumps. Biophys J. 2014;106(12):2555–2565. doi: 10.1016/j.bpj.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]