Identification and characterization of a putative Arabidopsis sphingolipid glycosyltranferase suggests that glycosylation of these crucial lipids affects cellulose content and plant defense signaling.
Abstract
Glycosylinositol phosphorylceramides (GIPCs) are a class of glycosylated sphingolipids found in plants, fungi, and protozoa. These lipids are abundant in the plant plasma membrane, forming ∼25% of total plasma membrane lipids. Little is known about the function of the glycosylated headgroup, but two recent studies have indicated that they play a key role in plant signaling and defense. Here, we show that a member of glycosyltransferase family 64, previously named ECTOPICALLY PARTING CELLS1, is likely a Golgi-localized GIPC-specific mannosyl-transferase, which we renamed GIPC MANNOSYL-TRANSFERASE1 (GMT1). Sphingolipid analysis revealed that the Arabidopsis thaliana gmt1 mutant almost completely lacks mannose-carrying GIPCs. Heterologous expression of GMT1 in Saccharomyces cerevisiae and tobacco (Nicotiana tabacum) cv Bright Yellow 2 resulted in the production of non-native mannosylated GIPCs. gmt1 displays a severe dwarfed phenotype and a constitutive hypersensitive response characterized by elevated salicylic acid and hydrogen peroxide levels, similar to that we previously reported for the Golgi-localized, GIPC-specific, GDP-Man transporter GONST1 (Mortimer et al., 2013). Unexpectedly, we show that gmt1 cell walls have a reduction in cellulose content, although other matrix polysaccharides are unchanged.
INTRODUCTION
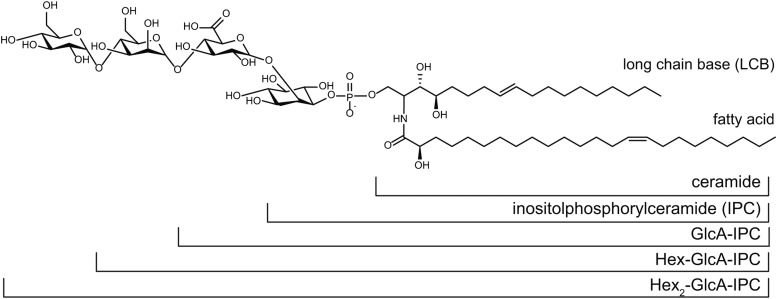
Glycosylinositol phosphorylceramides (GIPCs; Figure 1) and glucosylceramides are the major sphingolipid classes in the plasma membrane of plant cells. GIPCs have been estimated to make up 64% of total sphingolipids and, therefore, ∼25% of the plasma membrane lipids in the Arabidopsis thaliana leaf (Markham and Jaworski, 2007). Despite their abundance, the functions of GIPCs in plants are not well understood, although they have been proposed to participate in many important processes, including symbiosis (Perotto et al., 1995), pollen development (Rennie et al., 2014), defense against pathogens (Mortimer et al., 2013; Tartaglio et al., 2016), and membrane organization and trafficking (Mongrand et al., 2004; Cacas et al., 2016).
Figure 1.
Example GIPC Structure, Including the Nomenclature Used in This Work.
The most abundant GIPC in Arabidopsis callus, t18:1/h24:1, is shown, with the various sugar headgroup structures described in this article. It should be noted that although the final hexose is shown here as α-1,4-glucose, its identity is as yet unknown. The LCB nomenclature of t18:1 indicates that it is a trihydoxyl LCB (i.e., it has three hydroxyl groups), a single double bond, and 18 carbon atoms. An LCB with only two hydroxyl groups (dihydroxy LCB) would be referred to as d18:1.
GIPCs are found only in plants, fungi, and some protozoa (Sperling and Heinz, 2003; Sperling et al., 2005). The enzymes involved in GIPC biosynthesis have been well studied in the yeast Saccharomyces cerevisiae (Zäuner et al., 2010; Markham et al., 2013). The GIPC ceramide backbone is synthesized in the endoplasmic reticulum, starting with the condensation of serine and palmitoyl-CoA by serine palmitoyltransferase to produce 3-ketosphinganine. 3-Ketosphinganine is reduced to the long-chain base (LCB) sphinganine (d18:0) by 3-ketosphinganine reductase. The sphinganine can be used directly or modified by hydroxylation or desaturation for ceramide biosynthesis. The ceramide synthases in Arabidopsis have different substrate preferences. Dihydroxyl LCBs are primarily acylated by ceramide synthase I (CSI) for glucosylceramide biosynthesis. Trihydroxyl LCBs are primarily acylated with very-long-chain fatty acyl-CoAs by ceramide synthase II for both glucosylceramide and GIPC biosynthesis (Markham et al., 2013). For glucosylceramide biosynthesis, the glucosylation occurs in the endoplasmic reticulum and is catalyzed by glucosylceramide synthase (Hillig et al., 2003). For GIPC biosynthesis, the ceramide is trafficked to the Golgi, where a phosphorylinositol headgroup is added to form an inositol phosphorylceramide (IPC) by IPC synthase (Wang et al., 2008). The IPC core can then be glycosylated by various glycosyltransferases (GTs) to produce mature GIPCs. The biosynthesis of IPCs is well conserved between plants and fungi (Dunn et al., 2004). However, plant GIPCs are more highly glycosylated than those in S. cerevisiae, usually containing α-glucuronic acid (GlcA) linked to the IPC core structure, to which additional sugar units such as glucosamine (GlcN), N-acetyl-glucosamine (GlcNAc), mannose (Man), and arabinose (Ara) may be attached (Markham et al., 2006, 2013; Buré et al., 2011). In most Arabidopsis tissues, the major GIPC glycosylation consists of one hexose (Hex) attached to GlcA linked to IPC (Hex-GlcA-IPC), while in seeds, the dominant GIPC contains an amino sugar linked to the GlcA (Tellier et al., 2014). A recent report found that Arabidopsis pollen GIPCs also have a different glycosylation pattern, consisting of a complex array of N-acetyl-glycosylated GIPCs, including species with up to three pentose units (Luttgeharm et al., 2015). Other plant tissues, such as tobacco (Nicotiana tabacum) leaves, do not contain the Hex-GlcA-IPC found in Arabidopsis but instead the dominant structure consists of GlcNAc attached to the GlcA linked to IPC (Carter et al., 1958; Kaul and Lester, 1975; Hsieh et al., 1981; Buré et al., 2011). The significance of the glycan structural variation is not clear.
The site of GIPC glycosylation is the Golgi apparatus. The Golgi contains a large and diverse group of membrane-bound GTs responsible for the biosynthesis of noncellulosic cell wall polysaccharides and for the glycosylation of glycoproteins and GIPCs. It is not yet clear how this myriad of glycosylation reactions is controlled, and various hypotheses including spatio/temporal separation and substrate channeling have been proposed (Moore et al., 1991; Seifert et al., 2002; Mortimer et al., 2013; Oikawa et al., 2013). This complexity, combined with the notorious difficulty of predicting GT activity based on amino acid sequence and the lack of corresponding GTs in yeast, has meant that the use of bioinformatic approaches to predict the identity of GIPC GTs has been limited.
Recent work has identified the first two genes involved in plant GIPC glycosylation and demonstrated the importance of the glycan headgroup for plant growth and development (Mortimer et al., 2013; Rennie et al., 2014). INOSITOL PHOSPHORYLCERAMIDE GLUCURONOSYLTRANSFERASE1 (IPUT1) is responsible for transferring GlcA from UDP-GlcA to IPC (Rennie et al., 2014). Mutations in IPUT1 are not transmitted through the pollen, indicating that GIPCs are critical to pollen function. GOLGI LOCALIZED NUCLEOTIDE SUGAR TRANSPORTER1 (GONST1) is a GDP-sugar transporter that appears to specifically transport GDP-Man into the Golgi for use in GIPC glycosylation (Mortimer et al., 2013). gonst1 Arabidopsis plants have a dwarfed phenotype and a constitutive hypersensitive response, with elevated salicylic acid and hydrogen peroxide levels. gonst1 GIPCs lack mannosylation, and the dominant GIPC headgroup structure is GlcA-IPC. Previously it had been proposed that disruption to the free ceramide pool induces defense responses and programmed cell death (Wang et al., 2008), but the sphingolipid profile was unchanged in gonst1, suggesting that GIPC misglycosylation itself can trigger this phenotype. This, and other studies, have revealed that increased free-ceramide, increased salicylic acid, and cell death may not be directly linked, but, as has been discussed (Markham et al., 2013), there may be other factors involved. In order to investigate this further, it is important that additional members of the GIPC biosynthetic pathway are identified. Since there is a GIPC-specific GDP-Man transporter, an unidentified Golgi-localized GT is therefore required to transfer the Man onto the GlcA-IPC product.
Previously, a mutant in a member of CAZy GT family 64 (Lombard et al., 2014; At3g55830) named ectopically parting cells1 (epc1-1; later referred to in this report as gmt1-1 [gipc mannosyl-transferase 1-1]) was reported to have reduced cell-cell adhesion in hypocotyl and cotyledon tissues (Singh et al., 2005). The epc1-1 (gmt1-1) plants were also shown to have a severe phenotype characterized by reduced leaf expansion and defects in vascular development. A second publication described a new allele, epc1-2 (gmt1-2), which displayed similar growth phenotypes to epc1-1 (gmt1-1), but no cell adhesion defects were observed (Bown et al., 2007). However, the authors did report that the epc1-2 (gmt1-2) mutant exhibits an exaggerated response to exogenous abscisic acid but not to any other hormone tested. Analysis of the epc1 (gmt1) mutants in the two reports did not suggest a biochemical function for of EPC1/GMT1.
In this work, we investigated the function of EPC1/GMT1 and propose to rename it GIPC MANNOSYL-TRANSFERASE1 (GMT1) based on the evidence described here. Reanalysis of published epc1/gmt1 alleles, along with isolation of a novel allele, gmt1-3, revealed a defect in GIPC glycosylation. Heterologous expression of GMT1 in tobacco and yeast resulted in the production of Arabidopsis-like GIPCs in these systems. As with gonst1, the gmt1 mutants showed severely stunted growth and a constitutive defense response. We also discovered that gmt1 also has a reduction in cellulose quantity, although other cell wall components are unaffected. Based on these data, we propose that GMT1 is likely a mannosyltransferase (ManT) that glycosylates the major GIPC classes in Arabidopsis.
RESULTS
The Phenotype of gmt1 Resembles That of gonst1
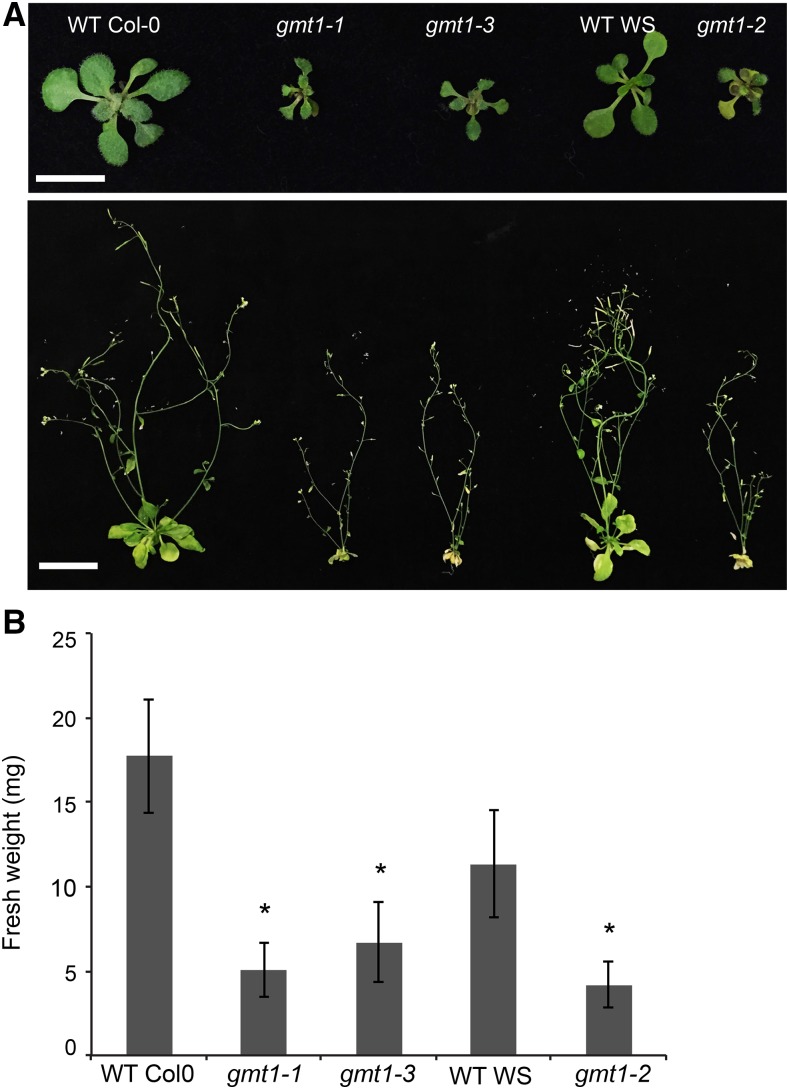
gonst1 mutants, which lack a GIPC-specific GDP-Man transporter, are dwarfed and exhibit a constitutive defense response. We noted that this phenotype is reminiscent of the phenotype reported for epc1 (hereafter referred to as gmt1) (Singh et al., 2005; Bown et al., 2007), suggesting that the GT might function in the same pathway as the transporter. We therefore hypothesized that GMT1 (EPC1) could be the GIPC-specific mannosyltransferase. To test this, three allelic homozygous T-DNA insertional mutants, gmt1-1 (Columbia-0 [Col-0] ecotype; previously described in Singh et al. [2005]), gmt1-2 (Wassilewskija [Ws] ecotype; previously described in Bown et al. [2007]), and gmt1-3 (Col-0; a new allele) were obtained and confirmed as transcriptional knockouts (Supplemental Figure 1). All three gmt1 allelic mutants have severely stunted growth (Figure 2A), with a fresh weight approximately half that of the wild type (Figure 2B). gmt1 plants can successfully complete a full life cycle, but only a small number of viable seeds are produced.
Figure 2.
Phenotype of gmt1.
(A) Top row: 15-d-old agar-grown wild-type and gmt1 plants. Bar = 1 cm. Bottom row: 6-week-old wild-type and gmt1 plants. Bar = 3 cm.
(B) Fresh weight of seedlings of 15-d-old, agar-grown wild type and gmt1; n = 6 to 16 individual plants grown simultaneously, ±sd. An asterisk indicates a significant difference from the wild type (Student’s t test, P < 0.01).
GMT1 is a member of CAZy family GT64 (Supplemental Figure 2). There are two other members in the GT64 family, At1g80290 and At5g04500, although conservation among the three members of the family at the amino acid level is relatively low at ∼26% (Supplemental Figures 2 and 3). The function of the other two genes remains unknown. Neither of the two genes can functionally compensate for the inactivation of GMT1 (Bown et al., 2007). The GT64 family described in the CAZy database (http://www.cazy.org/; Coutinho et al., 2003) includes members from a diverse range of species including humans and Drosophila melanogaster, Selaginella moellendorffii, Arabidopsis, and rice (Oryza sativa). However, while most animal proteins contain both a GT64 and GT47 domain, the 143 genes currently annotated as GT64 in the plant CAZyme database have only a GT64 domain (Edvardsson et al., 2010; Ekstrom et al., 2014).
GMT1 Is Golgi Localized
The GMT1 protein is predicted to be a type II membrane protein targeted to the secretory pathway. To investigate the subcellular localization of GMT1, the GMT1 coding sequence was fused to YFP at the C terminus under the control of the CaMV 35S promoter. This construct was cotransformed with the Golgi marker α-mannosidase-ECFP into onion epidermal cells by particle bombardment (Lao et al., 2014). The localization of the fusion proteins was examined by fluorescence confocal microscopy. Consistent with a previous report (Bown et al., 2007), the signal from the Golgi marker colocalizes with that from GMT1 (Supplemental Figure 2).
gmt1 GIPCs Have Reduced Glycosylation
Plant GIPCs have extensive structural complexity due to the various fatty acids, LCBs, and glycan headgroups from which they are made, with ∼200 species reported in Arabidopsis (Markham and Jaworski, 2007). The major GIPC structure in callus, as detected by mass spectrometry (MS), was identified as trihydroxyl C18 monounsaturated LCBs acylated to hydroxyl C24 monounsaturated LCBs (t18:1/h24:1), which is consistent with previous reports for callus (Mortimer et al., 2013) and leaves (Markham and Jaworski, 2007). The composition of the ceramide moiety (Supplemental Figure 4 and Supplemental Data Set 1) in the GIPCs and glucosylceramides of gmt1 was similar compared with the wild type, suggesting no major changes to the lipid moiety of the sphingolipids.
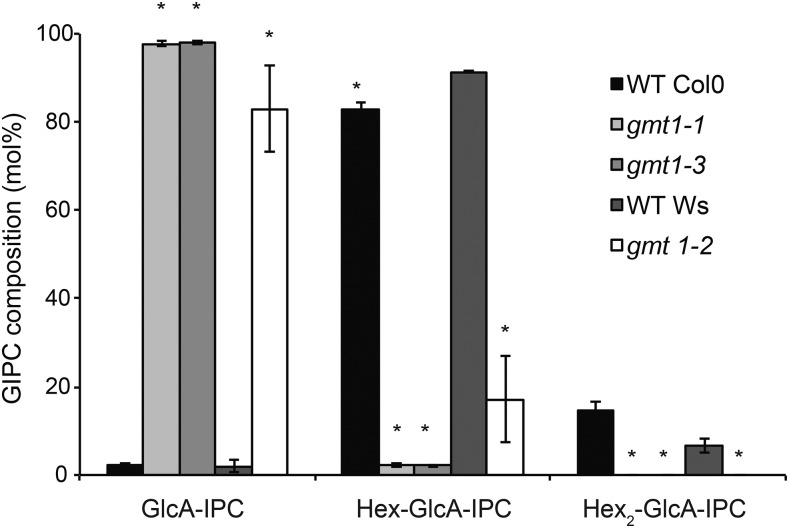
GIPCs extracted from Arabidopsis cells contain one to six sugar residues (Buré et al., 2011) linked to the GlcA-IPC core. However, in wild-type Arabidopsis callus, the dominant headgroup structure is Hex-GlcA-IPC (where the hexose is a Man, sometimes referred to as series A GIPCs), with Hex-Hex-GlcA-IPC (Hex2-GlcA-IPC or series B) the second most abundant (Buré et al., 2011; Mortimer et al., 2013) (Figure 1). In each series, the GIPC molecular species share the same glycosylation structure but contain many different ceramide structures (Supplemental Data Set 1). GIPCs were extracted from liquid-grown callus and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) multiple reaction monitoring as previously described (Mortimer et al., 2013). The amounts of GlcA-IPC and Hex2-GlcA-IPC species were estimated using the same quantitative parameter as the Hex-GlcA-IPC, since we do not have access to these as standards (the MS response factor is shown in Supplemental Data Set 2). The relative amounts of each series of GIPCs are shown in Figure 3. The wild-type membranes contain mostly Hex-GlcA-IPC (∼80% for Col-0 and ∼90% for Ws) and a minor percentage of Hex2-GlcA-IPC. By comparison, the dominant GIPC series in gmt1 was GlcA-IPC, with the amount of Hex-GlcA-IPC reduced to ∼2% for gmt1-1 and gmt1-3 and to ∼17% for gmt1-2. No Hex2-GlcA-IPC was detectable in any of the gmt1 lines.
Figure 3.
Analysis of gmt1 GIPCs.
GIPCs were isolated from wild-type and gmt1 callus total membrane preparations. The GIPCs were analyzed by MS and categorized according to the number of hexoses in the sugar moiety (Hexn-GlcA-IPC). Amounts were determined based on the peak areas of detected GIPCs; n = 3, ±sd. An asterisk indicates a significant difference from the wild type (Student’s t test, P < 0.01). See Supplemental Data Set 1 for the raw data used to prepare this figure.
gmt1 GIPCs Have Reduced Mannosylation
Since the identity of the hexose could not be determined directly by MS, the GIPC-containing fraction above was then further enriched by anion exchange chromatography, hydrolyzed with trifluoroacetic acid (TFA), and the monosaccharide composition of the headgroups determined by high-performance anion exchange chromatography coupled with pulsed amperometric detection (HPAEC-PAD) as previously described (Mortimer et al., 2013). The gmt1 mutants showed a significant reduction in Man compared with the wild type (Table 1; Supplemental Data Set 3). It should be noted that a large (and variable) amount of glucose (Glc) was detected, although this was not significantly different between the wild-type and gmt1 lines (Table 1). The amount of Ara became undetectable in gmt1, perhaps suggesting that pentoses are added to a Hex-GlcA-IPC. Alternatively, GMT1 could be functioning as an arabinosyltransferase. We therefore took a heterologous expression approach to testing GMT1 function.
Table 1. Monosaccharide Composition of an Enriched GIPC Fraction Isolated from Arabidopsis Wild-Type and gmt1 Callus.
Heterologous Expression of Arabidopsis GMT1 in a GlcA-IPC-Producing Yeast Strain Resulted in Hex-GlcA-IPC Biosynthesis
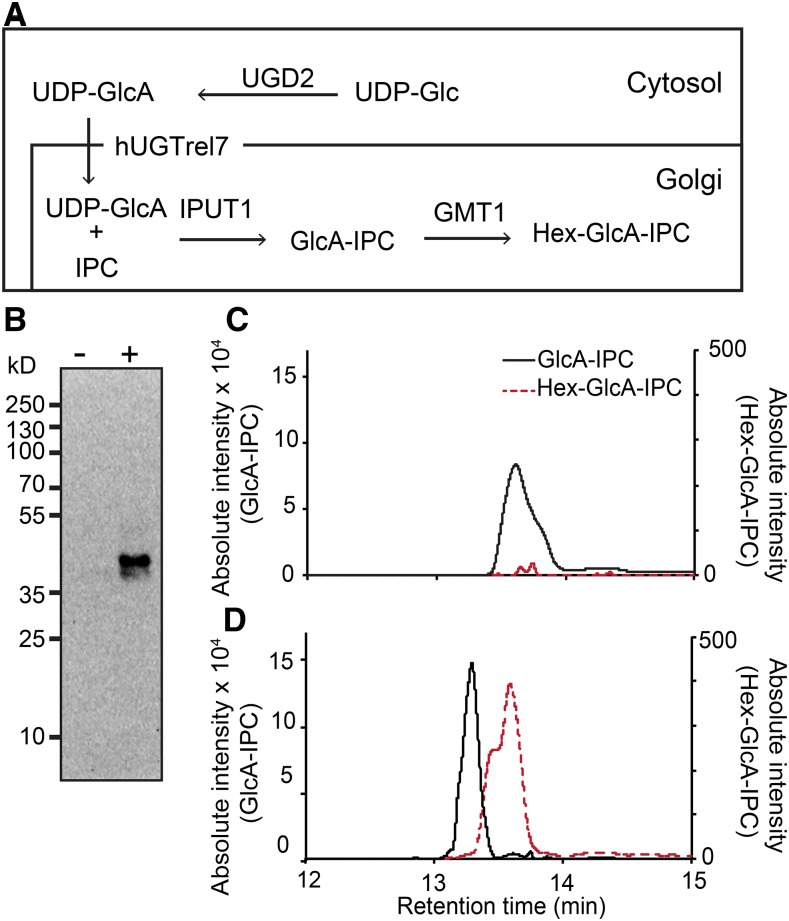
The major GIPC species in yeast are mannosylated IPCs (Obeid et al., 2002). The sur1Δ knockout S. cerevisiae mutant is able to synthesize and accumulate IPCs but is defective in the conversion of IPC to mannosylinositol phosphorylceramide. The pathway for biosynthesis of GlcA-IPC, which is the predicted substrate of GMT1, has been successfully assembled in sur1Δ yeast by expressing IPUT1, UDP-Glc Dehydrogenase 2 (UGD2), and human UDP-Galactose Transporter Relative 7 (hUGTrel7; a UDP-GlcA transporter) (Rennie et al., 2014). We additionally introduced GMT1 into this system to test its function, as outlined in Figure 4A. Lysates from sur1Δ yeast expressing GMT1 and the other three genes were analyzed by immunoblotting to ensure the GMT1 protein was expressed, as shown in Figure 4B.
Figure 4.
Heterologous Expression of GMT1 in GlcA-IPC-Synthesizing Yeast.
(A) The proposed pathway for producing the Hex-GlcA-IPC using yeast. UGD2, hUGTrel7, IPUT1 as previously reported (Rennie et al., 2014), and GMT1 were expressed in sur1Δ yeast.
(B) Immunoblot showing expression of the GMT1 protein in yeast expressing UGD2, hUGTrel7, and IPUT1 (−) and yeast expressing UGD2, hUGTrel7, IPUT1, and GMT1 (+). The predicted size of GMT1 is 37 kD.
(C) and (D) MS/MS chromatograms showing the production of GlcA-IPC (black line) and Hex-GlcA-IPC (red dashed line) from yeast expressing UGD2, hUGTrel7, and IPUT1 (C) and yeast expressing UGD2, hUGTrel7, IPUT1, and GMT1 (D).
Lipids from the GMT1-expressing yeast and the control yeast were extracted and analyzed by MS/MS. The production of GlcA-IPC and Hex-GlcA-IPC species with a t18:0-h26:0 ceramide backbone (the predominant S. cerevisiae IPC) was monitored (Figures 3C and 3D). The absolute intensity was used to estimate the amount of GIPC produced, since standards are not available. The level of GlcA-IPC detected in yeast in the absence or presence of GMT1 expression was comparable (Figures 4C and 4D). However, Hex-GlcA-IPC was detectable only in the GMT1-expressing yeast, albeit at a very low level (Figure 4D). This result indicates that GMT1 is able to add a hexose to the yeast-synthesized IPUT1 product GlcA-IPC.
An attempt was also made to demonstrate the in vitro activity of GMT1. Whilst we were able to express recombinant GMT1 protein in Escherichia coli and to purify a small amount as a soluble fraction, we were unable to identify GlcA-IPC hexosylation by MALDI-MS under the assay conditions tested (Supplemental Figure 5).
Heterologous Expression of Arabidopsis GMT1 in Tobacco BY2 Cells Resulted Hex-GlcA-IPC Biosynthesis
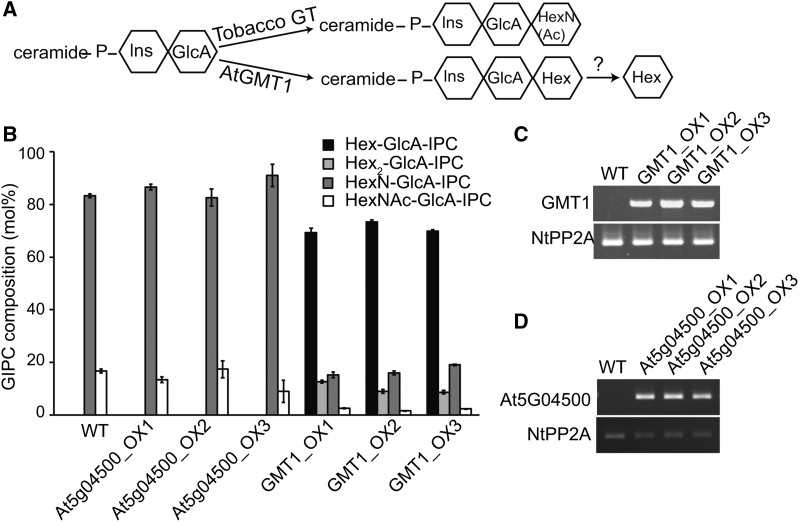
To complement the heterologous expression of GMT1 in yeast, we introduced the GMT1 protein into tobacco Bright Yellow 2 (BY2) cells. Unlike Arabidopsis, the major GIPCs in tobacco BY2 cells contain GlcN(Ac) linked to the GlcA-IPC core (Buré et al., 2011, 2014) (Figure 5A). GMT1 and a homolog from GT64, At5g05400 (Supplemental Figure 1), were placed under a constitutive promoter (CaMV 35S) and introduced into tobacco BY2 cells (Figures 5C and 5D). GIPCs extracted from BY2 cells expressing GMT1 and At5g05400 were analyzed by LC-MS/MS (Figure 5B; Supplemental Figure 6). Hex- and Hex2-GlcA-IPCs were not detected in wild-type cells or cells expressing At5g05400. However, GMT1-expressing cells contained ∼20% Hex-GlcA-IPCs and ∼2% Hex2-GlcA-IPCs. The presence of Hex-GlcA-IPCs demonstrates that when expressed in tobacco BY2 cells, the GMT1 protein is able to produce Arabidopsis-like, Hexn-GlcA-PCs. The presence of small amounts of Hex2-GIPCs can either be interpreted as GMT1 itself being able to further glycosylate Hex-GlcA-IPC to produce Hex2-GlcA-IPC, or alternatively, tobacco contains other GTs able to use Hex-GlcA-IPC as a substrate at a low rate.
Figure 5.
Heterologous Expression of Arabidopsis GMT1 in Tobacco BY2 Cells.
(A) The proposed mechanism for the production of Hex-GlcA-IPCs in BY2 cells expressing Arabidopsis GMT1.
(B) Arabidopsis GMT1 was expressed in tobacco BY2 cells under the control of the CaMV 35S promoter. GIPCs were isolated and characterized by LC-MS/MS and the data grouped by GIPC headgroup composition. A second member of the Arabidopsis GT64 family, At5g04500, was also transformed into BY2 cells as a control. Three independently transformed lines were analyzed for each transgene; n = 3, ±sd.
(C) and (D) RT-PCR showing expression of GMT1 (C) and At5G04500 (D), along with a control gene, tobacco PROTEIN PHOSPHATASE2A (NtPP2A). It should be noted that for clarity, the tobacco headgroup structure is shown as HexN(Ac), but it has been demonstrated to be GlcN(Ac) (Kaul and Lester, 1975; Hsieh et al., 1978).
Using this system, we now wanted identify the hexose that was added in the GMT1-expressing BY2 cells. As described above for Arabidopsis, we performed monosaccharide analysis on a TFA-hydrolyzed fraction of the crude GIPCs (Table 2). Wild-type BY2 cells have trace amounts of Man (0.07 ± 0.01 µg/mg), whereas the GMT1 overexpressing lines showed a large and significant increase (0.98 ± 0.20 to 1.45 ± 0.14 µg/mg) in Man. No significant difference in the amount of GlcA was recorded, indicating that the overall quantity of GlcA-IPC had not changed in the extracted crude GIPC fractions (Table 2). It should be noted that the HPAEC-PAD only detects GlcN, since the TFA hydrolyzes GlcNAc to GlcN.
Table 2. Monosaccharide Composition of a Crude GIPC Fraction Isolated from Tobacco BY2 Cells Overexpressing Arabidopsis GMT1.
| Monosaccharide (µg/mg DW) | Ara | GlcN | Gal | Glc | Man | GlcA |
|---|---|---|---|---|---|---|
| Wild type | 1.18 ± 0.37 | 2.14 ± 0.45 | 3.97 ± 1.41 | 12.77 ± 5.84 | 0.07 ± 0.01 | 0.99 ± 0.05 |
| GMT1_OX1 | 1.92 ± 0.92 | 2.98 ± 1.43 | 3.13 ± 1.94 | 18.48 ± 4.88 | 1.45 ± 0.14 | 1.00 ± 0.14 |
| GMT1_OX2 | 2.07 ± 0.55 | 3.18 ± 0.65 | 2.07 ± 0.33 | 11.02 ± 4.92 | 1.28 ± 0.43 | 0.86 ± 0.59 |
| GMT1_OX3 | 2.18 ± 0.15 | 3.16 ± 0.19 | 1.86 ± 0.29 | 11.82 ± 2.26 | 0.98 ± 0.20 | 0.62 ± 0.20 |
gmt1 Displays a Constitutive Immune Response
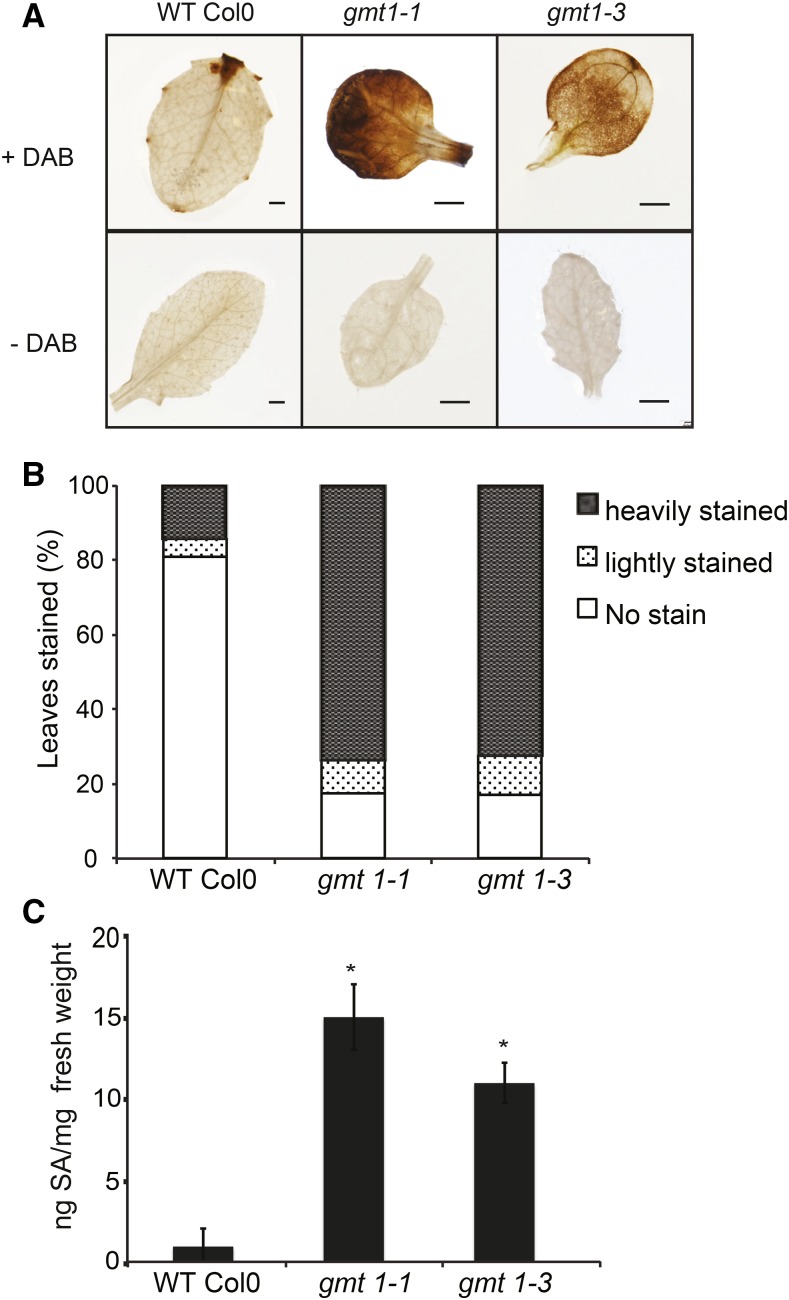
Both gonst1 and gmt1 plants exhibit an early senescence phenotype under normal growth conditions (Figure 2A), as does the pollen-specific rescued iput1 Arabidopsis plant (Tartaglio et al., 2016). Both gonst1 and iput1 display a constitutive defense response, as measured by elevated salicylic acid (SA) and H2O2 levels (Mortimer et al., 2013; Tartaglio et al., 2016), and the early senescence phenotype could be partially rescued by suppression of SA production (Mortimer et al., 2013). To test whether the link between GIPC misglycosylation and plant defense signaling was also seen in the gmt1 plants, we estimated in situ H2O2 production by 3,3-diaminobenzidine (DAB) staining. Darker brown precipitation in gmt1 leaves signified a greater level of constitutive H2O2 accumulation compared with the wild type (Figures 6A and 6B). The total SA was also quantified in leaves of wild-type and gmt1 plants by LC-MS/MS. gmt1-1 and gmt1-3 had ∼10-fold increase in SA accumulation compared with the wild type (Figure 6C).
Figure 6.
H2O2 and SA in gmt1 Leaves.
(A) H2O2 detection in 15-d-old, agar-grown leaves by DAB staining. Bar = 1 mm.
(B) Percentage and extent of leaves stained by DAB (n = 21 to 34 individual plants).
(C) Total SA content of 15-d-old, agar-grown leaves; n = 3, ±sd. An asterisk indicates a significant difference from the wild type (Student’s t test, P < 0.01).
Golgi-Synthesized Cell Wall Polysaccharides Are Unaffected in gmt1
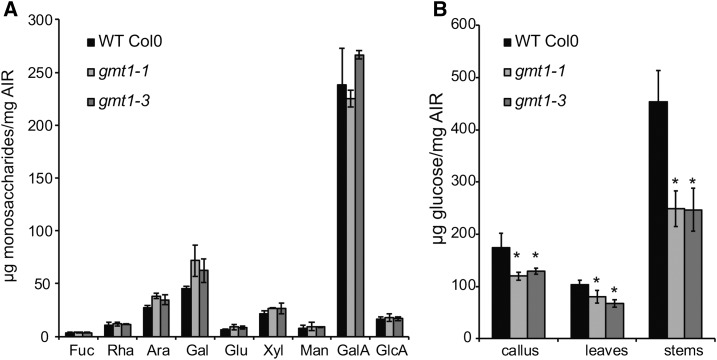
Previously, it was proposed that GMT1 plays a role in pectin biosynthesis (Singh et al., 2005; Bown et al., 2007). However, the previous studies analyzed a single T-DNA insertion line each and disagreed on the exact nature of the modification, which was ascribed to different growth conditions between the studies (Bown et al., 2007). Therefore, we collected three independently grown biological replicates (plants grown at three different times; more than one plant was collected each time and the material pooled) of gmt1-1, gmt1-2, and gmt1-3, and their respective wild-type ecotypes, and prepared destarched cell wall extracts (alcohol insoluble residue [AIR]) from liquid-grown callus, 15-d-old agar-grown leaves, and mature stems. The AIR was hydrolyzed with TFA to release noncellulosic sugars, and the monosaccharides were quantified by HPAEC-PAD (Figure 7A; Supplemental Table 1 and Supplemental Data Set 3). Under our growth conditions, no statistically significant difference was detected between the wild type and any of the three gmt1 mutants in any of the tissue types analyzed.
Figure 7.
Cell Wall Composition Analysis.
(A) Monosaccharide analysis of wild-type and gmt1 leaves; n = 3, ±sd. Destarched AIR was hydrolyzed with 2 M TFA, and the released monosaccharides were quantified by HPAEC-PAD.
(B) Crystalline cellulose content of gmt1 callus, leaves, and stems. Following sulfuric acid hydrolysis of the TFA-insoluble fraction of AIR, the Glc released was quantified by HPAEC-PAD; n = 3; ±sd. An asterisk indicates a significant difference from the wild type (Student’s t test, P < 0.01).
gmt1 Has Reduced Cellulose Content
We then further hydrolyzed the residual TFA insoluble fraction with sulfuric acid to estimate the crystalline cellulose content (Figure 7B, Table 3). Surprisingly, all gmt1 mutants showed a large decrease in cellulose contents for all three tissue types (∼22% for leaves, ∼31% for callus, and ∼45% for stem).
Table 3. Cellulose and Lignin Content of gmt1 and Wild-Type Stems.
| Lignin (µg/mg) | Cellulose (µg/mg) | |
|---|---|---|
| Wild-type Col-0 | 110.0 ± 12.4 | 453.7 ± 60.0 |
| gmt1-1 | 221.3 ± 38.0 | 248.7 ± 34.5 |
Stem cellulose content was estimated by sulfuric acid-hydrolysis of TFA-resistant AIR. The Glc released was quantified by HPAEC-PAD. The lignin content of stem tissue was estimated using the acetyl bromide method; n = 3, ±sd. Bold indicates significant difference from the wild type (Student’s t test, P < 0.01).
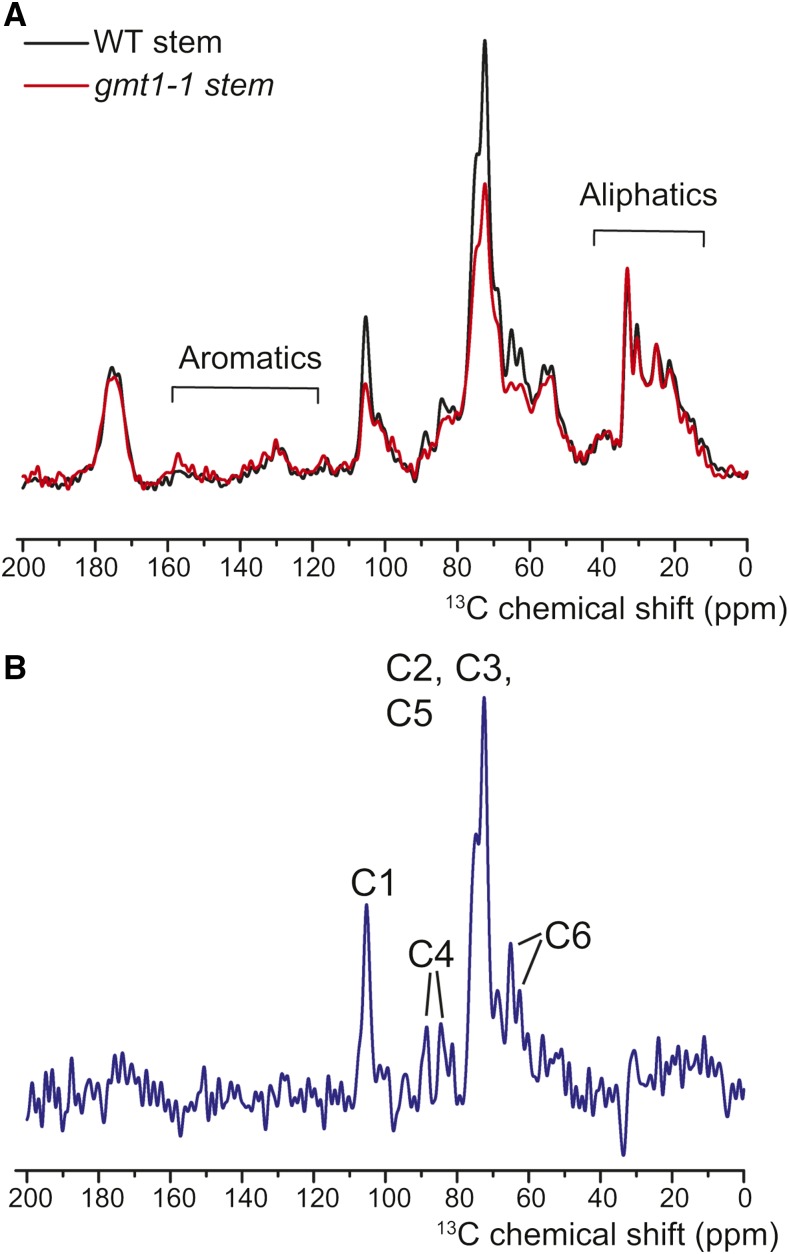
To further investigate this reduced level of crystalline cellulose, 1D 13C solid-state NMR was used to further examine the cell wall structure differences between the wild type and gmt1-1 (Figure 8A). A linear subtraction of gmt1-1 from wild-type spectra (Figure 8B) was enriched for signals from the carbons of Glc in cellulose, suggesting a specific reduction in the cellulose content of gmt1-1. These results are consistent with the results of the sulfuric acid hydrolysis and confirm that gmt1 has reduced quantities of crystalline cellulose.
Figure 8.
Solid-State 13C CP MAS NMR Spectra of Wild-Type and gmt1-1 Stems.
(A) 13C MAS solid-state NMR spectra of wild-type (black) and gmt1-1 (red) stems. As previously described (Zhang et al., 2016), the signal intensity was normalized to the aliphatic protein region (35 to 25 ppm) with the assumption that the gmt1-1 mutant does not show significant changes to proteins or waxes.
(B) The subtraction spectrum (wild type − gmt1-1) reveals specific decreases in the cellulose carbons in gmt1-1. Cn refers to the carbons in the cellulosic Glc, where C1 is the anomeric C.
gmt1 Has an Increase in Lignin Content
The solid-state NMR data also revealed a slight increase in the signal intensity in the lignin aromatic region (110 to 155 ppm) in gmt1 plants compared with the wild type (Figure 7A). It should be noted that the cross-polarization (CP) methods used here preferentially enhance signals from rigid species such as cellulose, as opposed to the more mobile lignin signals (Dick-Pérez et al., 2011), so changes to the lignin are less evident. Therefore, we estimated the total lignin content of stems of wild-type and gmt1 plants using the acetyl-bromide method (Foster et al., 2010). The lignin content of gmt1-1 was increased ∼50% compared with the wild type (Table 3), a phenotype that has been reported for other cellulose mutants (Caño-Delgado et al., 2003). It is interesting to note that the reduced level of crystalline cellulose in stems is accompanied by increases in lignin, but not by any compensatory changes to the matrix polysaccharides.
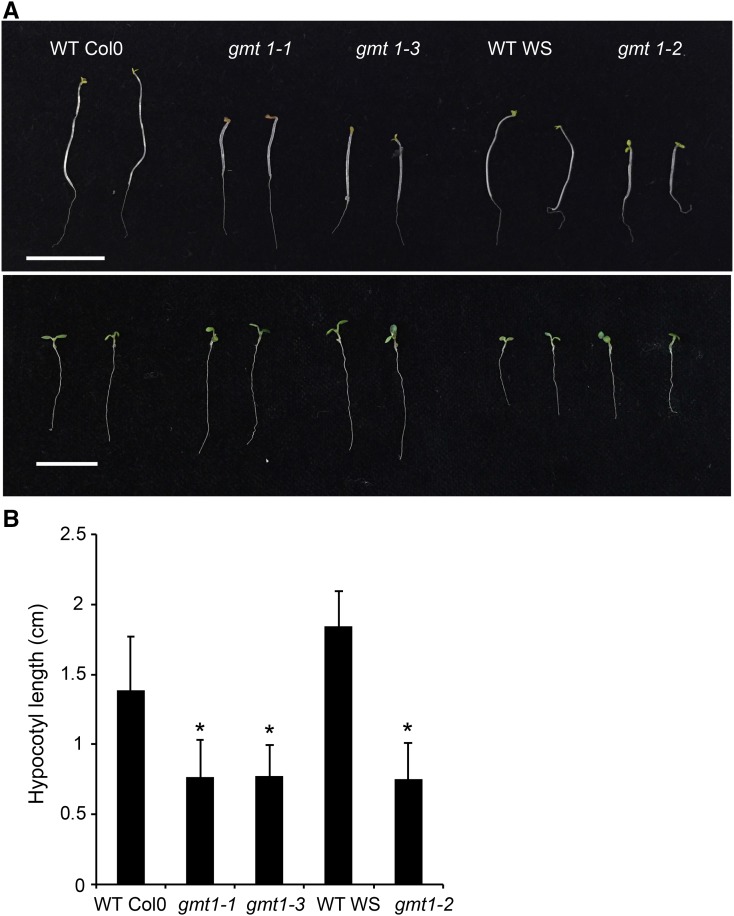
gmt1 Has Defective Hypocotyl Elongation
Reduced etiolated hypocotyl length is a common phenotype associated with defects in cellulose biosynthesis (Persson et al., 2007; Harris et al., 2012). Seedlings were germinated and grown in the dark or light for 5 d, and hypocotyl length measured (Figure 9A). While gmt1 showed a slight growth defect following 5 d in the light, there was a severe and significant effect on hypocotyl elongation in the dark of ∼50% reduction compared with the wild type (Figure 9B).
Figure 9.
Skotomorphogenic Response of gmt1.
(A) Five-day-old dark-grown (top) and light-grown (bottom) wild-type and gmt1 seedlings. Bar = 1 cm.
(B) Quantitation of etiolated hypocotyl length; n = 8 to 12 individual plants grown simultaneously, ±sd. An asterisk indicates a significant difference from the wild type (Student’s t test, P < 0.01).
DISCUSSION
A number of recent studies have now revealed that GIPCs are major components of the plant plasma membrane and that the nature of the glycan headgroup can vary by tissue and species (Cacas et al., 2013; Buré et al., 2014; Tellier et al., 2014). In this work, our goal was to identify proteins involved in GIPC glycosylation. Since the only known GIPC glycosylation mutants are either gametophytic lethal (iput1, a GIPC GlcAT; Rennie et al., 2014) or dwarfed with a constitutive defense response (gonst1, a GIPC-specific GDP-Man transporter; Mortimer et al., 2013), we screened the literature for GT mutants with severe phenotypes. gmt1 (epc1) was selected as a promising candidate for further investigation, as the reported growth phenotype was reminiscent of gonst1. We now conclude that GMT1 is specifically involved in GIPC glycosylation and likely functions as a GIPC-specific ManT.
We obtained both previously published mutant alleles (gmt1-1 [Singh et al., 2005] and gmt1-2 [Bown et al., 2007]), as well as a new null mutant allele (gmt1-3), and when grown side-by-side, all displayed the gonst1-like developmental morphology of short stature, early senescence, poor survival on soil, and poor seed set. Under our growth conditions, we did not detect the previously reported phenotypes of defective cell-cell adhesion or changes to the noncellulosic cell wall fraction, although we could confirm the Golgi localization of the YFP fusion protein.
When we isolated the GIPCs from gmt1, in all lines there was a specific loss of Hex-GlcA-IPC and Hex2-GlcA-IPC, with the gmt1 GIPC headgroups consisting almost entirely of GlcA-IPC. This biochemical phenotype was very similar to that reported for gonst1, although the proportion of Hex-GlcA-IPC loss was higher in gmt1. Since MS is used for the headgroup analysis, the identity of the hexoses terminal to the GlcA cannot be determined from these data. However, monosaccharide analysis of the sphingolipid enriched fraction of gmt1 showed a significant decrease in Man compared with the wild type. The levels of other monosaccharides were also reduced in the gmt1 mutant, including galactose, which has the same mass as Man. It has been demonstrated previously that the first hexose linked to the GlcA is likely Man (Mortimer et al., 2013); therefore, we propose that these changes are due to the loss of sugars that are terminal to the Man, such as those reported previously (Cacas et al., 2013; Buré et al., 2014), and it is expected that not all have been reported. Alternatively, they could derive from non-GIPCs in the analyzed fraction. The sugar composition of the enriched GIPC fraction from callus also revealed the presence of large and variable amount of Glc. The GIPC fraction isolated from wild-type BY2 cells also contained a very high level of Glc, although no Hex-GlcA-IPC was detected. Thus, we propose that the Glc is derived from unknown molecule(s) other than the GIPC headgroup. We were unable to determine the source of this Glc, but our current hypothesis is that it may come from certain unknown molecules that bound to the GIPCs during the enrichment process.
Enzymatic cleavage of the phosphate group would release the glycan headgroup from the ceramide, enabling the use of standard glycobiology methods to study the range of GIPC headgroups. Although it has been reported that Brassicacea extracts (from cabbage and Arabidopsis) have GIPC-specific phosphatase activity (Tanaka et al., 2013), as yet the gene encoding the enzyme responsible has not been cloned. Identification of this enzyme will be very beneficial for GIPC research, enabling NMR characterization of the released glycan.
Since loss of activity in a mutant is not sufficient to infer enzyme activity, we took several approaches to test enzyme activity in a heterologous system. First, we used a yeast system we had previously developed, in which the yeast synthesizes the proposed GMT1 substrate GlcA-IPC (Rennie et al., 2014). When GMT1 was expressed in this yeast strain, we could detect a Hex-GlcA-IPC form of this substrate, although the amount was relatively small. The reasons for the poor hexosylation of the GlcA-IPC are unknown, but it could be that the product is toxic to the cell, that there is not enough of the substrate available, or that the plant proteins show poor activity in yeast. Therefore, we took a second approach using a plant system. Tobacco BY2 cells do not produce a Hex-GlcA-IPC. Instead, GlcN(Ac) is linked to GlcA-IPC (Buré et al., 2011). This means that in the tobacco cells, there is native production of the proposed GMT1 substrate, prior to its glycosylation by the unknown tobacco GlcN(Ac) transferase. Expression of Arabidopsis GMT1 (but not a second member of the Arabidopsis GT64 family) resulted in the production of Arabidopsis-like Hex-GlcA-IPCs. Sugar composition analysis of these lines revealed a significant increase in Man. It should be noted that we did not detect a significant increase in Ara or other sugars. While large amounts of Glc were measured in the GMT1-overexpressing lines, it was also present in the untransformed BY2 lines, where Hex-GlcA-IPCs were not detected. There was an increase in total GIPC content in both gmt1 Arabidopsis plants and GMT1-overexpressing BY2 lines compared with the wild type, while a reduced GIPC content was measured in the iput1 mutant (Tartaglio et al., 2016). These results suggest that modification of GIPC headgroups might cause changes to GIPC accumulation. The reason is unknown, but may be due to feedback mechanisms which detect the abnormal GIPCs.
Our attempts to demonstrate GMT1 in vitro activity using recombinant GMT1 protein expressed in E. coli have thus far been unsuccessful. Plant glycosyltransferases are notoriously difficult to demonstrate in vitro activity for, for many proposed reasons including unknown cofactors and poor protein stability when heterologously expressed (Scheible and Pauly, 2004; Bencúr et al., 2005). However, we believe that the heterologous expression of GMT1 in both yeast and tobacco, as well as the mutant characterization, allows us to conclude that the most likely function of GMT1 is as a GIPC-specific ManT.
These data, along with the previously reported work on IPUT1 (Rennie et al., 2014; Tartaglio et al., 2016) and GONST1 (Mortimer et al., 2013), confirm that GIPCs are, as in yeast, glycosylated in the Golgi and that the biosynthetic pathway is somehow distinct from other glycosylation processes in the Golgi that use the same substrates, supporting a substrate channeling hypothesis. However, these data do not reveal the function of the GIPC headgroup. For this reason, we further investigated the phenotype of the gmt1 plants. Previously, it has been shown that gonst1 (Mortimer et al., 2013), pollen-rescued iput1 (Tartaglio et al., 2016), and erh1 (inositol phosphorylceramide synthase1) expressing RPW8 (a resistance gene) (Wang et al., 2008) display constitutive SA accumulation, hypersensitive lesion formation, and stunted growth, which we now report for the gmt1 mutant. The constitutive production of this defense hormone is likely upstream of the excessive production of reactive oxygen species (here, we measured the most stable species, H2O2), which leads to the cell death and senescence phenotype visible from the seedling stage. We therefore conclude that misglycosylation of GIPCs specifically induces a constitutive defense response. However, the signaling cascade leading to this induction remains unknown and is something we are continuing to investigate.
The degree of GIPC headgroup alteration is linked to the severity of the phenotypes observed. Complete loss of IPUT1, resulting in only an IPC core as the dominant GIPC, is a lethal mutation (Rennie et al., 2014) and affects pollen tube guidance (Tartaglio et al., 2016), whereas both gonst1 (Mortimer et al., 2013) and gmt1, which have GlcA-IPC as the dominant GIPC, have viable pollen. It should be noted that the pollen quality of these plants has not yet been explored. Seed yield is certainly lower than the wild type, but this could also be due to the poor overall health of the parent plants.
One unexpected finding from this study was the reduced quantity of cellulose in the gmt1 mutant and a concomitant increase in lignin. Cellulose is synthesized at the plasma membrane by the cellulose synthase (CESA) complex (CSC). This rosette of proteins takes UDP-Glc as a substrate from the cytosol and extrudes glucan chains to the exterior of the cell, which then assembles into the crystalline microfibril. The CSC is associated with a large number of other proteins. For example, cytosolic proteins, such as KORRIGAN and CSI1, directly interact with the CESA complex and affect the direction and velocity of the CSC (McFarlane et al., 2014). Extracellular proteins, including glycosylphosphatidylinositol-anchored proteins CTL1 and COBRA, also affect CSC motility. Glycosylphosphatidylinositol-anchored proteins are enriched in detergent-resistant membrane fractions (Borner et al., 2003), which are enriched in sphingolipids (including GIPCs) and sterols (Cacas et al., 2016). While CSCs have not been found in detergent-resistant membrane fractions, the sterol environment has previously been implicated in CSC function, since several sterol mutants are disrupted in cellulose biosynthesis, and like gmt1, no other changes to the cell wall were reported (Schrick et al., 2004). As expected, mutants in the biosynthesis of sphingolipids are lethal, and although RNAi lines have been made, these were not analyzed for cellulose content (Chen et al., 2006; Dietrich et al., 2008), something that may now be worth revisiting. The mechanism for how GIPCs may affect cellulose content is not clear. It is possible that the glycan headgroup assists in ordering proteins within lipid “microdomains” (Simons and Sampaio, 2011). It is also possible that the CSCs or associated proteins could interact with the GIPC headgroups, impeding their movement through the plasma membrane. Research trying to understand the constraints on movement of proteins within the plasma membrane has shown that the interactions of the protein with the cell wall are critical (Martinière et al., 2012). In the future, we will use fluorescently tagged CESA proteins to try to understand whether the insertion rate, velocity, or residence time of the CESA rosettes are affected in the gmt1 plants in order to try to test these hypotheses.
The recent identification of multiple mutants in GIPC glycosylation processes with severe but related phenotypes supports a critical role for GIPC headgroups in plant development and signaling. Future work should focus on linking these biochemical and physiological phenotypes to mechanistic roles for the glycan headgroup.
METHODS
Phylogenetic Analysis
GT64 protein sequences were downloaded from PlantCAZyme (Ekstrom et al., 2014) on February 10, 2015. Sequences were aligned using ClustalOmega using default settings (www.ebi.ac.uk; McWilliam et al., 2013) and exported in PHYLIP format. The PHYLIP package (v3.695) was used to produce a phylogeny (http://evolution.genetics.washington.edu/phylip.html; Felenstein, 2005). Specifically, the alignment was first resampled using seqboot (2000 replicates, otherwise default settings), followed by proml (maximum likelihood without a molecular clock) using an outgroup root (GUX1) and the not-rough analysis. Consense was used to build a rooted tree, which was visualized using drawgram. Arabidopsis (Arabidopsis thaliana) GUX1 was chosen as an outgroup since it is also a single-transmembrane, Golgi-localized GT (family 8; Mortimer et al., 2010).
Plant Material and Growth Conditions
Arabidopsis seeds were surface sterilized and sown on solid medium containing 0.5× Murashige and Skoog salts including vitamins and 1% (w/v) sucrose. Following stratification (48 h, 4°C, in the dark), plates were transferred to a growth room (22°C, 100 to 200 µmol m−2 s−1, 14 h light/10 h dark, 60% humidity). After 2 to 3 weeks, plants were transferred to Magenta boxes under the same conditions. Liquid callus cultures were derived from Arabidopsis roots and maintained as described previously (Prime et al., 2000). All experiments were performed on at least three independently grown biological replicates unless otherwise stated.
Mutant Identification
Seeds of T-DNA mutants in At3g55830 were obtained from the ABRC (gmt1-1, SAIL_A11_68; as described in Singh et al. [2005]; and gmt1-3, SAIL_303_A11) and a kind donation from Steven Jackson, University of Warwick (gmt1-2; Bown et al., 2007). Plants homozygous for the T-DNA insertion were identified by PCR (Phire Plant Direct PCR kit; Finnzymes), and transcriptional knockouts were identified by RT-PCR. Total RNA was extracted from wild-type and mutant plants using the RNeasy Plant Mini Kit (Qiagen). RNA was first treated with DNase I (Qiagen) and first-strand cDNA synthesis was performed using SuperScript II RT (Life Technologies). ACTIN7 was used as an internal control. All primers used in this study are listed in Supplemental Table 2.
Subcellular Localization
The construction of the pBullet expression clone, transient expression in onion cells, and confocal microscopy were performed exactly as described by Lao et al. (2014).
AIR Preparation
Samples were collected from inflorescence stems (6-week-old plants), young leaves (15-d-old plants), and liquid-grown callus tissue. The tissue was harvested into 96% ethanol and incubated for 30 min at 70°C to inactivate cell wall-degrading enzymes. The tissue was homogenized using a Retsch mixer mill and centrifuged. The pellet was washed with 100% ethanol and twice with a mixture of chloroform and methanol (2:1), followed by three successive washes with 65% (v/v), 80% (v/v), and 100% (v/v) ethanol. The pellet was air-dried overnight. The starch in the samples was degraded with α-amylase, amyloglucosidase, and pullulanase (Megazyme) as described previously (Harholt et al., 2006). The destarched residue was referred to as AIR.
Analysis of Cell Wall Monosaccharide Composition
AIR (5 mg) was hydrolyzed with fresh 2 M TFA at 121°C for 1 h. The supernatant was retained, dried under vacuum, and resuspended in 200 μL water. The TFA-insoluble material was washed with water and further hydrolyzed with 72% (v/v) sulfuric acid containing 10 µg myo-inositol for 1 h at room temperature. The sulfuric acid was then diluted to 1 M with water and the samples further incubated at 100°C for 3 h and neutralized with BaCO3 to provide the crystalline cellulose fraction. Samples were filtered through a 22-µm syringe filter and analyzed by HPAEC on an ICS-5000 instrument (Thermo Fisher Scientific) equipped with a CarboPac PA20 analytical anion exchange column (3 mm × 150 mm; Thermo Fisher Scientific), PA20 guard column (3 mm × 30 mm), borate trap, and a pulsed amperometric detector. In order to achieve full separation of all the major plant cell wall monosaccharides, we used a method modified from Mortimer et al. (2013). Following a 5-min equilibration, the samples were eluted using an isocratic gradient of 4 mM NaOH from 0 to 6 min, followed by a linear gradient of 4 mM NAOH to 1 mM NaOH from 6 to 19 min. At 19.1 min, the gradient was increased to 450 mM NaOH to elute the acidic sugars.
Determination of Acetyl Bromide Soluble Lignin
Lignin content was estimated from 6-week-old stem samples by the acetyl bromide method, with slight modification (Foster et al., 2010). Five mg of finely ground freeze-dried stem was thoroughly exacted with ethanol/toluene (1:1) until the extracts no longer absorbed UV light at 280 nm. The dried samples were placed into loosely capped glass tubes containing 1 mL of acetyl bromide/acetic acid (1:3) and incubated at 70°C for 30 min. The sample was then cooled in an ice bath and mixed with 0.9 mL of 2 M NaOH, 5 mL glacial acetic acid, and 0.1 mL 7.5 M hydroxylamine hydrochloride. The final volume was adjusted to 10 mL with glacial acetic acid and the absorbance measured at 280 nm using a SpectraMax M2 spectrometer (Molecular Devices). A blank control was used to correct for background absorbance. The amount of acetyl bromide soluble lignin was calculated according to the formula described by Foster et al. (2010). The coefficient used for Arabidopsis was 15.69.
Cell Wall Analysis by 13C Solid-State NMR
13C solid-state magic angle spinning NMR on wild-type and gmt1-1 air-dried stems was performed at 16.4 T on a Bruker Avance spectrometer at 15 kHz. A Bruker narrow bore H/C/N magic-angle-spinning (MAS) probe was used. The 13C chemical shift was set on adamantine (38.48 ppm relative to TMS for methylene signal). 1H to 13C CP and two-pulse phase modulation decoupling conditions were optimized using 13C-labeled glycine. A contact time of 1 ms and a pulse delay of 6 s were used.
GIPC Purification and Analysis
Approximately 1 g of lyophilized callus tissue was ground with a pestle and mortar and resuspended in 10 mL deionized water. Lipids were first extracted with 100 mL CHCl3/CH3OH/4 M NH4OH (9:7:2 v/v/v) with 0.2 M C2H3O2NH4 at 37°C and centrifuged for 1 min at 800g to separate the organic and aqueous phases (Kaul and Lester, 1975; Rennie et al., 2014). The top aqueous phase was collected, cooled to −20°C, and then centrifuged again. The bottom phase was collected as the crude GIPC fraction and dried under a N2 stream. The enrichment of GIPCs from the polar lipid fraction in the bottom phase was performed using a weak anion exchange SPE cartridge (Strata X-AW; Phenomenex) as described by Mortimer et al. (2013) with slight modification. The extracted lipid fractions were resuspended in 1 mL CHCl3/C2H5OH/18 M NH3/water (10:60:6:24 v/v/v/v) overnight at room temperature. After centrifugation (20,000g, 5 min), the supernatant was transferred to an AEX cartridge that had been equilibrated with CHCl3. The following wash steps were performed using one column volume (3 mL) to elute the non-GIPC fractions: CHCl3, CHCl3/CH3OH (2:1, 1:1, and 1:2 v/v), and CH3OH. After all the washing steps, the cartridge was dried overnight. Enriched GIPCs were eluted with 3 mL CHCl3/C2H5OH/18 M NH3/water (10:60:6:24 v/v/v/v) and dried under a N2 stream. To estimate the monosaccharide composition of GIPC headgroups, crude or enriched GIPC fractions were hydrolyzed with of 2 M TFA at 121°C for 1 h and analyzed with HPAEC-PAD. For the sphingolipidomics, total sphingolipids were extracted from dried (30 to 50 mg) or fresh (100 to 200 mg) plant materials and subjected to lipid profiling by LC-MS/MS as described previously (Nagano et al., 2014). GIPCs were quantified according to Ishikawa et al. (2016).
Cloning of Yeast Expression Vectors
The cloning and construction of yeast expression vectors was described previously (Rennie et al., 2014). Expression of UGD2 and GMT1 was accomplished using the yeast shuttle vector pDRf1-GW-PHXT7 described by Eudes et al. (2011). The UGD2 coding sequence was inserted into the Gateway site of this vector. The GMT1 coding sequence was amplified from the Arabidopsis GT clone collection (Lao et al., 2014) with primers including a C terminus HA tag sequence and was cloned into the multiple cloning site of the second yeast expression cassette in this vector using the NotI restriction site. Finally, the PHXT7 promoter in this cassette was replaced with PTEF1 using SacI and XmaI restriction sites in order to ensure that expression of GMT1 was similar to that of the other heterologous genes. The three vectors were cotransformed into yeast strain BY4742 using the Frozen-EZ Yeast Transformation II kit (Zymo Research) and plated on appropriate selective media. Yeast proteins were extracted and prepared for immunoblotting as described by Rennie et al. (2014). Protein blots were probed with a 1:1000 dilution of rat monoclonal anti-HA antibody (Sigma-Aldrich) followed by a 1:15,000 dilution of goat anti-rat IgG conjugated to horseradish peroxidase (Sigma-Aldrich). The transformed yeast strain was grown for 72 h in the selective medium for GIPC analysis. The yeast cells were subjected to lipid profiling by LC-MS/MS as described previously (Nagano et al., 2014).
Transformation of BY2 Cells
The coding sequences of GMT1 (At3g55830) and At5g04500 were cloned into pDONR207 (Invitrogen) and transferred to pH2GW7 (Karimi et al., 2002). Agrobacterium tumefaciens-mediated transformation of BY2 cells was performed as described previously (Kawai-Yamada et al., 2004). Hygromycin-resistant cells were collected for lipid analysis and RT-PCR. NtPP2A was used as a stable control gene (Schmidt and Delaney, 2010). The sugars and lipids of GIPC from BY2 cells were quantified as described above.
Histochemical Detection of H2O2
Detection of H2O2 was performed by endogenous peroxidase-dependent histochemical staining using DAB as described by Mortimer et al. (2013). Leaves of 15-d-old agar grown plants were submerged in 1 mL buffer (100 mM HEPES-KOH, pH 6.8) or 1 mg mL−1 DAB in buffer. After 4 min of vacuum infiltration, leaves were incubated at 22°C under a light intensity of 100 to 200 µmol m−2 s−1 for 6 h period. Leaves were cleared for 30 min in 96% (v/v) ethanol solution at 70°C and examined using a light microscope.
Quantitation of Total Leaf SA
For total SA determination, 500 mg leaves were frozen and ground in liquid nitrogen. The powder obtained was mixed with 1 mL 80% (v/v) methanol and mixed for 15 min at 70°C. This step was repeated four times. Pooled extracts were centrifuged and filtered through Amicon Ultra centrifugal filters (10,000 D molecular mass cutoff; EMD Millipore). Conjugated SA in the filtered extracts was dried and hydrolyzed in 1 n HCl at 95°C for 3 h. The mixture was subjected to three ethyl acetate partitioning steps. Ethyl acetate fractions were pooled, dried in vacuo, and resuspended in 50% (v/v) methanol. SA was quantified using HPLC-electrospray ionization-time-of-flight MS. The detailed running conditions were previously described (Eudes et al., 2013).
Expression of GMT1 in Escherichia coli and in Vitro Enzyme Assay
The GMT1 coding sequence was cloned into the pVP16 Gateway vector (Thermo-Fisher) containing an N terminus 8x His and maltose binding protein tag and transformed into E. coli One-Shot BL21 (DE3) competent cells (Thermo-Fisher). A single colony of the recombinant strain was inoculated into Luria-Bertani medium containing 50 µg/mL kanamycin and cultured at 37°C. An overnight treatment of 1 mM IPTG at 18°C was used for the induction of GMT1. Cells were harvested by centrifugation and lysed using sonication. The protein in the supernatant was purified by amylose resin (New England Biolabs). Purified protein fractions were boiled for 5 min and analyzed by 12% SDS-PAGE. Proteins were visualized by Coomassie Brilliant Blue staining and protein identity confirmed by MS. To assay for GlcA-IPC mannosyltransferase activity, each reaction contained 50 mM HEPES-KOH (pH 7.0), 5 mM MnCl2, 5 mM MgCl2, 1 mM DTT, 400 mM sucrose, 10 μg purified protein fractions, and 15 nmol GlcA-IPC enriched from gmt1 callus as an acceptor and 10 μM GDP-Man/UDP-Glc as a substrate in a total volume of 50 µL. Reactions were incubated at 21°C for 2 h, and the reaction was then stopped by adding CHCl3/CH3OH /4M NH4OH (9:7:2 v/v/v).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under accession numbers NP_191142.1 (At3g55830, GMT1) and NP_196070.2 (At5g04500).
Supplemental Data
Supplemental Figure 1. Summary of T-DNA lines used in this study.
Supplemental Figure 2. GMT1 localization and phylogeny.
Supplemental Figure 3. Sequence alignment of Arabidopsis GT64 members, including GMT1 (At3g55830).
Supplemental Figure 4. Sphingolipidomic analysis of wild-type Col-0 and gmt1-1 callus.
Supplemental Figure 5. Assay of GMT1 enzyme activity.
Supplemental Figure 6. Analysis of GIPCs from tobacco BY2 cells overexpressing Arabidopsis GMT1.
Supplemental Table 1. Monosaccharide composition of AIR prepared from callus and stems from 6-week-old plants.
Supplemental Table 2. Primers used in this study.
Supplemental Data Set 1. Sphingolipidomics and GIPC data.
Supplemental Data Set 2. Target list of Hex0-2 species in Arabidopsis.
Supplemental Data Set 3. Monosaccharide composition of cell wall and GIPC headgroups.
Supplementary Material
Acknowledgments
We thank Ramana Pidatala for assistance with callus culture, Mi-Yeon Lee for assistance with plant growth, Leanne Chan and Chris Petzold for protein sequencing, and Misato Ohtani and the Demura team for their support whilst J.C.M. was at RIKEN. This work was part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy (L.F., E.A.R., J.L., E.E.K.B., J.D.K., J.C.M., H.V.S.), by a RIKEN FPR fellowship (J.C.M.), and by JSPS KAKENHI 24010084 and 15K20909 to T.I. and 26292190 to M.K.-Y.
AUTHOR CONTRIBUTIONS
J.C.M. conceived the research program. J.C.M., L.F., T.I., and E.A.R. designed research. J.C.M., L.F., T.I., E.A.R., G.M.M., J.L., J.Y., A.Y.-L.T., E.E.K.B., and J.X. performed research. J.C.M., L.F., T.I., E.A.R., G.M.M., J.L., T.D., M.K.-Y., J.D.K., and H.V.S. analyzed the data. J.C.M. and L.F. wrote the article.
Glossary
- GIPC
glycosylinositol phosphorylceramide
- LCB
long-chain base
- IPC
inositol phosphorylceramide
- GT
glycosyltransferase
- GlcA
α-glucuronic acid
- GlcN
glucosamine
- GlcNAc
N-acetyl-glucosamine
- ManT
mannosyltransferase
- MS
mass spectrometry
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- TFA
trifluoroacetic acid
- HPAEC-PAD
high-performance anion exchange chromatography coupled with pulsed amperometric detection
- BY2
Bright Yellow 2
- SA
salicylic acid
- DAB
3,3-diaminobenzidine
- AIR
alcohol insoluble residue
- CP
cross-polarization
- CESA
cellulose synthase
- CSC
cellulose synthase complex
- MAS
magic-angle-spinning
Footnotes
Articles can be viewed without a subscription.
References
- Bencúr P., Steinkellner H., Svoboda B., Mucha J., Strasser R., Kolarich D., Hann S., Köllensperger G., Glössl J., Altmann F., Mach L. (2005). Arabidopsis thaliana β1,2-xylosyltransferase: an unusual glycosyltransferase with the potential to act at multiple stages of the plant N-glycosylation pathway. Biochem. J. 388: 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner G.H., Lilley K.S., Stevens T.J., Dupree P. (2003). Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 132: 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown L., Kusaba S., Goubet F., Codrai L., Dale A.G., Zhang Z., Yu X., Morris K., Ishii T., Evered C., Dupree P., Jackson S. (2007). The ectopically parting cells 1-2 (epc1-2) mutant exhibits an exaggerated response to abscisic acid. J. Exp. Bot. 58: 1813–1823. [DOI] [PubMed] [Google Scholar]
- Buré C., Cacas J.-L., Mongrand S., Schmitter J.-M. (2014). Characterization of glycosyl inositol phosphoryl ceramides from plants and fungi by mass spectrometry. Anal. Bioanal. Chem. 406: 995–1010. [DOI] [PubMed] [Google Scholar]
- Buré C., Cacas J.L., Wang F., Gaudin K., Domergue F., Mongrand S., Schmitter J.M. (2011). Fast screening of highly glycosylated plant sphingolipids by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 25: 3131–3145. [DOI] [PubMed] [Google Scholar]
- Cacas J.-L., Buré C., Furt F., Maalouf J.-P., Badoc A., Cluzet S., Schmitter J.-M., Antajan E., Mongrand S. (2013). Biochemical survey of the polar head of plant glycosylinositolphosphoceramides unravels broad diversity. Phytochemistry 96: 191–200. [DOI] [PubMed] [Google Scholar]
- Cacas J.-L., et al. (2016). Revisiting plant plasma membrane lipids in tobacco: A focus on sphingolipids. Plant Physiol. 170: 367–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño-Delgado A., Penfield S., Smith C., Catley M., Bevan M. (2003). Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 34: 351–362. [DOI] [PubMed] [Google Scholar]
- Carter H.E., Gigg R.H., Law J.H., Nakayama T., Weber E. (1958). Biochemistry of the sphingolipides. XI. Structure of phytoglycolipide. J. Biol. Chem. 233: 1309–1314. [PubMed] [Google Scholar]
- Chen M., Han G., Dietrich C.R., Dunn T.M., Cahoon E.B. (2006). The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell 18: 3576–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho P.M., Deleury E., Davies G.J., Henrissat B. (2003). An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328: 307–317. [DOI] [PubMed] [Google Scholar]
- Dick-Pérez M., Zhang Y., Hayes J., Salazar A., Zabotina O.A., Hong M. (2011). Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry 50: 989–1000. [DOI] [PubMed] [Google Scholar]
- Dietrich C.R., Han G., Chen M., Berg R.H., Dunn T.M., Cahoon E.B. (2008). Loss-of-function mutations and inducible RNAi suppression of Arabidopsis LCB2 genes reveal the critical role of sphingolipids in gametophytic and sporophytic cell viability. Plant J. 54: 284–298. [DOI] [PubMed] [Google Scholar]
- Dunn T.M., Lynch D.V., Michaelson L.V., Napier J.A. (2004). A post-genomic approach to understanding sphingolipid metabolism in Arabidopsis thaliana. Ann. Bot. (Lond.) 93: 483–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edvardsson E., Singh S.K., Yun M.-S., Mansfeld A., Hauser M.-T., Marchant A. (2010). The plant glycosyltransferase family GT64: in search of a function. In Annual Plant Reviews, Plant Polysaccharides: Biosynthesis and Bioengineering, Vol. 41, P. Ulvskov, ed (Wiley-Blackwell), pp. 285–303. [Google Scholar]
- Ekstrom A., Taujale R., McGinn N., Yin Y. (2014). PlantCAZyme: a database for plant carbohydrate-active enzymes. Database (Oxford) pii: bau079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eudes A., Baidoo E.E.K., Yang F., Burd H., Hadi M.Z., Collins F.W., Keasling J.D., Loqué D. (2011). Production of tranilast [N-(3′,4′-dimethoxycinnamoyl)-anthranilic acid] and its analogs in yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 89: 989–1000. [DOI] [PubMed] [Google Scholar]
- Eudes A., Juminaga D., Baidoo E.E., Collins F.W., Keasling J.D., Loqué D. (2013). Production of hydroxycinnamoyl anthranilates from glucose in Escherichia coli. Microb. Cell Fact. 12: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felenstein J. (2005). PHYLIP (Phylogeny Inference Package). (Seattle, WA: University of Washington). [Google Scholar]
- Foster C.E., Martin T.M., Pauly M. (2010). Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part I: lignin. J. Vis. Exp. pii: 1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J., Jensen J.K., Sørensen S.O., Orfila C., Pauly M., Scheller H.V. (2006). ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol. 140: 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D.M., et al. (2012). Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903V and CESA3T942I of cellulose synthase. Proc. Natl. Acad. Sci. USA 109: 4098–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillig I., Leipelt M., Ott C., Zähringer U., Warnecke D., Heinz E. (2003). Formation of glucosylceramide and sterol glucoside by a UDP-glucose-dependent glucosylceramide synthase from cotton expressed in Pichia pastoris. FEBS Lett. 553: 365–369. [DOI] [PubMed] [Google Scholar]
- Hsieh T.C., Lester R.L., Laine R.A. (1981). Glycophosphoceramides from plants. Purification and characterization of a novel tetrasaccharide derived from tobacco leaf glycolipids. J. Biol. Chem. 256: 7747–7755. [PubMed] [Google Scholar]
- Hsieh T.C., Kaul K., Laine R.A., Lester R.L. (1978). Structure of a major glycophosphoceramide from tobacco leaves, PSL-I: 2-deoxy-2-acetamido-D-glucopyranosyl(alpha1 leads to 4)-D-glucuronopyranosyl(alpha1 leads to 2)myoinositol-1-O-phosphoceramide. Biochemistry 17: 3575–3581. [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Ito Y., Kawai-Yamada M. (2016). Molecular characterization and targeted quantitative profiling of the sphingolipidome in rice. Plant J., http://dx.doi.org/10.1111/tpj.13281. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kaul K., Lester R.L. (1975). Characterization of inositol-containing phosphosphingolipids from tobacco leaves: Isolation and identification of two novel, major lipids: N-acetylglucosamidoglucuronidoinositol phosphorylceramide and glucosamidoglucuronidoinositol phosphorylceramide. Plant Physiol. 55: 120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai-Yamada M., Ohori Y., Uchimiya H. (2004). Dissection of Arabidopsis Bax inhibitor-1 suppressing Bax-, hydrogen peroxide-, and salicylic acid-induced cell death. Plant Cell 16: 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao J., et al. (2014). The plant glycosyltransferase clone collection for functional genomics. Plant J. 79: 517–529. [DOI] [PubMed] [Google Scholar]
- Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42: D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttgeharm K.D., Kimberlin A.N., Cahoon R.E., Cerny R.L., Napier J.A., Markham J.E., Cahoon E.B. (2015). Sphingolipid metabolism is strikingly different between pollen and leaf in Arabidopsis as revealed by compositional and gene expression profiling. Phytochemistry 115: 121–129. [DOI] [PubMed] [Google Scholar]
- Markham J.E., Jaworski J.G. (2007). Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21: 1304–1314. [DOI] [PubMed] [Google Scholar]
- Markham J.E., Li J., Cahoon E.B., Jaworski J.G. (2006). Separation and identification of major plant sphingolipid classes from leaves. J. Biol. Chem. 281: 22684–22694. [DOI] [PubMed] [Google Scholar]
- Markham J.E., Lynch D.V., Napier J.A., Dunn T.M., Cahoon E.B. (2013). Plant sphingolipids: function follows form. Curr. Opin. Plant Biol. 16: 350–357. [DOI] [PubMed] [Google Scholar]
- Martinière A., et al. (2012). Cell wall constrains lateral diffusion of plant plasma-membrane proteins. Proc. Natl. Acad. Sci. USA 109: 12805–12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane H.E., Döring A., Persson S. (2014). The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 65: 69–94. [DOI] [PubMed] [Google Scholar]
- McWilliam H., Li W., Uludag M., Squizzato S., Park Y.M., Buso N., Cowley A.P., Lopez R. (2013). Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 41: W597–W 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrand S., Morel J., Laroche J., Claverol S., Carde J.-P., Hartmann M.-A., Bonneu M., Simon-Plas F., Lessire R., Bessoule J.-J. (2004). Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 279: 36277–36286. [DOI] [PubMed] [Google Scholar]
- Moore P.J., Swords K.M., Lynch M.A., Staehelin L.A. (1991). Spatial organization of the assembly pathways of glycoproteins and complex polysaccharides in the Golgi apparatus of plants. J. Cell Biol. 112: 589–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer J.C., et al. (2013). Abnormal glycosphingolipid mannosylation triggers salicylic acid-mediated responses in Arabidopsis. Plant Cell 25: 1881–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer J.C., Miles G.P., Brown D.M., Zhang Z., Segura M.P., Weimar T., Yu X., Seffen K.A., Stephens E., Turner S.R., Dupree P. (2010). Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc. Natl. Acad. Sci. USA 107: 17409–17414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M., Ishikawa T., Ogawa Y., Iwabuchi M., Nakasone A., Shimamoto K., Uchimiya H., Kawai-Yamada M. (2014). Arabidopsis Bax inhibitor-1 promotes sphingolipid synthesis during cold stress by interacting with ceramide-modifying enzymes. Planta 240: 77–89. [DOI] [PubMed] [Google Scholar]
- Obeid L.M., Okamoto Y., Mao C. (2002). Yeast sphingolipids: metabolism and biology. Biochim. Biophys. Acta 1585: 163–171. [DOI] [PubMed] [Google Scholar]
- Oikawa A., Lund C.H., Sakuragi Y., Scheller H.V. (2013). Golgi-localized enzyme complexes for plant cell wall biosynthesis. Trends Plant Sci. 18: 49–58. [DOI] [PubMed] [Google Scholar]
- Perotto S., Donovan N., Drobak B.K., Brewin N.J. (1995). Differential expression of a glycosyl inositol phospholipid antigen on the peribacteroid membrane during pea nodule development. Mol. Plant Microbe Interact. 8: 560–568. [Google Scholar]
- Persson S., Paredez A., Carroll A., Palsdottir H., Doblin M., Poindexter P., Khitrov N., Auer M., Somerville C.R. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 15566–15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prime T.A., Sherrier D.J., Mahon P., Packman L.C., Dupree P. (2000). A proteomic analysis of organelles from Arabidopsis thaliana. Electrophoresis 21: 3488–3499. [DOI] [PubMed] [Google Scholar]
- Rennie E.A., et al. (2014). Identification of a sphingolipid α-glucuronosyltransferase that is essential for pollen function in Arabidopsis. Plant Cell 26: 3314–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible W.-R., Pauly M. (2004). Glycosyltransferases and cell wall biosynthesis: novel players and insights. Curr. Opin. Plant Biol. 7: 285–295. [DOI] [PubMed] [Google Scholar]
- Schmidt G.W., Delaney S.K. (2010). Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genomics 283: 233–241. [DOI] [PubMed] [Google Scholar]
- Schrick K., Fujioka S., Takatsuto S., Stierhof Y.-D., Stransky H., Yoshida S., Jürgens G. (2004). A link between sterol biosynthesis, the cell wall, and cellulose in Arabidopsis. Plant J. 38: 227–243. [DOI] [PubMed] [Google Scholar]
- Seifert G.J., Barber C., Wells B., Dolan L., Roberts K. (2002). Galactose biosynthesis in Arabidopsis: genetic evidence for substrate channeling from UDP-D-galactose into cell wall polymers. Curr. Biol. 12: 1840–1845. [DOI] [PubMed] [Google Scholar]
- Simons K., Sampaio J.L. (2011). Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 3: a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.K., Eland C., Harholt J., Scheller H.V., Marchant A. (2005). Cell adhesion in Arabidopsis thaliana is mediated by ECTOPICALLY PARTING CELLS 1--a glycosyltransferase (GT64) related to the animal exostosins. Plant J. 43: 384–397. [DOI] [PubMed] [Google Scholar]
- Sperling P., Heinz E. (2003). Plant sphingolipids: structural diversity, biosynthesis, first genes and functions. Biochim. Biophys. Acta 1632: 1–15. [DOI] [PubMed] [Google Scholar]
- Sperling P., Franke S., Lüthje S., Heinz E. (2005). Are glucocerebrosides the predominant sphingolipids in plant plasma membranes? Plant Physiol. Biochem. 43: 1031–1038. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Kida T., Imai H., Morishige J., Yamashita R., Matsuoka H., Uozumi S., Satouchi K., Nagano M., Tokumura A. (2013). Identification of a sphingolipid-specific phospholipase D activity associated with the generation of phytoceramide-1-phosphate in cabbage leaves. FEBS J. 280: 3797–3809. [DOI] [PubMed] [Google Scholar]
- Tartaglio V., Rennie E.A., Cahoon R., Wang G., Baidoo E., Mortimer J.C., Cahoon E.B., Scheller H.V. (2016). Glycosylation of inositol phosphorylceramide sphingolipids is required for normal growth and reproduction in Arabidopsis. Plant J., http://dx.doi.org/10.1111/tpj.13382 [DOI] [PubMed] [Google Scholar]
- Tellier F., Maia-Grondard A., Schmitz-Afonso I., Faure J.-D. (2014). Comparative plant sphingolipidomic reveals specific lipids in seeds and oil. Phytochemistry 103: 50–58. [DOI] [PubMed] [Google Scholar]
- Wang W., et al. (2008). An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in Arabidopsis. Plant Cell 20: 3163–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zäuner S., Ternes P., Warnecke D. (2010). Biosynthesis of sphingolipids in plants (and some of their functions). In Sphingolipids as Signaling and Regulatory Molecules, C. Chalfant and M. Del Poeta, eds (New York: Springer-Verlag; ), pp. 249–263. [DOI] [PubMed] [Google Scholar]
- Zhang Y., et al. (2016). Golgi-localized STELLO proteins regulate the assembly and trafficking of cellulose synthase complexes in Arabidopsis. Nat. Commun. 7: 11656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.