Organ-specific shade-regulated transcriptome analysis identifies genes whose differential expression may underlie organ-specific elongation and reveals a growth promotion gene expression signature.
Abstract
In response to neighbor proximity, plants increase the growth of specific organs (e.g., hypocotyls) to enhance access to sunlight. Shade enhances the activity of Phytochrome Interacting Factors (PIFs) by releasing these bHLH transcription factors from phytochrome B-mediated inhibition. PIFs promote elongation by inducing auxin production in cotyledons. In order to elucidate spatiotemporal aspects of the neighbor proximity response, we separately analyzed gene expression patterns in the major light-sensing organ (cotyledons) and in rapidly elongating hypocotyls of Arabidopsis thaliana. PIFs initiate transcriptional reprogramming in both organs within 15 min, comprising regulated expression of several early auxin response genes. This suggests that hypocotyl growth is elicited by both local and distal auxin signals. We show that cotyledon-derived auxin is both necessary and sufficient to initiate hypocotyl growth, but we also provide evidence for the functional importance of the local PIF-induced response. With time, the transcriptional response diverges increasingly between organs. We identify genes whose differential expression may underlie organ-specific elongation. Finally, we uncover a growth promotion gene expression signature shared between different developmentally regulated growth processes and responses to the environment in different organs.
INTRODUCTION
The light environment in dense vegetation is characterized by reduced PAR and hence represents a threatening situation to which plants respond with a variety of growth and developmental strategies (Casal, 2013). Broadly speaking, plants are classified into shade-tolerant and shade-avoiding species, which can be distinguished based on their response to shade signals (Gommers et al., 2013). In shade-avoiding species, neighboring plants, prior to direct shading, trigger growth responses aimed at reaching unfiltered sunlight in anticipation of an unfavorable environment (de Wit et al., 2012; Casal, 2013). The shade-avoidance syndrome comprises a variety of growth and developmental responses in several plant organs (Casal, 2013). Stems and hypocotyls elongate, petioles adopt a more upright position (hyponasty) and elongate, while leaf blade growth is often reduced and branching is inhibited (Kozuka et al., 2010; Casal, 2013; Reddy et al., 2013; de Wit et al., 2015). It is generally accepted that these organ-specific growth responses are the result of resource reallocation and are favorable to maximize growth and survival in an environment with limited light availability (Cagnola et al., 2012; Casal, 2013).
Foliar shade is characterized by the selective reduction of the red and blue wavebands absorbed by photosynthetic pigments, while other parts of the spectra, such as green and far red, are less depleted (Casal, 2013). Hence, under a canopy, there is a reduction in PAR and the red/far-red (R/FR) ratio. Such a reduction in the R/FR ratio also occurs during neighbor detection due to the reflection of FR light by neighboring leaves (Casal, 2013). The phytochrome (phy) photoreceptors play a crucial role in shade and neighbor perception, with phyB having a predominant function in the model plant Arabidopsis thaliana (Franklin and Quail, 2010). The reduction in blue light typical of true shade is sensed by the cryptochrome photoreceptors (Sellaro et al., 2010; Keller et al., 2011; Pedmale et al., 2016). In young seedlings, cotyledons are considered to be the primary site of low R/FR perception; nevertheless, several studies have shown that other parts of the plant can also sense this light cue (Morgan et al., 1980; Tanaka et al., 2002; Procko et al., 2014; Nito et al., 2015).
Signaling events triggered by shade are best understood following phyB-mediated detection of the reduced R/FR ratio (Casal, 2013). In the sun (high R/FR ratio), active phyB inhibits several elongation-promoting bHLH factors from the Phytochrome Interaction Family (PIF; de Wit et al., 2014; Leivar and Monte, 2014). Low R/FR releases phyB-mediated inhibition of several PIFs, leading to shade-induced elongation of hypocotyls and petioles (Lorrain et al., 2008; Keller et al., 2011; Leivar et al., 2012; Li et al., 2012; de Wit et al., 2015; Mizuno et al., 2015; Nozue et al., 2015). Shade-induced auxin biosynthesis in the cotyledons and young leaves followed by transport of the phytohormone into the hypocotyl is considered essential to trigger hypocotyl elongation (Steindler et al., 1999; Tao et al., 2008; Keuskamp et al., 2010; de Wit et al., 2014; Procko et al., 2014, 2016). In response to shade, PIF transcription factors directly regulate the expression of auxin biosynthesis and auxin response genes, thereby providing a link between environmental perception and the growth response (Hornitschek et al., 2012; Li et al., 2012; Hersch et al., 2014; Nozue et al., 2015).
Several lines of evidence, including analyses of genome-wide changes in gene expression, suggest that multiple hormones play roles during the shade-avoidance syndrome (Devlin et al., 2003; Sessa et al., 2005; Tao et al., 2008; Hornitschek et al., 2012; Leivar et al., 2012; Li et al., 2012; Crocco et al., 2015; Nito et al., 2015; Nozue et al., 2015; Das et al., 2016). In particular, brassinosteroids, gibberellic acid (GA), and jasmonic acid have been shown to function in growth regulation in shaded environments (Djakovic-Petrovic et al., 2007; Kozuka et al., 2010; Robson et al., 2010; Keller et al., 2011; Keuskamp et al., 2011; Cifuentes-Esquivel et al., 2013; Bou-Torrent et al., 2014; Moreno and Ballaré, 2014; Nozue et al., 2015; Das et al., 2016; Procko et al., 2016). Abscisic acid (ABA), cytokinins, and ethylene have also been linked to a subset of shade-regulated responses (Carabelli et al., 2007; Pierik et al., 2009; Reddy et al., 2013; Zheng et al., 2013; Pierik and Testerink, 2014). A recurrent problem with many transcriptional studies is that shade-regulated growth is most often measured in hypocotyls but is being correlated with gene expression analyzed in whole seedlings/plants. In such studies of whole seedlings, including both growing and nongrowing tissues, it is challenging to directly relate changes in gene expression with the growth response.
In order to characterize the transcriptional events leading to shade-induced hypocotyl elongation, we analyzed changes in transcript abundance at different time points during shade treatment in dissected Arabidopsis cotyledons and hypocotyls. We concentrated our analysis on these two organs because the former is considered to be the primary shade-sensing organ, while elongation is rapidly triggered in the hypocotyl (Tao et al., 2008; Cole et al., 2011; de Wit et al., 2014; Procko et al., 2014). Genetic experiments were designed based on interesting expression patterns, enabling us to test current models of shade-regulated hypocotyl elongation. Moreover, this data set reveals the expression signature of enhanced organ growth at the genome-wide level.
RESULTS
Selecting the Time Points for Shade-Induced Transcriptional Reprogramming
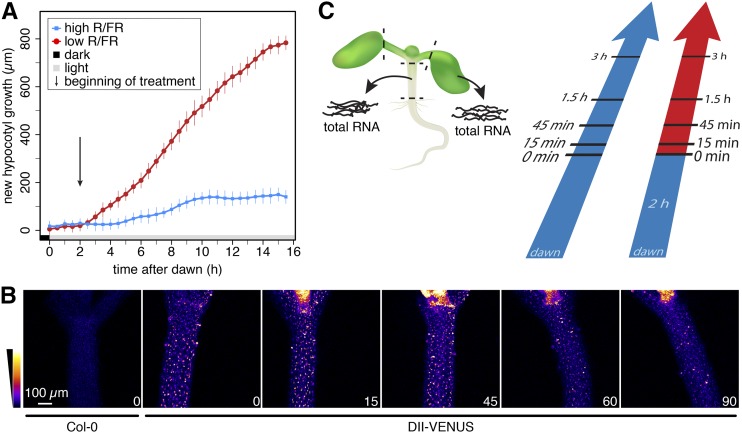
In order to select the time points of our gene expression analysis, we first determined the timing of shade-induced hypocotyl elongation. In response to additional FR light mimicking neighbor detection (low R/FR, referred to as shade), we observed enhanced hypocotyl elongation in Arabidopsis seedlings after approximately 1 h of shade treatment (Figure 1A; Supplemental Figure 1A). Given that shade-induced hypocotyl growth is likely due to an increase in auxin (indole-3-acetic acid [IAA]) concentration (Tao et al., 2008; Keuskamp et al., 2010; Hornitschek et al., 2012; Li et al., 2012; Procko et al., 2014), we indirectly determined auxin levels in the hypocotyl using the DII-VENUS auxin sensor (Brunoud et al., 2012). We observed a reduction of the DII-VENUS signal in the hypocotyl within 1 h, indicating increased auxin levels (Figure 1B). Importantly, in mutants that are impaired in shade-induced auxin production (pif7 and shade avoidance3 [sav3]) (Tao et al., 2008; Li et al., 2012), the DII-VENUS signal did not significantly decrease following shade treatment (Supplemental Figure 1B). We decided to concentrate on early time points during the response to shade for two reasons: (1) to capture the early signaling events believed to primarily occur in cotyledons and (2) to capture the transition from slow elongation to fast elongation, which occurs rapidly in hypocotyls (Cole et al., 2011) (Figure 1A). The aforementioned experiments prompted us to select 15 and 45 min time points to capture changes in transcript abundance occurring prior to an obvious change in hypocotyl growth and 90 and 180 min, when hypocotyls of the shade-treated seedlings grew faster than control seedlings. To correct for diurnally regulated gene expression, we collected hypocotyl and cotyledon samples for high and low R/FR-treated seedlings at each time point for investigation by RNA-seq analysis (Figure 1C).
Figure 1.
Experimental Design for the Organ-Specific Shade-Regulated Transcriptional Response.
(A) Growth analysis of 5-d-old Arabidopsis seedlings grown in long days and treated with high or low R/FR at ZT2. Additional hypocotyl growth of 26 and 20 seedlings in high and low R/FR, respectively, was recorded in 30-min time intervals. Measurements were smoothed and expressed as average. Error bar represent the 2× se of the mean prior to smoothing.
(B) DII-VENUS signal intensity in hypocotyls decreases within the first hour in low R/FR. Seedlings were grown as in (A). Similar image slices of representative seedlings are shown. The duration of the low R/FR treatment in minutes is given in the bottom right corner of each image.
(C) Schematic representation of sample preparation for subsequent RNA-seq analysis. Seedlings in both light conditions were separately sampled on day 6 after the indicated time in high (blue) or low (red) R/FR. Dashed lines represent the locations of the applied cuts.
The Shade-Induced Transcriptional Program Becomes More Organ-Specific with Time
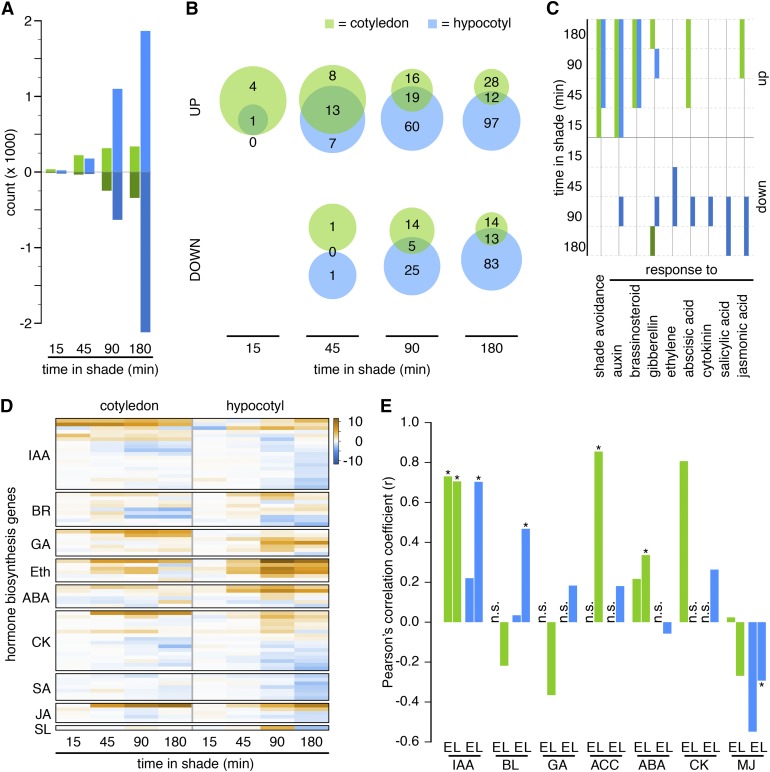
Using principle component (PC) analysis, we showed that PC1 separates samples according to the organ, while PC2 separates them according to time and treatment (shade or control) (Supplemental Figure 2A). Genes showing shade-regulated expression were defined using a 2-fold change threshold with an adjusted P value < 0.01. To validate our RNA-seq data, we performed quantitative RT-PCR (RT-qPCR) on 21 selected genes from an independent experiment (Supplemental Figure 1C). Using these stringent criteria, we found that a similar number of genes were shade-regulated in hypocotyls and cotyledons during the early time points (15 and 45 minutes). In contrast, at times when hypocotyl growth was significantly enhanced in shade-treated samples, many more genes were regulated in hypocotyls than in cotyledons (Figure 2A; Supplemental Data Sets 1 and 2). To compare our data with previous analyses performed at the level of whole seedlings, we selected the study from Li et al. (2012) because they also used RNA-seq. The overlap between both studies was particularly small between genes specifically regulated by shade in hypocotyls and the previous data set (Supplemental Figure 2C). This indicates that our strategy has a great potential to identify novel shade-regulated genes in hypocotyls, which contribute less to changes in RNA than cotyledons. In order to concentrate on processes regulated by shade rather than individual genes, we performed Gene Ontology (GO) enrichment analyses. The list of significantly enriched GO terms at each time point was analyzed separately for up- and downregulated genes in each organ (Supplemental Data Set 3). Among the early shade-induced genes, both organs responded similarly to the treatment, while later, the gene expression response became markedly organ specific (Figure 2B). This trend was less apparent when comparing individual genes (Supplemental Figure 2B), and it was specific for upregulated genes (Figure 2B).
Figure 2.
Regulation of Transcript Abundance Becomes More Organ Specific with Time.
(A) Number of significantly up- or downregulated genes in cotyledons and hypocotyls over time.
(B) Set analyses of enriched GO terms between organs at various time points.
(C) Absent/present analysis of selected GO terms. Solid boxes represent significant detection (weight ≤ 0.05). Green shades = cotyledon; blue shades = hypocotyl.
(D) Hierarchically clustered log fold change of shade-regulated genes involved in hormone biosynthetic pathways.
(E) Correlation analysis of the transcriptional responses to shade and hormone treatments (Goda et al., 2008). For each hormone and organ, an early time point comparing 45 min in shade and 60 min of hormone treatment (marked with E) and a late time point comparing 3 h of treatment in both experiments (marked with L) were investigated. n.s., not significantly overlapping gene lists (hypergeometric test: P < 2.5 × 10−4); asterisks indicate P < 0.05 for the correlation coefficient. Eth, ethylene; SL, strigolactone.
Shade Regulates the Expression of Multiple Hormone Pathways in Cotyledons and Hypocotyls
The importance of multiple hormones in shade-regulated morphogenesis prompted us to focus on GO terms associated with hormone responses (Tao et al., 2008; Hornitschek et al., 2012; Li et al., 2012; Bou-Torrent et al., 2014; Procko et al., 2014; Nozue et al., 2015). Consistent with the early role of auxin in this process, the term response to auxin was significantly overrepresented among genes upregulated by shade both in hypocotyls and cotyledons at all time points of our analysis (Figure 2C). The GO category response to brassinosteroid showed a similar pattern in both organs during the last three time points. GO terms associated with responses to other hormones were more organ specific. For example, response to ABA was selectively overrepresented among upregulated genes in cotyledons. The term response to jasmonic acid was overrepresented among upregulated genes in the cotyledons at 90 and 180 min, while in the hypocotyl, it was overrepresented among late downregulated genes, suggesting an opposite jasmonic acid gene expression pattern in cotyledons versus hypocotyls during shade treatment (Figure 2C).
In Arabidopsis, shade or low light leads to the rapid production of auxin and ethylene, while GA levels do not change significantly and levels of the brassinosteroid castasterone decrease (Vandenbussche et al., 2003; Tao et al., 2008; Bou-Torrent et al., 2014). Rapid auxin production has been linked to PIF-mediated YUCCA (YUC) expression (Hornitschek et al., 2012; Li et al., 2012), which prompted us to analyze the expression patterns of hormone biosynthetic genes in our data set. The expression of four YUC genes (YUC2, 5, 8, and 9) increased within 15 min of shade treatment, specifically in cotyledons, while YUC3 and YUC8 expression increased in hypocotyls later in response to shade (Figure 2D; Supplemental Data Set 2). Among brassinosteroid (BR) biosynthetic genes, PHYB ACTIVATION TAGGED SUPPRESSOR1 (BAS1), which is involved in BR catabolism (Turk et al., 2005), was the most robustly shade-induced gene. The BR biosynthetic gene DWARF4 (DWF4) was also moderately upregulated in hypocotyls, while in cotyledons, its expression was reduced at the later time points (Figure 2D; Supplemental Data Set 2) (Choe et al., 1998). Members of three GA biosynthetic gene families were shade regulated, including GIBBERELLIN3-OXIDASE (GA3ox) and GA20ox genes (encoding active GA-producing enzymes) and GA2ox genes (encoding GA-inactivating enzymes [Yamaguchi, 2008]). Interestingly, different members of the AtGA2ox and AtGA20ox families were shade induced in hypocotyls versus cotyledons (Figure 2D; Supplemental Figure 2D). In contrast, AtGA3ox1 was the only member of this family to be shade induced only in the hypocotyl. Multiple members of the 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE (ACS) family of ethylene biosynthetic genes were induced by shade, with more being induced in hypocotyls (ACS4, 5, 6, 8, and 11) than cotyledons (ACS4 and 8) (Figure 2D; Supplemental Data Set 2). Among the hormones typically considered to be growth inhibitory, two salicylic acid biosynthetic genes showed selective downregulation by shade in hypocotyls (ISOCHORISMATE SYNTHASE1 [ICS1]/SALICYLIC ACID INDUCTION DEFICIENT2 and ICS2) (Garcion et al., 2008), while expression of the ABA biosynthetic genes NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3 (NCED3) and NCED5 was particularly enhanced in hypocotyls (Nambara and Marion-Poll, 2005). Several members of the CYTOKININ OXIDASE (CKX) family, encoding enzymes inactivating cytokinin, were regulated by shade (Schmülling et al., 2003). Most prominently, CKX5 was strongly induced in cotyledons, whereas CKX4 and CKX7 expression was downregulated in both organs. These data show rapid transcriptional regulation of multiple hormone biosynthesis genes, with examples of organ specificity in several instances.
To analyze the potential effects of shade-regulated hormone levels further, we compared the effects of our shade treatment with the regulation of gene expression elicited by seven different phytohormones (1 and 3 h treatments) (Goda et al., 2008). For comparisons yielding a significant overlap between shade and hormone treatments, we correlated the expression patterns (Figure 2E, significant correlation coefficients are marked with an asterisk). Very high positive correlations were identified for auxin and shade in both cotyledons and hypocotyls. For brassinosteroids, we detected a significant positive correlation at the late time point, specifically in hypocotyls. In contrast, significant positive correlations were detected between ABA or 1-aminocyclopropane-1-carboxylic acid (ethylene precursor) treatments and shade only in cotyledons. Finally, we detected a significant negative correlation between methyl jasmonate and shade treatment, specifically in hypocotyls, suggestive of a downregulation of defense responses (Figure 2E). Taken together, these analyses suggested a strong overlap between shade and auxin treatments in both hypocotyls and cotyledons, while other hormones showed more organ-specific patterns (Figure 2E).
PIF Transcription Factors Directly Regulate Many Early Shade-Induced Genes in Both Organs
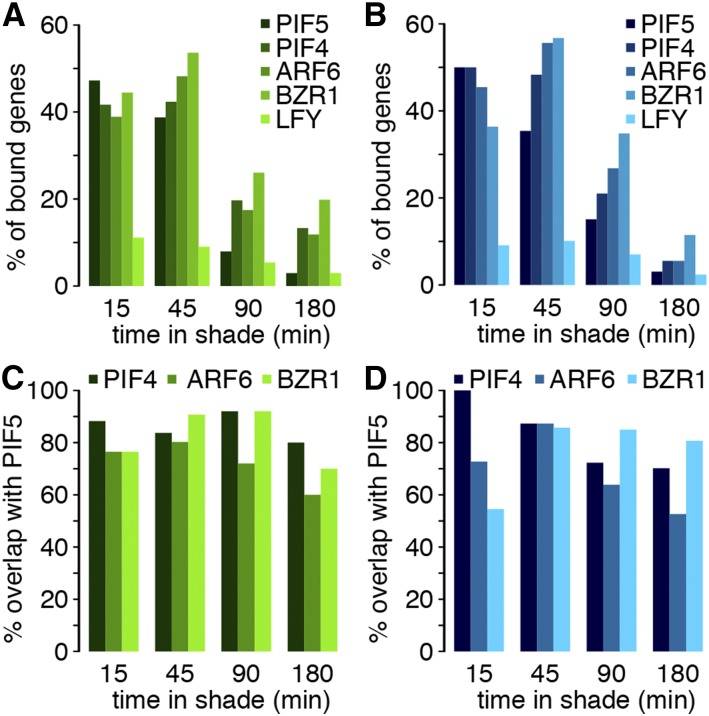
PIF transcription factors are considered to act early in this pathway by serving as a direct link between the phyB photoreceptor and the transcriptional program (Lorrain et al., 2008; Hornitschek et al., 2012; Leivar et al., 2012; Li et al., 2012). This model predicts that many early shade-regulated genes are direct PIF target genes. To address this question, we analyzed the overlap between the shade-regulated genes identified here and the PIF5 binding sites determined in shade-treated seedlings (Hornitschek et al., 2012). Given that PIF5 primarily acts as a transcriptional activator (Hornitschek et al., 2012), we separately analyzed shade-induced and -repressed genes. Strikingly, approximately half of the genes upregulated by shade in hypocotyls and cotyledons at 15 and 45 min were also bound by PIF5 (Figures 3A and 3B). This percentage strongly declined at later time points (Figures 3A and 3B). These data are consistent with an early involvement of PIF5 in the shade response. Moreover, given that many PIF target genes are themselves transcriptional regulators (Hornitschek et al., 2012; Oh et al., 2012; Pfeiffer et al., 2014), this finding suggests the involvement of other transcription factors later in the response. PIFs are known to share many target genes; for example, PIF4, PIF5, and PIF7 bind to the YUC8 promoter (Hornitschek et al., 2012; Li et al., 2012; Pfeiffer et al., 2014). We tested the degree of PIF binding overlap by reanalyzing genome-wide PIF4 ChIP (chromatin immunoprecipitation) data in etiolated seedlings (Oh et al., 2012). Although PIF4 ChIP and PIF5 ChIP were done using different biological materials, the comparison between shade-regulated gene expression and PIF4 or PIF5 binding sites both identified ∼50% of the early shade-regulated genes as bound by those PIFs (Figures 3A and 3B; Supplemental Data Set 4). Importantly, there was a very high overlap in binding sites, which is consistent with an earlier study that showed that both PIF4 and PIF5 bind to the same region of selected shade-regulated genes (Figures 3C and 3D) (Hornitschek et al., 2012). The degree of overlap between PIF binding and shade-regulated expression was similar in hypocotyls and cotyledons, suggesting that these PIFs directly regulate a large fraction of early shade-induced genes in both organs (Figures 3A and 3B). Interestingly, these patterns were specific for genes upregulated by shade, suggesting PIF regulation of early shade upregulated but not downregulated genes (Figure 3; Supplemental Figure 3).
Figure 3.
Many Early Shade-Regulated Genes in Both Hypocotyls and Cotyledons Are Likely Direct PIF Target Genes.
(A) and (B) Relative number of shade-induced direct target genes (DTG) in the cotyledon (A) and hypocotyl (B).
(C) and (D) Overlap between shade-induced DTG of PIF4, ARF6, and BZR1 relative to shade-induced DTG of PIF5 in the cotyledon and hypocotyl, respectively.
The overlap between shade, auxin, and brassinosteroid-induced expression patterns (Tao et al., 2008; Keuskamp et al., 2011; Das et al., 2016) (Figure 2), together with the high degree of chromatin co-occupancy of AUXIN RESPONSE FACTOR6 (ARF6), BRASSINAZOLE-RESISTANT1 (BZR1), and PIF4 (Oh et al., 2014), prompted us to characterize the binding sites of these transcription factors in the context of shade-regulated genes identified here. As a control, we compared these data with the binding sites of LEAFY (LFY) (Moyroud et al., 2011), a transcription factor not known to be required for this morphogenetic response. We found that many shade upregulated genes were bound by PIF5, PIF4, ARF6, and BZR1 (Figures 3A and 3B). As observed for PIF5 and PIF4, the fraction of genes upregulated by shade that is bound by ARF6 and BZR1 was very high early during the shade response but declined markedly later (Figures 3A and 3B). Moreover, we found a high overlap between ARF6- or BZR1- and PIF5-bound genes, indicating that all four transcription factors bind to the same shade-regulated genes (Figures 3C and 3D). In contrast, only a low fraction of shade-regulated genes was bound by LFY at all time points (Figures 3A and 3B; Supplemental Figure 3). Collectively, these data are consistent with an early role of PIF5, PIF4, ARF6, and BZR1 during the response to shade and the potential for the concerted control of shade upregulated genes by different classes of transcription factors.
Shade-Regulated Expression of a Subset of Auxin-Responsive Genes in the Hypocotyl Occurs Independently of Auxin Transport from the Cotyledons
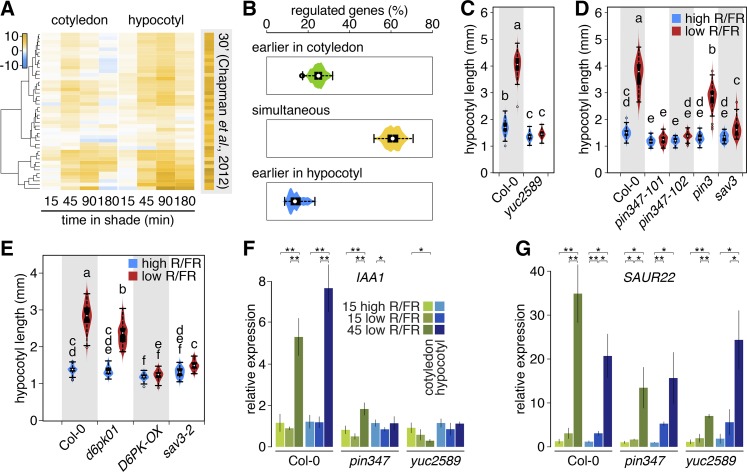
According to the current model of shade-induced hypocotyl elongation, shade leads to increased auxin production in the cotyledons, which is subsequently transported to the hypocotyl to promote growth (Tanaka et al., 2002; Tao et al., 2008; Keuskamp et al., 2010; Li et al., 2012; Procko et al., 2014). This suggests that following shade treatment, it might be possible to observe a temporal delay in the regulation of auxin-responsive genes between cotyledons and hypocotyls. To verify this prediction, we analyzed shade regulation of genes that rapidly (within 30 minutes) respond to picloram (a synthetic auxin) application in hypocotyls (Chapman et al., 2012). We detected expression of 46 (70%) picloram-regulated genes in our data set, among which 43 were regulated by shade. In the hypocotyl, the expression of all of these genes responded very similarly to shade and picloram application, revealing an astounding overlap between both treatments (Figure 4A). The expression pattern of this set of genes was also very similar in shade-treated cotyledons (Figure 4A). In both tissues, most of these genes were induced after 45 min of shade treatment. However, while in the hypocotyl, the expression levels of many of these genes continued to rise until 90 min; in the cotyledons, the peak of expression was typically at 45 min (Figure 4A). As this initial analysis did not provide evidence for an obvious temporal delay of auxin-regulated genes between hypocotyls and cotyledons, we tested this further by analyzing the expression of genes in response to a 2-h picloram application (Chapman et al., 2012). These genes displayed a more organ-specific shade response but did not provide evidence for an earlier auxin response in cotyledons than hypocotyls (Supplemental Figure 4A). In addition, we analyzed shade regulation of all the genes identified as being auxin regulated in two studies (Nemhauser et al., 2006; Chapman et al., 2012). The 539 auxin-regulated genes that are regulated by shade in both hypocotyls and cotyledons were categorized as follows: regulated simultaneously by shade in both organs, regulated first in the hypocotyl, and regulated first in the cotyledons. We then varied the P value and/or fold change criteria to redefine shade regulation. More than half of these genes showed simultaneous shade regulation in both organs, while ∼15% responded first in the hypocotyl and ∼25% responded first in the cotyledons (Figure 4B). The outcome of this analysis did not depend on the criteria selected to define shade regulation (Figure 4B; Supplemental Figure 4B). Hence, with the temporal resolution used here, we did not observe an obvious pattern of earlier shade regulation among auxin-regulated genes in cotyledons compared with the hypocotyl.
Figure 4.
The Shade Response of Auxin-Regulated Genes in Hypocotyls Depends on Cotyledon-Mediated Auxin Production as Well as Local Signals.
(A) Hierarchical clustering of picloram-inducible shade-regulated genes showing various response patterns within and between organs. Each gene is significantly enriched in at least one condition.
(B) Repeated classification of transcriptional responses to shade of auxin-regulated genes using various criteria of significance (1.5 ≤ fold change ≤ 5; 0.05 ≥ adjusted P ≥ 0.001).
(C) to (E) Hypocotyl elongation of yuc2 yuc5 yuc8 yuc9 (n > 100), pin (22 ≤ n ≤ 69), d6pk (n ≥ 50), and sav3-2 (n ≥ 28) mutant lines in high and low R/FR, respectively. Data are represented as box plots and distribution of measurements (violin plot). Different letters indicate significant differences (ANOVA; significance level = 0.01).
(F) and (G) Transcript levels of the auxin-inducible genes IAA1 and SAUR22, respectively, were analyzed by RT-qPCR in Col-0, pin3 pin4 pin7, and yuc2 yuc5 yuc8 yuc9. P value for significance of difference in mean (t test) is given above the bars. *P < 0.05 and **P < 0.01; n.s., not significant. Error bars indicate ± 2× se of three biological replicates prepared from separate pools of seedlings, with three technical repeats per biological replicate.
The importance of YUC genes in shade-induced hypocotyl elongation remains poorly understood, given that a quintuple yuc mutant only shows a moderate phenotype (Li et al., 2012). We detected upregulation of YUC2, YUC5, YUC8, and YUC9 in cotyledons after 15 min of shade treatment (Supplemental Figure 4C). We therefore tested shade-regulated hypocotyl elongation in a yuc2 yuc5 yuc8 yuc9 quadruple mutant and found that under our conditions, hypocotyl growth of this quadruple mutant was unresponsive to shade. This validates the importance of PIF-mediated YUC expression in this growth process (Figure 4C) (Hornitschek et al., 2012; Li et al., 2012; see also Nozue et al., 2015). These data also suggest that YUC-mediated auxin production in cotyledons triggers elongation of the hypocotyl. To test whether induction of a YUC gene in cotyledons can induce hypocotyl elongation, we used an inducible YUC3 line in which this gene is selectively expressed in photosynthetic tissues (Chen et al., 2014). Interestingly, following YUC3 induction, this line showed marked hypocotyl elongation, confirming that cotyledon-derived auxin is sufficient to trigger hypocotyl elongation (Supplemental Figure 4D). PIN-FORMED3 (PIN3) was previously proposed to be essential for shade-induced hypocotyl elongation (Keuskamp et al., 2010); however, under our growth conditions, a pin3 mutant showed a diminished but robust response to shade (Figure 4D). The expression of PIN3, PIN4, and PIN7 was induced following shade treatment (Supplemental Figure 4E). Moreover, shade-induced hypocotyl elongation was abolished in the pin3 pin4 pin7 triple mutant (Figure 4D). The activity of several PIN proteins requires phosphorylation by members of the D6 PROTEIN KINASE (D6PK) family (Zourelidou et al., 2014). The expression of D6PK and D6PKL1 was selectively upregulated in the hypocotyls of shade-treated seedlings (Supplemental Data Set 1). Therefore, we analyzed shade-regulated hypocotyl elongation in d6pk d6pkl1 double mutants and in plants ectopically expressing D6PK. The loss-of-function mutant displayed a reduced response to shade, and seedlings expressing D6PK driven by the 35S promoter were unresponsive to the treatment, indicating that D6PK proteins are important for shade-induced growth (Figure 4E). Hence, our expression data guided our genetic analyses of shade-induced hypocotyl elongation, providing strong support for the importance of YUC-mediated auxin production in the cotyledons and regulated PIN-mediated auxin transport in this process. Our data suggest that PIFs regulate more than auxin biosynthesis in response to shade. To test this idea, we compared the auxin sensitivity of the yuc2 yuc5 yuc8 yuc9 and pif4 pif5 pif7 triple mutants. Both mutants are similarly impaired in their response to low R/FR, but while the yuc2 yuc5 yuc8 yuc9 mutant showed a wild-type response to auxin, the response of pif4 pif5 pif7 was reduced, confirming that PIFs regulate more than auxin biosynthesis (Supplemental Figure 4F).
Our gene expression analysis identified auxin-regulated genes that were very rapidly shade induced in the hypocotyl (Figure 4; Supplemental Figure 4G). To determine whether the expression of such genes depended on YUC-mediated auxin production and/or PIN-mediated auxin transport, we used RT-qPCR to analyze gene expression in cotyledons and hypocotyls of dissected wild-type, pin3 pin4 pin7, and yuc2 yuc5 yuc8 yuc9 mutant seedlings. This analysis revealed different types of expression patterns (Figures 4F and 4G). Shade-induced IAA1 expression in cotyledons and hypocotyls was abolished in the pin3 pin4 pin7 and yuc2 yuc5 yuc8 yuc9 mutants. This indicates that shade induction of IAA1 is dependent on YUC-mediated IAA production and PIN-mediated IAA transport. The situation was different for SMALL AUXIN UP RNA22 (SAUR22), another typical early auxin-responsive gene (Chapman et al., 2012; Ren and Gray, 2015). SAUR22 induction was apparent after 15 min of shade and further increased at 45 min both in cotyledons and hypocotyls. Interestingly, SAUR22 induction remained robust in the pin3 pin4 pin7 and yuc2 yuc5 yuc8 yuc9 mutants, suggesting a local response in the hypocotyl to the shade signal (Figures 4F and 4G). Consistent with this hypothesis, we found that shade led to a small but significant reduction in the DII-VENUS signal in the hypocotyls of pin3 pin4 pin7 triple mutants (Supplemental Figure 4H). Collectively, our analysis of typical auxin-regulated genes suggests that shade controls gene expression both through distal (cotyledon-derived auxin) and local signals (Figure 4; Supplemental Figure 4). Very rapid and coordinated low R/FR-induced expression of members of the SAUR19 group (SAUR19-24) (Supplemental Figure 4G) was particularly interesting in this context because induction of the tested members still occurred in the pin and yuc mutants (Figure 4). To test their functional involvement in shade-induced hypocotyl elongation, we used an artificial microRNA line with downregulated expression of several members of the SAUR19 group (Spartz et al., 2012). Interestingly, the hypocotyl of these mutants was undistinguishable from the wild type in high R/FR, while in low R/FR, it was slightly but significantly shorter (Supplemental Figure 4I). This finding is consistent with the positive role of SAURs in elongation growth (Spartz et al., 2012, 2014). Finally, to test whether both the YUC-dependent and independent responses required the PIFs, we analyzed the expression of IAA1 and SAUR22 in pif4 pif5 pif7 hypocotyls and cotyledons and found that low R/FR induction of these genes was strongly attenuated in the pif triple mutant (Supplemental Figures 4J and 4K). Collectively, these data indicate that PIFs are required for the rapid shade induction of auxin related genes in both organs and that the local SAUR response in hypocotyls may contribute to shade-induced elongation.
Organ-Specific Hormonal Responses That May Underlie Selective Hypocotyl Growth Promotion
Previous studies have suggested that the organ-specific growth response elicited by shade might be due to a tissue-specific auxin response (de Wit et al., 2015). At a broad level, auxin response genes were upregulated both in cotyledons and hypocotyls (Figure 2; Supplemental Figure 5A and Supplemental Data Set 3; where the most significant GO term among genes upregulated in both tissues is: response to auxin). However, a careful inspection revealed interesting additional patterns. By comparing the correlation coefficient between rapid picloram-regulated genes and shade-regulated expression in cotyledons versus hypocotyls, we observed high correlation during the combined first two time points (15 and 45 min, r = 0.73), while this coefficient declined markedly for the last two time points (r = 0.51) (Chapman et al., 2012). The same analysis for two families of early auxin-responsive genes (SAUR and IAA) comprising many shade-regulated members showed similar patterns (Supplemental Figure 5). We conclude that for numerous early auxin response genes, shade led to similar expression patterns in cotyledons and hypocotyls after 15 and 45 min of treatment, while later the shade response, these patterns became more organ specific.
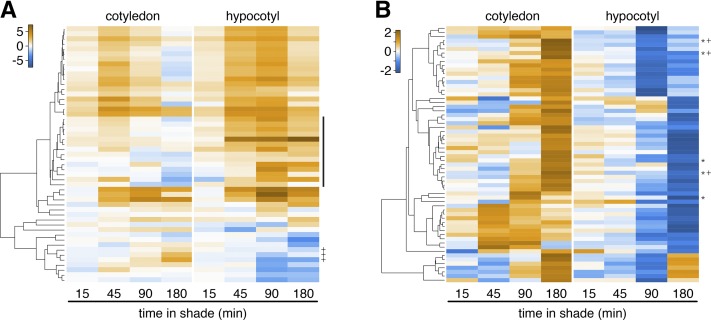
To extend this analysis, we classified the genes regulated by shade in both organs into four groups: upregulated in both organs, downregulated in both organs, upregulated in hypocotyls/downregulated in cotyledons, and upregulated in cotyledons/downregulated in hypocotyls (Figure 5B; Supplemental Figures 6A to 6C and Supplemental Data Set 5). Among the genes upregulated by shade in both organs, the GO terms response to auxin, response to brassinosteroid, shade avoidance, and polar auxin transport were heavily over represented (Supplemental Figure 6A and Supplemental Data Set 6). However, the most significantly overrepresented term among genes upregulated in cotyledons and downregulated in hypocotyls was response to auxin, with three SAUR genes contributing to this term (Figure 5B, marked with a +; Supplemental Data Sets 5 and 6). A closer inspection of the expression patterns of the SAUR genes also revealed that a number of these genes (middle cluster on Figure 5A) were significantly induced by shade in the hypocotyl, while the expression of these genes was rather repressed in cotyledons. Taken together, the analysis of shade-regulated auxin-responsive genes in cotyledons and hypocotyls revealed two interesting trends. First, the patterns of gene expression were more similar early than late during the shade treatment (Figures 4A and 5A; Supplemental Figure 5). Second, while overall, the majority of auxin-responsive genes were upregulated by shade in both hypocotyls and cotyledons, there were notable exceptions showing differential shade regulation in cotyledons versus hypocotyls (Figure 5; Supplemental Figure 5).
Figure 5.
Opposite Transcriptional Regulation by Shade of Several Early Auxin Response Genes in Hypocotyls versus Cotyledons.
Hierarchical clustering of transcriptional ratios of shade-regulated SAUR genes (A) and expression patterns (z-score) of cotyledon-induced and hypocotyl-repressed genes (B). The middle cluster in (A) is marked with a black bar. +, Three SAUR genes with opposite regulation direction in cotyledons and hypocotyls; *, genes contained in the GO category response to auxin.
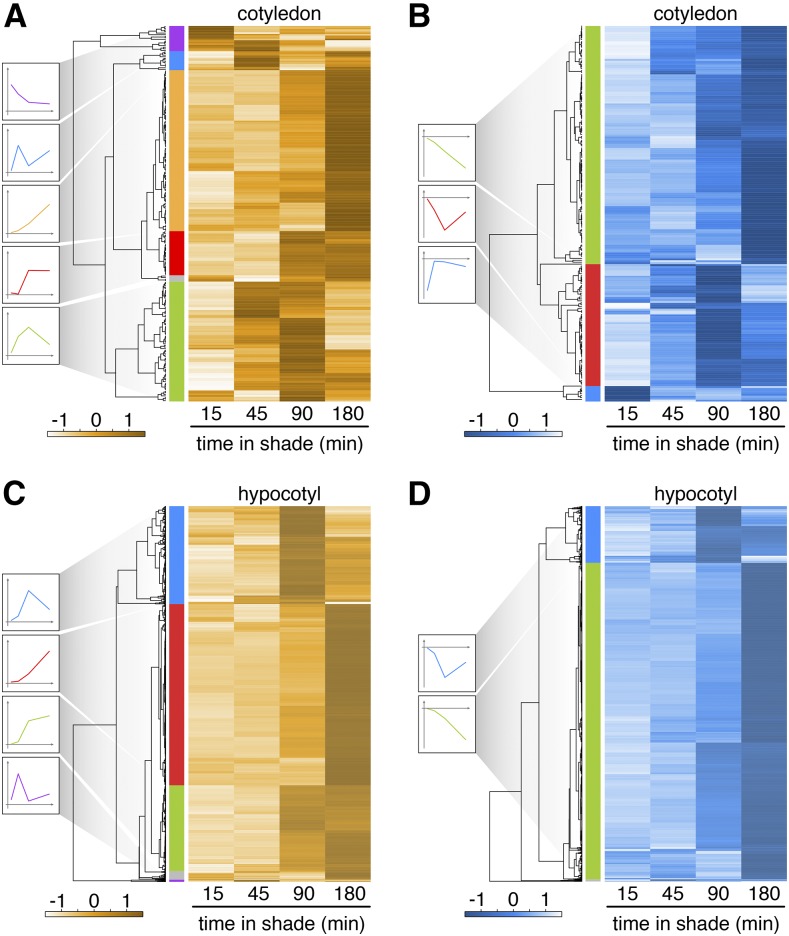
The organ-specific shade response may also be due to differential expression of other hormonal pathways involved in growth control. Examples for this were described above (Figures 2C and 2E). In addition, the GO terms response to jasmonic acid, response to ABA, and response to ethylene were enriched among genes upregulated in cotyledons and downregulated in the hypocotyl (Figure 5B; Supplemental Data Set 6). In order to investigate this more thoroughly, we performed separate analyses of genes regulated by shade in cotyledons and hypocotyls and grouped them as up- or downregulated. Each group was hierarchically clustered to identify different temporal response patterns, which are illustrated to the left of each cluster (Figure 6; Supplemental Data Set 7). An interesting trend that emerged by analyzing GO terms among genes that were upregulated in cotyledons is that several of these terms are associated with drought responses, including response to water deprivation, response to ABA, and hyperosmotic salinity response. This trend is consistent with the positive correlation between ABA-responsive genes and shade regulation in cotyledons (Figure 2E). These terms were particularly enriched among cotyledon-specific shade-regulated genes that were transiently induced by shade (peak at 90 min; Figure 6A, green cluster; Supplemental Data Set 7). Interestingly, the same GO terms were enriched in the transiently downregulated cluster of hypocotyl-specific shade-responsive genes (minimal expression after 90 min of shade; Figure 6D, blue cluster). Hence, our organ-specific transcriptome analysis reveals gene expression signatures pointing to distinct shade-elicited hormonal responses in hypocotyls versus cotyledons, which may explain selective growth enhancement in the hypocotyl (Figures 2, 5, and 6; Supplemental Figure 6).
Figure 6.
Hierarchical Clustering of Organ-Specific Shade-Regulated Genes.
Hierarchical clustering of normalized transcriptional ratios of cotyledon-specific ([A] and [B]) and hypocotyl-specific ([C] and [D]) upregulated ([A] and [C]) or downregulated ([B] and [D]) genes. Defined clusters are marked with various colors. Schematic regulatory patterns are shown for each cluster to the left.
Shade Treatment Leads to a Strong Genomic Signature of Enhanced Growth and Decreased Defense in the Hypocotyl
Due to the large number of shade-responsive genes in the hypocotyl, we will discuss the different clusters separately (Figures 6C and 6D; Supplemental Data Set 7). The expression of a small group of genes peaked at 45 min (Figure 6C, purple cluster). Notably, this group includes the gene ILI1 BINDING BHLH1, encoding a small HLH protein that was previously shown to regulate hypocotyl elongation (Bai et al., 2012; Ikeda et al., 2012). Among the genes whose expression peaked at 90 min (Figure 6C, blue cluster), several terms associated with hormone pathways were enriched (Supplemental Data Set 8). This group was also enriched in the terms response to mechanical stimulus and cellular glucan metabolic process, which are interesting in the context of a fast growing hypocotyl. A third cluster comprises genes that maintained high expression at 90 and 180 min of shade treatment (Figure 6C, green cluster). This group of genes was enriched in numerous GO categories that can be directly linked to the growth process, such as sterol biosynthetic process and many cell wall biosynthesis-associated terms (Supplemental Data Set 8). The expression of the fourth and largest group of genes peaked at 180 min (Figure 6C, red cluster; Supplemental Data Set 7). This group of genes was enriched in many GO terms, suggesting strong metabolic activity, including several terms related to protein biosynthesis but also protein degradation/modification (Supplemental Data Set 8). A comparative analysis of the shade regulation of ribosome subunit genes in hypocotyls versus cotyledons was quite revealing, as it showed robust and selective induction of these genes in hypocotyls (Supplemental Figure 6D). This gene cluster was also enriched in GO terms associated with cellular transport and cell wall remodeling terms (Supplemental Data Set 8). The overall list of GO terms and the timing at which they became enriched among shade upregulated genes in the hypocotyl are fully consistent with accelerated growth in this organ.
The genes that were downregulated in response to shade in the hypocotyl could be clustered into two main groups: a smaller group with lowest expression after 90 min and a large group with continued downregulation throughout shade treatment (Figure 6D, blue and green clusters, respectively). The first group was enriched in many GO terms related to hormones, including response to salicylic acid, response to ABA, response to jasmonic acid, and response to auxin. The term response to auxin among this list is somewhat surprising, given that it is also strongly enriched among genes upregulated in the hypocotyl following shade treatment (Figure 6C, blue cluster). This group of transiently downregulated genes comprised IAA28, ARF6, and five SAURs. The presence of the other hormone-associated terms is consistent with the growth-defense dilemma, as in rapidly growing plants, defense mechanisms are typically reduced (Moreno and Ballaré, 2014). This pattern is consistent with the negative correlation observed between shade-regulated expression in the hypocotyl and methyl jasmonate treatment (Figure 2E). Additional evidence for this hypothesis was also present among the list of genes with lowest expression after 180 min of shade treatment (Figure 6D, green cluster). Indeed, this list of genes was enriched in the terms glucosinolate biosynthetic process and defense response. We conclude that the analysis of gene expression in a rapidly growing organ provides striking genomic evidence for the opposite regulation of growth versus defense mechanisms. Of particular interest is the observation that this opposite regulation occurs selectively in the fast growing tissue (the hypocotyl) and is not the result of different regulatory processes occurring in different parts of the plant in response to a shade stimulus.
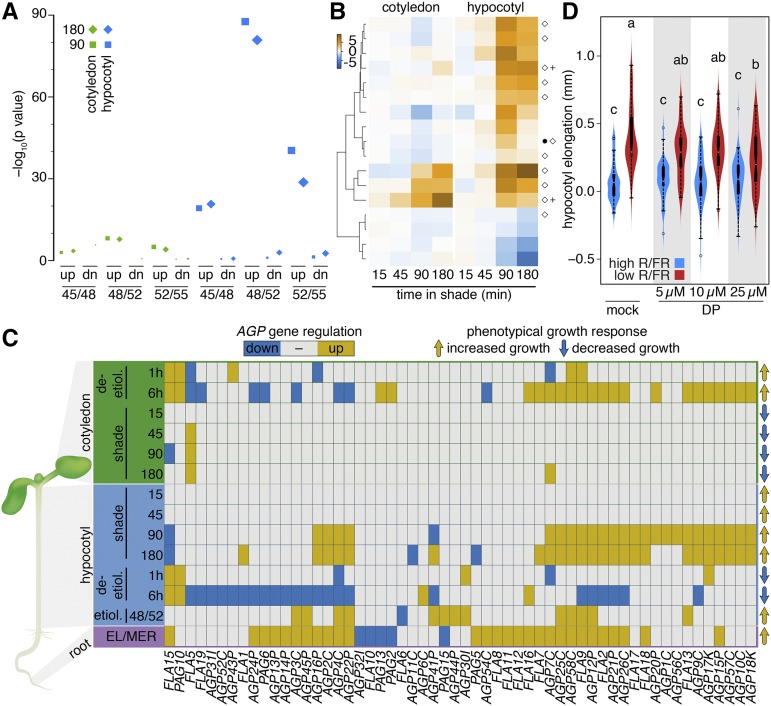
To test whether the gene expression signature observed in rapidly growing shade-treated hypocotyls is also observed in other situations leading to hypocotyl elongation, we compared our data with gene expression patterns in rapidly elongating etiolated hypocotyls (Pelletier et al., 2010). In this study, gene expression was compared among hypocotyls at 45 to 48 h, 48 to 52 h, and 52 to 55 h postgermination, with rapid hypocotyl elongation starting after 48 h (Pelletier et al., 2010). Interestingly, the shade expression patterns most significantly overlapped with the genes differentially regulated in etiolated seedlings between 48 and 52 h postgermination (Figure 7A). This high overlap in transcriptional profiles was specific for genes upregulated in hypocotyls, indicating that similar gene expression patterns underlie rapid hypocotyl elongation in green shade-treated seedlings and etiolated seedlings (Figure 7A). To test whether related expression patterns also underlie elongation in other organs, we identified genes differentially expressed between the root meristem and the early elongation zone (Supplemental Data Set 9; Wilson et al., 2015). Interestingly, among the genes upregulated in the root elongation zone, a large fraction (Supplemental Figure 7A, light green cluster) was also specifically upregulated in shade-treated hypocotyls at 180 min. GO analysis of this group of genes revealed the enrichment of response to brassinosteroid, which is consistent with a recent study showing enhance BR signaling in the root elongation zone (Chaiwanon and Wang, 2015) and the positive correlation between expression patterns induced by shade and brassinolide (BL) treatment (Figure 2E). In addition, we found multiple terms associated with cell elongation, such as sterol biosynthetic process, and several terms associated with cell wall remodeling (Supplemental Data Set 10).
Figure 7.
Similar Expression Profiles Underlie Rapid Growth in Different Conditions and Organs.
(A) Significance of the overlap between gene expression in etiolated and shade-treated seedlings. 45/48/52/55 correspond to hours in darkness as defined by Pelletier et al. (2010). The symbol size represents the log10-transformed gene count.
(B) Hierarchically clustered log fold change of shade-regulated members of the XTH family. Black circle, genes induced in rapidly growing etiolated seedlings; white diamond, genes induced in the root elongation zone; plus, PIF5 target gene.
(C) Transcriptional regulation of AGP genes in hypocotyls during shade avoidance, etiolation (Pelletier et al., 2010), and deetiolation (Sun et al., 2016) and in the root elongation zone (Wilson et al., 2015).
(D) Hypocotyl elongation on DMSO (mock) or different concentrations of DP (α,α-dipyridyl) during 1 day of treatment in high or low R/FR (38 ≤ n ≤ 65). Different letters indicate significantly different hypocotyl elongation (ANOVA; P value < 0.01).
Shade-regulated expression of many gene families encoding cell wall components or modifying enzymes has been observed before (Hornitschek et al., 2009; Keuskamp et al., 2011; Pedmale et al., 2016) (Supplemental Data Set 5). Notably, the expression of XTH (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE) genes regulated by PIFs and/or IAA and/or BL has been linked to growth promotion (Hornitschek et al., 2009; Sasidharan et al., 2010; Keuskamp et al., 2011; de Wit et al., 2015; Pedmale et al., 2016). Our organ-specific analysis allowed us to test whether shade-induced expression of XTH genes selectively occurred in the fast growing hypocotyl. Seventeen XTH genes were shade regulated under our experimental conditions. The expression of 10 of these was selectively induced in hypocotyls and can therefore be easily correlated with growth (Figure 7B). In contrast, the expression of some XTH genes was shade-induced in both hypocotyls and cotyledons (XTH15, 19, and 33), while the expression of four XTH genes specifically declined in the hypocotyls of shade-treated seedlings (e.g., XTH31 and 32). An inspection of cell wall-related gene families revealed that members of the arabinogalactan protein (AGP) gene family (Showalter et al., 2010) showed an expression pattern that was highly correlated with growth, with 26 members showing strong shade-induced expression in hypocotyls but not cotyledons (Figure 7C). We therefore analyzed the expression of this family during rapid hypocotyl elongation in the dark, in the root elongation zone, and during deetiolation based on a study in which hypocotyls and cotyledons were analyzed separately (Pelletier et al., 2010; Wilson et al., 2015; Sun et al., 2016). The pattern of AGP expression nicely correlated with growth in different organs both in the developmental context and in response to the environment (Figure 7C; Supplemental Data Set 11). Interestingly, nine of the AGPs that were selectively induced by shade in hypocotyls are also bound by PIF5, suggesting direct PIF regulation of AGP expression in hypocotyls (Supplemental Figure 7C, marked with a +). To test the functional importance of AGPs in shade-induced hypocotyl growth, we used α,α-dipyridyl, which interferes with the posttranslational hydroxyprolination of AGPs (Velasquez et al., 2011). Consistent with the notion that AGPs play a role in shade-regulated growth, this treatment led to reduced hypocotyl elongation, specifically in low R/FR-treated seedlings (Figure 7D). We conclude that our comparative analysis of gene expression in different organs and conditions triggering cell expansion revealed a surprisingly large overlap in gene expression signatures and identified members of the AGP family as particularly robust markers of growth (Figure 7).
DISCUSSION
Our analysis provided strong support for aspects of the current model of shade-induced hypocotyl elongation that had not been firmly established and identifies additional players in this pathway (Keuskamp et al., 2010; Won et al., 2011; Li et al., 2012; Procko et al., 2014) (Figure 4; Supplemental Figure 4). We showed that four YUC genes (YUC2, 5, 8, and 9) are shade induced within 15 min in cotyledons and that the corresponding quadruple mutant lacks shade-induced hypocotyl elongation (Figures 2D and 4C; Supplemental Figure 4C) (Nozue et al., 2015). Moreover, the induction of a YUC gene in green tissues is sufficient to promote hypocotyl elongation (Supplemental Figure 4D). Transport of cotyledon-derived auxin to the hypocotyl is required for the shade response (Steindler et al., 1999; Tanaka et al., 2002; Tao et al., 2008; Keuskamp et al., 2010; Procko et al., 2014). We confirm that shade leads to increased expression of PIN3 (Keuskamp et al., 2010), but also of PIN4 and PIN7 (Supplemental Figure 4). Moreover, our genetic analysis showed that all three PINs contribute to the shade response (Figure 4). In addition, we found that the expression of two genes encoding members of the D6PK protein kinase family is selectively induced in the hypocotyl. PIN3-mediated auxin transport depends on phosphorylation of this efflux carrier by D6PKs (Zourelidou et al., 2014). Interestingly, a mutant lacking the shade-regulated members of the D6PK family has a reduced hypocotyl response to shade, and plants ectopically expressing D6PK are severely defective for this response (Figure 4E). These findings identify D6PK proteins as regulators of shade-induced hypocotyl growth and show that the PIN transport system is transcriptionally regulated at multiple levels during shade avoidance.
Although shade-induced hypocotyl elongation requires an interaction between cotyledons and hypocotyls (see above), some shade responses occur in an organ-autonomous manner (Morgan et al., 1980; Procko et al., 2014; Nito et al., 2015; Zheng et al., 2016). The expression of members of the SAUR19 subfamily of early auxin-responsive genes (Chapman et al., 2012; Spartz et al., 2012) is very rapidly shade induced in hypocotyls, and SAUR22 induction still occurs in the pin3 pin4 pin7 and yuc2 yuc5 yuc8 yuc9 mutants, suggesting the involvement of a local response (Figure 4). The rapid induction of SAUR22 could be due to locally regulated auxin homeostasis (Zheng et al., 2016). Consistent with such a possibility, we found that DII-VENUS levels declined in the hypocotyls of shade-treated pin3 pin4 pin7 seedlings (Supplemental Figure 4H). Alternatively, the rapid induction of SAUR genes could be due to direct PIF-regulated expression, as ChIP-seq analysis showed that all members of the SAUR19 subfamily are bound by PIF5, that SAUR22 expression is reduced under low R/FR in the pif4 pif5 pif7 mutant, and that the expression of SAURs is reduced in pif mutants during deetiolation (Supplemental Figure 4K) (Hornitschek et al., 2012; Leivar et al., 2012; Sun et al., 2016). Irrespective of the mechanism underlying the regulation of SAUR genes in the hypocotyl, our results are consistent with a functional role for rapid shade-inducted SAUR19 subfamily expression (Supplemental Figure 4I). Such a mechanism may explain how local shade-simulating irradiation of white mustard (Sinapis alba) internodes stimulates growth within 15 min of treatment (Morgan et al., 1980). However, while it is possible that YUC-independent changes in hypocotyl gene expression contribute to the normal growth response, they are insufficient to lead to sustained growth (Figure 4C). One possibility is that the coincidence of a local hypocotyl signal and auxin coming from the cotyledons constitutes a coherent signal ensuring a coordinated growth response.
By comparing shade-regulated gene expression with ChIP data, we provide strong support for an early role of PIF transcription factors, specifically for upregulated genes (Figure 3; Supplemental Figure 3). Interestingly, the fractions of PIF-bound, shade-regulated genes in hypocotyls and cotyledons were similar, suggesting an early role of these transcription factors in both organs. We confirmed the importance of PIFs beyond shade-induced auxin production in cotyledons by comparing the auxin responsiveness of yuc2 yuc5 yuc8 yuc9 and pif4 pif5 pif7 mutants (Supplemental Figure 4F) (Nozue et al., 2011; Hornitschek et al., 2012; Hersch et al., 2014). Consistent with the additive function of PIF4 and PIF5 during shade avoidance (Lorrain et al., 2008; Hornitschek et al., 2012; Pfeiffer et al., 2014), both bound to a highly overlapping set of shade-induced genes (Figure 3). The rapid and strong overrepresentation of auxin-regulated genes among our shade-regulated gene list (Figures 2 and 4) prompted us to determine whether many of these genes are also bound by ARF transcription factors (Guilfoyle, 2015). By analyzing ARF6 ChIP-seq data (Oh et al., 2014), we showed that like PIF4 and PIF5, ARF6 bound to many early shade-induced genes and that this set was highly overlapping with those bound by the PIFs (Figure 3). These data indicate that many early shade-regulated genes are bound by PIFs and ARFs, showing that, as observed in other growth-stimulating conditions (Oh et al., 2014), many genes are bound by both classes of transcription factors in response to shade (Figure 3). Furthermore, consistent with the role of BL in multiple growth responses (Oh et al., 2014; Das et al., 2016; Procko et al., 2016), we also detected BZR1 binding to many rapidly shade-regulated genes (Figure 3).
Our gene expression analysis suggests that several hormones contribute to the organ-specific growth response elicited by shade. The more sustained shade induction of numerous auxin response genes in hypocotyls (Figure 4A) is consistent with the notion that an organ-specific auxin response contributes to selective hypocotyl elongation (Bou-Torrent et al., 2014; Hersch et al., 2014; de Wit et al., 2015; Sun et al., 2016). Hypocotyl-specific induction of many (but not all) SAUR genes is also consistent with this idea (Figure 5) (Spartz et al., 2014; Ren and Gray, 2015). Our data also support a role for BL in defining organ-specific growth. DWF4 expression was repressed in cotyledons but enhanced in hypocotyls late in the response to low R/FR (Figure 2). Moreover, the expression of BAS1, encoding a BL-inactivating enzyme, was selectively downregulated in hypocotyls (Figure 2). These expression patterns suggest a rise in BL levels in hypocotyls, which is consistent with the positive correlation between expression patterns triggered by BL and shade, specifically in hypocotyls (Figure 2E). The functional importance of DWF4 and BAS1 in this process was shown genetically, suggesting that local BL homeostasis contributes to selective hypocotyl elongation (Figure 2) (Das et al., 2016; Procko et al., 2016). Similarly, the hypocotyl-specific induction of AtGA3ox1 that we report here (Figure 2D; Supplemental Figure 2D and Supplemental Data Set 2), together with the requirement of AtGA3ox1 for normal shade-induced hypocotyl elongation (Yu et al., 2015), suggest a role for GA in selective growth enhancement of this organ. GA levels increase relatively late in response to shade (after 24 but not 4 h) (Bou-Torrent et al., 2014). An organ-specific analysis of GA levels would further elucidate the spatial and temporal aspects of GA-regulated growth in the context of shade avoidance. Finally, analysis of ABA-associated gene expression patterns suggests a role for this hormone in the organ-specific shade response (Figures 2 and 6). Terms associated with ABA and drought were selectively upregulated in cotyledons, while they were rather downregulated in the hypocotyl. Such a response pattern is compatible with the opposite growth responses of these organs, given that ABA is rather growth inhibiting. Tomato (Solanum lycopersicum) plants subjected to long-term shade treatments and the constitutively shade-avoiding phyB mutant (in Arabidopsis) have higher ABA levels in leaves than in hypocotyls, but it is currently unknown whether shade leads to rapid changes in ABA levels and what the consequences of such changes are (Cagnola et al., 2012; González et al., 2012).
Our organ-specific gene expression analysis showed that the response to low R/FR becomes increasingly more organ specific with time in the shade (Figure 2; Supplemental Figure 2). This conclusion can be visualized using enrichment maps of GO terms (Supplemental Figures 8 and 9). Moreover, when focusing on terms enriched in hypocotyls, this analysis revealed a temporally coordinated pattern consistent with rapid growth (Supplemental Figures 8 and 9). Our comparative gene expression analysis revealed similarities underlying growth processes in different organs and in different developmental or environmental contexts (Figure 7; Supplemental Figure 7). First, we observed a striking correlation between gene expression in etiolated seedlings at the time of rapid hypocotyl elongation and shade-induced expression in hypocotyls at the time of growth acceleration (Figure 7) (Pelletier et al., 2010). This includes upregulation of YUC8 and a number of auxin-responsive genes in both situations (Figure 7) (Pelletier et al., 2010). Hence, despite the limited importance attributed to auxin in etiolated hypocotyl elongation (Gray et al., 1998), the hypocotyl growth programs in etiolated seedlings and in shade-treated green seedlings appear to be more similar than previously anticipated (Figure 7; Supplemental Figure 7). The XTH family of cell wall-modifying enzymes is involved in shade responses (Keuskamp et al., 2011; de Wit et al., 2015; Pedmale et al., 2016). The expression of several members of this family is selectively induced in hypocotyls, which is consistent with their growth-promoting roles (Figure 7; Supplemental Figure 7). Moreover, the expression of several XTHs is also induced in rapidly growing etiolated hypocotyls and cells in the root elongation zone (Figure 7B). A remarkable pattern emerged by analyzing members of the AGP gene family. AGPs are extensively hydroxyprolinated glycoproteins found in the apoplasm but associated with the plasma membrane (Lamport et al., 2014). These proteins have been connected to a variety of growth and developmental processes (Velasquez et al., 2011; Lamport et al., 2014). It was hypothesized that AGPs play a signaling role involving pH-regulated Ca2+ release, an activity that might be controlled by auxin-mediated acidification of the apoplasm (Lamport et al., 2014). We identified 26 members of the AGP family showing selective shade induction in hypocotyls coinciding with the time of shade-promoted elongation (Figure 7C). Expression of several AGPs also increases at the time of rapid hypocotyl elongation in the dark and in the root elongation zone (Figure 7C). Of note, several AGP genes are upregulated in roots of a Brachypodium mutant with enhanced root cell elongation (Pacheco-Villalobos et al., 2016). Moreover, during deetiolation, the expression of numerous AGPs increases in the expanding cotyledon, while it decreases in hypocotyls where growth is rapidly inhibited (Figure 7C) (Sun et al., 2016). PIF5 binding sites were identified in the proximity of nine of the shade-induced AGPs, suggesting direct regulation of their expression by PIFs (Supplemental Figure 7C) (Hornitschek et al., 2012). Finally, we provide functional data supporting the importance of AGPs in shade-induced hypocotyl elongation (Figure 7D). Collectively, our comparative genomic analysis suggests that (1) multiple AGPs are robust growth-marker genes over a range of conditions and organs, and (2) many AGPs might be direct PIF target genes.
METHODS
Plant Growth and Light Conditions
We used the following Arabidopsis thaliana genotypes (cv Columbia-0): pin3 pin4 pin7, d6pk01, D6PK-OX (Willige et al., 2013), sav3-2 (Tao et al., 2008), pif7-1 (Li et al., 2012), DII-VENUS (Brunoud et al., 2012), pif4 pif5 pif7 (de Wit et al., 2015), amiD2/5/7/8/9 (Spartz et al., 2014), amiSAUR19/23/24 (Spartz et al., 2012), and iYUC3 (FRO6:XVE:YUC3; Chen et al., 2014). To generate the yuc2 yuc5 yuc8 yuc9 quadruple mutant, T-DNA and transposon insertion lines were obtained from the ABRC, the Cold Spring Harbor Laboratory, or GABI-Kat. Mutant yucca lines from Yunde Zhao were previously described (Chen et al., 2014). The yuc quadruple mutant was obtained by repeated crossing and PCR genotyping using the described primers. DII-VENUS in the pif7, sav3, and pin3 pin4 pin7 backgrounds were obtained by crossing. Plant growth, growth analysis, and microscopy are described in the Supplemental Methods.
RNA Extraction, RNA-Seq Library Preparation, and RT-qPCR
Arabidopsis Col-0 was grown for 5 d in long days at 21°C on one horizontal plate of ∼140 mm in diameter per sample. Each plate contained 25 mL 0.5× MS medium covered with a nylon mesh. On day 6 at ZT2, seedlings were either kept in high R/FR or transferred to low R/FR. After 0, 15, 45, 90, or 180 min in low R/FR, samples exposed to both light qualities were imbibed within 75 s in ice-cold 100% (v/v) acetone and subjected twice to ∼600 mbar below atmospheric pressure for 5 min while remaining on ice. The dissection of acetone-fixed seedlings was performed in 70% (v/v) 4°C cold ethanol under a binocular lens. Cotyledon and hypocotyl tissues from 50 seedlings per sample were collected in 100% (v/v) ethanol. For each time point and light condition, duplicates were prepared. Plant materials were manually ground using pestles, and total RNA was extracted using an RNeasy kit (Qiagen) with on-column DNA digestion, according to the manufacturer’s instructions. Finally, RNA samples were precipitated using 3 M NaOH (pH 5.2) and 100% (v/v) ethanol. The precipitate was visualized with glycogen and washed with 80% (v/v) ice-cold ethanol. Stranded libraries were prepared using 400 ng high-quality RNA according to the TruSeq protocol (Illumina). This included RNA purification steps using the Agencourt AMPure XP system (Beckman Coulter) and cDNA preparation using a mix of random and poly(A) primers. RNA-seq libraries were subsequently sequenced with the HiSeq 2500 System (Illumina) in the Lausanne Genome Technology Facility.
Growth conditions and the RNA preparation protocol used for RT-qPCR analysis were similar to those used for the RNA-seq experiment. For each experiment, equal amounts of RNA were reverse transcribed into cDNA with Superscript II reverse transcriptase (Invitrogen). RT-qPCR was performed in three biological replicates with material from separate pools of seedlings. For each biological replicate, three technical replicates were prepared (7900HT Applied Biosystems). Data were normalized against three reference housekeeping genes (ACT2, PEX4/UBC21, and YLS8) using Biogazelle qbase software (list of primers provided in Supplemental Table 1). Subsequent analyses were done in R (v. 3.3.1).
RNA-seq analysis and additional bioinformatics protocols are described in the Supplemental Methods.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AGP10C (At4g09030), AGP11C (At3g01700), AGP12P (At3g13520), AGP13P (At4g26320), AGP14P (At5g56540), AGP15P (At5g11740), AGP16P (At2g46330), AGP17K (At2g23130), AGP18K (At4g37450), AGP1C (At5g64310), AGP20P (At3g61640), AGP21P (At1g55330), AGP22P (At5g53250), AGP24P (At5g40730), AGP25C (At5g18690), AGP26C (At2g47930), AGP2C (At2g22470), AGP30I (At2g33790), AGP31I (At1g28290), AGP32I (At5g21160), AGP3C (At4g40090), AGP41P (At5g24105), AGP43P (At2g41905), AGP44P (At3g01730), AGP45P (At5g12880), AGP4C (At5g10430), AGP52C (At1g63530), AGP54C (At2g28440), AGP56C (At3g22070), AGP57C (At3g45230), AGP58C (At4g16980), AGP6C (At5g14380), AGP7C (At5g65390), AGP9C (At2g14890), AIF4 (At1g09250), ARF6 (At1g30330), bZIP52 (At1g06850), BZR1 (At1g75080), D6PK (At5g55910), D6PKL1 (At4g26610), FLA1 (At5g55730), FLA10 (At3g60900), FLA11 (At5g03170), FLA12 (At5g60490), FLA13 (At5g44130), FLA15 (At3g52370), FLA16 (At2g35860), FLA17 (At5g06390), FLA18 (At3g11700), FLA19 (At1g15190), FLA2 (At4g12730), FLA5 (At4g31370), FLA6 (At2g20520), FLA7 (At2g04780), FLA8 (At2g45470), FLA9 (At1g03870), GA20OX1/GA5 (At4g25420), GA20OX3 (At5g07200), GA2OX6 (At1g02400), GA2OX8 (At4g21200), GA3OX1/GA4 (At1g15550), HFR1 (At1g02340), IAA1 (At4g14560), IAA2 (At3g23030), IAA29 (At4g32280), KDR (At1g26945), LFY (At5g61850), MAKR6 (At5g52900), MAPKKK14 (At2g30040), PAG10 (At4g27520), PAG13 (At4g31840), PAG15 (At5g25090), PAG2 (At2g25060), PAG5 (At2g32300), PAG6 (At2g44790), PER3 (At3g08640), PIF4 (AT2G43010), PIF5 (AT3G59060), PIF7 (AT5G61270), PIN3 (At1g70940), PIN4 (At2g01420), PIN7 (At1g23080), PRE5 (At3g28857), RCI2A (At3g05880), RTFL17 (At1g13245), SAUR19 (At5g18010), SAUR20 (At5g18020), SAUR21 (At5g18030), SAUR22 (At5g18050), SAUR23 (At5g18060), SAUR24 (At5g18080), SAUR67 (At1g29510), TAA1/sav3 (AT1G70560), TCP15 (At1g69690), YUC2 (At4g13260), YUC3 (At1g04610), YUC5 (At5g43890), YUC8 (At4g28720), and YUC9 (At1g04180). Additional gene identifiers can be found in the Supplemental Data Set 1. RNA-seq data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus as GEO series GSE81202.
Supplemental Data
Supplemental Figure 1. Data validation and quantification.
Supplemental Figure 2. Transcriptional regulation in shade shows organ-specific traits.
Supplemental Figure 3. Binding of PIF4, PIF5, ARF6, BZR1, and LFY to shade-repressed genes in hypocotyls and cotyledons.
Supplemental Figure 4. Different types of shade regulation among typical auxin response genes.
Supplemental Figure 5. Shade regulation of the early auxin response in cotyledons versus hypocotyls.
Supplemental Figure 6. Response patterns of genes regulated by shade in both hypocotyls and cotyledons.
Supplemental Figure 7. A set of genes with similar expression patterns in the root elongation zone and shade-treated hypocotyls.
Supplemental Figure 8. Similarity networks of Gene Ontology terms become increasingly more organ specific with time in the shade.
Supplemental Figure 9. Similarity networks of Gene Ontology terms.
Supplemental Table 1. Primer sequences used for RT-qPCR.
Supplemental Data Set 1. Gene expression table of shade-regulated genes.
Supplemental Data Set 2. Significantly regulated genes in response to low R/FR.
Supplemental Data Set 3. GO analyses of significantly regulated genes in response to low R/FR.
Supplemental Data Set 4. ChIP analysis.
Supplemental Data Set 5. Clustered genes shade-regulated in both organs.
Supplemental Data Set 6. GO analysis of clustered genes shade-regulated in both organs.
Supplemental Data Set 7. Clustered genes organ-specifically shade-regulated.
Supplemental Data Set 8. GO analysis of clustered genes organ-specifically shade-regulated.
Supplemental Data Set 9. Gene expression of differentially regulated genes in the root elongation zone.
Supplemental Data Set 10. GO analysis of upregulated genes in the root and hypocotyl elongation zone.
Supplemental Data Set 11. AGP regulation under various conditions.
Supplementary Material
Acknowledgments
Funding in the Fankhauser lab comes from the University of Lausanne and the Swiss National Science Foundation (FNS 31003A_160326, Sinergia Grant CRSII3_154438, and SystemsX Grant PlantMechanix 51RT-0_145716). In the Maloof lab, this work was funded by National Science Foundation Grants DBI-0227103 and IOS-0923752, USDA NIFA Project CA-D-PLB-7226-H, and UC Davis intramural funds. We thank Anne-Sophie Fiorucci, Mieke de Wit, Vinicius Costa Galvao, and Micha Hersch for helpful comments on the manuscript. We thank our microscopy facility (CIF, http://cifweb.unil.ch/) for technical advice and the Lausanne Genome Technology Platform (LGTF) for advice on RT-qPCR and help with library generation and sequencing. We are grateful to Sylvain Pradervand, Sandra Calderon Copete (Lausanne Genome Technology Platform), and Frédéric Schütz for advice on statistics, initial data analysis, and sequence read mapping. We thank Yunde Zhao and Youfa Cheng (UCSD) for sharing yucca seed prior to publication and Bill Gray (University of Minnesota) for providing the SAUR ami-RNA lines. We thank Joanne Chory and Ullas Pedmale (The Salk Institute) for sharing unpublished information.
AUTHOR CONTRIBUTIONS
M.V.K. designed, acquired, analyzed, and interpreted data. M.T., L.A.P., F.S., and E.S.-S. acquired, analyzed, and interpreted data. P.M.-M. and J.M. provided important unpublished material for the experiments. I.X. analyzed and interpreted data. C.F. designed, analyzed, and interpreted data and wrote the manuscript with input from all authors.
Glossary
- R/FR
red/far-red
- GA
gibberellic acid
- ABA
abscisic acid
- IAA
indole-3-acetic acid
- GO
Gene Ontology
- BR
brassinosteroid
- ChIP
chromatin immunoprecipitation
- BL
brassinolide
Footnotes
Articles can be viewed without a subscription.
References
- Bai M.Y., Fan M., Oh E., Wang Z.Y. (2012). A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24: 4917–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Torrent J., Galstyan A., Gallemí M., Cifuentes-Esquivel N., Molina-Contreras M.J., Salla-Martret M., Jikumaru Y., Yamaguchi S., Kamiya Y., Martínez-García J.F. (2014). Plant proximity perception dynamically modulates hormone levels and sensitivity in Arabidopsis. J. Exp. Bot. 65: 2937–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoud G., Wells D.M., Oliva M., Larrieu A., Mirabet V., Burrow A.H., Beeckman T., Kepinski S., Traas J., Bennett M.J., Vernoux T. (2012). A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106. [DOI] [PubMed] [Google Scholar]
- Cagnola J.I., Ploschuk E., Benech-Arnold T., Finlayson S.A., Casal J.J. (2012). Stem transcriptome reveals mechanisms to reduce the energetic cost of shade-avoidance responses in tomato. Plant Physiol. 160: 1110–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli M., Possenti M., Sessa G., Ciolfi A., Sassi M., Morelli G., Ruberti I. (2007). Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev. 21: 1863–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J.J. (2013). Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64: 403–427. [DOI] [PubMed] [Google Scholar]
- Chaiwanon J., Wang Z.Y. (2015). Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr. Biol. 25: 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.J., Greenham K., Castillejo C., Sartor R., Bialy A., Sun T.P., Estelle M. (2012). Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS One 7: e36210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Dai X., De-Paoli H., Cheng Y., Takebayashi Y., Kasahara H., Kamiya Y., Zhao Y. (2014). Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 55: 1072–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S., Dilkes B.P., Fujioka S., Takatsuto S., Sakurai A., Feldmann K.A. (1998). The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10: 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Esquivel N., Bou-Torrent J., Galstyan A., Gallemi M., Sessa G., Salla Martret M., Roig-Villanova I., Ruberti I., Martinez-Garcia J.F. (2013). The bHLH proteins BEE and BIM positively modulate the shade avoidance syndrome in Arabidopsis seedlings. Plant J. 75: 989–1002. [DOI] [PubMed] [Google Scholar]
- Cole B., Kay S.A., Chory J. (2011). Automated analysis of hypocotyl growth dynamics during shade avoidance in Arabidopsis. Plant J. 65: 991–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocco C.D., Locascio A., Escudero C.M., Alabadí D., Blázquez M.A., Botto J.F. (2015). The transcriptional regulator BBX24 impairs DELLA activity to promote shade avoidance in Arabidopsis thaliana. Nat. Commun. 6: 6202. [DOI] [PubMed] [Google Scholar]
- Das D., St Onge K.R., Voesenek L.A., Pierik R., Sasidharan R. (2016). Ethylene- and shade-induced hypocotyl elongation share transcriptome patterns and functional regulators. Plant Physiol. 172: 718–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin P.F., Yanovsky M.J., Kay S.A. (2003). A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol. 133: 1617–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit M., Lorrain S., Fankhauser C. (2014). Auxin-mediated plant architectural changes in response to shade and high temperature. Physiol. Plant. 151: 13–24. [DOI] [PubMed] [Google Scholar]
- de Wit M., Ljung K., Fankhauser C. (2015). Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytol. 208: 198–209. [DOI] [PubMed] [Google Scholar]
- de Wit M., Kegge W., Evers J.B., Vergeer-van Eijk M.H., Gankema P., Voesenek L.A., Pierik R. (2012). Plant neighbor detection through touching leaf tips precedes phytochrome signals. Proc. Natl. Acad. Sci. USA 109: 14705–14710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakovic-Petrovic T., de Wit M., Voesenek L.A., Pierik R. (2007). DELLA protein function in growth responses to canopy signals. Plant J. 51: 117–126. [DOI] [PubMed] [Google Scholar]
- Franklin K.A., Quail P.H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61: 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcion C., Lohmann A., Lamodière E., Catinot J., Buchala A., Doermann P., Métraux J.P. (2008). Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 147: 1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H., et al. (2008). The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 55: 526–542. [DOI] [PubMed] [Google Scholar]
- Gommers C.M., Visser E.J., St Onge K.R., Voesenek L.A., Pierik R. (2013). Shade tolerance: when growing tall is not an option. Trends Plant Sci. 18: 65–71. [DOI] [PubMed] [Google Scholar]
- González C.V., Ibarra S.E., Piccoli P.N., Botto J.F., Boccalandro H.E. (2012). Phytochrome B increases drought tolerance by enhancing ABA sensitivity in Arabidopsis thaliana. Plant Cell Environ. 35: 1958–1968. [DOI] [PubMed] [Google Scholar]
- Gray W.M., Ostin A., Sandberg G., Romano C.P., Estelle M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 7197–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T.J. (2015). The PB1 domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell 27: 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch M., Lorrain S., de Wit M., Trevisan M., Ljung K., Bergmann S., Fankhauser C. (2014). Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 6515–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28: 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., Lopez-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., Xenarios I., Fankhauser C. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71: 699–711. [DOI] [PubMed] [Google Scholar]
- Ikeda M., Fujiwara S., Mitsuda N., Ohme-Takagi M. (2012). A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 24: 4483–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M.M., Jaillais Y., Pedmale U.V., Moreno J.E., Chory J., Ballare C.L. (2011). Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J. 67: 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuskamp D.H., Pollmann S., Voesenek L.A., Peeters A.J., Pierik R. (2010). Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc. Natl. Acad. Sci. USA 107: 22740–22744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuskamp D.H., Sasidharan R., Vos I., Peeters A.J., Voesenek L.A., Pierik R. (2011). Blue-light-mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J. 67: 208–217. [DOI] [PubMed] [Google Scholar]
- Kozuka T., Kobayashi J., Horiguchi G., Demura T., Sakakibara H., Tsukaya H., Nagatani A. (2010). Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol. 153: 1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport D.T., Varnai P., Seal C.E. (2014). Back to the future with the AGP-Ca2+ flux capacitor. Ann. Bot. (Lond.) 114: 1069–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E. (2014). PIFs: systems integrators in plant development. Plant Cell 26: 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Cohn M.M., Monte E., Al-Sady B., Erickson E., Quail P.H. (2012). Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24: 1398–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26: 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53: 312–323. [DOI] [PubMed] [Google Scholar]
- Mizuno T., Oka H., Yoshimura F., Ishida K., Yamashino T. (2015). Insight into the mechanism of end-of-day far-red light (EODFR)-induced shade avoidance responses in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 79: 1987–1994. [DOI] [PubMed] [Google Scholar]
- Moreno J.E., Ballaré C.L. (2014). Phytochrome regulation of plant immunity in vegetation canopies. J. Chem. Ecol. 40: 848–857. [DOI] [PubMed] [Google Scholar]
- Morgan D.C., O’Brien T., Smith H. (1980). Rapid photomodulation of stem extension in light-grown Sinapis alba L.: Studies on kinetics, site of perception and photoreceptor. Planta 150: 95–101. [DOI] [PubMed] [Google Scholar]
- Moyroud E., Minguet E.G., Ott F., Yant L., Posé D., Monniaux M., Blanchet S., Bastien O., Thévenon E., Weigel D., Schmid M., Parcy F. (2011). Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell 23: 1293–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56: 165–185. [DOI] [PubMed] [Google Scholar]
- Nemhauser J.L., Hong F., Chory J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475. [DOI] [PubMed] [Google Scholar]
- Nito K., Kajiyama T., Unten-Kobayashi J., Fujii A., Mochizuki N., Kambara H., Nagatani A. (2015). Spatial regulation of the gene expression response to shade in Arabidopsis seedlings. Plant Cell Physiol. 56: 1306–1319. [DOI] [PubMed] [Google Scholar]
- Nozue K., Harmer S.L., Maloof J.N. (2011). Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol. 156: 357–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K., Tat A.V., Kumar Devisetty U., Robinson M., Mumbach M.R., Ichihashi Y., Lekkala S., Maloof J.N. (2015). Shade avoidance components and pathways in adult plants revealed by phenotypic profiling. PLoS Genet. 11: e1004953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Bai M.Y., Arenhart R.A., Sun Y., Wang Z.Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco-Villalobos D., et al. (2016). The effects of high steady state auxin levels on root cell elongation in Brachypodium. Plant Cell 28: 1009–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale U.V., Huang S.S., Zander M., Cole B.J., Hetzel J., Ljung K., Reis P.A., Sridevi P., Nito K., Nery J.R., Ecker J.R., Chory J. (2016). Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier S., et al. (2010). A role for pectin de-methylesterification in a developmentally regulated growth acceleration in dark-grown Arabidopsis hypocotyls. New Phytol. 188: 726–739. [DOI] [PubMed] [Google Scholar]
- Pfeiffer A., Shi H., Tepperman J.M., Zhang Y., Quail P.H. (2014). Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant 7: 1598–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R., Testerink C. (2014). The art of being flexible: how to escape from shade, salt, and drought. Plant Physiol. 166: 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R., Djakovic-Petrovic T., Keuskamp D.H., de Wit M., Voesenek L.A. (2009). Auxin and ethylene regulate elongation responses to neighbor proximity signals independent of gibberellin and della proteins in Arabidopsis. Plant Physiol. 149: 1701–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C., Crenshaw C.M., Ljung K., Noel J.P., Chory J. (2014). Cotyledon-generated auxin is required for shade-induced hypocotyl growth in Brassica rapa. Plant Physiol. 165: 1285–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C., Burko Y., Jaillais Y., Ljung K., Long J.A., Chory J. (2016). The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev. 30: 1529–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S.K., Holalu S.V., Casal J.J., Finlayson S.A. (2013). Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiol. 163: 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Gray W.M. (2015). SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant 8: 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F., Okamoto H., Patrick E., Harris S.R., Wasternack C., Brearley C., Turner J.G. (2010). Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22: 1143–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R., Chinnappa C.C., Staal M., Elzenga J.T., Yokoyama R., Nishitani K., Voesenek L.A., Pierik R. (2010). Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases. Plant Physiol. 154: 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmülling T., Werner T., Riefler M., Krupková E., Bartrina y Manns I. (2003). Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J. Plant Res. 116: 241–252. [DOI] [PubMed] [Google Scholar]
- Sellaro R., Crepy M., Trupkin S.A., Karayekov E., Buchovsky A.S., Rossi C., Casal J.J. (2010). Cryptochrome as a sensor of the blue/green ratio of natural radiation in Arabidopsis. Plant Physiol. 154: 401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G., Carabelli M., Sassi M., Ciolfi A., Possenti M., Mittempergher F., Becker J., Morelli G., Ruberti I. (2005). A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 19: 2811–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter A.M., Keppler B., Lichtenberg J., Gu D., Welch L.R. (2010). A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 153: 485–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz A.K., Ren H., Park M.Y., Grandt K.N., Lee S.H., Murphy A.S., Sussman M.R., Overvoorde P.J., Gray W.M. (2014). SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz A.K., Lee S.H., Wenger J.P., Gonzalez N., Itoh H., Inze D., Peer W.A., Murphy A.S., Overvoorde P.J., Gray W.M. (2012). The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 70: 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler C., Matteucci A., Sessa G., Weimar T., Ohgishi M., Aoyama T., Morelli G., Ruberti I. (1999). Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126: 4235–4245. [DOI] [PubMed] [Google Scholar]
- Sun N., Wang J., Gao Z., Dong J., He H., Terzaghi W., Wei N., Deng X.W., Chen H. (2016). Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc. Natl. Acad. Sci. USA 113: 6071–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Nakamura S., Mochizuki N., Nagatani A. (2002). Phytochrome in cotyledons regulates the expression of genes in the hypocotyl through auxin-dependent and -independent pathways. Plant Cell Physiol. 43: 1171–1181. [DOI] [PubMed] [Google Scholar]
- Tao Y., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk E.M., et al. (2005). BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J. 42: 23–34. [DOI] [PubMed] [Google Scholar]
- Vandenbussche F., Vriezen W.H., Smalle J., Laarhoven L.J., Harren F.J., Van Der Straeten D. (2003). Ethylene and auxin control the Arabidopsis response to decreased light intensity. Plant Physiol. 133: 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez S.M., et al. (2011). O-glycosylated cell wall proteins are essential in root hair growth. Science 332: 1401–1403. [DOI] [PubMed] [Google Scholar]
- Willige B.C., Ahlers S., Zourelidou M., Barbosa I.C., Demarsy E., Trevisan M., Davis P.A., Roelfsema M.R., Hangarter R., Fankhauser C., Schwechheimer C. (2013). D6PK AGCVIII kinases are required for auxin transport and phototropic hypocotyl bending in Arabidopsis. Plant Cell 25: 1674–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.H., et al. (2015). Multi-omics analysis identifies genes mediating the extension of cell walls in the Arabidopsis thaliana root elongation zone. Front. Cell Dev. Biol. 3: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., Kasahara H., Kamiya Y., Chory J., Zhao Y. (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 18518–18523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251. [DOI] [PubMed] [Google Scholar]
- Yu J., Qiu H., Liu X., Wang M., Gao Y., Chory J., Tao Y. (2015). Characterization of tub4(P287L), a β-tubulin mutant, revealed new aspects of microtubule regulation in shade. J. Integr. Plant Biol. 57: 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Guo Y., Novak O., Chen W., Ljung K., Noel J.P., Chory J. (2016). Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat. Plants 2: 16025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Guo Y., Novák O., Dai X., Zhao Y., Ljung K., Noel J.P., Chory J. (2013). Coordination of auxin and ethylene biosynthesis by the aminotransferase VAS1. Nat. Chem. Biol. 9: 244–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou M., et al. (2014). Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. eLife 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.