Characterization of an Arabidopsis heat-sensitive mutant identified a chlorophyll dephytylase involved in steady-state chlorophyll turnover, revealing a pathway of chlorophyll metabolism.
Abstract
Chlorophyll turns over in green organs during photosystem repair and is salvaged via de- and rephytylation, but the enzyme involved in dephytylation is unknown. We have identified an Arabidopsis thaliana thylakoid protein with a putative hydrolase domain that can dephytylate chlorophyll in vitro and in vivo. The corresponding locus, CHLOROPHYLL DEPHYTYLASE1 (CLD1), was identified by mapping a semidominant, heat-sensitive, missense allele (cld1-1). CLD1 is conserved in oxygenic photosynthetic organisms, sharing structural similarity with pheophytinase, which functions in chlorophyll breakdown during leaf senescence. Unlike pheophytinase, CLD1 is predominantly expressed in green organs and can dephytylate chlorophyll in vitro. The specific activity is significantly higher for the mutant protein encoded by cld1-1 than the wild-type enzyme, consistent with the semidominant nature of the cld1-1 mutation. Supraoptimal CLD1 activities in cld1-1 mutants and transgenic seedlings led to the proportional accumulation of chlorophyllides derived from chlorophyll dephytylation after heat shock, which resulted in light-dependent cotyledon bleaching. Reducing CLD1 expression diminished thermotolerance and the photochemical efficiency of photosystem II under prolonged moderate heat stress. Taken together, our results suggest that CLD1 is the long-sought enzyme for removing the phytol chain from chlorophyll during its turnover at steady state within the chloroplast.
INTRODUCTION
Oxygenic photosynthesis is the predominant mechanism for converting solar energy into chemical energy to perpetuate most biological activities on Earth. In the photosynthetic apparatus, chlorophyll plays a pivotal role in harvesting light energy, which is used by the chlorophyll-containing multiprotein complexes called photosystems to produce the reducing power NADPH and generate a proton gradient across the thylakoid membrane, thereby driving ATP synthesis. Chlorophyll is a pigment consisting of two moieties: a chlorin ring containing a magnesium ion at its center and a long hydrophobic phytol chain, joined by an ester bond. Once formed through highly coordinated pathways (Kim et al., 2013), chlorophylls are tightly bound to the photosystems until the partial or complete dismantling of the photosynthetic machinery in response to environmental and developmental cues, during which they undergo turnover or breakdown.
Visible chlorophyll breakdown occurs during leaf senescence, seed maturation, and fruit ripening, when the green pigments are massively catabolized into nonphototoxic breakdown products via the PHEOPHORBIDE A OXYGENASE pathway (Hörtensteiner, 2013a). In contrast, chlorophyll turns over at steady state often without a visible change in chlorophyll content due to the balance of its biosynthesis and degradation (Matile et al., 1999). The pulse-chase experiment revealed constant chlorophyll turnover in the green leaves of barley (Hordeum vulgare; Raskin et al., 1995), rye (Secale cereale; Feierabend and Dehne, 1996), and Arabidopsis thaliana (Beisel et al., 2010), which were attributed to the turnover of D1 protein of the photosystem II (PSII) reaction center.
D1 protein is the most labile component of PSII and is subject to damage by irradiation (Mattoo et al., 1989; Aro et al., 1993). In the absence of light, D1 protein can be damaged under heat stress, probably due to the backflow of electrons from the stroma to PSII (Marutani et al., 2012). Repair of PSII requires the degradation of the D1 protein by proteases and the cotranslational insertion of a new D1 into the partially disassembled PSII core complex (Edelman and Mattoo, 2008; Komenda et al., 2012). Each D1 protein binds three chlorophyll a and one pheophytin a (Phein a), which is structurally the same as chlorophyll a without the magnesium ion. With the turnover of the D1 protein, the bound chlorophyll a and Phein a undergo concomitant turnover. Analysis of the fate of isotope-labeled chlorophylls in the cyanobacterium Synechocystis sp PCC6803 suggests that a substantial portion of the chlorophyll a released during PSII repair is recycled via de- and rephytylation steps (Vavilin and Vermaas, 2007). Recently, studies of an Arabidopsis mutant with reduced chlorophyll synthase (CHLG) suggested that CHLG is involved in chlorophyll recycling by catalyzing the rephytylation of chlorophyllide a (Chlide a) derived from chlorophyll dephytylation (Lin et al., 2014). The enzyme responsible for the dephytylation step has not been identified.
For more than a century, chlorophyllase (CLH) was the only protein shown to dephytylate chlorophylls and Pheins in vitro (Hörtensteiner, 2013b). With its enzymatic function, CLH has been proposed to be involved in chlorophyll turnover and breakdown (Jacob-Wilk et al., 1999; Tsuchiya et al., 1999; Morita et al., 2009). However, genetic studies of Arabidopsis CLH mutants do not support this proposition (Schenk et al., 2007; Lin et al., 2014; Zhang et al., 2014). Instead of dephytylation of chlorophyll a, chlorophyll breakdown in leaf senescence is mainly initiated by removing the magnesium ion from chlorophyll a to yield Phein a by Mg-dechelatase (Shimoda et al., 2016). Phein a is then dephytylated by pheophytinase (PPH), generating pheophorbide (Pheide) a for further degradation via the PHEOPHORBIDE A OXYGENASE pathway (Morita et al., 2009; Schelbert et al., 2009; Guyer et al., 2014). In contrast to CLH, recombinant PPH can dephytylate Pheins but not chlorophylls (Morita et al., 2009; Schelbert et al., 2009). Thus, the enzyme responsible for removing the phytol chain from chlorophyll has remained elusive. Recently, PPH was found to be dispensable for chlorophyll breakdown during Arabidopsis seed maturation (Zhang et al., 2014) and tomato (Solanum lycopersicum) fruit ripening (Guyer et al., 2014), indicating the existence of an alternative dephytylating enzyme(s).
Several intermediates of chlorophyll metabolism are photosensitizers that can induce oxidative stress (Rüdiger, 2009), so their accumulation must be avoided under all conditions. Genetic mutants that conditionally accumulate these intermediates are useful in deciphering the complex and highly regulated pathways in chlorophyll metabolism (Kusaba et al., 2013). Here, we provide genetic and biochemical evidence that the Arabidopsis AT5G38520 locus encodes a chloroplast protein with chlorophyll dephytylation activity. The gene was identified by characterizing and mapping an Arabidopsis EMS-mutagenized allele, named defect in long-term acquired thermotolerance3-1 (dlt3-1), which causes a heat-sensitive phenotype in seedlings (Wu et al., 2013). dlt3-1 is a semidominant allele of AT5G38520 caused by a missense mutation that produces a protein with a G193D substitution. AT5G38520 encodes a conserved protein sharing peptide sequence similarity with PPH and other α/β-hydrolases. Similar to CLH, the recombinant AT5G38520 protein showed substantial chlorophyll/Phein dephytylating activity, and the G193D enzyme was much more active than the wild type based on the kinetic characterization. In seedlings harboring the dlt3-1 allele, Chlide a and b levels highly accumulated after heat shock (HS) treatment. These results suggest that AT5G38520 encodes a novel “chlorophyllase.” Because the protein is structurally distinct from the known CLH, hereafter it is named chlorophyll dephytylase and its encoding locus CHLOROPHYLL DEPHYTYLASE1 (CLD1). Suppression of CLD1 expression by artificial microRNA reduced PSII efficiency in transgenic seedlings exposed to prolonged moderate high temperature, which suggests that chlorophyll turnover via de- and rephytylation is essential for maintaining the homeostasis of the photosystem under heat stress. The finding of CLD1 opens an opportunity for further understanding the mechanistic details of chlorophyll turnover at steady state and its role in photosystem repair.
RESULTS
Cloning of CLD1, Which Encodes an α/β-Hydrolase Superfamily Protein of Unknown Function
In a previous effort to identify new thermotolerance components in Arabidopsis, several dlt mutant alleles were isolated, and the genetic identities of dlt1-1, dlt1-2, and dlt4-1 (or chlg1-1) were revealed (Wu et al., 2013; Lin et al., 2014). In this study, we characterized and cloned the dlt3-1 allele, which we renamed cld1-1 based on its molecular function identified in this study. The homozygous mutant plants are referred to hereafter as cld1-1 or cld1-1/cld1-1.
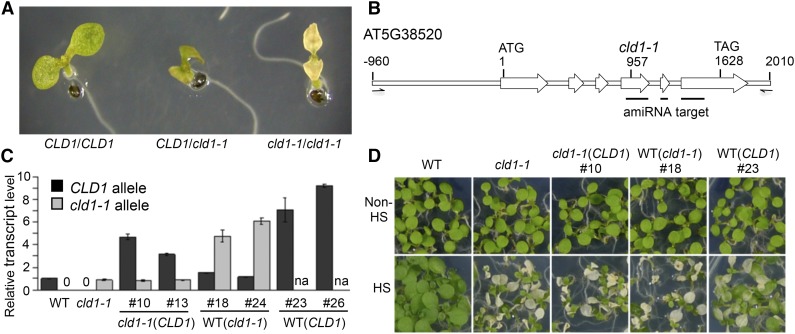
Under normal conditions, cld1-1 mutant plants did not differ from the wild type in growth and development. However, after HS at 40°C for 1 h, the mutant seedlings showed a cotyledon bleaching phenotype after a 3-d recovery (fast bleaching) following the HS treatment, whereas the wild-type cotyledons remained green (Figure 1A). Heterozygous F1 seedlings (CLD1/cld1-1) from a cross between the wild type (Col-0) and cld1-1 showed slow cotyledon bleaching (Figure 1A). Similar results were obtained in the F1 seedlings of the cross between the wild type (Landsberg erecta [Ler]) and cld1-1 (Supplemental Figure 1). After HS treatment, an F2 offspring population showed an approximate 1:2:1 ratio of nonbleaching:slow-bleaching:fast-bleaching phenotype that passed a χ2 test (Supplemental Table 1), which indicates that cld1-1 is a semidominant allele. The nonbleaching seedlings, which should carry homozygous CLD1 allele, were further collected for bulked segregant analysis and map-based cloning with molecular markers.
Figure 1.
cld1-1 Is a Semidominant Allele of AT5G38520 that Causes a Heat-Sensitive Phenotype.
(A) Phenotypes of 5-d-old Arabidopsis seedlings with homozygous or heterozygous cld1-1 allele after treatment at 40°C for 1 h (HS) plus 3-d recovery under normal growth condition. The representative seedlings showed nonbleaching (CLD1/CLD1), slow-bleaching (CLD1/cld1-1), and fast-bleaching (cld1-1/cld1-1) phenotypes. Without HS, the wild-type and mutant cotyledons were all green and are not shown here.
(B) Diagram of CLD1 gene between −960 to 2010 bp is shown with the translational start site (ATG) set as +1 and stop codon (TAG) at 1628. The white arrows indicate the exons. The mutation site in cld1-1 is indicated at position 957 with a G-to-A transition. The black lines underneath indicate the target regions of artificial microRNA. The half arrows, which are not to scale, indicate the forward and reverse primers for PCR amplification of CLD1 genomic DNA for complementation test.
(C) Relative CLD1 or cld1-1 transcript levels in 5-d-old seedlings of different transgenic lines normalized to that of the wild type or cld1-1 mutant, respectively. Data are means ± sd of three replicates. na, not analyzed.
(D) Phenotyping of representative transgenic lines with or without HS treatment. The transgenic lines are labeled as cld1-1(CLD1), cld1-1 transformed with ectopic CLD1 genomic DNA; WT(cld1-1), the wild type transformed with ectopic cld1-1 genomic DNA; WT(CLD1), the wild type transformed with ectopic CLD1 genomic DNA. Numbers after the symbol “#” indicate the line designations derived from independent transformation events. Seedlings in the same row were grown on the same plate and reorganized for presentation.
Following the map-based cloning of CLD1, a mutated site was subsequently identified in cld1-1 as a G-to-A transition in exon 4 of locus AT5G38520 (Figure 1B; Supplemental Figure 2). Two protein-coding models of AT5G38520 were predicted in TAIR as being due to a putative alternative splicing site between exons 3 and 4. The AT5G38520.1 model is the primary, if not the only, version supported by the cDNA sequences compiled by the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/cgi-bin/tdnaexpress) as well as the cloning results obtained in this study. The protein was annotated as an α/β-hydrolase superfamily protein of unknown function and containing 362-amino acid residues with a predicted transit peptide for chloroplast localization (Supplemental Figure 3). The mutation results in the replacement of Gly-193 by Asp (G193D).
To examine whether the missense mutation caused the bleaching phenotype, we conducted complementation tests by transforming the wild-type CLD1 genomic DNA into cld1-1/cld1-1 mutant and the cld1-1 genomic DNA into the wild-type (Col-0) background to yield transgenic lines designated as cld1-1 (CLD1) and WT (cld1-1), respectively. Homozygous lines of the transgenic plants with a single T-DNA insertion event were isolated for analysis. The transgenes, including ∼1 kb of its native promoter, tended to be overexpressed in all transgenic plants (Figure 1C), suggesting that an upstream negative cis-element is missing in the construct. Phenotyping results showed that the CLD1 transgene could not rescue the bleaching of cld1-1/cld1-1 cotyledons, whereas the cld1-1 transgene conferred a heat-induced bleaching phenotype to cotyledons with a wild-type background (Figure 1D). As a control, transgenic plants with CLD1 transgene overexpressed in the wild-type background showed no or slow bleaching of cotyledons (Figure 1D). These results confirmed that cld1-1 is a semidominant allele causing the heat-sensitive phenotype.
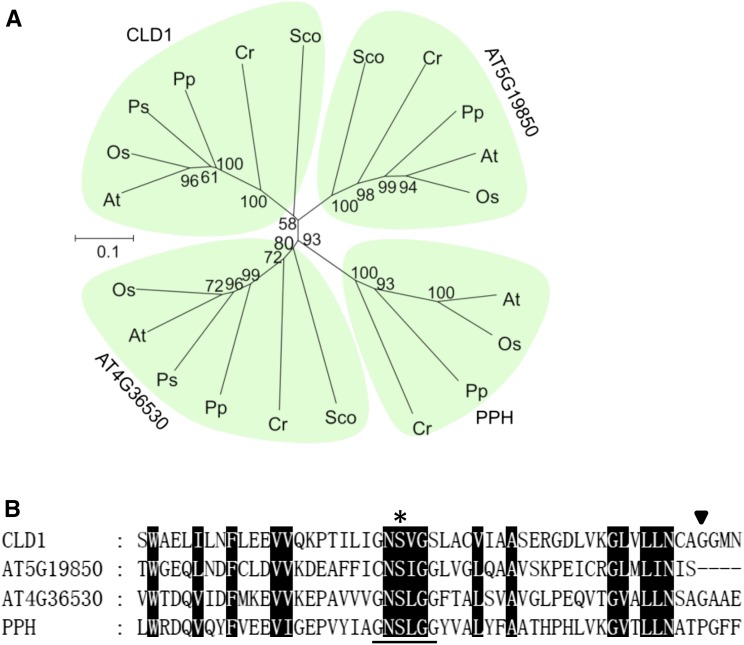
CLD1 Is Conserved in Oxygenic Photosynthetic Organisms, Sharing Sequence Similarity with PPH
To explore the molecular function of CLD1, we used the amino acid sequence of CLD1 without the putative transit peptide for a BLASTP search of the National Center for Biotechnology Information database. Homologous sequences of CLD1 (E-value < 1e−63) were found in green organisms from cyanobacteria, algae, moss, gymnosperms, and angiosperms, suggesting that CLD1 is a plant lineage-specific protein (Karpowicz et al., 2011). Many of these homologs are annotated as PPH-like proteins or putative α/β-hydrolases. PPH is involved in chlorophyll breakdown in senescent leaves in Arabidopsis (Schelbert et al., 2009) and rice (Oryza sativa; Morita et al., 2009). Phylogenetic analysis showed that CLD1s and PPHs form different clades (Figure 2A). In Arabidopsis, two putative α/β-hydrolases encoded by AT4G36530 and AT5G19850 share similarity with CLD1 and PPH (Supplemental Figure 4), thereby forming two additional clades. The phylogenetic tree suggests that CLD1 is an ancient protein that evolved early in the divergence of the green lineages. Although Arabidopsis CLD1 and PPH share only ∼27% protein sequence identity (Supplemental Figure 4), CLD1 contains a short peptide sequence very similar to the core sequence of the PPH motif (G-N-S-[LIV]-G-G; Figure 2B), proposed to be the active site of the enzyme (Schelbert et al., 2009). Gly-193 of CLD1, which is mutated into an Asp residue in cld1-1, is adjacent to the PPH motif and is conserved in all CLD1 orthologs (Supplemental Figure 3) but not PPH and the protein encoded by AT5G19850 (Figure 2B; Supplemental Figure 4).
Figure 2.
Phylogenetic Relationship of CLD1 Homologs and the Conserved PPH Motif.
(A) Phylogenetic tree of CLD1 homologous proteins of representative species from plants, algae, and cyanobacteria analyzed using MEGA6. Bootstrap values indicated as the percentage of 1000 replicates are provided at the branches. The scale bar represents replacements per unit distance per site. At, Arabidopsis; Os, Oryza sativa; Ps, Picea sitchensis; Pp, Physcomitrella patens; Cr, Chlamydomonas reinhardtii; Sco, Synechococcus sp PCC 7002.
(B) Alignment of the peptide sequences surrounding the PPH motif of CLD1 homologs in Arabidopsis. Asterisk, putative active site; black triangle, the mutation site in cld1-1; black box, conserved amino acid residue. The PPH motif sequence is underlined.
CLD1 Is Mainly Expressed in Green Tissues and Light-Grown Seedlings
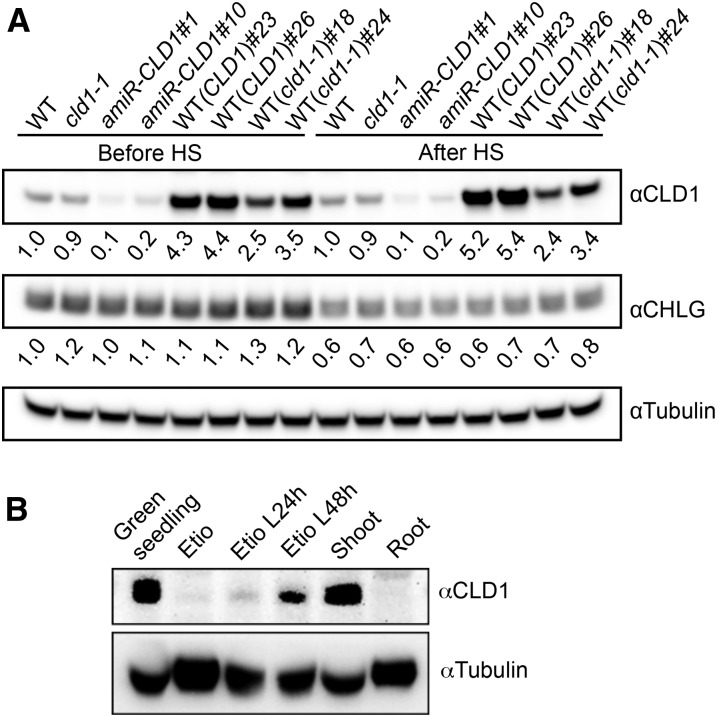
The expression profile of CLD1 was analyzed by examining the public Arabidopsis transcriptome database. CLD1 transcripts are more abundant in green tissues such as shoot apex, cotyledon, nonsenescent leaves, and immature seeds but not roots (Supplemental Figure 5A). This expression profile differs from that of PPH, which is mainly expressed in senescent leaves (Supplemental Figure 5B) and mature seeds. To examine whether the CLD1 protein matches its transcript level, we generated an antiserum against the Arabidopsis CLD1, which recognized a 36-kD protein in the crude extract of wild-type seedlings on immunoblot (Supplemental Figure 6A). This 36-kD protein corresponds to mature CLD1 without the predicted transit peptide (Mr 35,297). To confirm this notion, we generated two independent transgenic lines expressing an artificial microRNA targeting CLD1 (amiR-CLD1) because we found no available T-DNA insertion mutant of CLD1. In both amiR-CLD1 lines, the intensity of the 36-kD band substantially decreased (Supplemental Figure 6A), consistent with the reduction of CLD1 transcripts (Supplemental Figure 6B). This result indicates that the 36-kD protein is CLD1. We observed a good agreement between the CLD1 protein levels and the CLD1 transcript levels in different lines (Figure 3A), suggesting that the expression of CLD1 is primarily regulated at the transcriptional level.
Figure 3.
Immunoblot Analysis of CLD1 Protein.
(A) Immunoblots of CLD1 and CHLG in the total proteins extracted from 5-d-old seedlings before and after treatment at 40°C for 1 h. The band intensities of CLD1 and CHLG in each sample were quantified with ImageJ software and normalized to that of tubulin, which serves as the loading control. The normalized CLD1 and CHLG band intensities in each sample were divided by the corresponding values in the wild type and shown underneath each band. amiR-CLD1#1 and #10 are independent transgenic lines expressing artificial microRNA targeting CLD1. Other line designations are described in Figure 1.
(B) Immunoblots of total proteins extracted from the shoot and root, whole 5-d-old green seedlings, or 4-d-old etiolated seedlings (Etio) without or with illumination for 24 or 48 h (L24 h, L48 h). In each lane, 70 μg of protein was loaded.
Furthermore, immunoblot analysis showed that CLD1 was present in the shoots but not the roots of the light-grown seedlings (Figure 3B). The protein was more abundant in light-grown seedlings than etiolated seedlings, and the level of CLD1 increased after shining light on the etiolated seedlings (Figure 3B). These results agree with the transcript profile of CLD1, which is mainly expressed in green tissues (Supplemental Figure 5).
CLD1 Is Located in Chloroplasts and Associated with Thylakoid
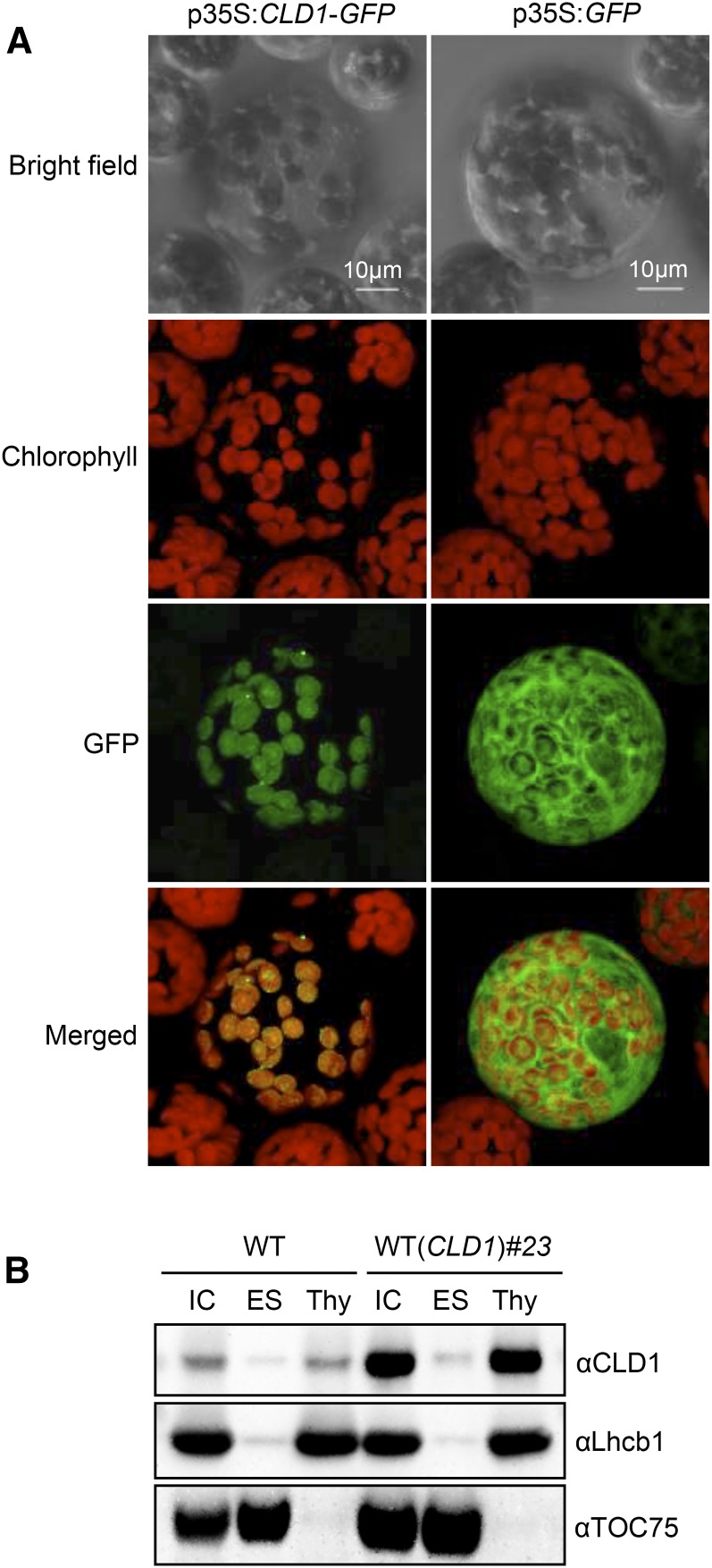
CLD1 contains a putative transit peptide at the N terminus, suggesting that CLD1 is a chloroplast protein. To confirm this, we transiently expressed the full-length CLD1 cDNA fused to the 5′-end of the reporter gene encoding GFP in Arabidopsis protoplasts. Figure 4A shows that the fluorescence of the CLD1-GFP fusion protein entirely overlapped with that of chlorophylls, with the GFP protein mainly in the cytosol, which confirms that CLD1 is localized within the chloroplast.
Figure 4.
Subcellular Localization of CLD1.
(A) CaMV35S promoter-driven CLD1-GFP (left panel) and GFP only (right panel) constructs were transiently expressed in Arabidopsis protoplasts.
(B) Intact chloroplasts (IC) isolated from 4-week-old plants were disrupted and further separated into envelope/stroma (ES) and thylakoid (Thy) fractions for immunoblot analysis. TOC75 and Lhcb1 were markers for ES and Thy fractions, respectively. Proteins equivalent to 5 μg of chlorophyll were loaded in each lane.
Fractionation of different chloroplastic compartments followed by immunoblot analysis revealed that mature CLD1 protein in the wild-type plants was present in the thylakoid fraction, but not in the envelope or stroma (Figure 4B). Overexpressed CLD1 in a transgenic line was also mainly associated with the thylakoid (Figure 4B). These results are consistent with the curated location of CLD1 reported in The Plant Proteome Database (Sun et al., 2009).
Recombinant CLD1 Dephytylates Chlorophyll a, Chlorophyll b, and Phein a in Vitro
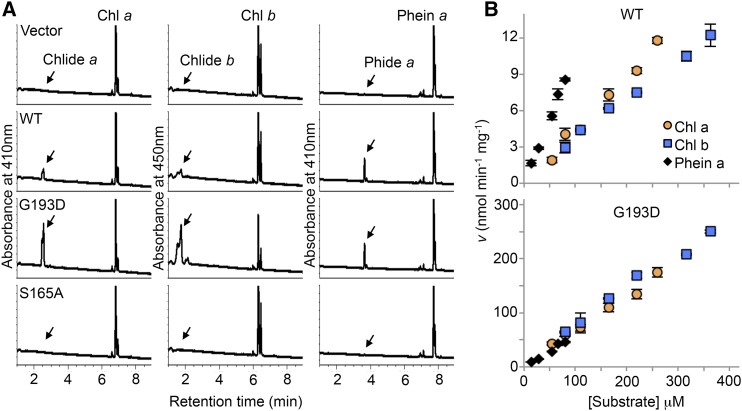
The sequence similarity with PPH prompted us to speculate that CLD1 has dephytylation activity on Phein a or similar compounds such as chlorophyll a and b. To verify this, we produced the wild-type and G193D mutant enzymes without the predicted transit peptide in Escherichia coli cells. Another mutant protein (S165A) was engineered by replacing the conserved Ser residue in the PPH motif with Ala. A cell line transformed with the empty expression vector was generated as a negative control. The recombinant CLD1 proteins were readily produced in all cell lines except the vector control but primarily present in the insoluble pellet fractions after centrifugation of the crude extracts (Supplemental Figure 7A). The G193D protein was present in the supernatant at a lower level than the wild-type and S165A proteins probably because of inefficient folding in a heterologous expression system.
Incubation of the supernatants containing the wild-type and G193D enzymes with chlorophyll a, chlorophyll b, and Phein a produced substantial amounts of Chlide a, Chlide b, and Pheide a, respectively, whereas no dephytylase activity was detected in the sample containing the S165A protein or of the vector control (Figure 5A). This result indicates that the recombinant CLD1 can act as a dephytylase in hydrolyzing chlorophylls and Phein a in vitro. Analysis of the activity under various temperatures at 25, 30, 35, and 40°C showed that the wild-type enzyme was most active at 35°C, and G193D at 30 to 35°C (Supplemental Figure 8A). We observed a considerable reduction in the activity of both enzymes at 40°C compared with that at lower temperatures. Acetone, which was used to solubilize the hydrophobic substrates, inhibited CLD1 activity remarkably. In the presence of 10% acetone in the reaction mixtures, over 80% reduction of chlorophyll dephytylase activity was observed for the wild-type and mutant CLD1 compared with that with 5% acetone (Supplemental Figure 8B). The activity was totally abolished by 20% of acetone.
Figure 5.
In Vitro Chlorophyll Dephytylase Activity of Recombinant CLD1 Expressed in E. coli Cells.
(A) HPLC trace of chlorophyll a (Chl a), chlorophyll b (Chl b), or Phein a after being incubated with the recombinant wild-type or mutated CLD1 proteins (G193D and S165A) in reaction mixtures contained 55 μM of each substrate and 5% acetone. The reactions were conducted at 35°C for 60 min.
(B) The effect of substrate concentration on the initial velocity of the wild type (upper panel) and G193D enzyme (lower panel). The substrate of indicated concentration was incubated with 0.4 or 0.08 μg of wild-type or G193D protein, respectively, at 35°C for 30 min. The reactions were stopped by adding acetone to the reaction mixtures, and the dephytylated products were quantified by HPLC. The velocity (v) is expressed as nmol of dephytylated product formed per min per mg CLD1 protein. Data are means ± sd from three replicates.
To further investigate the effect of the cld1-1 mutation, we attempted to determine the Km and Vmax of the wild-type and G193D enzymes in crude extracts. For determination of Vmax or specific activity, CLD1 protein levels were quantified by immunoblotting with purified histidine-tagged CLD1 protein as standard (Supplemental Figure 7B). The kinetic analysis was conducted by incubating the enzymes with varying concentrations of chlorophyll a, chlorophyll b, and Phein a at 35°C in the presence of 5% acetone for 30 min, within which the initial velocity was maintained (Supplemental Figure 8C). Under this assay condition, both the wild-type and mutant enzymes were not saturated at the highest concentration achievable for each substrate because the initial velocities with these three substrates remained linear (Figure 5B). The G193D mutation results in 15-, 20-, and 5.4-fold enhancement in dephytylation of chlorophyll a, chlorophyll b, and Phein a, respectively, which is consistent with the semidominant nature of the cld1-1 allele. At lower substrate concentrations, dephytylation of Phein a proceeded faster than that of chlorophyll a and chlorophyll b with the wild-type enzyme. However, this difference was not as noticeable for the G193D enzyme.
Supraoptimal CLD1 Activity Leads to Chlide a and b Accumulation in Arabidopsis Seedlings after Heat Shock
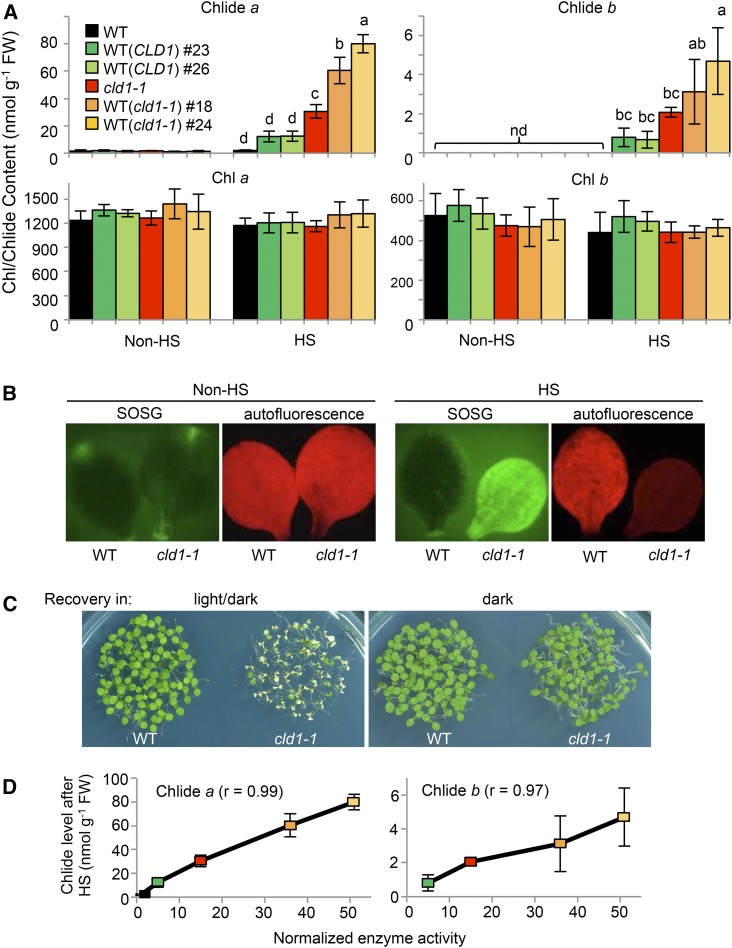
The results of in vitro activity assay and subcellular localization suggested that CLD1 is involved in chlorophyll dephytylation in chloroplasts. The cld1-1 and transgenic lines with supraoptimal CLD1 activities would probably accumulate higher levels of Chlides than wild-type plants. To test this hypothesis, we analyzed the levels of Chlides in young green seedlings before and after HS treatment. Under normal conditions, the wild-type seedlings contained little Chlide a, < 2 nmol g−1 fresh weight, with no Chlide b detected. Chlide a and b levels did not differ between the wild type and cld1-1 mutant and transgenic lines ectopically expressing the cld1-1 allele in the wild-type background, WT (cld1-1)#18 and #24 (Figure 6A). However, immediately after HS at 40°C for 1 h, Chlide a and b levels were significantly increased in cld1-1, up to 30 and 2 nmol g−1 fresh weight, respectively, with no significant change in the wild type. Chlide a and b levels were further increased in the transgenic lines WT (cld1-1)#18 and #24, up to 80 and 4 nmol g−1 fresh weight, respectively (Figure 6A). Overexpression of the mutant protein did not cause a massive degradation of chlorophylls in the transgenic lines because total chlorophyll levels did not significantly differ from that of the wild type (Figure 6A).
Figure 6.
Heat-Induced Accumulation of Chlide a and b Is Associated with Supraoptimal CLD1 Activities, Resulting in Singlet Oxygen Formation and Light-Dependent Cotyledon Bleaching.
(A) Levels of Chlides and chlorophylls extracted from pooled 5-d-old seedlings before and after treatment at 40°C for 1 h (HS). The line designations are as described in Figure 1. Data are means ± sd of three independent experiments. One-way ANOVA with Tukey’s method was applied to show the significant differences at P < 0.01 in pairwise comparison and classified with the letter a, b, c, or d. ANOVA tables can be found in Supplemental Figure 10. Column without any letter indicates no significant difference from that of the wild type before HS.
(B) Detection of singlet oxygen in wild-type and cld1-1 seedlings. After HS, the singlet oxygen level was analyzed by incubating the cut cotyledons with 10 μM SOSG solution under light for 4 h at 22°C. Non-heat-treated samples were used as the control. Images with false color were obtained under a fluorescence microscope. Green indicates the accumulation of singlet oxygen, and red indicates the autofluorescence of chlorophyll.
(C) Heat-induced cotyledon bleaching in cld1-1 is light-dependent. Five-d-old seedlings were treated at 40°C for 1 h and recovered for 2 d at 22°C under 16-h/8-h light-dark cycle (100 μmole m−2 s−1) and dark. The pictures were obtained right after recovery.
(D) Correlation of levels of heat-induced Chlide a and b versus the normalized chlorophyll dephytylase activities in indicated lines. The enzyme activity in each line was calculated by multiplying the protein level shown in Figure 3A and the normalized reaction rate determined in Figure 5B. The activity in the wild type is assigned to 1. The color codes are as in (A). Pearson’s correlation coefficients (R) are shown.
Because the accumulation of Chlides in cells could lead to the production of singlet oxygen when exposed to light, we stained cotyledons of the wild-type and cld1-1 seedlings with the fluorescence probe singlet oxygen sensor green (SOSG). Only the heat-treated cld1-1 cotyledons emitted strong fluorescence (Figure 6B), which indicates the surge of singlet oxygen in the mutant after HS treatment. Moreover, cotyledon bleaching of cld1-1 was induced during recovery after HS treatment under a light/dark cycle condition but not in the dark (Figure 6C). These results are consistent with the heat-induced accumulation of phototoxic Chlides in the mutant.
The levels of Chlide a and b in the transgenic lines overexpressing wild-type CLD1, WT (CLD1)#23 and #26, were also significantly increased after HS treatment (Figure 6A), so the accumulation of Chlides was not simply due to the G193D mutation of CLD1 but to the supraoptimal CLD1 activities expressed. When the normalized chlorophyll dephytylase activities were plotted against the levels of Chlides present in different lines after HS treatment, we observed a good correlation between these two variables (see correlation coefficients in Figure 6D). Although reduced CHLG level can also result in accumulation of Chlide a (Lin et al., 2014), the levels of CHLG in cld1-1 and transgenic lines were comparable to that of the wild type before and after HS treatment (Figure 3A). Of note, the level of CHLG was reduced ∼40% by HS treatment in all lines tested, with that of CLD1 (both the wild type and the G193D mutant) largely unchanged (Figure 3A). These results suggest that CLD1 dephytylates chlorophyll a and b in vivo and that supraoptimal CLD1 activity led to the accumulation of Chlides after HS treatment. Interestingly, the ratio of heat-induced Chlide a and b was ∼15 to 1, which is much higher than the chlorophyll a and b ratio present in the lines with supraoptimal CLD1 activities (Figure 6A), so chlorophyll a is the primary target subjected to dephytylation by CLD1. However, we could not exclude the possibility that Chlide b came from the oxidation of Chlide a catalyzed by CAO (Tanaka and Tanaka, 2011).
Despite CLD1 being able to hydrolyze Phein a in vitro, we did not observe accumulation of Pheide a in all the lines tested before or after the HS treatment, and the levels of Phein a were also not significantly different between the wild type and the mutants (Supplemental Figure 11A). This observation suggests that alteration of CLD1 activity does not affect the turnover of Phein a in vivo.
Accumulation of Chlide a Is Further Enhanced in cld1-1 chlg-1 Double Mutants after Heat Shock
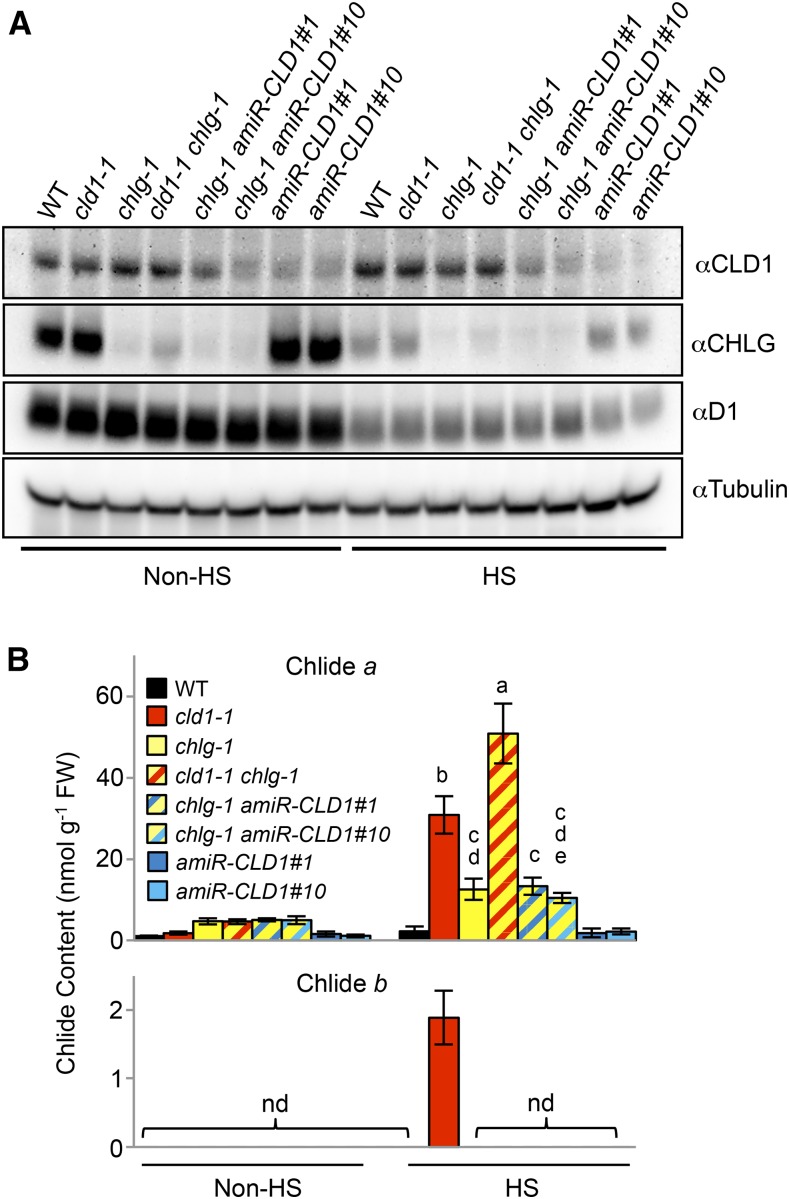
The accumulation of Chlides is likely a result of the increased ratio of de- and rephytylation reactions catalyzed by CLD1 and CHLG, respectively. The missense mutation in chlg-1 reduces ∼90% of CHLG protein and results in heat-induced Chlide a accumulation (Lin et al., 2014). To further examine the effect of varying CLD1/CHLG ratio on Chlide accumulation, we produced the cld1-1 chlg-1 double mutant (Figure 7A; Supplemental Figure 9A). After HS treatment, the cld1-1 chlg-1 double mutant accumulated a significantly higher level of Chlide a than each of the single mutants (Figure 7B). Intriguingly, cld1-1 chlg-1 did not accumulate Chlide b like cld1-1, despite having a higher Chlide a level. The absence of Chlide b in cld1-1 chlg-1 was probably due to the substantially reduced chlorophyll b level (∼30% of the wild-type level; Supplemental Figure 9B) in the double mutant, in which chlorophyll b concentration might be below the threshold for CLD1 action.
Figure 7.
Effect of Varied CLD1/CHLG Ratio on Heat-Induced Chlide Accumulation.
Immunoblots of CLD1 and CHLG proteins (A) and Chlide a and b (B) amounts in 5-d-old seedlings before and after treatment at 40°C for 1 h (HS). The data are means ± sd of three independent experiments. One-way ANOVA with Tukey method was used to examine significant differences at P < 0.01 in pairwise comparison and classified by a, b, c, d, or e. ANOVA tables can be found in Supplemental Figure 10. nd, not detected.
To determine whether suppressed CLD1 expression in a chlg-1 background would reduce the accumulation of Chlides, we crossed the amiR-CLD1 lines with chlg-1 to generate the amiR-CLD1#1 chlg-1 and amiR-CLD1#10 chlg-1 double mutants. However, these double mutants did not show significantly decreased Chlide a level after HS treatment compared with chlg-1 (Figure 7B). Levels of CLD1 were higher in the double mutants amiR-CLD1#1 chlg-1 and amiR-CLD1#10 chlg-1 than amiR-CLD1#1 and #10 (Figure 7A), respectively, so the effect of artificial microRNA was reduced in the double mutants. The level of CLD1 in the double mutants may be high enough to trigger the accumulation of Chlide a when the level of CHLG is reduced.
Unsaturated Geranylgeraniol Chlorophyll b Level Increased in Seedlings with Supraoptimal CLD1 Activity
While analyzing chlorophylls and related pigments, we detected substantial increases of chlorophyll b conjugated with unsaturated geranylgeraniol side chains, i.e., geranylgeraniol chlorophyll b (chlorophyll bGG), dihydrogeranylgeraniol chlorophyll b (chlorophyll bDHGG), and tetrahydrogeranylgeraniol chlorophyll b (chlorophyll bTHGG), in all lines with supraoptimal CLD1 activity under normal conditions (Supplemental Figure 11A). HS treatment did not further increase their abundance. Although the level of phytyl chlorophyll b (chlorophyll bphy) was comparable in the wild type and these lines (Figure 6A), total chlorophyll b level with unsaturated geranylgeraniols increased from <1% in the wild type to >5% in cld1-1 and the two WT (cld1-1) transgenic lines. A minor but significant increase in levels of chlorophyll bGG, chlorophyll bDHGG, and chlorophyll bTHGG was present in transgenic lines with an ectopic copy of CLD1 in the wild-type background. Intriguingly, the levels of chlorophyll a with unsaturated geranylgeraniols did not differ as substantially as those found in b-type pigments between the wild type and the mutants. The amount of geranylgeranyl reductase (GGR), which catalyzes the subsequent reduction of chlorophyll bGG to chlorophyll bphy, was not affected in cld1-1 and transgenic lines (Supplemental Figure 11B), so the accumulation of unsaturated chlorophyll bGG was not due to reduced GGR in the plants with supraoptimal CLD1 activity. Moreover, the mutants lil3:1 and lil3:2, which also accumulate high levels of chlorophyll bGG, bDHGG, and bTHGG (Tanaka et al., 2010), did not show any heat-sensitive phenotype (Supplemental Figure 11C). These results indicate that the heat-sensitive phenotype in cld1-1 and WT (cld1-1) transgenic lines was not due to the accumulation of chlorophyll b with unsaturated side chains. Our observations suggest that CLD1 activity higher than the wild-type level somehow perturbs the function of GGR directly or indirectly. The asymmetric effects on the accumulation of chlorophyll a and chlorophyll b with unsaturated side chains suggest that the perturbation on GGR is mainly due to its reduction of the geranylgeranyl side chain of chlorophyll instead of free GGPP.
Suppression of CLD1 Expression Reduces the Maximum Quantum Efficiency of PSII Photochemistry and Thermotolerance under Chronic Heat Stress
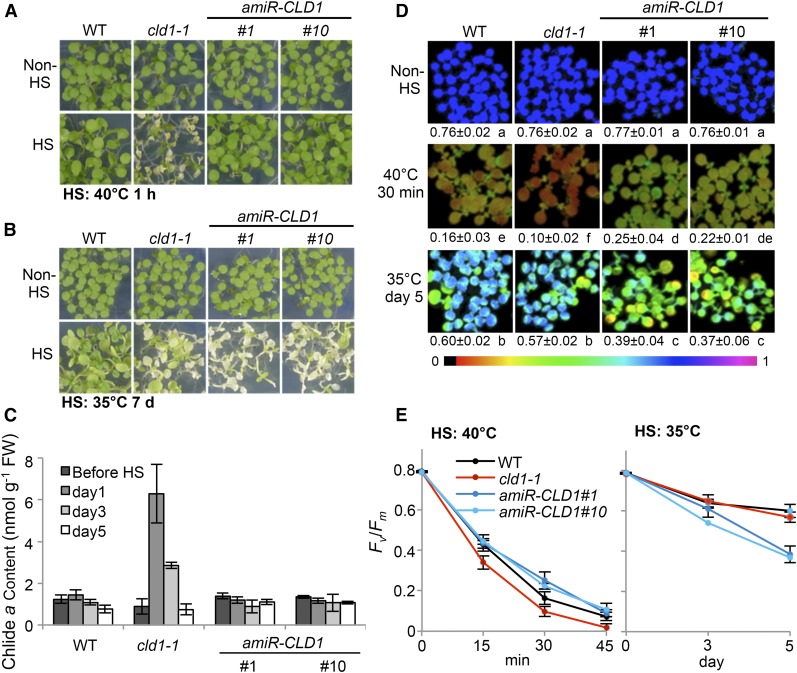
Our in vitro and in vivo results strongly suggest that CLD1 is a novel chlorophyll dephytylase involved in chlorophyll turnover at steady state. However, the biological significance of this chlorophyll dephytylation activity is not clear. HS treatment at 40°C for 1 h induced cotyledon bleaching in cld1-1 but no visible damage to the amiR-CLD1#1 and #10 lines compared with the wild type (Figure 8A), which indicates that a wild-type level of CLD1 is not required for tolerating a short exposure to severely high temperature (acute heat stress). Since plants often experience moderately high temperatures, which could also damage photosystems (Murata et al., 2007), we examined the response of the amiR-CLD1 lines to prolonged moderate heat stress (chronic heat stress at 33 to 35°C for 7 d). Surprisingly, these transgenic lines with reduced CLD1 level were more sensitive than the wild type to chronic heat stress, showing bleached cotyledons, retarded growth, and reduced viability during recovery (Figure 8B). The cld1-1 mutant showed a heat-sensitive phenotype under the same conditions but not as severe as the amiR-CLD1 lines. Similar to the case of acute heat stress, Chlide a accumulated in cld1-1 under the chronic heat stress condition to a higher level than in the wild type and the amiR-CLD1 lines (Figure 8C).
Figure 8.
Suppression of CLD1 Reduces Maximum Quantum Efficiency of PSII and Thermotolerance under Chronic Heat Stress.
(A) and (B) Phenotypes of 5-d-old seedlings with or without HS treatment under two different HS regimes. Seedlings in the same treatment were grown on the same plate. Lower panels in (A) and (B) were obtained after 3- and 7-d recovery, respectively.
(C) Chlide a level before and after treatment with different duration at 33 to 35°C. Samples were collected before the light turned on. Data are means ± sd of three independent experiments.
(D) Maximum quantum efficiency of PSII of seedlings before and after exposure to different HS regimes. The false color represents the Fv/Fm value from 0 (black) to 1 (magenta). Mean Fv/Fm value ± sd of five independent experiments is indicated below each representative image. One-way ANOVA with Tukey’s method was used to analyze significant differences in pairwise comparison at P < 0.01 and classified with a, b, c, d, e, or f. ANOVA tables can be found in Supplemental Figure 10.
(E) Time course of Fv/Fm change at 40 or 35°C.
Since heat stress can damage PSII, we estimated the PSII activities of seedlings of different Arabidopsis lines by measuring the maximum quantum efficiency of PSII photochemistry (Fv/Fm) after acute or chronic heat stress treatment. A decline in Fv/Fm reflects the damage of PSII (Murchie and Lawson, 2013). Figure 8D shows that without heat stress, the Fv/Fm values were similar between the wild type, cld1-1, and amiR-CLD1 lines. A marked decrease in Fv/Fm was observed in all lines after HS treatment at 40°C for 30 min, but the value for cld1-1 was significantly lower than that of the wild type and amiR-CLD1 lines (Figures 8D and 8E). In contrast, after chronic heat stress treatment for 5 d, the Fv/Fm values were significantly lower for the amiR-CLD1 lines than the wild type, with the value of cld1-1 only slightly less than that of the wild type (Figures 8D and 8E). The results are consistent with the phenotypes shown in Figures 8A and 8B. The alteration in Fv/Fm and bleaching phenotype were quite specific for heat stress because we did not observe these phenomena under osmotic, salt, or high-light stress (Supplemental Figure 12).
DISCUSSION
The cleavage of the phytol chain occurs at the early stage of chlorophyll catabolism, including chlorophyll breakdown and turnover (Hörtensteiner, 2013b). Finding the enzymes responsible for the dephytylation reaction is necessary for understanding these issues. An activity-based approach had led to the cloning of CLH genes encoding chlorophyllases from citrus and Chenopodium album (Jacob-Wilk et al., 1999; Tsuchiya et al., 1999), and a reverse genetic approach led to the identification of PPH from Arabidopsis (Schelbert et al., 2009). In this study, we serendipitously identified a new dephytylase, CLD1, from Arabidopsis by a forward genetic approach initially aiming to identify thermotolerance components (Wu et al., 2013). Isolation of the cld1-1 mutant is critical in shedding light on the biological function of CLD1 because the semidominant allele confers a heat-sensitive phenotype that facilitated mapping and cloning of the mutant gene.
CLD1 Is a “Chlorophyllase” Distinct from CLH
CLD1 belongs to the α/β-hydrolase superfamily that also includes CLH and PPH (Schelbert et al., 2009). The overall sequence identity between these proteins is low (<30%), but they catalyze similar dephytylation reactions on chlorophylls or chlorophyll derivatives. In contrast to PPH, which removes the phytol chain from Phein a but not chlorophylls (Schelbert et al., 2009), CLD1 can dephytylate chlorophyll a and Phein a in vitro (Figure 5), similar to CLHs (McFeeters et al., 1971; Schelbert et al., 2009). The structural basis underlying the difference between CLD1 and PPH in substrate specificity is unclear and worthy of further investigation. Similar to the case in PPH (Schelbert et al., 2009) and CLH (Tsuchiya et al., 2003), mutation of the putative catalytic serine residue in CLD1 abolished the enzyme activity (Figure 5B), which suggests that CLD1 is also a serine-type hydrolase.
The natural enzyme capable of dephytylating chlorophylls in vivo has been enigmatic (Hörtensteiner, 2013b). Now, at least two enzymes, CLH and CLD1, are capable of cleaving the ester bond of chlorophyll in vitro. Based on this common catalytic activity, CLD1 can be referred to as a “chlorophyllase.” However, it differs remarkably from the typical chlorophyllases (CLHs) in enzyme behavior. The Km values of CLHs for chlorophyll a were reported to range from 1.76 to 103 μM depending on the source of the protein (McFeeters et al., 1971; Trebitsh et al., 1993; Tsuchiya et al., 1997; Fang et al., 1998; Okazawa et al., 2006). However, the reaction rate of CLD1 was not near saturation even with 260 μM of chlorophyll a (Figure 5B), suggesting that CLD1 has a much lower apparent affinity for chlorophyll a than does CLH. Since CLD1 activity is substantially diminished in the presence of >7% acetone (Supplemental Figure 8B), we were unable to obtain the Vmax for CLD1 with higher chlorophyll a concentrations. The mode of inhibition of CLD1 by acetone remains unclear. Of note, CLH apparently is tolerant to high acetone concentration as the enzymes from sugar beet (Beta vulgaris), snap bean (Phaseolus vulgaris), C. album, and Arabidopsis were routinely assayed in reaction mixtures with ≥20% acetone (Bacon and Holden, 1970; Fang et al., 1998; Tsuchiya et al., 1999).
Another significant difference between CLD1 and CLH is their catalytic efficiency. The specific activities of CLHs purified from citrus peel and mature leaves of C. album are 174 and 145 μmol chlorophyll a hydrolyzed min−1 mg−1 protein, respectively (Trebitsh et al., 1993; Tsuchiya et al., 1997). The specific activities of Arabidopsis recombinant CLH1 and CLH2 were shown to be similar to that of C. album recombinant CLH expressed in E. coli (Tsuchiya et al., 1999). These numbers are about four orders of magnitude higher than the highest specific activity determined for wild-type CLD1 (∼12 nmol chlorophyll a hydrolyzed min−1 mg−1 protein; Figure 5B). This magnitude of difference suggests that CLD1 and CLH must be spatially separated when they are coexpressed in the same cell at least in Arabidopsis. The idea is supported by the finding that CLD1 is localized within the chloroplasts (Figure 4), and CLH isoforms are not (Schenk et al., 2007). Recent work showed that Arabidopsis CLH1, the major isoform, is localized to the tonoplast and the endoplasmic reticulum, and artificially mistargeting CLH1 into chloroplasts causes Chlide accumulation and cell death (Hu et al., 2015), which suggests detrimental effects of the constitutive presence of CLH in chloroplasts. From these observations, we infer that CLD1 catalyzes a dephytylation reaction functionally distinct from that catalyzed by CLH. CLH was shown to be involved in chlorophyll breakdown in postharvest citrus fruits and broccoli (Brassica oleracea var italica) florets (Azoulay Shemer et al., 2008; Chen et al., 2008) but not in senescent leaves and maturing seeds of Arabidopsis (Schenk et al., 2007; Zhang et al., 2014). The exact role of CLH in chlorophyll catabolism remains controversial, but it was shown to function in defense against pathogens and herbivores, which may disrupt the integrity of chloroplasts and render accessibility of chlorophyll to CLH (Kariola et al., 2005; Hu et al., 2015).
CLD1 Is Involved in Chlorophyll Turnover at Steady State
Because of its localization and in vitro chlorophyll dephytylase activity, CLD1 is a better candidate than CLH for catalyzing chlorophyll dephytylation within intact chloroplasts. Being located to the thylakoid would place CLD1 in proximity to its substrates. Because massive breakdown of chlorophylls does not occur before the initiation of senescence (Pruzinská et al., 2007) and CLD1 is highly expressed in green tissues (Supplemental Figure 5) and induced in greening seedlings (Figure 3B), CLD1 is likely mainly, if not solely, involved in chlorophyll turnover rather than chlorophyll breakdown. This notion is consistent with the result that reduction or increase of CLD1 activity did not affect chlorophyll breakdown in dark-induced senescent leaves (Supplemental Figure 13).
The cld1-1 allele encodes a mutant enzyme with specific activity significantly higher than that of the wild-type enzyme (Figure 5B), which is consistent with the semidominant heat-sensitive phenotype caused by the mutant allele (Figure 1A). The heat-induced cotyledon bleaching of cld1-1 seedlings could be attributed to the accumulation of Chlide a (Figure 6A), which is potent in generating reactive oxygen species light-dependently (Lin et al., 2014). Because the HS treatment was performed in the dark, the Chlide a level accumulated in the mutant seedlings was unlikely derived from the de novo light-dependent reduction of protochlorophyllide, which is catalyzed by NADPH:protochlorophyllide oxidoreductase (Rüdiger, 2009). Hence, the accumulation of Chlide a should result from the dephytylation of chlorophyll a catalyzed by CLD1 in vivo. This notion is strongly supported by the good correlation of normalized CLD1 activities and levels of Chlides in Arabidopsis seedlings after HS treatment (Figure 6D). Interestingly, supraoptimal CLD1 activities did not result in the accumulation of Chlides under normal conditions (Figure 6A), which suggests that chlorophyll turnover rate may not be solely determined by CLD1 activity. There may be other factors involved in regulating CLD1 activity or facilitating the accessibility of it substrates in response to HS treatment. HS temperature apparently is unlikely to be a direct factor in activating CLD1, as the enzyme in vitro is substantially inhibited at 40°C (Supplemental Figure 8A). The low apparent affinity of CLD1 for chlorophyll a might be overcome by interacting with certain chlorophyll binding proteins that enrich chlorophyll a via substrate channeling (Miles et al., 1999). It would be of interest to identify CLD1 interacting proteins to verify this hypothesis.
The accumulation of Chlide b was substantial but much lower than Chlide a in the seedlings with supraoptimal CLD1 activities. The Chlide a and b ratios in the heat-treated mutant seedlings were much higher than the ratio of chlorophyll a and b (Figure 6A), which is consistent with previous reports that chlorophyll turnover mainly affects chlorophyll a (Feierabend and Dehne, 1996; Beisel et al., 2010). Chlorophyll b is present in the peripheral antennae, light-harvesting complex I (LHCI), and LHCII, and in the interface between PSI core complex and LHCI (Mazor et al., 2015; Qin et al., 2015). Thus, heat stress may also stimulate the turnover of chlorophyll b released from LHCs or PSI complex. Damage of PSI occurred under stress conditions (Alboresi et al., 2009; Sonoike, 2011), and turnover of chlorophyll b bound to these protein complexes via dephytylation by CLD1 is plausible. However, interconversion of chlorophyll/Chlide a and chlorophyll/Chlide b could occur in the chlorophyll cycle (Tanaka and Tanaka, 2011), so the origin of the accumulated Chlide b in heat-treated cld1-1 is uncertain at this moment. More effort is needed to address this question.
CLD1 and CHLG Form a Chlorophyll Salvage Cycle
Previous studies suggested that a chlorophyll salvage pathway via de- and rephytylation steps are involved in recycling chlorophyll a during its turnover at steady state, but the enzyme responsible for the dephytylation step remained unknown (Vavilin and Vermaas, 2007; Lin et al., 2014). The results of this work suggest that CLD1 plays this role, so we propose a modified scheme of chlorophyll metabolism incorporating the function of CLD1 (Figure 9). In this model, CLD1 and CHLG form a salvage cycle in recycling the chlorin rings and phytol tails (not shown in the scheme) of chlorophylls. Reuse of the phytol tails for chlorophyll synthesis has been reported previously (Ischebeck et al., 2006; Vavilin and Vermaas, 2007). We reason that an appropriate ratio of CLD1/CHLG should be maintained to avoid accumulation of the phototoxic intermediates such as Chlide a and b. In the wild type, the CHLG level is sufficient to convert Chlides back to chlorophylls when CLD1 is at the wild-type level. However, when CLD1 activity is abnormally high, such as in cld1-1 and transgenic plants overexpressing CLD1 (mutant or wild-type form), the CHLG level could not match the increased activity of CLD1, which results in accumulation of Chlides. This notion is consistent with our earlier findings that the reduction of CHLG in the chlg-1 mutant leads to the heat-induced accumulation of Chlide a (Lin et al., 2014) and that the level of Chlide a is much higher in the cld1-1 chlg-1 double mutant than individual single mutants (Figure 7B). Intriguingly, downregulation of CLD1 in the chlg-1 background did not reduce the level of Chlide a (Figure 7B) probably due to the reduced effect of CLD1 gene silencing in the double mutant (Figure 7A). Alternatively, the other three CLD1 homologs (Figure 2A) may share a redundant function in dephytylating chlorophylls. Recent studies of tomato PPH proposed a similar view on the functional redundancy of PPH homologs (Lira et al., 2016). Characterization of the CLD1 homologs encoded by AT4G36530 and AT5G19850 is under way. Our preliminary data showed that at least one of these homologs catalyzes chlorophyll dephytylation reaction in vitro like CLD1, which suggests the existence of additional chlorophyll dephytylase.
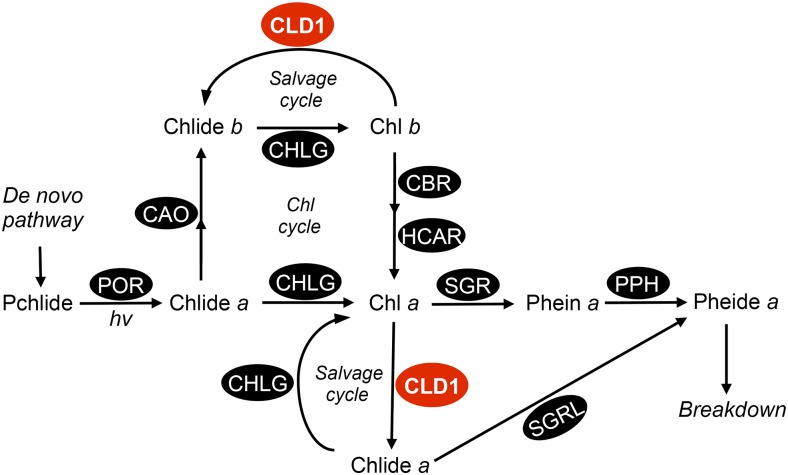
Figure 9.
A Simplified Scheme of Chlorophyll Metabolism with CLD1 in Chlorophyll Turnover.
Chlide a, the precursor of chlorophyll a, can be produced from a de novo pathway via protochlorophyllide (Pchlide) or from dephytylation of chlorophyll a catalyzed by CLD1 during chlorophyll turnover. The Chlide b accumulated in cld1-1 could result from chlorophyll b dephytylation or oxidation of Chlide a, which is not yet certain. POR, protochlorophyllide oxidoreductase; CHLG, chlorophyll synthase; CAO, Chlide a oxidase; CBR, chlorophyll b reductase; HCAR, 7-hydroxymethyl chlorophyll a reductase; MCS, metal chelating substance; SGR, STAY-GREEN (chlorophyll a Mg-dechelatase); SGRL, STAY-GREEN-LIKE (Chlide/chlorophyll a Mg-dechelatase).
Recently, STAY-GREEN LIKE (SGRL) was shown to encode a protein with in vitro activity of Chlide a dechelatase (Shimoda et al., 2016), suggesting that CLD1 could also be involved in chlorophyll degradation (Figure 9). However, chlorophyll degradation via CLD1 and SGRL is not a primary route in sequestering chlorophyll a under heat stress. Otherwise, the accumulation of Chlide a in cld1-1 would not have been observed. Hence, we think that CLD1 is primarily involved in recycling chlorophyll a with CHLG via the salvage cycle. The physiological significance of recycling chlorophylls via this salvage cycle is unclear. One possibility is that it is essential for reassembling of the recycled chlorophyll molecules into the photosystem protein-pigment complex (Vavilin and Vermaas, 2007). Alternatively, chlorophyll dephytylation is required for the degradation of D1 protein in repairing PSII (Nixon et al., 2010). However, we did not observe a difference in D1 levels between the wild-type and transgenic plants with reduced CLD1 after heat stress (Figure 7A). Again, this finding could be due to the functional redundancy conferred by the CLD1 homologs, which awaits further investigation.
CLD1 Plays a Role in Thermotolerance to Moderately High Temperature
Plants have evolved different thermotolerance responses that require the functions of many components, including transcription factors, protein chaperones, and enzymes (Yeh et al., 2012). Heat stress is well known to damage photosystems and cause photoinhibition (Allakhverdiev et al., 2008). So far, only a small number of components have been shown to protect photosystems against heat stress. Adjustment of lipid unsaturation of the thylakoid membrane plays an important role in the thermotolerance of photosystems (Routaboul et al., 2012). Rubisco activase is required for the thermotolerance of the effective quantum yield of PSII (Salvucci, 2008). In maize (Zea mays), phytoene synthase PSY1 is involved in carotenogenesis in the dark and heat stress tolerance (Li et al., 2008). A chloroplastic small HSP was shown to protect PSII against heat stress (Heckathorn et al., 1998; Neta-Sharir et al., 2005). Isoprene formation in certain plant species is a well-known feature in increasing the thermostability of thylakoid membranes (Behnke et al., 2007; Sharkey et al., 2008; Velikova et al., 2011). Here, we provide genetic evidence indicating that CLD1 is required for thermotolerance to moderately high temperature (Figure 8). This result suggests that chlorophyll turnover mediated by CLD1 is a protective measure in maintaining the homeostasis of PSII under chronic heat stress. CLD1-mediated chlorophyll turnover may be involved in the repair of photodamage, which is induced in PSII by moderate heat stress (Murata et al., 2007). Although PSII turnover and repair has been intensively investigated (Järvi et al., 2015), whether and how chlorophyll turnover and salvage is required for these processes has not been studied because of the complex metabolism of the pigment. The identification of CLD1 and the related mutants generated in this study would provide an opportunity to gain better insight into this issue. Of note, suppression of CLD1 expression seemed not to affect the tolerance of PSII to acute heat stress in Arabidopsis seedlings (Figures 8D and 8E). Intriguingly, the maximum quantum efficiency of PSII was less affected by HS at 40°C for 30 min in the CLD1 silencing lines than the wild type. This observation suggests that CLD1 acts as not only a negative regulator of PSII function under acute heat stress but also as a positive regulator of PSII under chronic heat stress. XRN4, an RNA exonuclease, operates as a negative and positive regulator of thermotolerance during acute and chronic heat stress, respectively (Merret et al., 2013; Nguyen et al., 2015). Plants probably feature a tradeoff in adapting to different heat stress conditions.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana EMS mutants cld1-1 (dlt3-1) and chlg-1 (dlt4-1) were isolated by a forward genetic approach as described (Wu et al., 2013). The pph-1 mutant (SALK_000095) was obtained from Stefan Hörtensteiner, University of Zürich, Switzerland. The mutants lil3:1 and lil3:2 were Ds transposon-tagged mutants of the Nossen ecotype (Tanaka et al., 2010) obtained from Ayumi Tanaka and Ryouichi Tanaka, Hokkaido University, Japan. The transgenic lines WT (cld1-1), WT (CLD1), and amiR-CLD1 were generated as described below. Seeds were sterilized, sown on a plate containing 0.8% agar with half-strength Murashige-Skoog medium and 1% sucrose (medium A), and imbibed for 3 d at 4°C in the dark. Then plates were incubated at 22°C with a 16-h/8-h light-dark cycle (100 μmole m−2 s−1 light intensity, white light, LED bulbs) until the indicated growth stage for stress treatments. HS treatment involved a water bath at 40°C for 1 h in the dark (Charng et al., 2006). Chronic heat stress was applied to Arabidopsis seedlings as described (Hu et al., 2012). For salt and osmotic stress treatments, 5-d-old seedlings grown under the normal condition on medium A were transferred to medium A containing NaCl or mannitol at the indicated concentrations for another 3 d of growth under a 16-h/8-h light-dark cycle.
Map-Based Cloning of cld1-1 and Complementation Test
Map-based cloning of cld1-1 involved the F2 population from the cross between cld1-1 (in the Col-0 background) and the wild type (in Ler background). Seedlings with a nonbleaching phenotype were collected for DNA extraction and molecular marker mapping as described (Jander, 2006). The mutated site in cld1-1 was located on chromosome 5 within the region between the markers PHYC.2 and ciw9, and fine mapping narrowed down the locus to the region flanked by the markers CER457644 and CER458106 (Supplemental Figure 2); the genes within this region were examined by sequencing. For the complementation of cld1-1, genomic DNA of Arabidopsis CLD1 or cld1-1 was amplified by PCR with the primers YP011 and YP012 (Supplemental Table 2). The 2970-bp PCR fragment encompassing CLD1 or cld1-1 genomic DNA with the 5′-end starting at 959 bp upstream of the transcriptional start site (Figure 1B) was cloned into pCR8/GW/TOPO (Invitrogen), sequenced to verify no unwanted mutation, and subcloned into the binary vector pBGW,0 (Karimi et al., 2002) for Agrobacterium tumefaciens-mediated transformation of Arabidopsis. Transformation and selection of transgenic plants were performed as described (Charng et al., 2007).
Phylogenetic Analysis
The sequence of Arabidopsis CLD1 peptide with 362 amino acids (AT5G38520.1) was extracted from TAIR (http://www.arabidopsis.org). Homologs of CLD1 in Arabidopsis and other species were identified by a BLASTP search of the databases National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi) or the Joint Genome Institute (https://phytozome.jgi.doe.gov/pz/portal.html#!search). The peptide sequences (see below for accession numbers) without predicted chloroplast transit peptide were aligned using the MEGA6 ClustalW program (Tamura et al., 2013), then used for generation of a phylogenetic tree by the neighbor-joining method. The transit peptide was predicted using ChloroP 1.1 (Emanuelsson et al., 2007). A text file of the alignment can be found in Supplemental File 1.
Artificial MicroRNA-Mediated Suppression of CLD1 Expression
Suppression of CLD1 expression in transgenic plants was achieved by an artificial microRNA-mediated method (Schwab et al., 2006). Briefly, a portion of CLD1 cDNA (Figure 1B) was amplified by PCR with the primers YP021 and YP022 (Supplemental Table 2). The amplified 435-bp DNA fragments were cloned into the binary vector pHELLGATE2 (Helliwell and Waterhouse, 2003) to yield pHG2-amiR-CLD1 for the Agrobacterium-mediated transformation of Arabidopsis. Expression of the artificial microRNA was driven by the constitutive CaMV35S promoter in pHELLGATE2. Transformation and selection of transgenic plants were performed as previously described (Lin et al., 2014).
RNA Extraction and Quantitative RT-PCR
Total RNA extraction, cDNA preparation, and quantitative RT-PCR were performed as previously described (Liu and Charng, 2013). Expression of the CLD1 and cld1-1 alleles was analyzed by quantitative RT-PCR with allele-specific primers (Supplemental Table 2). The expression of CLD1 or cld1-1 was normalized to that of ACTIN2 by subtracting the cycle threshold (CT) value of ACTIN2 from the CT value of CLD1 or cld1-1. The amplification efficiency (E) of PCR reactions using different primer sets was >0.95 (Peirson et al., 2003).
Protein Extraction and Immunoblotting
Total proteins were extracted from Arabidopsis seedlings and used for immunoblotting as described (Charng et al., 2006). Protein content was quantified using the DC protein assay kit (Bio-Rad) with a standard curve of BSA. For immunoblot analysis, total protein was loaded to an SDS-PAGE minigel (NuPAGE 4-12% BisTris gel system; Invitrogen) for separation and then transferred to a nitrocellulose membrane for immunodetection using chemiluminescence reagent (Western Lighting Plus-ECL; Perkin-Elmer). The polyclonal antiserum against Arabidopsis CLD1 was produced by immunizing rabbits with a synthetic peptide (N′-IGERSKKWKWKGEYSVNY; Supplemental Figure 3) in a service provided by LTK Biotechnology (http://www.ltk.com.tw/). The antibodies against CHLG, D1, Lhcb1, and tubulin were described by Lin et al. (2014), and the antibodies against GGR were described by Tanaka et al. (1999). Anti-TOC75 antibody (AS06 150, 2000× dilution) was from Agrisera. Chemiluminescent signals were visualized with FluorChem HD2 (Alpha Innotech) and quantified using ImageJ (National Institutes of Health). The membrane was stained with Amido black (0.1%, w/v) following immunodetection to ensure equal protein loading.
Subcellular Localization of CLD1 Protein
The full-length cDNA sequence of CLD1 was amplified by RT-PCR with the primers YP051 and YP052 (Supplemental Table 2). CLD1 cDNA was ligated into the vector pCR8/GW/TOPO for sequencing confirmation and subsequent subcloning into pMDC83 for generating a chimeric gene with C-terminal in-frame fusion to GFP as described (Hu et al., 2012). The resulting construct was used to transform Arabidopsis protoplasts to determine the subcellular localization of the CLD1-GFP fusion protein as described (Yoo et al., 2007; Wu et al., 2009). For the GFP control, pMDC83 was used for the transient transformation. The fluorescence signal of GFP and autofluorescence of chlorophylls were analyzed under a Zeiss LSM 510 Meta confocal microscope with an appropriate filter set.
Fractionation of chloroplastic compartments was performed as described (Chu and Li, 2011). Briefly, leaves of 4-week-old plants were collected, and chloroplasts were isolated and partitioned into envelope/stroma and thylakoid fractions. Protein in the intact chloroplast, envelope/stroma, or thylakoid was extracted for immunoblotting as previously described.
Analysis of Chlorophyll/Phein and Chlide/Pheide
Chlorophyll and related pigments were extracted from seedlings and analyzed by HPLC as described (Lin et al., 2014). Highly purified chlorophyll a and b and Phein a were purchased from Sigma-Aldrich or Wako. Standard Chlide a, Chlide b, and Pheide a were generated from dephytylation of chlorophyll a and b and Phein a catalyzed by recombinant AtCLH1 as previously described (Lin et al., 2014). The identities of chlorophylls with unsaturated side chains was confirmed according to their retention time and absorption spectrum in comparison with the extra peaks present in lil3:2.
Singlet Oxygen Detection
Cotyledons of 5-d-old seedlings were cut and stained with 10 μM aqueous solution of SOSG (Invitrogen) at 22°C for 4 h in a transparent humid-box under light (100 μmole m−2 s−1). Fluorescence images were captured under a Z1 microscope (Zeiss) with a 488/525-nm excitation/emission filter set.
Production of Recombinant CLD1 and in Vitro Assay of Chlorophyll Dephytylase Activity
The cDNA of CLD1 or cld1-1 without the chloroplast transit peptide-encoding sequence (111 bp) was amplified by RT-PCR with the primers YP061 and YP062 (Supplemental Table 2). PCR products were cloned into the vector pGEM-T Easy (Promega) for sequencing confirmation and subcloning into pET32a to produce recombinant proteins in Escherichia coli (BL21DE3) cells (New England Biolabs). The construct for encoding the S165A mutant enzyme was generated by a PCR-based site-directed mutagenesis method (Hemsley et al., 1989) with the pET32a plasmid containing wild-type CLD1 cDNA as a template and primers YP071 and YP072 (Supplemental Table 2). The recombinant proteins were induced by adding 1 mM isopropyl β-d-1-thiogalactopyranoside to the cell culture, which was further incubated at 28°C for ∼3 h with shaking at 200 rpm until the OD600 value reached 1.8. The cells were pelleted by centrifugation and resuspended in 0.1 M MOPS buffer (pH 7.0). Cells were lysed by adding lysozyme (1 mg/mL) and DNase (0.1 mg/mL) to the suspension. After sonication, insolubles in the total lysate were spun down at 12,000 rpm for 30 min at 4°C, and the supernatant was used for enzyme activity assay. Dephytylase activity was measured by adding 5 μL soluble lysates to 45 μL reaction solution containing 0.1 M MOPS, 5% acetone, 0.5% Triton X-100, and the indicated amount of chlorophyll a, chlorophyll b, or Phein a. The reaction was incubated at indicated temperature in the dark for 30 min to 3 h. After adding 200 μL of 100% acetone to the reaction mixture and vortexing briefly to stop the reaction, the solution was centrifuged at 12,000 rpm for 5 min, and the supernatant analyzed by HPLC to measure Chlide a, Chlide b, and Pheide a.
For quantification of CLD1 protein level in crude extracts, (His)6-CLD1 fusion protein was produced and purified from E. coli cells harboring a pET28 construct containing CLD1 cDNA fragment. (His)6-CLD1 protein was purified through HisTrap HP column (GE 17-524-01) according to the manufacturer’s protocol. The purified (His)6-CLD1 was used as the standard for immunoblot analysis.
Measurement of Maximum Quantum Efficiency of PSII
Chlorophyll fluorescence emission of 5-d-old seedlings grown on the plate was monitored with a fluorescence imaging system (Imaging-PAM M-Series, MAXI version; Walz). The Fv/Fm values were calculated from the chlorophyll fluorescence emitted from seedlings with or without 20 min dark adaptation using ImageWin (Walz). The values obtained from the sample with or without dark adaptation did not differ substantially in this study. Fv/Fm was presented as the mean of ∼20 to 30 seedlings in each measurement.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: CLD1 and orthologs (Arabidopsis, AT5G38520.1; Oryza sativa, NP_001055569.1; Picea sitchensis, ABK26500.1; Physcomitrella patens, Pp1s90_2V6; Chlamydomonas reinhardtii, Cre16.g664350; Synechococcus sp PCC 7002, WP_041443291.1); AT4G36530 and orthologs (O. sativa, EAZ24328.1; P. sitchensis, ABK22395.1; P. patens, XP_001762523.1; Chlamydomonas, Cre10.g456150; Synechococcus sp PCC 7002, WP_012307877.1); AT5G19850 and orthologs (O. sativa, EAZ40401.1; P. patens, XP_001785779.1; C. reinhardtii, Cre12.g558550; Synechococcus sp PCC 7002, WP_041443820.1); PPH and orthologs (Arabidopsis, AT5G13800; O. sativa, NP_001057593.1; P. patens, XP_001761725.1; C. reinhardtii, XP_001702982.1).
Supplemental Data
Supplemental Figure 1. cld1-1 Is a Semidominant Mutation Revealed by Genetic Crossing and Complementation Test.
Supplemental Figure 2. Map-Based Cloning of cld1-1.
Supplemental Figure 3. Alignment of the Amino Acid Sequences of CLD1 Orthologs from Different Species.
Supplemental Figure 4. Alignment of the Amino Acid Sequences of Arabidopsis CLD1 Homologs.
Supplemental Figure 5. Abundance of CLD1 mRNA in Different Tissues and Developmental Stages in Arabidopsis.
Supplemental Figure 6. Immunoblot and Quantitative RT-PCR Analyses of CLD1 Expression in amiR-CLD1 Lines.
Supplemental Figure 7. Recombinant CLD1 Expressed in E. coli Cells and Its Quantification.
Supplemental Figure 8. Effect of Temperature and Acetone on CLD1 Activity.
Supplemental Figure 9. Phenotypes of Arabidopsis Seedlings with Varied CLD1/CHLG Ratios in Response to Heat Shock and Their Chlorophyll Content.
Supplemental Figure 10. ANOVA Tables for Figures 6 to 8.
Supplemental Figure 11. Accumulation of Chlorophylls with Unsaturated Side Chains and Effect on Cotyledon Bleaching Phenotype.
Supplemental Figure 12. Growth and PSII Maximum Efficiency of cld1-1 and Two amiCLD1 Lines under Osmotic, Salt, and High Light Stresses.
Supplemental Figure 13. Chlorophyll Breakdown in Dark-Induced Senescence Leaves.
Supplemental Table 1. Phenotypic Segregation of F2 Seedlings from the Cross between cld1-1 (Col) and the Wild Type (Ler) after Heat Shock Treatment.
Supplemental Table 2. List of Oligonucleotide Primers Used in This Study.
Supplemental File 1. Text File of the Alignment for Phylogenetic Analysis in Figure 2.
Supplementary Material
Acknowledgments
We thank Ayumi Tanaka and Ryouichi Tanaka for providing lil3:1 and lil3:2 seeds and the antibody against GGR and Stefan Hörtensteiner for providing pph-1 seeds. We thank the technical service of Lin-Yun Kuang, Shu-Chen Shen, and Fu-Hui Wu from the core facilities of Academia Sinica and Ching-Ju Tseng for assistance in experiments. We also thank The Small Molecule Metabolomics Core Facility, Institute of Plant and Microbial Biology, and the Scientific Instrument Center, Academia Sinica, for lending the UPLC system. This work was supported by the Foresight Project of Academia Sinica (Grant AS-97-FP-L19-1) and the Ministry of Science and Technology of the Republic of China (Grant 103-2311-B-001-011-MY3 and 104-2923-B-001-001-MY3).
AUTHOR CONTRIBUTIONS
Y.-P.L. and Y.-y.C. designed the experiments. Y.-P.L. and M.-C.W. performed the research. Y.-P.L and Y.-y.C. analyzed data and wrote the article.
Glossary
- Pheide
pheophorbide
- PPH
pheophytinase
- HS
heat shock
- SOSG
singlet oxygen sensor green
- GGR
geranylgeranyl reductase
- CLH
chlorophyllases
Footnotes
Articles can be viewed without a subscription.
References
- Alboresi A., Ballottari M., Hienerwadel R., Giacometti G.M., Morosinotto T. (2009). Antenna complexes protect photosystem I from photoinhibition. BMC Plant Biol. 9: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allakhverdiev S.I., Kreslavski V.D., Klimov V.V., Los D.A., Carpentier R., Mohanty P. (2008). Heat stress: an overview of molecular responses in photosynthesis. Photosynth. Res. 98: 541–550. [DOI] [PubMed] [Google Scholar]
- Aro E.-M., Virgin I., Andersson B. (1993). Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143: 113–134. [DOI] [PubMed] [Google Scholar]
- Azoulay Shemer T., Harpaz-Saad S., Belausov E., Lovat N., Krokhin O., Spicer V., Standing K.G., Goldschmidt E.E., Eyal Y. (2008). Citrus chlorophyllase dynamics at ethylene-induced fruit color-break: a study of chlorophyllase expression, posttranslational processing kinetics, and in situ intracellular localization. Plant Physiol. 148: 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon M.F., Holden M. (1970). Chlorophyllase of sugar-beet leaves. Phytochemistry 9: 115–125. [Google Scholar]
- Behnke K., Ehlting B., Teuber M., Bauerfeind M., Louis S., Hänsch R., Polle A., Bohlmann J., Schnitzler J.-P. (2007). Transgenic, non-isoprene emitting poplars don’t like it hot. Plant J. 51: 485–499. [DOI] [PubMed] [Google Scholar]
- Beisel K.G., Jahnke S., Hofmann D., Köppchen S., Schurr U., Matsubara S. (2010). Continuous turnover of carotenes and chlorophyll a in mature leaves of Arabidopsis revealed by 14CO2 pulse-chase labeling. Plant Physiol. 152: 2188–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng Y.Y., Liu H.C., Liu N.Y., Hsu F.C., Ko S.S. (2006). Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol. 140: 1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng Y.Y., Liu H.C., Liu N.Y., Chi W.T., Wang C.N., Chang S.H., Wang T.T. (2007). A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 143: 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-F.O., Lin C.-H., Kelkar S.M., Chang Y.-M., Shaw J.-F. (2008). Transgenic broccoli (Brassica oleracea var. italica) with antisense chlorophyllase (BoCLH1) delays postharvest yellowing. Plant Sci. 174: 25–31. [Google Scholar]
- Chu C.-C., Li H.-m. (2011). Determining the location of an Arabidopsis chloroplast protein using in vitro import followed by fractionation and alkaline extraction. Methods Mol. Biol. 774: 339–350. [DOI] [PubMed] [Google Scholar]
- Edelman M., Mattoo A.K. (2008). D1-protein dynamics in photosystem II: the lingering enigma. Photosynth. Res. 98: 609–620. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953–971. [DOI] [PubMed] [Google Scholar]
- Fang Z., Bouwkamp J.C., Solomos T. (1998). Chlorophyllase activities and chlorophyll degradation during leaf senescence in non-yellowing mutant and wild type of Phaseolus vulgaris L. J. Exp. Bot. 49: 503–510. [Google Scholar]
- Feierabend J., Dehne S. (1996). Fate of the porphyrin cofactors during the light-dependent turnover of catalase and of the photosystem II reaction-center protein D1 in mature rye leaves. Planta 198: 413–422. [Google Scholar]
- Guyer L., Hofstetter S.S., Christ B., Lira B.S., Rossi M., Hörtensteiner S. (2014). Different mechanisms are responsible for chlorophyll dephytylation during fruit ripening and leaf senescence in tomato. Plant Physiol. 166: 44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckathorn S.A., Downs C.A., Sharkey T.D., Coleman J.S. (1998). The small, methionine-rich chloroplast heat-shock protein protects photosystem II electron transport during heat stress. Plant Physiol. 116: 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C., Waterhouse P. (2003). Constructs and methods for high-throughput gene silencing in plants. Methods 30: 289–295. [DOI] [PubMed] [Google Scholar]
- Hemsley A., Arnheim N., Toney M.D., Cortopassi G., Galas D.J. (1989). A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 17: 6545–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S. (2013a). Update on the biochemistry of chlorophyll breakdown. Plant Mol. Biol. 82: 505–517. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. (2013b). The pathway of chlorophyll degradation: Catabolites, enzymes and pathway regulation. In Plastid Development in Leaves during Growth and Senescence, B. Biswal and K. Krupinska, eds (Dordrecht, The Netherlands: Springer; ), pp. 363–392. [Google Scholar]
- Hu C., Lin S.Y., Chi W.T., Charng Y.Y. (2012). Recent gene duplication and subfunctionalization produced a mitochondrial GrpE, the nucleotide exchange factor of the Hsp70 complex, specialized in thermotolerance to chronic heat stress in Arabidopsis. Plant Physiol. 158: 747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Makita S., Schelbert S., Sano S., Ochiai M., Tsuchiya T., Hasegawa S.F., Hörtensteiner S., Tanaka A., Tanaka R. (2015). Reexamination of chlorophyllase function implies its involvement in defense against chewing herbivores. Plant Physiol. 167: 660–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T., Zbierzak A.M., Kanwischer M., Dörmann P. (2006). A salvage pathway for phytol metabolism in Arabidopsis. J. Biol. Chem. 281: 2470–2477. [DOI] [PubMed] [Google Scholar]
- Jacob-Wilk D., Holland D., Goldschmidt E.E., Riov J., Eyal Y. (1999). Chlorophyll breakdown by chlorophyllase: isolation and functional expression of the Chlase1 gene from ethylene-treated citrus fruit and its regulation during development. Plant J. 20: 653–661. [DOI] [PubMed] [Google Scholar]
- Jander G. (2006). Gene identification and cloning by molecular marker mapping. In Arabidopsis Protocols, J. Salinas and J.J. Sanchez-Serrano, eds (Totowa, NJ: Humana), pp. 115–126. [DOI] [PubMed] [Google Scholar]
- Järvi S., Suorsa M., Aro E.-M. (2015). Photosystem II repair in plant chloroplasts--Regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim. Biophys. Acta 1847: 900–909. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kariola T., Brader G., Li J., Palva E.T. (2005). Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell 17: 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz S.J., Prochnik S.E., Grossman A.R., Merchant S.S. (2011). The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J. Biol. Chem. 286: 21427–21439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Schlicke H., Van Ree K., Karvonen K., Subramaniam A., Richter A., Grimm B., Braam J. (2013). Arabidopsis chlorophyll biosynthesis: an essential balance between the methylerythritol phosphate and tetrapyrrole pathways. Plant Cell 25: 4984–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J., Sobotka R., Nixon P.J. (2012). Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr. Opin. Plant Biol. 15: 245–251. [DOI] [PubMed] [Google Scholar]
- Kusaba M., Tanaka A., Tanaka R. (2013). Stay-green plants: what do they tell us about the molecular mechanism of leaf senescence. Photosynth. Res. 117: 221–234. [DOI] [PubMed] [Google Scholar]
- Li F., Vallabhaneni R., Yu J., Rocheford T., Wurtzel E.T. (2008). The maize phytoene synthase gene family: overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance. Plant Physiol. 147: 1334–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-P., Lee T.Y., Tanaka A., Charng Y.Y. (2014). Analysis of an Arabidopsis heat-sensitive mutant reveals that chlorophyll synthase is involved in reutilization of chlorophyllide during chlorophyll turnover. Plant J. 80: 14–26. [DOI] [PubMed] [Google Scholar]
- Lira B.S., Rosado D., Almeida J., de Souza A.P., Buckeridge M.S., Purgatto E., Guyer L., Hörtensteiner S., Freschi L., Rossi M. (2016). Pheophytinase knockdown impacts carbon metabolism and nutraceutical content under normal growth conditions in tomato. Plant Cell Physiol. 57: 642–653. [DOI] [PubMed] [Google Scholar]
- Liu H.C., Charng Y.Y. (2013). Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol. 163: 276–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marutani Y., Yamauchi Y., Kimura Y., Mizutani M., Sugimoto Y. (2012). Damage to photosystem II due to heat stress without light-driven electron flow: involvement of enhanced introduction of reducing power into thylakoid membranes. Planta 236: 753–761. [DOI] [PubMed] [Google Scholar]
- Matile P., Hörtensteiner S., Thomas H. (1999). Chlorophyll degradation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 67–95. [DOI] [PubMed] [Google Scholar]
- Mattoo A.K., Marder J.B., Edelman M. (1989). Dynamics of the photosystem II reaction center. Cell 56: 241–246. [DOI] [PubMed] [Google Scholar]
- Mazor Y., Borovikova A., Nelson N. (2015). The structure of plant photosystem I super-complex at 2.8 Å resolution. eLife 4: e07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFeeters R.F., Chichester C.O., Whitaker J.R. (1971). Purification and properties of chlorophyllase from Ailanthus altissima (Tree-of-Heaven). Plant Physiol. 47: 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merret R., Descombin J., Juan Y.T., Favory J.J., Carpentier M.C., Chaparro C., Charng Y.Y., Deragon J.M., Bousquet-Antonelli C. (2013). XRN4 and LARP1 are required for a heat-triggered mRNA decay pathway involved in plant acclimation and survival during thermal stress. Cell Reports 5: 1279–1293. [DOI] [PubMed] [Google Scholar]
- Miles E.W., Rhee S., Davies D.R. (1999). The molecular basis of substrate channeling. J. Biol. Chem. 274: 12193–12196. [DOI] [PubMed] [Google Scholar]
- Morita R., Sato Y., Masuda Y., Nishimura M., Kusaba M. (2009). Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 59: 940–952. [DOI] [PubMed] [Google Scholar]
- Murata N., Takahashi S., Nishiyama Y., Allakhverdiev S.I. (2007). Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta 1767: 414–421. [DOI] [PubMed] [Google Scholar]
- Murchie E.H., Lawson T. (2013). Chlorophyll fluorescence analysis: a guide to good practice and understanding some new applications. J. Exp. Bot. 64: 3983–3998. [DOI] [PubMed] [Google Scholar]
- Neta-Sharir I., Isaacson T., Lurie S., Weiss D. (2005). Dual role for tomato heat shock protein 21: protecting photosystem II from oxidative stress and promoting color changes during fruit maturation. Plant Cell 17: 1829–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A.H., Matsui A., Tanaka M., Mizunashi K., Nakaminami K., Hayashi M., Iida K., Toyoda T., Nguyen D.V., Seki M. (2015). Loss of Arabidopsis 5′-3′ exoribonuclease AtXRN4 function enhances heat stress tolerance of plants subjected to severe heat stress. Plant Cell Physiol. 56: 1762–1772. [DOI] [PubMed] [Google Scholar]
- Nixon P.J., Michoux F., Yu J., Boehm M., Komenda J. (2010). Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. (Lond.) 106: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazawa A., Tango L., Itoh Y., Fukusaki E., Kobayashi A. (2006). Characterization and subcellular localization of chlorophyllase from Ginkgo biloba. Z. Naturforsch., C, J. Biosci. 61: 111–117. [DOI] [PubMed] [Google Scholar]
- Peirson S.N., Butler J.N., Foster R.G. (2003). Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 31: e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A., Anders I., Aubry S., Schenk N., Tapernoux-Lüthi E., Müller T., Kräutler B., Hörtensteiner S. (2007). In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19: 369–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., Suga M., Kuang T., Shen J.-R. (2015). Photosynthesis. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 348: 989–995. [DOI] [PubMed] [Google Scholar]
- Raskin V.I., Fleminger D., Marder J.B. (1995). Integration and turnover of photosystem II pigment. In Photosynthesis: from light to biosphere, Mathis P., ed (Amsterdam, The Netherlands: Kluwer; ), pp. 945–948. [Google Scholar]
- Routaboul J.M., Skidmore C., Wallis J.G., Browse J. (2012). Arabidopsis mutants reveal that short- and long-term thermotolerance have different requirements for trienoic fatty acids. J. Exp. Bot. 63: 1435–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdiger W. (2009). Regulation of the late steps of chlorophyll biosynthesis. In Tetrapyrroles, M. Warren and A. Smith, eds (New York: Springer; ), pp. 263–273. [Google Scholar]
- Salvucci M.E. (2008). Association of Rubisco activase with chaperonin-60beta: a possible mechanism for protecting photosynthesis during heat stress. J. Exp. Bot. 59: 1923–1933. [DOI] [PubMed] [Google Scholar]
- Schelbert S., Aubry S., Burla B., Agne B., Kessler F., Krupinska K., Hörtensteiner S. (2009). Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21: 767–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk N., Schelbert S., Kanwischer M., Goldschmidt E.E., Dörmann P., Hörtensteiner S. (2007). The chlorophyllases AtCLH1 and AtCLH2 are not essential for senescence-related chlorophyll breakdown in Arabidopsis thaliana. FEBS Lett. 581: 5517–5525. [DOI] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey T.D., Wiberley A.E., Donohue A.R. (2008). Isoprene emission from plants: why and how. Ann. Bot. (Lond.) 101: 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda Y., Ito H., and Tanaka A. (2016). Arabidopsis STAY-GREEN, Mendel's green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 28: 2147–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoike K. (2011). Photoinhibition of photosystem I. Physiol. Plant. 142: 56–64. [DOI] [PubMed] [Google Scholar]
- Sun Q., Zybailov B., Majeran W., Friso G., Olinares P.D.B., van Wijk K.J. (2009). PPDB, the plant proteomics database at Cornell. Nucleic Acids Res. 37: D969–D974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R., Tanaka A. (2011). Chlorophyll cycle regulates the construction and destruction of the light-harvesting complexes. Biochim. Biophys. Acta 1807: 968–976. [DOI] [PubMed] [Google Scholar]
- Tanaka R., Oster U., Kruse E., Rüdiger W., Grimm B. (1999). Reduced activity of geranylgeranyl reductase leads to loss of chlorophyll and tocopherol and to partially geranylgeranylated chlorophyll in transgenic tobacco plants expressing antisense RNA for geranylgeranyl reductase. Plant Physiol. 120: 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R., Rothbart M., Oka S., Takabayashi A., Takahashi K., Shibata M., Myouga F., Motohashi R., Shinozaki K., Grimm B., Tanaka A. (2010). LIL3, a light-harvesting-like protein, plays an essential role in chlorophyll and tocopherol biosynthesis. Proc. Natl. Acad. Sci. USA 107: 16721–16725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh T., Goldschmidt E.E., Riov J. (1993). Ethylene induces de novo synthesis of chlorophyllase, a chlorophyll degrading enzyme, in Citrus fruit peel. Proc. Natl. Acad. Sci. USA 90: 9441–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T., Ohta H., Masuda T., Mikami B., Kita N., Shioi Y., Takamiya K.-i. (1997). Purification and characterization of two isozymes of chlorophyllase from mature leaves of Chenopodium album. Plant Cell Physiol. 38: 1026–1031. [Google Scholar]
- Tsuchiya T., Ohta H., Okawa K., Iwamatsu A., Shimada H., Masuda T., Takamiya K. (1999). Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: finding of a lipase motif and the induction by methyl jasmonate. Proc. Natl. Acad. Sci. USA 96: 15362–15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T., Suzuki T., Yamada T., Shimada H., Masuda T., Ohta H., Takamiya K. (2003). Chlorophyllase as a serine hydrolase: identification of a putative catalytic triad. Plant Cell Physiol. 44: 96–101. [DOI] [PubMed] [Google Scholar]
- Vavilin D., Vermaas W. (2007). Continuous chlorophyll degradation accompanied by chlorophyllide and phytol reutilization for chlorophyll synthesis in Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1767: 920–929. [DOI] [PubMed] [Google Scholar]
- Velikova V., Várkonyi Z., Szabó M., Maslenkova L., Nogues I., Kovács L., Peeva V., Busheva M., Garab G., Sharkey T.D., Loreto F. (2011). Increased thermostability of thylakoid membranes in isoprene-emitting leaves probed with three biophysical techniques. Plant Physiol. 157: 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.-H., Shen S.-C., Lee L.-Y., Lee S.-H., Chan M.-T., Lin C.-S. (2009). Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.Y., Juan Y.T., Hsu Y.H., Wu S.H., Liao H.T., Fung R.W., Charng Y.Y. (2013). Interplay between heat shock proteins HSP101 and HSA32 prolongs heat acclimation memory posttranscriptionally in Arabidopsis. Plant Physiol. 161: 2075–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh C.H., Kaplinsky N.J., Hu C., Charng Y.Y. (2012). Some like it hot, some like it warm: phenotyping to explore thermotolerance diversity. Plant Sci. 195: 10–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang W., Liu T., Ren G., Hörtensteiner S., Zhou Y., Cahoon E.B., Zhang C. (2014). Chlorophyll degradation: the tocopherol biosynthesis-related phytol hydrolase in Arabidopsis seeds is still missing. Plant Physiol. 166: 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.