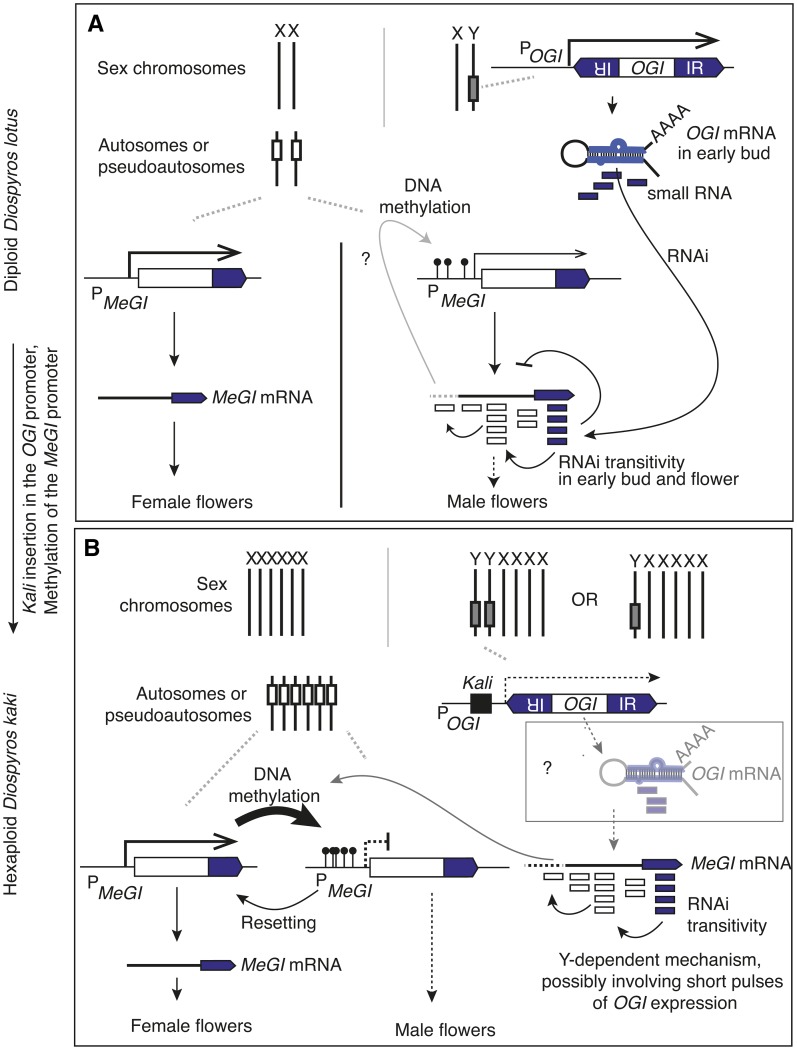
The transition from diploidy to polyploidy in cultivated persimmon is associated with an added layer of regulation in the form of gene methylation, allowing for a reversible sex-determination system.
Abstract
Epigenetic regulation can add a flexible layer to genetic variation, potentially enabling long-term but reversible cis-regulatory changes to an allele while maintaining its DNA sequence. Here, we present a case in which alternative epigenetic states lead to reversible sex determination in the hexaploid persimmon Diospyros kaki. Previously, we elucidated the molecular mechanism of sex determination in diploid persimmon and demonstrated the action of a Y-encoded sex determinant pseudogene called OGI, which produces small RNAs targeting the autosomal gene MeGI, resulting in separate male and female individuals (dioecy). We contrast these findings with the discovery, in hexaploid persimmon, of an additional layer of regulation in the form of DNA methylation of the MeGI promoter associated with the production of both male and female flowers in genetically male trees. Consistent with this model, developing male buds exhibited higher methylation levels across the MeGI promoter than developing female flowers from either monoecious or female trees. Additionally, a DNA methylation inhibitor induced developing male buds to form feminized flowers. Concurrently, in Y-chromosome-carrying trees, the expression of OGI is silenced by the presence of a SINE (short interspersed nuclear element)-like insertion in the OGI promoter. Our findings provide an example of an adaptive scenario involving epigenetic plasticity.
INTRODUCTION
Epigenetic variation potentially enables long-term but reversible cis-regulatory changes to take place in an allele while maintaining its DNA sequence. Like mutations based on DNA changes, epialleles occur in wild plant populations and may evolve neutrally or adaptively, carrying the added advantage of simple reversibility (Chen, 2007; Kaeppler et al., 2000; Manning et al., 2006). There are very few cases where metastable or programmed variable epigenetic states inferred to be adaptive have been documented. Yet, epialleles offer a natural switch system that can prove particularly well suited in specific situations. Epigenetic switching might also be particularly advantageous for trees, whose persistent meristems can traverse large temporal and spatial distances.
Diploid persimmon tree (Diospyros lotus) is dioecious, i.e., each tree bears exclusively male or female flowers. Dioecy is found in ∼6% of plant species and is thought to have evolved independently multiple times (Ming et al., 2007). Most dioecious species carry sex chromosomes, as is the case for persimmon, in which males are heterogametic (XY) and females carry two copies of the X chromosome (Akagi et al., 2014b). In diploid persimmon, we previously established that sex determination is governed by a pair of genes called OGI (Japanese for male tree) and MeGI (Japanese for female tree) (Akagi et al., 2014a). OGI is a Y-encoded pseudogene that produces a noncoding hairpin-forming RNA, ultimately producing small RNA (smRNA) molecules that target the homologous MeGI gene (Akagi et al., 2014a). MeGI is an autosomal gene that encodes a homeodomain transcription factor regulating flower development and anther fertility in a dosage-dependent fashion (Akagi et al., 2014a). MeGI is a homolog of the barley (Hordeum vulgare) Vrs1 gene, previously identified because it controls lateral flower development and inflorescence development in barley flowers (Komatsuda et al., 2007). In male persimmon trees, OGI smRNAs trigger transitive expression of MeGI smRNAs (smMeGI), which in turn act to repress MeGI expression in developing male flowers. While the diploid persimmon D. lotus is dioecious, the closely related species Diospyros kaki contains mostly autohexaploid trees, with varieties that are either fully female or monoecious, i.e., each tree bears both male and female flowers. With the exception of a few female cultivars that occasionally bear male flowers (Akagi et al., 2014b; Yonemori et al., 1993), all monoecious trees tested so far carry at least one copy of the Y chromosome (presence of the Y-specific OGI gene), while the female-only trees do not (Akagi et al., 2014b).
In this study, we investigated the mechanisms of flower sex determination in monoecious D. kaki and compared the results to our findings about sex determination in D. lotus. We found that the MeGI gene undergoes alternative epigenetic states in monoecious D. kaki, adding a layer of epigenetic regulation to the genetic system and leading to the formation of male and female flower branches on genetically male but developmentally monoecious individuals.
RESULTS
To begin evaluating the mechanism of flower sex regulation in monoeocious hexaploid D. kaki, we compared the developmental patterns of male and female flowers of dioecious D. lotus and monoecious D. kaki and found that, aside from differing in overall flower size, they were almost identical (Supplemental Figure 1). This provided a tractable system to investigate the molecular mechanisms that allow male and female flowers to be present on Y-carrying trees of hexaploid persimmon.
The Sex Determination Pathway Is Mostly Conserved in Hexaploid Persimmon
In diploid D. lotus, we previously determined that the OGI pseudogene produces smRNAs that target the MeGI autosome and that these smRNAs result in transitive expression of smRNAs from MeGI, repressing its own expression (Akagi et al., 2014a). This results in a reduction in MeGI expression levels in developing male flowers compared with developing female flowers. To investigate if similar events occur in hexaploid D. kaki, we compared three types of flowers: (1) female flowers from female trees (individuals lacking OGI and Y-specific sequence entirely and bearing exclusively female flowers), (2) female flowers from monoecious trees, and (3) male flowers from monoecious trees.
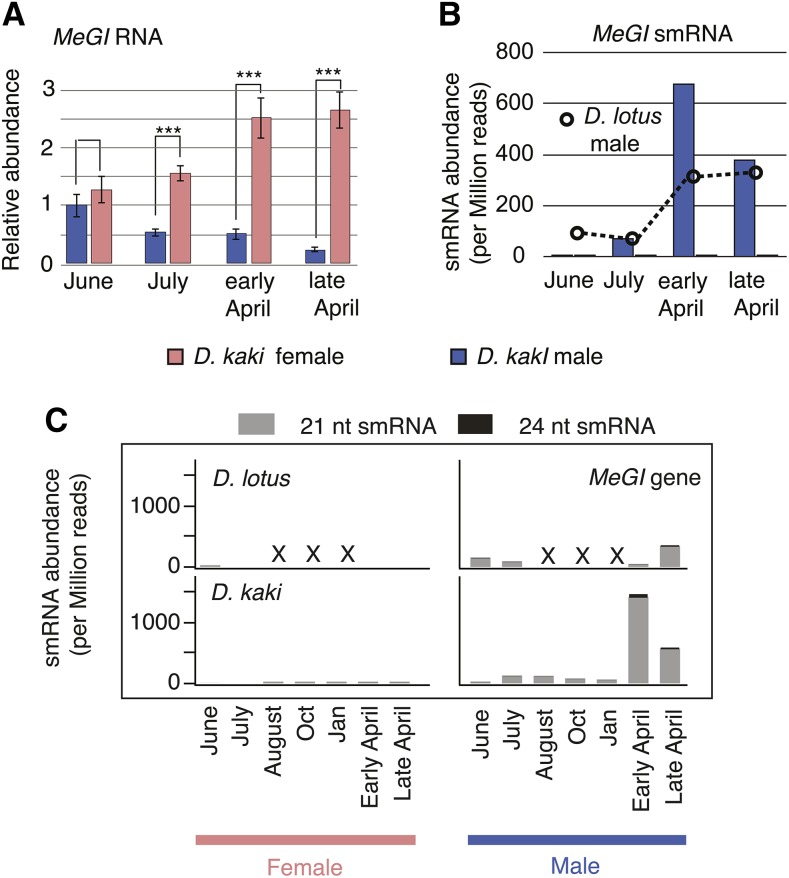
We observed that the production of smMeGI and associated repression of MeGI were conserved and specific to male flowers in monoecious D. kaki (Figures 1A and 1B). The developmental pattern observed was consistent with that observed in D. lotus male flowers, except that smMeGI expression began slightly earlier in D. lotus compared with D. kaki. The pattern of smRNA accumulation on the MeGI transcript was conserved as well (Supplemental Figure 2). As previously observed in diploid D. lotus, MeGI was targeted by 21- and 22-nucleotide smRNA but not by 24-nucleotide smRNA (Figure 1C; Supplemental Table 1). Finally, we investigated sequence variation within the MeGI alleles in Taishu, a hexaploid D. kaki cultivar used in this study. Between the six copies of this gene, so far, we only found three single nucleotide polypmorphic sites in the genic region, none of which were located on the regions of the gene targeted by the smRNA peaks (T. Akagi and T. Kawai, unpublished data). These results suggest that all six copies of MeGI are equally targeted by the smRNA population.
Figure 1.
Comparison of MeGI RNA and Small RNA Expression in Diploid and Hexaploid Persimmon.
(A) RT-PCR analysis of the expression of MeGI RNA during primordia formation and flower development in monoecious D. kaki. Male buds/flowers exhibited significantly lower MeGI expression than female buds/flowers (***P < 0.001, paired Student’s t test), which is consistent with previous observations in D. lotus (Akagi et al., 2014a). The average of two to four biological replicates is represented by the height of the bars; sd values are indicated as well.
(B) Expression of MeGI smRNA during primordia formation and flower development, in monoecious D. kaki, as well as in male flowers of dioecious D. lotus.
(C) The 21- and 24-nucleotide smMeGI abundance in male and female buds/flowers of dioecious D. lotus and monoecious D. kaki throughout the year. “X” indicates a missing data point.
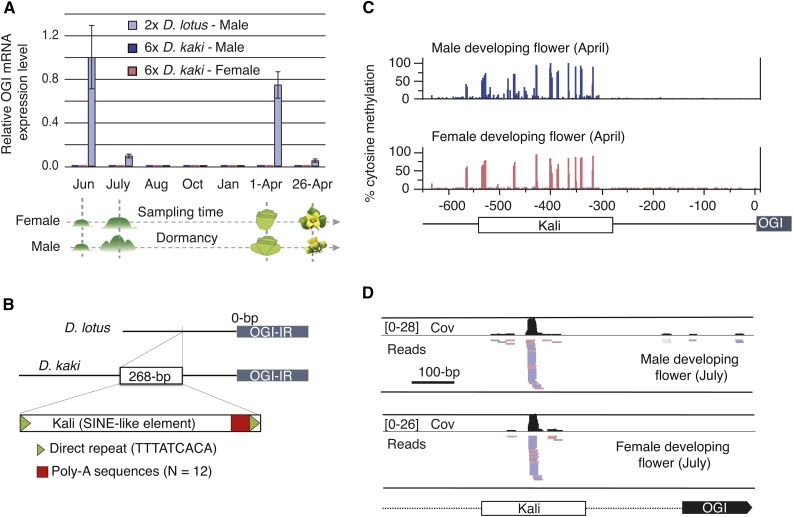
We next investigated the expression of the sex-linked pseudogene OGI. In male diploid D. lotus, two peaks of OGI expression were detected: one at the onset of primordia development (June) and the other at the early flower development stage (April 1st). As expected, OGI expression was completely undetectable in female flowers of hexaploid persimmon (female or monoecious D. kaki trees; Figure 2A). Surprisingly, OGI expression was undetectable in the male flowers as well, at least at the developmental stages and tissues that we sampled (Figure 2A). This is particularly puzzling because the OGI sequences from hexaploid D. kaki and diploid D. lotus are >96% similar, and a hairpin structure thought to be involved in smRNA production (Akagi et al., 2014a) is conserved in D. kaki as well, suggesting that the D. kaki OGI gene is functional.
Figure 2.
Characterization of OGI RNA and Small RNA Expression.
(A) RT-PCR analysis of the expression of OGI mRNA during primordia formation and flower development in monoecious D. kaki and dioecious D. lotus. The average of two to four biological replicates is represented by the height of the bars; sd values are indicated as well.
(B) Schematic structure of the OGI promoter region and the Kali insertion.
(C) Cytosine methylation levels across the OGI promoter in developing flowers from hexaploid D. kaki cv Taishu. Each bar represents the methylation level of one cytosine residue in either the sense or antisense strand. The position of the SINE element relative to the start codon of the OGI pseudogene is represented at the bottom.
(D) The 24-nucleotide small RNA accumulation on the Kali SINE-like insertion in the OGI promoter. For both samples, the coverage track is shown in black above the smRNA mapping tracks. Nearly all smRNAs that mapped to the Kali SINE-like region were 24 nucleotides long (Supplemental Figure 4). Mapped reads are shown in different colors depending on their mapping quality, with unambiguously mapped reads shown in pink (forward mapped reads) or blue (reversely mapped reads) and ambiguously mapped reads shown in gray.
OGI Expression Is Repressed by a SINE-Like Insertion
Investigating the sequence context around the OGI pseudogene provided a potential way to explain the lack of OGI expression in developing male flowers. Indeed, compared with its counterpart in diploid D. lotus, the OGI promoter region in hexaploid D. kaki contains a 268-bp insertion that exhibits high homology to a SINE (short interspersed nuclear element)-like retrotransposon element (Figure 2B), which we named Kali (after the Hindu goddess). PCR analysis of OGI promoter sequences from multiple D. kaki cultivars revealed that this insertion is present in all monoecious D. kaki cultivars tested, but not in other Diospyros species (Supplemental Figure 3). We identified at least six Kali-like SINEs in the hexaploid D. kaki genome, which could be derived from a D. kaki-specific burst of this SINE family (Supplemental Figure 4).
To investigate if this element might be involved in the regulation of the OGI transcript, we next documented smRNA accumulation on OGI and its promoter, including the SINE-like insertion. We observed a strong accumulation of 24-nucleotide smRNA on the Kali element and on a short fragment of the OGI promoter upstream of the Kali element (Figure 2D). We could not determine if this 24-nucleotide smRNA was produced from Kali itself or from one or several of the related sequences, due to high nucleotide similarity between Kali and the identified Kali-like SINEs (Supplemental Figure 4A). This smRNA accumulation was visible in both male and female flowers from monoecious D. kaki. Consistent with the absence of the Kali element in D. lotus, no 24-nucleotide smRNA accumulation was detected in diploid D. lotus at any stage or any location of the OGI promoter (Supplemental Figures 5 and 6). Conversely, the strong peak of OGI 21-nucleotide smRNA on the OGI gene body was not visible in either male or female developing flowers in D. kaki (Supplemental Figure 5). The Kali element also exhibited strong cytosine methylation along its entire length in both male and female developing buds and flowers, leaves, and stems (Figure 2C; Supplemental Figure 7). Taken together, these observations are consistent with a repressive role of the Kali element on the expression of OGI in hexaploid D. kaki.
Sex Determination Is Associated with MeGI Promoter Methylation
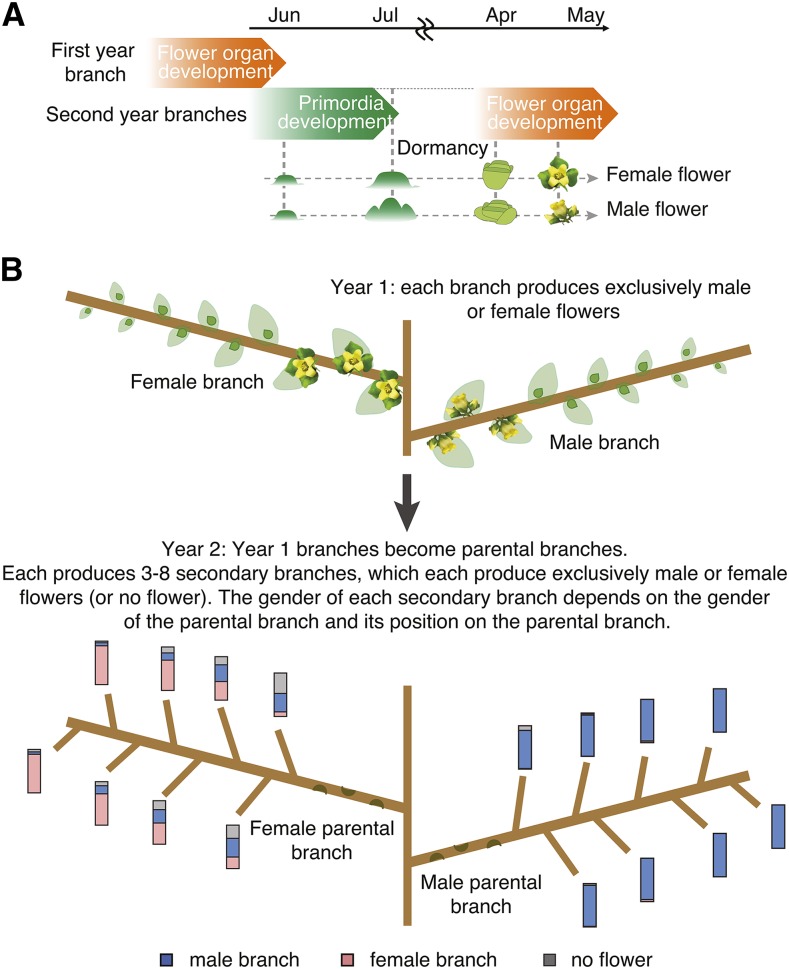
The apparent lack of expression of the OGI gene suggests the existence of an alternative mechanism leading to differential expression of MeGI in male versus female developing buds and flowers and in the presence of different flower sexes within a genetically male tree. To characterize this process, we recorded the pattern of male and female flower formation in multiple D. kaki individuals over multiple years. The results are summarized in Figure 3. In short, each year, each branch bears flowers of a single type (male or female), as well as developing buds of unknown sex. The subsequent year, these branches become parental branches and the buds develop into new branches, each bearing only male or female flowers. Consistent with previous observations (Yonemori et al., 1993), the future gender of each bud could be predicted, sometimes with high accuracy, based on the combination of the gender and length of its parental branch and its position on the parental branch (Figure 3). Specifically, buds located at the very ends of female branches and buds located on male branches had a posterior probability >92.5% (see Figure 3, Methods, and Supplemental Table 2) to develop into female and male branches, respectively. On the other hand, more proximal buds located on female branches have almost equal probability of developing into male or female branches. This knowledge was used for all analyses involving undifferentiated buds. More importantly, the observed association between the sexuality of parental and filial branches suggested a potential role for epigenetic regulation in sex determination.
Figure 3.
Male and Female Flower Development and Transitions in Monoecious D. kaki.
(A) Annual cycle of bud/flower development. The initial development of floral buds occurs in June, at which point primordial development starts and male and female primordia become distinguishable. Consistent with flower development in diploid D. lotus (Akagi et al., 2014a), female primordia exhibit a simple “solitary” structure, while male primordia exhibit trifurcated architecture (Supplemental Figure 1). Buds then enter dormancy until April of the next year, at which point flowers start to develop.
(B) Trends in male and female flower development in the next annual cycle (see Methods). Apical buds on branches carrying female flowers most often develop into female flowers (>80%), while medial buds on those same branches can be male or female. On the other hand, buds from male parent branches strongly tend to develop into male branches irrespective of their position on the parental branch (>95%). For each branch, the relative percentages of male (blue), female (pink), and branches with no flowers (gray) are represented as stacked bars.
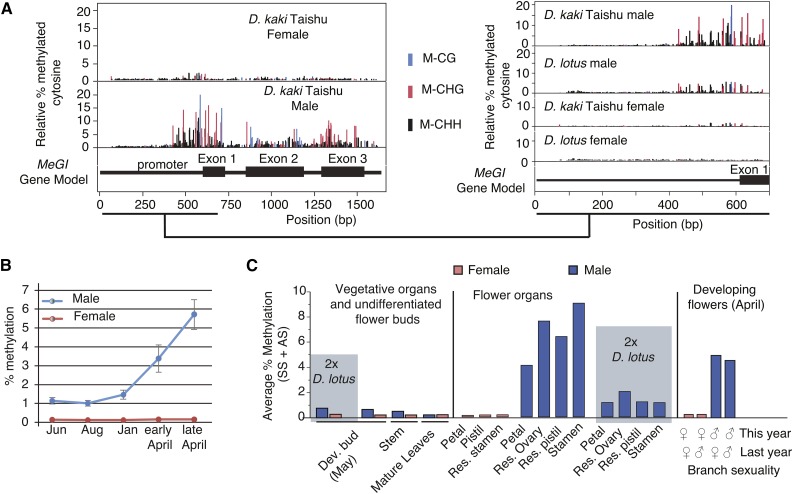
We thus investigated the role of cytosine methylation in the regulation of the MeGI gene. Sex-specific methylation was visible in all three methylation contexts (CG, CHG, and CHH; Figure 4, Supplemental Figure 8 and Supplemental Table 3) across the length of the MeGI promoter and genic regions (Figure 4; Supplemental Figure 7). Although we did not detect high methylation levels across these regions, methylation levels were clearly higher in male tissue compared with female tissue, especially in the 250-bp immediately upstream of the MeGI start codon (Figure 4A). It is possible that we observed an overall low level of methylation because MeGI expression is restricted to a subset of the cells within developing buds and the overall methylation level was diluted in our samples of entire developing buds. This is consistent with our preliminary observation that MeGI expression is limited to meristems in developing buds (T. Akagi and R. Tao, unpublished data). At all developmental stages, male developing buds or flowers exhibited stronger methylation than the corresponding female tissue (Figures 4A and 4B). This difference in methylation status increased with developmental stage and was most extreme in late developing flowers (late April; Figure 4B; Supplemental Figure 8), which is consistent with the observed differential expression of the MeGI RNA transcripts (Figure 1A). Cytosine methylation in the CHH sequence context is thought to be most relevant to gene silencing. Consistently, the stronger signal of CHH methylation in male flower buds corresponded with the reduction in MeGI expression. Cytosine methylation was specific to flower organs (Figure 4C) and was fully male-specific. This remained true in cases of gender reversal, i.e., when parental and filial branches carried flowers of the opposite sex, in which case the methylation status of the developing flower matched that of the current year (Figure 4C, right panel). Importantly, this observed male-specific methylation level in hexaploid D. kaki was higher than that observed in diploid D. lotus (Figure 4C). This is consistent with the hypothesis that a stronger methylation signal might be required to maintain the silenced state of MeGI in the absence of continual reinforcement by OGI in hexaploid D. kaki.
Figure 4.
DNA Methylation on the MeGI Promoter.
(A) Variation in DNA methylation levels across the MeGI gene and promoter region in developing male and female flowers of monoecious D. kaki (left panel) and across the MeGI promoter region in both diploid dioecious D. lotus and hexaploid monoecious D. kaki (right panel). Different colors represent the different sequence contexts (CH, CHG, and CHH). The gene model is shown at the bottom of each panel. Methylation data values at each position were normalized based on the control gene MatK (see Methods).
(B) Average DNA methylation values across the MeGI promoter, in male and female buds and flowers of monoecious D. kaki. The percentages of methylated cytosine at all cytosine positions were averaged over the 300 bp upstream of the start codon. Gray bars indicate sd values.
(C) Male organ predominant methylation patterns. The percentage of methylated cytosine at all cytosine positions was averaged over the 200 bp upstream of the start codon. Methylation is visible in male organs but not in female organs (middle panel). Methylation is also consistent with the gender of the current year’s flowers, even if the previous year’s flowers were of the opposite gender (four right-most bars). All samples are from monoecious hexaploid D. kaki, cv Taishu, with the exception of the bars in the two blue boxes, which represent diploid D. lotus samples, as indicated. Methylation data values at each position were normalized based on the control gene MatK (see Methods), with the exception of the last four bars (developing flower samples), for which samples were no longer available to measure methylation percentages on the MatK locus.
These results link methylation of the MeGI promoter with sex-specific repression of the MeGI gene, indicating that both de novo methylation and demethylation occur in the cases of sex reversal. To investigate how early these events occur, we sampled single developing buds (June). Specifically, we sampled buds located at different positions on parental female branches and investigated the methylation status of the MeGI promoter in these buds. We found that cytosine methylation on the MeGI promoter in these developing buds was tightly associated with the ratio of male-to-female flowers expected to develop from these buds. Specifically, buds located at the ends of female branches were not methylated, while approximately half of the proximal developing buds carried cytosine methylation and the other half did not (Supplemental Figure 9). Taken together with the lack of MeGI promoter methylation in female flower buds, a parsimonious explanation is that the shoot apical meristem of the primary female branch forms inflorescence meristems in which the MeGI promoter is not methylated. Some of the inflorescence buds reset, resulting in early deposition of methylation marks on MeGI promoters. Subsequently, these form male branches. The well-known persistence of epialleles might explain the low conversion rate of male into female branches.
Treatment with Methylation Inhibitors Can Result in Altered Flower Development
Given the potential role of DNA methylation in sex determination of developing branches, we investigated the effect of the nonspecific methylation inhibitor zebularine on sex determination. Flowers from female D. lotus (diploid) trees were unaffected by the treatment (Supplemental Table 3B). Treatment of developing flowers from male D. lotus trees produced no phenotypic change in 60% of the treated flowers, intermediate changes in 30% (Supplemental Figure 10H), and distinctly inhibited stamen and pollen development in 10% of flowers but did not promote pistil development (Supplemental Table 3B and Supplemental Figure 10). Zebularine application to developing monoecious D. kaki male buds and flowers had no effect in the case of D. kaki cultivar Fudegaki (Supplemental Figure 11A and Supplemental Table 4A). In Zenjimaru and Taishu (male-biased monoecious cultivars), zebularine treatment promoted pistil development, resulting in semihermaphroditic flowers in 59/71 and 20/32 treated flowers, respectively (Figure 5A; Supplemental Figure 11A and Supplemental Table 4). Zebularine also inhibited anther development, resulting in a smaller proportion of pollen grains exhibiting normal pollen tube growth (Figure 5B; Supplemental Figure 11B). Female flowers from the same monoecious trees treated with zebularine did not exhibit any distinct changes. Consistent with the role of zebularine in MeGI regulation, smMeGI expression levels were also reduced in zebularine-treated male flowers, from 133 to 35 reads per kilobase per million mapped reads in control and zebularine-treated flowers, respectively (Supplemental Figure 11D). Comparison of the level of cytosine methylation in the semifeminized and wild-type male flowers in these three cultivars showed that semifeminized flowers tended to exhibit reduced cytosine methylation levels, while unaffected flowers did not exhibit any significant decrease in methylation level (Supplemental Figure 10C). While D. kaki displayed a variable response to zebularine treatment, the phenotypic and molecular responses of Zenjimaru were thus associated with both the repression of male organs and the promotion of female organs, the two elements being necessary to switch gender. Mechanistically, these results are consistent with the hypothesis that, in male flowers, methylation of the MeGI promoter promotes smMeGI production to repress MeGI expression, which in turns results in male flower development. It is possible that at least some of the variation observed in the response to zebularine stems from variable penetration of zebularine depending on tissue type, developmental stage, and/or cultivar.
Figure 5.
Effect of the Methylation Inhibitor Zebularine on Flower Development.
Feminization of male flowers in monoecious D. kaki, cv Zenjimaru, after zebularine treatment compared with control male flowers.
(A) Male flowers exhibited elongated styles and formed normal ovules including immature embryos. At, anther; Em, embryo; Ov, ovule; Pn, pistinode; Sg, stigma; St, style. Bars = 2 mm. For both the zebularine-treated and control flowers, images on the right are close-up photographs of the samples on the left.
(B) Pollen germination was reduced in zebularine-treated male flowers, in comparison to the control male flowers (P < 0.00001, Student’s t test). Each mean value was calculated from five flowers, which each contained data from 200 to 300 pollen grains. Standard errors are indicated.
DISCUSSION
We set out to investigate the mechanisms underlying sex determination in persimmons; more specifically, we wanted to understand how a tree that is genetically male can produce both male and female flowers. While flower development and most of the molecular pathway leading to sex determination are conserved between the two species (Figure 1), we found two notable differences. First, OGI expression remained undetectable in developing male flowers of D. kaki, potentially due to the insertion of a SINE-like element in its promoter (Figure 2). Second, we found a tight association between sex determination and methylation of the MeGI promoter (Figure 4), which exhibits 5′ cytosine methylation and repression of MeGI expression in buds that form male flowers, but not in the rest of the plant body or in buds that form female flowers. Confirming this hypothesis, we observed that treatment of developing male flowers with a methylation inhibitor could result in feminized flowers (Figure 5).
A Model for Sex Determination in Monoecious Persimmon
This epigenetic sex regulation mechanism stands in contrast to genetic determination in the closely related diploid persimmon (Figure 6). Our results suggest that monoecy in hexaploid D. kaki and the observed strong association between the sexuality of a branch and that of its parental branch is based on the presence of transitive smMeGI and DNA methylation on the MeGI promoter. In this model, the methylation signal present on the MeGI promoter can activate smMeGI production (Figure 6). This is consistent with the observation that MeGI promoter methylation is observed very early in bud development (June), prior to any observation of smMeGI production. Based on the observation that the smRNAs targeting the MeGI promoter are 21 nucleotides long, this regulation may involve the noncanonical RNA-dependent DNA methylation pathway and the action of Pol II (RNA polymerase II) and RDR6 (RNA-Dependent RNA Polymerase 6) (Matzke and Mosher, 2014; Matzke et al., 2015).
Figure 6.
Summary and Evolutionary Model of Sex Determination in Diospyros.
(A) Genetic sex determination in diploid D. lotus. Heterogametic individuals are always male. The Y chromosome carries OGI, a male-specific gene whose inverted repeat transcript results in a hairpin RNA processed into small 21-nucleotide RNA. The OGI small RNA triggers RNAi and possibly also methylation on the homologous target MeGI, resulting in suppression of the feminizing factor.
(B) Epigenetic sex determination in genetic males of D. kaki. The presence, in the OGI promoter, of the SINE element Kali and the associated near inactivation of OGI expression allow for a more flexible reversible regulation of MeGI expression and the production of both male and female flowers in genetically male trees.
In most D. kaki cultivars/accessions, the first flowers produced by monoecious trees are female, suggesting that the default state is for the MeGI promoter to be active and unmethylated. This state changes in some developing buds, in which methylation marks are deposited on the MeGI promoter and which will eventually develop into branches bearing male flowers. This suggests that de novo methylation occurs in these specific buds, followed by maintenance of this methylation so that all flowers produced by these buds are male and most branches produced in subsequent years from these male branches are also male. Occasional sex reversal from male to female could originate from spontaneous demethylation of the MeGI promoter, as suggested by the results of our zebularine treatment.
Lack of OGI Expression in Monoecious D. kaki
High DNA methylation levels were observed across the Kali SINE insertion within the OGI 5′-promoter region. The accumulation of 24-nucleotide small RNA was observed as well, probably triggered by the application of these methylation marks. Both of these observations are consistent with the action of the canonical RNA-dependent DNA methylation pathway, involving the action of Pol IV (Matzke et al., 2015). Identification of the corresponding 26- to 45-nucleotide precursor Pol IV and RDR2-dependent RNAs (Blevins et al., 2015; Zhai et al., 2015) would confirm this hypothesis. The switch from expression of OGI as a Pol II transcript in D. lotus to repression via a Pol IV transcript in D. kaki could be due to the presence of the Kali SINE element in the promoter of OGI. This type of switch from posttranscriptional gene silencing to transcriptional gene silencing of transposable elements has been observed previously (Borges and Martienssen, 2015), although in this case, it is associated with the regulation of a major functional pathway and possibly critical to the switch from dioecy to monoecy (see below).
The identity of the signal that triggers smMeGI production and the switch from female to male branches remains unclear (Figure 4). Although we have been unable to detect any substantial OGI expression in D. kaki so far, OGI expression remains an obvious candidate, since spikes in OGI mRNA and smRNA expression are observed in diploid D. lotus during male floral bud development, and given the known repressive action of OGI on MeGI expression (Akagi et al., 2014a). Additionally, the presence of the Y chromosome (and the OGI gene) is associated with male flowers in a wide variety of hexaploid D. kaki cultivars. It is thus possible that rare and short developmental expression of OGI suffices to activate transitive RNAi on the MeGI promoter. Alternatively, another Y-encoded regulator may exist that remains to be uncovered.
What Is the Origin of This Additional Level of Regulation in D. kaki?
Epigenetic regulation adds a flexible layer to genetic variation, potentially enabling long-term but reversible cis-regulatory changes in an allele while maintaining its DNA sequence. While the exact nature of the triggers that result in sex reversal remains elusive, it is possible that this epigenetic plasticity is advantageous in the D. kaki hexaploid background. In the diploid D. lotus, sex reversal is virtually never observed and tree sex is completely genetically linked the presence of the OGI gene (Akagi et al., 2014a). Indeed, while lower but detectable levels of MeGI promoter methylation are present in diploid male persimmon, female flowers are virtually never produced from trees carrying OGI, consistent with full penetrance of the action of OGI RNA, either through constant reinforcement of MeGI promoter methylation or via a separate mechanism. It is possible that the potential involvement of an epigenetic control mechanism for flower sex determination in D. kaki is associated with autopolyploidization, the selection of specific plants for selfing, or domestication. Indeed, although we could not detect signs of ongoing selection across this region (Tajima’s D = −0.358, Fu and Li’s D = −0.162, Fu and Li’s F = −0.252), complete conservation of the Kali SINE insertion in the OGI promoter region of all D. kaki varieties tested suggests a historical bottleneck, possibly under natural selection or, alternatively, from domestication or historical breeding for optimal ratios of male to female flowers for fruit production. Investigation of the mechanism for sex determination in other polyploid Diospyros species will help distinguish between these possibilities. So far, our results provide evidence that sexuality can be epigenetically regulated in a tree species, enabling mechanistic exploration of epigenetic mechanisms affecting reproductive traits.
METHODS
Plant Materials
For observation of the sex determination patterns and discriminant analysis of sexuality in monoecious Diospyros kaki, eight monoecious cultivars (Supplemental Table 5), all planted in the Kyoto University orchard (Kyoto, Japan, 35°03’, 135°78’ L/L), were used. For expression analyses, mixed buds from seven cultivars (Supplemental Table 5) were harvested on June 9 to 13, July 9 to 13, August 9 to 13, and October 10, 2014 and on January 10, April 1 to 4, and April 23 to 26, 2015. These time periods correspond to the male/female primordia (June-July), dormancy (August-January), and male/female flower developmental stages (April). To compare expression patterns in dioecious and monoecious male/female buds/flowers, mix male and female buds of Diospyros lotus cv Kunsenshi were also sampled at the same developmental stages in 2012 to 2014. To detect DNA methylation levels on the MeGI promoter in monoecious D. kaki, male and female buds/flowers from D. kaki cv Taishu were harvested at similar stages. Developing flowers from two male (L1 and L8) and two female (L18 and L20) individuals of the D. lotus KK population (Akagi et al., 2014b) were also sampled on April 26 to compare the DNA methylation status with that of D. kaki. To characterize the Kali SINE-like sequence in the OGI promoter region, 79 D. kaki cultivars, 2 D. lotus, 1 D. glaucifolia, 1 D. virginiana, and 2 D. mespiliformis were tested by PCR analysis (Supplemental Table 5). For zebularine treatment, D. kaki monoecious cvs Zenjimaru, Taishu, and Fudegaki, and male and female D. lotus cv Kunsenshi were used. In the Kyoto University orchard, cv Zenjimaru was maintained in 40-liter planting pots. For the zebularine experiment, potted trees were utilized. All other cultivars were planted in the ground.
Flower Sex Determination Patterns in Monoecious D. kaki Cultivars
A total of 919 buds from eight monoecious D. kaki cultivars (Supplemental Table 5) were observed to assess the relationship between environmental variables, parental branch gender, and buds in 2014 to 2015. A previous report (Yonemori et al., 1993) suggested that flower gender in monoecious cultivars is affected by three main parameters: (1) the gender of the parental branch, (2) the length of the parental branch, and (3) bud positions. A multiple discriminant model was thus constructed using these three parameters, with data from the 919 buds, and using the lda function in the R MASS library. At least 20 buds were assessed for each parameter. Note that male parental branches normally only bear two to five buds. The numbers of buds in the 6th to 8th positions on male parental branches were thus smaller. Based on the discriminant model obtained (Supplemental Table 2), apical buds developing on long (>40 cm) female parental branches were used as developing female buds (posterior probability > 92.5% in MDA; Supplemental Table 2). Any buds developing on short (<12 cm) male parental branches were used as developing male buds (posterior probability >95% in MDA; Supplemental Table 2). The results obtained from the quantitative PCR and smRNA-seq analyses were consistent with these assessments.
smRNA-Seq Libraries
Total RNA was extracted from pools of seven to nine developing buds using the Plant RNA Purification Reagent (Invitrogen) and purified by phenol/chloroform extraction. The small RNA fraction was concentrated from total RNA using a mirVana miRNA Isolation kit (Life Technologies). Approximately 80 to 120 ng of small RNA was subjected to library construction using a NEBNext Small RNA Library Prep Set (NEB), according to the manufacturer’s instructions. PCR enrichment reactions were performed using 12 to 15 cycles of amplification, followed by filtration of approximately >200-bp fragments using AMPure (AMPure:reaction = 1.1:1 v/v) and a final cleanup step (AMPure:reaction = 3: 1 v/v) to remove self-ligated adapter dimers. Library quality and quantity were assessed using an Agilent Bioanalyzer (Agilent Technologies) and a Qubit fluorometer (Invitrogen). The constructed libraries were sequenced using Illumina’s HiSeq 2500/4000 sequencer (50-bp single-end reads).
Bisulfite PCR Sequencing Library
Genomic DNA was extracted using the CTAB method (Akagi et al., 2014b) and purified by phenol/chloroform extraction. Two micrograms of genomic DNA was subjected to bisulfite treatment using a MethylEasy Xceed Rapid DNA bisulfite modification kit (TaKaRa) to deaminate nonmethylated cytosine (C) residues into uracil (U) residues. The bisulfite-treated sequences of the MeGI 5′ promoter and genic regions were amplified by TaKaRa EpiTaq HS (TaKaRa) using sense- and antisense-specific primer sets (Supplemental Table 5). The resulting amplicons were subjected to end repair, A-base overhanging, and adaptor ligation, as described in the mRNA-seq library construction section in a previous report (Akagi et al., 2014a). The eluted DNA was enriched by PCR using Phusion 2X HF master mix (NEB) or PrimeSTAR Max DNA polymerase (TaKaRa), with the following PCR conditions: 30 s at 95°C; 8 to 12 cycles of 10 s at 95°C, 30 s at 65°C, and 30 to 60 s at 72°C, and a final extension step of 1 min at 72°C. Enriched libraries were purified with AMPure (1:1 v/v AMPure:reaction). Libraries were sequenced on the Illumina MiSeq 2500 sequencer (150-bp paired-end or 250-bp paired reads). For deeper sequencing (coverage >50,000) of the amplicons, a fragmentation step was added, according to the previous report (Akagi et al., 2014a), and the resulting libraries were sequenced on the Illumina HiSeq 2500/4000 sequencer (50-bp single or 150-bp paired reads).
Sequencing and Initial Read Processing
All Illumina sequencing was conducted at the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, and the raw sequencing reads were processed using custom Python scripts developed in the Comai laboratory and available online (http://comailab.genomecenter.ucdavis.edu/index.php/Barcoded_data_preparation_tools), as previously described (Akagi et al., 2014a). Briefly, reads were split based on index information and trimmed for quality (average Phred sequence quality >20 over a 5-bp sliding window) and adaptor sequence contamination. Next, a read length cutoff of 35 bp was applied to mRNA reads, while a cutoff of 19 bp was applied to smRNA reads. All samples used to generate Illumina sequences are listed in Supplemental Tables 6 and 7.
Expression Profiling
cDNA was synthesized from 500 ng of total RNA using ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO), and 4-fold diluted cDNA was used as a template for quantitative RT-PCR (qPCR) analysis. qPCR analysis of MeGI and OGI expression (primers in Supplemental Table 8) was performed using SYBR Green Master Mix and a LightCycler 480 (Roche). PCR was performed under the following conditions: 95°C for 3 min followed by 45 cycles of 95°C for 10 s, 57 to 59°C for 5 s, and 72°C for 15 s. Gene-specific amplification was confirmed by melting curve analysis. The average values of two independently synthesized cDNAs (technical replicates) were used for each cultivar (biological replicates). For the reference gene, primer pairs specific to Actin (AB473616) were used under the same PCR conditions as above. Statistic significance of differential MeGI expression between male and female buds/flowers from seven D. kaki cultivars (Supplemental Table 5) was assessed by paired Student’s t test (n = 7 × 2).
For smRNA analysis, smRNA-seq reads were mapped to the previously assembled D. lotus transcriptome (Akagi et al., 2014a), except that the OGI and MeGI transcripts were replaced with full-length MeGI and OGI genomic sequences including the surrounding regions, which were sequenced from cv Taishu (D. kaki) and cv Kunsenshi-male (D. lotus) for mapping of D. kaki and D. lotus reads, respectively. Mapping was performed using BWA (Li and Durbin, 2009) and allowing no nucleotide mismatches. Read counts per contig statistics were generated from the aligned .SAM files using a custom R script, which recorded the number of smRNAs of different sizes, and mapping to different parts of the OGI and MeGI genomic sequences. Expression levels were recorded as reads per million reads. The coverage and distribution of the smRNA reads were visualized as the sum of the sense and antisense strand reads using Integrative Genomics Viewer (IGV) version 2.3 (Robinson et al., 2011).
Detection of Methylated Cytosine Residue
The bisulfite PCR sequencing reads were mapped to the modified MeGI and OGI sequences, where all the cytosine residues were altered to thymine residues. The mapping was performed using the BWA aligner (Li and Durbin, 2009), allowing up to 10-, 25-, and 35-bp nucleotide mismatches for SE50, PE150, and PE250 reads, respectively. The informative SNPs in the read sequences were called using the mpileup tool in SAMtool (Li et al., 2009). The frequency of thymine-to-cytosine modifications was calculated at each of the cytosine positions present in the original sequence, with coverage values ranging from 50 to 100,000.
To normalize the conversion effect in bisulfite treatment, the matK region from chloroplast DNA of Diospyros (GU471729) was also amplified in each bisulfite treated-sample (see Supplemental Table 8 for primer information), followed by Illumina sequencing analysis, as described above. Generally, >99.5% of Cs were converted to Us in our analyses. The detected conversion ratios were used for normalization of methylation levels among the samples, according to Masser et al. (2015). When methylation data are shown as a mean per region, the normalized percentage of methylation at each cytosine was recorded and these values were averaged across the length of the assayed region.
One of the representative methylation residues in the MeGI promoter, located 26-bp upstream from the start codon, was converted to a cleaved amplified polymorphic sequence marker recognized by HpyCH4 IV and used as a proxy for methylation status in single developing buds (Supplemental Figure 9). The bisulfite-treated sequences of the MeGI 5′ promoter region were amplified by TaKaRa EpiTaq HS using primers MeGI-SenseProm-bis-F1 and MeGI-SenseProm-bis-F1 (Supplemental Table 8), followed by HpyCH4 IV digestion.
Analyses of the Kali SINE-Like Insertion in the OGI Promoter
The primer pairs for DNA sequence analysis of the 5′ promoter region of OGI were designed based on D. lotus BAC sequences (Akagi et al., 2014a). PCR was performed using PrimeSTAR GXL DNA polymerase (TaKaRa). Primer pairs OGI-prom2F and OGI-spR (Supplemental Table 8) consistently amplified the full genomic region including the 5′ promoter of OGI in a wide variety of Diospyros species. The amplicons from D. lotus, D. virginiana, D. mespiliformis, and D. glaucifolia male individuals, and from 12 OGI-carrying monoecious D. kaki cultivars (Supplemental Table 5), were purified and either directly sequenced or cloned into pGEM T Easy vector (Promega) followed by sequencing analysis. The nucleotide sequences were aligned using MAFFT version 7 (Katoh and Standley, 2013) with the L-INS-i model. The resulting alignment of the Kali SINE-like sequences from the D. kaki cultivars was subjected to DnaSP 5.1 (Librado and Rozas, 2009) to calculate the index of evolutionary selection (Tajima’s D, and Fu and Li’s neutrality tests). PCR analyses using primer pairs specific to Kali and surrounding sequences (see Supplemental Figure 3 for the exact locations of the primers and Supplemental Table 8 for primer sequences) were conducted to examine the conservation of this insertion and to detect sequences homologous to Kali in 79 D. kaki cultivars, D. lotus male and female individuals, D. virginiana male, D. mespiliformis male, and D. glaucifolia male individuals.
The Kali-like SINEs from the D. kaki genome were isolated from random genomic Illumina reads (PE150) from cultivar Taishu. The Illumina reads mapped to the Kali element, allowing up to 20-bp mismatches, were assembled using CLC (CLC bio). The nucleotide sequences of the Kali-like SINEs in D. kaki and the corresponding sequences from D. lotus and D. virginiana were aligned using MAFFT version 7 with the L-INS-i model and subjected to maximum likelihood phylogenetic analysis using MEGA v5.0, and a GTR model with 1000 replications for bootstrap. All sites, including missing and gap data, were used for the construction of the phylogenic trees. The alignment is shown in Supplemental File 1. Bootstrap values are shown as percentages on the branches of the phylogenetic tree (Supplemental Figure 4).
Treatment with the Demethylating Agent Zebularine
To test the effect of demethylation on flower sexuality in planta, zebularine (TGI) was applied to male and female branches of monoecious D. kaki cultivars, Zenjimaru and Fudegaki, and to male and female dioecious D. lotus cultivars Kunsenshi. Every 2 to 3 d, 100 mL of 1 mM zebularine solution or water (control) including 0.01% Tween 20 was sprayed onto 10 to 15 male and female branches carrying ∼20 to 120 flowers. In D. kaki, the treatments were applied from March 24, 2015 and March 23, 2016 (immediately after bud burst for Zenjimaru and before bud burst for Fudegaki), until May 5, 2015 and April 29, 2016, when androecia and gynoecia in both cultivars were almost mature. In D. lotus, treatments were applied from March 20, 2015 (immediately after bud burst for both male and female individuals) until May 5, 2015. Pollen tube germination was assessed six hours after placing the pollen grains on 15% sucrose/0.005% boric acid/1.0% agarose medium at 25°C. Pollen germination frequency was recorded as average percentages, using batches of 200 to 300 pollen grain from five flowers (biological replicates) from the same branch. Pollen grains exhibiting pollen tubes longer than 3 times the length of the grain were counted as “germinated.” Pistil length and pollen germination ratio were measured in fully mature flowers, i.e., 1 to 2 days after the petals had completely opened. Total RNA was extracted from pools of three developing flowers harvested on April 26, 2015, using the Plant RNA Purification Reagent (Invitrogen), and purified by phenol/chloroform extraction, as described above. From the total RNA extracted, the small RNA fraction was concentrated and subjected to smRNA-seq analysis as described above. To compare methylation levels between samples, genomic DNA was extracted from six feminized and nonfeminized flowers from the zebularine-treated sections, on May 3 to 4, 2016, using the CTAB method. Two micrograms of genomic DNA was subjected to bisulfite treatment, followed by PCR amplification of the MeGI promoter region. The amplicons were subjected to Illumina sequencing, which was used to calculate the cytosine methylation ratio, as described above.
Accession Numbers
All sequence data generated in the context of this manuscript have been deposited in the appropriate DDBJ database: Illumina reads for smRNA-seq and bisulfite amplicon sequencing were deposited in the Short Read Archive database (BioProject ID PRJDB4334), and the OGI promoter genomic sequences from D. kaki cultivar Taishu were submitted to GenBank (ID LC120366).
Supplemental Data
Supplemental Figure 1. Comparison of the developmental stages of D. lotus and D. kaki flowers.
Supplemental Figure 2. Pattern of smRNA accumulation on the MeGI transcript.
Supplemental Figure 3. Conservation of D. kaki-specific SINE insertions in the OGI 5′ region.
Supplemental Figure 4. Analysis of Kali-like sequences in the Diospyros genome.
Supplemental Figure 5. Small RNAs targeting the OGI gene and promoter elements.
Supplemental Figure 6. 24-Nucleotide small RNA accumulation on the Kali SINE-like insertion in the OGI promoter.
Supplemental Figure 7. DNA methylation across the OGI promoter.
Supplemental Figure 8. Seasonal DNA methylation on the MeGI promoter in male buds/flowers.
Supplemental Figure 9. DNA methylation of the MeGI promoter in single developing buds.
Supplemental Figure 10. Phenotypic effects of zebularine treatment on male D. lotus flowers.
Supplemental Figure 11. Effect of zebularine treatment on male D. kaki flowers.
Supplemental Table 1. Small-MeGI expression levels in male buds/flowers of cv Taishu throughout the year.
Supplemental Table 2. Multiple discriminant analysis of the pattern of bud/flower sexuality based on biological and environmental variables.
Supplemental Table 3. DNA methylation status of the MeGI promoter throughout the year.
Supplemental Table 4. Flower phenotypes after zebularine treatment in D. kaki and D. lotus.
Supplemental Table 5. List of plant materials.
Supplemental Table 6. mRNA-seq and smRNA-seq sample information.
Supplemental Table 7. Bisulfite amplicon sequencing sample information.
Supplemental Table 8. Primer sequences.
Supplemental File 1. Alignment used to produce the phylogenetic tree shown in Supplemental Figure 4.
Supplementary Material
Acknowledgments
We thank Meric Lieberman (UC Davis Genome Center) for bioinformatics support, Ayaka Sugimoto (Graduate School of Agriculture, Kyoto University) for experimental support, and Akihiko Sato, Atsushi Kono, and Noriyuki Onoue (Grape and Persimmon Research Station, NIFTS/NARO) for preliminary tests of gender determination patterns of D. kaki cultivars. Some of this work was performed at the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 OD018174 Instrumentation Grant. This work was supported by PRESTO, Japan Science and Technology Agency (to T.A.), by a Grant-in-Aid for Young Scientists (A) (no. 26712005 to T.A.), by Challenging Exploratory Research (no. 15K14654 to T.A. and no. 26660025 to R.T.) from the JSPS, by National Science Foundation IOS award under Grant 1457230 (to I.M.H. and L.C.), and by Grant-in-Aid for Scientific Research on Innovative Areas No. J16H06471 to T.A. (from JSPS).
AUTHOR CONTRIBUTIONS
T.A., I.M.H., L.C., and R.T. conceived the study. T.A. and I.M.H. designed most of the experiments. T.A. and T.K. performed the experiments. T.A. and I.M.H. analyzed the data. T.A., I.M.H., and L.C. drafted the manuscript. All authors interpreted data, edited the manuscript, and approved the final manuscript.
Glossary
- smRNA
small RNA
- SINE
short interspersed nuclear element
Footnotes
Articles can be viewed without a subscription.
References
- Akagi T., Henry I.M., Tao R., Comai L. (2014a). Plant genetics. A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science 346: 646–650. [DOI] [PubMed] [Google Scholar]
- Akagi T., Kibe T., Morimura H., Kajita K., Tsujimoto T., Nishiyama S., Kawai T., Yamane H., Tao R. (2014b). Development of molecular markers associated with sexuality in Diospyros lotus L. and their application in D. kaki Thunb. J. Jpn. Soc. Hortic. Sci. 76: 214–221. [Google Scholar]
- Blevins T., Podicheti R., Mishra V., Marasco M., Wang J., Rusch D., Tang H., Pikaard C.S. (2015). Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 4: e09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F., Martienssen R.A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16: 727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J. (2007). Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 58: 377–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeppler S.M., Kaeppler H.F., Rhee Y. (2000). Epigenetic aspects of somaclonal variation in plants. Plant Mol. Biol. 43: 179–188. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsuda T., et al. (2007). Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl. Acad. Sci. USA 104: 1424–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Manning K., Tör M., Poole M., Hong Y., Thompson A.J., King G.J., Giovannoni J.J., Seymour G.B. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38: 948–952. [DOI] [PubMed] [Google Scholar]
- Masser D.R., Stanford D.R., Freeman W.M. (2015). Targeted DNA methylation analysis by next-generation sequencing. J. Vis. Exp. 96: e52488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M.A., Kanno T., Matzke A.J. (2015). RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 66: 243–267. [DOI] [PubMed] [Google Scholar]
- Matzke M.A., Mosher R.A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15: 394–408. [DOI] [PubMed] [Google Scholar]
- Ming R., Wang J., Moore P.H., Paterson A.H. (2007). Sex chromosomes in flowering plants. Am. J. Bot. 94: 141–150. [DOI] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemori K., Sugiura A., Tanaka K., Kameda K. (1993). Floral ontogeny and sex determination in monoecious-type persimmons. J. Am. Soc. Hortic. Sci. 118: 293–297. [Google Scholar]
- Zhai J., et al. (2015). A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 163: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.