The transcription factor ARF2 represses HAK5 expression under K-sufficient conditions, and low-K-induced ARF2 phosphorylation relieves this repression in order to enhance HAK5-mediated high-affinity K+ uptake.
Abstract
Potassium (K+) plays crucial roles in plant growth and development. In natural environments, K+ availability in soils is relatively low and fluctuating. Transcriptional regulation of K+ transporter genes is one of the most important mechanisms in the plant’s response to K+ deficiency. In this study, we demonstrated that the transcription factor ARF2 (Auxin Response Factor 2) modulates the expression of the K+ transporter gene HAK5 (High Affinity K+ transporter 5) in Arabidopsis thaliana. The arf2 mutant plants showed a tolerant phenotype similar to the HAK5-overexpressing lines on low-K+ medium, whose primary root lengths were longer than those of wild-type plants. High-affinity K+ uptake was significantly increased in these plants. ARF2-overexpressing lines and the hak5 mutant were both sensitive to low-K+ stress. Disruption of HAK5 in the arf2 mutant abolished the low-K+-tolerant phenotype of arf2. As a transcriptional repressor, ARF2 directly bound to the HAK5 promoter and repressed HAK5 expression under K+ sufficient conditions. ARF2 can be phosphorylated after low-K+ treatment, which abolished its DNA binding activity to the HAK5 promoter and relieved the inhibition on HAK5 transcription. Therefore, HAK5 transcript could be induced, and HAK5-mediated high-affinity K+ uptake was enhanced under K+ deficient conditions. The presented results demonstrate that ARF2 plays important roles in the response to external K+ supply in Arabidopsis and regulates HAK5 transcription accordingly.
INTRODUCTION
Potassium (K+) is an essential macronutrient for plant growth and development. It participates in many physiological processes in living plant cells, e.g., electrical neutralization, enzyme activation, stomata movement, membrane potential maintenance, and osmotic regulation (Clarkson and Hanson, 1980). It also promotes photosynthesis, starch synthesis, and transport of assimilation products, which determines crop yield and quality in agricultural production (Pettigrew, 2008; Zörb et al., 2014). As a major nutrient ion, K+ constitutes 2 to 10% of the plant’s dry weight, whose concentration in the cytoplasm is relatively stable at ∼100 mM (Leigh and Wyn Jones, 1984; Walker et al., 1996). However, K+ concentration in soils is relatively low and fluctuating, varying from micro- to millimolar (Schroeder et al., 1994; Maathuis, 2009). Thus, plants must possess multiple K+ transport systems with different K+ affinities and transport activities to absorb adequate amounts of K+ from the soil. An early investigation revealed that dual mechanisms of K+ absorption exist in plants (Epstein et al., 1963). Mechanism 1 is responsible for high-affinity K+ uptake under low K+ concentrations (below 0.2 mM), while mechanism 2 contributes to low-affinity K+ uptake at higher K+ concentrations (over 0.5 mM). Plants can sense the fluctuation of external K+ concentrations and switch between these two mechanisms to absorb K+ more efficiently (Maathuis and Sanders, 1997; Ashley et al., 2006; Wang and Wu, 2013; Chérel et al., 2014).
Over the past decade, an increased number of plant K+ transporters and K+ channels have been identified, which have different K+ affinities and operate in different K+ uptake mechanisms (Véry and Sentenac, 2003; Véry et al., 2014). In general, K+ channels mediate low-affinity K+ uptake, and K+ transporters conduct high-affinity K+ uptake. Plants employ K+ channels or K+ transporters in response to external K+ concentrations and regulate K+ absorption in root cells (Maathuis and Sanders, 1997). In Arabidopsis, the K+ channel AKT1 from the Shaker family has been identified as one of the most important K+ uptake components in roots (Hirsch et al., 1998). AKT1 is involved in not only low-affinity ([K+]ext > 1 mM) but also high-affinity ([K+]ext < 100 μM) K+ uptake (Hirsch et al., 1998; Spalding et al., 1999; Xu et al., 2006). Another important component, High Affinity K+ transporter 5 (HAK5), belonging to KUP/HAK/KT family, has been characterized as a high-affinity K+ transporter (Ahn et al., 2004; Gierth et al., 2005). AKT1 and HAK5 constitute the main K+ absorption systems in Arabidopsis roots (Gierth et al., 2005; Pyo et al., 2010).
Transcriptional regulation is an important mechanism in the plant’s response to low-K+ stress (Ashley et al., 2006; Wang and Wu, 2013; Chérel et al., 2014). In Arabidopsis thaliana, the HAK5 transcript is remarkably induced by K+ deficiency in order to enhance high-affinity K+ uptake (Shin and Schachtman, 2004; Gierth et al., 2005). HAK5 is considered as a marker gene in the response to K+ deficiency in Arabidopsis. A previous report showed that a transcription factor, RAP2.11 (Related to AP2 11), positively regulates HAK5 transcription under low-K+ stress (Kim et al., 2012). It was also suggested that HAK5 may be modulated by multiple transcription factors (Hong et al., 2013).
Auxin response factors (ARFs) are important transcription factors that play crucial roles in regulating the expression of auxin response genes (Tiwari et al., 2003; Guilfoyle and Hagen, 2007). Twenty-three members constitute the ARF gene family in Arabidopsis. According to their functions, these ARFs are divided into two classes: transcriptional activators and transcriptional repressors (Guilfoyle and Hagen, 2007). Many investigations have revealed the essential roles of ARFs in plant growth and development as well as in stress adaptation (Hardtke and Berleth, 1998; Nemhauser et al., 2000; Ellis et al., 2005; Okushima et al., 2005a, 2005b; Wang et al., 2011; Yang et al., 2013). In this study, we demonstrate that ARF2 participates in the regulation of K+ uptake. It functions as a transcriptional repressor and modulates HAK5 transcription in the response to low-K+ stress in Arabidopsis.
RESULTS
The arf2 Mutant Is Tolerant to Low-K+ Stress
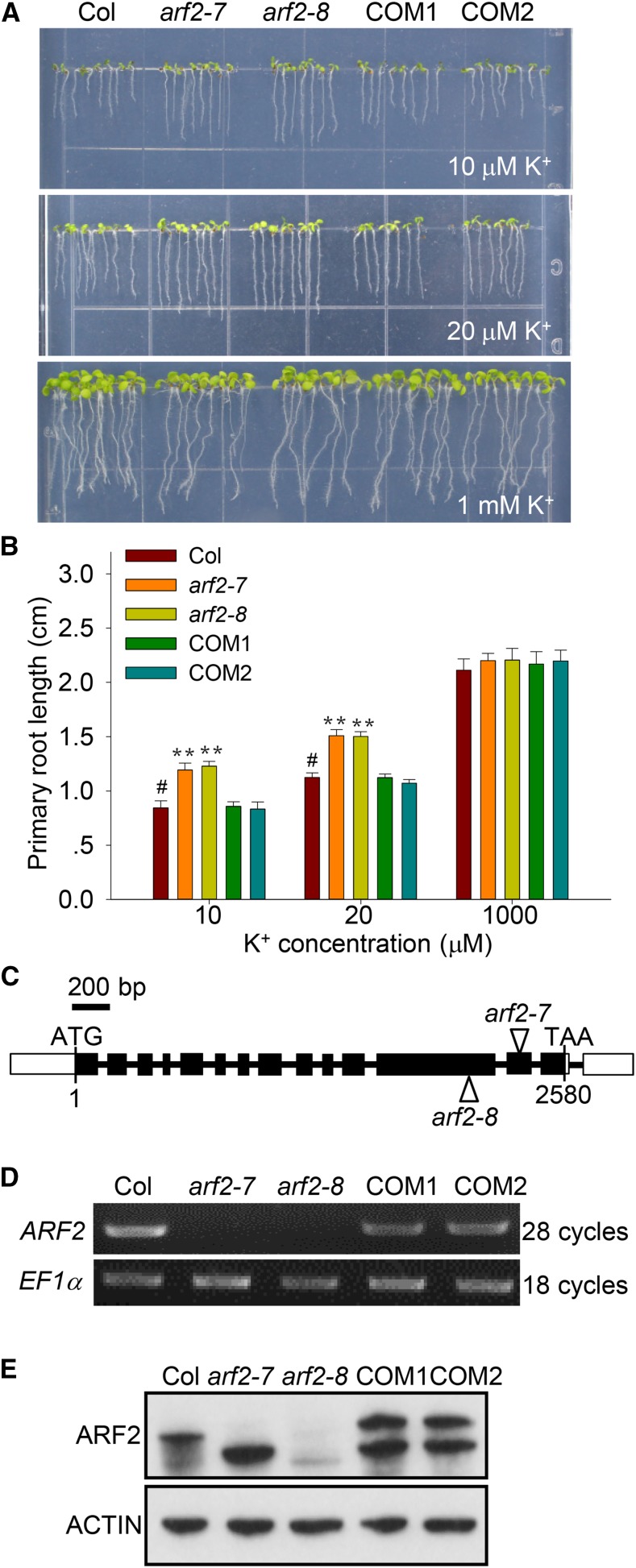
AKT1 and HAK5 are the two major components mediating high-affinity K+ uptake in Arabidopsis roots (Gierth et al., 2005; Pyo et al., 2010). Their single mutant and double mutant exhibit root growth defects at extremely low-K+ concentrations (10 μM K+). Previous reports showed that CIPK23 (CBL-interacting protein kinase 23) and KC1 (K+ rectifying channel 1) are two important regulators of AKT1; the transcription of both CIPK23 and KC1 are induced after low-K+ treatment (Shin and Schachtman, 2004; Gierth et al., 2005; Xu et al., 2006). In addition, HAK5 transcription is also elevated after K+ starvation (Shin and Schachtman, 2004; Gierth et al., 2005). To investigate the mechanism of transcriptional regulation in the response to low-K+ stress in Arabidopsis, we analyzed the promoter regions of these genes and identified some phytohormone-related cis-elements. Then, 15 mutants of phytohormone-related transcription factors, whose low-K+ phenotypes were determined, were collected from the ABRC, NASC (Nottingham Arabidopsis Stock Centre), and other research groups. We found that the mutant with a defect in transcription factor ARF2 showed a root growth advantage under low-K+ conditions (Figure 1A). The primary root lengths of two allelic mutants of ARF2 (arf2-7 and arf2-8) were longer than those of wild-type plants (Col), when they were directly germinated on low-K+ medium (10 and 20 μM K+) for 7 d (Figures 1A and 1B). However, the root growth advantage of arf2 did not appear under K+ sufficient conditions (1 mM K+) (Figures 1A and 1B). The ARF2 transcripts in these two T-DNA insertion mutants were both disrupted (Figures 1C and 1D), although a truncated ARF2 protein accumulated in arf2-7 (Figure 1E). ARF2 contains three domains, a DNA binding domain (160 to 293 amino acids), a repression domain (290 to 372 amino acids), and a C-terminal dimerization domain (CTD; 716 to 818 amino acids) (http://www.ebi.ac.uk/interpro/protein/Q94JM3). The truncated proteins in arf2-7 and arf2-8 may contain DNA binding domains, but lose CTD domains (Figures 1C). A previous study has revealed that the CTD domain stabilizes the dimerization of ARF DNA binding domains and facilitates ARF DNA binding activity in vivo (Boer et al., 2014). Therefore, both arf2-7 and arf2-8 used in this study should be loss-of-function mutants.
Figure 1.
The arf2 Mutant Is Tolerant to Low-K+ Stress.
(A) Phenotype comparison among the wild type (Col), arf2 mutants (arf2-7 and arf2-8), and complementation lines arf2-7/ProARF2:ARF2-Flag (COM1 and COM2). Seedlings were germinated and grown on low-K+ (10 and 20 μM) medium or K+-sufficient (1 mM) medium for 7 d.
(B) Primary root length of various plants tested in (A). Data are means ± se (n = 15). Student’s t test (**P < 0.01) was used to analyze statistical significance, and “#” represents control.
(C) Diagram of ARF2. Filled boxes and lines represent exons and introns, respectively. Triangles indicate the T-DNA insertion sites in arf2-7 and arf2-8.
(D) RT-PCR analyses of ARF2 expression in various plants. EF1α was used as a control.
(E) Immunoblot analyses of ARF2 protein levels in various plants. ACTIN was used as a control.
We constructed the complementation lines arf2-7/ProARF2:ARF2-Flag (COM1 and COM2), in which the transcription and protein levels of ARF2 were both restored (Figures 1D and 1E). The primary root lengths of these two independent transgenic lines (COM1 and COM2) were both reverted to wild-type lengths (Figures 1A and 1B).
The results of K+ content measurement indicated that the K+ contents in arf2 mutants was significantly increased compared with the wild-type and complementation lines, on low-K+ (10 and 20 μM) medium but not on K+ sufficient (1 mM) medium (Figure 2A). We then performed a 86Rb+ uptake assay to test the high-affinity K+ uptake capacity among various plants. Seven-day-old seedlings were starved of K+ for 3 d prior to the addition of 86Rb+, so that high-affinity K+ uptake was activated. Then, the 86Rb+ uptake rate was measured at 0.5 and 1 h. The results confirmed that the high-affinity Rb+ (K+) uptake capacity of arf2 mutants was markedly enhanced compared with wild-type plants (Figure 2B). These results suggest that ARF2 regulates primary root growth under low-K+ conditions by modulating high-affinity K+ uptake in Arabidopsis roots.
Figure 2.
High-Affinity K+ Uptake Is Enhanced in arf2 Mutants.
The K+ content (A) and 86Rb+ uptake rate (B) of the wild type, arf2 mutants, and complementation lines. Data are means ± se (n = 3). Student’s t test (**P < 0.01) was used to analyze statistical significance, and “#” represents control.
(A) Seedlings were germinated and grown on low-K+ medium or K+-sufficient medium for 7 d.
(B) Seven-day-old seedlings were starved of K+ for 3 d. Then, 86Rb+ uptake rates were measured at the indicated times (0.5 and 1 h).
ARF2-Overexpressing Lines Are Sensitive to Low-K+ Stress
ARF2-overexpressing lines were also constructed, in which the ARF2 coding sequence was driven by the 35S promoter. We obtained two independent transgenic lines (ARF2 OE1 and OE2). An RT-qPCR assay confirmed that the ARF2 transcripts were increased in both lines (Supplemental Figure 1A). Phenotype analysis showed that ARF2 OE1 and OE2 were sensitive to low-K+ stress, exhibiting primary roots that were shorter than those of wild-type plants (Supplemental Figures 1B and 1C). However, these two overexpressing lines had shorter primary roots and more lateral roots and root hairs compared with wild-type plants under K+ sufficient conditions (Supplemental Figure 1B). This abnormal root architecture might be due to the constitutive overexpression of ARF2.
We thus constructed inducible ARF2-overexpressing lines (ARF2 OE3 and OE4) driven by a β-estradiol (E2) inducible promoter (Schlücking et al., 2013). A GFP tag was also fused to the C terminus of ARF2 in ARF2 OE3 and OE4 to monitor the expression and localization of ARF2. As shown in Figure 3A, without E2 in the medium, ARF2 OE3 and OE4 did not show any phenotype difference compared with wild-type plants, both on low-K+ and K+-sufficient medium (Figures 3A and 3C). Upon application of E2 (10 μM), the ARF2 transcript was induced (Figure 3B), and ARF2-GFP proteins were expressed and targeted to the nucleus of the root epidermis and root hairs (Supplemental Figure 2). After application of E2 in the medium, ARF2 OE3 and OE4 exhibited shorter primary roots than did wild-type plants, only under low-K+ conditions (Figures 3A and 3C). Although the primary root lengths of all plants became shorter on K+-sufficient medium after application of E2, there was no phenotype difference between wild-type and ARF2-overexpressing plants (Figures 3A and 3C). Furthermore, the K+ contents in ARF2 OE3 and OE4 were significantly reduced on low-K+ medium, only when E2 was added in the medium (Figure 3D). However, ARF2 OE3 and OE4 did not show any difference in K+ content with wild-type plants under K+ sufficient conditions (Figure 3E). Together, these results demonstrated that ARF2 is a negative regulator involved in the response to low-K+ stress in Arabidopsis.
Figure 3.
ARF2-Overexpressing Lines Are Sensitive to Low-K+ Stress.
(A) Phenotype comparison between wild-type (Col) and inducible ARF2-overexpressing lines (OE3 and OE4). Seedlings were germinated and grown on low-K+ medium or K+-sufficient medium with or without 10 μM β-estradiol (E2) for 7 d.
(B) RT-qPCR analyses of ARF2 expression in various plants treated with or without β-estradiol (E2). Data are means ± se (n = 3). Student’s t test (**P < 0.01) was used to analyze statistical significance, and “#” represents control.
(C) to (E) Primary root length (C) and K+ content ([D] and [E]) of various plants tested in (A). Data are shown as means ± se. Student’s t test (*P < 0.05 and **P < 0.01) was used to analyze statistical significance, and “#” represents control. Sample number (n) was 15 in (C) and 3 in (D) and (E).
We noticed that the induced ARF2-GFP signal was only observed in the root epidermis and root hairs (Supplemental Figure 2). This is because E2 was added in the medium and can easily enter the outer layer of root tissues (epidermis and root hairs), but barely enters the inner layer (cortex and endodermis), where ARF2-GFP is thus not induced.
ARF2 Negatively Regulates HAK5 Expression
Phenotype analysis and K+ content measurement suggested that ARF2 negatively regulates high-affinity K+ uptake in Arabidopsis roots. Since ARF2 has been reported as a transcription factor that represses the transcription of target genes (Guilfoyle and Hagen, 2007; Wang et al., 2011), we hypothesized that ARF2 inhibits the expression of genes that control high-affinity K+ uptake in Arabidopsis roots, such as HAK5, AKT1, KC1, CIPK23, and CBL1/9/10. After detecting the expression of these relevant genes, we found that only HAK5 showed a difference in expression between the wild type and arf2 mutants (Figure 4A; Supplemental Figure 3). The transcript of HAK5 was markedly increased in arf2 mutants, especially under low-K+ conditions (Figure 4A), while HAK5 expression was significantly repressed in ARF2 OE3 and OE4 after application of E2 (Figure 4B). In addition, HAK5 expression was reduced in ARF2 OE1 and OE2 (Supplemental Figure 1D).
Figure 4.
ARF2 Negatively Regulates HAK5 Expression.
(A) and (B) RT-qPCR analyses of HAK5 expression in arf2 mutants (A) and inducible ARF2-overexpressing lines (B). Data are means ± se (n = 3). Student’s t test (**P < 0.01) was used to analyze statistical significance, and “#” represents control.
(A) Seedlings were germinated and grown on K+-sufficient (1 mM) medium for 5 d and then transferred to low-K+ (10 μM) medium or K+-sufficient (1 mM) medium for 12 h.
(B) Seedlings were germinated and grown on low-K+ medium or K+-sufficient medium with or without β-estradiol (E2) for 7 d.
(C) GUS activity measurement in tobacco leaves after transient expression of ProHAK5:GUS and Pro35S:ARF2. LUC was used as an internal control. Data are means ± se (n = 6). Student’s t test (**P < 0.01) was used to analyze statistical significance, and “#” represents control.
(D) Luminescence observation in ProHAK5:LUC transgenic plants. Seedlings were germinated and grown on low-K+ medium or K+-sufficient medium for 7 d. Luminescence intensity was indicated using pseudo color.
(E) Quantification of luminescence intensity shown in (D). Data are means ± se (n = 6 to 8). Student’s t test (**P < 0.01) was used to analyze statistical significance, and “#” represents control.
To further test whether ARF2 directly regulated HAK5 expression, we performed transient expression experiments in tobacco (Nicotiana benthamiana) leaves. When Pro35S:ARF2 and ProHAK5:GUS were coexpressed in tobacco leaves, the GUS activity was significantly reduced (Figure 4C). This suggests that ARF2 inhibits HAK5 promoter activity. Using the ProHAK5:LUC Arabidopsis transgenic line, we also tested the expression level of HAK5 in vivo. As shown in Figures 4D and 4E, the arf2 mutant emitted stronger luminescence than control plants, especially under low-K+ conditions. These results demonstrate that ARF2 negatively modulates HAK5 expression in response to low-K+ stress in Arabidopsis.
ARF2 Directly Binds to the HAK5 Promoter
As a transcription factor, ARF2 contains a conserved DNA binding domain at the N terminus that can bind to auxin-responsive elements (AuxREs) (Ulmasov et al., 1997; Guilfoyle and Hagen, 2007). Promoter sequence analysis showed that several AuxREs are located within the HAK5 promoter region. According to their positions, we designated three fragments (P1 to P3) within the HAK5 promoter, each of which contains at least one AuxRE (Figure 5A). Using an ARF2 antibody, we conducted a chromatin immunoprecipitation (ChIP) assay to detect the DNA binding activity of ARF2 to the HAK5 promoter in vivo. The arf2-8 mutant was used as a negative control. ChIP-qPCR results indicated that ARF2 binds to the P1 and P3 fragments but not to the P2 fragment (Figure 5B).
Figure 5.
ARF2 Directly Binds to the HAK5 Promoter.
(A) Diagram of the HAK5 promoter region. Gray lines represent the positions of AuxREs in the HAK5 promoter. Black lines indicate three fragments (P1 to P3) containing AuxREs in the HAK5 promoter that were used in the ChIP-qPCR assay.
(B) ChIP-qPCR analyses of ARF2 DNA binding activity to the HAK5 promoter. Chromatin was isolated from 7-d-old seedlings grown on K+-sufficient medium. Chromatin was immunoprecipitated with ARF2 antibody (anti-ARF2). Then, the three fragments (P1 to P3) were tested using qPCR. The ratios of immunoprecipitated DNA over the input DNA were calculated and presented as percentage of input (% IP). Chromatin without ARF2 antibody was used as a negative control. Data are means ± se (n = 3).
(C) EMSA to analyze the DNA binding activity of ARF2 N-terminal to P1 and P3 fragments in the HAK5 promoter. The purified CKS-ARF2-N-His proteins were incubated with P1 or P3 probes labeled with biotin. An excess of unlabeled probes was added to compete with biotin-labeled probes. ARF2-N can bind to P1 and P3 fragments, but not interact with the fragments with mutated AuxREs. The AuxREs were labeled in red. CKS-His protein was used as a negative control. The positions of hysteresis bands and free labeled probes are indicated with arrows.
Furthermore, ARF2 DNA binding activity was also validated in vitro using an EMSA (electrophoresis mobility shift assay). The N terminus of ARF2 (ARF2-N), which contains a DNA binding domain, was fused with CKS (CTP:CMP-3-deoxy-d-mannooctulosonate cytidylyltransferase synthetase) protein and His tag. The CKS-ARF2-N-His protein was expressed in Escherichia coli and purified from the soluble fraction using His antibody. EMSA results showed that ARF2-N can directly bind to P1 and P3 probes labeled with biotin (Figure 5C). This binding activity was gradually reduced by the addition of unlabeled competitive probes (Figure 5C). In addition, when the AuxREs within P1 and P3 were mutated, ARF2-N no longer interacted with these two fragments (Figure 5C). As a negative control, CKS-His protein alone did not bind to the HAK5 promoter (Figure 5C). Taking all these data together, we demonstrated that ARF2 directly binds to the AuxREs within the HAK5 promoter region, suggesting that ARF2 regulates HAK5 transcription.
ARF2 Participates in K+ Uptake by Regulating HAK5 Expression
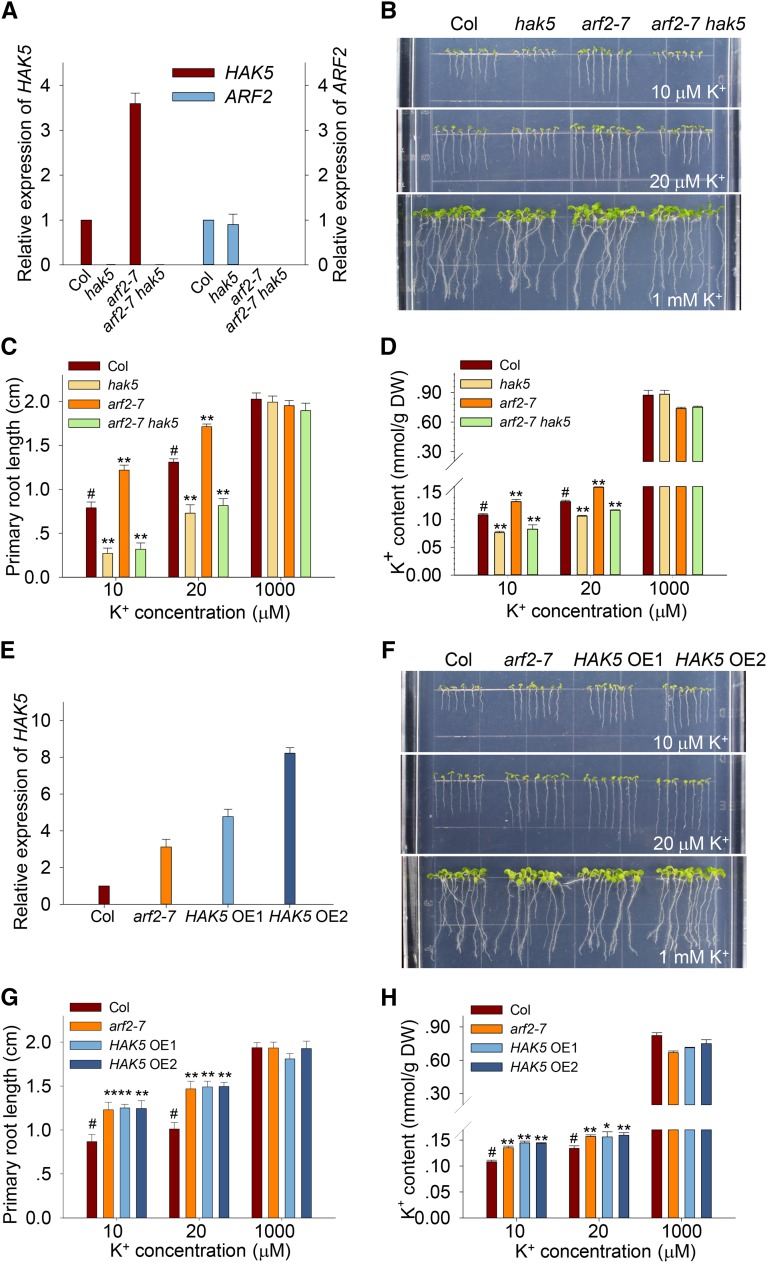
Since ARF2 can bind to the HAK5 promoter and repress HAK5 expression, we further investigated this regulation in planta by constructing arf2 hak5 double mutant and HAK5-overexpressing lines. The arf2-7 single mutant was crossed with the hak5 single mutant to generate the arf2-7 hak5 double mutant, in which the transcripts of both ARF2 and HAK5 are disrupted (Figure 6A). Phenotype analysis indicated that the low-K+-tolerant phenotype of arf2-7 was entirely abolished in the arf2-7 hak5 double mutant, whose primary root length was as short as that of the hak5 single mutant under low-K+ conditions (Figures 6B and 6C). These results suggest that HAK5 is epistatic to ARF2. The K+ content of the arf2-7 hak5 double mutant was also as low as that of the hak5 single mutant on low-K+ medium, even though the arf2-7 single mutant showed a high K+ content (Figure 6D). These results indicate that the low-K+-tolerant phenotype of arf2 relies on HAK5 function.
Figure 6.
ARF2 Participates in K+ Uptake by Regulating HAK5 Expression.
(A) RT-qPCR verification of ARF2 and HAK5 expression in various plants as indicated. Seedlings were germinated and grown on K+-sufficient (1 mM) medium for 7 d, and then total RNA of plants was extracted and used for RT-qPCR analysis.
(B) to (D) Root phenotype (B), primary root length (C), and K+ content (D) of various plants as indicated. Seedlings were germinated and grown on low-K+ medium or K+-sufficient medium for 7 d.
(E) RT-qPCR analysis of HAK5 expression in the arf2 mutant and HAK5-overexpressing lines.
(F) to (H) Root phenotype (F), primary root length (G), and K+ content (H) of arf2 mutant and HAK5-overexpressing lines. Data are means ± se. Sample numbers (n) were 15 in (C) and (G) and 3 in (D) and (H). Student’s t test (*P < 0.05 and **P < 0.01) was used to analyze statistical significance, and “#” represents control.
In addition, we determined the phenotype and K+ content of HAK5-overexpressing lines. These overexpressing lines were generated by transformation of the HAK5 coding sequence driven by the HAK5 native promoter into the hak5 mutant. Two independent transgenic lines (HAK5 OE1 and OE2) were selected by detecting the HAK5 expression level (Figure 6E). They exhibited a low-K+-tolerant phenotype similar to that of the arf2-7 single mutant, whose primary root lengths were longer than those of wild-type plants under low-K+ conditions (Figures 6F and 6G). Furthermore, the arf2-7 mutant and HAK5-overexpressing lines accumulated more K+ ions in plants compared with wild-type plants under low-K+ conditions (Figure 6H). These results demonstrate that the low-K+-tolerant phenotype of arf2 is due to the elevation of HAK5 expression.
The K+ content measurement was consistent with the results of the 86Rb+ uptake assay. As shown in Figure 7, both the arf2-7 mutant and the HAK5-overexpressing lines had higher Rb+ (K+) uptake rates than did wild-type plants within the high-affinity range. By contrast, the Rb+ (K+) uptake rates in the hak5 mutant and arf2-7 hak5 double mutant were significantly reduced (Figure 7). These data indicate that ARF2 modulates HAK5-mediated high-affinity K+ uptake.
Figure 7.
Rb+ Uptake Rates Are Increased in arf2 Mutant and HAK5-Overexpressing Lines.
Seven-day-old seedlings were starved of K+ for 3 d. Then 86Rb+ uptake rates were measured at the indicated times (0.5 and 1 h). Data are means ± se (n = 3). Student’s t test (*P < 0.05 and **P < 0.01) was used to analyze statistical significance, and “#” represents control.
Overexpression of HAK5 Stimulates Root Hair Elongation under Low-K+ Conditions
A previous report showed that low-K+ treatment induces root hair elongation (Jung et al., 2009). In this study, we observed that both root hair length and density were increased in wild-type plants, when plants were directly germinated on low-K+ medium (Figure 8). This increase in root hair length and density is likely an adaptive morphological alteration in response to low-K+ stress in Arabidopsis. Furthermore, we found that the root hair length of HAK5-overexpressing lines was longer than that of wild-type plants only under low-K+ conditions (Figure 8). According to the results presented above, HAK5 expression was elevated in arf2 mutants (Figure 4A). It is conceivable that arf2-7 showed longer root hairs, similar to HAK5-overexpressing lines (Figure 8). By contrast, the hak5 mutant exhibited extremely short root hairs on low-K+ medium (Figure 8). In addition, the long root hair phenotype of the arf2-7 mutant was entirely abolished in the arf2-7 hak5 double mutant (Figure 8). It should be noticed that the root hair differences only occurred on low-K+ medium, but not on K+ sufficient medium (Figure 8). The root hair phenotype analyses further confirmed that HAK5 is the downstream target of ARF2. The elevated expression of HAK5 could contribute to root hair elongation under low-K+ conditions.
Figure 8.
The arf2 Mutant and HAK5-Overexpressing Lines Show Longer Root Hairs under Low-K+ Conditions.
(A) Root hair phenotype of various plants as indicated. Seedlings were germinated and grown on low-K+ medium or K+-sufficient medium for 4 d. Bars = 1 mm.
(B) Diagram of root hairs in primary roots. Root hair length was measured in the upper region (2 mm) of the root hair differentiation zone (RHDZ).
(C) Root hair length of various plants tested in (A). Data are means ± se (n = 120 from eight individual plants for each bar). Student’s t test (**P < 0.01) was used to analyze statistical significance, and “#” represents control.
Phosphorylation of ARF2 Relieves the Repression of HAK5 Transcription under Low-K+ Conditions
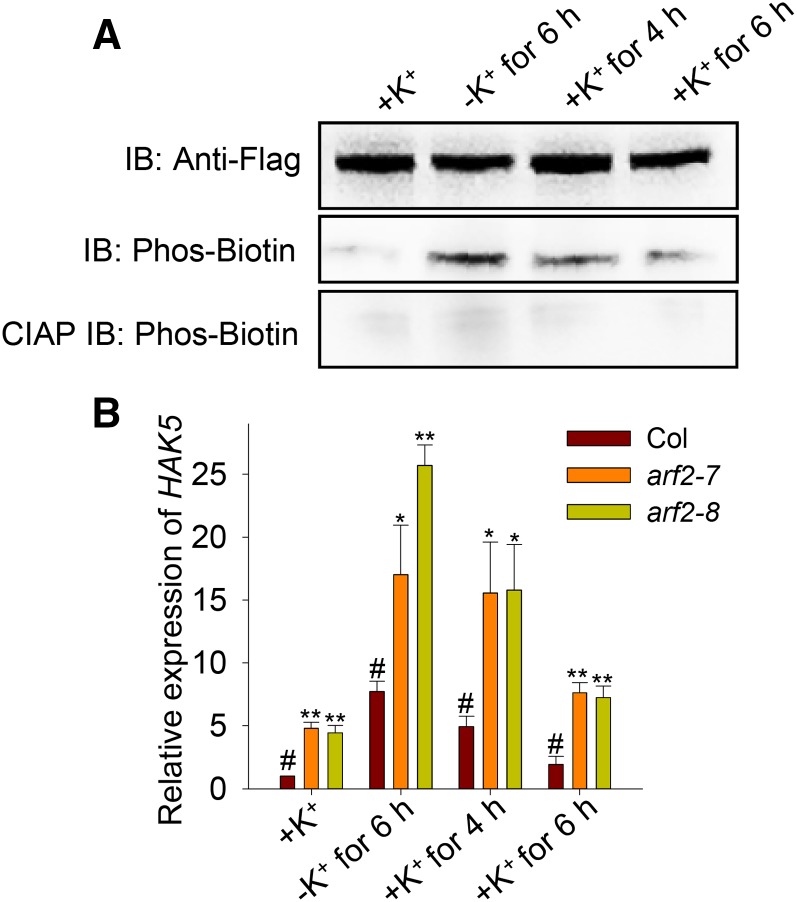
Consistent with previous reports (Shin and Schachtman, 2004; Gierth et al., 2005; Rubio et al., 2014), we also observed that HAK5 expression was induced by low-K+ treatment (Supplemental Figure 4A). Since ARF2 is a negative regulator of HAK5, we hypothesized that low-K+ treatment might relieve the repression of HAK5 transcription by reducing ARF2 transcription or ARF2 activity. RT-qPCR analysis revealed that ARF2 transcription was not affected after low-K+ treatment (Supplemental Figure 4B). Therefore, it is possible that the DNA binding activity of ARF2 declines under low-K+ conditions. A ChIP-qPCR assay confirmed that ARF2 no longer bound to the P1 and P3 fragments within the HAK5 promoter after low-K+ treatment (Figure 9A). How does this binding activity disappear? A previous report showed that phosphorylation can block ARF2 DNA binding activity (Vert et al., 2008). In this study, the arf2-7/ProARF2:ARF2-Flag transgenic line (COM1) was used to detect the ARF2 phosphorylation level in planta using the Phos-biotin method (Kinoshita-Kikuta et al., 2007). As shown in Figure 9B, low-K+ treatment triggers the rapid phosphorylation of ARF2 within several hours. After addition of CIAP (calf intestine alkaline phosphatase), this low-K+-induced ARF2 phosphorylation vanished (Figure 9B). These results indicate that low-K+ treatment leads to ARF2 phosphorylation and then may remove ARF2 repression from the HAK5 promoter.
Figure 9.
Phosphorylation of ARF2 Relieves the Repression of HAK5 Transcription under Low-K+ Conditions.
(A) ChIP-qPCR analyses of ARF2 DNA binding activity to P1 and P3 fragments in the HAK5 promoter with or without low-K+ treatment. Wild-type seedlings were germinated and grown on K+-sufficient medium for 5 d and then transferred to K+-sufficient medium or low-K+ medium for 6 h. Data are means ± se (n = 3).
(B) ARF2 can be phosphorylated after low-K+ treatment. arf2-7/ProARF2:ARF2-Flag (COM1) transgenic plants were germinated and grown on K+-sufficient medium for 5 d and then transferred to K+-sufficient medium or low-K+ medium for 2, 4, and 6 h. Total proteins of the plants at different time points were extracted. ARF2 proteins were immunoprecipitated using anti-Flag mAb agarose. Phosphorylated ARF2 proteins were detected by immunoblotting (IB) using biotinylated Phos-tag. CIAP can dephosphorylate ARF2, which was used as a control.
(C) GUS activity measurement in tobacco leaves after transient expression of ProHAK5:GUS and Pro35S:ARF2 with point mutations (S689A and S689D). ARF2S689D can relieve the repression of HAK5 transcription. Data are means ± se (n = 6). Student’s t test (**P < 0.01) was used to analyze statistical significance, and “#” represents control.
(D) Ser-689 on ARF2 is phosphorylated after low-K+ treatment. Wild-type ARF2 peptide (ARF2501-700) and mutated peptide (ARF2501-700 S689A) were expressed in E. coli and purified, respectively. Wild-type plants were germinated and grown on K+-sufficient medium for 5 d and then transferred to K+-sufficient medium or low-K+ medium for 2, 4, and 6 h. Total proteins were extracted from wild-type plants at the indicated time points. The purified ARF2 peptides were incubated with the total proteins for 40 min. Then the phosphorylation status of ARF2 peptides was tested using Phos-tag SDS-PAGE. ARF2 peptides were detected by immunoblot analysis using His antibody. CIAP can dephosphorylate ARF2 peptides, which were used as a control.
(E) EMSA to analyze the DNA binding activity of ARF2S689A (SA) and ARF2S689D (SD) to P1 and P3 fragments. The purified CKS-ARF2-His proteins were incubated with P1 or P3 probes labeled with biotin. An excess of unlabeled probes was added to compete with biotin-labeled probes. The CKS-His protein was used as a negative control. The positions of hysteresis bands and free labeled probes are indicated with arrows.
To further investigate the role of ARF2 phosphorylation in the regulation of HAK5 transcription, we tried to identify the phosphorylation sites on the ARF2 sequence. Phosphoproteomics analysis revealed three Ser residues (Ser-533, Ser-689, and Ser-840) on ARF2 that are phosphorylated in the response to abiotic stresses in Arabidopsis (http://phosphat.uni-hohenheim.de/). Then, we examined the roles of these Ser residues in HAK5 regulation. These three Ser residues were mutated to Ala or Asp in order to mimic the dephosphorylation or phosphorylation status, respectively, of ARF2. Then, the activities of mutated ARF2 were determined in tobacco leaves using a GUS/LUC assay. The results indicated that mutations on Ser-533 and Ser-840 did not affect ARF2 activity in repressing HAK5 transcription (Supplemental Figure 5). However, ARF2S689D significantly relieves the repression on HAK5 transcription (Figure 9C), suggesting that Ser-689 may be crucial for ARF2 DNA binding activity.
Using the Phos-tag method, we detected the phosphorylation status of Ser-689 in the response to low-K+ stress in Arabidopsis. As shown in Figure 9D, the ARF2 peptide (ARF2501-700) containing Ser-689 showed low phosphorylation levels under K+ sufficient conditions, while those phosphorylation levels were markedly enhanced after low-K+ treatment. However, the mutated ARF2501-700 S689A can hardly be phosphorylated under low-K+ conditions (Figure 9D). These results indicate that Ser-689 is an important phosphorylation site on ARF2 that responds to low-K+ stress.
Furthermore, we tested the DNA binding activity of ARF2S689A and ARF2S689D to the HAK5 promoter using the EMSA method. Here, the full-length ARF2 protein was expressed and purified. EMSA results indicated that full-length ARF2 can also bind to the P1 and P3 fragments within the HAK5 promoter (Figure 9E). This binding activity was reduced by the addition of unlabeled competitive probes (Figure 9E). The S689A mutation did not affect ARF2 binding activity to P1 and P3 (Figure 9E). However, ARF2S689D no longer interacted with P1, whose binding activity to P3 was also significantly impaired (Figure 9E). All these data demonstrated that low-K+-induced phosphorylation of ARF2 abolishes its DNA binding activity to the HAK5 promoter and removes the repression of HAK5 transcription. Therefore, HAK5 could be rapidly induced under low-K+ stress.
Conversely, the phosphorylated ARF2 induced by low-K+ stress can be rapidly dephosphorylated again when K+ was resupplied (Figure 10A). We also found that the phosphorylation status of ARF2 corresponded to HAK5 expression levels (Figure 10). The dephosphorylated ARF2 repressed HAK5 transcription again after K+ was resupplied (Figure 10B). Disruption of ARF2 relieved this repression of HAK5 transcription (Figure 10B). These results indicated that the regulation of phosphorylation status of ARF2 is dependent on the external K+ level, which directly determines the expression level of HAK5.
Figure 10.
Potassium Level Controls ARF2 Phosphorylation Status and HAK5 Expression.
(A) ARF2 phosphorylation status in wild-type plants under different K+ conditions. arf2-7/ProARF2:ARF2-Flag (COM1) transgenic plants were germinated and grown on K+-sufficient medium (1 mM K+) for 5 d and then transferred to low-K+ medium (10 μM K+) for 6 h. After that, the seedlings were transferred back to K+-sufficient medium (1 mM K+) for 4 and 6 h. Total proteins of the plants at different time points were extracted. ARF2 proteins were immunoprecipitated using anti-Flag mAb agarose. Phosphorylated ARF2 proteins were detected by immunoblotting (IB) using biotinylated Phos-tag. CIAP can dephosphorylate ARF2, which was used as a control.
(B) HAK5 expression in wild-type plants and arf2 mutants under different K+ conditions described in (A). Total RNA of the plants at different time points was extracted and used for RT-qPCR analyses. Transcript level of HAK5 was quantified.
DISCUSSION
Transcriptional Regulation of HAK5 by ARF2 May Not Rely on Auxin Signaling
ARFs are an important family of transcription factors that regulate the expression of auxin response genes (Tiwari et al., 2003; Guilfoyle and Hagen, 2007). Some arf mutants exhibit obvious auxin-related phenotypes, such as arf3, arf5, arf7, and arf7 arf19 (Hardtke and Berleth, 1998; Nemhauser et al., 2000; Okushima et al., 2005b; Schruff et al., 2006). In this study, we also wondered if the transcriptional regulation of HAK5 by ARF2 is dependent on auxin signaling. Phenotype analysis indicated that the arf2 mutant did not show any phenotypic difference in root growth compared with the wild type under K+-sufficient conditions (Supplemental Figure 6). Application of 1-naphthylacetic acid or 1-N-naphthylphthalamic acid can change the root architecture in both the wild type and arf2 mutant; however, there was still no phenotype difference between these lines (Supplemental Figure 6). This result indicates that the arf2 mutant exhibits a normal response to exogenous auxin, similar to the wild type, which was also observed previously (Okushima et al., 2005a). On low-K+ medium, the tolerant phenotype of the arf2 mutant was not affected after 1-naphthylacetic acid or 1-N-naphthylphthalamic acid was added to the medium (Supplemental Figure 6), suggesting that the low-K+-tolerant phenotype of arf2 is not dependent on auxin signaling. A previous report also revealed that ARF2 may not participate in auxin signaling at the young seedling stage (Okushima et al., 2005a). Taken together, we consider that ARF2-mediated HAK5 transcriptional regulation may not rely on auxin signaling, even though the arf2 mutant exhibits pleiotropic developmental phenotypes at the adult stage (Okushima et al., 2005a).
ARF2 has been found to be involved in many phytohormone signaling pathways. It can function as a negative regulator in ABA-mediated seed germination and primary root growth (Wang et al., 2011). ARF2 is also regulated by BIN2 (BRASSINOSTEROID-INSENSITIVE2) kinase and involved in brassinosteroid (BR) signaling (Vert et al., 2008). In addition, ARF2 proteins are degraded by ethylene signaling in a HLS1-dependent manner, which suggests that ARF2 participates in ethylene and light signaling pathways that control hypocotyl bending (Li et al., 2004). All these reports demonstrated that, as an auxin response factor, ARF2 has diverse roles in multiple phytohormone signaling pathways and may function as an important connecting node in the plant’s response to different stresses or stimuli. As a transcriptional repressor, ARF2 might function as a general “brake” that controls plant growth and development.
ARF2 Maintains the Low Expression Level of HAK5 under K+-Sufficient Conditions
HAK5 transcription is induced by low-K+ stress, which is one of the most important mechanisms in the adaptation to low-K+ stress in Arabidopsis (Ahn et al., 2004; Armengaud et al., 2004; Shin and Schachtman, 2004; Gierth et al., 2005). In this study, we demonstrated that ARF2 is a negative regulator of HAK5 transcription. Why do plants need negative regulation of HAK5? HAK5 is a high-affinity K+ transporter (Km ∼15 to 24 μM) and conducts K+ uptake only at the low K+ concentration range (Gierth et al., 2005; Pyo et al., 2010). HAK5-mediated K+ uptake constitutes a high energetic cost (Maathuis and Sanders, 1997). By contrast, AKT1 has a lower affinity than HAK5 for K+ uptake (Gierth et al., 2005; Pyo et al., 2010). AKT1-mediated K+ uptake is more energy efficient at a range of higher K+ concentrations (Maathuis and Sanders, 1997; Hirsch et al., 1998; Spalding et al., 1999). Under K+-sufficient conditions (millimolar level), HAK5-mediated K+ uptake would be energetically expensive and wasteful, which should be shut down and replaced by the low-affinity K+ uptake system mainly mediated by AKT1 (Maathuis and Sanders, 1997). Therefore, the HAK5 transcript should be maintained at extremely low levels under K+ sufficient conditions, and ARF2 plays a crucial role in the process.
In this study, we found that HAK5 repression was impaired in the arf2 mutant under K+ sufficient conditions (Figures 4A and 10B). Furthermore, when plants were transferred from low-K+ medium to K+-sufficient medium, HAK5 transcript was rapidly reduced in wild-type plants. However, HAK5 expression in the arf2 mutant cannot decline as rapidly as in the wild type (Figure 10B). It has been demonstrated that the physiological role of ARF2 is maintaining the low expression level of HAK5 under K+-sufficient conditions in order to save energy. Since HAK5 transcription is induced by low-K+ stress, there must exist other transcription factors that positively regulate HAK5 expression under low-K+ conditions (Kim et al., 2012; Hong et al., 2013). These transcription factors are also important regulators of HAK5 expression and thus merit further investigation.
After resupply of K+, HAK5 transcript levels steadily decreased in arf2 mutants, although the expression levels were still higher than in wild-type plants (Figure 10B). Some other negative regulators that repress HAK5 transcription might exist. Alternatively, the positive regulators may be removed from HAK5 promoter after K+ resupply, thereby reducing HAK5 expression.
Phosphorylation Is an Important Regulatory Mechanism for ARF2 Function
In plants, phosphorylation-mediated regulation of transcription factors is a common mechanism, which has been revealed in many signaling pathways. In the BR signaling pathway, the protein kinase BIN2 phosphorylates two transcription factors, BZR1 (BRASSINAZOLE-RESISTANT1) and BZR2/BRI1-EMS-SUPPRESSOR1, when BR levels are low. Phosphorylated BZR1/2 lose DNA binding activity and cannot regulate target genes. In the presence of BR, BZR1/2 are dephosphorylated and then bind to BR response elements in the promoters of BR target genes to regulate their expression (Wang et al., 2012). In the ABA signaling pathway, transcription factor RAV1 represses the expression of ABI3, ABI4, and ABI5. Protein kinases SnRK2s are activated by the ABA signal and then phosphorylate RAV1, which removes RAV1 repression on ABI5 transcription (Feng et al., 2014). Here, we demonstrated that low-K+ stress triggers a rapid phosphorylation of ARF2 (Figure 9B). Conversely, phosphorylated ARF2 can be dephosphorylated again when K+ is resupplied (Figure 10A). The phosphorylation status of ARF2 directly determines the expression level of HAK5 (Figure 10B). In addition, a previous report has shown that the protein kinase BIN2 from the BR signaling pathway can phosphorylate ARF2, which also results in loss of ARF2 DNA binding and repression activities (Vert et al., 2008). Phosphorylation might be a universal mechanism in ARF2-mediated stress responses. Future studies should aim to identify the protein kinases and protein phosphatases that modulate ARF2.
The results presented in this study reveal a regulatory mechanism of HAK5 transcription that is controlled by the transcription factor ARF2. As shown in Figure 11, we propose a working model to illustrate this mechanism. Under K+ sufficient conditions, ARF2 binds to the AuxREs within the HAK5 promoter and represses HAK5 transcription to maintain a low expression level of HAK5. When plants are subjected to low-K+ stress, ARF2 is rapidly phosphorylated and loses DNA binding activity. ARF2 is removed from the HAK5 promoter, which relieves the repression on HAK5 transcription. Then some other transcription factors (such as RAP2.11) bind to the HAK5 promoter and activate HAK5 transcription. Finally, the elevated HAK5 expression enhances HAK5-mediated high-affinity K+ uptake, which helps plants survive under K+-deficient conditions.
Figure 11.
Working Model of HAK5 Transcriptional Regulation by ARF2.
Under K+-sufficient conditions, ARF2 binds to AuxREs within the HAK5 promoter and represses HAK5 transcription. When plants are subjected to low-K+ stress, ARF2 is rapidly phosphorylated and removed from the HAK5 promoter, which relieves the repression on HAK5 transcription. Then some other transcription factors (such as RAP2.11) bind to the HAK5 promoter and activate HAK5 transcription. The T-bar represents transcriptional repression, and the arrow represents transcriptional activation.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana mutants (Columbia ecotype) used in this study, including arf2-7 (CS24601), arf2-8 (SALK_108995), and hak5 (SALK_005604), were obtained from the ABRC or the NASC.
For plant hybridization and seed harvesting, Arabidopsis plants were grown in a potting soil mixture (rich soil:vermiculite = 2:1, v/v) and kept in growth chambers at 22°C with illumination (fluorescent lamps) at 120 μmol m−2 s−1 under long-day (16 h light/8 h darkness) conditions. The relative humidity was ∼70% (±5%). For the low-K+ phenotype assay, seeds were surface sterilized and treated at 4°C in darkness for 4 d. Then, the seeds were placed on K+-sufficient (1 mM) or low-K+ (10 and 20 μM) medium containing 0.5% (w/v) sucrose and 0.7% (w/v) agarose (USB) and grown at 22°C with illumination at 100 μmol m–2 s–1 under long-day conditions, unless otherwise indicated.
Generation of Arabidopsis Transgenic Plants
The ARF2 coding sequence fused with the Flag tag was cloned into the pCAMBIA1305 vector driven by an ARF2 native promoter and was transformed into the arf2-7 mutant to obtain complementation lines (COM1 and COM2). The ARF2 coding sequence was cloned into the pCAMBIA1300 vector driven by the 35S promoter and transformed into wild-type (Columbia ecotype) plants to obtain constitutive ARF2-overexpressing lines (ARF2 OE1 and OE2). The ARF2 coding sequence was cloned into the SUPERR:sXVE:GFPC:Bar vector driven by an inducible promoter. This construct was transformed into wild-type plants to obtain inducible ARF2-overexpressing lines (ARF2 OE3 and OE4). The HAK5 coding sequence was cloned into the pCAMBIA1391 vector driven by the HAK5 native promoter. This construct was transformed into the hak5 mutant to obtain the HAK5-overexpressing lines (HAK5 OE1 and OE2). Arabidopsis plants were transformed with Agrobacterium tumefaciens (strain GV3101) using the floral dip method (Clough and Bent, 1998). The T4 homozygous transgenic plants were used for the phenotype test.
Root Phenotype Observation and K+ Content Measurement
The K+-sufficient medium used for phenotype analysis was prepared by modifying the Murashige and Skoog medium. NH4NO3 and KNO3 were removed, and KH2PO4 was replaced by NH4H2PO4. The medium contained 1.5 mM MgSO4·7H2O, 1.25 mM NH4H2PO4, 2.99 mM Ca(NO3)2·4H2O, 0.1 mM MnSO4·4H2O, 5 μM KI, 0.1 μM CuSO4·5H2O, 0.1 mM H3BO5, 0.1 μM CoCl·6H2O, 0.03 mM ZnSO4·7H2O, 10 μM Na2MO4·2H2O, 0.1 mM FeSO4·7H2O, and 0.1 mM Na2EDTA. For low-K+ medium, the NH4H2PO4 was replaced by H3PO4. Different K+ concentrations were supplemented with KCI. Seeds were germinated and grown on K+-sufficient (1 mM) or low-K+ (10 and 20 μM) medium for 4 or 7 d and then photographs were taken. ImageJ software was used to measure root hair length and primary root length. The 7-d-old seedlings grown on K+-sufficient or low-K+ medium were harvested and used for K+ content measurement. The dry weight was measured as biomass. Samples were treated in a muffle furnace at 300°C for 1 h and 575°C for 5 h. The ashes were dissolved and diluted in 0.1 M HCl. The K+ concentrations were measured using 4100-MP AES (Agilent).
GUS/LUC Assay
The transient GUS/LUC assay was performed as described previously (Feng et al., 2014). The constructs Pro35S:ARF2 and ProHAK5:GUS in the pCAMBIA1381 vector were transfected into Agrobacterium (strain GV3101) separately. Then, the bacterial fluid was injected into tobacco (Nicotiana benthamiana) leaves using syringes. Three days later, the GUS activity was measured using methyl umbelliferyl glucuronide (Sigma-Aldrich), and luciferase (LUC) activity was used as an internal control. The GUS/LUC ratio was used to determine the ARF2 binding activity to the HAK5 promoter.
EMSA
EMSA was conducted using a LightShift Chemiluminescent EMSA Kit (Pierce) following the manufacturer’s protocol. The recombinant proteins CKS-ARF2-N-His, CKS-ARF2-His, and CKS-His were expressed and purified from Escherichia coli. The QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies) was used to generate CKS-ARF2S689A-His and CKS-ARF2S689D-His constructs. Three fragments (P1 to P3) in the HAK5 promoter were amplified by PCR and labeled with or without 5′-biotin. The unlabeled fragments were used as competitors, and CKS-His protein was used as a negative control.
ChIP-qPCR Assay
The ChIP-qPCR assay was performed as described previously (Feng et al., 2014). Mouse polyclonal ARF2 antibody was generated against His-tagged recombinant ARF2 peptides (residues 1 to 60 and 395 to 627) purified from E. coli. The 5-d-old seedlings were transferred to K+-sufficient (1 mM) or low-K+ (10 μM) medium for 6 h. Then the plants were harvested and used for ChIP assays.
86Rb+ Uptake Assay
86Rb+ isotope was used as a tracer to analyze K+ uptake. Seven-day-old seedlings were transferred from K+-sufficient medium to low-K+ medium for 3 d. The fresh weight of 12 individual seedlings was measured. Then, the plants were incubated in 1.5 mL uptake solution (5 mM MES and 0.2 mM CaCl2) that was K+-free and supplemented with 100 μM RbCl containing 0.005 μCi/nmol 86Rb+ (Amersham Biosciences). At the end of the uptake period, plants were blotted with tissue paper and immediately washed two times for 30 min in 4°C washing solution (K+-free and supplemented with 1.75 mm nonradiolabeled RbCl). The radioactivity in plants was measured with a scintillation counter (Perkin-Elmer 1450 MicroBeta TriLux) after the addition of 1 mL scintillation liquid.
RT-qPCR Analysis
Total RNA was extracted from Arabidopsis seedlings using TRIzol reagent (Invitrogen) and then treated with DNase I (Takara) to eliminate genomic DNA contamination. The cDNA was synthesized using SuperScript II RNase H2 reverse transcriptase (Invitrogen) and Radom Hexamer Primer (Promega). Quantitative real-time PCR was conducted using Power SYBR Green PCR Master Mix (Applied Biosystems; P/N 4368577) on a 7500 Real-Time PCR System (Applied Biosystems) following the manufacturer’s protocols. The amplification reactions were performed in a total volume of 20 μL, which contained 1 μL cDNA, 7 μL double-distilled water, 2 μL forward and reverse primers (1 μM), and 10 μL SYBR Green premix. The PCR was programmed as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The Actin2/8 gene was used as an internal standard for normalization of the test gene expression levels. The fluorescence signal was obtained during the PCR annealing step. Each RT-qPCR analysis contained three biological replicates. Each replicate contained ∼60 to 80 individual plants.
Luminescence Observation
ProHAK5:LUC Arabidopsis transgenic plants were germinated and grown on K+-sufficient or low-K+ medium for 7 d. The seedlings were sprayed with luciferin solution (0.5 mg/mL) evenly and pretreated in darkness for 20 min to avoid false signals. Then, luminescence was observed using a Lumazone CCD camera. The luminescence intensity was analyzed using WinView32 software and shown using pseudo color.
ARF2 Phosphorylation in Vivo
The arf2-7/ProARF2:ARF2-Flag (COM1) transgenic plants were germinated and grown on K+-sufficient medium for 5 d and then transferred to K+-sufficient medium or low-K+ medium for 2, 4, and 6 h. Total proteins of the plants at different time points were extracted. ARF2-Flag proteins were separated by 10% SDS-PAGE and immunoprecipitated using anti-Flag mAb agarose (Sigma-Aldrich; lot no. SLBJ8441V). Phosphorylated ARF2 proteins were detected by immunoblotting using biotinylated Phos-tag as described previously (Kinoshita-Kikuta et al., 2007). CIAP (Takara) was used to dephosphorylate ARF2 proteins.
ARF2 Peptide Phosphorylation in Vitro
The 1501 to 2100 bp of ARF2 coding sequence fused with His tag was cloned into the pTrc-CKS vector. The point mutations of ARF2 were generated using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies). Then, wild-type ARF2 peptide and mutated ARF2 peptide were expressed in E. coli and purified. Arabidopsis wild-type plants were germinated and grown on K+-sufficient medium for 5 d and then transferred to K+-sufficient medium or low-K+ medium for 2, 4, and 6 h. Total proteins were extracted from wild-type plants at the indicated time points. Then, 100 μg of the total proteins was incubated with 2 μg of ARF2 peptides in phosphorylation buffer with 10 μM ATP for 40 min at 22°C. The phosphorylated and dephosphorylated ARF2 peptides were separated using 8% Phos-tag SDS-PAGE (NARD AAL-107) following the manufacturer’s protocol. The peptides were detected by immunoblotting using His antibody. CIAP (Takara) was used to dephosphorylate ARF2 peptides.
Accession Numbers
Sequence data for the genes described in this article can be found in the Arabidopsis TAIR database (https://www.arabidopsis.org/index.jsp) under the following accession numbers: At5g62000 for ARF2, At4g13420 for HAK5, At2g26650 for AKT1, At1g30270 for CIPK23, At4g32650 for KC1, At4g17615 for CBL1, At5g47100 for CBL9, and At4g33000 for CBL10.
Supplemental Data
Supplemental Figure 1. Phenotype Analysis of ARF2-Overexpressing Lines.
Supplemental Figure 2. ARF2 Proteins Are Targeted to the Nucleus in the Root Epidermis and Root Hairs.
Supplemental Figure 3. Expression Analyses of Potassium Relevant Genes in arf2 Mutants.
Supplemental Figure 4. Expression Analyses of HAK5 and ARF2 in Response to Low-K+ Stress.
Supplemental Figure 5. GUS/LUC Assay in Tobacco Leaves to Test the Activity of Mutant ARF2 Proteins.
Supplemental Figure 6. Phenotype Test of arf2 Mutant in Response to NAA or NPA.
Supplemental Table 1. Primer Sequences and Enzyme Sites Used in This Study.
Supplementary Material
Acknowledgments
We thank Zhizhong Gong and Yan Guo (China Agricultural University) for kindly providing us with the arf2-7 mutant and pCAMBIA1305-Flag vector, respectively. We thank Jörg Kudla (Universität Münster, Germany) and Ryoung Shin (RIKEN Plant Science Center, Japan) for providing the SUPERR:sXVE:GFPc:Bar vector and ProHAK5:LUC transgenic line, respectively. This work was supported by grants from the National Key Research and Development Program of China (2016YFD0100700) and the National Natural Science Foundation of China (31270306, 31570243, and 31421062).
AUTHOR CONTRIBUTIONS
S.Z. and Y.W. designed the research. S.Z., M.-L.Z., and T.-L.M. performed research and analyzed data. S.Z. and Y.W. wrote and revised the article.
Glossary
- CTD
C-terminal dimerization domain
- AuxRE
auxin-responsive element
- EMSA
electrophoresis mobility shift assay
- CIAP
calf intestine alkaline phosphatase
- BR
brassinosteroid
- ChIP
chromatin immunoprecipitation
Footnotes
Articles can be viewed without a subscription.
References
- Ahn S.J., Shin R., Schachtman D.P. (2004). Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol. 134: 1135–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud P., Breitling R., Amtmann A. (2004). The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol. 136: 2556–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley M.K., Grant M., Grabov A. (2006). Plant responses to potassium deficiencies: a role for potassium transport proteins. J. Exp. Bot. 57: 425–436. [DOI] [PubMed] [Google Scholar]
- Boer D.R., Freire-Rios A., van den Berg W.A., Saaki T., Manfield I.W., Kepinski S., López-Vidrieo I., Franco-Zorrilla J.M., de Vries S.C., Solano R., Weijers D., Coll M. (2014). Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156: 577–589. [DOI] [PubMed] [Google Scholar]
- Chérel I., Lefoulon C., Boeglin M., Sentenac H. (2014). Molecular mechanisms involved in plant adaptation to low K(+) availability. J. Exp. Bot. 65: 833–848. [DOI] [PubMed] [Google Scholar]
- Clarkson D.T., Hanson J.B. (1980). The mineral nutrition of higher plants. Annu. Rev. Plant Physiol. 31: 239–298. [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Ellis C.M., Nagpal P., Young J.C., Hagen G., Guilfoyle T.J., Reed J.W. (2005). AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132: 4563–4574. [DOI] [PubMed] [Google Scholar]
- Epstein E., Rains D.W., Elzam O.E. (1963). Resolution of dual mechanisms of potassium absorption by barley roots. Proc. Natl. Acad. Sci. USA 49: 684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C.Z., Chen Y., Wang C., Kong Y.H., Wu W.H., Chen Y.F. (2014). Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 80: 654–668. [DOI] [PubMed] [Google Scholar]
- Gierth M., Mäser P., Schroeder J.I. (2005). The potassium transporter AtHAK5 functions in K(+) deprivation-induced high-affinity K(+) uptake and AKT1 K(+) channel contribution to K(+) uptake kinetics in Arabidopsis roots. Plant Physiol. 137: 1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T.J., Hagen G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10: 453–460. [DOI] [PubMed] [Google Scholar]
- Hardtke C.S., Berleth T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17: 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch R.E., Lewis B.D., Spalding E.P., Sussman M.R. (1998). A role for the AKT1 potassium channel in plant nutrition. Science 280: 918–921. [DOI] [PubMed] [Google Scholar]
- Hong J.P., Takeshi Y., Kondou Y., Schachtman D.P., Matsui M., Shin R. (2013). Identification and characterization of transcription factors regulating Arabidopsis HAK5. Plant Cell Physiol. 54: 1478–1490. [DOI] [PubMed] [Google Scholar]
- Jung J.Y., Shin R., Schachtman D.P. (2009). Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21: 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.J., Ruzicka D., Shin R., Schachtman D.P. (2012). The Arabidopsis AP2/ERF transcription factor RAP2.11 modulates plant response to low-potassium conditions. Mol. Plant 5: 1042–1057. [DOI] [PubMed] [Google Scholar]
- Kinoshita-Kikuta E., Aoki Y., Kinoshita E., Koike T. (2007). Label-free kinase profiling using phosphate affinity polyacrylamide gel electrophoresis. Mol. Cell. Proteomics 6: 356–366. [DOI] [PubMed] [Google Scholar]
- Leigh R.A., Wyn Jones R.G. (1984). A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol. 97: 1–13. [Google Scholar]
- Li H., Johnson P., Stepanova A., Alonso J.M., Ecker J.R. (2004). Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev. Cell 7: 193–204. [DOI] [PubMed] [Google Scholar]
- Maathuis F.J.M. (2009). Physiological functions of mineral macronutrients. Curr. Opin. Plant Biol. 12: 250–258. [DOI] [PubMed] [Google Scholar]
- Maathuis F.J.M., Sanders D. (1997). Regulation of K+ absorption in plant root cells by external K+: interplay of different plasma membrane K+ transporters. J. Exp. Bot. 48: 451–458. [DOI] [PubMed] [Google Scholar]
- Nemhauser J.L., Feldman L.J., Zambryski P.C. (2000). Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127: 3877–3888. [DOI] [PubMed] [Google Scholar]
- Okushima Y., Mitina I., Quach H.L., Theologis A. (2005a). AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J. 43: 29–46. [DOI] [PubMed] [Google Scholar]
- Okushima Y., et al. (2005b). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettigrew W.T. (2008). Potassium influences on yield and quality production for maize, wheat, soybean and cotton. Physiol. Plant. 133: 670–681. [DOI] [PubMed] [Google Scholar]
- Pyo Y.J., Gierth M., Schroeder J.I., Cho M.H. (2010). High-affinity K(+) transport in Arabidopsis: AtHAK5 and AKT1 are vital for seedling establishment and postgermination growth under low-potassium conditions. Plant Physiol. 153: 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio F., Fon M., Ródenas R., Nieves-Cordones M., Alemán F., Rivero R.M., Martínez V. (2014). A low K+ signal is required for functional high-affinity K+ uptake through HAK5 transporters. Physiol. Plant. 152: 558–570. [DOI] [PubMed] [Google Scholar]
- Schlücking K., Edel K.H., Köster P., Drerup M.M., Eckert C., Steinhorst L., Waadt R., Batistič O., Kudla J. (2013). A new β-estradiol-inducible vector set that facilitates easy construction and efficient expression of transgenes reveals CBL3-dependent cytoplasm to tonoplast translocation of CIPK5. Mol. Plant 6: 1814–1829. [DOI] [PubMed] [Google Scholar]
- Schroeder J.I., Ward J.M., Gassmann W. (1994). Perspectives on the physiology and structure of inward-rectifying K+ channels in higher plants: biophysical implications for K+ uptake. Annu. Rev. Biophys. Biomol. Struct. 23: 441–471. [DOI] [PubMed] [Google Scholar]
- Schruff M.C., Spielman M., Tiwari S., Adams S., Fenby N., Scott R.J. (2006). The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133: 251–261. [DOI] [PubMed] [Google Scholar]
- Shin R., Schachtman D.P. (2004). Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc. Natl. Acad. Sci. USA 101: 8827–8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding E.P., Hirsch R.E., Lewis D.R., Qi Z., Sussman M.R., Lewis B.D. (1999). Potassium uptake supporting plant growth in the absence of AKT1 channel activity: Inhibition by ammonium and stimulation by sodium. J. Gen. Physiol. 113: 909–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.B., Hagen G., Guilfoyle T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868. [DOI] [PubMed] [Google Scholar]
- Véry A.A., Sentenac H. (2003). Molecular mechanisms and regulation of K+ transport in higher plants. Annu. Rev. Plant Biol. 54: 575–603. [DOI] [PubMed] [Google Scholar]
- Véry A.A., Nieves-Cordones M., Daly M., Khan I., Fizames C., Sentenac H. (2014). Molecular biology of K+ transport across the plant cell membrane: what do we learn from comparison between plant species? J. Plant Physiol. 171: 748–769. [DOI] [PubMed] [Google Scholar]
- Vert G., Walcher C.L., Chory J., Nemhauser J.L. (2008). Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc. Natl. Acad. Sci. USA 105: 9829–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D.J., Leigh R.A., Miller A.J. (1996). Potassium homeostasis in vacuolate plant cells. Proc. Natl. Acad. Sci. USA 93: 10510–10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hua D., He J., Duan Y., Chen Z., Hong X., Gong Z. (2011). Auxin Response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet. 7: e1002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wu W.H. (2013). Potassium transport and signaling in higher plants. Annu. Rev. Plant Biol. 64: 451–476. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Bai M.Y., Oh E., Zhu J.Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46: 701–724. [DOI] [PubMed] [Google Scholar]
- Xu J., Li H.D., Chen L.Q., Wang Y., Liu L.L., He L., Wu W.H. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360. [DOI] [PubMed] [Google Scholar]
- Yang J., Tian L., Sun M.X., Huang X.Y., Zhu J., Guan Y.F., Jia Q.S., Yang Z.N. (2013). AUXIN RESPONSE FACTOR17 is essential for pollen wall pattern formation in Arabidopsis. Plant Physiol. 162: 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zörb C., Senbayram M., Peiter E. (2014). Potassium in agriculture--status and perspectives. J. Plant Physiol. 171: 656–669. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.