Abstract
Acidity is a serious limitation to plant production on many of the world's agricultural soils. Toxic aluminium (Al) cations solubilized by the acidity rapidly inhibit root growth and limit subsequent uptake of water and nutrients. Recent work has shown that the ALMT1 gene of wheat (Triticum aestivum) encodes a malate transporter that is associated with malate efflux and Al tolerance. We generated transgenic barley (Hordeum vulgare) plants expressing ALMT1 and assessed their ability to exude malate and withstand Al stress. ALMT1 expression in barley conferred an Al-activated efflux of malate with properties similar to those of Al-tolerant wheat. The transgenic barley showed a high level of Al tolerance when grown in both hydroponic culture and on acid soils. These findings provide additional evidence that ALMT1 is a major Al-tolerance gene and demonstrate its ability to confer effective tolerance to acid soils through a transgenic approach in an important crop species.
Acid soils cover some 40% of the Earth's arable land and represent a major limitation to plant production (1). The main constraint to plant growth on these soils is the aluminum (Al) that is solubilized by the acidity into the toxic Al3+ cation. Al toxicity is manifest by inhibition of root growth resulting in poor uptake of water and nutrients (2). Plant production on acid soils can be maintained by neutralizing the acidity with lime (CaCO3) and through the use of Al-tolerant plant species. Lime can take decades to correct acidity at depth, and many important crop and pasture species lack sufficient Al tolerance within their germplasm to allow effective breeding for this character. Genetic engineering provides an opportunity to enhance the Al tolerance of sensitive species through the overexpression of endogenous genes or by the expression of foreign genes. Toward this end, the Al tolerance of canola (Brassica napus) (3), Arabidopsis thaliana (4), tobacco (Nicotiana tabacum) (5), and alfalfa (Medicago sativum) (6) have been reported to be enhanced by increasing organic acid biosynthesis through overexpression of citrate synthase or malate dehydrogenase genes derived from plants or bacteria. Other strategies have sought to increase Al tolerance by overexpression of genes associated with stress responses (7-9). In some cases, manipulation of organic acid biosynthesis has led to increased secretion of organic acids from roots, and the increased Al tolerance was attributed to the ability of organic acids to chelate and detoxify Al3+. However, the increases in Al tolerance have at best been modest or, as in the case of a Psuedomonas aeuriginosa citrate synthase gene, not easily reproducible (10). Whereas an enhanced ability to secrete organic acids from roots might need to be linked to the biosynthesis of organic acids, the transport of these molecules to the external medium appears to be a rate-limiting step (11).
The Al-activated efflux of organic acid anions from roots is now a well established mechanism that is proposed to be used by a range of Al-tolerant plants (11, 12). This mechanism for Al tolerance has been thoroughly studied in wheat (Triticum aestivum). A pair of near-isogenic lines that differ in tolerance at a single genetic locus (13) was used to show that Al-activated efflux of malate from root apices was greater in the tolerant genotype than the sensitive genotype (14). The “malate hypothesis” proposes that the secreted malate binds Al into a nontoxic form and protects the root apex from damage. Subsequently, Ryan et al. (15) and Zhang et al. (16) identified an Al-activated anion channel permeable to malate present on the plasma membrane of apical root cells of wheat and speculated that the Al-tolerance gene encoded this channel. Recently, Sasaki et al. (17) cloned a gene (ALMT1) encoding a protein with properties consistent with it being the Al-activated channel. The ALMT1 protein is membrane-bound, the gene cosegregates with Al tolerance, and the expression of ALMT1 in Xenopus oocytes, rice (Oryza sativa), and tobacco cells confers an Al-activated efflux of malate. Furthermore, ALMT1 expression in tobacco cells grown in suspension culture conferred an enhanced ability to recover from an 18-h exposure to Al stress, providing additional evidence that ALMT1 is the Al-tolerance gene of wheat. However, ALMT1 did not confer enhanced Al tolerance to rice plants despite conferring an Al-activated efflux of malate. The inability of ALMT1 to confer Al tolerance to rice was attributed to the already high endogenous Al tolerance of this species (17).
To establish whether ALMT1 is capable of conferring Al tolerance to intact plants, we chose to express this gene in barley (Hordeum vulgare). Barley is an economically important crop in many parts of the world and is among the most Al-sensitive of the cereal crops (18). Here we show that expressing ALMT1 in barley confers an Al-activated efflux of malate that is associated with increased Al tolerance both in hydroponic culture and acid soil. These findings provide additional evidence that ALMT1 is a major Al-tolerance gene and demonstrate the potential of using this gene to enhance the acid-soil tolerance of important crop species.
Materials and Methods
Plant Materials. The barley cultivar Golden Promise was transformed by Agrobacterium tumifaciens as described in ref. 19. Primary transformants (T0) were maintained as clonal populations by taking tillers and growing them separately. In this way, up to 20 individual plants were maintained for each transgenic line. The vector controls were transformed with an empty binary vector, whereas WT plants were derived from plants that had progressed through tissue culture but were not transformed. The clonal plants were maintained by hydroponic culture in a nutrient solution that contained 500 μM KNO3, 500 μM CaCl2, 500 μM NH4NO3, 150 μM MgSO4, 100 μM KH2PO4, 2 μM Fe:EDTA, 11 μM H3BO3, 2 μM MnCl2, 0.35 μM ZnCl2, and 0.2 μM CuCl2 adjusted to pH 5.5. Several T0 plants of each line were transferred to soil and grown to maturity to produce seeds of the T1 generation. Seeds collected from T1 plants (T2 generation) were germinated and grown in hydroponic culture by using Al screening solution (same solution as above except that it contained 10 μMKH2PO4 and 2 μM FeCl3 instead of Fe:EDTA and was adjusted to pH 4.3) with AlCl3 added to 10 μM. In this way, lines homozygous for ALMT1 and sister lines azygous for ALMT1 (null segregants) were identified and confirmed independently by either their level of hygromycin resistance or by using PCR to amplify the ALMT1 gene.
Gene Constructs. The coding region of the ALMT1-1 (referred to as ALMT1 in the text; GenBank accession no. AB081803) gene was amplified by using RT-PCR from polyA+ RNA as described in ref. 17. The resulting fragment was digested with SalI and NotI and cloned into the plasmid pTH2 (20) by replacing the GFP sequence to yield pTH-ALMT1-1. After the ALMT1 coding region was verified by sequencing, plasmid pTH-ALMT1-1 was digested with SalI and NotI, and the ALMT1-1 fragment was blunted by end-filling and then inserted into the SmaI site of pWUbi (21). The orientation with respect to the ubiquitin promoter was verified, and then pWUbi was digested with NotI to excise the fragment containing the ubiquitin promoter, the ALMT1 coding region, and the terminator. This fragment was then inserted into the NotI site of the binary vector pWBVec8 (21), the orientation with respect to the selectable marker was verified, and the plasmid was introduced into Agrobacterium by triparental mating. Southern blots were performed by using the procedures described in ref. 17.
Real-Time Quantitative RT-PCR. Total RNA was prepared with an RNeasy minikit (Qiagen) from 20 root tips (3 mm long) collected from the various genotypes with three biological replicates for each line. The RNA extraction included an on-column DNase step to degrade any contaminating genomic DNA. cDNA was prepared from total RNA (2 μg) as described in ref. 22, except that the final elution from the spin column was diluted to 100 μl. Levels of ALMT1 and control gene expressions were determined by real-time quantitative RT-PCR on Rotor-Gene 2000 or 3000 Real Time Cyclers (Corbett Research, Sydney). One-tenth dilutions were used as a template for the quantitative RT-PCR reaction in a total volume of 10 μl as follows: 5 μl of SYBR Green JumpStart Taq ReadyMix (Sigma), 0.5 μl of primer mix (50:50 mix of forward and reverse primers at 10 pmol/μl each), and 4.5 μl of template. The primers 5′-CGTGAAAGCAGCGGAAAGCC-3′ and 5′-CCCTCGACTCACGGTACTAACAACG-3′ were used for amplification of ALMT1 transcript; 5′-AACAAGACTGCTTTCACCAC-3′ and 5′-TCTCAGAAAGCTCACGGTAG-3′ were used for amplification of a proton-pump transcript from barley (GenBank accession no. AY136627); 5′-AACAAGACTGCTTTCACCAC-3′ and 5′-TCTCAGAGAGCTCACGGTAG-3′ were used for amplification of a proton-pump transcript from wheat (GenBank accession no. AY543630); 5′-GAAGGACATCTTCACGGCGATC-3′ and 5′-CACGGCCATGAAGAAGAAGC-3′ were used for amplification of the wheat phosphate transporter transcript PT-1 (GenBank accession no. AF110180); 5′-GAAGGACATCTTCACGGCGATC-3′ and 5′-CACCGCCATGAAGAAGAATC-3′ were used for amplification of the barley phosphate transporter transcript Pht1-6 (GenBank accession no. AF543198); and 5′-CTGATCTTCTGTGAAGGGT-3′ and 5′-TGATAGAACTCGTAATGGGC-3′ for amplification of both the wheat and barley 28S ribosomal transcripts (GenBank accession no. AY049041). Cycling conditions were as follows: 5 min at 94°C, followed by 45 cycles of 15 s at 94°C, 15 s at 55°C, and 20 s at 72°C. At the end of the cycling, the samples were incubated at 40°C for 5 min, then at 55°C for 1 min followed by a melting curve program (55-99°C in 1° increments with a 5-s hold at each temperature).
Malate and K+ Efflux. Malate efflux from root segments was assayed by using modification methods described in ref. 23. Unless stated otherwise, four 3-mm-long root apices were incubated with shaking in 1 ml of 0.2 mM CaCl2 (pH 4.3) for ≈1 h. The apices were then rinsed with the CaCl2 solution three times and replaced with 1 ml of either the same solution (control) or treatment solution (0.2 mM CaCl2 with added treatment at pH 4.3). After an incubation that typically lasted for 1 h, the solution was removed and dried, and the residue was resuspended in 200 μl of buffer used for the spectrographic assay of malate (0.25 M glycine/0.20 M hydrazine/2.7 mM NAD adjusted to pH 9). A subsample (100 μl) was transferred to a microcuvette, and the increase in absorbance at 340 nm measured 6 min after addition of 1 μl of malate dehydrogenase (5 mg/ml) was used to calculate the amount of malate by comparison to a standard curve. For K+ efflux, samples from treatments that had not been concentrated were analyzed by atomic absorption with procedures described in ref. 23.
Al Tolerance. Plants in hydroponic culture were assessed for Al tolerance by using the Al screening solution described above supplemented with a range of AlCl3 concentrations. Soil experiments used an acid subsoil (10- to 40-cm layer, CaCl2 extracted pH of 3.9, water-extracted pH of 5.2, and 40 μg/g of CaCl2-extractable Al) derived from Chiltern in Australia (24) and a nonallophanic andosol obtained from the Field Science Center at Tohoku University (Sendai, Japan). In the first experiment, pregerminated seeds, where the longest root was 8-35 mm, were planted in 300 g of either unamended Chiltern soil or Chiltern soil mixed with 0.75 g of CaCO3 per kg of soil (water-extracted pH of 6.0). Each pot contained a single plant with each combination of genotype and treatment consisting of eight replicates. The experiment was set up as four blocks in a greenhouse with two replicates of each genotype/treatment placed randomly per block. The pots were weighed daily and watered to either 323 g (acid soil) or 316 g (amended soil). After 4 days of growth, the seedlings were removed and the longest root was remeasured. In the second experiment, pregerminated seeds, where the longest root was 5-26 mm, were planted in either 140 g of moistened acidic andosol (water-extracted pH of 4.5) or control medium consisting of 80 g of peat moss (Primemix TKS1, Sakata Seed, Yokohama, Japan; water-extracted pH of 6.2) moistened with 50 ml of water and fertilized with nutrients (N, 100-140 mg/ml; P, 80-120 mg/ml; K, 130-190 mg/liter). Each pot contained a single plant with each combination of genotype and treatment consisting of six replicates. The experiment was set up as three blocks in a growth cabinet with two replicates of each genotype/treatment placed randomly per block. The growth cabinet was set at 25°C, and lighting was adjusted to 150-200 μmol of photons per m2 perswith a 14-h photoperiod. The pots were weighed daily and watered (to 140 g for the acid soil and 130 g for the control medium), and the roots were measured after 4 days of growth.
Results
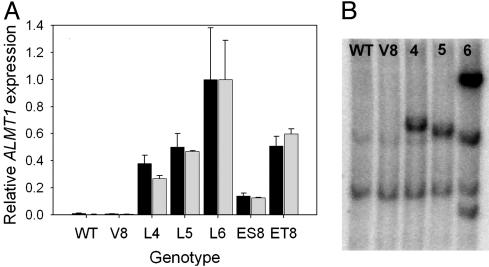
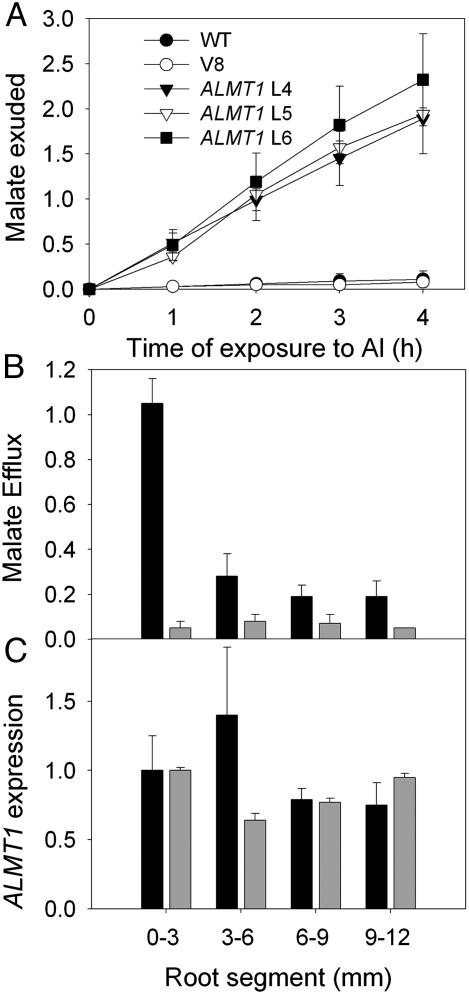
Transgenic Barley Expressing ALMT1 Shows an Al-Activated Malate Efflux. A. tumefaciens transformation was used to introduce the ALMT1 gene under the control of the ubiquitin promoter into barley. From 25 primary transformants, the three highest-expressing lines were selected for detailed analysis. Analysis of root apices by real-time quantitative RT-PCR verified ALMT1 expression in these lines to a level comparable with an Al-tolerant wheat when either a phosphate transporter or proton-pump gene was used to normalize expression (Fig. 1A). Southern blots showed the presence of a single insertion (lines 4 and 5) or multiple insertions (line 6) of the gene, whereas a band common to all genotypes indicated the presence of a related gene in the barley genome (Fig. 1B). Primary transformants (T0) were maintained by clonal propagation of tillers that allowed a homogenous population of plants to be maintained for each line. All three ALMT1 lines showed an Al-activated malate efflux from root apices that was absent from either WT plants or plants transformed with the vector alone (Fig. 2A). Efflux was maintained from the excised root apices over at least 4 h of Al exposure.
Fig. 1.
Expression of ALMT1 in primary transformants of barley. (A) Real-time quantitative RT-PCR was used to assess ALMT1 expression in root apices of three independent transgenic barley lines (lines 4-6, denoted by L4-L6) transformed with the ALMT1 coding region and the near-isogenic wheat lines ET8/ES8 that differ in Al tolerance (13). The controls consisted of nontransformed WT plants and plants transformed with the empty vector (V8). Expression is expressed relative to phosphate transporter (black columns) or proton-pump (gray columns) control genes and is shown as a proportion of the highest expressing transgenic line (arbitrarily set to 1.0). The error bars show ±SEM (n = 3). (B) Southern blot analysis of the transgenic barley lines expressing ALMT1 (lanes 4-6) and control plants (WT, wild type; V8, plasmid control). Genomic DNA was digested with HindIII, and the filter was probed with the ALMT1 coding region.
Fig. 2.
ALMT1 expression confers Al-activated malate efflux from barley roots. (A) Al-activated malate efflux from root apices of transgenic barley expressing ALMT1 (lines 4-6 denoted by L4-L6) and control lines consisting of WT plants and plants transformed with the empty vector (V8). Excised root apices were incubated with 50 μM Al solution in basal solution (0.2 mM CaCl2, pH 4.3) and solutions were changed after every hour. Efflux is expressed as nmol per apex per h, and error bars show ±SEM (n = 3) of the cumulative exudation of malate into the medium. Excised root apices incubated in basal solution exuded little or no malate for all genotypes. (B) Al-activated malate efflux from root segments of transgenic barley line 5 expressing ALMT1. The measurements indicate the distance from the end of the root apex. Excised root segments were incubated for 1 h with 50 μM Al solution in basal solution (0.2 mM CaCl2, pH 4.3; black columns) or in basal solution only (gray columns). Error bars show ±SEM (n = 3). (C) Relative ALMT1 expression in root segments of transgenic barley line 5. RNA from excised root segments was extracted, transcribed to cDNA, and analyzed for ALMT1 expression by real-time quantitative RT-PCR. Expression is expressed relative to phosphate-transporter (black columns) or proton-pump (gray columns) genes and is shown as a proportion of the 0- to 3-mm segment (arbitrarily set to 1.0). Error bars show ±SEM (n = 3).
The root apex (≈3 mm) represents the most Al-sensitive part of the root (25) and is the region that specifically possesses the Al-activated efflux of malate in Al-tolerant wheat (23). Because ALMT1 expression in the transgenic barley was under the control of the ubiquitin promoter with a high level of constitutive expression throughout the plant, we also determined the level of Al-activated malate efflux from more mature root segments. Although older root segments showed an Al-activated efflux of malate, it was considerably smaller than that observed from the root apex (Fig. 2B). ALMT1 was expressed in all root segments analyzed for transgenic line 5 when normalized to the expression of two other transporter genes (Fig. 2C). A similar pattern was found when ALMT1 expression was normalized to the expression of ribosomal RNA, but the reproducibility of the data was less reliable presumably because of the large difference in expression between the two genes (data not shown).
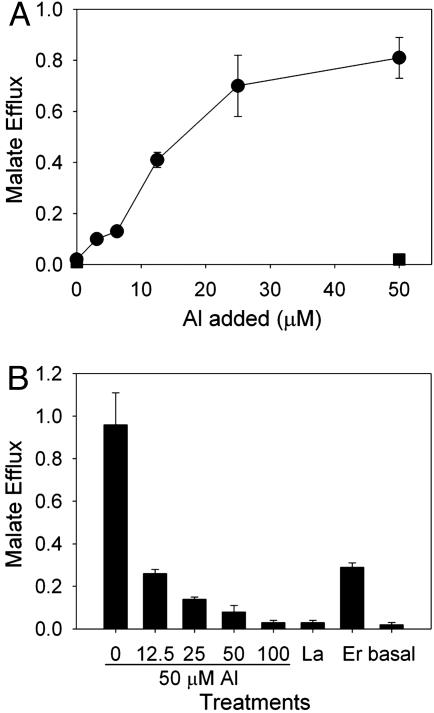
Malate efflux from the roots of line 5 of the ALMT1 transgenics, a line with a single insert, was characterized in detail. The efflux responded to Al concentration in a dose-responsive manner, plateauing at ≈25 μM added Al (Fig. 3A). Malate efflux was inhibited by the anion channel blocker niflumic acid (Fig. 3B). Lanthanum (La) was ineffective in activating malate efflux, and the rare earth element erbium (Er) was capable of eliciting malate efflux, albeit at a lower level than observed for Al (Fig. 3B). Previously, Kataoka et al. (26) had found that Er, along with a range of other rare earth elements, was capable of eliciting malate efflux from Al-tolerant wheat. Al-activated malate efflux from root apices of transgenic barley expressing ALMT1 was accompanied by the efflux of K+. Net K+ effluxes from excised apices of line 5 were calculated by subtracting corresponding values obtained for the vector-only control line, resulting in an efflux of 0.4 ± 0.3 nmol of K+ per apex per h in the absence of Al and an efflux of 5.2 ± 1.0 nmol of K+ per apex per h in the presence of 50 μM Al (means ± SE; n = 4).
Fig. 3.
Properties of malate efflux from root apices of transgenic barley expressing ALMT1. (A) Effect of Al concentration on malate efflux. Excised root apices from ALMT1 line 5 (solid circles) were exposed to Al treatments in basal solution for 1 h. Efflux is expressed as nmol per apex per h and error bars show ±SEM (n = 3). The filled squares show efflux from root apices of the control line transformed with the empty vector. (B) Effects of niflumic acid in combination with Al (0-100 μM niflumic acid with the concentration denoted above the bar labeled 50 μM Al), La (50 μM), Er (50 μM), and basal solution (0.2 mM CaCl2, pH 4.3) on malate efflux from root apices of line 5. Excised root apices from line 5 were exposed to the various treatments in basal solution for 1 h. Error bars show ±SEM (n = 3).
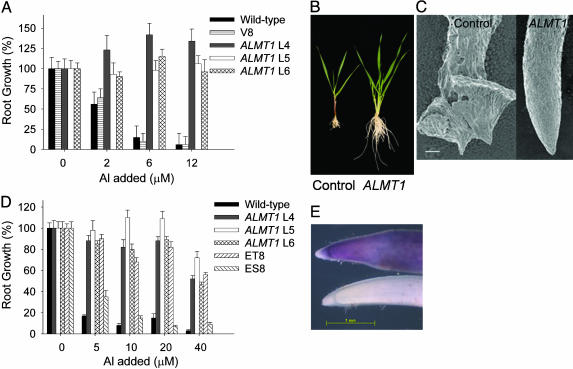
Expression of ALMT1 in Barley Confers Enhanced Al Tolerance. Al tolerance can be assessed by determining root elongation of plants grown in hydroponic culture in the continual presence of added Al. All three barley T0 lines expressing ALMT1 showed robust root growth in hydroponic culture at Al concentrations that severely inhibited roots of control plants (Fig. 4 A and B). Root apices of the ALMT1 plants were unaffected by the Al, whereas those of the controls were severely damaged and malformed (Fig. 4C). Similarly, homozygous T2 lines expressing ALMT1 were Al tolerant in hydroponic culture, compared with the WT parental line (Fig. 4D) or azygous sister lines (data not shown). Azygous sister lines derived from the same transformation events that generated the ALMT1-expressing lines provide ideal controls because they are plants that have experienced the same tissue culture conditions during the transformation procedure. The level of tolerance in hydroponic culture was comparable with ET8, the Al-tolerant wheat line that is the original source of the ALMT1 gene. Hematoxylin staining of roots previously exposed to Al solutions is another method commonly used to assess Al tolerance in cereals (27). Hematoxylin forms a purple-red complex with Al and provides an indirect measure of noncomplexed Al in root apices, with the intensity of staining correlated with sensitivity to Al in wheat. Root apices of control plants became stained with hematoxylin, suggesting that they had accumulated Al, whereas those of the ALMT1 transgenics remained clear (Fig. 4E). Some swelling also was apparent behind the tip of the control line, a typical early symptom of Al toxicity.
Fig. 4.
ALMT1 confers Al tolerance to barley grown in hydroponic culture. (A) Root elongation of T0 (primary transgenics; lines 4-6 denoted by L4-L6) barley lines grown in hydroponic culture. Plants were grown for 10 days in nutrient solution that contained a range of Al concentrations. Control lines included WT and a transgenic line transformed with the vector only (V8). Elongation of the longest root over 5 days is expressed as a percentage of the minus-Al treatment, and error bars show ±SEM (n = 4). Root lengths of plants grown in the absence of Al were as follows: WT, 103 ± 14 mm; vector-only control, 103 ± 14 mm; ALMT1 line 4, 99 ± 12 mm; ALMT1 line 5, 122 ± 12 mm; and ALMT1 line 6, 118 ± 8 mm. (B) Effect of 3 μM Al on growth over 10 days of the T0 generation of the control (empty vector) line and ALMT1 line 5. (C) Scanning electron micrograph showing the effect of Al (3 μM) on the morphology of the root apex from the control line (empty vector) and ALMT1 line 5 grown for 10 days. (Scale bar, 100 μm.) (D) Root elongation of T2 homozygous barley lines grown in hydroponic culture. For each transgenic line, a sister line azygous for ALMT1 derived from the same transformation event was developed (lines 4-6 denoted by L4-L6). The wheat lines ET8 and ES8 are near-isogenic lines that differ in Al tolerance. Elongation of the longest root over 5 days is expressed as a percentage of the minus-Al treatment, and error bars show ±SEM (n = 7). Root lengths of plants grown in the absence of Al were as follows: WT, 54 ± 3 mm; ALMT1 line 4, 62 ± 4 mm; ALMT1 line 5, 46 ± 3 mm; ALMT1 line 6, 60 ± 4 mm; ET8, 66 ± 3 mm; and ES8, 74 ± 4. (E) Roots stained with hematoxylin. The bottom root is derived from the T2 homozygous ALMT1 line 5, and the top root is from its azygous sister line. Roots were exposed to 10 μM in nutrient solution for 24 h before being stained according to the procedures described in ref. 13. (Scale bar, 1 mm.)
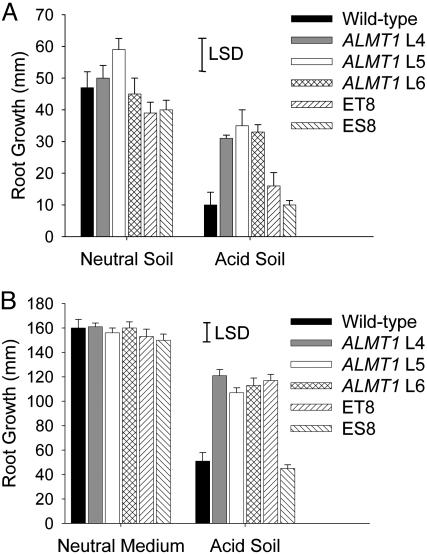
To determine whether the Al tolerance of the transgenic barley expressing ALMT1 was apparent in soil, the homozygous lines were grown on two different acid soils. The first was a subsoil that contained high concentrations of soluble Al. ALMT1-expressing transgenics clearly showed better root growth in the acid soil than WT controls (Fig. 5A) or their azygous sister lines (data not shown). When the soil was neutralized with CaCO3, root growth of all barley lines was similar regardless of genotype. The toxicity of the soil was apparent on the ET8/ES8 wheat lines where root growth of the Al-tolerant ET8 line was inhibited on the unamended soil to such an extent that it did not differ significantly from the sensitive ES8 line. When the lines were grown on a second acidic soil, the homozygous lines expressing ALMT1 showed better root growth than did WT barley, and ET8 wheat grew significantly better than ES8 (Fig. 5B).
Fig. 5.
ALMT1 enhances root growth of barley on acid soils. (A) Seedlings of the various T2 homozygous ALMT1-expressing lines of barley (lines 4-6 denoted by L4-L6), WT barley, and the near-isogenic wheat lines ET8/ES8 were grown on an unamended acid soil obtained from Chiltern, Australia, and the same soil neutralized with CaCO3. After 4 days, the longest root of each seedling was measured and root growth over 4 days calculated [error bars denote ±SEM (n = 6-8), and the least significant difference (LSD) at P < 0.05 is shown for the interactions of root length with treatments and genotypes]. (B) Seedlings of the various T2 homozygous ALMT1-expressing lines of barley (lines 4-6 denoted by L4-L6), WT barley (WT), and the near-isogenic wheat lines ET8/ES8 were grown on an acid soil obtained from Tohoku, Japan, and a neutral medium consisting of fertilized peat moss. After 4 days, the longest root of each seedling was measured [error bars denote ±SEM (n = 6), and the LSD at P < 0.05 is shown for the interactions of root length with treatments and genotypes].
Discussion
The barley cultivar used in this study (cv. Golden Promise) is very susceptible to Al toxicity and does not possess an Al-activated efflux of malate. The generation of transgenic Golden Promise expressing ALMT1 conferred an Al-activated malate efflux that was accompanied by an Al-tolerance phenotype that was at least as effective as Al-tolerant wheat. These data provide evidence that ALMT1 is capable of conferring Al tolerance to intact plants and further support the notion that ALMT1 is a major gene for Al tolerance in wheat. In addition, they provide further support for the “malate hypothesis” as a tolerance mechanism in wheat. Similar to Al-tolerant wheat genotypes (23, 26), malate efflux in the transgenic barley showed an Al dose-response, was inhibited by the anion channel-blocker niflumic acid, was activated by Er, and was accompanied by K+ efflux. The malate efflux from root apices of the transgenic barley (≈1 nmol per apex per h) is comparable with that found in Al-tolerant wheat genotypes (23), resulting in a correspondingly similar level of Al tolerance in hydroponic culture but an apparent greater level of tolerance in a severely acidic soil (Fig. 5A). These features are consistent with ALMT1 encoding an Al-gated anion channel that is specifically permeable to malate. The finding that a single gene was able to confer an Al-activated malate efflux to a similar level found in Al-tolerant genotypes of wheat suggests that the biosynthesis of malate is not a rate-limiting step for its efflux from root apices of barley, as concluded previously for Al-sensitive wheat (23). Although there is evidence that overexpressing genes involved in organic acid biosynthesis can increase the content of organic acid anions and their subsequent efflux from roots of some species, it appears that transport of organic acid anions across the plasma membrane is the major factor that limits efflux in species such as barley and wheat.
The ALMT1 gene was expressed under the control of the ubiquitin promoter that has previously been shown to confer high-level constitutive expression of transgenes in both meristimatic and mature regions of roots (28). By contrast, transcripts for ALMT1 in wheat under the control of its native promoter are restricted to the apical 2-3 mm of the root that coincides with the region of malate efflux (17). The level of ALMT1 transcripts when the gene was expressed under the control of the ubiquitin promoter was similar in mature and apical segments when two other genes encoding transporters were used as reference genes (Fig. 2C). However, when it is taken into account that root apices contain 5- to 10-fold more total RNA than the older segments, the absolute transcript level is considerably greater in root apices. Root apices are particularly susceptible to Al toxicity, and restricting malate efflux to this tissue, and only in the presence of Al, ensures that the metabolic cost to the Al-tolerant wheat plant is minimized. Similarly, malate efflux from the transgenic barley expressing ALMT1 was largely restricted to the apical 3 mm of roots, although some efflux was detectable in more mature segments. The lower efflux from more mature regions of the root might be due to the lower level of ALMT1 transcripts relative to the root apex, as discussed above, assuming that the ALMT1 protein is continuously turned over. Alternatively, it might represent an inability of more mature segments to replenish malate pools as effectively as the root apex. In any case, despite our use of a constitutive promoter to express ALMT1, the resulting phenotype mimics the situation in Al-tolerant wheat and avoids potential metabolic costs to the plant associated with a high level of malate efflux from all root tissues.
Although barley shows some variation in Al tolerance, it is the most sensitive of the cereal crops with Al-tolerant genotypes, being considerably less tolerant than Al-tolerant wheat genotypes. In barley, Al tolerance is associated with an Al-activated efflux of citrate (18). Whereas citrate forms a stronger complex with Al than malate and protects roots more effectively (13), citrate efflux from barley roots is much less than the malate efflux found from Al-tolerant wheat. Zhao et al. (18) showed that, for a range of cereal genotypes, a strong correlation exists between Al tolerance and a value that takes into account both the level of organic acid efflux and the ability of the organic acid to chelate Al. Mapping of genes for Al tolerance in barley has revealed a major gene on chromosome 4H (29, 30) in a region corresponding to that found for an Al-tolerance gene in wheat on chromosome 4DL. In view of the similar physiology of Al tolerance of the two species, coupled with a similar genetic location for a major gene encoding tolerance, it is possible that a barley orthologue of ALMT1 encodes Al tolerance and that the protein is permeable to citrate instead of malate. A fragment that cross-hybridizes with ALMT1 in genomic DNA (Fig. 1B) may indicate such an orthologous gene in barley.
We have demonstrated the utility of a single gene to confer a high level of Al tolerance to an important agricultural species and demonstrated its effectiveness on acid soils. Unlike genes for disease or insect resistance that can be overcome by mutations in the attacking organism, genes for abiotic stresses such as Al are not prone to breakdown in a similar fashion. However, continual use of Al-tolerant germplasm without neutralizing the acidity could exacerbate soil acidity through continued removal of alkaline produce, such as grain, and could lead to increased concentrations of toxic Al that may eventually overcome the protection conferred by genes such as ALMT1. For this reason, the effective management of acid soils combines the application of lime with genotypes tolerant of acid soils. For instance, studies have shown that when grown on acid soils that have been limed, Al-tolerant wheat still out-yields Al-sensitive wheat when the acidity is at depth (31, 32). Furthermore, although Al is generally the major toxic metal of acid soils, Mn toxicity also can occur in these soils. The recent isolation of genes that confer Mn tolerance (33) and their use in combination with ALMT1 and liming practices could provide additional options for managing acid soils. This strategy has the potential of allowing normally sensitive crop species, such as barley, to be grown effectively on a wide range of acid soils.
Acknowledgments
We thank Dr. Richard Culvenor (Commonwealth Scientific and Industrial Research Organization Plant Industry) for providing the Chiltern soil, Dr. Masahiko Saigusa and Dr. Toyoaki Ito (Tohoku University) for providing the acidic andosol, Dr. Syuntaro Hiradate (National Institute for Agro-Environmental Sciences, Japan) for technical comments, and Ms. Michiyo Ariyoshi for experimental assistance.
Author contributions: E.D., P.R.R., Y.Y., T.S., and H.M. designed research; E.D., P.R.R., D.M.H., and T.S. performed research; E.D., P.R.R., D.M.H., Y.Y., T.S., and H.M. analyzed data; and E.D., P.R.R., D.M.H., Y.Y., T.S., and H.M. wrote the paper.
References
- 1.von Uexküll, H. R. & Mutert, E. (1995) Plant Soil 171, 1-15. [Google Scholar]
- 2.Kochian, L. V. (1995) Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 237-260. [Google Scholar]
- 3.Anoop, V. M., Basu, U., McCammon, M. T., McAlister-Henn, L. & Taylor, G. J. (2003) Plant Physiol. 132, 2205-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koyama, H., Kawamura, A., Kihara, T., Hara, T., Takita, E. & Shibata, D. (2000) Plant Cell Physiol. 41, 1030-1037. [DOI] [PubMed] [Google Scholar]
- 5.de la Fuente, J. M., Ramírez-Rodríguez, V., Cabrera-Ponce, J. L. & Herrera-Estrella, L. (1997) Science 276, 1566-1568. [DOI] [PubMed] [Google Scholar]
- 6.Tesfaye, M., Temple, S. J., Allan, D. L., Vance, C. P. & Samac, D. A. (2001) Plant Physiol. 127, 1836-1844. [PMC free article] [PubMed] [Google Scholar]
- 7.Basu, U., Good, A. G. & Taylor, G. J. (2001) Plant Cell Environ. 24, 1269-1278. [Google Scholar]
- 8.Ezaki, B., Gardner, R. C., Ezaki, Y. & Matsumoto, H. (2000) Plant Physiol. 122, 657-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sivaguru, M., Ezaki, B., He, Z. H., Tong, H., Osawa, H., Baluska, F., Volkmann, D. & Matsumoto, H. (2003) Plant Physiol. 132, 2256-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delhaize, E., Hebb, D. M. & Ryan, P. R. (2001) Plant Physiol. 125, 2059-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan, P. R., Delhaize, E. & Jones, D. L. (2001) Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 527-560. [DOI] [PubMed] [Google Scholar]
- 12.Ma, J. F., Ryan, P. R. & Delhaize, E. (2001) Trends Plant Sci. 6, 273-278. [DOI] [PubMed] [Google Scholar]
- 13.Delhaize, E., Craig, S., Beaton, C. D., Bennet, R. J., Jagadish, V. C. & Randall, P. J. (1993) Plant Physiol. 103, 685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delhaize, E., Ryan, P. R. & Randall, P. J. (1993) Plant Physiol. 103, 695-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan, P. R., Skerrett, M., Findlay, G. P., Delhaize, E. & Tyerman, S. D. (1997) Proc. Natl. Acad. Sci. USA 94, 6547-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang, W.-H., Ryan, P. R. & Tyerman, S. D. (2001) Plant Physiol. 125, 1459-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasaki, T., Yamamoto, Y., Ezaki, B., Katsuhara, M., Ahn, S. J., Ryan, P. R., Delhaize, E. & Matsumoto, H. (2004) Plant J. 37, 645-653. [DOI] [PubMed] [Google Scholar]
- 18.Zhao, Z., Ma, J. F., Sato, K. & Takeda, K. (2003) Planta 217, 794-800. [DOI] [PubMed] [Google Scholar]
- 19.Tingay, S., McElroy, D., Kalla, R., Fieg, S., Wang, M., Thornton, S. & Brettell, R. (1997) Plant J. 11, 1369-1376. [Google Scholar]
- 20.Chiu, W.-I., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H. & Sheen, J. (1996) Curr. Biol. 6, 325-330. [DOI] [PubMed] [Google Scholar]
- 21.Wang, M.-B., Li, Z.-Y., Upadhyaya, N. & Waterhouse, P. M. (1998) Acta Hort. 461, 401-405. [Google Scholar]
- 22.Schenk, P. M., Kazan, K., Wilson, I., Anderson, J. P., Richmond, T., Somerville, S. C. & Manners, J. M. (2000) Proc. Natl. Acad. Sci. USA 97, 11655-11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan, P. R., Delhaize, E. & Randall, P. J. (1995) Planta 196, 103-110. [Google Scholar]
- 24.Culvenor, R. A., Wood, J. T., Avery, A. L., Dempsey, W., McDonald, S. E., Ronnfeldt, G. & Veness, P. E. (2004) Aust. J. Agric. Res. 55, 681-692. [Google Scholar]
- 25.Ryan, P. R., DiTomaso, J. M. & Kochian, L. V. (1993) J. Exp. Bot. 44, 437-446. [Google Scholar]
- 26.Kataoka, T., Stekelenburg, A., Nakanishi, T. M., Delhaize, E. & Ryan, P. R. (2001) Plant Cell Environ. 25, 453-460. [Google Scholar]
- 27.Polle, E., Konzak, C. F. & Kittrick, J. A. (1978) Crop Sci. 18, 823-827. [Google Scholar]
- 28.Schunmann, P. H. D., Surin, B. & Waterhouse, P. M. (2003) Funct. Plant Biol. 30, 453-460. [DOI] [PubMed] [Google Scholar]
- 29.Tang, Y., Sorrells, M. E., Kochian, L. V. & Garvin, D. F. (2000) Crop Sci. 40, 778-782. [Google Scholar]
- 30.Raman, H., Moroni, J. S., Sato, K., Read, B. J. & Scott, B. J. (2002) Theor. Appl. Genet. 105, 458-464. [DOI] [PubMed] [Google Scholar]
- 31.Scott, B. J., Conyers, M. K., Poile, G. J. & Cullis, B. R. (1997) Aust. J. Agric. Res. 48, 843-854. [Google Scholar]
- 32.Tang, C., Diatloff, E., Rengel, Z. & McGann, B. (2001) Plant Soil 236, 1-10. [Google Scholar]
- 33.Delhaize, E., Kataoka, T., Hebb, D. M., White, R. G. & Ryan, P. R. (2003) Plant Cell 15, 1131-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]