Abstract
Various chemotherapies and radiation therapies are useful for killing cancer cells mainly by inducing DNA double-strand breaks (DSBs). Uncovering the molecular mechanisms of DSB repair processes is crucial for developing next-generation radiotherapies and chemotherapeutics for human and animal cancers. XRCC4 plays a critical role in Ku-dependent nonhomologous DNA-end joining (NHEJ) in human cells, and is one of the core NHEJ factors. The localization of core NHEJ factors, such as human Ku70 and Ku80, might play a crucial role in regulating NHEJ activity. Recently, companion animals, such as canines, have been proposed to be a good model in many aspects of cancer research. However, the localization and regulation mechanisms of core NHEJ factors in canine cells have not been elucidated. Here, we show that the expression and subcellular localization of canine XRCC4 changes dynamically during the cell cycle. Furthermore, EYFP-canine XRCC4 accumulates quickly at laser-microirradiated DSB sites. The structure of a putative human XRCC4 nuclear localization signal (NLS) is highly conserved in canine, chimpanzee and mouse XRCC4. However, the amino acid residue corresponding to the human XRCC4 K210, thought to be important for nuclear localization, is not conserved in canine XRCC4. Our findings might be useful for the study of the molecular mechanisms of Ku-dependent NHEJ in canine cells and the development of new radiosensitizers that target XRCC4.
Keywords: canine, companion animal, DNA damage, Ku, XRCC4
Resistance to radiation or chemotherapeutics is a common clinical problem when treating people and pets with cancer. The lethal effect of photon radiation, such as X-ray, carbon ion radiation and various chemotherapeutics, on cancer cells is primarily caused by DNA double-strand breaks (DSBs). Nonhomologous DNA-end joining (NHEJ) is a major DSB repair pathway in animal and human cells, and might repair most DSB produced by radiation and chemotherapeutics in cancer and normal cells [4, 22]. Therefore, to elucidate the molecular mechanisms of the DSB repair process, especially NHEJ, it is important to develop next-generation radiotherapies and novel chemotherapeutics for animal and human cancer.
NHEJ does not require an intact template and might be independent of the cell cycle [4, 22]. NHEJ starts with the binding of Ku, which consists of Ku70 and Ku80, to DSB ends. The control mechanism for subcellular localization and/or heterodimerization of Ku70 and Ku80 might play a key role in regulating the physiological functions of Ku [11, 13]. NHEJ requires other core factors including Artemis, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), DNA Ligase IV, XRCC4-like factor (XLF; also called Cernunnos or NHEJ1) and X-ray repair cross-complementing protein 4 (XRCC4) [4, 22]. Recently, PAralog of XRCC4 and XLF (PAXX; also called C9orf142 or XLS) was suggested as a new factor involved in NHEJ [2, 25, 31]. Human Ku70 and Ku80 accumulate at micro-laser-induced DSB sites following irradiation, and heterodimerization of Ku70 and Ku80 might be essential for the recruitment of some core NHEJ factors including DNA-PKcs, XLF and XRCC4 [12, 15, 22, 26]. However, dimerization of Ku70 and Ku80 is not required to recruit another core NHEJ factor, Artemis, or the HR-related protein Rad52, to DSB sites [15, 17]. The mechanism by which recruitment of core NHEJ factors to DSB sites occurs in canine cells has not been clarified.
The XRCC4/DNA Ligase IV complex is crucial for the final ligation of DSBs through NHEJ [22]. The XRCC4 homodimer stabilizes DNA Ligase IV by binding to a single molecule [29]. It was experimentally observed that absence of XRCC4 results in an impaired ability to repair DSB in rodent cells making it an indispensable factor in NHEJ [7, 21, 22]. Recently, some groups have reported that missense and truncation mutations in XRCC4 cause a human disease including primordial dwarfism [1, 3, 8, 24, 27, 28]. Expectedly, XRCC4-defective patient fibroblasts showed radiosensitivity and diminished DSB repair capacity.
Companion animals, such as canines, are proposed to be a good cancer model for both veterinary and human medical cancer research [10, 30]. The localization of a core NHEJ factor, such as XRCC4, might play a critical role in regulating NHEJ. Thus, it is important to elucidate the mechanism of XRCC4 accumulation at DSB sites. To our knowledge, there are currently no reports on the localization of XRCC4 of companion animal species including the canine. Here, we examined the expression and subcellular localization of canine XRCC4 in canine cells. Additionally, we cloned XRCC4 cDNA from a beagle dog testis library and examined whether canine XRCC4 accumulates at DNA damage sites quickly after laser-microirradiation.
MATERIALS AND METHODS
Cloning of canine XRCC4: Oligonucleotide primers to amplify canine XRCC4 cDNA from male Beagle dog cDNA library (Biochain, Newark, CA, U.S.A.) were designed based on the predicted XRCC4 genomic sequence of female Boxer dog, Canis lupus familiaris (XM_546040.1). EcoR1 and BamH1 restriction enzyme sites were appended on the 5’ end of the sense and antisense primers, respectively. PCR amplification with sense (5′-CGAATTCGATGGAGAGAAAAGTAAGCAGAA-3′) and antisense (5′-CGGATCCTTAAATCTCATCAAAGAGGTCTT-3′) primers was carried out for 30 cycles in a Thermal Cycler PC-700 (ASTEC, Fukuoka, Japan) using LA Taq polymerase (Takara Bio Inc., Otsu, Japan). After pre-denaturing (94°C for 5 min), each cycle consisted of denaturation at 94°C for 1 min, annealing at 56°C for 1 min and extension at 72°C for 1 min, followed by a final extension (4 min). Other PCR primers used in this study were as follows: F1: 5′-TTTCGGAGGAGGCCGGAAGT-3′, R1: 5′-ATCGGCTTCTTGGGAAATCT-3′, F2: 5′-GCGGATGCAAAAAAATCTTGGAAC-3′, and R2: 5′-GAAGATAGATAAGTAAATGGGACACGG-3′. The PCR product was subcloned into pCR4-TOPO vector (Invitrogen, Carlsbad, CA, U.S.A.), and the nucleotide sequences was determined by sequencing.
Cell lines, cultures and transfections: A Madin-Darby canine kidney cell line (MDCK) (HSRRB, Osaka, Japan), a human cervical carcinoma cell line (HeLa) (Riken Cell Bank, Tsukuba, Japan), human colon cancer cell line (HCT116) (Riken Cell Bank), a murine embryonic fibroblast cell line NIH3T3 (Riken Cell Bank) and a murine lung epithelial cell line (Ku70 +/− MLE) previously established [15] were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS). XRCC4 cDNA from the pCR4-canine XRCC4 was subcloned into the EcoRI and BamHI sites of pEYFP-C1 to produce the in-frame fusion gene. pEYFP-canine XRCC4 or pEYFP-C1 was transiently transfected in cells using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol. Cells were cultured for 2 days and then monitored under an FV300 confocal laser scanning microscope (Olympus, Tokyo, Japan) as previously described [12, 15, 16].
Immunofluorescence staining: Immunofluorescence staining was conducted as previously described [12, 14, 15] with the following modifications. Briefly, the fixed cells were blocked using a blocking solution and then incubated for 30 min at room temperature with mouse anti-γH2AX monoclonal antibody (JBW301) (Upstate Biotechnology Inc., Charlottesville, VA, U.S.A.) or goat anti-XRCC4 polyclonal antibody (C-20) (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.). After washing with PBS, detection of each protein was performed using Alexa fluor 488- or 568-conjugated secondary antibodies (Molecular Probes, OR, U.S.A.).
Immunoblotting: The extraction of total cell lysates and Western blot analysis were conducted as described previously [14, 19] with the following modifications. The membranes were blocked in Blocking One (Nacalai Tesque, Kyoto, Japan) for 30 min. The following antibodies were used: goat anti-XRCC4 polyclonal antibody (C-20), rabbit anti-GFP polyclonal antibody (FL) (Santa Cruz Biotechnology) or mouse anti-β-actin monoclonal antibody (Sigma, St. Louis, MO, U.S.A.). The anti-XRCC4 and anti-GFP antibodies, were diluted in Signal Enhancer HIKARI (Nacalai Tesque). In accordance with the manufacturer’s instructions, the binding to each protein was detected using a Select Western blotting detection system (GE Healthcare Bio-Sci. Corp.) and visualized using the ChemiDoc XRS system (Bio-Rad, Hercules, CA, U.S.A.).
Local DNA damage induction using laser and cell imaging: Local DNA damage induction using laser and subsequent cell imaging was conducted as described previously [12, 15, 17]. Briefly, local DSB were induced using a 5–30% power scan (for 1 s) from a 405 nm laser. Images of living or fixed cells expressing EYFP-canine XRCC4 proteins or EYFP alone were obtained using an FV300 confocal scanning laser microscopy system (Olympus).
RESULTS
Expression and subcellular localization of canine XRCC4: We examined the expression and subcellular localization of XRCC4 in canine cells. Originally, there have been reports on the expression of XRCC4 in human and mouse cells [21]. First, we examined the expression of XRCC4 in the canine cell line MDCK, the two human cell lines HeLa and HCT116, and the two mouse cell lines NIH3T3 and Ku70 +/− MLE by Western blotting using the anti-XRCC4 antibody. As shown in Fig. 1A, a signal of canine XRCC4 as well as human and mouse XRCC4 was detected. These results demonstrate that XRCC4 is expressed in canine cells. In addition, our data showed that the electrophoretic mobility of canine XRCC4 was different from that of human and mouse XRCC4, suggesting the possibility that the structure and/or post-translational modifications of canine XRCC4 are different from those of human and/or mouse XRCC4. Next, immunofluorescence analysis was performed in MDCK cells to examine the subcellular localization of XRCC4 in canine cells (Fig. 1B and 1C). Indirect immunofluorescence staining using an anti-XRCC4 antibody showed that fluorescence was weaker in mitotic cells than in interphase cells. During interphase, fluorescence was detected in the nucleoplasm and was excluded from the nucleolus. During mitosis, fluorescence was detected throughout the cytoplasm of MDCK cells, but not localized to mitotic chromosomes. These results suggest that the expression and localization of canine XRCC4 change dynamically during the cell cycle.
Fig. 1.
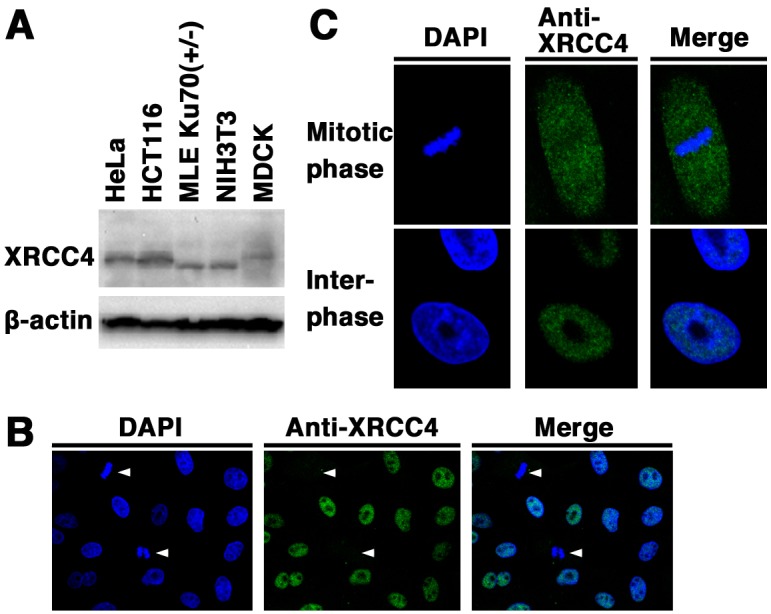
Expression and subcellular localization of XRCC4 in canine cells. (A) Total cell lysates from the canine cell line (MDCK), two human cell lines (HeLa and HCT116), and two mouse cell lines (NIH3T3 and Ku70 +/− MLE) were analyzed by Western blotting using an anti-XRCC4 antibody or an anti-β-actin antibody. (B, C) Subcellular localization of XRCC4 in canine (MDCK) cells. The cells were fixed and stained with an anti-XRCC4 antibody. Nuclear DNA was counterstained with DAPI. The stained cells were analyzed by confocal laser microscopy. Left panel, DAPI image; center panel, XRCC4 image; right panel, merged image. Arrowheads indicate mitotic phase cells (B). The images shown are a representative example for interphase cells or mitotic phase cells (C).
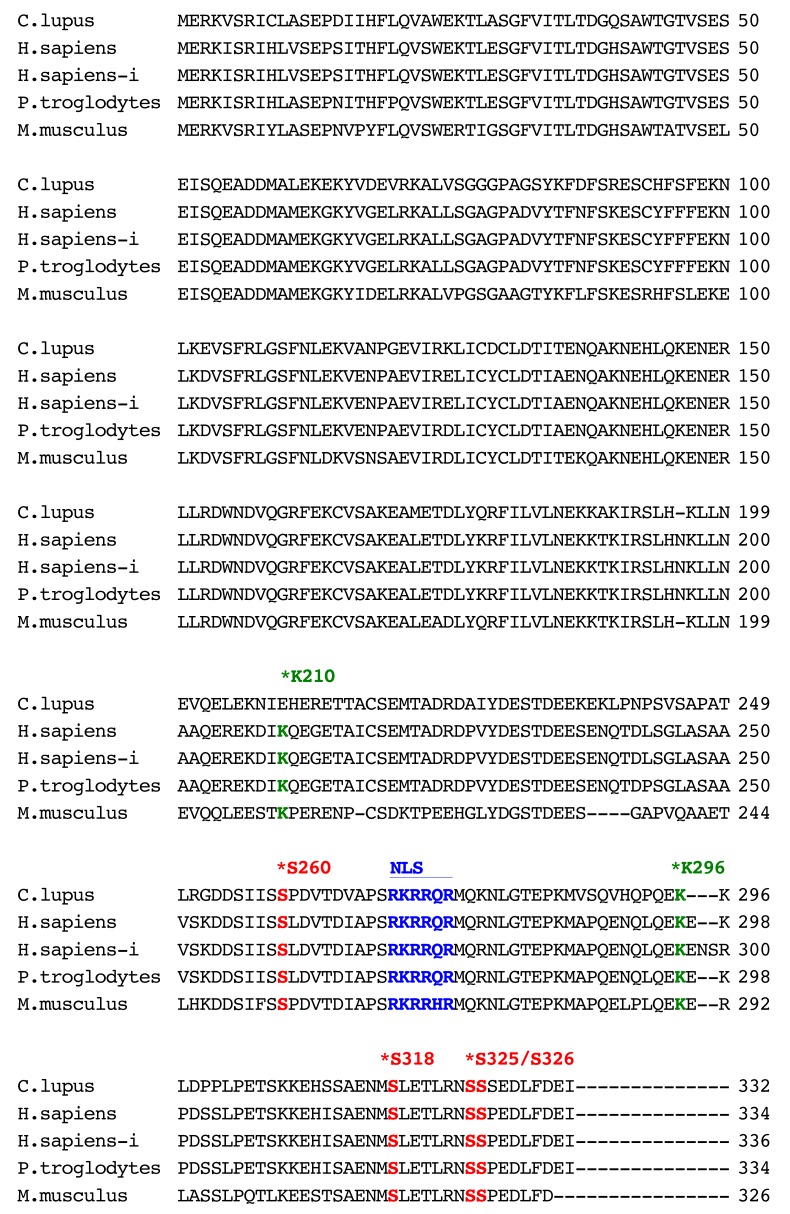
Sequence analysis of canine XRCC4: The canine XRCC4 cDNA was cloned and sequenced from a beagle dog testis library. We isolated a 999-nucleotide open-reading-frame encoding a protein of 332 amino acids (Fig. 2). The cDNA sequence obtained from the Beagle dog was identical to that predicted sequence from a female Boxer dog genomic sequence (XM_546040.1). The obtained canine sequence has been submitted to the DDBJ/ENA/NCBI database [accession number LC168634]. Human XRCC4 was originally identified as a protein which consists of 334 amino acids [21]. As shown in Fig. 2, there is another splice variant form encoding human XRCC4 isoform of 336 amino acids (labeled H. sapiens-i). Comparative analysis of XRCC4 sequences showed that canine XRCC4 had 79.3%, 78.6%, 79.6% and 73.3% amino acid identity with human (H. sapiens: AAC50339.1), human isoform (H. sapiens-i: NP_071801.1), chimpanzee (XP_001148110.2) and mouse (NP_082288.1), respectively. Post-translational modifications of DNA repair proteins including phosphorylation, ubiquitylation and SUMOylation, might play an important role in the regulation of various DNA repair pathways. We found that the DNA-PK major phosphorylation sites (S260, S318, S325 and S326) and a polyubiquitylation site (K296) of human XRCC4 [20, 32, 34] are evolutionarily conserved in canine, chimpanzee and mouse XRCC4. It is reported that human XRCC4 has a putative nuclear localization signal (NLS) sequence, RKRRQR, at amino acid positions 270–275 [21]. We found that the putative NLS sequence is conserved in canine XRCC4, as well as in chimpanzee and mouse. Human XRCC4 is modified with the small ubiquitin-like modifier SUMO at Lys210, and this SUMOylation is important for the nuclear localization of XRCC4 [33]. Interestingly, the amino acid sequence corresponding to the Lys210 in human XRCC4 is not conserved in canine XRCC4 (Fig. 2).
Fig. 2.
Amino acid sequences of XRCC4 from canine (Canis lupus familiaris, GenBank accession number: LC168634), human (Homo sapiens, GenBank accession number: AAC50339.1), human isoform (H.sapience-i) (Homo sapiens, GenBank accession number: NP_071801.1), chimpanzee (Pan troglodytes, GenBank accession number: XP_001148110.2) and mouse (Mus musculus, GenBank accession number: NP_082288.1) species. The location of a putative nuclear localization signal (NLS) sequence in human XRCC4 [21]. Stars mark the location of the SUMO modification site (K210), polyubiquitylation site (K296) and DNA-PK phosphorylation sites (S260, S318, S325 and S326) in the human sequences (AAC50339.1)) [20, 32, 34].
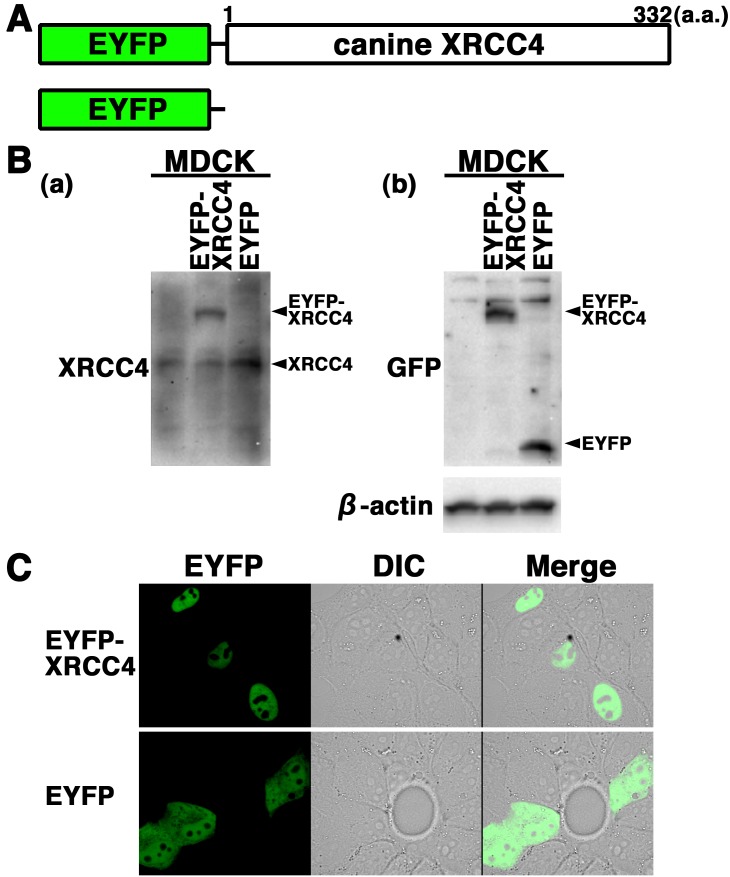
EYFP-canine XRCC4 accumulates quickly at laser-microirradiation-induced DSBs: To further examine the expression and localization of XRCC4 in living canine cells, we generated cells transiently expressing EYFP-canine XRCC4 in MDCK cells. The expression vector pEYFP-C1 containing canine XRCC4 (pEYFP-canine XRCC4) was transfected into MDCK cells (Fig. 3A). Western blotting using anti-XRCC4 and anti-GFP antibodies showed that EYFP-canine XRCC4 was expressed in the transfectants (Fig. 3B). Confocal laser microscopy showed that EYFP-canine XRCC4 was localized to the nuclei of living interphase cells in EYFP-canine XRCC4 transfectants (Fig. 3C). EYFP, used as a control, was distributed throughout the cell excluding the nucleolus in EYFP transfectants (Fig. 3C).
Fig. 3.
Subcellular localization of EYFP-canine XRCC4 in living canine cells. (A) Schematics of EYFP-canine XRCC4 chimeric protein and control protein (EYFP). (B) EYFP-canine XRCC4 was expressed in canine (MDCK) cells, and the expression of EYFP-canine XRCC4 was examined by Western blotting using the anti-XRCC4, anti-GFP or anti-β-actin antibody. (C) Imaging of living EYFP-canine XRCC4-transfected cells. Living MDCK cells transiently expressing EYFP-canine XRCC4 or EYFP were analyzed by confocal laser microscopy. EYFP images for the same cells are shown alone (left panel) or merged (right panel) with differential interference contrast images (DIC) (center panel).
We next examined whether EYFP-canine XRCC4 accumulates quickly at 405 nm laser-induced DSB sites in canine cells (Fig. 4A). EYFP-canine XRCC4, but not EYFP alone, accumulated at the microirradiated sites in living MDCK cells (Fig. 4B). To determined whether EYFP-canine XRCC4 accumulated at 405 nm laser-induced DSB sites, we immunostained cells with an antibody that detects γH2AX, a marker of DSBs. EYFP-canine XRCC4 colocalized with γH2AX at microirradiated sites in MDCK cells (Fig. 4C). To investigate the temporal dynamics of XRCC4 localization, we performed time-lapse imaging of EYFP-canine XRCC4 transfected MDCK cells. We observed EYFP-canine XRCC4 accumulation at the microirradiated sites 5 sec after irradiation, and the intensity of the EYFP signal increased quickly at the microirradiated sites (Fig. 4D). These results reveal that after irradiation, EYFP-canine XRCC4 quickly accumulates and forms foci at laser-induced DSBs in living cells.
Fig. 4.
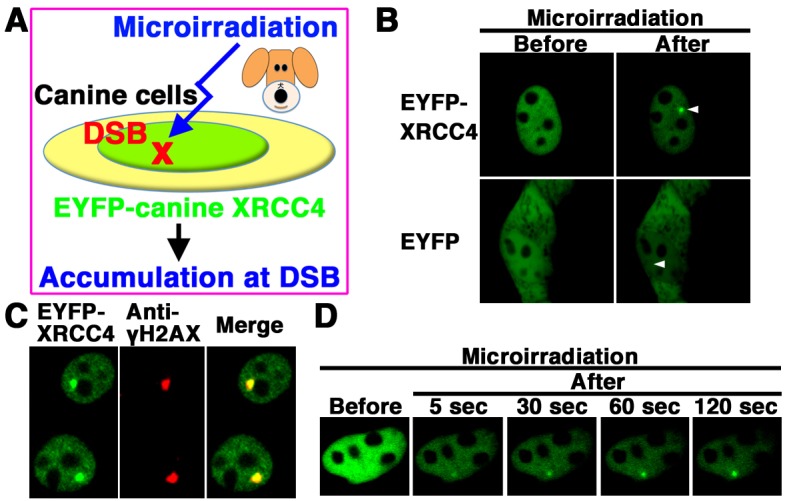
EYFP-canine XRCC4 accumulated quickly at DSBs induced by laser microirradiation. (A) The localization and accumulation of EYFP-canine XRCC4 at DSBs induced by 405 nm laser irradiation were examined. (B) Imaging of living EYFP-canine XRCC4-transfected MDCK cells before (left panel) and after (right panel) microirradiation. Upper panel, EYFP-canine XRCC4-transfected cells; Lower panel, EYFP-transfected cells. Arrowheads indicate the microirradiated sites. (C) Immunostaining of microirradiated EYFP-canine XRCC4-transfected cells with anti-γH2AX antibody. Cells were fixed and stained with anti-γH2AX antibody at 5 min postirradiation. Left panel, EYFP-canine XRCC4; Center panel, γH2AX image; Right panel, merged image. (D) Time-dependent EYFP-canine XRCC4 accumulation in living cells (5-120 s) after irradiation.
DISCUSSION
To develop next-generation chemoradiotherapies and chemotherapeutics for cancers is important to understand the molecular mechanisms of DNA repair process including NHEJ. XRCC4 plays a critical role in the final ligation step of NHEJ at DSB [24]. The regulation of XRCC4 localizations might play a key role in regulating NHEJ activity. However, there have been no reports on the expression and localization of XRCC4 in canine cells. Here, we examined the expression and subcellular localization of canine XRCC4. We found that the expression and localization of canine XRCC4 change dynamically during the cell cycle. Canine XRCC4 was cloned from a beagle dog testis library, and sequence alignment indicated that the putative NLS sequence of human XRCC4 is conserved in canine, chimpanzee and mouse XRCC4. Transient expression of EYFP-tagged canine XRCC4 showed that canine XRCC4 was localized within the nucleus of interphase cells and accumulated at laser-irradiated DSB sites in canine MDCK cells. These observations clarify the regulation mechanism of XRCC4 localization and further our understanding of the molecular mechanisms of Ku-dependent NHEJ in canine cells.
Human XRCC4 contains many post-translational modification sites. Human XRCC4 is a nuclear phosphoprotein that is phosphorylated by DNA-PK in response to radiation in vivo [23, 34]. Recently, Zhang et al. (2016) reported that DNA-PK phosphorylates XRCC4 at S325/S326, which promotes the binding of XRCC4 to FBXW7 [34]. SCFFBXW7 E3 ligase then promotes polyubiquitylation of XRCC4 at K296 via K63 linkage for enhanced association with the Ku70/Ku80 complex to facilitate NHEJ repair. Sequence alignments indicated that DNA-PK major phosphorylation sites of human XRCC4 (S260, S318, S325 and S326) and a polyubiquitylation site (K296) are evolutionarily conserved in canine, chimpanzee and mouse XRCC4. However, the equivalent of the human XRCC4 residue K210, which is modified with SUMO, is not conserved in canine XRCC4. We speculate that phosphorylation and ubiquitylation, but not SUMOylation, post-translational modifications of canine XRCC4 play a critical role in the regulation of canine NHEJ, although further studies need to confirm this. Further studies would clarify the relative importance of different post-translational modifications in canine NHEJ
The nuclear localization of human XRCC4 is regulated by the XRCC4 NLS (amino acids 270-275), SUMO modification at K210 and regulation by DNA Ligase IV [5, 21, 33]. Yurchenko et al. (2006) showed that SUMOylation at K210 is necessary and sufficient for the nuclear localization of XRCC4 [33]. However, Fukuchi et al. (2015) have recently reported that mutation of the SUMOylation site (K210) had no effect on the nuclear localization of human XRCC4 [6]. Additionally, they indicated that the amino acid sequence surrounding K210 in human XRCC4 is divergent even among mammalian species, e.g., E. caballus and B. Taurus. Here, we showed that canine XRCC4 is localized to the nuclei of interphase cells and that localization of canine XRCC4 changes dynamically during the cell cycle. Additionally, we showed that the structure of the putative human XRCC4 NLS is highly conserved in canine XRCC4. However, the amino acid sequence corresponding to the K210 of human XRCC4 is not conserved in canine XRCC4. While the role of the SUMOylation site (K210) of XRCC4 remains controversial in human cells, we speculate that SUMOylation might not be important for the nuclear localization of canine XRCC4. Altogether, we consider that the structure of a putative XRCC4 NLS might be vital for the nuclear localization of canine XRCC4.
The veterinary/human medical collaboration has resulted in improved oncological outcomes for both canine and human [30]. An attractive approach to chemotherapy is the design of drugs that target core DNA repair factors including NHEJ [9]. Additionally, NHEJ is an attractive target for strategies aimed at increasing the sensitivity of tumors to anticancer treatments including various radiotherapies. The regulation mechanism controlling the localization of core NHEJ factors, including Ku, plays a critical role in controlling NHEJ activity [11, 18]. Here, we demonstrated that EYFP-canine XRCC4 accumulation at micro-laser induced DSBs began immediately after irradiation. This is a first report examining the localization and accumulation of core NHEJ factors at DSB sites in living cells of companion animal species, such as canine. Our findings provide the foundation for further studies to develop drugs using XRCC4, and other core NHEJ factors, as target molecules in both canine and human.
Acknowledgments
This work was supported in part by JSPS KAKENHI Grant Number JP26450438.
REFERENCES
- 1.Bee L., Nasca A., Zanolini A., Cendron F., d’Adamo P., Costa R., Lamperti C., Celotti L., Ghezzi D., Zeviani M.2015. A nonsense mutation of human XRCC4 is associated with adult-onset progressive encephalocardiomyopathy. EMBO Mol. Med. 7: 918–929. doi: 10.15252/emmm.201404803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Craxton A., Somers J., Munnur D., Jukes-Jones R., Cain K., Malewicz M.2015. XLS (c9orf142) is a new component of mammalian DNA double-stranded break repair. Cell Death Differ. 22: 890–897. doi: 10.1038/cdd.2015.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Bruin C., Mericq V., Andrew S. F., van Duyvenvoorde H. A., Verkaik N. S., Losekoot M., Porollo A., Garcia H., Kuang Y., Hanson D., Clayton P., van Gent D. C., Wit J. M., Hwa V., Dauber A.2015. An XRCC4 splice mutation associated with severe short stature, gonadal failure, and early-onset metabolic syndrome. J. Clin. Endocrinol. Metab. 100: E789–E798. doi: 10.1210/jc.2015-1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Downs J. A., Jackson S. P.2004. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 5: 367–378. doi: 10.1038/nrm1367 [DOI] [PubMed] [Google Scholar]
- 5.Francis D. B., Kozlov M., Chavez J., Chu J., Malu S., Hanna M., Cortes P.2014. DNA Ligase IV regulates XRCC4 nuclear localization. DNA Repair (Amst.) 21: 36–42. doi: 10.1016/j.dnarep.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuchi M., Wanotayan R., Liu S., Imamichi S., Sharma M. K., Matsumoto Y.2015. Lysine 271 but not lysine 210 of XRCC4 is required for the nuclear localization of XRCC4 and DNA ligase IV. Biochem. Biophys. Res. Commun. 461: 687–694. doi: 10.1016/j.bbrc.2015.04.093 [DOI] [PubMed] [Google Scholar]
- 7.Gao Y., Sun Y., Frank K. M., Dikkes P., Fujiwara Y., Seidl K. J., Sekiguchi J. M., Rathbun G. A., Swat W., Wang J., Bronson R. T., Malynn B. A., Bryans M., Zhu C., Chaudhuri J., Davidson L., Ferrini R., Stamato T., Orkin S. H., Greenberg M. E., Alt F. W.1998. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell 95: 891–902. doi: 10.1016/S0092-8674(00)81714-6 [DOI] [PubMed] [Google Scholar]
- 8.Guo C., Nakazawa Y., Woodbine L., Björkman A., Shimada M., Fawcett H., Jia N., Ohyama K., Li T. S., Nagayama Y., Mitsutake N., Pan-Hammarström Q., Gennery A. R., Lehmann A. R., Jeggo P. A., Ogi T.2015. XRCC4 deficiency in human subjects causes a marked neurological phenotype but no overt immunodeficiency. J. Allergy Clin. Immunol. 136: 1007–1017. doi: 10.1016/j.jaci.2015.06.007 [DOI] [PubMed] [Google Scholar]
- 9.Jekimovs C., Bolderson E., Suraweera A., Adams M., O’Byrne K. J., Richard D. J.2014. Chemotherapeutic compounds targeting the DNA double-strand break repair pathways: the good, the bad, and the promising. Front. Oncol. 4: 86. doi: 10.3389/fonc.2014.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khanna C., Lindblad-Toh K., Vail D., London C., Bergman P., Barber L., Breen M., Kitchell B., McNeil E., Modiano J. F., Niemi S., Comstock K. E., Ostrander E., Westmoreland S., Withrow S.2006. The dog as a cancer model. Nat. Biotechnol. 24: 1065–1066. doi: 10.1038/nbt0906-1065b [DOI] [PubMed] [Google Scholar]
- 11.Koike M.2002. Dimerization, translocation and localization of Ku70 and Ku80 proteins. J. Radiat. Res. (Tokyo) 43: 223–236. doi: 10.1269/jrr.43.223 [DOI] [PubMed] [Google Scholar]
- 12.Koike M., Koike A.2008. Accumulation of Ku80 proteins at DNA double-strand breaks in living cells. Exp. Cell Res. 314: 1061–1070. doi: 10.1016/j.yexcr.2007.11.014 [DOI] [PubMed] [Google Scholar]
- 13.Koike M., Awaji T., Kataoka M., Tsujimoto G., Kartasova T., Koike A., Shiomi T.1999. Differential subcellular localization of DNA-dependent protein kinase components Ku and DNA-PKcs during mitosis. J. Cell Sci. 112: 4031–4039. [DOI] [PubMed] [Google Scholar]
- 14.Koike M., Shiomi T., Koike A.2001. Dimerization and nuclear localization of Ku proteins. J. Biol. Chem. 276: 11167–11173. doi: 10.1074/jbc.M010902200 [DOI] [PubMed] [Google Scholar]
- 15.Koike M., Yutoku Y., Koike A.2011. Accumulation of Ku70 at DNA double-strand breaks in living epithelial cells. Exp. Cell Res. 317: 2429–2437. doi: 10.1016/j.yexcr.2011.07.018 [DOI] [PubMed] [Google Scholar]
- 16.Koike M., Yutoku Y., Koike A.2011. Accumulation of p21 proteins at DNA damage sites independent of p53 and core NHEJ factors following irradiation. Biochem. Biophys. Res. Commun. 412: 39–43. doi: 10.1016/j.bbrc.2011.07.032 [DOI] [PubMed] [Google Scholar]
- 17.Koike M., Yutoku Y., Koike A.2013. The C-terminal region of Rad52 is essential for Rad52 nuclear and nucleolar localization, and accumulation at DNA damage sites immediately after irradiation. Biochem. Biophys. Res. Commun. 435: 260–266. doi: 10.1016/j.bbrc.2013.04.067 [DOI] [PubMed] [Google Scholar]
- 18.Koike M., Yutoku Y., Koike A.2014. Impact of amino acid substitutions in two functional domains of Ku80: DNA-damage-sensing ability of Ku80 and survival after irradiation. J. Vet. Med. Sci. 76: 51–56. doi: 10.1292/jvms.13-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koike M., Yutoku Y., Koike A.2015. Dynamic changes in subcellular localization of cattle XLF during cell cycle, and focus formation of cattle XLF at DNA damage sites immediately after irradiation. J. Vet. Med. Sci. 77: 1109–1114. doi: 10.1292/jvms.14-0516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee K. J., Jovanovic M., Udayakumar D., Bladen C. L., Dynan W. S.2004. Identification of DNA-PKcs phosphorylation sites in XRCC4 and effects of mutations at these sites on DNA end joining in a cell-free system. DNA Repair (Amst.) 3: 267–276. doi: 10.1016/j.dnarep.2003.11.005 [DOI] [PubMed] [Google Scholar]
- 21.Li Z., Otevrel T., Gao Y., Cheng H. L., Seed B., Stamato T. D., Taccioli G. E., Alt F. W.1995. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell 83: 1079–1089. doi: 10.1016/0092-8674(95)90135-3 [DOI] [PubMed] [Google Scholar]
- 22.Mahaney B. L., Meek K., Lees-Miller S. P.2009. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 417: 639–650. doi: 10.1042/BJ20080413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumoto Y., Suzuki N., Namba N., Umeda N., Ma X. J., Morita A., Tomita M., Enomoto A., Serizawa S., Hirano K., Sakaia K., Yasuda H., Hosoi Y.2000. Cleavage and phosphorylation of XRCC4 protein induced by X-irradiation. FEBS Lett. 478: 67–71. doi: 10.1016/S0014-5793(00)01800-7 [DOI] [PubMed] [Google Scholar]
- 24.Murray J. E., van der Burg M., IJspeert H., Carroll P., Wu Q., Ochi T., Leitch A., Miller E. S., Kysela B., Jawad A., Bottani A., Brancati F., Cappa M., Cormier-Daire V., Deshpande C., Faqeih E. A., Graham G. E., Ranza E., Blundell T. L., Jackson A. P., Stewart G. S., Bicknell L. S.2015. Mutations in the NHEJ component XRCC4 cause primordial dwarfism. Am. J. Hum. Genet. 96: 412–424. doi: 10.1016/j.ajhg.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochi T., Blackford A. N., Coates J., Jhujh S., Mehmood S., Tamura N., Travers J., Wu Q., Draviam V. M., Robinson C. V., Blundell T. L., Jackson S. P.2015. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 347: 185–188. doi: 10.1126/science.1261971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynolds P., Botchway S. W., Parker A. W., O’Neill P.2013. Spatiotemporal dynamics of DNA repair proteins following laser microbeam induced DNA damage - when is a DSB not a DSB? Mutat. Res. 756: 14–20. doi: 10.1016/j.mrgentox.2013.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosin N., Elcioglu N. H., Beleggia F., Isgüven P., Altmüller J., Thiele H., Steindl K., Joset P., Rauch A., Nürnberg P., Wollnik B., Yigit G.2015. Mutations in XRCC4 cause primary microcephaly, short stature and increased genomic instability. Hum. Mol. Genet. 24: 3708–3717. [DOI] [PubMed] [Google Scholar]
- 28.Shaheen R., Faqeih E., Ansari S., Abdel-Salam G., Al-Hassnan Z. N., Al-Shidi T., Alomar R., Sogaty S., Alkuraya F. S.2014. Genomic analysis of primordial dwarfism reveals novel disease genes. Genome Res. 24: 291–299. doi: 10.1101/gr.160572.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sibanda B. L., Critchlow S. E., Begun J., Pei X. Y., Jackson S. P., Blundell T. L., Pellegrini L.2001. Crystal structure of an Xrcc4-DNA ligase IV complex. Nat. Struct. Biol. 8: 1015–1019. doi: 10.1038/nsb725 [DOI] [PubMed] [Google Scholar]
- 30.Withrow S. J., Wilkins R. M.2010. Cross talk from pets to people: translational osteosarcoma treatments. ILAR J. 51: 208–213. doi: 10.1093/ilar.51.3.208 [DOI] [PubMed] [Google Scholar]
- 31.Xing M., Yang M., Huo W., Feng F., Wei L., Jiang W., Ning S., Yan Z., Li W., Wang Q., Hou M., Dong C., Guo R., Gao G., Ji J., Zha S., Lan L., Liang H., Xu D.2015. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat. Commun. 6: 6233. doi: 10.1038/ncomms7233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu Y., Wang W., Ding Q., Ye R., Chen D., Merkle D., Schriemer D., Meek K., Lees-Miller S. P.2003. DNA-PK phosphorylation sites in XRCC4 are not required for survival after radiation or for V(D)J recombination. DNA Repair (Amst.) 2: 1239–1252. doi: 10.1016/S1568-7864(03)00143-5 [DOI] [PubMed] [Google Scholar]
- 33.Yurchenko V., Xue Z., Sadofsky M. J.2006. SUMO modification of human XRCC4 regulates its localization and function in DNA double-strand break repair. Mol. Cell. Biol. 26: 1786–1794. doi: 10.1128/MCB.26.5.1786-1794.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Q., Karnak D., Tan M., Lawrence T. S., Morgan M. A., Sun Y.2016. FBXW7 facilitates nonhomologous end-joining via K63-linked polyubiquitylation of XRCC4. Mol. Cell 61: 419–433. doi: 10.1016/j.molcel.2015.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]