Abstract
To determine the occurrence and genetic diversity of Sapelovirus A (SV-A) in diarrhea and non-diarrhea feces of Korean pigs, 110 specimens from different age groups of pigs in the same farm were analyzed by RT-nested PCR. SV-As were detected in 60% of both diarrhea and non-diarrhea specimens regardless of age groups with primer pairs for 2C region, in which all diarrhea samples were co-infected by other enteric pathogens. Phylogenetical analysis of partial VP1 region showed that our strains and several other Korean strains belonged to cluster I, distinct from some strains reported in Korea and other countries. These data indicate that genetically distinct SV-As are frequently detected in Korean pigs irrespective of diarrhea and age.
Keywords: diarrhea, genetic diversity, occurrence, porcine, Sapelovirus A
Sapelovirus A (SV-A), a member of new genus Sapelovirus within the family Picornaviridae, has typical picornavirus genome organization of 5′ untranslated region (UTR)-L-VP4-VP2-VP3-VP1-2A-2B-2C-3A-3B-3C-3D-3′ UTR [1, 11]. However, the structural genome features within SV-A strains are different, i.e., the cis-acting RNA element (CRE) in the 2C coding region and kissing domain in the 3′UTR between the recent Korean and Chinese strains, and past English and Chinese strains are different [11]. Likewise, SV-A strains isolated from the different countries or continents or even in the same country have antigenic diversities [2]. These differences in antigenic and genomic features could influence the virulence and/or pathogenicity of SV-A strains isolated in the recent and past decades from different regions or countries [11].
SV-As can cause many symptoms ranging from asymptomatic to symptomatic infections including diarrhea, pneumonia, polioencephalomyelitis and reproductive disorders [2]. Although only a limited number of epidemiological studies on SV-A infections in different countries have been reported [3,4,5, 8, 14], the reported prevalence has ranged from 36.6% in the Czech Republic [8] to 69.2% in Japan [5]. A recent Spanish study has demonstrated that SV-A is detected in the fecal samples from asymptomatic pigs and its infection has an age predilection to postweaning pigs [10, 14, 15]. In Korea, SV-A infections are common in pigs with diarrhea [12]. However, the occurrence and genetic diversity of SV-As in fecal samples from asymptomatic pigs and age predilection in both diarrhea and asymptomatic pigs are largely unknown. Therefore, the aim of this study was to determine the occurrence and genetic diversity of SV-As as well as its age predilections in both diarrhea and healthy Korean pigs from different age groups.
To determine the exact occurrence of SV-A infections across all age groups and their association with diarrhea, 110 fecal specimens from diarrheic or non-diarrheic pigs from previously SV-A positive farm [12] were collected during the winter of 2011 (Table 1). The ages and number of pigs (age of pigs at sample collection; no. of collected samples [diarrheic/non-diarrheic]) tested were as follows: suckling (≤3 weeks; n=30 [15/15]), weaned (≤8 weeks; n=20 [10/10]), grower (≤20 weeks; n=20 [10/10]), finisher (≥20 weeks; n=20 [10/10]) and sows (≥1 year; n=20 [10/10]). To detect SV-A RNA in each fecal sample, RT-nested PCR assays with primer pairs specific to VP1 or 2C regions were performed with extracted RNA [12]. In addition, other common enteric pathogens including viruses and bacteria were screened by RT-nested PCR, PCR or specific agar media [12]. To determine the genetic diversity of SV-A in each age group, nested PCR products for partial VP1 (542 bp) region were purified using GenClean II kit (BIO 101, LaJolla, CA, U.S.A.). DNA sequencing was carried out using an automated DNA sequencer (ABI system 3700; Applied Biosystem, Foster City, CA, U.S.A.) [12]. Using DNA Basic module (DNAsis MAX, Alameda, CA, U.S.A.), the partial nucleotide sequences of VP1 (500 bp, devoid of primer pair sequences) were compared to those of other known SV-A sequences. Phylogenetic analyses based on nucleotide alignments were conducted using MEGA 6 software package [13]. Genetic distances between our strains and other reference strains were calculated using Kimura-2 correction parameter at the nucleotide level. Phylogenetic trees were constructed using neighbor-joining method with 1,000 bootstrap replicates. Data analysis was performed with Fisher’s exact test at confidence level of 95% (P<0.05) using SPSS Statistics version 20 for Windows (SPSS, Chicago, IL, U.S.A.).
Table 1. Detection rates of Sapelovirus A in pigs with diarrhea or non-diarrhea in different age groups by RT-nested PCR assays with primer pairs specific for VP1 and 2C regions.
| Age group | RT-nested PCR assays specific for | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| VP1 region | 2C region | ||||||||
| Diarrhea | Non-diarrhea | Total | P valuea) | Diarrhea | Non-diarrhea | Total | P value | ||
| Suckling (≤3 weeks; n=30) | 1/15 (7%) | 0/15 (0%) | 1/30 (3%) | NS | 10/15 (67%) | 9/15 (60%) | 19/30 (63%) | NS | |
| Weaned (≤8 weeks; n=20) | 0/10 (0%) | 1/10 (10%) | 1/20 (5%) | NS | 4/10 (40%) | 4/10 (40%) | 8/20 (40%) | NS | |
| Grower (≤20 weeks; n=20) | 3/10 (30%) | 1/10 (10%) | 4/20 (20%) | NS | 6/10 (60%) | 7/10 (70%) | 13/20 (65%) | NS | |
| Finisher (≥20 weeks; n=20) | 0/10 (0%) | 4/10 (40%) | 4/20 (20%) | 0.0253 | 7/10 (70%) | 8/10 (80%) | 15/20 (75%) | NS | |
| Sows (≥1 year; n=20) | 1/10 (10%) | 1/10 (10%) | 2/20 (10%) | NS | 6/10 (60%) | 5/10 (50%) | 11/20 (55%) | NS | |
| Total | 5/55 (9%) | 7/55 (13%) | 12/110 (11%) | NS | 33/55 (60%) | 33/55 (60%) | 66/110 (60%) | NS | |
a) Data analysis was performed by Fisher’s exact test with confidence limits of 95% (P<0.05) using SPSS Statistics version 20 for Windows (SPSS). NS: Statistically no significant.
The RT-nested PCR assays revealed different sensitivities for SV-As depending on primer pairs (Table 1). The detection rate by RT-nested PCR assay with primer pairs specific to SV-A 2C region was 60%, whereas VP1 region-targeting RT-nested PCR assay showed 11% positive rate. The 60% detection rate was similar to that reported in Japan [5]. SV-As were highly detected throughout all age groups (Table 1). This indicates that SV-A infections in Korea have no age predilections, unlike that in Spanish pigs [14]. Importantly, no significant difference in SV-A infections between diarrhea and non-diarrhea groups among all age groups was observed. SV-As are known to be able to cause asymptomatic and symptomatic infection in the field and experimental pigs [2, 3, 9, 10]. Our results also confirmed that SV-As could be detected in both diarrhea and non-diarrhea fecal samples of Korean pigs.
Of the 55 diarrhea fecal samples from each age group, single infection of SV-A was not detected. However, 33 samples (60%) tested as positive for SV-A were also positive for other common enteric pathogens (Table 2). Interestingly, coinfection status of SV-A with other enteric pathogens in each age group was quite different (Table 2). For example, occurrence of porcine enterovirus infection prevailed in suckling piglets (80%), whereas porcine teschovirus was most frequently detected in grower pigs (100%). The remaining 22 fecal samples (40%) tested as negative for SV-A were found to be positive for other common enteric pathogens (Table 2). Therefore, SV-A also co-infected with other enteric pathogens in Korean pigs, similar to the findings of previous reports regarding the status of coinfection of other enteric viruses in pigs with diarrhea [6, 8].
Table 2. Summary of enteric pathogens present in the samples obtained from pigs with diarrhea.
| Enteric pathogens presenta) | No. of samples (%) | ||||||
|---|---|---|---|---|---|---|---|
| Suckling | Weaned | Grower | Finisher | Sows | Total | ||
| SV-A plus other pathogens (%) | 10 (100%) | 4 (100%) | 6 (100%) | 7 (100%) | 6 (100%) | 33 (100%) | |
| PKV | 5 (50%) | 0 (0%) | 1 (17%) | 1 (14%) | 4 (67%) | 11 (33%) | |
| PEV | 8 (80%) | 3 (75%) | 4 (67%) | 3 (43%) | 2 (33%) | 20 (61%) | |
| PTV | 1 (10%) | 3 (75%) | 6 (100%) | 2 (29%) | 2 (33%) | 14 (42%) | |
| PSaV | 0 (0%) | 0 (0%) | 1 (17%) | 2 (29%) | 2 (33%) | 5 (15%) | |
| PRVA | 9 (90%) | 2 (50%) | 3 (50%) | 2 (29%) | 6 (100%) | 20 (61%) | |
| PRVB | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| PRVC | 1 (10%) | 3 (75%) | 1 (17%) | 1 (14%) | 3 (50%) | 9 (27%) | |
| EPEC | 0 (0%) | 0 (0%) | 0 (0%) | 1 (14%) | 0 (0%) | 1 (3%) | |
| C, perfringens | 3 (30%) | 1 (25%) | 0 (0%) | 0 (0%) | 0 (0%) | 4 (12%) | |
| B. hyodysenteriae | 0 (0%) | 1 (25%) | 1 (17%) | 0 (0%) | 0 (0%) | 2 (6%) | |
| B. pilosicoli | 1 (10%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (3%) | |
| SV-A negative but other pathogens (%) | 5 (100%) | 6 (100%) | 4 (100%) | 3 (100%) | 4 (100%) | 22 (100%) | |
| PKV | 3 (60%) | 2 (33%) | 0 (0%) | 0 (0%) | 2 (50%) | 7 (32%) | |
| PEV | 3 (60%) | 6 (100%) | 3 (75%) | 1 (33%) | 2 (50%) | 15 (68%) | |
| PTV | 0 (0%) | 5 (83%) | 3 (75%) | 2 (67%) | 3 (75%) | 13 (59%) | |
| PSaV | 0 (0%) | 0 (0%) | 0 (0%) | 3 (100%) | 0 (0%) | 3 (14%) | |
| PRVA | 4 (80%) | 2 (33%) | 3 (75%) | 1 (33%) | 3 (75%) | 13 (59%) | |
| PRVB | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (25%) | 1 (5%) | |
| PRVC | 2 (40%) | 4 (67%) | 1 (25%) | 0 (0%) | 1 (25%) | 8 (36%) | |
| EPEC | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| C, perfringens | 0 (0%) | 1 (17%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (5%) | |
| B. hyodysenteriae | 1 (10%) | 1 (17%) | 0 (0%) | 0 (0%) | 1 (25%) | 3 (14%) | |
| B. pilosicoli | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
a) SV-A: Sapelovirus A; PTV: Porcine teschovirus; PEV: Porcine enterovirus; PKV: Porcine kobuvirus; PRVA-C: Porcine groups A, B and C rotaviruses; PSaV: Porcine sapovirus; EPEC: Enteropathogenic Escherichia coli; C. perfringens: Clostridium perfringens; B. hyodysenteriae: Brachyspira hyodysenteriae; B. pilosicoli: Brachyspira pilosicoli.
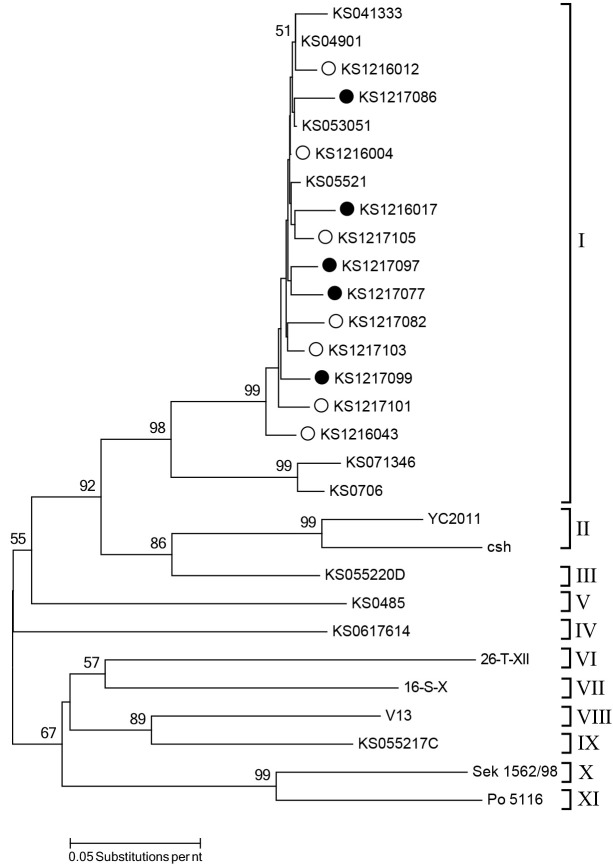
Sequence comparison of VP1 region is valuable for the determination of genetic relationships among picornaviruses [7]. In this study, the nucleotide sequences of the partial VP1 region (500 nucleotides) among SV-A strains circulated in the same farm shared high nucleotide identities (96.1–99.2%), regardless of diarrhea (data not shown). However, these strains showed high genetic diversities with reported strains circulating in Korea, sharing 69.2 to 99.8% of nucleotide identities. These results suggest that SV-As have evolved in different pathways, resulting in distinct genetic properties [12]. The hypervariability of VP1 nucleotide sequences of SV-As could also influence the genotype diversity [12]. Based on the phylogenetic analysis of the VP1 region, SV-As could be classified into 11 clusters [12]. The current strains identified in this study, regardless of diarrhea, and several other Korean strains belonged to cluster I. They were distinct from other strains reported in Korean or other countries (Fig. 1). These data could support the hypothesis that different genetic diversity of SV-As can result in various genotypes or clusters [12].
Fig. 1.
Phylogenetic tree based on the partial nucleotide sequences of VP1 region. This tree was constructed using the neighbor-joining method with 1,000 bootstrap replicates using MEGA6 [13]. The Korean strains detected in the pigs with diarrhea are indicated by solid circles, whereas the Korean strains detected in the non-diarrhea feces are indicated by empty circles.
In conclusion, SV-A infections are common in both diarrhea and non-diarrhea feces of pigs without age predilection. These SV-As that are genetically distinct from other known strains in Korea and other countries can co-infect pigs with other enteric pathogens. The virulence, pathogenesis and host ranges of these Korean SV-A strains distinct from other known strains remain to be determined in future studies.
ETHICAL STATEMENT.
This article does not contain any animal experiments performed by any of the authors.
Acknowledgments
This study was supported by the Bio-industry Technology Development Program (315021–04) through the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (iPET) funded by the Ministry of Agriculture, Food and Rural Affairs, and Korea Basic Science Institute grant (C33730), Republic of Korea. Chonnam National University provided funding to SIP (2012–2013).
REFERENCES
- 1.Adams M. J., Lefkowitz E. J., King A. M., Bamford D. H., Breitbart M., Davison A. J., Ghabrial S. A., Gorbalenya A. E., Knowles N. J., Krell P., Lavigne R., Prangishvili D., Sanfaçon H., Siddell S. G., Simmonds P., Carstens E. B.2015. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2015). Arch. Virol. 160: 1837–1850. doi: 10.1007/s00705-015-2425-z [DOI] [PubMed] [Google Scholar]
- 2.Alexandersen S., Knowles N. J., Dekker A., Belsham G. J., Zhang Z., Koenen F.2012. Picornaviruses. pp. 587–620. In: Diseases of Swine, 10th ed. (Zimmerman, J. J., Karriker, L. A., Ramirez, A., Schwartz, K. J. and Stevenson, G. W. eds), Wiley-Blackwell, Sussex. [Google Scholar]
- 3.Donin D. G., de Arruda Leme R., Alfieri A. F., Alberton G. C., Alfieri A. A.2014. First report of Porcine teschovirus (PTV), Porcine sapelovirus (PSV) and Enterovirus G (EV-G) in pig herds of Brazil. Trop. Anim. Health Prod. 46: 523–528. doi: 10.1007/s11250-013-0523-z [DOI] [PubMed] [Google Scholar]
- 4.Dunne H. W., Gobble J. L., Hokanson J. F., Kradel D. C., Bubash G. R.1965. Porcine reproductive failure associated with a newly identified “SMEDI” group of picorna viruses. Am. J. Vet. Res. 26: 1284–1297. [PubMed] [Google Scholar]
- 5.Honda E., Hattori I., Oohara Y., Taniguchi T., Ariyama K., Kimata A., Nagamine N., Kumagai T.1990. Sero- and CPE-types of porcine enteroviruses isolated from healthy and diarrheal pigs: possible association of CPE type II with diarrhea. Jpn. J. Vet. Sci. 52: 85–90. doi: 10.1292/jvms1939.52.85 [DOI] [PubMed] [Google Scholar]
- 6.Kim H. J., Park S. I., Ha T. P., Jeong Y. J., Kim H. H., Kwon H. J., Kang M. I., Cho K. O., Park S. J.2010. Detection and genotyping of Korean porcine rotaviruses. Vet. Microbiol. 144: 274–286. doi: 10.1016/j.vetmic.2010.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oberste M. S., Maher K., Kilpatrick D. R., Pallansch M. A.1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73: 1941–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prodělalová J.2012. The survey of porcine teschoviruses, sapeloviruses and enteroviruses B infecting domestic pigs and wild boars in the Czech Republic between 2005 and 2011. Infect. Genet. Evol. 12: 1447–1451. doi: 10.1016/j.meegid.2012.04.025 [DOI] [PubMed] [Google Scholar]
- 9.Schock A., Gurrala R., Fuller H., Foyle L., Dauber M., Martelli F., Scholes S., Roberts L., Steinbach F., Dastjerdi A.2014. Investigation into an outbreak of encephalomyelitis caused by a neuroinvasive porcine sapelovirus in the United Kingdom. Vet. Microbiol. 172: 381–389. doi: 10.1016/j.vetmic.2014.06.001 [DOI] [PubMed] [Google Scholar]
- 10.Sibalin M.1963. An investigation and characterization of enterovirus strains in Swedish pigs. II. Pathogenicity tests and serological properties. Acta Vet. Scand. 4: 332–355. [Google Scholar]
- 11.Son K. Y., Kim D. S., Kwon J., Choi J. S., Kang M. I., Belsham G. J., Cho K. O.2014. Full-length genomic analysis of Korean porcine Sapelovirus strains. PLOS ONE 9: e107860. doi: 10.1371/journal.pone.0107860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Son K. Y., Kim D. S., Matthijnssens J., Kwon H. J., Park J. G., Hosmillo M., Alfajaro M. M., Ryu E. H., Kim J. Y., Kang M. I., Cho K. O.2014. Molecular epidemiology of Korean porcine sapeloviruses. Arch. Virol. 159: 1175–1180. doi: 10.1007/s00705-013-1901-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vilar M. J., Peralta B., García-Bocanegra I., Simon-Grifé M., Bensaid A., Casal J., Segalés J., Pina-Pedrero S.2016. Distribution and genetic characterization of Enterovirus G and Sapelovirus A in six Spanish swine herds. Virus Res. 215: 42–49. doi: 10.1016/j.virusres.2016.01.019 [DOI] [PubMed] [Google Scholar]
- 15.Yamanouchi K., Bankowski R. A., Howarth J. A.1965. Physical and biological properties of the Chico strain of porcine enterovirus. J. Infect. Dis. 115: 345–355. doi: 10.1093/infdis/115.4.345 [DOI] [PubMed] [Google Scholar]