Abstract
Transforming growth factor β (TGFβ) is one of few known negative regulators of hematopoiesis, yet the mechanisms by which it affects cell cycle arrest and stem cell quiescence are poorly understood. Induction of the cyclin-dependent kinase inhibitors, p15INK4b (p15) and p21WAF1 (p21) is important for TGFβ-mediated cytostasis in epithelial cells but not in hematopoietic cells. Using primary human hematopoietic cells and microarray analysis, we identified p57KIP2 (p57) as the only cyclin-dependent kinase inhibitor induced by TGFβ. Up-regulation of p57 mRNA and protein occurs before TGFβ-induced G1 cell cycle arrest, requires transcription, and is mediated via a highly conserved region of the proximal p57 promoter. The up-regulation of p57 is essential for TGFβ-induced cell cycle arrest in these cells, because two different small interfering RNAs that prevent p57 up-regulation block the cytostatic effects of TGFβ on human hematopoietic cells. Reduction of basal p57 expression by this approach also allows hematopoietic cells to proliferate more readily in the absence of TGFβ. p57 is a putative tumor suppressor gene whose expression is frequently silenced by promoter hypermethylation in hematologic malignancies. Our studies identify a molecular pathway by which TGFβ mediates its cytostatic effects on human hematopoietic cells and suggests an explanation for the frequent silencing of p57 expression.
Keywords: transcription control, negative regulators, cyclin-dependent kinase inhibitor
Transforming growth factor β (TGFβ) has pleiotropic effects on diverse cell types and tissues, and dysregulated TGFβ signal transduction is an important, and common, property of malignant cells, including those of hematopoietic origin (1-3). TGFβ maintains hematopoietic stem cells in a quiescent undifferentiated state, whereas lineage-restricted progenitor cells are generally less responsive to its cytostatic effects (4). Early hematopoietic progenitors produce TGFβ, and neutralization of autocrine TGFβ by monoclonal antibodies can recruit early progenitor cells into the cell cycle and initiate their differentiation (5). Whereas primary cultures of normal cells are highly sensitive to the cytostatic action of TGFβ, most malignant cells and leukemia-derived cell lines are resistant to this effect (5). Despite the frequency with which mutations in the TGFβ signaling pathway arise in certain solid tumors, they are exceedingly uncommon in leukemias (6, 7), which suggests that other mechanisms dysregulate this pathway in hematologic malignancies.
Although TGFβ is one of few negative regulators of hematopoiesis, the mechanism by which this effect is mediated is not well understood. In epithelial cells, TGFβ can trigger cell cycle arrest by up-regulating cytostatic proteins such as the cyclin-dependent kinase inhibitors (CDKI), p15 and p21, while down-modulating growth-stimulating proteins, notably c-Myc and Id family members.
Determining how TGFβ affects hematopoietic cell cycle arrest and stem cell quiescence has proven difficult, because neither p21 nor p27KIP1 (p27) appears to be required for the inhibition of hematopoietic stem cell or progenitor cell proliferation by TGFβ (8). Using gene expression profiling, we have identified p57 as an immediate-early target of TGFβ in normal human CD34+ hematopoietic progenitor/stem cells. Using small interfering RNA (siRNA), we found that p57 is required for TGFβ-mediated cell cycle arrest of hematopoietic cells, and that the basal expression of p57 restrains the proliferation of these cells. Expression of p57 is silenced in the malignant cells of 30-55% of patients with acute lymphoblastic leukemia, acute myelogenous leukemia, and B cell lymphomas, suggesting that p57 could function as a tumor suppressor in these diseases (9-12). Our results provide mechanistic insight into the activity of p57 and its role as a suppressor of hematopoietic cell transformation.
Materials and Methods
Plasmids and Cloning. Cytomegalovirus (CMV) promoter-based expression plasmids for Smad3, Smad4, the constitutively active TGFβ type I receptor, TβR-I(T204D), and the p57 promoter-regulated luciferase reporter plasmids, -2191KIP2, -1550KIP2, -595KIP2, and -165KIP2, have been described (13-15). Further experimental details are provided in Supporting Text, which is published as supporting information on the PNAS web site.
Cell Culture. The acute myelogenous leukemia-derived cell line, M091 was kindly provided by M. Okabe (16), and HeLa cells were obtained from the American Type Culture Collection (ATCC). Primary human cord blood-derived CD34+ hematopoietic progenitors (CB-CD34) were maintained in Iscove's modified Dulbecco's medium supplemented with 20% BIT 9500 (StemCell Technologies, Vancouver), 100 ng/ml FLT3 ligand (FLT3L), 100 ng/ml stem cell factor, and 20 ng/ml IL-6. Kirin (Tokyo), Amgen (Thousand Oaks, CA), and Novartis (Basel) kindly provided these cytokines.
Gene Expression Analysis. Total RNA was isolated from CB-CD34 or M091 cells 0, 2, and 4 h after stimulation with TGFβ. Biotin-labeled cRNA was synthesized and hybridized to Affymetrix U133A (CB-CD34) or U95Av2 (M091) arrays by using standard protocols (Affymetrix, Santa Clara, CA). Data processing was performed by using mas 5.0 software (Affymetrix) and the genespring software package (Silicon Genetics, Redwood City, CA).
p57 Expression Studies. Total RNA was purified from CB-CD34 or M091 cells before or after treatment with 200 pM TGFβ1 for the indicated times. Semiquantitative analysis of expression was assessed visually after PCR was performed by using primers specific for p57 or hypoxanthine phosphoribosyltransferase. Quantitative PCR was performed by using the ABI 7700 instrument and reagents (Applied Biosystems) and primer/probe sets designed for the indicated transcripts. Actinomycin D (10 μg/ml), cycloheximide (5 μg/ml), or diluent control (0.2% DMSO) were added 15 min before TGFβ addition, when indicated.
To discriminate among the three known isoforms of p57, PCR amplification of M091 and CB-CD34 cell-derived cDNA was performed by using the KIP2 Pan-Isoform primers that bracket intron 1 and generate a 381-bp fragment from the A1 transcript (and from genomic DNA), a 246-bp fragment from the A2 transcript, and a 215-bp fragment from the A3 transcript. A second set of primers, KIP2 Isoform-A1A2, amplify a 309-bp fragment from the A1 and A2 transcripts and from genomic DNA but do not amplify the A3 transcript (because the sense primer is located within an intron for the A3 transcript).
Assessment of p57 Imprinting. Genomic DNA was purified from single-donor cord-blood mononuclear cells, and total RNA was obtained from CB-CD34 cells before and after TGFβ treatment. PCR was used to amplify the PAPA repeat region of exon 2, which is known to contain several polymorphisms (GenBank accession no. XM_006479). Either genomic DNA or cDNA generated from CB-CD34 cells obtained from the same single donor were used as template.
Western Blot Analysis. Lysates from M091 cells, untreated or treated with TGFβ for the indicated times, were used for Western blotting with standard procedures and antibodies, as indicated.
Proliferation and Cell Cycle Analysis. Cell proliferation was determined either by counting or by using the WST-1 reagent (Amersham Pharmacia). Univariate cell cycle analysis was performed by using ethanol-fixed cells stained with propidium iodide in buffer containing RNase A. DNA content was assessed by flow cytometry, and cell cycle analysis was performed by using the multicycle software package (Phoenix Flow Systems, San Diego).
Transcription Reporter Assays. HeLa cells were plated in 24-well plates at a density of 105 cells per well. When the cells had reached 80-90% confluence, they were transfected in triplicate, with each well receiving 100 μl of OptiMem I (Invitrogen) containing 1.5 μl of Lipofectamine 2000, 250 ng of KIP2-luciferase reporter construct, and 600 ng of some combination of the pCMV5- or pCMV5-based Flag-Smad3, Flag-Smad4, and TβR-I(T204D) expression plasmids, as well as 50 ng of pEGFP-C3 (BD-Clontech) and 25 ng of CMV-RNL (Renilla luciferase) (Promega). Forty-eight hours after transfection, cell lysates were prepared for analysis by using the Dual-luciferase system. Firefly luciferase activity was normalized to RNL activity as the first step of data analysis. Subsequent normalization to an intraexperiment reference activity was performed, as indicated.
siRNA-Mediated Knockdown of Gene Expression. Annealed double-stranded siRNA (Dharmacon, Lafayette, CO) was introduced into logarithmically growing M091 cells by electroporation by using a Genepulser electroporator (Bio-Rad).
Results
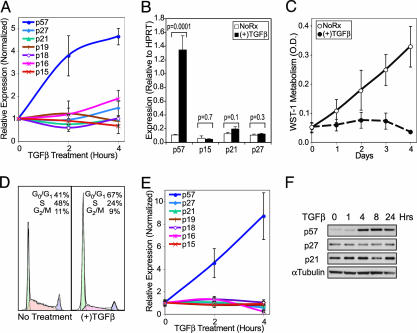
TGFβ Induces p57 Expression in Hematopoietic Cells. To determine how TGFβ induces the cytostasis of hematopoietic cells, we used microarrays to define the transcription response of primary human CD34+ umbilical cord blood progenitor/stem cells (CB-CD34) to treatment with TGFβ (200 pM) for 2 and 4 h. TGFβ increased the expression of 31 probe sets, and it decreased the expression of 46 probe sets (of 21,608 probe sets on the Affymetrix U133A chip) by a least 1.5-fold in each of four independent experiments (see Supporting Text). Although several well established TGFβ gene responses, including induction of SMAD6 and SMAD7 and repression of c-MYC (17-19), were identified, TGFβ did not affect the expression of p21 or p15 in any data set. However, we did observe the brisk and robust up-regulation of p57 mRNA. This finding is in marked contrast to the induction of p21 and/or p15 typically observed in epithelial cells, which is believed to play a critical role in mediating the cytostatic effects of TGFβ (20, 21). TGFβ did not affect the expression of any other CDKI (p21, p15, p27, p16INK4a, p18INK4c, and p19INK4d) in CB-CD34 (Fig. 1A). Using quantitative RT-PCR to confirm these findings, we found that TGFβ induced significant up-regulation of p57 mRNA (6.6- to 20-fold induction) but had no effect on the expression of p15, p21, or p27 mRNA in CB-CD34 (Fig. 1B) or in M091 cells (not shown).
Fig. 1.
TGFβ causes G1 cell cycle arrest and up-regulates p57 mRNA and protein. (A) p57 mRNA is rapidly up-regulated by TGFβ in primary human hematopoietic progenitor cells. The average signal for the various CDKI probe sets is shown normalized to the expression before stimulation with TGFβ. (B) Quantitative RT-PCR demonstrates strong up-regulation of p57 mRNA 4 h after exposure of CB-CD34 to TGFβ. There is no statistically significant regulation of other CDKIs by TGFβ (Student's t test). Expression of the indicated mRNA is reported relative to the level of hypoxanthine phosphoribosyltransferase expression. (C) Proliferation rates of M091 cells treated or untreated with TGFβ (200 pM) are shown. (D) Representative cell cycle profiles of M091 cells treated with TGFβ (200 pM) for 24 h, or left untreated (Left). (E) Only p57 mRNA is up-regulated by TGFβ in M091 cells. Expression array data are presented as described for A. (F) Western blot analysis of p57, p27, and p21 protein expression using lysates from M091 cells stimulated with TGFβ for the indicated periods of time. p15 is not detectable by Western blot at any time. α-Tubulin staining was used as a control for protein loading. All error bars represent the standard errors of measurements from three to four independent experiments.
Although most leukemia cell lines are resistant to TGFβ-induced cell cycle arrest (unpublished observations), we found that M091 cells, derived from a patient with an undifferentiated acute myelogenous leukemia, mimic the sensitivity of normal primary hematopoietic progenitor cells to the cytostatic effects of TGFβ and arrest in G1 with an EC50 of 4 pM (Fig. 1 C and D and data not shown). p57 mRNA and protein are rapidly up-regulated by TGFβ in these cells, whereas expression of other members of the CIP/KIP and INK4 families is unaffected (Fig. 1 E and F). The strong up-regulation of p57 precedes the G1 arrest of the cells and persists for at least 30 h (Fig. 1F and data not shown).
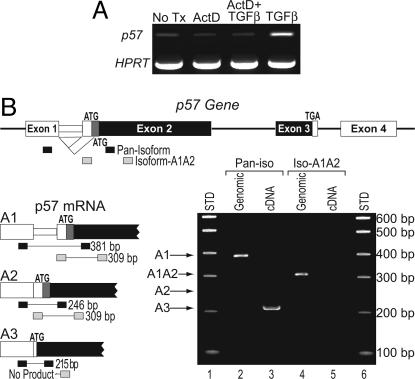
Hematopoietic Cells Express a Single p57 Isoform, and Its Regulation by TGFβ Is Transcriptional. We used the transcription inhibitor, actinomycin D, to assess whether de novo transcription was required for the rapid up-regulation of p57 mRNA by TGFβ. Pretreatment of M091 cells with actinomycin D before TGFβ completely abrogated the TGFβ-induced up-regulation of p57 mRNA (Fig. 2A), strongly suggesting a transcriptional mechanism of regulation. Consistent with this, TGFβ did not alter the half-life of p57 mRNA (data not shown). Furthermore, new protein synthesis is not required for TGFβ to induce p57 transcription, because pretreatment of M091 cells with cycloheximide does not prevent the robust induction of p57 mRNA by TGFβ (data not shown).
Fig. 2.
TGFβ-induced up-regulation of p57 involves increased transcription of the A3 isoform. (A) M091 cells were pretreated with actinomycin D (lanes 2 and 3) or DMSO alone (lanes 1 and 4) and then stimulated with TGFβ (200 pM) (lanes 3 and 4) for 90 min. The relative expression of p57 mRNA was assessed by RT-PCR. Amplification of hypoxanthine phosphoribosyltransferase mRNA is shown to demonstrate equal RNA input. (B) The organization of the p57 gene is shown (Upper). The translated portion of each exon is colored black if it is invariant or dark gray if it is isoform specific. The 5′ end of the three known isoforms of p57 are presented (A1, A2, and A3) (Lower Left). The locations of the PCR primers used for this analysis are represented by black (the Pan-Isoform Amplimer Set) or gray bars (the Isoform-A1A2 Amplimer Set). Amplification of genomic DNA by these primers yields 381- and 309-bp fragments, respectively. PCR products generated from genomic DNA (lanes 2 and 4) or cDNA (lanes 3 and 5) templates are shown (Lower Right). Only the A3 transcript is detected in CB-CD34 cells (and M091 cells; data not shown).
Three p57 mRNAs can be generated from the primary transcript by alternative splicing of intron 1 (Fig. 2B) (22, 23). The A1 and A2 transcripts encode p57 proteins that are 12 aa longer than that coded by the A3 transcript, due to their use of an upstream initiation codon not found in the A3 transcript. There is sparse information regarding the relative expression of these variant transcripts or the extent to which their expression is tissue restricted. Using isoform-specific PCR primers, a single band corresponding to the p57 A3 transcript was amplified from primary human hematopoietic progenitor cell cDNA (Fig. 2B, lane 3). We observed no expression of the A1 or A2 p57 isoforms in untreated or TGFβ-treated CB-CD34 cells (or M091 cells) (Fig. 2B, lane 5, and data not shown).
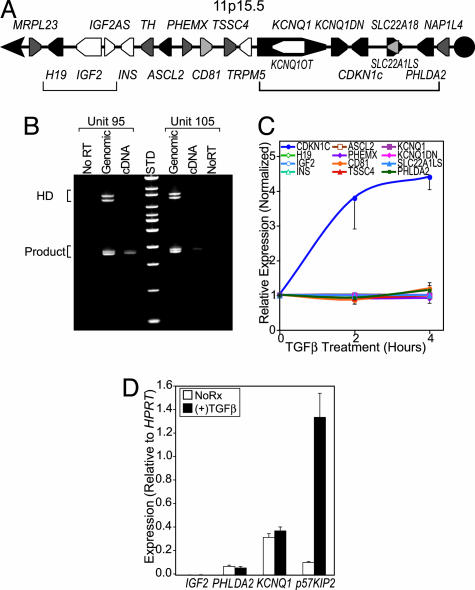
TGFβ Induces Monoallelic Up-Regulation of p57 but Does Not Affect Expression of Coimprinted Genes. The gene encoding p57, CDKN1c, is located within an imprinted region of chromosome 11p15.5 (Fig. 3A), and it is expressed from the maternal allele in most tissues (24). To determine whether p57 expression is monoallelic (i.e., imprinted) in primary CD34+ hematopoietic progenitors, we used two different cord-blood samples (units 95 and 105) that possessed informative size polymorphisms (12-bp deletions) and single-nucleotide polymorphisms within the PAPA repeat region of p57. When genomic DNA was used as a template, the fragment amplified from the PAPA repeat region formed heteroduplexes. However, amplification of the CB-CD34 cDNAs generated only a single band (Fig. 3B), demonstrating that only one allele is expressed. Monoallelic expression, which implies genomic imprinting, was confirmed by sequencing at least 10 clones derived from each PCR product (not shown).
Fig. 3.
TGFβ induces monoallelic up-regulation of p57 but does not regulate other genes in the imprinted region of chromosome 11p15.5. (A) The genomic organization of a 1-Mb imprinted cluster of genes on chromosome 11p15.5 is shown. Maternally and paternally imprinted genes are represented as pink or blue-filled pentagons, respectively. Nonimprinted genes within the region are shown in black, and those for which the imprinting status is unknown are filled with white. The brackets correspond to the two clusters of genes that are coordinately imprinted and are under the control of independent regulatory elements. (B) The PAPA repeat region was amplified from two individual units of cord blood by using genomic DNA and cDNA made from CB-CD34 cells stimulated with TGFβ for 4 h as templates. Heteroduplexes are seen for the amplified genomic fragments but not when the cDNA is amplified. The doublet seen using genomic DNA but not with the cDNA is due to different allele lengths. (C) p57 mRNA, but not that of other imprinted genes on chromosome 11p15.5, is rapidly up-regulated by TGFβ in CB-CD34. The average signal for the various probe sets is shown normalized to the expression before stimulation with TGFβ. (D) Quantitative RT-PCR analysis of PHLDA2, KCNQ1, IGF2, and CDKN1c (p57) gene expression before and 4 h after exposure of CB-CD34 to TGFβ. Expression of the indicated mRNA is reported relative to the expression of the hypoxanthine phosphoribosyltransferase reference transcript. All error bars represent the standard errors of measurements from three to four independent experiments.
Chromosome 11p15.5 contains two imprinted clusters of genes that are under the control of distinct imprint control regions, and CDKN1c is the parent member of an imprinted subdomain that includes the PHLDA2, KCNQ1, KCNQ1OT, and SLC22A18 genes (Fig. 3A) (24, 25). Genes that are coordinately imprinted are sometimes coordinately expressed in a tissue-specific manner (26-28). We queried our microarray data from both primary CB-CD34 and the M091 cell line to determine whether TGFβ regulates other genes within the imprinted regions of 11p15.5. The expression of IGF2, INSULIN, ASCL2, TSSC4, and BWRT was not detectable above the microarray baseline at any point (i.e., called “absent” in Affymetrix mas 5 software), and none of the expressed genes were regulated by TGFβ (Fig. 3C; data for M091 not shown). We confirmed these findings with quantitative RT-PCR for representative genes in both clusters of imprinted genes (Fig. 3D). These results indicate that TGFβ acts through p57-specific regulatory elements.
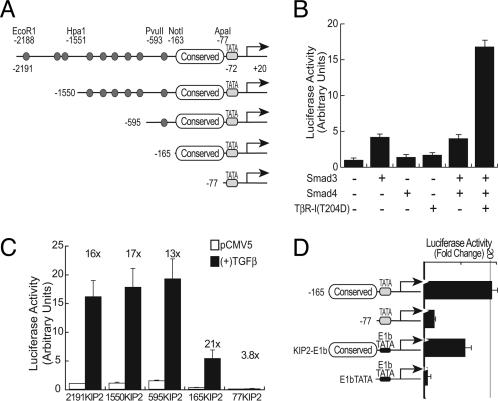
TGFβ May Regulate p57 Expression Through a Highly Conserved Region of Its Promoter. Distant elements important for the genomic imprinting of p57 and for its tissue-specific expression have been recently identified (26, 27, 29, 30). However, the regulatory elements and transcription-modulating proteins that act at the proximal promoter have not been elucidated. To develop a model system with which to identify the TGFβ-responsive region(s) of the p57 promoter, we introduced luciferase reporter gene constructs into a variety of leukemia-derived cell lines. For unknown reasons, however, we observed no inducible luciferase activity in these cell lines. Other TGFβ-regulated constructs, such as the Igα-class switch element, which is normally expressed only in B cells, faithfully respond to TGFβ in both mesenchymal and epithelial cells (31, 32). Therefore, we resorted to HeLa cells to test the activity of the p57 promoter constructs. Using this approach, we observed consistent induction of the p57 promoter by TGFβ mediators and subsequently found that HeLa cells express p57 at a low level that can be modestly up-regulated by exogenous TGFβ (data not shown). We used HeLa cells for our reporter gene assays.
Using a series of luciferase reporter constructs driven by various portions of the p57 promoter, we found that the -2191KIP2 reporter plasmid (Fig. 4A), which contains p57 promoter sequences from -2191 to +20, was activated 4-fold by Smad3 expression and 17-fold by the coexpression of Smad3, Smad4, and TβR-I(T204D) (Fig. 4B). Smad4, or the TβR-I(T204D) receptor alone, had minimal effects on the reporter activity. Using truncation mutant plasmids, we identified a region just upstream of the TATA box (extending from -165 to -77) as being critical for TGFβ-mediated activation (Fig. 4C). This region is highly conserved in the mouse, rat, and human promoters, and when placed in front of a heterologous minimal promoter, it is sufficient to confer TGFβ-responsiveness (Fig. 4D). Thus, TGFβ induces transcription of p57, but not other genes, in the imprinted region of 11p15.5 and could do so through a highly conserved region (-165 to -77) of the p57 promoter.
Fig. 4.
Activation of the p57 promoter by TGFβ signaling requires a highly conserved region upstream of the TATA-box. (A) Schematic diagram of the p57 promoter indicating nine canonical Smad-binding elements (CAGAC, represented as small ovals) and the restriction sites used for generating the truncation mutations used for this analysis. Also indicated is a GC-rich region located between -165 and -77 that is highly conserved between the human, rat, and murine p57 genes. (B) The full-length -2191KIP2 reporter was cotransfected into HeLa cells in combination with Smad3, Smad4, and TβR-I(T204D), as indicated. Relative luminescence units were normalized -2191KIP2 reporter activity when cotransfected with pCMV5 alone. (C) The indicated p57 reporter constructs were transfected into HeLa cells with either pCMV5 or TGFβ mediators [Smad3, Smad4, and TβR-I(T204D)]. Relative luminescence units were normalized as described for A. The numbers above each set of bars indicate the fold increase in reporter activity induced by the cotransfection of the TGFβ mediators. (D) The p57 promoter constructs used for these experiments are diagrammed (Left). The indicated p57 reporter constructs were transfected into HeLa cells with either pCMV5 or TGFβ mediators [Smad3, Smad4, and TβR-I(T204D)]. (Right) The fold increase in reporter activity with cotransfection of the TGFβ mediators compared to the reporter activity in the presence of pCMV5. All error bars represent the standard errors of measurements from two to four independent experiments each carried out in triplicate.
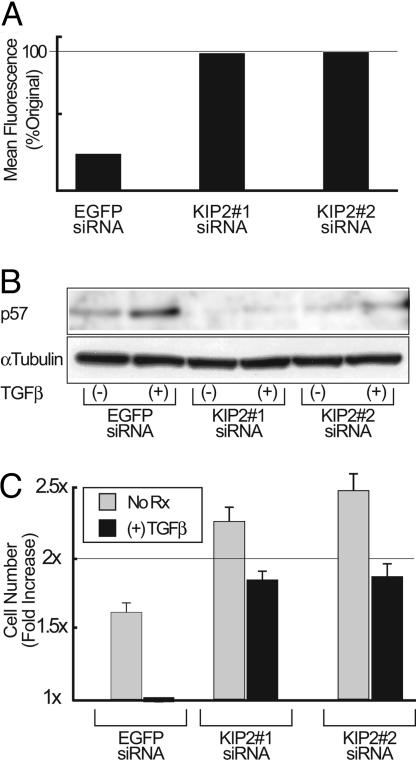
Basal p57 Expression Restrains Hematopoietic Cell Proliferation, and Its Up-Regulation Is Required for TGFβ-Mediated Cytostasis. To evaluate the primacy of p57 as the immediate early target of TGFβ that leads to cell cycle arrest of hematopoietic cells, we generated siRNAs to specifically knockdown its expression. We generated three different siRNAs, two specific for the p57 transcript and a third siRNA directed against enhanced GFP (EGFP) to serve as a control. Using cells that constitutively express EGFP, we demonstrated that the EGFP siRNA, but neither of the p57-specific siRNAs, knocked down EGFP expression (Fig. 5A). Conversely, both of the p57-specific siRNAs blocked p57 up-regulation, whereas the EGFP-specific siRNA did not (Fig. 5B). Cells transfected with the anti-EGFP siRNA proliferate normally and undergo TGFβ-induced cell cycle arrest in a normal fashion (Fig. 5C). In contrast, cells transfected with either of the siRNAs directed against p57 are no longer arrested by TGFβ and, in fact, proliferate more briskly in the absence of TGFβ than the parental cell line. Thus, p57 up-regulation is required for the TGFβ-mediated cell cycle arrest of hematopoietic progenitor cells, and basal p57 expression appears to restrain the proliferation of these cells.
Fig. 5.
Up-regulation of p57 is required for the TGFβ-induced cell cycle inhibition of hematopoietic cells. M091 were transfected with siRNA directed against either EGFP or p57 mRNA. (A) The effect of the indicated siRNA on EGFP expression was assessed by flow cytometry by using M091-MIGR1 cells that constitutively express EGFP from the MSCV LTR. Presented is the mean fluorescence normalized to untransfected control cells (not shown). (B) M091 cells were transfected with the indicated siRNA, and split into equal portions, half of which were treated with TGFβ (200 pM) for 4 h before Western blot. The α-tubulin control is shown to demonstrate equal protein loading in all lanes. (C) The fold change in M091 cell numbers after transfection with the indicated siRNA is shown in the presence or absence of TGFβ (200 pM) for 24 h. Error bars represent the standard error of three independent experiments.
Discussion
Hematopoiesis consists of a series of highly tuned branching processes through which progenitor/stem cells undergo self renewal, proliferation, and differentiation. This process, which continues throughout the lifetime of an organism, gives rise to all mature blood cell lineages. An intimate relationship exists between cell cycle regulation and hematopoietic progenitor and stem cell function. Murine studies show that CDKIs p18INK4c and p21 are important for normal hematopoietic stem cell quiescence (33, 34), whereas p27 regulates the pool of more mature progenitors (8, 35). Little is known of the function performed by other CDKIs in immature hematopoietic cells.
Using CD34-positive primary human hematopoietic progenitor/stem cells, we have identified p57 as the only CDKI rapidly and robustly up-regulated by TGFβ. CDKIs that are often seen to be up-regulated by TGFβ in epithelial and neural progenitor cells (e.g., p15, p27, and p21) are not immediate targets of this cytokine in hematopoietic cells, and neither p21 nor p27 is necessary for TGFβ to affect cytostasis in immature hematopoietic cells (8, 34, 35). p15 does not appear to be an early target of TGFβ in CB-CD34+ cells (or in the M091 cells at any time), suggesting that the delayed up-regulation of p15 sometimes seen in these cells in response to TGFβ (36, 37) is a consequence of earlier events. The strong TGFβ-mediated up-regulation of p57 that we observed in immature human hematopoietic cells is not seen in many epithelial cell types (18), further demonstrating that critical aspects of TGFβ-signaling are cell type specific.
Although we demonstrate transcriptional up-regulation of p57 by TGFβ in hematopoietic cells, proteasome-mediated degradation of p57 has been shown to be triggered by TGFβ in serum-starved rat osteoblastic cells (38). This tissue-specific regulation of p57 is not entirely surprising, because osteoblasts are one of very few cell types that are induced to proliferate by TGFβ. TGFβ-mediated up-regulation of p57 protein in hematopoietic cells persists for at least 1 day, suggesting that proteasome-dependent degradation of this CDKI is not triggered by TGFβ in hematopoietic cells and further demonstrating important differences in the tissue-specific regulation of p57.
Expression of p57 is tightly controlled, but little is known of its physiological regulators. We were able to demonstrate that the genomic imprint of p57 extends to primitive hematopoietic cells and is unaffected by TGFβ. Under some circumstances, genes that are coordinately imprinted are also coregulated, perhaps because of the presence of shared enhancers (26, 28). This appears to be the case for the induction of p57 and KCNQ1 by the p53 family member, p73β (28). In contrast, TGFβ appears to mediate its effects through tissue-specific p57 promoter/enhancer elements.
The regulation of p57 by TGFβ is transcriptional, and we used luciferase reporter gene constructs derived from various portions of the human p57 promoter to further define this transcriptional regulation. Neither exogenous TGFβ nor the cotransfection of components of the TGFβ pathway induced luciferase expression in hematopoietic cell lines. However, p57 promoter activity was robustly stimulated in HeLa cells. This disparity could relate to technical differences in the methodologies used to transfect suspension cells and adherent cells, but it could also indicate that the transcriptional regulation of p57 expression in hematopoietic cells is more complex than our studies have addressed. We found that the region of the p57 promoter extending from -165 to -77 is critical for TGFβ-mediated regulation of this gene in HeLa cells. Several lines of evidence suggest that this region serves an important role in the regulation of p57 expression. There is 91% sequence identity within this region of the mouse, rat, and human p57 promoters. Furthermore, CpG methylation of this element, but not of more upstream regions of the p57 promoter (10), is associated with the epigenetic silencing of p57 expression in hematopoietic and nonhematopoietic malignancies. Furthermore, this “proximal” region of the p57 promoter indirectly mediates the down-regulation of p57 by EWS-FLI1 Ewing's sarcoma-associated fusion transcription factor (15), which is known to interfere with TGFβ signaling (39). Although Smad3 stimulates p57 reporter gene activation, there is neither a consensus Smad-binding element nor an E-box located between -165 and -77 of the p57 promoter. It is possible that a TGFβ-activated Smad complex contacts this region with the cooperation of a sequence specific DNA-binding cofactor. The region does contain putative binding sites for several transcription factors (e.g., Sp1, YY1, and RUNX1) that are known to interact with Smads as well as binding sites for other transcription factors known to play important roles in hematopoiesis (including C/EBPα, EGR1, WT-1, ICSBP, and NRF1). Whether any of these proteins is involved in the regulation of p57 expression in hematopoietic cells remains to be elucidated.
We used siRNA to knock-down expression of p57 and observed marked reduction in the cytostatic response to TGFβ, indicating that p57 is a critical downstream mediator of TGFβ in these hematopoietic cells. M091 cells proliferate more rapidly after p57 levels are knocked down with siRNA. We have twice observed spontaneous silencing of p57 expression by promoter hypermethylation (data not shown) after M091 cells were maintained in continuous cell culture for 10-14 months. Both of these sublines proliferated more rapidly than the parental M091 cell line, implicating reduced p57 expression in the biological selection of more rapidly growing hematopoietic cells. As we had observed with siRNA knockdown of p57 expression, both of the M091 sublines that had acquired silencing of p57 expression through promoter hypermethylation lost much of their cytostatic response to TGFβ.
p57 is a putative tumor-suppressor gene with an established role in the cancer-predisposing Beckwith-Wiedemann Syndrome as well as a postulated role in several hematopoietic malignancies (9-11, 40-44). Hematopoietic stem and progenitor cells possess a vast proliferative capacity, making it surprising that the p57 gene is imprinted, with one allele silenced since early embryogenesis. In vitro experiments have demonstrated that p57 can suppress oncogene-mediated transformation of primary cells (41), suggesting that interference with the expressed p57 allele could serve as a “second hit” during oncogenesis. Indeed, this may occur during the transformation of chronic myelogenous leukemia from the indolent chronic phase to the aggressive blast crisis during which the expression of p57, but not other CDKIs, is silenced (12).
Despite the many effects TGFβ exerts on cells, cancer cells most commonly acquire the ability to escape from TGFβ-mediated cytostasis (3). TGFβ-induced cell cycle arrest of hematopoietic cells depends upon the up-regulation of the CDKI, p57. This suggests that the frequent silencing of p57 expression seen in myeloid and lymphoid hematopoietic malignancies significantly contributes to their transformation.
Supplementary Material
Acknowledgments
We thank Dr. O. Delattre for p57 reporter plasmids, Dr. M. Okabe for the M091 cells, Jin Zhang for technical assistance, and Dr. Agnes Viale and the Sloan-Kettering Institute Genomics Core Laboratory for assistance and many helpful discussions. This work was funded by a Specialized Center of Research Grant from the Leukemia & Lymphoma Society (to S.D.N.), a Leukemia & Lymphoma Society Fellowship (to J.M.S.), and National Institutes of Health R01 Grant DK52621 (to S.D.N.). J.M.S. is a recipient of a Ruth L. Kirschstein National Research Service Award and has been supported by a Charles A. Dana Fellowship and a Mortimer Lacher Fellowship. P.B. is a fellow of the Leukemia Research Foundation and is also sponsored by the Charles H. Revson Foundation.
Author contributions: J.M.S. and S.D.N. designed research; J.M.S. and P.B. performed research; J.M.S., J.M., and S.D.N. analyzed data; J.M.S., J.M., and S.D.N. wrote the paper.
Abbreviations: TGFβ, transforming growth factor β; CDKI, cyclin-dependent kinase inhibitors; siRNA, small interfering RNA; CMV, cytomegalovirus; EGFP, enhanced GFP.
Data deposition: Microarray data are available in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo.
References
- 1.Fynan, T. M. & Reiss, M. (1993) Crit. Rev. Oncog. 4, 493-540. [PubMed] [Google Scholar]
- 2.Siegel, P. M. & Massagué, J. (2003) Nat. Rev. Cancer 3, 807-820. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan, D. & Weinberg, R. A. (2000) Cell 100, 57-70. [DOI] [PubMed] [Google Scholar]
- 4.Ohta, M., Greenberger, J. S., Anklesaria, P., Bassols, A. & Massagué, J. (1987) Nature 329, 539-541. [DOI] [PubMed] [Google Scholar]
- 5.Fortunel, N. O., Hatzfeld, A. & Hatzfeld, J. A. (2000) Blood 96, 2022-2036. [PubMed] [Google Scholar]
- 6.Imai, Y., Kurokawa, M., Izutsu, K., Hangaishi, A., Maki, K., Ogawa, S., Chiba, S., Mitani, K. & Hirai, H. (2001) Oncogene 20, 88-96. [DOI] [PubMed] [Google Scholar]
- 7.Xu, J. & Attisano, L. (2000) Proc. Natl. Acad. Sci. USA 97, 4820-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, T., Shen, H., Rodrigues, N., Stier, S. & Scadden, D. T. (2001) Blood 98, 3643-3649. [DOI] [PubMed] [Google Scholar]
- 9.Shen, L., Toyota, M., Kondo, Y., Obata, T., Daniel, S., Pierce, S., Imai, K., Kantarjian, H. M., Issa, J.-P. J. & Garcia-Manero, G. (2003) Blood 101, 4131-4136. [DOI] [PubMed] [Google Scholar]
- 10.Kikuchi, T., Toyota, M., Itoh, F., Suzuki, H., Obata, T., Yamamoto, H., Kakiuchi, H., Kusano, M., Issa, J. P., Tokino, T. & Imai, K. (2002) Oncogene 21, 2741-2749. [DOI] [PubMed] [Google Scholar]
- 11.Li, Y., Nagai, H., Ohno, T., Yuge, M., Hatano, S., Ito, E., Mori, N., Saito, H. & Kinoshita, T. (2002) Blood 100, 2572-2577. [DOI] [PubMed] [Google Scholar]
- 12.Iolascon, A., Della Ragione, F., Giordani, L., Serra, A., Saglio, G. & Faienza, M. F. (1998) Haematologica 83, 771-777. [PubMed] [Google Scholar]
- 13.Wotton, D., Lo, R. S., Lee, S. & Massagué, J. (1999) Cell 97, 29-39. [DOI] [PubMed] [Google Scholar]
- 14.Wieser, R., Wrana, J. L. & Massagué, J. (1995) EMBO J. 14, 2199-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dauphinot, L., De Oliveira, C., Melot, T., Sevenet, N., Thomas, V., Weissman, B. E. & Delattre, O. (2001) Oncogene 20, 3258-3265. [DOI] [PubMed] [Google Scholar]
- 16.Okabe, M., Kunieda, Y., Shoji, M., Nakane, S., Kurosawa, M., Tanaka, J., Hansen, S. R. & Asaka, M. (1995) Leuk. Res. 19, 933-943. [DOI] [PubMed] [Google Scholar]
- 17.Afrakhte, M., Moren, A., Jossan, S., Itoh, S., Sampath, K., Westermark, B., Heldin, C. H., Heldin, N. E. & ten Dijke, P. (1998) Biochem. Biophys. Res. Commun. 249, 505-511. [DOI] [PubMed] [Google Scholar]
- 18.Kang, Y., Chen, C. R. & Massagué, J. (2003) Mol. Cell 11, 915-926. [DOI] [PubMed] [Google Scholar]
- 19.Chen, C. R., Kang, Y., Siegel, P. M. & Massagué, J. (2002) Cell 110, 19-32. [DOI] [PubMed] [Google Scholar]
- 20.Reynisdottir, I., Polyak, K., Iavarone, A. & Massagué, J. (1995) Genes Dev. 9, 1831-1845. [DOI] [PubMed] [Google Scholar]
- 21.Hannon, G. J. & Beach, D. (1994) Nature 371, 257-261. [DOI] [PubMed] [Google Scholar]
- 22.Tokino, T., Urano, T., Furuhata, T., Matsushima, M., Miyatsu, T., Sasaki, S. & Nakamura, Y. (1996) Hum. Genet. 97, 625-631. [DOI] [PubMed] [Google Scholar]
- 23.Lee, M. H., Reynisdottir, I. & Massagué, J. (1995) Genes Dev. 9, 639-649. [DOI] [PubMed] [Google Scholar]
- 24.Matsuoka, S., Thompson, J. S., Edwards, M. C., Bartletta, J. M., Grundy, P., Kalikin, L. M., Harper, J. W., Elledge, S. J. & Feinberg, A. P. (1996) Proc. Natl. Acad. Sci. USA 93, 3026-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, M. P., DeBaun, M. R., Mitsuya, K., Galonek, H. L., Brandenburg, S., Oshimura, M. & Feinberg, A. P. (1999) Proc. Natl. Acad. Sci. USA 96, 5203-5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cleary, M. A., van Raamsdonk, C. D., Levorse, J., Zheng, B., Bradley, A. & Tilghman, S. M. (2001) Nat. Genet. 29, 78-82. [DOI] [PubMed] [Google Scholar]
- 27.John, R. M., Ainscough, J. F., Barton, S. C. & Surani, M. A. (2001) Hum. Mol. Genet. 10, 1601-1609. [DOI] [PubMed] [Google Scholar]
- 28.Blint, E., Phillips, A. C., Kozlov, S., Stewart, C. L. & Vousden, K. H. (2002) Proc. Natl. Acad. Sci. USA 99, 3529-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mager, J., Montgomery, N. D., de Villena, F. P. & Magnuson, T. (2003) Nat. Genet. 33, 502-507. [DOI] [PubMed] [Google Scholar]
- 30.Fitzpatrick, G. V., Soloway, P. D. & Higgins, M. J. (2002) Nat. Genet. 32, 426-431. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, Y. & Derynck, R. (2000) J. Biol. Chem. 275, 16979-16985. [DOI] [PubMed] [Google Scholar]
- 32.Jakubowiak, A., Pouponnot, C., Berguido, F., Frank, R., Mao, S., Massagué, J. & Nimer, S. D. (2000) J. Biol. Chem. 275, 40282-40287. [DOI] [PubMed] [Google Scholar]
- 33.Yuan, Y., Shen, H., Franklin, D. S., Scadden, D. T. & Cheng, T. (2004) Nat. Cell Biol. 6, 436-442. [DOI] [PubMed] [Google Scholar]
- 34.Cheng, T., Rodrigues, N., Shen, H., Yang, Y., Dombkowski, D., Sykes, M. & Scadden, D. T. (2000) Science 287, 1804-1808. [DOI] [PubMed] [Google Scholar]
- 35.Cheng, T., Rodrigues, N., Dombkowski, D., Stier, S. & Scadden, D. T. (2000) Nat. Med. 6, 1235-1240. [DOI] [PubMed] [Google Scholar]
- 36.Dao, M. A., Hwa, J. & Nolta, J. A. (2002) Blood 99, 499-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dao, M. A., Taylor, N. & Nolta, J. A. (1998) Proc. Natl. Acad. Sci. USA 95, 13006-13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishimori, S., Tanaka, Y., Chiba, T., Fujii, M., Imamura, T., Miyazono, K., Ogasawara, T., Kawaguchi, H., Igarashi, T., Fujita, T., et al. (2001) J. Biol. Chem. 276, 10700-10705. [DOI] [PubMed] [Google Scholar]
- 39.Hahm, K. B., Cho, K., Lee, C., Im, Y. H., Chang, J., Choi, S. G., Sorensen, P. H., Thiele, C. J. & Kim, S. J. (1999) Nat. Genet. 23, 222-227. [DOI] [PubMed] [Google Scholar]
- 40.Matsuoka, S., Edwards, M. C., Bai, C., Parker, S., Zhang, P., Baldini, A., Harper, J. W. & Elledge, S. J. (1995) Genes Dev. 9, 650-662. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe, H., Pan, Z. Q., Schreiber-Agus, N., DePinho, R. A., Hurwitz, J. & Xiong, Y. (1998) Proc. Natl. Acad. Sci. USA 95, 1392-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hatada, I., Ohashi, H., Fukushima, Y., Kaneko, Y., Inoue, M., Komoto, Y., Okada, A., Ohishi, S., Nabetani, A., Morisaki, H., et al. (1996) Nat. Genet. 14, 171-173. [DOI] [PubMed] [Google Scholar]
- 43.Maher, E. R. & Reik, W. (2000) J. Clin. Invest. 105, 247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, P., Liegeois, N. J., Wong, C., Finegold, M., Hou, H., Thompson, J. C., Silverman, A., Harper, J. W., DePinho, R. A. & Elledge, S. J. (1997) Nature 387, 151-158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.