ABSTRACT
Hosts utilize macroautophagy/autophagy to clear invading bacteria; however, bacteria have also developed a specific mechanism to survive by manipulating the host cell autophagy mechanism. One pathogen, Legionella pneumophila, can hinder host cell autophagy by using the specific effector protein RavZ that cleaves phosphatidylethanolamine-conjugated LC3 on the phagophore membrane. However, the detailed molecular mechanisms associated with the function of RavZ have hitherto remained unclear. Here, we report on the biochemical characteristics of the RavZ-LC3 interaction, the solution structure of the 1:2 complex between RavZ and LC3, and crystal structures of RavZ showing different conformations of the active site loop without LC3. Based on our biochemical, structural, and cell-based analyses of RavZ and LC3, both distant flexible N- and C-terminal regions containing LC3-interacting region (LIR) motifs are important for substrate recognition. These results suggest a novel mechanism of RavZ action on the phagophore membrane and lay the groundwork for understanding how bacterial pathogens can survive autophagy.
KEYWORDS: ATG4B, crystal structure, LC3, Legionella pneumophila, RavZ, SAXS, xenophagy
Introduction
Autophagy, a process by which a cell's own organelles or proteins undergo bulk degradation by lysosomes (or vacuoles in fungi), is tightly regulated to maintain a balance between the synthesis and degradation of cellular products.1,2 Initially, autophagy was presumed to be a nonspecific, bulk process; however, selective autophagy has also been studied extensively.3,4 Selective autophagy requires specific autophagy receptors for specific types of cargo, in that receptor molecules are present for target recognition and removal of various cellular structures, including protein aggregates, various cellular organelles, and intracellular pathogens.5-8 In order to maintain the selectivity of the process, autophagic receptors that simultaneously bind target substrates and the key autophagic molecule Atg8 (LC3 in mammals) are required.3 In addition to its role in the proteasomal degradation system, ubiquitination has been found to play a key role in autophagy by marking specific targets for autophagy-mediated degradation.9 To date, 5 ubiquitin (Ub)-binding autophagic receptors, NBR1, SQSTM1/p62, OPTN (optineurin), CALCOCO2/NDP52 and TOLLIP, are known to interact with LC3 through the so-called LC3-interacting region (LIR) motif.8,10-15
Although a basal level of autophagy occurs in most cells, autophagy activity can increase when specific events occur, such as bacterial infection.16,17 When bacteria invade cells, hosts implement autophagy, in combination with an immune response, to remove the pathogen.16,18 The process involving autophagic elimination of intracellular pathogens is referred to as ‘xenophagy.’19,20 Recent research suggests that the upregulation of autophagy for certain infectious diseases can be utilized as a therapeutic strategy.21 However, the precise mechanisms involved in xenophagy are not fully understood. To survive xenophagy, certain contagious intracellular bacterial pathogens have evolved mechanisms to counter or avoid this process. Three strategies have been identified as being employed by bacteria to avoid host cell autophagy, and include bacterial evasion, inhibition, and subversion.17,19 For example, Legionella pneumophila has a unique mechanism of autophagy inhibition that involves use of an effector protein, RavZ.22 Legionella secretes RavZ protein, via the Dot/Icm type IV secretion system, into the infected cells.22,23,24 The RavZ protein localizes onto the phagophore membrane (the precursor of the autophagosome) and then cleaves the C-terminal region of phosphatidylethanolamine (PE) -conjugated Atg8-family proteins such as LC3 and GABARAP.17,22 Although Atg8-family members possess Ub folding structural domains, they do not possess the Gly-Gly sequence conserved among ubiquitin-like (Ubl) modifiers, but do contain a conserved aromatic residue (phenylalanine or tyrosine)-Gly sequence at the C terminus.14
The intracellular LC3 processing enzyme, ATG4B (Atg4 in yeast), cleaves nascent LC3 to expose the C-terminal glycine, which then allows for conjugation to PE and participation of the mature LC3 in the phagophore elongation steps. ATG4B is also involved in the recycling of LC3 by specific deconjugation of the PE moiety.25 In contrast, the cleavage of LC3–PE by RavZ occurs between the conserved C-terminal glycine and aromatic amino acid in Atg8-family members.22 Cleaved LC3 bearing the aromatic residue at the C terminus is unable to participate in the phagophore elongation step.22 Thus, the cysteine protease RavZ might possess a catalytic mechanism similar to ATG4B, although there are clear differences between the 2 enzymes. The molecular mechanism involving the recognition of LC3 by RavZ, and in particular the recognition of membrane-anchored LC3–PE, remains unknown. During preparation of this manuscript the structure of RavZ was published, and largely focused on the membrane targeting activity of RavZ.26 Here, we report on a wealth of structural, biochemical, biophysical, and cell biology data we have acquired, and use it to propose a novel mechanism of RavZ action. In contrast to ATG4B, different conformations of the active site loop of RavZ without LC3 were found in our structures, and the critical role of the flexible N- and C-terminal regions of RavZ were also investigated. Furthermore, we show that RavZ forms a 1:2 complex with 2 LC3 molecules (we are referring to the LC3B isoform throughout the manuscript) through interaction of the LIR motifs located on the N- and C-terminal regions. These results lay the groundwork for understanding how bacterial pathogens survive autophagy. Furthermore, our acquired structural information may contribute toward the successful design and implementation of antimicrobial drugs that target specific bacteria.
Results
RavZ forms a complex with 2 molecules of LC3
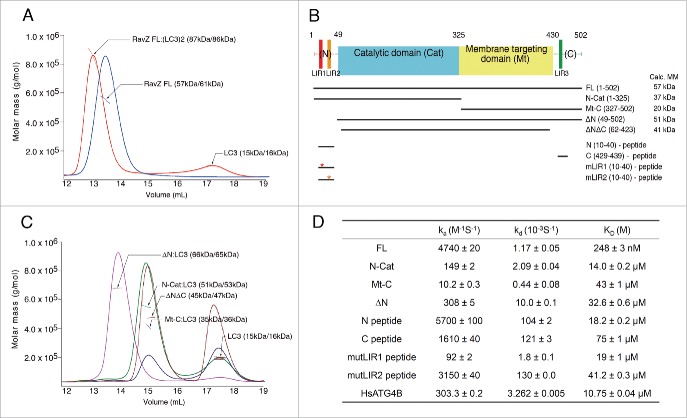
RavZ cleaves LC3–PE to expose the C-terminal phenylalanine, but is unable to cleave unconjugated LC3.19 After RavZ and LC3 were purified separately, a mixture comprising the 2 proteins was subjected to size exclusion chromatography with multi-angle light scattering (SEC-MALS). The 2 proteins formed a tight complex and, interestingly, the molecular mass (MM) determined by MALS was 86 kDa, which is substantially higher than expected because the MM of wild-type (WT) full-length RavZ (FL; residues 1–502) and LC3 is 57 and 15 kDa, respectively (Fig. 1A). This result indicated that RavZ could interact with 2 LC3 molecules simultaneously. This was intriguing since a similar molecule, ATG4B, forms a 1:1 complex with LC3 in solution (Fig. S1). In order to identify the LC3-binding regions of RavZ, several different constructs of RavZ were generated (Fig. 1B). We deleted the N-terminal (ΔN; residues 49–502), C-terminal (N-Cat; residues 1–325), and both terminal regions (ΔNΔC; residues 62–423) of RavZ to examine whether these regions are critical for LC3 binding (Fig. 1C). SEC-MALS experiments with these deletion mutants were also performed. As shown in Fig. 1C, the calculated MM of the ΔN and N-Cat mutants was 66 and 51 kDa, respectively, suggesting the presence of a 1:1 complex. Moreover, the ΔNΔC mutant, comprising deletions of both N- and C-terminal regions, exhibited no binding to LC3. The MALS data clearly showed that the experimentally determined MM was the same as expected from the calculated MM of the ΔNΔC mutant without LC3 (Fig. 1C). The mutant comprising the membrane-targeting domain (Mt) with the C-terminal region (Mt-C; residues 327–502) also formed a 1:1 complex with LC3. These results clearly demonstrate that each N- and C-terminal region of RavZ binds one molecule of LC3 independently.
Figure 1.
Interaction between RavZ and LC3. (A) The RavZ-LC3 complex (red line) and RavZ alone (blue line) analyzed by SEC-MALS. The horizontal line represents the measured MM. Each species is indicated by an arrow with experimental (MALS) and theoretically calculated (Calc) molar mass values shown in parentheses (MALS/Calc). This shows the 1:2 binding stoichiometry between RavZ and LC3. (B) Domain architecture and RavZ constructs in this study. Three potential LIR motifs in the primary sequence are numbered sequentially: LIR1, LIR2 and LIR3. The abbreviations of each construct and the calculated molecular weights are shown. (C) The SEC-MALS results with deletion mutants of RavZ in the presence of LC3. ΔN (magenta line), N-Cat (green line) and Mt-C (blue line) in the presence of LC3 elute earlier and their MMs determined by SEC-MALS are 15-kDa greater than the calculated MMs of the free mutants, indicating 1:1 complex formation. The ΔNΔC mutant comprising deletion of the flexible N- and C-terminal regions (brown line) is unable to form a complex with LC3. Excess LC3 with MM of 15 kDa elutes later. (D) Summary of SPR data. The binding of RavZ and its deletion mutants to immobilized LC3 was measured, and the binding of ATG4B was also measured as a test case (See Fig. S1A and S2 for the SPR sensorgrams). The equilibrium dissociation constant (KD) was obtained by dividing the dissociation rate constant (kd) by the association rate constant (ka).
Next, we measured the binding affinity of RavZ WT in addition to that of the above mutants with LC3 using surface plasmon resonance (SPR) (Fig. 1D and Fig. S2). The dissociation constant (KD) between RavZ FL and LC3 was 248 nM, which is very tight, and that between the deletion mutants of RavZ and LC3 was reduced by an order of magnitude. As expected, the KD between the ΔNΔC mutant and LC3 could not be measured. The binding affinity between the N-terminal region of RavZ and LC3 was higher than that between the C-terminal region and LC3, and interestingly the single second LIR (LIR2) peptide had a similar binding affinity to LC3 as the intact N-terminal peptide containing the 2 LIRs, LIR1 and LIR2 (Fig. 1D). Although there are 2 LIR motifs in the N-terminal region, 2 molecules of LC3 cannot bind to this region simultaneously based on the SEC-MALS results, and the SRP data clearly showed that the LIR2 peptide had stronger binding affinity to LC3 than the LIR1 peptide. RavZ containing only the N-terminal LIRs, N-Cat, had similar binding affinity to LC3 as the single LIR2 peptide or N-terminal peptide, and the almost 50-fold stronger binding affinity between RavZ and LC3 might simply reflect a synergistically increased chance of the interaction. In contrast to the mutant peptides of the N-terminal region that still have significant binding affinity (Fig. 1D), the mutant peptide of the C-terminal region essentially lost binding affinity (KD of 15 mM; data not shown).
Interestingly, the KD between LC3 and ATG4B, which has only one LIR motif, was 10.75 μM, approximately 40-fold lower than that between LC3 and RavZ (Fig. 1D and Fig. S1B). This value is comparable with the data obtained using RavZ mutants (ΔN and N-Cat), which have 1:1 binding stoichiometry with LC3. The affinity measurement is consistent with previous SEC-MALS data, and the strong binding affinity between RavZ and LC3 can be accounted for by the synergistic binding of LC3 using separate N- and C-terminal regions of RavZ. Therefore, we concluded that RavZ forms a 1:2 complex with LC3 using its N- and C-terminal regions, and interacts with LC3 much more tightly than ATG4B.
The LC3-binding N- and C-terminal regions of RavZ are structurally flexible
Three crystal structures of RavZ were refined at 2.74, 2.81 and 2.54 Å resolution to an Rfree of 0.287, 0.227 and 0.213, respectively (Table 1). The N-terminal 48 residues and C-terminal 71 residues were not visible in the electron density map from crystal form-I and -II using RavZ FL, suggesting that these regions have high flexibility. The structure determined using crystal form-III with the ΔN construct also showed no electron density at the C-terminal region, even with different crystal packing. Interestingly, the invisible regions were critical for LC3 binding, as shown in Fig. 1B. The overall structure of RavZ (residues 49–431 and nearly the same as ΔNΔC) comprised 2 separate domains (Fig. 2A and 2B), an N-terminal α/β-fold (residues 49–325) and a C-terminal α-helical domain (residues 326–431). The N-terminal domain showed a well-conserved folding pattern typical of Ubl deconjugating proteases (Fig. S3). Specifically, 7 α-helices and 9 β-strands formed a distinct globular domain, and α5 was a central helix that was surrounded by the other α-helices and β-strands in a manner similar to a hand grabbing a stick (Fig. 2A). This domain includes the catalytic triad (His176, Asp197 and Cys258) and thus defines the catalytic domain. The catalytic domain of RavZ showed high structural similarity (Z-score > 9) to XopD from Xanthomonas (PDB ID: 2OIX),27 SENP8/Den1 in complex with NEDD8 (PDB ID: 1XT9),28 human SENP2 protease (PDB ID: 1TH0),29 SENP3/Ulp1 (PDB ID: 2HL8),30 and unexpected structural differences with human ATG4B (PDB ID: 2CY7).31 However, the catalytic triads of these Ubl deconjugating proteases were all quite similar (Fig. S4).
Table 1.
Data collection and refinement statistics.
| FL – form I | FL – form II | FL SeMet (SAD) – form II | ΔN – form III | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | I422 | F222 | F222 | P6322 |
| Cell dimensions | ||||
| a, b, c (Å) | 222.0, 222.0, 71.3 | 72.2, 312.8, 313.7 | 72.2, 312.8, 313.7 | 219.3, 219.3, 65.9 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 120 |
| Peak | ||||
| Wavelength (Å) | 1.0000 | 1.0000 | 0.9794 | 0.9000 |
| Resolution (Å) | 39.3–2.74 (2.84–2.74) | 43.6–2.81 (2.92–2.81) | 30.0–3.5 (3.56–3.5) | 41.4–2.54 (2.64–2.54) |
| Rmerge | 0.051 (0.422) | 0.066 (0.448) | 0.105 (0.244) | 0.084 (0.455) |
| I / σI | 20.3 (4.7) | 43.6 (2.3) | 29.6 (6.0) | 47.6 (7.1) |
| Completeness (%) | 97.9 (98.1) | 97.7 (94.7) | 99.2 (97.6) | 99.7 (100) |
| Redundancy | 10.6 (10.0) | 5.0 (3.1) | 9.2 (5.9) | 9.1 (9.2) |
| Refinement | ||||
| Resolution (Å) | 2.74 | 2.81 | 2.54 | |
| No. reflections | 23,216 | 42,554 | 30,865 | |
| Rwork / Rfree | 0.224/0.287 | 0.187/0.227 | 0.178/0.213 | |
| No. atoms | 3,071 | 5,873 | 3,067 | |
| Protein | 3,038 | 5,873 | 3,002 | |
| Water | 33 | – | 65 | |
| B-factors | 78.02 | 95.50 | 62.09 | |
| Protein | 78.30 | 95.50 | 62.26 | |
| Water | 52.54 | – | 53.94 | |
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.011 | 0.011 | 0.009 | |
| Bond angles (°) | 1.34 | 1.49 | 1.10 | |
| Ramachandran plot (%) | ||||
| Favored | 96.1 | 95.6 | 96.8 | |
| Allowed | 3.7 | 3.7 | 3.2 | |
| Outliers | 0.2 | 0.7 | 0 | |
| MolProbity | ||||
| Clashscore | 28.24 | 19.82 | 6.86 |
Figure 2.
Structure of RavZ. (A) Ribbon diagram of RavZ from L. pneumophila. The catalytic (residues 49–325) and membrane targeting (residues 326–423) domains are colored aquamarine and yellow, respectively. The catalytic site is indicated by an arrow. The N-terminal 48 and C-terminal 71 residues are missing and shown as red and green dots, respectively. The secondary structural elements are labeled sequentially. (B) Schematic representation of full-length RavZ. The invisible N-terminal and C-terminal regions in the crystal structure are represented by a red and green dotted line, respectively. The putative LC3-binding sites, LIR1, LIR2 and LIR3, are marked. (C) Superposition of 2 crystal structures (crystal form I and III) showing different loop conformations near the active site. This view is re-oriented from Fig. 2A to show the catalytic triad (approximately 110° and 30° along the horizontal and vertical axes, respectively). The overall structure is virtually identical except for the active site region. (D) A close-up view of the active site, which is highlighted as a transparent box in panel (C). The active site is covered by the flexible internal loop in the closed conformation of crystal form I (cyan), whereas it is widely accessible in the open conformation of crystal form III (blue). Catalytic triad residues (His176, Asp197 and Cys258) are presented as stick models. The interacting atoms in the catalytic triad are lined green and yellow for the closed and open conformations, respectively. The movement of the flexible loop is indicated by a double-headed red arrow.
The rest of the molecule was made up of a small helical domain comprising all 5 α-helices (Fig. 2A). Although 2 bacterial proteins, YvqK protein (PDB ID: 1RTY)32 and hypothetical protein TA0546 (PDB ID: 1NOG),33 showed high structural similarity (Z score > 6) with this helical domain, its role is not immediately apparent. Horenkamp et al. reported that this helical region is a targeting domain for phagophore membranes by the binding of phosphatidylinositol 3-phosphate (PtdIns3P).26 Thus, the helical domain is referred to as a membrane targeting (Mt) domain; full-length RavZ is divided into 4 domains, comprising an N-terminal LC3 binding, catalytic, membrane targeting, and C-terminal LC3 binding domains (Fig. 2B).
The active site loop of RavZ shows different conformations without LC3
Three different crystal forms of RavZ, I, II and III, were obtained and the structures compared. Structures derived from 2 of our 3 crystal forms (form I and II) were virtually identical, although the crystal packing differed (Table 1), and our crystal form I and a previously reported structure26 have the same crystal packing, even though different constructs were used for the crystallization. The structure in the other crystal form (form III) with the ΔN mutant, however, showed a notable difference (Fig. 2C). Although the C-terminal region was invisible in both crystal forms, several residues including the N-terminal histidine tag were visible in crystal form III. Furthermore, there was a clear difference in the loop region covering the active site (Fig. 2D). The loop (residues 250–256) also showed high flexibility and 2 different conformations, most probably due to different crystal packing, the behavior of which might differ from the conformational regulation suggested by the ATG4B structures.25 The regulatory loop of free ATG4B covers the active site and consequently catalytic Cys74 is buried, whereas the regulatory loop undergoes a conformational change to form a groove to accommodate the C-terminal region of LC3 upon complex formation. In contrast, the corresponding loop in RavZ showed an open conformation without LC3 complex formation (Fig. 2C), suggesting that the loop covering the active site is also very flexible and that the regulation of RavZ differs from that of ATG4B.
The catalytic triad of RavZ was also similar to other Ubl cysteine proteases (Fig. S4). The Nϵ2 atom of His176 was located 3.3 Å from the Oγ atom of Ser258 (Cys258), and the Nδ1 atom of His176 was located 2.7 Å from the Oδ1 atom of Asp197 in the conformation of crystal form I and II of RavZ, whereas the corresponding distances were 4.1 and 2.4 Å, respectively, in the conformation of crystal form III of RavZ (Fig. 2D and Fig S4A). The orientation of the Oγ atom of Ser258 was rotated out from the catalytic triad and was located at a relatively longer distance. The exact meaning of this rotation is not yet clear, and may be revealed following examination of the complex structure between RavZ and LC3–PE.
Both LIR motifs in the N- and C-terminal region of RavZ are critical for LC3 binding
In an effort to identify the exact binding sites of LC3, the sequence of RavZ was analyzed. It is well known that the conserved sequence motif W-x-x-L (where ‘x’ is any amino acid residue) is utilized for specific interactions with Atg8-family proteins, including LC3.10,14 The W-site is sometimes substituted with Tyr or Phe, and the L-site with Ile or Val.10 Three potential LIR motifs, 16F-x-x-L19, 29F-x-x-L32 and 435F-x-x-I438, referred to as LIR1, LIR2 and LIR3, respectively, were identified (Fig. 3A and Fig. S5). Compared to the WT LIR motif, the F-type frequently contains acidic residues at the N-terminal side of the motif.10 All 3 potential LIR motifs in RavZ have similar characteristics to the F-type LIR. Interestingly, the first and second LIRs were located close within the primary sequence at the N-terminal region of RavZ, and the third LIR3 was located at the C-terminal region of RavZ (Fig. S5). The functional LIR must be a short linear motif arising from an intrinsically disordered region,34 and all aforementioned potential LIRs possess a high PONDR-FIT score and the regions are all disordered in our crystal structures (Fig. 2B and Fig S5).
Figure 3.
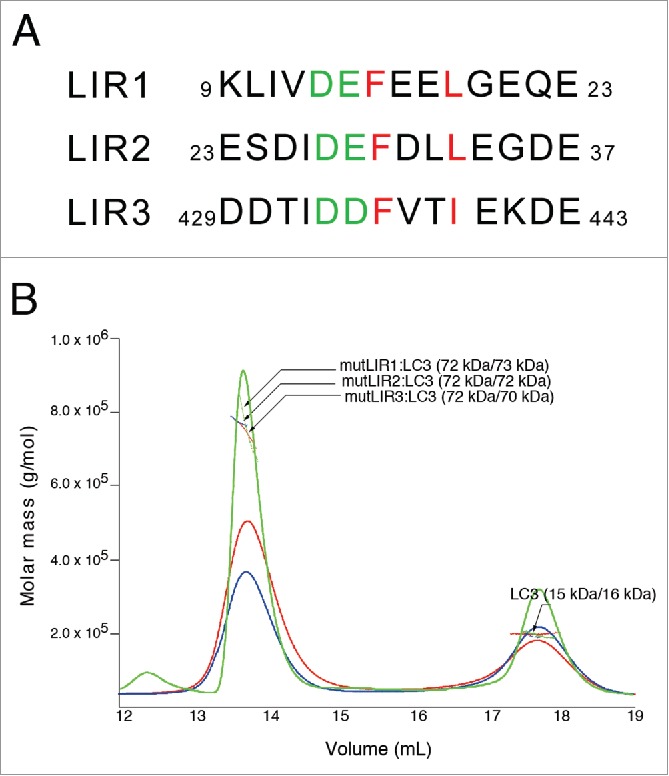
Sequence alignment of LIR motifs of RavZ and solution behavior of the mutants. (A) Sequence alignment of 3 potential RavZ LIR motifs, LIR1, LIR2 and LIR3. The critical hydrophobic phenylalanine and leucine (or isoleucine) residues in the F-x-x-L(I) motif are colored red and the preceding 2 acidic residues are colored green. (B) SEC-MALS results confirming the importance of the LIR motifs. Green, red and blue profiles represent mutLIR1, mutLIR2 and mutLIR3, respectively, in the presence of LC3, and the corresponding horizontal lines represent the measured data obtained by MALS. Each species is indicated by an arrow with experimental (MALS) and theoretically calculated (Calc) molar mass values shown in parentheses (MALS/Calc). All LIR mutants form a 1:1 complex and, taken together with the MALS results using deletion constructs (See Fig. 1C), both LIR1 and LIR2 motifs participate in the binding of one LC3, while LIR3 binds to one LC3, independently.
To further delineate these potential LIR motifs, we generated a series of double-mutant constructs comprising mutLIR1: F16A L19A; mutLIR2: F29A L32A; and mutLIR3: F435A I438A. We purified each mutant protein and performed SEC-MALS experiments with LC3. As noted in the above section, RavZ FL formed a complex with 2 molecules of LC3 (Fig. 1A), however, all double mutants (mutLIR1, mutLIR2 and mutLIR3) in the presence of LC3 generated 1:1 complexes (Fig. 3B), as shown by ΔN and ΔC with LC3 (Fig. 1B). It is intriguing that 2 N-terminal LIRs are able to bind 1 LC3, and that any mutation in 1 LIR motif affects the binding affinity of the other LIR. These data clearly show that RavZ forms a complex with 2 LC3 molecules, one LC3 molecule interacting with N-terminal LIR motifs and the other using a C-terminal LIR motif, LIR3. Compared with ATG4B, which only has 1 LIR motif at its N-terminal region (7Y-x-x-L10), utilization of separate LIR motifs at the N- and C-terminal regions of RavZ accounts for its higher affinity to LC3 with a 1:2 binding stoichiometry (Fig. 1D), and also suggests a different mode of action on the phagophore membrane, which will be discussed later.
The LC3-binding N- and C-terminal regions of RavZ are critical for inhibiting autophagy
To confirm the above in vitro data of RavZ, various mutant constructs were generated for the monitoring of autophagy flux using green fluorescent protein (GFP)-tagged LC3, a well-known autophagy marker.34-36 Autophagy was induced by the addition of rapamycin, and punctate GFP-LC3 fluorescent signals indicating recruitment of LC3 protein to phagophores and/or its presence on autophagosomal membranes were detected in the cytosol (Fig. 4). Overexpression of WT RavZ FL removed almost all of the GFP puncta in the cells, indicating that LC3 was immediately cleaved from the phagophores or autophagosomes. As a control, the inactive C258S mutant was tested and the results showed clear accumulation of autophagosomes in cells due to the absence of RavZ enzymatic activity (Fig. 4A).
Figure 4.
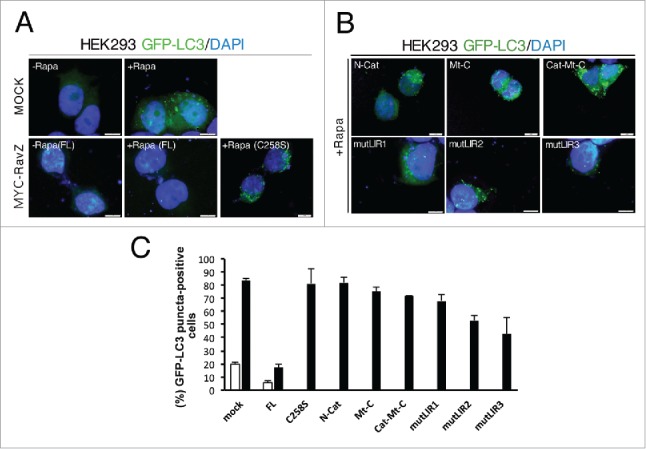
Accumulation of autophagosomes in RavZ mutant cells. (A) HEK293 cells were transduced with GFP-LC3. After 24 h of infection, cells were transfected with expression vectors for MYC-tagged RavZ WT (FL) and C258S inactive mutant as a control. After 24 h of transfection, cells were left untreated (-Rapa) or treated (+Rapa) with 500 nM rapamycin for 12 h. Cells were then fixed, permeabilized, and stained with DAPI. Representative images of GFP-LC3 (green) and DAPI (blue) fluorescence are shown. Scale bar: 10 μm. (B) The same experiments with RavZ variants (N-Cat, Mt-C, Cat-Mt-C, mutLIR1, mutLIR1 and mutLIR1). (C) The fluorescence intensity of GFP-LC3 in the cytoplasm of > 50 cells for each experimental group was quantified and expressed as a percentage relative to the control group. Data represent the means ± SEM (standard error of mean) from 3 independent experiments.
Use of RavZ deletion mutants such as N-term, C-term, or both, resulted in the accumulation of autophagosomes in cells to levels very similar to those observed with use of the catalytically inactive mutant (Fig. 4B and 4C). We also performed the same experiments with point mutants, mutLIR1, mutLIR2 and mutLIR3, which possess a defect in LC3-binding (Fig. 3A). Based on our in vitro data, all double mutants formed a 1:1 complex with LC3 (Fig. 3B), and therefore must have a lower binding affinity compared with RavZ FL. Quantification of the fluorescence data indicated that a milder effect was shown by the point mutants compared with the deletion mutants (Fig. 4C), with different effects being observed with some of the point mutants. Similar results were also obtained using western blot analyses of LC3 turnover by examination of the LC3-I and LC3-II bands (Fig. S6). This result is consistent with those of our biochemical assays and structural analyses. In conclusion, the flexible N- and C-terminal regions in RavZ, and in particular the LIR motifs, are important for the recognition of LC3–PE conjugates and play a critical role in blocking autophagy in cells.
Solution structure of the RavZ-LC3 complex
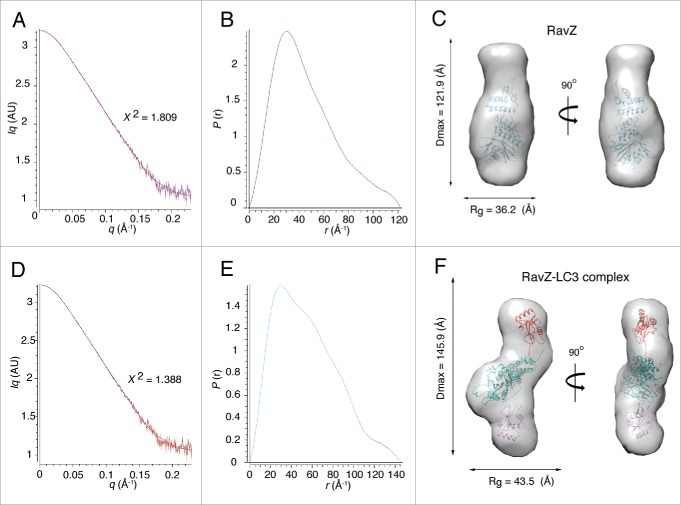
Although it is necessary to determine the complex structure between RavZ and LC3 to confirm that 2 independent LC3 molecules bind to the LIR motifs located on the flexible N- and C-terminal regions of RavZ, the crystal was not available for high-resolution structure determination. Instead, we performed a structural study in solution using small-angle X-ray scattering (SAXS). Two SAXS data were obtained using RavZ alone and RavZ in complex with LC3 (Fig. 5 and Fig. S7). These data fit well with the theoretically calculated model based on the Guinier plot (Fig. 5A and Fig. 5D). For the free RavZ model, Rg and Dmax values were calculated to be 36.20 ± 2.4 and 121.9 Å, respectively (Fig. 5B). The molecular mass derived from the estimated Porod volume was very similar to the calculated MM (Table S1). The molecular envelope matched well with the crystal structure of RavZ (Fig. 5C). Because the regulatory domain was smaller than the catalytic domain, the apo RavZ model resembles a gourd-shaped bottle or snowman, and the empty region must be the location of the invisible N- and C-terminal domains containing the LIR motifs.
Figure 5.
Structure of the 1:2 complex between RavZ and LC3. (A) Scattering curve of RavZ alone, (B) Distance distribution function, P(r), of RavZ alone and (C) Molecular envelope of RavZ alone. The high-resolution crystal structure of RavZ was fitted into the low resolution molecular envelope generated by the SAXS data by rigid body docking. (D) Scattering curve of the RavZ-LC3 complex, (E) Distance distribution function, P(r), of the RavZ-LC3 complex and (F) Molecular envelope of the RavZ-LC3 complex. The 1:2 LC3-RavZ-LC3 model was generated using Chimera (UCSF). Two molecules of LC3 are located in the direction of the invisible N- and C-terminal regions of RavZ.
The SAXS data of the RavZ and LC3 complex yielded Rg and Dmax values of 43.47 ± 0.38 and 145.9 Å, respectively (Fig. 5E). The calculated molecular mass derived from the estimated Porod volume was very close to that of the 1:2 complex between RavZ and LC3. The molecular envelope of the RavZ and LC3 complex showed a twisted Z-shaped conformation that had an extra space toward the invisible N- and C-terminal direction of RavZ (Fig. 5F). This additional space was large enough to accommodate an LC3 molecule at both sides, and thus we generated the initial 1:2 complex model considering the previous complex structure of the Ubl protease-Ubl as well as the HsATG4B-LC3 complex (Fig. S8). The final rigid-body fitting of the initial model was matched well with the SAXS molecular envelope (Fig. 5F), and a plausible mode of action of RavZ on the phagophore membrane can be suggested (Fig. 6) and will be discussed below.
Figure 6.
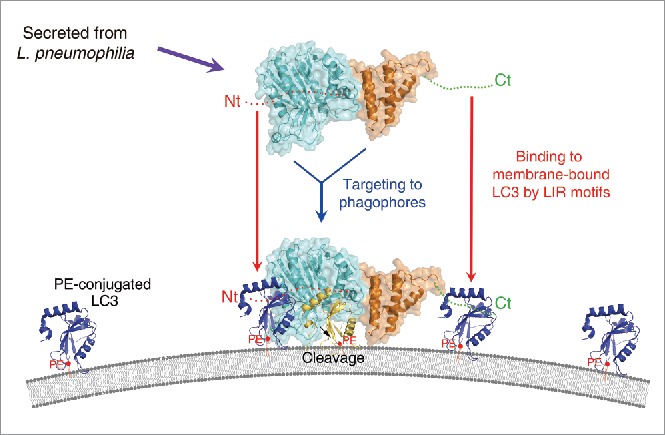
A plausible model for the mode of action of RavZ. The RavZ protein is secreted from L. pneumophila and then targeted to the phagophore or autophagosomal membrane. The catalytic domain of RavZ possessing cysteine protease activity is colored cyan and the membrane-targeting domain possessing binding affinity for the PtdIns3P-rich membrane23 is colored orange. The LIR motif containing the N- and C-terminal regions are indicated as red and green dots, and labeled as Nt and Ct, respectively. PE-conjugated and membrane-anchored LC3 is shown as a blue ribbon and PE is shown as a red ball with tails. Cleaved LC3 (by RavZ) is shown in yellow. RavZ possesses higher binding affinity for LC3 than ATG4B because it has 2 independent LIR motifs at the N- and C-terminal regions (Nt and Ct, respectively). We propose that RavZ achieves greater binding affinity for membrane-bound LC3 molecules (blue ribbon) by using the N- and C-terminal LIR motifs, and this event is critical for correct orientation to facilitate cleavage of the LC3–PE substrate generating a product that cannot be reconjugated to PE (yellow ribbon). Using this elegant and superior mechanism with RavZ as the key player, L. pneumophila has evolved an effective survival mechanism against host cell autophagy.
Discussion
Legionella pneumophila invades and replicates inside macrophages in humans, and causes Legionnaires' disease and Pontiac fever.37 This bacterium has evolved to survive in host cells, and one survival mechanism involves the inhibition of host cell autophagy by irreversible deconjugation of LC3 using the effector RavZ.17,22 Therefore, the RavZ enzyme competes with the host protease ATG4B, which catalyzes the maturation of LC3 for autophagosome formation and recycling of LC3 for continued autophagy.38,39 Taking into account a previously reported crystal structure, ATG4B is likely to interact with LC3 via 2 different regions of ATG4B, namely LC3(S), at which substrate LC3 binds to the active site of ATG4B, and LC3(N), at which nonsubstrate LC3 binds to the N-terminal LIR motif of ATG4B (Fig. S8B). A conformational change of the N-terminal tail upon LC3 binding allows for recognition of the C-terminal tail of LC3 by the catalytic residues of ATG4B. Indeed, LC3 is an enzymatic product of ATG4B and the 1:1 binding stoichiometry between ATG4B and LC3 in solution must be the result of the interaction between the LIR motif of ATG4B and LC3, with a dissociation constant KD of 10.75 μM (Fig. S1B). Compared with ATG4B, several key and immediate questions are raised concerning the mechanism of RavZ, including: 1) Why does RavZ cleave PE-conjugated LC3 in contrast to ATG4B, which cleaves cytosolic LC3 precursor in addition to LC3–PE on the autophagosomal membrane? 2) What are the structural similarities and differences of RavZ and ATG4B, such as they relate to conformational changes of the regulatory loop and N-terminal tail, and cleavage site specificity? 3) Why is RavZ targeted only to the phagophore membrane when high curvature and PtdIns3P concentrations are not the unique feature of these structures? Although some definitive answers await high-resolution complex structure determinations between RavZ and LC3–PE, our data can explain many of these issues in a reasonable manner.
It is natural that RavZ from L. pneumophila possesses higher affinity for the LC3 substrate than ATG4B for the purpose of destroying the host cell autophagy system. The binding affinity of RavZ to LC3 is approximately 40 times higher than that of ATG4B (Fig. 1D), and this is mainly due to the binding stoichiometry. Our SEC-MALS, mutagenesis and SAXS data clearly show that RavZ interacts with 2 molecules of LC3 with separate and flexible N- and C-terminal regions containing LIR motifs. As shown in Fig. 1D, the RavZ mutants containing only one LIR motif show a comparable dissociation constant with ATG4B. Therefore, we concluded that the stronger binding affinity of RavZ to LC3 originates from the synergistic binding avidity and that secreted RavZ from L. pneumophila efficiently removes the LC3 competing with ATG4B from the phagophore membrane. As shown in the crystal structure of the ATG4B-LC3 complex, ATG4B has 2 binding sites, one located within the N-terminal LIR motif and the other within the active site (Fig. S8B). The product LC3 does not bind to the active site, although LC3 binds to the LIR motif forming a 1:1 complex (Fig. S1A). The same scenario may occur where RavZ binds to 2 molecules of LC3 using the LIR motifs and a single substrate form of LC3, LC3–PE, binds to the active site of RavZ, but is not matured as well as precursor LC3.22 For this reason, a 1:2 complex of RavZ-LC3 was observed in solution (Fig. 1A).
As noted above, specific binding to the PE moiety and specific peptide bond cleavage between the aromatic (phenylalanine or tyrosine) and glycine residues can only be understood following high-resolution RavZ:LC3–PE complex structure determinations. However, our crystal structures clearly show that there is conformational flexibility of the loop covering the active site (Fig. 2D). The open active site of RavZ was observed using a construct comprising deletion of the N-terminal region (ΔN mutant) (Fig. 1B). Two LIRs, LIR1 and LIR2, accommodate only one LC3 with a short 9-residue gap in between and, more intriguingly, a double mutation in either the LIR1 or LIR2 region resulted in complete loss of binding affinity to the LC3 molecule, implying that the motifs somehow communicate with each other (Fig. 3B), although the isolated peptides behaved differently (Fig. 1D). We speculate that the 2 invisible LIRs may play a role in regulating the activity of RavZ. Furthermore, our low resolution RavZ-LC3 complex structure obtained by SAXS experiments shows that the LC3 molecules bind to the flexible N- and C-terminal regions of RavZ, forming an overall twisted Z-shaped conformation (Fig. 5F and S8C). We also determined the crystal structures of LC3 in complex with LIR1, LIR2 and LIR3 peptides independently (data not shown), but the results were still not clear enough to understand how the N- and C-terminal region regulate the activity of RavZ.
One of the unique structural features of RavZ is the inserted α-helical domain followed by the catalytic domain (Fig. 2A). In a recent report, the role of this domain was elucidated as comprising a PtdIns3P-binding module which, and in combination with a helix in the catalytic domain, effects targeting to the phagophore membrane.26 However, neither high curvature nor the PtdIns3P concentration are unique features of phagophores or autophagosomes, and the manner by which RavZ specifically localizes on these membranes remains unclear. We now have a plausible model to address this question. The full-length RavZ molecule consists of several distinct domains, and each domain possesses unique features for the efficient inhibition of host cell autophagy. Primarily, RavZ is directed to the PtdIns3P-rich membrane with correct orientation via the helical Mt domain and a helix in the catalytic domain.26 The N- and C-terminal regions of RavZ bind to the membrane-anchored LC3s independently with high avidity using the LIR motifs. The binding of LC3 molecules to both N- and C-terminal LIR regions may be coupled with access of substrate PE-conjugated LC3 to the active site of RavZ (Fig. 6). Once RavZ cleaves the peptide bond between the aromatic and glycine residues, the LC3 product cannot be utilized in the autophagy pathway. It is tempting to speculate that cleaved LC3 still possesses binding affinity to LIR motifs, which are present in numerous autophagic molecules including autophagy receptors and the autophagy core machinery.8,10,14,40 Therefore, the RavZ-generated product has a significant effect not only on the function of ATG4B, but also on the entire autophagy system in the host. In an evolutionary context, L. pneumophila has evolved this superior mechanism, which effects survival in host cells.
Materials and methods
Protein sample preparation
Full-length and shorter constructs of RavZ from Legionella were amplified by PCR. The amplified PCR products were treated with restriction enzymes BamHI and HindIII for full-length RavZ, and BamHI and XhoI for shorter constructs, and were then inserted into modified pMAL-c2X and pET-Duet vectors (Novagen,171254), respectively. Full-length genes of ATG4B and LC3B from Homo sapiens were amplified by PCR. The resultant plasmids were transformed into BL21(DE3) cells. The QuikChange site-directed mutagenesis method (Stratagene) was used to prepare constructs for the purpose of expressing RavZ double-mutants possessing LIR motif disruptions F16A L19A, F29A L32A and F435A I438A. Protein expression was induced by the addition of 0.5 mM IPTG at 18°C for 18 h. Cells were harvested by centrifugation, resuspended in 20 mM Tris-HCl, pH 8.0 buffer containing 100 mM NaCl, and then disrupted by sonication. The cell lysate was loaded onto a HisTrapTM column (GE Healthcare, 11–0004-58), and then eluted using a liner gradient of imidazole (0∼500 mM). The affinity tags were cleaved using thrombin or TEV protease (lab made) by overnight incubation at 4°C, and RavZ and ATG4B constructs were further purified by ion-exchange column chromatography using a HiTrapTM Q HP column (GE Healthcare, 17–5156-01). LC3 was further purified by cation-exchange column chromatography using a HiTrapTM SP HP column (GE Healthcare, 17–5054-01). Finally, proteins were loaded onto a HiLoadTM 16/600 SuperdexTM 200 gel filtration column (GE Healthcare, 28989335) pre-equilibrated with 20 mM HEPES (Sigma, H3375), pH 7.0 buffer containing 100 mM NaCl.
Crystallization and structure determination
Both RavZ FL and the ΔN mutant were concentrated to approximately 20 mg/ml and crystallized at 22°C by the hanging drop vapor diffusion method. The best crystal of FL (C258S mutant; crystal form I) was obtained by mixing 1 µl of protein with 1 µl of reservoir solution containing 1.7–2.0 M NaCl, 20–22% (w/v) PEG3350, 0.1 M magnesium chloride, and 0.1 M imidazole, pH 6.2–6.5 (Rigaku, 1009539). Interestingly, different crystal shapes (C258S mutant; crystal form II) were also obtained using the same crystallization conditions. The crystals of ΔN (C258S mutant; crystal form III) were obtained in 2.9–3.2 M sodium acetate trihydrate, pH 6.8–7.0 (Hampton Research, HR2–144). Both crystals were frozen in liquid nitrogen and cryoprotected in the presence of 20% (v/v) ethylene glycol.
Using crystal form I, diffraction data to 2.74 Å resolution were collected at beamline BL17A, Photon Factory, Japan. For phasing, single-wavelength anomalous dispersion (SAD) data using crystal form II were collected with a Se-peak wavelength (0.9794 Å) at beamline 5C, Pohang Accelerator Laboratory, South Korea. The highest resolution data to 2.54 Å were collected using crystal form III at beamline BL44XU, Spring-8, Japan. The data sets were integrated and scaling was performed using HKL2000 software.41 Statistics for the collected data are listed in Table 1. Using the Se-SAD data set, initial phases were obtained and improved using the SOLVE/RESOLVE module in PHENIX,42 resulting in approximately 80% of the initial model being built automatically. The model was rebuilt manually using COOT43 and refinement was performed using PHENIX.42 The structures of different crystal forms were determined by molecular replacement using the refined RavZ model of crystal form II. All 3 structures obtained using different crystal forms show nearly the same length of respective amino acid residues since the N- and C-terminal regions are invisible, even though RavZ FL was crystallized. All final models of RavZ were validated using MolProbity.44 Statistics for the refinement are also shown in Table 1. Structure comparisons were performed using DALI (http://ekhidna.biocenter.helsinki.fi/dali_server/),45 and all structural figures were drawn using PyMOL (http://www.pymol.org/).
Size-exclusion chromatography with multi-angle light scattering
SEC-MALS experiments were performed using a fast protein liquid chromatography system (GE Health care) connected to a Wyatt MiniDAWN TREOS MALS instrument and a Wyatt Optilab rEX differential refractometer.46 A Superdex 200 10/300 GL (GE Health care, 17–5175-01) gel filtration column pre-equilibrated with 20 mM HEPES, pH 7.0, 100 mM NaCl, 1 mM TCEP (Gold Bio, 51805–45-9) was normalized using ovalbumin. Complexes and individual proteins, prepared separately by the methods described earlier, were injected (1–3 mg/ml, 0.5 ml) at a flow rate of 0.5 ml/min. Data were analyzed using the Zimm model for static light scattering data fitting and represented using an EASI graph with a UV peak in the ASTRA V software (Wyatt).
Surface plasmon resonance
All SPR experiments were conducted at 22°C on a Reichert SR7500DC BIAcore 2000 instrument at Korea Basic Science Institute using a buffer comprised of 20 mM HEPES, pH 7.0, 150 mM NaCl, 1 mM DTT. Initially, LC3 was immobilized onto the PEG chip according to the manufacturer's instructions. Various concentrations of RavZ or mutants (10–62.5 nM) were then injected at 30 μl/min over the chip. The response of RavZ or mutants was calculated by subtracting that of the blank flow cell. The same experiments were performed with ATG4B for comparison. For further confirmation, full-length RavZ was immobilized onto the PEG chip described above and the same experiment was performed using various concentrations of LC3. All experiments were each performed 5 times. Data were calculated using Scrubber2 and Clamp software.
Small-angle X-ray scattering
Solutions of the RavZ and RavZ-LC3 complex were prepared in gel filtration buffer comprised of 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1mM DTT. The protein concentration of RavZ and RavZ-LC3 was 10 and 20 mg/ml, respectively. Scattering data of the RavZ and RavZ-LC3 complex were collected at beamline BL10C at Photon Factory and beamline 4C at Pohang Accelerator Laboratory. Details of the experimental parameters are shown in Table S1. Briefly, the scattering images from proteins at various concentrations were reduced into 2D data via circular integration. Preliminary analysis of the 2D data with PRIMUS (ATSAS program suite) provided the radius of gyration (Rg), Porod volume, and experimental molecular weight.47 Ab initio modeling and averaging of these models were performed using DAMMIF and DAMAVER, respectively. Rigid body modeling of the crystallographic structure on dummy-atom models was computed using the Situs program suite.48
Fluorescence analysis of autophagy
The full-length Legionella pneumophila RavZ gene containing a C-terminal MYC tag was cloned into the pCMV-5α vector (Agilent Tech., 211174) using the BamHI and XhoI restriction sites. The RavZ mutants, C258S, F435A I438A, N-Cat (residues 1–325), Mt-C (residues 328–502) and ΔN (residues 49–502), were generated by PCR-mediated site-directed mutagenesis using the same restriction enzymes. HEK293T or COS-7 cells were cultured under a humidified atmosphere of 5% (v/v) CO2 at 37°C in Dulbecco's modified Eagle's medium (Gibco, 11885–084) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone, SH30071). For DNA transfection, cells were transfected for 2 d with appropriate vectors using polyethyleneimine (Sigma-Aldrich, 764647). For fluorescence analyses, HEK293T or COS-7 cells were transduced for 24 h with GFP-LC3 according to the manufacturer's protocol (PremoTM Autophagy Sensor LC3B-FP [BacMam 2.0] system, Invitrogen). For autophagy induction, 500 nM rapamycin (Sigma-Aldrich, R0395) was added to the cells. The cells were fixed, permeabilized, stained with 4′,6-diamidino-2-phenylindole (DAPI; 5 µg/ml), and then examined by fluorescence microscopy. Fluorescence images were acquired using an Olympus BX53 fluorescence microscope equipped with an Olympus DP70 digital camera. The fluorescence intensity of GFP-LC3 in the perinuclear region of more than 50 cells in each group was quantified using Image Gauge V4.0 software (Fujifilm). Percentages of LC3 puncta-positive cells were calculated from 3 independent assays.
Immunoblot analysis
HEK293 cells were transfected with pCMV5α vectors for MYC-tagged RavZ variants (WT and its mutants). After 24 h of transfection, the cells were left untreated or treated with 1 μM rapamycin for 4 h. Cells were then lysed in lysis buffer (1% triton X-100 [Sigma, T8787], 150 mM NaCl, 5 mM EGTA, 20 mM Tris-HCl, pH 7.4, 0.5 % sodium deoxycholate [Sigma, D6750], 12 mM β-glycerophosphate, 1 mM phenylmethyl sulfonyl fluoride, 2 μg/ml aprotinin [Sigma, A1153], 2 μg/ml leupeptin [Sigma, L2884]). Cell lysates were centrifuged at 12,000 × g for 20 min at 4°C, and the resulting supernatant fractions were subjected to SDS-PAGE. For immunoblot analysis, proteins in polyacrylamide gel were transferred onto a polyvinylidene difluoride membrane (ThermoFisher, LC2002). After blocking with 5% bovine serum albumin (Sigma, A2153), the membrane was incubated overnight with appropriate primary antibodies at 4°C and immunoreactive bands were visualized using horseradish peroxidase-conjugated secondary antibodies (Sigma, RABHRP1) and enhanced chemiluminescence reagents (Thermo, NCI4080KR). Rabbit polyclonal antibody to LC3B was obtained from Cell Signaling Technology (2773S) and mouse monoclonal antibody to GAPDH was from Santa Cruz Biotechnology (sc-32233).
Accession codes
Atomic coordinates and structure factor files have been deposited in the Protein Data Bank under the following accession codes: 5IO3, 5IZV and 5HZY for crystal form I, II and III, respectively.
Supplementary Material
Abbreviations
- ΔN
N-terminal deletion mutant (residues 49–502)
- ΔNΔC
both termini deletion mutant (residues 62–423)
- Cat
catalytic domain of RavZ
- DAPI
4′,6-diamidino-2-phenylindole
- FL
full-length RavZ (residues 1–502)
- GFP
green fluorescent protein
- KD
dissociation constant
- LIR
LC3-interacting region
- MM
molecular mass
- Mt
membrane targeting
- Mt-C
Mt domain with the C-terminal region (residues 327–502)
- mutLIR1
F16A L19A mutant
- mutLIR2
F29A L32A mutant
- mutLIR3
F435A I438A
- N-Cat
C-terminal deletion mutant (residues 1–325)
- PE
phosphatidylethanolamine
- PtdIns3P
phosphatidylinositol 3-phosphate
- SAD
single-wavelength anomalous dispersion
- SAXS
small-angle X-ray scattering
- SEC-MALS
size exclusion chromatography with multi-angle light scattering
- SPR
surface plasmon resonance
- Ub
ubiquitin
- Ubl
ubiquitin-like
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the staff at beamline 5C, Pohang Accelerator Laboratory, Korea, beamline BL17A, Photon Factory, and beamline BL44XU, Spring-8, Japan for help with the X-ray data collection. We also thank the staff at beamline 4C, Pohang Accelerator Laboratory, Korea, and beamline BL10C, Photon Factory, Japan for help with the SAXS data collection.
Funding
This work was supported by National Research Foundation of Korea (NRF) grants from the Korean government (BRL grant: No. 2015041919, NRF-2011–0028168 and International Cooperation Program: No. 2015K2A2A6002008) and by the Basic Science Research Program through the NRF, funded by the Ministry of Education (NRF-2013R1A6A3A04063675 to YOJ).
References
- [1].Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 2009; 10:458-67; PMID:19491929; http://dx.doi.org/ 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- [2].Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000; 290:1717-21; PMID:11099404; http://dx.doi.org/ 10.1126/science.290.5497.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Svenning S, Johansen T. Selective autophagy. Essays Biochem 2013; 55:79-92; PMID:24070473; http://dx.doi.org/ 10.1042/bse0550079 [DOI] [PubMed] [Google Scholar]
- [4].Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 2014; 16:495-501; PMID:24875736; http://dx.doi.org/ 10.1038/ncb2979 [DOI] [PubMed] [Google Scholar]
- [5].von Muhlinen N, Thurston T, Ryzhakov G, Bloor S, Randow F. NDP52, a novel autophagy receptor for ubiquitin-decorated cytosolic bacteria. Autophagy 2010; 6:288-9; PMID:20104023; http://dx.doi.org/ 10.4161/auto.6.2.11118 [DOI] [PubMed] [Google Scholar]
- [6].Lim J, Lachenmayer ML, Wu S, Liu W, Kundu M, Wang R, Komatsu M, Oh YJ, Zhao Y, Yue Z. Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS Genet 2015; 11:e1004987; PMID:25723488; http://dx.doi.org/ 10.1371/journal.pgen.1004987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131-45; PMID:17580304; http://dx.doi.org/ 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- [8].Kim BW, Kwon H, Song HK. Structure biology of selective autophagy receptors. BMB Rep 2016; 49:73-80; PMID:26698872; http://dx.doi.org/ 10.5483/BMBRep.2016.49.2.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shaid S, Brandts CH, Serve H, Dikic I. Ubiquitination and selective autophagy. Cell Death Differ 2013; 20:21-30; PMID:22722335; http://dx.doi.org/ 10.1038/cdd.2012.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Birgisdottir AB, Lamark T, Johansen T. The LIR motif - crucial for selective autophagy. J Cell Sci 2013; 126:3237-47; PMID:23908376 [DOI] [PubMed] [Google Scholar]
- [11].Kim BW, Hong SB, Kim JH, Kwon do H, Song HK. Structural basis for recognition of autophagic receptor NDP52 by the sugar receptor galectin-8. Nat Communications 2013; 4:1613; PMID:23511477; http://dx.doi.org/ 10.1038/ncomms2606 [DOI] [PubMed] [Google Scholar]
- [12].Lu K, Psakhye I, Jentsch S. A new class of ubiquitin-Atg8 receptors involved in selective autophagy and polyQ protein clearance. Autophagy 2014; 10:2381-2; PMID:25470352; http://dx.doi.org/ 10.4161/15548627.2014.981919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lu K, Psakhye I, Jentsch S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell 2014; 158:549-63; PMID:25042851; http://dx.doi.org/ 10.1016/j.cell.2014.05.048 [DOI] [PubMed] [Google Scholar]
- [14].Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. Febs Letters 2010; 584:1379-85; PMID:20083108; http://dx.doi.org/ 10.1016/j.febslet.2010.01.018 [DOI] [PubMed] [Google Scholar]
- [15].Jacomin AC, Samavedam S, Promponas V, Nezis IP. iLIR database: A web resource for LIR motif-containing proteins in eukaryotes. Autophagy 2016; 12(10):1-9; PMID:18428766; http://dx.doi.org/ 10.1002/0471250953.bi0505s14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Boyle KB, Randow F. The role of ‘eat-me’ signals and autophagy cargo receptors in innate immunity. Curr Opin Microbiol 2013; 16:339-48; PMID:23623150; http://dx.doi.org/ 10.1016/j.mib.2013.03.010 [DOI] [PubMed] [Google Scholar]
- [17].Choy A, Roy CR. Autophagy and bacterial infection: an evolving arms race. Trends Microbiol 2013; 21:451-6; PMID:23880062; http://dx.doi.org/ 10.1016/j.tim.2013.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 2009; 10:1215-21; PMID:19820708; http://dx.doi.org/ 10.1038/ni.1800 [DOI] [PubMed] [Google Scholar]
- [19].Alvarez VE, Kosec G, Sant Anna C, Turk V, Cazzulo JJ, Turk B. Blocking autophagy to prevent parasite differentiation: a possible new strategy for fighting parasitic infections? Autophagy 2008; 4:361-3; PMID:18212533; http://dx.doi.org/ 10.4161/auto.5592 [DOI] [PubMed] [Google Scholar]
- [20].Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 2005; 120:159-62; PMID:15680321 [DOI] [PubMed] [Google Scholar]
- [21].Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, et al.. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 2013; 494:201-6; PMID:23364696; http://dx.doi.org/ 10.1038/nature11866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Choy A, Dancourt J, Mugo B, O'Connor TJ, Isberg RR, Melia TJ, Roy CR. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 2012; 338:1072-6; PMID:23112293; http://dx.doi.org/ 10.1126/science.1227026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science 1998; 279:873-6; PMID:9452389; http://dx.doi.org/ 10.1126/science.279.5352.873 [DOI] [PubMed] [Google Scholar]
- [24].Segal G, Purcell M, Shuman HA. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Natl Acad Sci U S A 1998; 95:1669-74; PMID:9465074; http://dx.doi.org/ 10.1073/pnas.95.4.1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J 2009; 28:1341-50; PMID:19322194; http://dx.doi.org/ 10.1038/emboj.2009.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Horenkamp FA, Kauffman KJ, Kohler LJ, Sherwood RK, Krueger KP, Shteyn V, Roy CR, Melia TJ, Reinisch KM. The legionella anti-autophagy effector RavZ targets the autophagosome via PI3P- and curvature-sensing motifs. Dev Cell 2015; 34:569-76; PMID:26343456; http://dx.doi.org/ 10.1016/j.devcel.2015.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chosed R, Tomchick DR, Brautigam CA, Mukherjee S, Negi VS, Machius M, Orth K. Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases. J Biol Chem 2007; 282:6773-82; PMID:17204475; http://dx.doi.org/ 10.1074/jbc.M608730200 [DOI] [PubMed] [Google Scholar]
- [28].Reverter D, Wu K, Erdene TG, Pan ZQ, Wilkinson KD, Lima CD. Structure of a complex between Nedd8 and the Ulp/Senp protease family member Den1. J Mol Biol 2005; 345:141-51; PMID:15567417; http://dx.doi.org/ 10.1016/j.jmb.2004.10.022 [DOI] [PubMed] [Google Scholar]
- [29].Reverter D, Lima CD. A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure 2004; 12:1519-31; PMID:15296745; http://dx.doi.org/ 10.1016/j.str.2004.05.023 [DOI] [PubMed] [Google Scholar]
- [30].Xu Z, Lam LS, Lam LH, Chau SF, Ng TB, Au SW. Molecular basis of the redox regulation of SUMO proteases: a protective mechanism of intermolecular disulfide linkage against irreversible sulfhydryl oxidation. FASEB J 2008; 22:127-37; PMID:17704192; http://dx.doi.org/ 10.1096/fj.06-7871com [DOI] [PubMed] [Google Scholar]
- [31].Sugawara K, Suzuki NN, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J Biol Chem 2005; 280:40058-65; PMID:16183633; http://dx.doi.org/ 10.1074/jbc.M509158200 [DOI] [PubMed] [Google Scholar]
- [32].Forouhar F, Kuzin A, Seetharaman J, Lee I, Zhou W, Abashidze M, Chen Y, Yong W, Janjua H, Fang Y, et al.. Functional insights from structural genomics. J Struct Funct Genomics 2007; 8:37-44; PMID:17588214; http://dx.doi.org/ 10.1007/s10969-007-9018-3 [DOI] [PubMed] [Google Scholar]
- [33].Saridakis V, Yakunin A, Xu X, Anandakumar P, Pennycooke M, Gu J, Cheung F, Lew JM, Sanishvili R, Joachimiak A, et al.. The structural basis for methylmalonic aciduria. The crystal structure of archaeal ATP:cobalamin adenosyltransferase. J Biol Chem 2004; 279:23646-53; PMID:15044458; http://dx.doi.org/ 10.1074/jbc.M401395200 [DOI] [PubMed] [Google Scholar]
- [34].Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016; 12:1-222; PMID:26799652; http://dx.doi.org/ 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19:5720-8; PMID:11060023; http://dx.doi.org/ 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kim JH, Hong SB, Lee JK, Han S, Roh KH, Lee KE, Kim YK, Choi EJ, Song HK. Insights into autophagosome maturation revealed by the structures of ATG5 with its interacting partners. Autophagy 2015; 11:75-87; PMID:25484072; http://dx.doi.org/ 10.4161/15548627.2014.984276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jules M, Buchrieser C. Legionella pneumophila adaptation to intracellular life and the host response: clues from genomics and transcriptomics. FEBS Lett 2007; 581:2829-38; PMID:17531986; http://dx.doi.org/ 10.1016/j.febslet.2007.05.026 [DOI] [PubMed] [Google Scholar]
- [38].Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 2000; 151:263-76; PMID:11038174; http://dx.doi.org/ 10.1083/jcb.151.2.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem 2003; 278:51841-50; PMID:14530254; http://dx.doi.org/ 10.1074/jbc.M308762200 [DOI] [PubMed] [Google Scholar]
- [40].Popelka H, Klionsky DJ. Analysis of the native conformation of the LIR/AIM motif in the Atg8/LC3/GABARAP-binding proteins. Autophagy 2015; 11(12):2153-9; PMID:26565669; http://dx.doi.org/ 10.1080/15548627.2015.1111503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode In: Carter J CW, Sweet RM, eds. Methods Enzymol New York: Academic Press; 1997:307-26. [DOI] [PubMed] [Google Scholar]
- [42].Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010; 66:213-21; PMID:20124702; http://dx.doi.org/ 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126-32; PMID:15572765; http://dx.doi.org/ 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- [44].Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 2010; 66:12-21; PMID:20057044; http://dx.doi.org/ 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Holm L, Kaariainen S, Wilton C, Plewczynski D. Using Dali for structural comparison of proteins. Curr Protoc Bioinformatics 2006; Chapter 5:Unit 5; PMID:18428766; http://dx.doi.org/ 10.1002/0471250953.bi0505s14 [DOI] [PubMed] [Google Scholar]
- [46].Hong SB, Kim BW, Lee KE, Kim SW, Jeon H, Kim J, Song HK. Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8. Nat Struct Mol Biol 2011; 18:1323-30; PMID:22056771; http://dx.doi.org/ 10.1038/nsmb.2165 [DOI] [PubMed] [Google Scholar]
- [47].Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HDT, Konarev PV, Svergun DI. New developments in the ATSAS program package for small-angle scattering data analysis. J Applied Crystallography 2012; 45:342-50; PMID:25484842; http://dx.doi.org/ 10.1107/S0021889812007662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wriggers W, Chacon P. Using Situs for the registration of protein structures with low-resolution bead models from X-ray solution scattering. J Applied Crystallography 2001; 34:773-6; http://dx.doi.org/ 10.1107/S0021889801012869 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.