Abstract
The detection of nucleic acid biomarkers for point-of-care (POC) diagnostics is currently limited by technical complexity, cost, and time constraints. To overcome these shortcomings, we have combined loop-mediated isothermal amplification (LAMP), programmable toehold-mediated strand exchange signal transduction, and standard pregnancy test strips. The incorporation of an engineered hCG-SNAP fusion reporter protein (human chorionic gonadotropin-O6-alkylguanine-DNA alkyltransferase) led to LAMP-to-hCG signal transduction on low-cost, commercially available pregnancy test strips. Our assay reliably detected as few as 20 copies of Ebola virus templates in both human serum and saliva and could be adapted to distinguish a common melanoma-associated SNP allele (BRAF V600E) from the wild-type sequence. The methods described are completely generalizable to many nucleic acid biomarkers, and could be adapted to provide point-of-care diagnostics for a range of pathogens.
Keywords: hCG, SNAP, LAMP, toehold-mediated strand exchange, pregnancy test strips
Graphical abstract
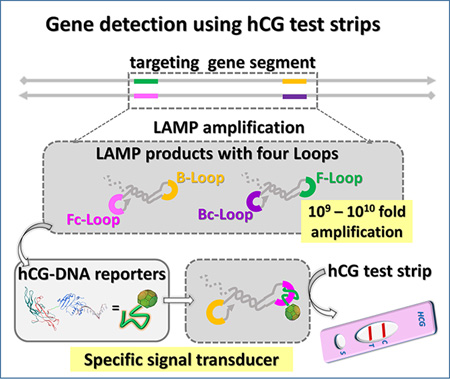
By engineering an hCG-SNAP fusion reporter protein (human chorionic gonadotro-pin-O6-alkylguanine-DNA alkyltransferase), we have successfully synthesized the hCG-DNA conjugate and further coupled isothermal amplification assays to strand exchange nucleic acid transducers and thereby generated hCG signals that can be directly read by off-the-shelf pregnancy test strips.
With recent advancements in medical diagnostics, it has become increasingly possible to quickly and accurately diagnose multiple disease states using nucleic acid or protein biomarkers.[1–3] However, these methods generally require complex instrumentation and are often restricted to medical diagnostic laboratories. Accurate, reliable assay methods that can be used in diverse or resource-limited locations are attractive alternatives for the early identification of pathogens.[4, 5] Only a few point-of-care (POC) products are commercially available, but these are potentially of great utility for establishing and expanding new diagnostics approaches.[6–8]
As an example, the already successful personal glucose meter (PGM)[9] was recently modified to quantify non-glucose targets via aptamer- or DNA-zyme-mediated conformational changes[10, 11] or sequence specific strand exchange[12] that in turn led to the release of invertase. Our laboratory has further enhanced the sensitivity of nucleic acid analyte detection with PGMs by coupling loop-mediated isothermal amplification (LAMP) reactions to strand exchange signal transduction.[13, 14] Unfortunately, the use of glucometers in POC diagnostics is problematic for many biomarkers because blood or urine samples are often infused with background glucose.
These limitations have inspired us to attempt a more generalizable approach to molecular diagnostics in which we transduce isothermal amplification to commercially available and easy-to-use pregnancy test strips.[15] The key innovation allowing signal readouts to be readily adapted from one off-the-shelf device (PGM) to another very different one (pregnancy test kits) is once again the remarkable plasticity of strand exchange signal transduction. The human chorionic gonadotropin (hCG) analyte that is already sensitively detected by commercial pregnancy test kits was site-specifically conjugated to a DNA oligonucleotide, thereby allowing transduction via strand exchange into both capture (signal-off) and release (signal-on) of hcG in the context of the lateral flow dipstick (LFD) assay. The reagents prove to be stable for months, making this an excellent option for POC diagnostics in low-resource settings. While lateral flow dipstick (LFD) assays have previously been adapted to sequence detection,[16, 17] the use of strand exchange probes, has greatly increased the sensitivity, specificity, and reliability of detection.
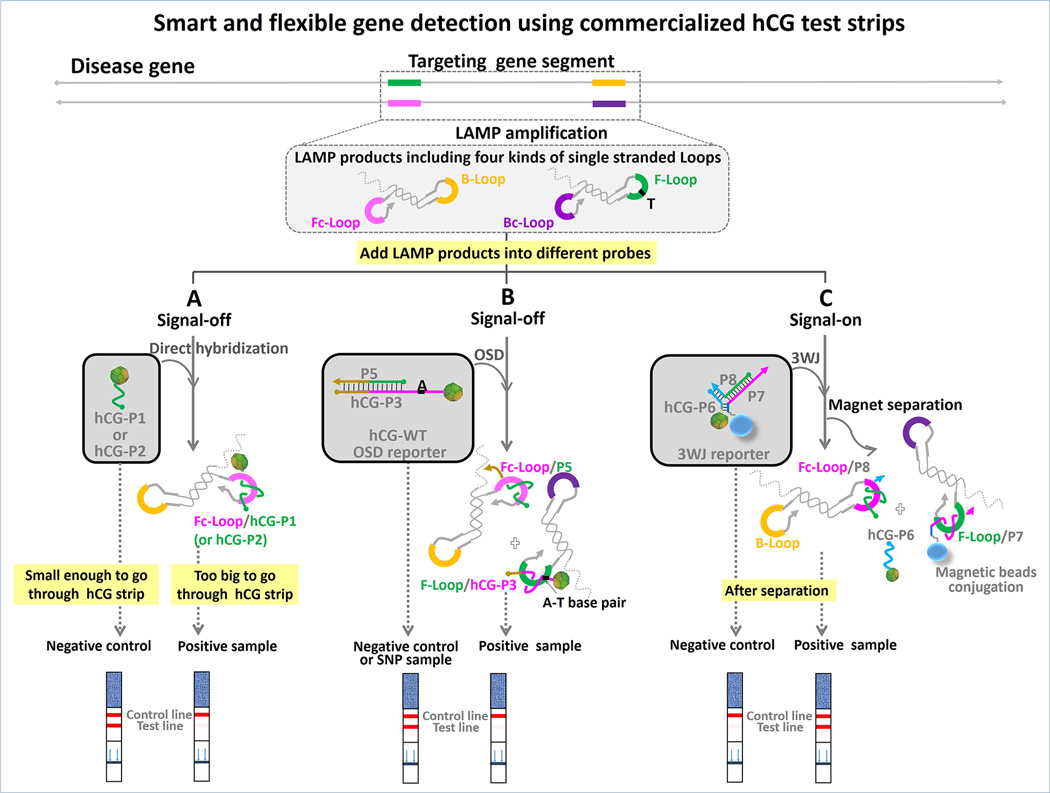
Adapting an isothermal amplification assay (LAMP) to detection via common pregnancy test strips consisted of three steps (Scheme 1): i) LAMP amplification; ii) capture of a hCG-DNA conjugate by the LAMP amplicon; and iii) readout via a pregnancy strip. LAMP produces concatameric amplicons that contain multiple different single strand loop sequences (between F2-F1 (F-Loop), F2c-F1c (Fc-Loop), B2-B1 (B-Loop), and B2c-B1c (Bc-Loop), respectively).[13,18,19] Because the LAMP amplicon is too large to migrate in the literal flow test trip, capture of the hCG-oligo conjugate by one or more of these loop sequences (herein the loop between F2c-F1c, Fc-Loop) results in a loss of signal, which can in turn be interpreted as a positive detection of the nucleic acid analyte. Additional variations on this robust theme are also shown, in which strand exchange reactions discriminate between SNPs (Scheme 1B) or produce positive signals (Scheme 1C). Each of these could be readily implemented in the overall simplistic context of applying molecular amplification reactions to a pregnancy test strip.
Scheme 1.
Three different strategies for detection of ZEBOV or wild-type BRAF (WT-BRAF) LAMP amplicons using pregnancy test strips. (A) “Signal-off” direct hybridization strategy. (B) “Signal-off” OSD strategy for discriminating mismatches. (C) “Signal-on” 3WJ strategy.
To implement this general strategy for antigen detection on a pregnancy test strip we needed to site-specifically conjugate DNA to hCG. We therefore designed a series of C- terminal hCGα- and hCGβ-SNAP-tag fusion proteins, expressed them in Expi293 cells, and tested their ability to bind a panel of commercial pregnancy test strips. The C-terminal fusion of the thermostable SNAP-tag to hCGβ provided the most robust signal in our initial screening, and all further assay optimizations were conducted with this fusion (hCGβ-SNAP) (Figure S1, in Supporting Information (SI)). Further assay of the fusion protein via ELISA revealed that the hCG-SNAP protein had a EC50 (half maximal effective concentration) value 1.0 nM, while the EC50 for commercially-obtained unmodified hCG was 46 nM (Figure S2, in SI). The hCGβ-SNAP fusion could be labeled with a benzylguanine-labeled oligonucleotide with > 90% efficiency, and purification using anion exchange chromatography eliminated unlabeled fusions (Figure S3, in SI). Probe stability was assessed following lyophilization and storage at room temperature. A particularly robust probe for Ebola virus was stored for at least 90 days without large signal deviations (Figure S4, in SI).
We then attempted to determine if yes / no signal-off detection could be reliably obtained for important biomedical targets such as the Zaire Ebolavirus VP30 genome (ZEBOV) and the melanoma-related oncogene biomarker BRAF. For the direct hybridization assay (Scheme 1A), a series of tubes containing 10 µL aliquots of hCG-P1 probe (18 nM, designed for ZEBOV LAMP amplicons; all seuqences described in Table S1 in SI) or hCG-P2 probe (8 nM, designed for BRAF LAMP amplicons) were mixed with 30 µL LAMP products. The hybridization reactions proceeded for 35 min at 25 °C. Subsequently, the pregnancy testing strips were dipped into the reaction solutions and pictures were taken after 2 minutes by cell phone (iPhone 6 plus, Apple, CA, USA). The LAMP amplicons cannot migrate on the strip after hybridization to the hCG-oligo probes, and thus only a single control line is obtained for a positive result (positive control, PC). Products that do not contain target sequence (negative controls, NCs) should not hybridize with the hCG-DNA probes, which can then flow to the test line. Thus, the appearance of signals at both the test and control lines indicate a negative result. Using the strategy described in Scheme 1A, we could definitively detect synthetic templates for these targets (Figure 1A and 1B). Further, following optimization of probe concentration (Figure S5, in SI) and hybridization times (Figure S6, in SI) we could detect as few as 20 copies of the ZEBOV templates and 2×103 copies of BRAF templates. The ZEBOV template spiked into human saliva (15%; Figure 2A) and human serum (10%; Figure 2B) also gave definitive results without any loss in signal intensity or specificity, even without pre-treatment. Electrophoretic analyses of comparable reactions also showed signals (Figure S7, in SI). While only 2.5 min of hybridization time was sufficient to distinguish between positive and negative signals, a final incubation time of 35 min was chosen because of its superior signal-to-noise ratio. With a 1.5 h LAMP preparation and reaction time, the total time for this assay is about 2.5 h.
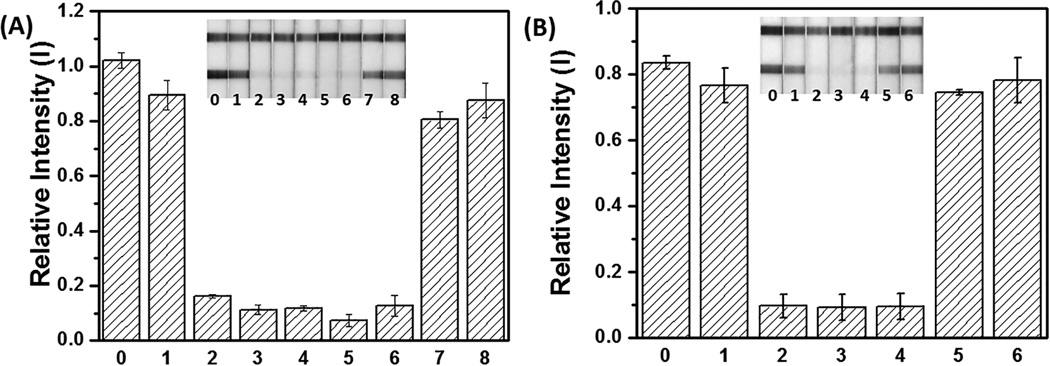
Figure 1.
Sensitive and specific detection of ZEBOV DNA plasmid and WT-BRAF PCR product using LAMP to hCG transduction. The LAMP reaction proceeded at 65 °C for 1.5 hours. (A) The hCG strip (inset) responses to negative control without template (strip 1), different amounts of ZEBOV templates (strip 2: 20 copies; strips 3: 200 copies; strip 4: 2×103 copies; strip 5: 2×104 copies; strip 6: 2×105 copies), side products (strip 7), and non-target WT-BRAF LAMP products (strip 8) are quantitated. Strip 0 is hCG-P1 in 1× iso buffer. (B) The hCG strip (inset) responses to negative control without template (strip 1), different amounts of WT-BRAF templates (strip 2: 2×103 copies; strip 3: 2×104 copies; strip 4: 2×105 copies), side products (strip 5), and non-target ZEBOV LAMP products (strip 6). Strip 0 is hCG-P2 in 1× iso buffer. Note: The side products are likely caused by the formation and extension of primer dimers.[14]
Figure 2.
Sensitive and selectivity of synthetic ZEBOV DNA plasmid in 15% human saliva (A) and in 10% human serum (B) using LAMP to hCG transduction. The hCG strip (inset) responses to different amounts of target (ZEBOV) templates and off-target BRAF templates. Strip 1: negative control without templates; strip 2: 20 copies; strips 3: 200 copies; strip 4: 2×103 copies; strip 5: 2×104 copies of ZEBOV templates; and strip 6: 2×106 copies of BRAF templates.
Nucleic acid amplification assays often rely on the ability to discriminate single nucleotide polymorphisms (SNPs) in samples, in order to discern exactly what variant of a gene or organism is present. We have previously shown that so-called oligonucleotide strand displacement (OSD) probes can be used for SNP detection,[14] and have now adapted OSD probes to our pregnancy test kit transduction assay (Scheme 1B). The hybridization of the reporter to the correct target loop is initiated at the single-stranded toehold and then proceeds via branch migration, leading to dissociation of the shorter signal strand.
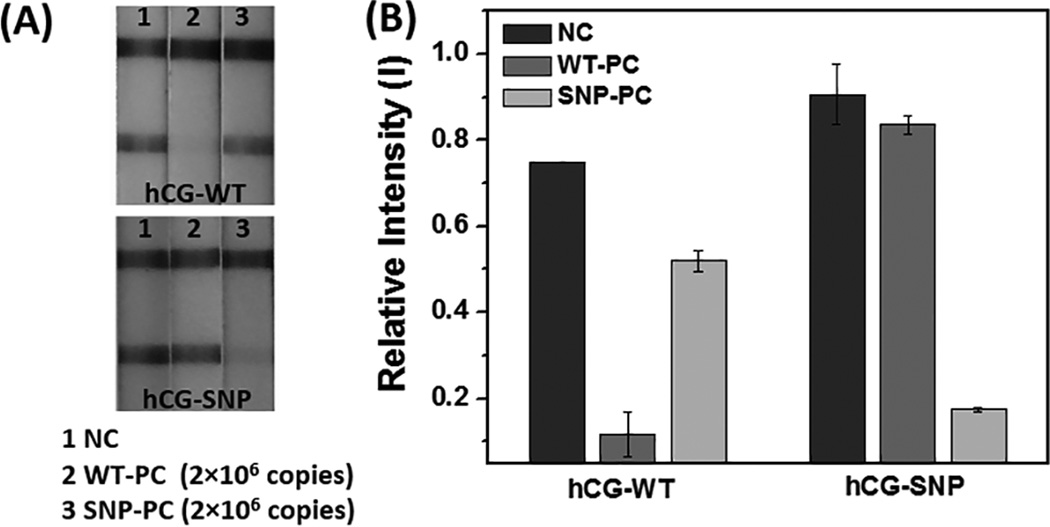
The V600E mutation of the BRAF gene is a melanoma-associated biomarker[20] and LAMP primers that incorporated either the wild-type BRAF sequence or the V600E BRAF (SNP-BRAF) T-to-A transversion mutation in the loop region were used to discriminate wild-type and mutant alleles. In parallel, hCG-labeled OSD reporters (hCG-WT and hCG-SNP) were designed that would be activated by the amplified wild-type and SNP-BRAF loop sequences, respectively. LAMP-OSD-to-hCG strip transduction was carried out with 0 and 2×106 copies of the WT-BRAF gene and SNP-BRAF. When the hCG-WT reporter was introduced, no band appeared on the test line for the WT-BRAF LAMP amplicons, as expected (Figure 3). In contrast, when the hCG-SNP reporter specific for the V600E allele was used, the mutant gene could be detected.
Figure 3.
Distinguishing single nucleotide polymorphisms (SNP) using strand displacement (OSD) probes. (A) The hCG strip responses to both WT- and SNP-BRAF templates with either hCG-WT or hCG-SNP reporters. (B) The bar-graph presents the quantitation of (B) using Image J software.
To achieve “signal-on” detection (a target shows a positive pregnancy test kit result) a new strand exchange probe was designed. The hCG conjugate (hCG-P6) was hybridized to a mediator strand (termed P8) to create a hemiduplex that could orderly be hybridized to a biotinylated strand (termed P7), creating a three-way junction (3WJ). The 3WJ was immobilized on streptavidin-modified magnetic beads (Scheme 1C). A toehold region on P7 can hybridize to LAMP amplicons via the target-specific loop region between F1 and F2 (F loop) and strand displace the P6:P8 hemiduplex. The released toehold region on P8 can hybridize to its complement on the Fc loop and thus displace the hCG-P6 reporter, allowing it to enter the lateral flow strip (Figure S8, in SI).
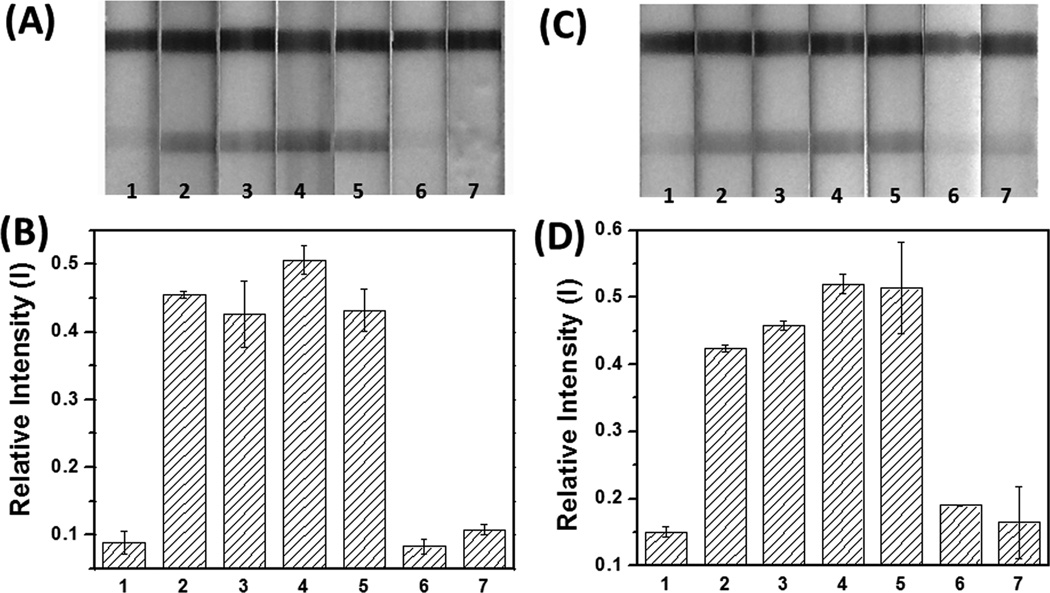
In order to demonstrate the potential utility of this design, we attempted to detect surrogate Ebola virus (ZEBOV) amplicons. LAMP reactions (30 µL) were mixed with 1 µL loaded magnetic beads (0.5 mg/mL) and incubated for 30 min before magnetic separation, and supernatant was run on individual pregnancy test strips. Figure 4A and 4B show a positive signal compared with negative controls (isothermal amplification buffer without ZEBOV templates, WT-BRAF templates with ZEBOV primers and non-cognate WT-BRAF LAMP products). Remarkably, as few as 20 copies of ZEBOV produced an obvious, easy-to-read positive signal on the test line of the pregnancy test strips. This was true even when the entire process was conducted with LAMP reactions in 5% human serum (Figures 4C and 4D).
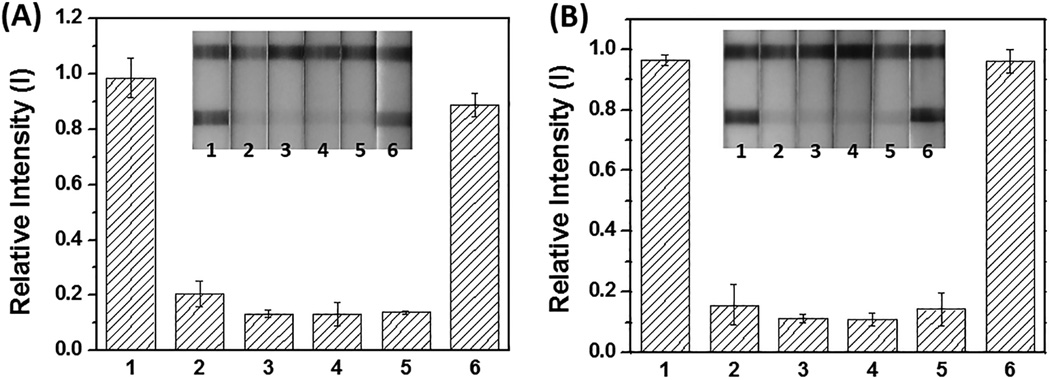
Figure 4.
Sensitive and selective detection of ZEBOV in buffer (A and B) and in 5% human serum (C and D). (A) and (C) The hCG strip responses to different amounts of target (ZEBOV) templates, and to off-target BRAF template and LAMP products. Strip 1: negative control; strip 2: 20 copies; strips 3: 200 copies; strip 4: 2×103 copies; strip 5: 2×104 copies of ZEBOV; strip 6: 5×106 copies of BRAF template amplified with ZEBOV primers; strip 7: 5×106 copies of BRAF LAMP product. (B) and (D) The bar-graphs represent the quantitation of (A) and (C), respectively, using Image J software.
We have coupled isothermal amplification assays to strand exchange nucleic acid transducers and thereby generated hCG signals for direct interpretation with off-the-shelf pregnancy test strips. These advances were abetted by protein engineering efforts that yielded a single attachment point for oligonucleotide probes to hCG. By capturing hCG-DNA probes (Scheme 1A), as few as 20 copies of ZEBOV virus nucleic acid biomarkers could be detected after a 1.5 h isothermal amplification reaction and a YES-or-NO answer for presence of ZEBOV virus could be obtained in buffer (Figure 1A), 15% human saliva (Figure 2A) and 10% human serum (Figure 2B). Similarly, as few as 2×103 copies of a melanoma-relevant BRAF sequence could be detected using a pregnancy test kit (Figure 1B). This limit of detection (LOD) is physiologically relevant, but higher than our previous work with this biomarker (20 copies)[14] because we limited ourselves to a shorter, more clinically relevant LAMP reaction time. Finally, we have also demonstrated the storage of lyophilized hCG-DNA probes at room temperature for at least 90 days without losing activity.
The sensitivity of these assays comes primarily from the powerful signal amplification properties of LAMP, while selectivity comes from both the LAMP primers and the novel strand exchange reactions. Strand exchange reactions can be engineered for specificity, and previous work has demonstrated that toehold design can significantly modulate the kinetics of strand displacement.[21, 22] We have previously shown that even a single mismatch between a LAMP amplicon and a toehold sequence can greatly reduce strand displacement and squelch background.[14] We reconfigured hCG capture to be dependent upon the initiation of strand exchange (Scheme 1B), and wild-type and BRAF V600E could be detected by their respective, matched probes, but not by mismatched probes (Figure 3). The discrimination afforded by mismatches with the short toehold sequence may be further amplified because of competition between the OSD reporter and the Fc loop for the F loop (since the Fc loop is complementary to the F loop; Scheme 1B).[14]
To reconfigure strand exchange reporters for “signal-on” detection with pregnancy test strips, we engineered a kinetically trapped three-way junction reporter (Scheme 1C), and assembled it on magnetic beads bearing a longer, toehold-containing strand (P7). Interaction of the F loop with P7 and the Fc loop with P8 results in the release (rather than capture) of a short oligonucleotide-hCG conjugate. Using this signal-on strategy the definitive detection of as few as 20 copies of the ZEBOV surrogate could be read relative to negative controls, including in LAMP reactions conducted in 5% human serum (Figure 4). The signal-on strategy incorporated some of the same design features used for allele discrimination, since toehold initiation must first occur for the overall strand exchange reaction to proceed.
The advantage of using nucleic acid strand exchange in analytical methods is that it can be readily transferred between platforms. By focusing on molecular transduction rather than on the development of a wholly new diagnostic platform it proved possible to develop rapid tests that could potentially be directly used for public health monitoring.
Supplementary Material
Acknowledgments
This study was supported by the William and Melinda Gates Foundation (OPP1028808), the National Institutes of Health (U54EB015403), the National Security Science and Engineering Faculty Fellowship (FA9550-10-1-0169), the Welch Foundation (F-1654) and the Cancer Prevention Research Institute of Texas (RP140315).
References
- 1.Wong DT. J. Am. Dent. Assoc. 2006;137:313–321. doi: 10.14219/jada.archive.2006.0180. [DOI] [PubMed] [Google Scholar]
- 2.Cheng MMC, Cuda G, Bunimovich YL, Gaspari M, Heath JR, Hill HD, Mirkin CA, Nijdam AJ, Terracciano R, Thundat T, Ferrari M. Curr. Opin. Chem. Biol. 2006;10:11–19. doi: 10.1016/j.cbpa.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Vo-Dinh T, Cullum B. Fresenius J. Anal. Chem. 2000;366:540–551. doi: 10.1007/s002160051549. [DOI] [PubMed] [Google Scholar]
- 4.Turner APF. Chem. Soc. Rev. 2013;42:3184–3196. doi: 10.1039/c3cs35528d. [DOI] [PubMed] [Google Scholar]
- 5.Urdea M, Penny LA, Olmsted SS, Giovanni MY, Kaspar P, Shepherd A, Wilson P, Dahl CA, Buchsbaum S, Moeller G, Hay Burgess DC. Nature. 2006;444(Suppl 1):73–79. doi: 10.1038/nature05448. [DOI] [PubMed] [Google Scholar]
- 6.Daar AS, Thorsteinsdottir H, Martin DK, Smith AC, Nast S, Singer PA. Nat. Genet. 2002;32:229–232. doi: 10.1038/ng1002-229. [DOI] [PubMed] [Google Scholar]
- 7.Tram K, Kanda P, Salena BJ, Huan S, Li Y. Angew. Chem. Int. Ed. 2014;53:12799–12802. doi: 10.1002/anie.201407021. Angew. Chem. 2014, 126, 13013–13016. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Z, Guan Z, Jia S, Lei Z, Lin S, Zhang H, Ma Y, Tian Z-Q, Yang CJ. Angew. Chem. Int. Ed. 2014;53:12503–12507. doi: 10.1002/anie.201405995. Angew. Chem. 2014, 126, 12711 –12715. [DOI] [PubMed] [Google Scholar]
- 9.Montagnana M, Caputo M, Giavarina D, Lippi G. Clin. Chim. Acta. 2009;402:7–13. doi: 10.1016/j.cca.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Xiang Y, Lu Y. Nat. Chem. 2011;3:697–703. doi: 10.1038/nchem.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu C, Lan T, Shi H, Lu Y. Anal. Chem. 2015;87:7676–7682. doi: 10.1021/acs.analchem.5b01085. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Z, Xiang Y, Tong A, Lu Y. Anal. Chem. 2014;86:3869–3875. doi: 10.1021/ac4040554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du Y, Hughes RA, Bhadra S, Jiang YS, Ellington AD, Li B. Sci. Rep. 2015;5:11039. doi: 10.1038/srep11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang YS, Bhadra S, Li B, Wu YR, Milligan JN, Ellington AD. Anal. Chem. 2015;87:3314–3320. doi: 10.1021/ac504387c. [DOI] [PubMed] [Google Scholar]
- 15.Davies RJ, Eapen SS, Carlisle SJ. Hoboken: Wiley; 2008. [Google Scholar]
- 16.Khunthong S, Jaroenram W, Arunrut N, Suebsing R, Mungsantisuk I, Kiatpathomchai W. J. Virol. Methods. 2013;188:51–56. doi: 10.1016/j.jviromet.2012.11.041. [DOI] [PubMed] [Google Scholar]
- 17.Yongkiettrakul S, Jaroenram W, Arunrut N, Chareanchim W, Pannengpetch S, Suebsing R, Kiatpathomchai W, Pornthanakasem W, Yuthavong Y, Kongkasuriyachai D. Parasitol. Int. 2014;63:777–784. doi: 10.1016/j.parint.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Nagamine K, Kuzuhara Y, Notomi T. Biochem. Biophys. Res. Commun. 2002;290:1195–1198. doi: 10.1006/bbrc.2001.6334. [DOI] [PubMed] [Google Scholar]
- 19.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Nucleic Acids Res. 2000;28 doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, Moses TY, Hostetter G, Wagner U, Kakareka J, Salem G, Pohida T, Heenan P, Duray P, Kallioniemi O, Hayward NK, Trent JM, Meltzer PS. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 21.Chen SX, Zhang DY, Seelig G. Nat. Chem. 2013;5:782–789. doi: 10.1038/nchem.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang DY, Chen SX, Yin P. Nat. Chem. 2012;4:208–214. doi: 10.1038/nchem.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.