Abstract
Iron-sulfur (Fe-S) clusters are inorganic cofactors that are fundamental to several biological processes in all three kingdoms of life. In most organisms, Fe-S clusters are initially assembled on a scaffold protein, ISCU, and subsequently transferred to target proteins or to intermediate carriers by a dedicated chaperone/co-chaperone system. The delivery of assembled Fe-S clusters to recipient proteins is a crucial step in the biogenesis of Fe-S proteins, and, in mammals, it relies on the activity of a multiprotein transfer complex that contains the chaperone HSPA9, the co-chaperone HSC20 and the scaffold ISCU. How the transfer complex efficiently engages recipient Fe-S target proteins involves specific protein interactions that are not fully understood. This mini review focuses on recent insights into the molecular mechanism of amino acid motif recognition and discrimination by the co-chaperone HSC20, which guides Fe-S cluster delivery.
Introduction
The chemical properties unique to transition metal ions enable them to participate in several processes that are essential to life by engaging directly in biochemical reactions. A central functional property of transition metals is the propensity to undergo one-electron transfer processes under physiological conditions. This property arises from the mid-range, one-electron reduction potentials of metals such as iron, copper, cobalt, nickel, manganese, and molybdenum. Iron-sulfur (Fe-S) clusters are ancient, ubiquitous cofactors composed of iron and inorganic sulfur. The combination of the chemical reactivity of iron and sulfur, together with many variations of cluster composition, oxidation states and protein environments, enables Fe-S clusters to participate in numerous biological processes (Figure 1A). The entire energy metabolism of the cell depends on electron transfer processes fulfilled by Fe-S clusters. Long-range electron transfers through distances of 10–14 Å link Fe-S centers of multimeric complexes involved in electron transfer pathways that catalyze ATP synthesis and processes essential to the global biogeochemical cycles of N, C, O, S, and H, some of which involve highly elaborate Fe-S prosthetic groups, such as those found in the molybdenum (Mo), vanadium (V), and iron only (Fe) nitrogenases that fix atmospheric N2 into ammonia1, in the [Ni–Fe] and [Fe–Fe] hydrogenases, responsible for hydrogen production and consumption2, and in the [Ni–Fe] and [Mo-Fe] carbon monoxide dehydrogenases (CODH) that carry out a reversible, biological water-gas shift reaction3–5.
Figure 1. Fe-S proteins are critical to several metabolic pathways.
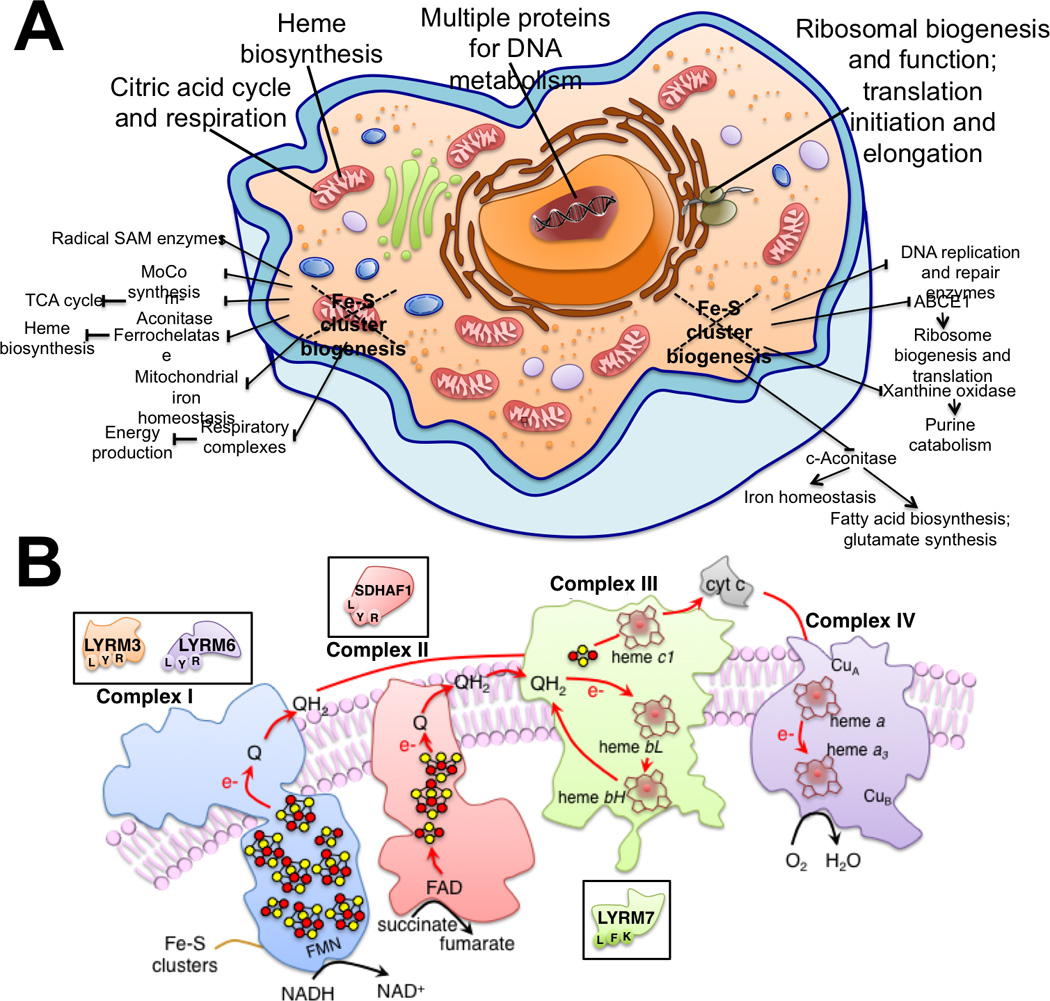
While the importance of the Fe-S enzymes mitochondrial aconitase and succinate dehydrogenase for the citric acid cycle has long been known, as well as the dependence of respiratory chain complexes I–III on the presence of Fe-S cofactors, the recent identification of new Fe-S proteins has revealed novel roles of Fe-S clusters in ribosomal biogenesis and in translation, DNA metabolism and repair (Panel A). (B) Schematic representation of mitochondrial respiratory chain complexes I-IV. LYR motif proteins are involved in the assembly of complex II, namely SDHAF1, and III (LYRM7). Two subunits of complex I are also LYR motif proteins, LYRM3 and LYRM6.
In 1951, researchers reported on the first Fe-S cluster dependent biochemical reaction: reduction of methaemoglobin by the crude or unwashed preparations of chloroplasts from a variety of plants6. In the following years, the use of electron paramagnetic resonance (EPR) imaging techniques enormously advanced the field, and EPR signals emanating from nonheme iron were first obtained for succinate dehydrogenase (SDH) in 19607, 8. By the mid-1960s, additional Fe-S proteins of bacterial origin became the subject of intensive studies, and they were shown to contain complexes of iron and acidic-labile sulphide in the form of two- and four-iron clusters9.
Underpinning the requirement of Fe-S proteins in the most crucial biological processes is the ability of Fe-S cofactors to undergo redox reactions, which are significantly influenced by the proteinaceous environment surrounding the cluster. Generally, the stability of the cluster decreases as its overall oxidation level increases. In addition to their capacity to undergo one-electron redox reactions, Fe-S clusters also carry out Lewis acid-based catalysis, such as in aconitase10–12. The cytosolic isoform of aconitase is a bifunctional enzyme (iron regulatory protein 1, IRP1/ cytosolic aconitase, ACO1), involved in regulation of cellular iron homeostasis13–15. Moreover, an astonishing array of diverse and biochemically challenging reactions is catalyzed by the radical S-adenosylmethionine (SAM) enzymes, which are involved in thermodynamically unfavorable reactions and require redox-active [Fe4–S4] conserved clusters for activity16. Several nucleic acid processing enzymes, such as DNA polymerases, glycosylases, helicases and primases require Fe-S clusters for their function17. Moreover, Fe-S clusters are present in DNA repair enzymes (Fanconi anemia group J protein, FANCJ, and Xeroderma pigmentosum group D protein, XPD)18, 19.
The inherent reactivity of Fe-S clusters towards a variety of redox-active species, including small redox-active compounds, reactive oxygen and reactive nitrogen species has been exploited during the evolution of bacterial sensors, which utilize the reactivity of Fe-S clusters to detect biologically relevant molecules (e.g., drugs, O2, NO)20, 21 and coordinate the cell’s response to oxidative and nitrosative stress conditions. However, this also means that Fe-S clusters are prone to degradation. Any of the reactions in which Fe-S cluster cofactors are involved may result in a change in the redox state of the cluster, cluster conversion, or even complete cluster loss. Moreover, interaction of Fe-S cofactors with strongly coordinating species may destroy the cluster by abstracting iron/sulfide or displacing thiolate ligands from the cluster22, 23.
While the cell usually strives to protect its complement of iron–sulfur proteins from the destructive effect of reactive species, Fe-S cofactors become particularly prone to damage when removed from the protective environment of the cell, a step that is necessary to characterize properties and functions of Fe-S proteins, but that increases the probability of Fe-S cluster destruction before characterization. It is, in fact, possible that previous failures to detect Fe-S clusters in numerous mammalian enzymes may have resulted from difficulties with overexpression or purification of intact cofactors24. Therefore, progress in the field of Fe-S protein biogenesis depends critically on the availability of methods for investigating structures and chemical properties of Fe-S enzymes, while preserving the integrity of their essential prosthetic groups25. An array of techniques and methods has been developed to enable researchers to obtain intact and biologically active Fe–S proteins in pure and stable forms. In addition, methods for the generation of highly concentrated samples containing Fe-S clusters isotopically labelled with 57Fe for Mössbauer and related spectroscopies have enabled researchers to identify novel mammalian Fe-S proteins, and to uncover new biological functions for Fe-S clusters in cellular metabolism26.
Bioinformatics approaches have dramatically expanded biological knowledge and enabled investigators to focus on particular candidates in their experimental work. Sophisticated bioinformatics tools were applied to the radical S-adenosylmethionine (SAM) superfamily to overcome the obstacles in the analysis of such a large number of enzymes which link different biosynthetic pathways. Radical SAM enzymes contain [Fe4-S4] cluster cofactors and are involved in an astonishing array of reactions that impact numerous cellular processes, including transcription, translation, gene regulation, the biosynthesis of several essential metabolites and complex metallo-cofactors, and signal transduction16, 27, 28. The term radical SAM was coined in a powerful bioinformatics approach, which used iterative profile methods to identify high homology and distinguishing features among the five founding members of the superfamily (biotin synthase, lipoyl synthase, lysine 2,3-aminomutase, and the activases for pyruvate-formate lyase and class III ribonucleotide reductases) and over 600 other uncharacterized proteins29. Iterative profile methods can greatly extend the power of sequence homology searches30, 31. While a strong match in a BLAST search can be used to infer protein homology, the weaker similarity detectable by an iterative profile method may uncover a more distant relationship and provide evidence for a conserved fold in the protein structure30. The most remarkable feature of radical SAM enzymes is a Cx3Cx2C motif, which ligates a [Fe4-S4] cluster. The fourth iron of the cluster was later found to be coordinated by the amino and carboxyl groups of SAM32, 33, allowing an electronic configuration that favors cleavage of SAM into methionine and 5’-deoxyadenosyl radical34, 35. More recently, a bioinformatics analysis conducted by the Structure Function Linkage Database (SFLD) expanded the number of radical SAM enzymes to almost 114,000 unique sequences in all organisms and over 60 distinct enzymatic reactions36. These approaches have linked functionally diverse enzymes based on conserved functional attributes such as a step or part of the reaction that they catalyze, creating the exciting possibility that the roles of novel Fe-S enzymes will be elucidated in the chemistry of numerous biological pathways36. Several other methods can facilitate identification of novel Fe-S proteins in mammalian cells.
This minireview focuses on recent advances achieved on the identification of molecular features that guide selection of specific subsets of nascent Fe-S recipient proteins by the co-chaperone HSC20. Short interaction modules, known as peptide motifs, determine biomolecular interactions in several cellular processes. Studies conducted in our lab allowed the identification of the tripeptide Leu-Tyr-Arg (LYR) as the molecular signature that mediates direct binding to the co-chaperone HSC20 and guides delivery of nascent clusters to succinate dehydrogenase subunit B, SDHB37. Here, we discuss the potential relevance of LYR motifs in proteins that were found to directly interact with HSC20, some of which are essential for the assembly and function of mitochondrial respiratory complexes I-III37, 38.
Fe-S cluster biogenesis: the assembly step
Biogenesis of Fe-S clusters is an elaborate process that involves formation of large dedicated multimeric complexes. In the 1980’s the synergistic work of chemists, biophysicists, biochemists and geneticists helped characterize the cellular mechanisms involved in in vivo assembly of Fe-S clusters in bacteria, and subsequent studies revealed that the basic components of the Fe-S biogenesis machinery are highly conserved throughout evolution, often functioning in large supra-molecular complexes39–41. Many details of how Fe-S cofactors are assembled and ligated to cellular proteins in the nucleus, cytosol and mitochondria of mammalian cells remain unclear.
In bacteria, the Fe-S biogenesis machinery supplies Fe-S clusters to recipient proteins in a single cellular compartment20, 42, whereas in mammalian cells the synthesis and distribution of Fe-S clusters are more complicated. The mammalian proteins involved in initial assembly of Fe-S clusters have been detected not only in mitochondria, but also in the cytosol and nucleus43, 44. In contrast, most of the early components of the biogenesis pathway have not been found in the cytosolic or nuclear compartments of the model organism Saccharomyces cerevisiae, in which the original building blocks of Fe-S clusters for cytosolic and nuclear proteins are proposed to derive from the mitochondrial machinery45. An additional level of complication arises from the presence of Fe-S clusters of different nuclearities that need to be delivered to specific recipients by a set of diverse intermediate carriers46. Most frequently, Fe-S proteins contain the rhomboid [Fe2-S2], the cuboidal [Fe3-S4], and the cubane [Fe4-S4] clusters5. However more complicated forms have been characterized that also harbor other metal ions5. Cysteine residues of the polypeptide chain typically ligate Fe-S clusters, but other amino acids, such as serine, arginine, and histidine, may also function as ligands. For instance two cysteines and two histidines coordinate the [Fe2-S2] cluster of the Rieske protein of respiratory chain complex III. Central to the initial assembly of Fe-S clusters is a cysteine desulfurase (NFS1 in mammals, Nfs1 in yeast, and IscS in bacteria), which abstracts sulfur from free cysteine and forms a persulfide intermediate on its active site cysteine (Figure 2). Two distinct conformational changes have been suggested to assist this initial step in Fe-S cluster biogenesis47, 48. One change exposes the buried substrate-binding sites thereby promoting cysteine binding, and it depends upon binding to NFS1 of the allosteric regulator of the cysteine desulfurase activity, frataxin (FXN). A second change that brings the bound substrate cysteine and the active site cysteine into close proximity for persulfide formation has been proposed to be mediated by binding of a small accessory protein, ISD11 (also known as LYRM4 in humans)49 to NFS1. Subsequently, the persulfide sulphur is transferred from NFS1 to the main scaffold protein ISCU, which provides the cysteine ligands to coordinate the nascent cluster50. In the bacterial IscU/IscS co-crystal structure, the ISCU ortholog binds to the cysteine desulfurase so that it orients the cysteine-rich active site in close proximity to the persulfide delivery loop of NFS151. Tight structural coupling of NFS1 and ISCU seems reasonable to preserve the efficiency of the pathway and to curb the toxicity that release of free sulphide would cause within the cell52, 53. A recent characterization of the thermodynamic, structural and electronic properties of Fe(II) bound to the scaffold protein Isu1 from S. cerevisiae, showed that the C-terminal helix of the ISCU scaffold protein, with its large number of conserved, surface exposed acidic and basic residues, is a potential candidate for the site of initial iron binding to ISCU52. The X-ray crystallographic structure of the E. coli apoIscU–IscS complex shows that each IscU molecule interacts with one subunit of the IscS dimer, leading to a 2:2 stoichiometry51, whereas, to date, no eukaryotic structures of the core complex are available. Using a co-expression approach, the mammalian recombinant ternary complex formed by NFS1, ISD11 and ISCU was biochemically and biophysically characterized, and a quaternary complex that formed when frataxin and two key substrates, iron and cysteine were added was analysed54. The mass of the ternary complex suggested formation of a homodimeric complex in which each unit had a 1:2:1 NFS1:ISD11:ISCU stoichiometry. Frataxin, which was shown to interact with the preformed ternary complex55, was proposed to bind in a ratio of 1:2:1:1 in the quaternary complex (NFS1:ISD11:ISCU:FXN)54. The source of iron for the nascent cluster remains to be identified, but proposed donors include frataxin56, 57, which has acidic surface patches to which iron binds with low affinity, or a complex of glutathione and glutaredoxin58. Electrons, needed to achieve the final electronic configurations of Fe-S clusters, are provided by ferredoxins (FDX1/2) and ferredoxin reductase (FDXR)59, 60. Ferredoxin has also been shown to facilitate reductive coupling of two distinct [Fe2-S2] clusters into a single [Fe4-S4] cluster on bacterial IscU in vitro61.
Figure 2. Fe-S cluster biogenesis in mammalian cells: a schematic of the main steps.
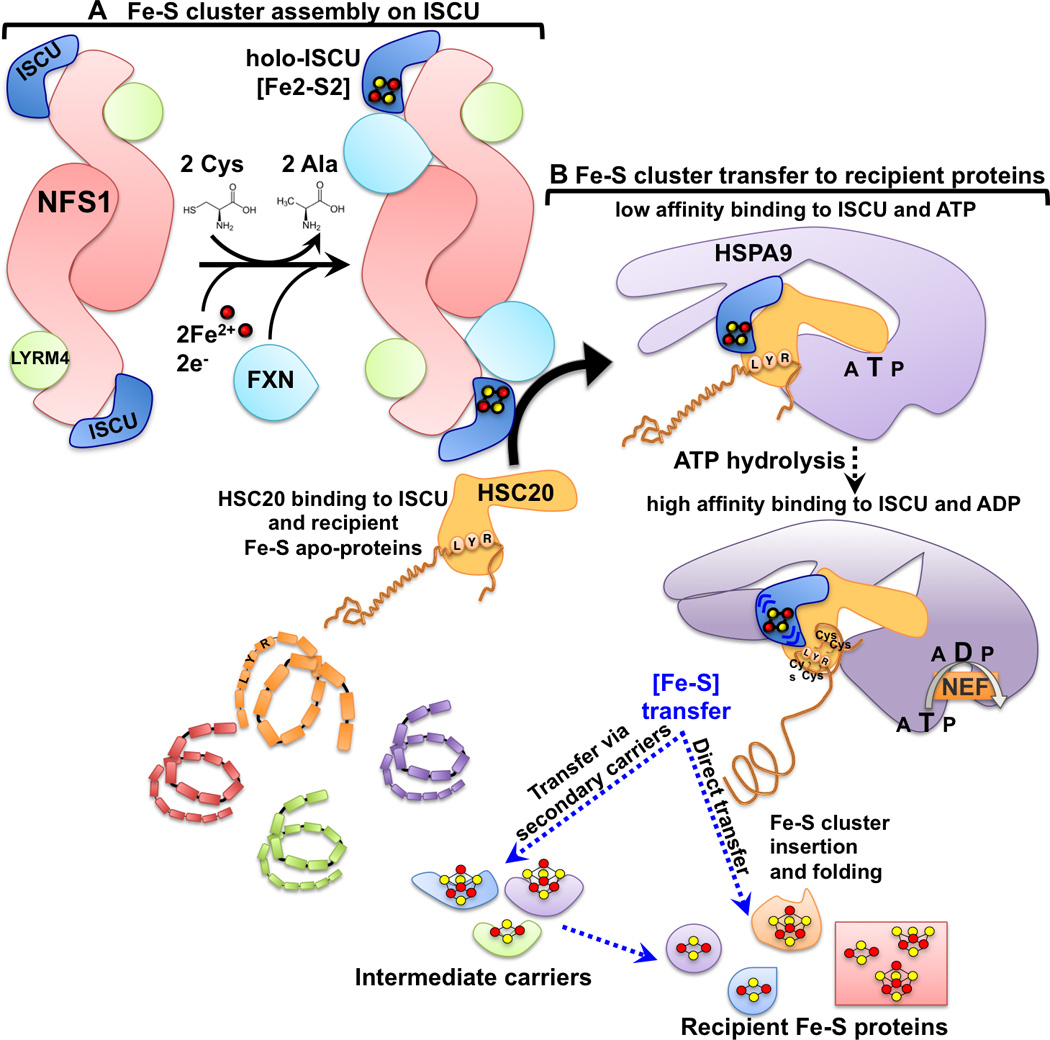
Nascent Fe-S clusters are initially assembled on the main scaffold protein ISCU. A cysteine desulfurase NFS1 forms a dimer to which monomers of the primary scaffold ISCU are proposed to bind at either end. LYRM4 is a structural component of the core complex in eukaryotes and it is required for the activity of NFS1, which, aided by its cofactor pyridoxal phosphate, provides sulfur, removed from cysteine, for the nascent cluster. Frataxin (FXN) is part of the core complex, potentially binding in a pocket-like region between NFS1 and ISCU. The cluster assembles upon ISCU when iron is provided together with the reducing equivalents needed to generate the final electronic configuration of the cluster. Cluster transfer from ISCU to recipient apoproteins is assisted by a dedicated chaperone/cochaperone (HSPA9/HSC20) system that facilitates cluster release from the primary scaffold ISCU and transfer to recipient apoproteins or to intermediate carriers, which then target specific recipients. The cochaperone HSC20 interacts with ISCU through a patch of hydrophobic amino acid residues in the C-terminus, and with the chaperone HSPA9 through the N-terminal domain (J-domain). The J-domain of HSC20 contains a conserved His (H), Pro (P), Asp (D) tripeptide essential for activation of the ATPase activity of HSPA9. The energy derived from activation of HSPA9 drives a conformational rearrangement in the substrate-binding domain of the chaperone, which is proposed to propagate a conformational change to its binding partner, ISCU. The conformational change is thought to facilitate release of the cluster from the main scaffold, ISCU, and allow protected transfer directly or via intermediate carriers to specific subsets of Fe-S recipient proteins. A nucleotide exchange factor (NEF) exchanges ADP with ATP and completes the ATPase cycle of the chaperone. The Leu-Tyr-Arg (LYR) motif present in recipient Fe-S proteins (i.e. SDHB) is a binding site for HSC20, which selects LYR-containing proteins from amongst the full spectrum of proteins in the proteome and forms a complex with holo-ISCU and the chaperone HSPA9. Upon hydrolysis of ATP, and concomitant with release of the cluster from ISCU, the Fe-S cluster is transferred into the domain of the recipient protein, which contains the Fe-S cluster cysteinyl ligands that can rapidly tether and protect the cluster, while driving folding of the primary peptide sequence.
Fe-S cluster biogenesis: transfer to recipient proteins
A specialized chaperone/co-chaperone system has evolved in bacteria (HscA/HscB) and in fungi (Ssq1/Jac1) to facilitate transfer of Fe-S clusters, newly assembled on the main scaffold ISCU, to Fe-S recipient proteins or to intermediate carriers, which then target specific clients62. Most eukaryotes utilize a multifunctional mitochondrial HSP70 (mtHSP70) that works together with a specialized co-chaperone, the ortholog of HscB/Jac163, 64. In vivo studies using a dominant negative mutant of the bacterial scaffold IscU (IscUD39A in Azotobacter vinelandii, or IscUD35A in Archaeglobus fulgidus), which remained trapped in a complex with the sulfur donor IscS, and contained a nascent Fe-S cluster65, 66, showed that genetic approaches could be used to uncouple cluster assembly and release steps. Fe-S cluster release and transfer is an ATP dependent process67, which occurs upon binding to IscU of the dedicated chaperone/co-chaperone system.
In mammalian cells, the cochaperone known as HSC2068 (HscB in bacteria63, and Jac1 in S. cerevisiae69) binds to the scaffold protein ISCU, and forms a complex with its chaperone partner, HSPA9 (HscA in bacteria, and Ssq1 in S. cerevisiae), a member of the HSP70 heat shock protein family37, 70. HSP70 homologs use the energy released by hydrolysis of ATP to drive conformational changes and refolding of target proteins71. The C-terminal domain of the cochaperone is directly responsible for binding the scaffold protein ISCU, with three highly conserved non-contiguous hydrophobic residues being of crucial importance for the HSC20-ISCU interaction46, 72–74. The Fe-S cluster transfer machinery is highly conserved among different organisms75, and cochaperones dedicated to Fe-S cluster biogenesis share common features73, 76, 77. A conserved structural core consists of two domains arranged in a L-shaped fold. The N-terminal J-domain, which contains an invariant histidine, proline, aspartate (HPD) motif, is responsible for stimulating the ATPase activity of the HSP70 cognate chaperone37, 76, 78, 79, whereas the C-terminal domain forms a three-helix bundle. The N-terminus of the human co-chaperone shows distinctive features compared to the specialized DnaJ type III proteins of bacteria and fungi, in that it contains, downstream of the mitochondrial targeting sequence (residues 1–2680), an additional domain, which harbors two CxxC modules (C41/C44 and C58/C61) that were found to coordinate a zinc ion in vitro77. The physiological relevance of the unique N-terminal domain of HSC20 remains to be elucidated, though it may facilitate dimerization of HSC20.
Recent studies with the purified components of the bacterial Fe-S transfer complex (HscA, HscB, IscU) have provided important insights into the molecular mechanism by which modulation of the ATPase activity of the chaperone HscA by the co-chaperone HscB is coupled to Fe-S cluster transfer from the scaffold protein IscU to a recipient Fe-S apo-protein81. Common to all HSP70 proteins is a highly conserved nucleotide-binding domain (NBD) that binds to and hydrolyzes ATP82, 83. The specialized substrate-binding domain (SBD) of HscA uniquely recognizes a central LPPVK motif in IscU as a substrate84–87. Similar to other HSP70s, nucleotide binding to the NBD of HscA allosterically regulates substrate binding affinity in the SBD67. The co-chaperone HscB binds to and delivers [Fe2-S2]-IscU to the SBD of HscA; together, IscU and HscB synergistically enhance HscA’s ATPase activity nearly 1000-fold63, 67. Two slowly interconverting states of IscU were shown to preferentially interact with HscA or HscB88–90: HscB binds preferentially to the more structured (S) state of IscU, which harbors a [Fe2-S2] cluster, whereas HscA binds preferentially the disordered (D) state of IscU, which forms after release of the cluster. Activation of the ATPase activity of HscA by the co-chaperone HscB, leads to hydrolysis of ATP to ADP in the NBD of HscA, driving a conformational change in the SBD of HscA through the HscA’s interdomain linker. The substrate-binding domain of HscA encloses the LPPVK peptide of IscU, which stabilizes the interaction and facilitates the transition of IscU from the S- to the D-state, which may promote release of the [Fe2-S2] cluster from holo-IscU81, 88. A similar mechanism has been proposed to assist biogenesis of Fe-S clusters in mammalian cells70. Inactivation of the HscA gene leads to diminished cellular growth rates and activities of Fe-S proteins, including SDH and aconitase91, 92, and inhibition of the ATPase activity of HscA via an ATPase deficient mutant, HscAT212V, abolishes [Fe2-S2] cluster transfer in vitro93. Mutations in the co-chaperone HSC20 and in its orthologs cause defects in Fe-S protein activities37, mitochondrial iron accumulation, and reduced mitochondrial respiration in human cell lines68, and multiple model systems, including yeast79, 94 and fly95. Additionally, recent studies in yeast confirm the central role of energy release coupled to conformational changes for Fe-S cluster biogenesis, as ATP consumption facilitates transition from the initial assembly of an Fe-S cluster on the main scaffold protein Isu by the core complex (NFS1-ISD11-FXN-ISCU), to the chaperone-cochaperone assisted transfer of the cluster to recipient proteins96.
In multiple cellular processes, co-chaperones are thought to determine the substrate specificity of their cognate chaperones64, 71, 73. The C-terminal domains of co-chaperones can selectively bind target substrates97, 98, facilitate refolding of denatured proteins, and enhance cell viability99, 100. Cochaperones serve a dual function in Fe-S cluster biogenesis: they guide ISCU to the substrate binding domain of the HSP70 cognate chaperone, and activate the ATPase activity of the chaperone, thereby driving a conformational change that might facilitate cluster release from ISCU and delivery to the final acceptor apoprotein or to intermediate carriers, which ultimately donate their clusters to specific recipients62, 63. The mechanism by which recipient Fe-S proteins are selected by the transfer machinery from amongst the full complement of human protiens has recently been partially characterized37. Interestingly, clinical and biochemical investigations of several newly described human diseases caused by mutations in NFU1, BOLA3, IBA57 or ISCA2 suggest that transfer of Fe-S clusters downstream of the holo-ISCU/chaperone/co-chaperone complex depends on selective pathways. The distinctive phenotypes associated with the various disease gene mutations reveal our lack of knowledge about how discrete Fe-S proteins are targeted in order to incorporate their vital prosthetic groups46, 101–111.
Recent progress: identification of molecular features that guide selection of recipient Fe-S proteins by the Fe-S transfer complex
Recent studies were conducted to try to elucidate how iron sulfur clusters are transferred from the initial scaffold protein, ISCU, to Fe-S clients, focusing on the human co-chaperone HSC2037 for two main reasons. Most eukaryotes, including humans, have a single multifunctional mitochondrial chaperone of the HSP70 family, which participates in diverse cellular functions by virtue of its ability to interact with an array of different J proteins (also known as HSP40s or co-chaperones) (for reviews, see71, 112–114). Therefore, HSPA9, a human HSP70 family member, would have not been the ideal prey to use to screen the human proteome for potential interacting Fe-S proteins, due to its promiscuous ATP-dependent substrate binding recognition activity. Co-chaperones are thought to play the crucial role of driving the functional specificity of HSP70s64, 71, 73, mostly through their C-terminal domains97–100, which makes the HSC20 co-chaperone a perfect candidate to use to search for specific interacting partners. Importantly, there is only one highly conserved HSC20 homologue in the human genome68, which also made HSC20 a good candidate to use in screening for target Fe-S recipient proteins. Since Fe-S clusters are highly sensitive to degradation by oxidants, the role of the co-chaperone may be to facilitate protected transfer of the cluster downstream of the holo-ISCU scaffold by directly binding recipient Fe-S apo-proteins. Studies in our lab identified several HSC20 binding partners in a stringent yeast two-hybrid (Y2H) screen, a method used to reveal direct molecular interactions between pairs of proteins37. Notably, multiple individual HSC20 interacting clones identified in the screen encoded succinate dehydrogenase subunit b (SDHB), which is the Fe-S cluster containing subunit of complex II. The interaction between HSC20 and SDHB was further validated in vivo in mammalian cells37. SDHB contains three clusters of different nuclearities; [Fe2-S2], [Fe4-S4] and [Fe3-S4]115, which are deeply buried within the mature protein and that likely need to be inserted during folding. Complex II is situated at the nexus of two essential energy-generating processes of the cell, the citric acid cycle and mitochondrial oxidative phosphorylation. In eukaryotes, mutations in each of the four nuclear-encoded succinate-coQ oxidoreductase (SQR) genes (SDHA, SDHB, SDHC, SDHD) or associated accessory factors (SDHAF1, SDHAF2) account for a variety of clinical phenotypes, including optic atrophy, myopathy, encephalopathy116, 117, and tumor formation118–120. Assembly of CII is a multistep process that depends upon incorporation of several prosthetic groups, including flavin adenine dinucleotide (FAD), covalently attached to SDHA, three Fe-S clusters coordinated by SDHB and an heme b moiety bound by the SDHC/SDHD membrane anchors121.
To identify the potential motifs that mediate Fe-S cluster transfer through binding to the co-chaperone HSC20, the SDHB primary sequence was subdivided into multiple peptides, which were cloned into Y2H prey constructs to screen 50 to 100 subclones of varying sizes for their ability to interact with HSC20. In recent years, investigators have focused increasingly on molecular interactions mediated by compact modules that are typically less than ten amino acid residues in length and are often found within intrinsically disordered regions of polypeptides. On average, binding motifs are 6–7 amino acids in length, and only 3–4 core positions confer specificity122. Because the binding surface area is small, these motifs bind with low affinity and engage in interactions that are transient and readily modulated. It has been suggested that over one million such motifs are present in the human proteome, and that they extend the functional capabilities of eukaryotes well beyond known structural domains123, influencing protein processing, localization, degradation, and transient participation in multiprotein complexes. Three independent binding sites for HSC20 were identified on SDHB, and two of these were iterations of the LYR motif. One SDHB peptide that interacted with HSC20 (residues 238–258 of SDHB) contained the distinctive motif, LYR, while the other (residues 35–52 of SDHB) contained a closely related sequence, IYR. Notably, the two native L(I)YR consensus sequences in SDHB are located in unstructured loops of the crystal structure. One of the L(I)YR motifs appears in SDHB near the N-terminus, proximal to the first cysteines that ligate the [Fe2-S2] cluster, whereas the second is closer to the C-terminus and the cysteinyl ligands of the [Fe4-S4] and [Fe3-S4] clusters, in positions where binding of the chaperone-cochaperone transfer apparatus can guide release of the cluster from holo-ISCU into the distal Fe-S binding sites of SDHB. Mutagenesis of the LYR or IYR tripeptide eliminated binding of motif-containing peptides to HSC2037. Interestingly, the amino acid residues of the two L(I)YR motifs were highly conserved in SDHB throughout evolution, including bacteria and yeast37. Homology searches indicated that the key features of this distinctive sequence, which mediated binding to HSC20, included having a hydrophobic residue in position 1, followed by an aromatic residue (Y or F) at position 2, and a positive residue, R or K, at position 3. Substitution of Arg46 into Gln in the IYR sequence was identified as a critical cause of renal cancer124, and substitutions of the IYR, LYR and cysteines important in ligating Fe-S clusters were identified as frequent causal mutations in rare tumors such as paragangliomas and gastrointestinal stromal tumors.
The LYR motif family is an annotated family present only in eukaryotes (Conserved Domains Accession: cl05087)125, which lists eleven proteins in humans characterized by having the LYR tripeptide close to their N-terminus. Members of the LYR family are SDHAF1 (LYRM8), a bona fide succinate dehydrogenase complex assembly factor117, ISD11 (LYRM4), a component of the initial Fe-S assembly complex126, LYRM7, a complex III assembly factor, and two subunits of respiratory chain complex I known as NDUFA6 and NDUFB9 (also annotated as LYRM6 and LYRM3, respectively)127 (Figure 1B). Several LYR motif proteins do not have a known function, such as LYRM2, LYRM5, and LYRM9, whereas others have been only partially characterized in yeast, such as ACN9 (acetate non-utilizing protein 9)128, or FMC1 (formation of mitochondrial complexes protein 1)129. Interestingly, human ISD11 and LYRM7 share high sequence homology with their yeast orthologues, Isd11 and Mzm1, respectively, and they perform similar functions126, 130, whereas human SDHAF1 has low similarity to the yeast ortholog Sdh6 and failed to restore respiration in a Sdh6 null background (ΔSdh6) yeast strain117.
SDHAF1, a member of the LYR motif family, assists Fe-S cluster incorporation into SDHB
SDHAF1 (also known as LYRM8) was shown to be important for SDH activity and for assembly of the holo-complex in fibroblasts117. Homozygous mutations in SDHAF1 cause a distinctive early-onset leukoencephalopathy in which accumulations of lactate and succinate in the white matter are associated with selective loss of SDH activity117, 118, 131; however the molecular role of SDHAF1 mutations in disease pathogenesis remained for some time undefined. A recent investigation used biochemical and functional approaches on cell lines derived from patients with SDHAF1 mutations to characterize the role of this accessory factor in the assembly of Complex II38. SDHAF1 was shown to recruit the Fe-S transfer complex to the C-terminus of SDHB through direct binding of its N-terminal LYR motif to the co-chaperone HSC20. The region L53-R65 of SDHAF1, enriched in arginine residues, was found to be involved in the interaction with SDHB at three binding sites containing multiple aromatic amino acids. Binding of SDHAF1 to SDHB and recruitment of the HSC20-HSPA9-holo-ISCU complex by its first LYR motif was shown to facilitate Fe-S cluster incorporation into SDHB. In vivo iron radiolabeling of SDHB showed defective Fe-S cluster incorporation into the protein synthesized in SDHAF1-deficient patient-derived cells. SDHB levels were drastically reduced in patient cells, where lack of functional SDHAF1 led to impaired biogenesis of SDHB, which was rapidly degraded by the mitochondrial protease, LONP138.
Potential role of LYR motif proteins in Fe-S cluster biogenesis
The functional significance of the common LYR motif was completely unknown when the Pfam clan Complex1_LYR-like superfamily (CL0491) was built by P. C. Coggill132. Based on bioinformatics analyses, the motif was defined as a tripeptide in which the first position was generally an aliphatic hydrophobic amino acid such as isoleucine or leucine, the second was an aromatic amino acid such as tyrosine or phenylalanine, and the third was a positively charged arginine or lysine. The tripeptide was followed by a conserved phenylalanine separated from LYR by about twenty-five residues127. The LYR motif was present in several other proteins selected in the Y2H screen as interacting partners of HSC20 (i.e. EPRS, HELZ, SPC25, GTF2E2, ETFA), and also in LYRM7, the Rieske Fe-S protein chaperone of complex III130 and an annotated member of the LYR family127. Overall, co-immunoprecipitation and Y2H studies indicated that LYR motifs present in known recipient Fe-S proteins (i. e. SDHB) or in accessory factors (i. e. SDHAF1, LYRM7) mediated an interaction with the co-chaperone HSC20, and guided insertion of Fe-S clusters into the Fe-S subunit of respiratory chain complex II, and, possibly, into complex III37. In fact, LYRM7, which has been characterized as a complex III assembly factor, binds UQCRFS1, the Rieske Fe-S protein of complex III130, and engages the HSC20- HSPA9-ISCU complex37, suggesting that the LYRM7-HSC20 interaction might guide the insertion of the [Fe2-S2] cluster into UQCRFS1. LYR containing subunits are present in mitochondrial complex I. The role of the complex I subunit LYRM6 (also known as NDUFA6 and as NB4M in the yeast Yarrowia lipolytica) has been recently characterized133. Chromosomal deletion of NB4M in Y. lipolytica or mutagenesis of the LYR tripeptide and of a conserved downstream phenylalanine into alanines caused loss of the [Fe4-S4] N2 cluster, which is the last Fe-S center in the ascending electron transfer sequence of the matrix arm of complex I with the highest redox potential, and totally abrogated the ubiquinone reductase activity of complex I, despite the fact that all the central subunits of the complex were intact. This finding suggests that NB4M (NDUFA6) is required for proper incorporation of the N2 [Fe4-S4] center, and for the function of complex I. Notably, NDUFS8, which ligates two [Fe4-S4] clusters, contains a conserved LYR motif in a non-canonical position, at its C-terminus. As only 14 of the 44 complex I subunits have a catalytic function, some of the accessory or supernumerary subunits may contribute to the assembly and stability of the complex, and the LYR-containing subunits may aid insertion of some of the Fe-S clusters.
A second consensus sequence, KKx(6–10)KK, was identified in the screening of the SDHB peptides that interacted with HSC20 through its C-terminal domain. Interestingly, human glutaredoxin 5 (GLRX5) has a similar pattern of lysines at its C-terminus (K139K140(x10)K151K152) and was found to interact with HSC20 in vivo37. The yeast ortholog Grx5 interacts with Ssq1, the mitochondrial chaperone dedicated to Fe-S cluster biogenesis134.
Molecular features of peptides containing the LYR motif that affect binding to HSC20
The function of the LYR and KKx(6–10)KK motifs may be very dependent on their molecular context and structural location37. Other features may define which LYR proteins engage HSC20 and the transfer apparatus, and these features will likely be defined as more LYR and Fe-S proteins are studied. Interestingly, there are two other examples in which a small peptide motif functions in the Fe-S transfer complex. The first is the well-known role of the tripeptide His, Pro, Asp (HPD) of HSC20 in activating the ATPase activity of its partner chaperone79. A second example is the role of the tripeptide Pro, Val, Lys (PVK) of ISCU in binding to the chaperone as part of the activation of the Fe-S transfer cycle 85, 87, 135. Experimental work will be needed to establish what other features are required for LYR motif-containing proteins to engage in binding with the Fe-S transfer complex. Upon inspection of the two motifs in SDHB and of the LYR motifs in SDHAF1 and LYRM7 that mediate an interaction with HSC20 (as assessed by co-immunoprecipitation of endogenous human proteins), it can be noticed that all four of these motifs appear in a peptide context in which a large hydrophobic residue, Phe, or two smaller hydrophobic amino acids, Val and Leu, are present just upstream of the LYR. For SDHB, the sequence F-X-IYR and F-X-LYR are highly conserved for the first and second LYR motifs, respectively. In LYRM7, the sequence VL-X-LFK is highly conserved, and in SDHAF1 (LYRM8), the sequence VLSLYR is highly conserved. Thus, it is possible that a small hydrophobic patch near the N-terminal side of LYR enhances binding to HSC20. A combination of informatics analyses and experimental tests will likely elucidate all the features of a functional LYR motif. In the years ahead, it will be interesting to see whether identification of LYR motifs through informatics can lead to discovery of unrecognized human Fe-S proteins24.
Intermediate carriers and secondary scaffolds that deliver Fe-S clusters to recipient proteins
A crucial although still largely uncharacterized process is the transfer of Fe-S clusters downstream of the main scaffold protein ISCU through secondary carriers. The co-chaperone HSC20 also binds to glutaredoxin 537, which was shown to interact with Ssq1, the chaperone dedicated to Fe-S cluster biogenesis in S. cerevisiae134. Glutaredoxin 5 has been characterized as an intermediate carrier involved in the transfer of Fe-S clusters downstream of the scaffold protein ISCU, in yeast134, 136–138, zebrafish139, and humans140–142. In eukaryotes, monothiol glutaredoxins localize to mitochondria or chloroplasts and have been implicated in Fe-S cluster biogenesis, whereas multidomain Glrxs (i.e. yeast Grx3/4 and human GLRX3) localize to the cytosolic/nuclear compartment and are proposed to play dual roles in iron trafficking and regulation as well as in Fe-S cluster biogenesis143–147. The crystal structure of human glutaredoxin 5 revealed the presence of a [Fe2-S2] cluster coordinated by a glutaredoxin homodimer and two glutathione molecules, which were held in place by non covalent interactions within the binding pocket of each GLRX5 monomer and by covalent linkages to the cluster148. Moreover, in this structure, two [Fe2-S2]-bridged homodimers interacted to form a tetramer. The current working model to describe the role of glutaredoxins in Fe-S cluster biogenesis proposes that they transiently accept a [Fe2-S2] cluster and engage the chaperone/co-chaperone transfer complex in order to facilitate insertion of the cluster into final acceptor apoproteins134, 149. An alternative cluster transfer mechanism hypothesizes that a [Fe2-S2] cluster coordinated by four glutathione molecules is exchanged with the cellular Fe-S cluster biogenesis components, including glutaredoxins150. Glutaredoxins have been linked by bioinformatics analyses, Y2H and affinity purification studies to another family of widely distributed proteins, the BolA-like proteins, both in prokaryotes and in eukaryotes, including plants151–155. BolA proteins were initially identified in E. coli with the observation that their overexpression promoted a switch from rod-shaped to rounded cell morphologies156. The Grx3/Grx4 proteins are involved in iron regulation in yeast by forming a complex with the BolA-like protein, Fra2, and ligating a [Fe2-S2] cluster coordinated by the Grx active site cysteine of Grx3/Grx4, a conserved histidine in Fra2 (His103), and a cysteine from glutathione144, 157, 158. Mammals have three BOLA orthologs, named BOLA1, BOLA2, and BOLA3, of unknown functions. Recently, mutations in human BOLA3 have been discovered to cause multiple mitochondrial dysfunctions syndrome 2101, 104, caused by defects in the biogenesis of the Fe-S cluster enzyme lipoic acid synthase (LIAS). Human BOLA3 has two isoforms that result from alternative splicing; both are predicted to localize to mitochondria (NM_001035505.1 and NM_212552.2) and isoform 2 differs because it lacks exon 3.
Human NFU1 has been recently characterized as a late acting factor specifically required for biogenesis of lipoic acid synthase, rather than as an alternative scaffold acting in parallel to ISCU, based on the fact that mutations in NFU1 are associated mainly with defective lipoic acid (LA) metabolism101, 102, 104, whereas other Fe-S proteins, such as aconitase, are not affected. The A-type scaffolds are thought to support the biogenesis of a distinct subset of Fe-S proteins. The isc and suf operons of bacteria encode two A-type proteins IscA and SufA, respectively, which were found to be involved in the biogenesis of some [Fe4-S4] proteins, such as aconitase and LIAS50, 159. An additional member of this family, ErpA, is specifically involved in isoprenoid biosynthesis160. The human genome encodes two A-type proteins, named ISCA1 and ISCA2, related to S. cerevisiae Isa1 and Isa2, respectively, thought to be necessary for biogenesis of several [Fe4-S4], but not [Fe2-S2], enzymes161. Consistent with the proposed role of ISCA1/ISCA2 and IBA57 in the biogenesis of [Fe4-S4] clusters for a subset of mitochondrial proteins, a mutation affecting the stability of IBA57 was found in two patients with severe myopathy and encephalopathy associated with compromised activities of the lipoic acid dependent enzymes, indicating defective maturation of LIAS109. A loss of function mutation in IBA57 was recently identified in a patient affected by infantile leukodystrophy with lack of protein lipoylation and decreased complex I and II activities162. In most cases, the conclusion that an intermediate carrier protein has specific downstream recipients is based on broad inferences rather than on direct experimental data. It should always be considered that mutations in secondary Fe-S cluster carriers that affect putative recipients may not directly interact with the affected Fe-S protein, and Fe-S proteins are sensitive to several cellular conditions that could indirectly affect their function.
A potential error in assigning roles of proteins in Fe-S cluster delivery may derive from giving a protein the function of an intermediate Fe-S cluster carrier solely based on the fact that its dysfunction leads to a defect in an Fe-S protein or protein complex. For instance, the mitochondrial P-loop NTPase IND1 (Fe-S protein required for NADH dehydrogenase) was proposed to function as an Fe-S cluster carrier for mitochondrial complex I, based on the fact that its deletion caused decreased complex I activity163, 164. Recent studies of IND1 in Arabidopsis thaliana (INDH) were able to unravel primary from secondary phenotypes caused by inactivating mutations in INDH, and demonstrated that INDH has a primary role in mitochondrial translation165. Impairment of complex I likely was due to the fact that seven of its subunits are encoded by the mitochondrial genome and translated in the mitochondrial matrix. In summary, it appears that virtually all mammalian Fe-S clusters are initially assembled on the main scaffold protein ISCU. Recipient Fe-S proteins can then acquire their clusters directly from ISCU in a chaperone/co-chaperone mediated transfer process, or indirectly from secondary carriers, which likely acquired their initial clusters from ISCU and that then deliver these clusters to only a subset of Fe-S proteins with which they specifically interact. Perhaps, the use of co-immunoprecipitation techniques may help to identify direct interactions between secondary carriers and Fe-S clients, although interpretation of these results can still be confounded by the presence of large multimeric complexes, in which two proteins can be found to interact even though they do not directly bind to one another. While the early steps in the biogenesis of Fe-S proteins are nearly universally shared, later steps may confer great specificity, and several molecular processes based on specific interactions allow for proper incorporation of Fe-S clusters into the correct recipient. One such mechanism is based on selection of substrates among the human proteome by the co-chaperone HSC20 which specifically recognizes a short interaction module in Fe-S apo-proteins37, 38. An additional level of specificity arises from the presence of secondary carriers, which seem to selectively target subsets of Fe-S recipient proteins.
Conclusions
During the last decade, Fe-S proteins have been identified at the core of ribosomal function and translation as well as of DNA metabolism. These discoveries were unexpected, and they were made possible by a combination of different approaches. The anaerobic preparation of crystals allowed detection of two [Fe4-S4] clusters in the bacterial ABCE1, an ABC cassette protein with a central although incompletely defined role in mRNA translation and ribosomal function166. Recently, an alternative approach for detecting Fe-S proteins through their physical interaction with the cytosolic Fe-S biogenesis machinery was developed, which linked essential enzymes involved in DNA metabolism and repair to MMS19, a protein that when mutated in yeast causes DNA instability, and to the cytosolic Fe-S biogenesis machinery167, 168. Bioinformatics approaches have accelerated identification of novel Fe-S proteins based on homology studies36. An alternative method was to identify specific motifs that are recognized by the Fe-S transfer machinery37. The cochaperone HSC20 binds to LYR motifs present in Fe-S recipient proteins or their binding partners. The discovery of the role of the LYR motif in engaging the Fe-S transfer complex has the potential to enhance the identification of novel Fe-S proteins. There are probably many more mammalian Fe-S proteins that have not been recognized so far because of the instability of Fe-S clusters and the previous lack of identifying sequence elements.
Acknowledgments
This work was supported by the intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, MD.
References
- 1.Hu Y, Ribbe MW. Nitrogenases-A Tale of Carbon Atom(s) Angew Chem Int Ed Engl. 2016 doi: 10.1002/anie.201600010. [DOI] [PubMed] [Google Scholar]
- 2.Peters JW, Schut GJ, Boyd ES, Mulder DW, Shepard EM, Broderick JB, King PW, Adams MW. [FeFe]- and [NiFe]-hydrogenase diversity, mechanism, and maturation. Biochim Biophys Acta. 2015;1853:1350–1369. doi: 10.1016/j.bbamcr.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 3.Drennan CL, Heo J, Sintchak MD, Schreiter E, Ludden PW. Life on carbon monoxide: X-ray structure of Rhodospirillum rubrum Ni-Fe-S carbon monoxide dehydrogenase. Proc Natl Acad Sci U S A. 2001;98:11973–11978. doi: 10.1073/pnas.211429998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sazanov LA. A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat Rev Mol Cell Biol. 2015;16:375–388. doi: 10.1038/nrm3997. [DOI] [PubMed] [Google Scholar]
- 5.Beinert H, Holm RH, Munck E. Iron-sulfur clusters: nature's modular, multipurpose structures. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 6.Davenport HE, Hill R, Whatley FR. A natural factor catalyzing reduction of methaemoglobin by isolated chloroplasts. Proc R Soc Lond B Biol Sci. 1952;139:346–358. doi: 10.1098/rspb.1952.0017. [DOI] [PubMed] [Google Scholar]
- 7.Beinert H, Sands RH. Studies on Succinic and Dpnh Dehydrogenase Preparations by Paramagnetic Resonance (Epr) Spectroscopy. Biochem Biophys Res Commun. 1960;3:41–46. [Google Scholar]
- 8.Sands RH, Beinert H. Studies on Mitochondria and Submitochondrial Particles by Paramagnetic Resonance (Epr) Spectroscopy. Biochem Biophys Res Commun. 1960;3:47–52. [Google Scholar]
- 9.Lovenberg W. Iron-sulfur proteins. New York: Academic Press; 1973. [Google Scholar]
- 10.Beinert H, Kennedy MC. Aconitase, a two-faced protein: enzyme and iron regulatory factor. FASEB J. 1993;7:1442–1449. doi: 10.1096/fasebj.7.15.8262329. [DOI] [PubMed] [Google Scholar]
- 11.Flint DH, Allen RM. Iron-Sulfur Proteins with Non redox Functions. Chem Rev. 1996;96:2315–2334. doi: 10.1021/cr950041r. [DOI] [PubMed] [Google Scholar]
- 12.Beinert H, Kennedy MC, Stout CD. Aconitase as Iron-Sulfur Protein, Enzyme, and Iron-Regulatory Protein. Chem Rev. 1996;96:2335–2374. doi: 10.1021/cr950040z. [DOI] [PubMed] [Google Scholar]
- 13.Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nature chemical biology. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 14.Volz K. The functional duality of iron regulatory protein 1. Current opinion in structural biology. 2008;18:106–111. doi: 10.1016/j.sbi.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of Mammalian iron metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 16.Landgraf BJ, McCarthy EL, Booker SJ. Radical S-Adenosylmethionine Enzymes in Human Health and Disease. Annu Rev Biochem. 2016;85:485–514. doi: 10.1146/annurev-biochem-060713-035504. [DOI] [PubMed] [Google Scholar]
- 17.White MF, Dillingham MS. Iron-sulphur clusters in nucleic acid processing enzymes. Current opinion in structural biology. 2012;22:94–100. doi: 10.1016/j.sbi.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nature chemical biology. 2012;8:125–132. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudolf J, Makrantoni V, Ingledew WJ, Stark MJ, White MF. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Molecular cell. 2006;23:801–808. doi: 10.1016/j.molcel.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 20.Mettert EL, Kiley PJ. How Is Fe-S Cluster Formation Regulated? Annu Rev Microbiol. 2015;69:505–526. doi: 10.1146/annurev-micro-091014-104457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crack JC, Green J, Thomson AJ, Le Brun NE. Iron-sulfur clusters as biological sensors: the chemistry of reactions with molecular oxygen and nitric oxide. Acc Chem Res. 2014;47:3196–3205. doi: 10.1021/ar5002507. [DOI] [PubMed] [Google Scholar]
- 22.Bobrik MA, Que L, Jr, Holm RH. Synthetic analogs of the active sites of iron-sulfur proteins. IV. Ligand substitution reactions of the tetranuclear clusters (Fe4S4(SR)4)2. J Am Chem Soc. 1974;96:285–287. doi: 10.1021/ja00808a064. [DOI] [PubMed] [Google Scholar]
- 23.Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A. 2009;106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rouault TA. Iron-sulfur proteins hiding in plain sight. Nat Chem Biol. 2015;11:442–445. doi: 10.1038/nchembio.1843. [DOI] [PubMed] [Google Scholar]
- 25.Maly T, Zwicker K, Cernescu A, Brandt U, Prisner TF. New pulsed EPR methods and their application to characterize mitochondrial complex I. Biochim Biophys Acta. 2009;1787:584–592. doi: 10.1016/j.bbabio.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Pandelia ME, Lanz ND, Booker SJ, Krebs C. Mossbauer spectroscopy of Fe/S proteins. Biochim Biophys Acta. 2015;1853:1395–1405. doi: 10.1016/j.bbamcr.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Broderick JB, Duffus BR, Duschene KS, Shepard EM. Radical S-adenosylmethionine enzymes. Chem Rev. 2014;114:4229–4317. doi: 10.1021/cr4004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Challand MR, Driesener RC, Roach PL. Radical S-adenosylmethionine enzymes: mechanism, control and function. Nat Prod Rep. 2011;28:1696–1721. doi: 10.1039/c1np00036e. [DOI] [PubMed] [Google Scholar]
- 29.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neuwald AF, Liu JS, Lipman DJ, Lawrence CE. Extracting protein alignment models from the sequence database. Nucleic Acids Res. 1997;25:1665–1677. doi: 10.1093/nar/25.9.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walsby CJ, Ortillo D, Yang J, Nnyepi MR, Broderick WE, Hoffman BM, Broderick JB. Spectroscopic approaches to elucidating novel iron-sulfur chemistry in the "radical-Sam" protein superfamily. Inorg Chem. 2005;44:727–741. doi: 10.1021/ic0484811. [DOI] [PubMed] [Google Scholar]
- 33.Dowling DP, Vey JL, Croft AK, Drennan CL. Structural diversity in the AdoMet radical enzyme superfamily. Biochim Biophys Acta. 2012;1824:1178–1195. doi: 10.1016/j.bbapap.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cosper NJ, Booker SJ, Ruzicka F, Frey PA, Scott RA. Direct FeS cluster involvement in generation of a radical in lysine 2,3-aminomutase. Biochemistry. 2000;39:15668–15673. doi: 10.1021/bi0022184. [DOI] [PubMed] [Google Scholar]
- 35.Nicolet Y, Amara P, Mouesca JM, Fontecilla-Camps JC. Unexpected electron transfer mechanism upon AdoMet cleavage in radical SAM proteins. Proc Natl Acad Sci U S A. 2009;106:14867–14871. doi: 10.1073/pnas.0904385106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akiva E, Brown S, Almonacid DE, Barber AE, 2nd, Custer AF, Hicks MA, Huang CC, Lauck F, Mashiyama ST, Meng EC, Mischel D, Morris JH, Ojha S, Schnoes AM, Stryke D, Yunes JM, Ferrin TE, Holliday GL, Babbitt PC. The Structure-Function Linkage Database. Nucleic Acids Res. 2014;42:D521–D530. doi: 10.1093/nar/gkt1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maio N, Singh A, Uhrigshardt H, Saxena N, Tong WH, Rouault TA. Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell Metab. 2014;19:445–457. doi: 10.1016/j.cmet.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maio N, Ghezzi D, Verrigni D, Rizza T, Bertini E, Martinelli D, Zeviani M, Singh A, Carrozzo R, Rouault TA. Disease-Causing SDHAF1 Mutations Impair Transfer of Fe-S Clusters to SDHB. Cell Metab. 2016;23:292–302. doi: 10.1016/j.cmet.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brigle KE, Newton WE, Dean DR. Complete nucleotide sequence of the Azotobacter vinelandii nitrogenase structural gene cluster. Gene. 1985;37:37–44. doi: 10.1016/0378-1119(85)90255-0. [DOI] [PubMed] [Google Scholar]
- 40.Dean DR, Brigle KE. Azotobacter vinelandii nifD- and nifE-encoded polypeptides share structural homology. Proc Natl Acad Sci U S A. 1985;82:5720–5723. doi: 10.1073/pnas.82.17.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dos Santos PC, Johnson DC, Ragle BE, Unciuleac MC, Dean DR. Controlled expression of nif and isc iron-sulfur protein maturation components reveals target specificity and limited functional replacement between the two systems. J Bacteriol. 2007;189:2854–2862. doi: 10.1128/JB.01734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Py B, Barras F. Genetic approaches of the Fe-S cluster biogenesis process in bacteria: Historical account, methodological aspects and future challenges. Biochim Biophys Acta. 2015;1853:1429–1435. doi: 10.1016/j.bbamcr.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 43.Tong WH, Rouault TA. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 2006;3:199–210. doi: 10.1016/j.cmet.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Rouault TA. Mammalian iron-sulphur proteins: novel insights into biogenesis and function. Nat Rev Mol Cell Biol. 2015;16:45–55. doi: 10.1038/nrm3909. [DOI] [PubMed] [Google Scholar]
- 45.Lill R, Dutkiewicz R, Freibert SA, Heidenreich T, Mascarenhas J, Netz DJ, Paul VD, Pierik AJ, Richter N, Stumpfig M, Srinivasan V, Stehling O, Muhlenhoff U. The role of mitochondria and the CIA machinery in the maturation of cytosolic and nuclear iron-sulfur proteins. Eur J Cell Biol. 2015;94:280–291. doi: 10.1016/j.ejcb.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 46.Maio N, Rouault TA. Iron-sulfur cluster biogenesis in mammalian cells: New insights into the molecular mechanisms of cluster delivery. Biochim Biophys Acta. 2015;1853:1493–1512. doi: 10.1016/j.bbamcr.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pandey A, Gordon DM, Pain J, Stemmler TL, Dancis A, Pain D. Frataxin directly stimulates mitochondrial cysteine desulfurase by exposing substrate-binding sites, and a mutant Fe-S cluster scaffold protein with frataxin-bypassing ability acts similarly. J Biol Chem. 2013;288:36773–36786. doi: 10.1074/jbc.M113.525857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoon H, Knight SA, Pandey A, Pain J, Zhang Y, Pain D, Dancis A. Frataxin-bypassing Isu1: characterization of the bypass activity in cells and mitochondria. Biochem J. 2014;459:71–81. doi: 10.1042/BJ20131273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pandey A, Golla R, Yoon H, Dancis A, Pain D. Persulfide formation on mitochondrial cysteine desulfurase: enzyme activation by a eukaryote-specific interacting protein and Fe-S cluster synthesis. Biochem J. 2012;448:171–187. doi: 10.1042/BJ20120951. [DOI] [PubMed] [Google Scholar]
- 50.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 51.Shi R, Proteau A, Villarroya M, Moukadiri I, Zhang L, Trempe JF, Matte A, Armengod ME, Cygler M. Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 2010;8:e1000354. doi: 10.1371/journal.pbio.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodrigues AV, Kandegedara A, Rotondo JA, Dancis A, Stemmler TL. Iron loading site on the Fe-S cluster assembly scaffold protein is distinct from the active site. Biometals. 2015;28:567–576. doi: 10.1007/s10534-015-9846-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bridwell-Rabb J, Fox NG, Tsai CL, Winn AM, Barondeau DP. Human frataxin activates Fe-S cluster biosynthesis by facilitating sulfur transfer chemistry. Biochemistry. 2014;53:4904–4913. doi: 10.1021/bi500532e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Colin F, Martelli A, Clemancey M, Latour JM, Gambarelli S, Zeppieri L, Birck C, Page A, Puccio H, Ollagnier de Choudens S. Mammalian frataxin controls sulfur production and iron entry during de novo Fe4S4 cluster assembly. J Am Chem Soc. 2013;135:733–740. doi: 10.1021/ja308736e. [DOI] [PubMed] [Google Scholar]
- 55.Schmucker S, Martelli A, Colin F, Page A, Wattenhofer-Donze M, Reutenauer L, Puccio H. Mammalian frataxin: an essential function for cellular viability through an interaction with a preformed ISCU/NFS1/ISD11 iron-sulfur assembly complex. PLoS One. 2011;6:e16199. doi: 10.1371/journal.pone.0016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Gakh O, Smith DYt, Isaya G. Oligomeric yeast frataxin drives assembly of core machinery for mitochondrial iron-sulfur cluster synthesis. J Biol Chem. 2009;284:21971–21980. doi: 10.1074/jbc.M109.011197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ranatunga W, Gakh O, Galeano BK, Smith DYt, Soderberg CA, Al-Karadaghi S, Thompson JR, Isaya G. Architecture of the Yeast Mitochondrial Iron-Sulfur Cluster Assembly Machinery: The subcomplex formed by the iron donor, Yfh1 protein, and the scaffold, Isu1 protein. J Biol Chem. 2016;291:10378–10398. doi: 10.1074/jbc.M115.712414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qi W, Cowan JA. Mechanism of glutaredoxin-ISU [2Fe-2S] cluster exchange. Chem Commun (Camb) 2011;47:4989–4991. doi: 10.1039/c0cc05079b. [DOI] [PubMed] [Google Scholar]
- 59.Shi Y, Ghosh M, Kovtunovych G, Crooks DR, Rouault TA. Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis. Biochim Biophys Acta. 2012;1823:484–492. doi: 10.1016/j.bbamcr.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sheftel AD, Stehling O, Pierik AJ, Elsasser HP, Muhlenhoff U, Webert H, Hobler A, Hannemann F, Bernhardt R, Lill R. Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc Natl Acad Sci U S A. 2010;107:11775–11780. doi: 10.1073/pnas.1004250107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chandramouli K, Unciuleac MC, Naik S, Dean DR, Huynh BH, Johnson MK. Formation and properties of [4Fe-4S] clusters on the IscU scaffold protein. Biochemistry. 2007;46:6804–6811. doi: 10.1021/bi6026659. [DOI] [PubMed] [Google Scholar]
- 62.Bonomi F, Iametti S, Morleo A, Ta D, Vickery LE. Facilitated transfer of IscU-[2Fe2S] clusters by chaperone-mediated ligand exchange. Biochemistry. 2011;50:9641–9650. doi: 10.1021/bi201123z. [DOI] [PubMed] [Google Scholar]
- 63.Vickery LE, Cupp-Vickery JR. Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation. Crit Rev Biochem Mol Biol. 2007;42:95–111. doi: 10.1080/10409230701322298. [DOI] [PubMed] [Google Scholar]
- 64.Pukszta S, Schilke B, Dutkiewicz R, Kominek J, Moczulska K, Stepien B, Reitenga KG, Bujnicki JM, Williams B, Craig EA, Marszalek J. Co-evolution-driven switch of J-protein specificity towards an Hsp70 partner. EMBO reports. 2010;11:360–365. doi: 10.1038/embor.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raulfs EC, O'Carroll IP, Dos Santos PC, Unciuleac MC, Dean DR. In vivo iron-sulfur cluster formation. Proc Natl Acad Sci U S A. 2008;105:8591–8596. doi: 10.1073/pnas.0803173105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marinoni EN, de Oliveira JS, Nicolet Y, Raulfs EC, Amara P, Dean DR, Fontecilla-Camps JC. (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer. Angew Chem Int Ed Engl. 2012;51:5439–5442. doi: 10.1002/anie.201201708. [DOI] [PubMed] [Google Scholar]
- 67.Hoff KG, Silberg JJ, Vickery LE. Interaction of the iron-sulfur cluster assembly protein IscU with the Hsc66/Hsc20 molecular chaperone system of Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:7790–7795. doi: 10.1073/pnas.130201997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uhrigshardt H, Singh A, Kovtunovych G, Ghosh M, Rouault TA. Characterization of the human HSC20, an unusual DnaJ type III protein, involved in iron-sulfur cluster biogenesis. Hum Mol Genet. 2010;19:3816–3834. doi: 10.1093/hmg/ddq301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Craig EA, Marszalek J. A specialized mitochondrial molecular chaperone system: a role in formation of Fe/S centers. Cellular and molecular life sciences : CMLS. 2002;59:1658–1665. doi: 10.1007/PL00012493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai K, Frederick RO, Kim JH, Reinen NM, Tonelli M, Markley JL. Human mitochondrial chaperone (mtHSP70) and cysteine desulfurase (NFS1) bind preferentially to the disordered conformation, whereas co-chaperone (HSC20) binds to the structured conformation of the iron-sulfur cluster scaffold protein (ISCU) J Biol Chem. 2013;288:28755–28770. doi: 10.1074/jbc.M113.482042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fuzery AK, Tonelli M, Ta DT, Cornilescu G, Vickery LE, Markley JL. Solution structure of the iron-sulfur cluster cochaperone HscB and its binding surface for the iron-sulfur assembly scaffold protein IscU. Biochemistry. 2008;47:9394–9404. doi: 10.1021/bi800502r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ciesielski SJ, Schilke BA, Osipiuk J, Bigelow L, Mulligan R, Majewska J, Joachimiak A, Marszalek J, Craig EA, Dutkiewicz R. Interaction of J-protein co-chaperone Jac1 with Fe-S scaffold Isu is indispensable in vivo and conserved in evolution. J Mol Biol. 2012;417:1–12. doi: 10.1016/j.jmb.2012.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fuzery AK, Oh JJ, Ta DT, Vickery LE, Markley JL. Three hydrophobic amino acids in Escherichia coli HscB make the greatest contribution to the stability of the HscB-IscU complex. BMC Biochem. 2011;12:3. doi: 10.1186/1471-2091-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barras F, Loiseau L, Py B. How Escherichia coli and Saccharomyces cerevisiae build Fe/S proteins. Adv Microb Physiol. 2005;50:41–101. doi: 10.1016/S0065-2911(05)50002-X. [DOI] [PubMed] [Google Scholar]
- 76.Cupp-Vickery JR, Vickery LE. Crystal structure of Hsc20, a J-type Co-chaperone from Escherichia coli. J Mol Biol. 2000;304:835–845. doi: 10.1006/jmbi.2000.4252. [DOI] [PubMed] [Google Scholar]
- 77.Bitto E, Bingman CA, Bittova L, Kondrashov DA, Bannen RM, Fox BG, Markley JL, Phillips GN., Jr Structure of human J-type co-chaperone HscB reveals a tetracysteine metal-binding domain. J Biol Chem. 2008;283:30184–30192. doi: 10.1074/jbc.M804746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Knieszner H, Schilke B, Dutkiewicz R, D'Silva P, Cheng S, Ohlson M, Craig EA, Marszalek J. Compensation for a defective interaction of the hsp70 ssq1 with the mitochondrial Fe-S cluster scaffold isu. J Biol Chem. 2005;280:28966–28972. doi: 10.1074/jbc.M503031200. [DOI] [PubMed] [Google Scholar]
- 79.Voisine C, Cheng YC, Ohlson M, Schilke B, Hoff K, Beinert H, Marszalek J, Craig EA. Jac1, a mitochondrial J-type chaperone, is involved in the biogenesis of Fe/S clusters in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2001;98:1483–1488. doi: 10.1073/pnas.98.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shan Y, Cortopassi G. HSC20 interacts with frataxin and is involved in iron-sulfur cluster biogenesis and iron homeostasis. Hum Mol Genet. 2012;21:1457–1469. doi: 10.1093/hmg/ddr582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alderson TR, Kim JH, Cai K, Frederick RO, Tonelli M, Markley JL. The specialized Hsp70 (HscA) interdomain linker binds to its nucleotide-binding domain and stimulates ATP hydrolysis in both cis and trans configurations. Biochemistry. 2014;53:7148–7159. doi: 10.1021/bi5010552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Silberg JJ, Hoff KG, Tapley TL, Vickery LE. The Fe/S assembly protein IscU behaves as a substrate for the molecular chaperone Hsc66 from Escherichia coli. J Biol Chem. 2001;276:1696–1700. doi: 10.1074/jbc.M009542200. [DOI] [PubMed] [Google Scholar]
- 83.Vickery LE, Silberg JJ, Ta DT. Hsc66 and Hsc20, a new heat shock cognate molecular chaperone system from Escherichia coli. Protein Sci. 1997;6:1047–1056. doi: 10.1002/pro.5560060511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tapley TL, Vickery LE. Preferential substrate binding orientation by the molecular chaperone HscA. J Biol Chem. 2004;279:28435–28442. doi: 10.1074/jbc.M400803200. [DOI] [PubMed] [Google Scholar]
- 85.Hoff KG, Ta DT, Tapley TL, Silberg JJ, Vickery LE. Hsc66 substrate specificity is directed toward a discrete region of the iron-sulfur cluster template protein IscU. J Biol Chem. 2002;277:27353–27359. doi: 10.1074/jbc.M202814200. [DOI] [PubMed] [Google Scholar]
- 86.Hoff KG, Cupp-Vickery JR, Vickery LE. Contributions of the LPPVK motif of the iron-sulfur template protein IscU to interactions with the Hsc66-Hsc20 chaperone system. J Biol Chem. 2003;278:37582–37589. doi: 10.1074/jbc.M305292200. [DOI] [PubMed] [Google Scholar]
- 87.Cupp-Vickery JR, Peterson JC, Ta DT, Vickery LE. Crystal structure of the molecular chaperone HscA substrate binding domain complexed with the IscU recognition peptide ELPPVKIHC. J Mol Biol. 2004;342:1265–1278. doi: 10.1016/j.jmb.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 88.Kim JH, Tonelli M, Frederick RO, Chow DC, Markley JL. Specialized Hsp70 chaperone (HscA) binds preferentially to the disordered form, whereas J-protein (HscB) binds preferentially to the structured form of the iron-sulfur cluster scaffold protein (IscU) J Biol Chem. 2012;287:31406–31413. doi: 10.1074/jbc.M112.352617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim JH, Tonelli M, Kim T, Markley JL. Three-dimensional structure and determinants of stability of the iron-sulfur cluster scaffold protein IscU from Escherichia coli. Biochemistry. 2012;51:5557–5563. doi: 10.1021/bi300579p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim JH, Tonelli M, Markley JL. Disordered form of the scaffold protein IscU is the substrate for iron-sulfur cluster assembly on cysteine desulfurase. Proc Natl Acad Sci U S A. 2012;109:454–459. doi: 10.1073/pnas.1114372109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Johnson DC, Unciuleac MC, Dean DR. Controlled expression and functional analysis of iron-sulfur cluster biosynthetic components within Azotobacter vinelandii. J Bacteriol. 2006;188:7551–7561. doi: 10.1128/JB.00596-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tokumoto U, Takahashi Y. Genetic analysis of the isc operon in Escherichia coli involved in the biogenesis of cellular iron-sulfur proteins. J Biochem. 2001;130:63–71. doi: 10.1093/oxfordjournals.jbchem.a002963. [DOI] [PubMed] [Google Scholar]
- 93.Bonomi F, Iametti S, Morleo A, Ta D, Vickery LE. Studies on the mechanism of catalysis of iron-sulfur cluster transfer from IscU[2Fe2S] by HscA/HscB chaperones. Biochemistry. 2008;47:12795–12801. doi: 10.1021/bi801565j. [DOI] [PubMed] [Google Scholar]
- 94.Kim R, Saxena S, Gordon DM, Pain D, Dancis A. J-domain protein, Jac1p, of yeast mitochondria required for iron homeostasis and activity of Fe-S cluster proteins. The Journal of biological chemistry. 2001;276:17524–17532. doi: 10.1074/jbc.M010695200. [DOI] [PubMed] [Google Scholar]
- 95.Uhrigshardt H, Rouault TA, Missirlis F. Insertion mutants in Drosophila melanogaster Hsc20 halt larval growth and lead to reduced iron-sulfur cluster enzyme activities and impaired iron homeostasis. Journal of biological inorganic chemistry : JBIC : a publication of the Society of Biological Inorganic Chemistry. 2013;18:441–449. doi: 10.1007/s00775-013-0988-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Majewska J, Ciesielski SJ, Schilke B, Kominek J, Blenska A, Delewski W, Song JY, Marszalek J, Craig EA, Dutkiewicz R. Binding of the chaperone Jac1 protein and cysteine desulfurase Nfs1 to the iron-sulfur cluster scaffold Isu protein is mutually exclusive. J Biol Chem. 2013;288:29134–29142. doi: 10.1074/jbc.M113.503524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Perales-Calvo J, Muga A, Moro F. Role of DnaJ G/F-rich domain in conformational recognition and binding of protein substrates. J Biol Chem. 2010;285:34231–34239. doi: 10.1074/jbc.M110.144642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Szabo A, Korszun R, Hartl FU, Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- 99.Lee S, Fan CY, Younger JM, Ren H, Cyr DM. Identification of essential residues in the type II Hsp40 Sis1 that function in polypeptide binding. J Biol Chem. 2002;277:21675–21682. doi: 10.1074/jbc.M111075200. [DOI] [PubMed] [Google Scholar]
- 100.Li J, Sha B. Structure-based mutagenesis studies of the peptide substrate binding fragment of type I heat-shock protein 40. Biochem J. 2005;386:453–460. doi: 10.1042/BJ20041050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cameron JM, Janer A, Levandovskiy V, Mackay N, Rouault TA, Tong WH, Ogilvie I, Shoubridge EA, Robinson BH. Mutations in iron-sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes. American journal of human genetics. 2011;89:486–495. doi: 10.1016/j.ajhg.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Navarro-Sastre A, Tort F, Stehling O, Uzarska MA, Arranz JA, Del Toro M, Labayru MT, Landa J, Font A, Garcia-Villoria J, Merinero B, Ugarte M, Gutierrez-Solana LG, Campistol J, Garcia-Cazorla A, Vaquerizo J, Riudor E, Briones P, Elpeleg O, Ribes A, Lill R. A fatal mitochondrial disease is associated with defective NFU1 function in the maturation of a subset of mitochondrial Fe-S proteins. American journal of human genetics. 2011;89:656–667. doi: 10.1016/j.ajhg.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ferrer-Cortes X, Font A, Bujan N, Navarro-Sastre A, Matalonga L, Arranz JA, Riudor E, del Toro M, Garcia-Cazorla A, Campistol J, Briones P, Ribes A, Tort F. Protein expression profiles in patients carrying NFU1 mutations. Contribution to the pathophysiology of the disease. Journal of inherited metabolic disease. 2013;36:841–847. doi: 10.1007/s10545-012-9565-z. [DOI] [PubMed] [Google Scholar]
- 104.Haack TB, Rolinski B, Haberberger B, Zimmermann F, Schum J, Strecker V, Graf E, Athing U, Hoppen T, Wittig I, Sperl W, Freisinger P, Mayr JA, Strom TM, Meitinger T, Prokisch H. Homozygous missense mutation in BOLA3 causes multiple mitochondrial dysfunctions syndrome in two siblings. Journal of inherited metabolic disease. 2013;36:55–62. doi: 10.1007/s10545-012-9489-7. [DOI] [PubMed] [Google Scholar]
- 105.Nizon M, Boutron A, Boddaert N, Slama A, Delpech H, Sardet C, Brassier A, Habarou F, Delahodde A, Correia I, Ottolenghi C, de Lonlay P. Leukoencephalopathy with cysts and hyperglycinemia may result from NFU1 deficiency. Mitochondrion. 2014;15:59–64. doi: 10.1016/j.mito.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 106.Baker PR, 2nd, Friederich MW, Swanson MA, Shaikh T, Bhattacharya K, Scharer GH, Aicher J, Creadon-Swindell G, Geiger E, MacLean KN, Lee WT, Deshpande C, Freckmann ML, Shih LY, Wasserstein M, Rasmussen MB, Lund AM, Procopis P, Cameron JM, Robinson BH, Brown GK, Brown RM, Compton AG, Dieckmann CL, Collard R, Coughlin CR, 2nd, Spector E, Wempe MF, Van Hove JL. Variant non ketotic hyperglycinemia is caused by mutations in LIAS, BOLA3 and the novel gene GLRX5. Brain : a journal of neurology. 2014;137:366–379. doi: 10.1093/brain/awt328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Invernizzi F, Ardissone A, Lamantea E, Garavaglia B, Zeviani M, Farina L, Ghezzi D, Moroni I. Cavitating leukoencephalopathy with multiple mitochondrial dysfunction syndrome and NFU1 mutations. Frontiers in genetics. 2014;5:412. doi: 10.3389/fgene.2014.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tonduti D, Dorboz I, Imbard A, Slama A, Boutron A, Pichard S, Elmaleh M, Vallee L, Benoist JF, Ogier H, Boespflug-Tanguy O. New spastic paraplegia phenotype associated to mutation of NFU1. Orphanet journal of rare diseases. 2015;10:13. doi: 10.1186/s13023-015-0237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ajit Bolar N, Vanlander AV, Wilbrecht C, Van der Aa N, Smet J, De Paepe B, Vandeweyer G, Kooy F, Eyskens F, De Latter E, Delanghe G, Govaert P, Leroy JG, Loeys B, Lill R, Van Laer L, Van Coster R. Mutation of the iron-sulfur cluster assembly gene IBA57 causes severe myopathy and encephalopathy. Hum Mol Genet. 2013;22:2590–2602. doi: 10.1093/hmg/ddt107. [DOI] [PubMed] [Google Scholar]
- 110.Lossos A, Stumpfig C, Stevanin G, Gaussen M, Zimmerman BE, Mundwiller E, Asulin M, Chamma L, Sheffer R, Misk A, Dotan S, Gomori JM, Ponger P, Brice A, Lerer I, Meiner V, Lill R. Fe/S protein assembly gene IBA57 mutation causes hereditary spastic paraplegia. Neurology. 2015;84:659–667. doi: 10.1212/WNL.0000000000001270. [DOI] [PubMed] [Google Scholar]
- 111.Al-Hassnan ZN, Al-Dosary M, Alfadhel M, Faqeih EA, Alsagob M, Kenana R, Almass R, Al-Harazi OS, Al-Hindi H, Malibari OI, Almutari FB, Tulbah S, Alhadeq F, Al-Sheddi T, Alamro R, AlAsmari A, Almuntashri M, Alshaalan H, Al-Mohanna FA, Colak D, Kaya N. ISCA2 mutation causes infantile neurodegenerative mitochondrial disorder. Journal of medical genetics. 2015;52:186–194. doi: 10.1136/jmedgenet-2014-102592. [DOI] [PubMed] [Google Scholar]
- 112.Wadhwa R, Taira K, Kaul SC. An Hsp70 family chaperone, mortalin/mthsp70/PBP74/Grp75: what, when, and where? Cell Stress Chaperones. 2002;7:309–316. doi: 10.1379/1466-1268(2002)007<0309:ahfcmm>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Daugaard M, Rohde M, Jaattela M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 114.Alderson TR, Kim JH, Markley JL. Dynamical Structures of Hsp70 and Hsp70-Hsp40 Complexes. Structure. 2016 doi: 10.1016/j.str.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Singer TP, Johnson MK. The prosthetic groups of succinate dehydrogenase: 30 years from discovery to identification. FEBS Lett. 1985;190:189–198. doi: 10.1016/0014-5793(85)81282-5. [DOI] [PubMed] [Google Scholar]
- 116.Birch-Machin MA, Taylor RW, Cochran B, Ackrell BA, Turnbull DM. Late-onset optic atrophy, ataxia, and myopathy associated with a mutation of a complex II gene. Annals of neurology. 2000;48:330–335. [PubMed] [Google Scholar]
- 117.Ghezzi D, Goffrini P, Uziel G, Horvath R, Klopstock T, Lochmuller H, D'Adamo P, Gasparini P, Strom TM, Prokisch H, Invernizzi F, Ferrero I, Zeviani M. SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nature genetics. 2009;41:654–656. doi: 10.1038/ng.378. [DOI] [PubMed] [Google Scholar]
- 118.Jain-Ghai S, Cameron JM, Al Maawali A, Blaser S, MacKay N, Robinson B, Raiman J. Complex II deficiency--a case report and review of the literature. American journal of medical genetics. Part A. 2013;161A:285–294. doi: 10.1002/ajmg.a.35714. [DOI] [PubMed] [Google Scholar]
- 119.Ricketts CJ, Shuch B, Vocke CD, Metwalli AR, Bratslavsky G, Middelton L, Yang Y, Wei MH, Pautler SE, Peterson J, Stolle CA, Zbar B, Merino MJ, Schmidt LS, Pinto PA, Srinivasan R, Pacak K, Linehan WM. Succinate dehydrogenase kidney cancer: an aggressive example of the Warburg effect in cancer. The Journal of urology. 2012;188:2063–2071. doi: 10.1016/j.juro.2012.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Saxena N, Maio N, Crooks DR, Ricketts CJ, Yang Y, Wei MH, Fan TW, Lane AN, Sourbier C, Singh A, Killian JK, Meltzer PS, Vocke CD, Rouault TA, Linehan WM. SDHB-Deficient Cancers: The Role of Mutations That Impair Iron Sulfur Cluster Delivery. J Natl Cancer Inst. 2016;108 doi: 10.1093/jnci/djv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hagerhall C, Hederstedt L. A structural model for the membrane-integral domain of succinate: quinone oxidoreductases. FEBS letters. 1996;389:25–31. doi: 10.1016/0014-5793(96)00529-7. [DOI] [PubMed] [Google Scholar]
- 122.Tompa P, Davey NE, Gibson TJ, Babu MM. A million peptide motifs for the molecular biologist. Molecular cell. 2014;55:161–169. doi: 10.1016/j.molcel.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 123.Babu MM, Kriwacki RW, Pappu RV. Structural biology. Versatility from protein disorder. Science. 2012;337:1460–1461. doi: 10.1126/science.1228775. [DOI] [PubMed] [Google Scholar]
- 124.Ricketts C, Woodward ER, Killick P, Morris MR, Astuti D, Latif F, Maher ER. Germline SDHB mutations and familial renal cell carcinoma. J Natl Cancer Inst. 2008;100:1260–1262. doi: 10.1093/jnci/djn254. [DOI] [PubMed] [Google Scholar]
- 125.Angerer H. Eukaryotic LYR Proteins Interact with Mitochondrial Protein Complexes. Biology (Basel) 2015;4:133–150. doi: 10.3390/biology4010133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shi Y, Ghosh MC, Tong WH, Rouault TA. Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Human molecular genetics. 2009;18:3014–3025. doi: 10.1093/hmg/ddp239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Angerer H. The superfamily of mitochondrial Complex1_LYR motif-containing (LYRM) proteins. Biochemical Society transactions. 2013;41:1335–1341. doi: 10.1042/BST20130116. [DOI] [PubMed] [Google Scholar]
- 128.Dennis RA, McCammon MT. Acn9 is a novel protein of gluconeogenesis that is located in the mitochondrial intermembrane space. European journal of biochemistry/FEBS. 1999;261:236–243. doi: 10.1046/j.1432-1327.1999.00267.x. [DOI] [PubMed] [Google Scholar]
- 129.Lefebvre-Legendre L, Vaillier J, Benabdelhak H, Velours J, Slonimski PP, di Rago JP. Identification of a nuclear gene (FMC1) required for the assembly/stability of yeast mitochondrial F(1)-ATPase in heat stress conditions. J Biol Chem. 2001;276:6789–6796. doi: 10.1074/jbc.M009557200. [DOI] [PubMed] [Google Scholar]
- 130.Sanchez E, Lobo T, Fox JL, Zeviani M, Winge DR, Fernandez-Vizarra E. LYRM7/MZM1L is a UQCRFS1 chaperone involved in the last steps of mitochondrial Complex III assembly in human cells. Biochimica et biophysica acta. 2013;1827:285–293. doi: 10.1016/j.bbabio.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ohlenbusch A, Edvardson S, Skorpen J, Bjornstad A, Saada A, Elpeleg O, Gartner J, Brockmann K. Leukoencephalopathy with accumulated succinate is indicative of SDHAF1 related complex II deficiency. Orphanet J Rare Dis. 2012;7:69. doi: 10.1186/1750-1172-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Angerer H, Radermacher M, Mankowska M, Steger M, Zwicker K, Heide H, Wittig I, Brandt U, Zickermann V. The LYR protein subunit NB4M/NDUFA6 of mitochondrial complex I anchors an acyl carrier protein and is essential for catalytic activity. Proc Natl Acad Sci U S A. 2014;111:5207–5212. doi: 10.1073/pnas.1322438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Uzarska MA, Dutkiewicz R, Freibert SA, Lill R, Muhlenhoff U. The mitochondrial Hsp70 chaperone Ssq1 facilitates Fe/S cluster transfer from Isu1 to Grx5 by complex formation. Mol Biol Cell. 2013;24:1830–1841. doi: 10.1091/mbc.E12-09-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dutkiewicz R, Schilke B, Cheng S, Knieszner H, Craig EA, Marszalek J. Sequence-specific interaction between mitochondrial Fe-S scaffold protein Isu and Hsp70 Ssq1 is essential for their in vivo function. The Journal of biological chemistry. 2004;279:29167–29174. doi: 10.1074/jbc.M402947200. [DOI] [PubMed] [Google Scholar]
- 136.Muhlenhoff U, Gerber J, Richhardt N, Lill R. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 2003;22:4815–4825. doi: 10.1093/emboj/cdg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Belli G, Polaina J, Tamarit J, De La Torre MA, Rodriguez-Manzaneque MT, Ros J, Herrero E. Structure-function analysis of yeast Grx5 monothiol glutaredoxin defines essential amino acids for the function of the protein. J Biol Chem. 2002;277:37590–37596. doi: 10.1074/jbc.M201688200. [DOI] [PubMed] [Google Scholar]
- 138.Rodriguez-Manzaneque MT, Tamarit J, Belli G, Ros J, Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell. 2002;13:1109–1121. doi: 10.1091/mbc.01-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wingert RA, Galloway JL, Barut B, Foott H, Fraenkel P, Axe JL, Weber GJ, Dooley K, Davidson AJ, Schmid B, Paw BH, Shaw GC, Kingsley P, Palis J, Schubert H, Chen O, Kaplan J, Zon LI, Tubingen Screen C. Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature. 2005;436:1035–1039. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
- 140.Rouhier N, Couturier J, Johnson MK, Jacquot JP. Glutaredoxins: roles in iron homeostasis. Trends Biochem Sci. 2010;35:43–52. doi: 10.1016/j.tibs.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li H, Outten CE. Monothiol CGFS glutaredoxins and BolA-like proteins: [2Fe-2S] binding partners in iron homeostasis. Biochemistry. 2012;51:4377–4389. doi: 10.1021/bi300393z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Camaschella C, Campanella A, De Falco L, Boschetto L, Merlini R, Silvestri L, Levi S, Iolascon A. The human counterpart of zebrafish shiraz shows sideroblastic-like microcytic anemia and iron overload. Blood. 2007;110:1353–1358. doi: 10.1182/blood-2007-02-072520. [DOI] [PubMed] [Google Scholar]
- 143.Muhlenhoff U, Molik S, Godoy JR, Uzarska MA, Richter N, Seubert A, Zhang Y, Stubbe J, Pierrel F, Herrero E, Lillig CH, Lill R. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 2010;12:373–385. doi: 10.1016/j.cmet.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kumanovics A, Chen OS, Li L, Bagley D, Adkins EM, Lin H, Dingra NN, Outten CE, Keller G, Winge D, Ward DM, Kaplan J. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J Biol Chem. 2008;283:10276–10286. doi: 10.1074/jbc.M801160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ojeda L, Keller G, Muhlenhoff U, Rutherford JC, Lill R, Winge DR. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J Biol Chem. 2006;281:17661–17669. doi: 10.1074/jbc.M602165200. [DOI] [PubMed] [Google Scholar]
- 146.Pujol-Carrion N, Belli G, Herrero E, Nogues A, de la Torre-Ruiz MA. Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J Cell Sci. 2006;119:4554–4564. doi: 10.1242/jcs.03229. [DOI] [PubMed] [Google Scholar]
- 147.Banci L, Ciofi-Baffoni S, Gajda K, Muzzioli R, Peruzzini R, Winkelmann J. N-terminal domains mediate [2Fe-2S] cluster transfer from glutaredoxin-3 to anamorsin. Nat Chem Biol. 2015;11:772–778. doi: 10.1038/nchembio.1892. [DOI] [PubMed] [Google Scholar]
- 148.Johansson C, Roos AK, Montano SJ, Sengupta R, Filippakopoulos P, Guo K, von Delft F, Holmgren A, Oppermann U, Kavanagh KL. The crystal structure of human GLRX5: iron-sulfur cluster co-ordination, tetrameric assembly and monomer activity. Biochem J. 2011;433:303–311. doi: 10.1042/BJ20101286. [DOI] [PubMed] [Google Scholar]
- 149.Brancaccio D, Gallo A, Mikolajczyk M, Zovo K, Palumaa P, Novellino E, Piccioli M, Ciofi-Baffoni S, Banci L. Formation of [4Fe-4S] clusters in the mitochondrial iron-sulfur cluster assembly machinery. J Am Chem Soc. 2014;136:16240–16250. doi: 10.1021/ja507822j. [DOI] [PubMed] [Google Scholar]
- 150.Qi W, Li J, Chain CY, Pasquevich GA, Pasquevich AF, Cowan JA. Glutathione complexed Fe-S centers. J Am Chem Soc. 2012;134:10745–10748. doi: 10.1021/ja302186j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huynen MA, Spronk CA, Gabaldon T, Snel B. Combining data from genomes, Y2H and 3D structure indicates that BolA is a reductase interacting with a glutaredoxin. FEBS Lett. 2005;579:591–596. doi: 10.1016/j.febslet.2004.11.111. [DOI] [PubMed] [Google Scholar]
- 152.Couturier J, Jacquot JP, Rouhier N. Evolution and diversity of glutaredoxins in photosynthetic organisms. Cellular and molecular life sciences : CMLS. 2009;66:2539–2557. doi: 10.1007/s00018-009-0054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, Jr, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 155.Couturier J, Wu HC, Dhalleine T, Pegeot H, Sudre D, Gualberto JM, Jacquot JP, Gaymard F, Vignols F, Rouhier N. Monothiol glutaredoxin-BolA interactions: redox control of Arabidopsis thaliana BolA2 and SufE1. Molecular plant. 2014;7:187–205. doi: 10.1093/mp/sst156. [DOI] [PubMed] [Google Scholar]
- 156.Aldea M, Garrido T, Hernandez-Chico C, Vicente M, Kushner SR. Induction of a growth-phase-dependent promoter triggers transcription of bolA, an Escherichia coli morphogene. EMBO J. 1989;8:3923–3931. doi: 10.1002/j.1460-2075.1989.tb08573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Li H, Mapolelo DT, Dingra NN, Naik SG, Lees NS, Hoffman BM, Riggs-Gelasco PJ, Huynh BH, Johnson MK, Outten CE. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry. 2009;48:9569–9581. doi: 10.1021/bi901182w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li H, Mapolelo DT, Dingra NN, Keller G, Riggs-Gelasco PJ, Winge DR, Johnson MK, Outten CE. Histidine 103 in Fra2 is an iron-sulfur cluster ligand in the [2Fe-2S] Fra2-Grx3 complex and is required for in vivo iron signaling in yeast. J Biol Chem. 2011;286:867–876. doi: 10.1074/jbc.M110.184176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tan G, Lu J, Bitoun JP, Huang H, Ding H. IscA/SufA paralogues are required for the [4Fe-4S] cluster assembly in enzymes of multiple physiological pathways in Escherichia coli under aerobic growth conditions. Biochem J. 2009;420:463–472. doi: 10.1042/BJ20090206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Loiseau L, Gerez C, Bekker M, Ollagnier-de Choudens S, Py B, Sanakis Y, Teixeira de Mattos J, Fontecave M, Barras F. ErpA, an iron sulfur (Fe S) protein of the A-type essential for respiratory metabolism in Escherichia coli. Proc Natl Acad Sci U S A. 2007;104:13626–13631. doi: 10.1073/pnas.0705829104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sheftel AD, Wilbrecht C, Stehling O, Niggemeyer B, Elsasser HP, Muhlenhoff U, Lill R. The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe-4S] protein maturation. Mol Biol Cell. 2012;23:1157–1166. doi: 10.1091/mbc.E11-09-0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Debray FG, Stumpfig C, Vanlander AV, Dideberg V, Josse C, Caberg JH, Boemer F, Bours V, Stevens R, Seneca S, Smet J, Lill R, van Coster R. Mutation of the iron-sulfur cluster assembly gene IBA57 causes fatal infantile leukodystrophy. Journal of inherited metabolic disease. 2015;38:1147–1153. doi: 10.1007/s10545-015-9857-1. [DOI] [PubMed] [Google Scholar]
- 163.Bych K, Kerscher S, Netz DJ, Pierik AJ, Zwicker K, Huynen MA, Lill R, Brandt U, Balk J. The iron-sulphur protein Ind1 is required for effective complex I assembly. EMBO J. 2008;27:1736–1746. doi: 10.1038/emboj.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sheftel AD, Stehling O, Pierik AJ, Netz DJ, Kerscher S, Elsasser HP, Wittig I, Balk J, Brandt U, Lill R. Human ind1, an iron-sulfur cluster assembly factor for respiratory complex I. Mol Cell Biol. 2009;29:6059–6073. doi: 10.1128/MCB.00817-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wydro MM, Sharma P, Foster JM, Bych K, Meyer EH, Balk J. The evolutionarily conserved iron-sulfur protein INDH is required for complex I assembly and mitochondrial translation in Arabidopsis [corrected] The Plant cell. 2013;25:4014–4027. doi: 10.1105/tpc.113.117283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Karcher A, Schele A, Hopfner KP. X-ray structure of the complete ABC enzyme ABCE1 from Pyrococcus abyssi. J Biol Chem. 2008;283:7962–7971. doi: 10.1074/jbc.M707347200. [DOI] [PubMed] [Google Scholar]
- 167.Gari K, Leon Ortiz AM, Borel V, Flynn H, Skehel JM, Boulton SJ. MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science. 2012;337:243–245. doi: 10.1126/science.1219664. [DOI] [PubMed] [Google Scholar]
- 168.Stehling O, Vashisht AA, Mascarenhas J, Jonsson ZO, Sharma T, Netz DJ, Pierik AJ, Wohlschlegel JA, Lill R. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science. 2012;337:195–199. doi: 10.1126/science.1219723. [DOI] [PMC free article] [PubMed] [Google Scholar]


