Abstract
Pregnant women with diabetes may have underlying beta cell dysfunction due to mutations/rare variants in genes associated with Maturity Onset Diabetes of the Young (MODY). MODY gene screening would reveal those women genetically predisposed and previously unrecognized with a monogenic form of diabetes for further clinical management, family screening and genetic counselling. However, there are minimal data available on MODY gene variants in pregnant women with diabetes from India. In this study, utilizing the Next generation sequencing (NGS) based protocol fifty subjects were screened for variants in a panel of thirteen MODY genes. Of these subjects 18% (9/50) were positive for definite or likely pathogenic or uncertain MODY variants. The majority of these variants was identified in subjects with autosomal dominant family history, of whom five were in women with pre-GDM and four with overt-GDM. The identified variants included one patient with HNF1A Ser3Cys, two PDX1 Glu224Lys, His94Gln, two NEUROD1 Glu59Gln, Phe318Ser, one INS Gly44Arg, one GCK, one ABCC8 Arg620Cys and one BLK Val418Met variants. In addition, three of the seven offspring screened were positive for the identified variant. These identified variants were further confirmed by Sanger sequencing. In conclusion, these findings in pregnant women with diabetes, imply that a proportion of GDM patients with autosomal dominant family history may have MODY. Further NGS based comprehensive studies with larger samples are required to confirm these finding
Introduction
Diabetes mellitus(DM) has evolved into a global epidemic and as Asian countries have undergone an economic, social and nutritional transition, type 2 diabetes mellitus and gestational diabetes mellitus (GDM) have increased exponentially[1]. GDM is defined as “carbohydrate intolerance of any degree of severity with an onset or first recognition during pregnancy”[2]. The prevalence of GDM ranges from 3.8 to 21%, worldwide. In India, it may affect around 200,000 pregnant women annually[2,3]. In a significant proportion of women, glucose intolerance during gestation appears to resolve after delivery. However, up to 50% of these women develop type 2 diabetes within five years after parturition[4]. Women who develop GDM early (<20 weeks) may have an additional underlying deficit, this may fall into one of three major categories: pre-existing insulin resistance, autoimmune induced disorders and monogenic disease like MODY[5]. With an increasing incidence of early onset diabetes and a high prevalence of GDM[3], there could be a significant proportion of MODY which may be misdiagnosed among pregnant women in India. Traditionally, MODY has been thought to account for 2% of diabetes and results from mutations in one of the thirteen genes that have been reported till date[6]. In addition, MODY gene screening would reveal not only those genetically predisposed women, but also their offspring who are at a 50% risk of inheriting the mutation and subsequently may enable the ability to predict the potential response to sulphonylureas (SU) in some patients[7] and aid in identifying the commodities in some forms of MODY[8].
The standard approach for diagnosis of MODY includes sequential screening of the first three common MODY genes which include hepatocyte nuclear factor 1alpha (HNF1A), hepatocyte nuclear factor 4 alpha (HNF4A) and Glucokinase (GCK)[9]. In addition, studies on MODY in pregnant women with diabetes were largely limited to the screening of GCK, HNF1A and HNF4A genes[10]. However, with the advent of next-generation sequencing(NGS) technology, there has been a significant improvement in the speed and scalability of sequencing[11]. Utilizing NGS based strategy, we have recently reported a wider spectrum of MODY mutations involving NEUROD1 and PDX1 in patients with young onset diabetes (<30years) in India[12]. Recognizing the presence and pattern of MODY genetic component in pregnant women with diabetes, will help in the identification of susceptible individuals, which in turn, may enable preventive and therapeutic measures for both the mother and the developing foetus[13]. In the present study using a similar NGS based approach[12], we have screened for pathogenic variants in a panel of 13 MODY genes in pregnant women with diabetes.
Methods
In the present cross-sectional observational study, we have recruited women with pregnancy complicated by hyperglycemia, from September 2012 to December 2013. Written informed consent was obtained from all study participants and the study was approved by the Institutional Review Board (IRB), Christian Medical College, Vellore (IRB Min.No.7998/2012). All pregnant women with any degree of glucose intolerance with an age of onset of disease ≤ 35 years and a body mass index (BMI) ≤ 30kg/m2 were included. Though MODY has been traditionally perceived to present before the age of 25 years, the diagnosis of diabetes may be delayed beyond 35 years[14,15]. Further, many MODY subjects in a previous study at our center had a higher BMI of up to 30kg/m2 with phenotypic characteristics that were typical of MODY and had a reduced insulin secretion when compared to subjects with type 2 diabetes and normal controls[12]. Those with an age of onset of diabetes >35 years or a body mass index (BMI) >30kg/m2, type 1 diabetes mellitus (T1DM) assigned based on the presence of glutamic acid decarboxylase (GAD)antibody and/or low C-peptide levels (<0.6ng/ml), secondary diabetes or fibrocalcific pancreatic diabetes were excluded from the study. We selected control subjects from a homogenous population of Dravidian ethnic origin (South Indian, Tamil speaking). All the control subjects had a normal OGTT with an HbA1c <5.7. Further, these subjects were without any known family history of diabetes. The body mass index was calculated from either the pre-conceptional weight or the weight recorded at the initial visit within 8 weeks of gestation. The biochemical assays were performed on a Hitachi 912 auto analyzer using reagents from Roche Diagnostics (Mannheim, Germany), GAD antibodies were measured with the radioimmunoassay technique (Bio-Rad iMark™ microplate ELISA reader, intra-assay; CV:5%). The glycosylated haemoglobin (HbA1c) was measured by the ion-exchange base High Performance Liquid Chromatography (HPLC) assay method (Bio-Rad Variant II turbo glycated haemoglobin (HbA1c) analyzer; CV: 2.6%). Based on the International Association of the Diabetes and Pregnancy Study Groups (IADPSG) classification[16] of the hyperglycemic disorders complicating pregnancy, our patients were classified into three groups.
1. Gestational Diabetes Mellitus [GDM]
The diagnosis of GDM is made when any of the following plasma glucose values are exceeded after a 75-g oral glucose tolerance test (OGTT) any time during pregnancy,
Fasting: ≥92 mg/dl (5.1 mmol/l)
1 h: ≥180 mg/dl (10.0 mmol/l)
2 h: ≥153 mg/dl (8.5 mmol/l)
[NOTE: All women not previously diagnosed with diabetes underwent a 75-g oral glucose tolerance test (OGTT) at 24–28 weeks of gestation. The OGTT was performed in the morning after an overnight fast of at least 8 hours. As per the IADPSG Consensus Panel OGTT was not routinely performed before 24–28 weeks gestation, but a fasting plasma glucose value (lower than those diagnostic of diabetes) in early pregnancy (≥ 5.1mmol/l) was also classified as GDM].
2. OvertDabetes Mellitus [Overt-GDM]
Glucose values / HbA1c that were diagnostic of diabetes using the standard American Diabetes Association (ADA) Criteria[16] at the first visit any time after conception or at 24–28 weeks of gestation during the one step 75-g oral glucose tolerance test (OGTT) were categorised as “overt-GDM”. Women who meet the criteria for diabetes are classified as a separate category named“overt diabetes in pregnancy” by the IADPSG criteria and represent the "highest at risk" GDM cohort, who have an increased risk of congenital abnormalities and diabetes related complications[17].
3. Pre-Gestational Diabetes Mellitus [PRE-GDM]
Pregnant women who were known to have diabetes prior to conception.
Statistical analysis
Depending on the selection criteria for screening, the prevalence of MODY in pregnant women with diabetes has been reported to range from 1% to 80%(10). This wide variation in previous studies on the prevalence of MODY mutations may be a result of highly variable inclusion criteria for GDM in order to increase the yield of the test. In this study, we have included a hybrid of patients with and without strong family histories, so a moderate prevalence was hypothesized and hence a 40% "median cumulative prevalence" was assumed and sample size was determined. Therefore, with a median cumulative prevalence of 40% and a 12 to 15% precision, 50 pregnant women were screened with a 13 MODY gene panel (calculated using the N-master software, www.nmaster.cmc-biostatistics.ac.in).
Continuous variables were described using means and standard deviation (SD). For variables with skewed distributions, medians with inter-quartile ranges were used. All categorical variables were summarized by using frequencies and related percentages. The association between pathogenic variants and other covariates (BMI, family history of diabetes) were tested by using Fischer’s exact test, the independent two sample t-test or Chi-square test. The data were analyzed using SPSS version 17.0.
NGS based MODY gene sequencing
Utilizing our recently published NGS based 2GDMODY protocol(13), we have screened 50 pregnant women with diabetes for a comprehensive panel of 13 MODY genes [HNF1A, HNF4A, GCK, PDX1, HNF1B, NEUROD1, KLF11, CEL, PAX4, INS, BLK, ABCC8,KCNJ11]. Sanger sequencing was performed to confirm all the identified variants of interest. Furthermore, sequencing data from 110 subjects with normal glucose tolerance was utilized for variant interpretation.
Bioinformatics Analysis
Sequencing was performed on an Ion torrent PGM using the Ion PGM™ 200 Sequencing Kit (Ion Torrent, Life Technologies), utilizing 314 chips (multiplex 4 samples) and 316 chips (multiplex 10–15 samples). The generated sequencing data was mapped to the human genome reference hg19. The coverage analysis was calculated using Torrent Coverage Analysis and potential pathogenic variants were identified using the Torrent Variant Caller and DNA Star software (DNASTAR, Madison, WI, USA). The Human Gene Mutation Database (HGMD®Professional 2012.4), was utilized to classify the identified variants as reported or novel. Further, the population sequencing database EXAC [18] was explored to validate the novel variants identified adhering to the latest guidelines by the American College of Medical Genetics19]. The in-silico analysis for likelihood of pathogenicity was performed for all the novel variants using PolyPhen-2[20], Sorting Intolerant From Tolerant (SIFT)[21] and Mutation taster [22] and for sequence conservation with Genomic Evolutionary Rate Profiling (GERP)[23].
Results
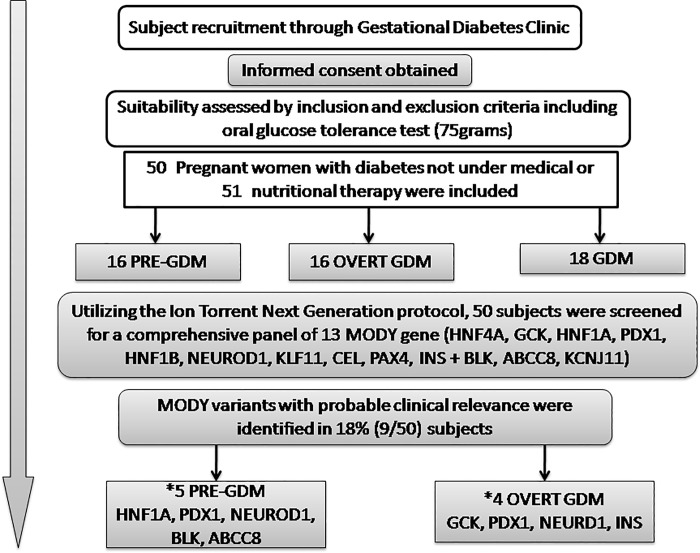
In the present study 50 subjects were screened, of whom 16 (32%) were with pre-gestational diabetes, 18 (36%) with gestational diabetes and overt diabetes in pregnancy was present in 16 (32%) subjects (Fig 1). There was no consanguinity in this study group and all members were of the Dravidian (south India) ethnicity, the site of recruitment being in the southern most Indian state of Tamil Nadu. The baseline characteristics of these subjects are summarized in Table 1. The mean age of the subjects was 29 years, mean BMI was 27.47kg/m2 with 37% (n = 17) below 25kg/m2. The median age of diagnosis of diabetes in the pre-GDM group was 22 months prior to conception (except in subject MG101 who was diagnosed at the age of 10 years and was negative for GAD antibodies). The median gestational age of diagnosis in the overt GDM and GDM group was 16 and 20 weeks respectively. The mean HbA1c of the subjects in the pre-GDM group was 8.2% and 7.4% in the overt-GDM group. Overall, 54% (n = 27) of the subjects had a three generation family history of diabetes, who predominantly (n = 20) were in the pre-GDM and overt-GDM groups. Postpartum, patients with overt diabetes in pregnancy (n = 16) who underwent an oral glucose tolerance test and/or HbA1c after 12 weeks post-partum, 25% (n = 4) had diabetes or 31% (n = 5) had impaired glucose tolerance and the rest (44%) were with normal glucose tolerance (n = 7). This was similar to the study by Wong et.al., (17) where about 27% had diabetes and 33% had impaired glucose tolerance with the remaining 40% returning to normal glucose tolerance post-partum. Among those with GDM (n = 18), 88% (n = 16) reverted to normal glucose tolerance with 12% (n = 2) having impaired glucose tolerance.
Fig 1. Detailed diagrammatic algorithm of the study with flow chart of NGS screening of the subject.
Table 1. Baseline characteristics of the study subjects.
| PARAMETERS | TOTAL | GDM | OVERT | PREGDM | p-value |
|---|---|---|---|---|---|
| Number of subjects | 50 | 18 | 16 | 16 | 0.102 |
| Age (years)mean ± SD | 29.1 ± 5.1 | 28.2 ± 4.8 | 27.3 ± 4.8 | 31.4 ± 5.1 | 0.905 |
| Age At Diagnosis (years) mean ± SD | 28.3 ± 4.7 | 28.2± 4.8 | 27.7 ± 5.1 | 28.5 ± 6.8 | NA |
| Average Week of detection /Duration (Median & range) | 20 weeks (8 to 32) | 16 Weeks (5 to 32) | #22 months (1 to 60) | ||
| * BMI (kg/m2) | 27.47 ± 3.82 | 26 ± 3.84 | 29.0 ± 2.56 | 26.4 ± 4.46 | 0.218 |
| 18.5–23 | 7 [14%] | 2 (11.1%) | 1(6.2%) | 4(25%) | 0.496 |
| 23.1–24.9 | 11[22%] | 5(27.8%) | 1(6.2%) | 1 (6.2%) | |
| 25–29.9 | 25 [50%] | 7(38.9%) | 10 (62.5%) | 8(50%) | |
| 30–34.9 | 7[14%] | 4 (22.2%) | 4(25%) | 3(18.7%) | |
| 3 generation family history of diabetes | 27 [54%] | 7(11.1%) | 9(56.2%) | 11(68.8%) | 0.213 |
| Number of pregnancies | |||||
| Primigravida | 24 (48%) | 11 (61.1%) | 7 (43.8%) | 6 (37.5%) | |
| Multigravida (≥ G2) | 26 (52%) | 7 (38.9%) | 9 (56.2%) | 10 (62.5%) | |
| Previous pregnancies | |||||
| Previous GDM | 5 (20%) | 2 | 1 | 1 | NS |
| Previous miscarriages | 13 (50%) | 6 | 5 | 2 | |
| Previous congenital anomaly | 1 (3.8%) | 0 | 0 | 1 | |
| Previous LGA | 4 (15.4%) | 1 | 1 | 2 | |
| Previous LSCS | 9 (34.6%) | 4 | 4 | 1 | |
| Glycaemic profile | |||||
| Fasting plasma glucose (mg%)[mean± sd] | 125 ± 35 | 98±13 | 150±24 | 130 ± 38 | <0.0001 |
| 2hour post meal Fasting plasma glucose (mg%) (mean± sd) | 186 ± 58 | 147±23 | 213±51 | 206 ± 70 | <0.003 |
| HbA1c (%) mean± sd | 7.18±1.51 | 5.5±0.5 [n = 13] | 7.4±1.2 [n = 13] | 8.2±1.3 [n = 15] | <0.0001 |
*Calculated based on preconception weight / weight recorded at the visit in first trimester.
# All except for one subject who had diabetes for 16 years.LGA / macrosomia = Birth weight of > 90th percentile (≥ 3.50 kg) in the neonates. BMI: Body Mass Index, N/S: not significant, N/A: not applicable. Multigravida (≥G2): a woman that is or has been pregnant for at least a second time. LSCS:
Next Generation Sequencing based MODY screening
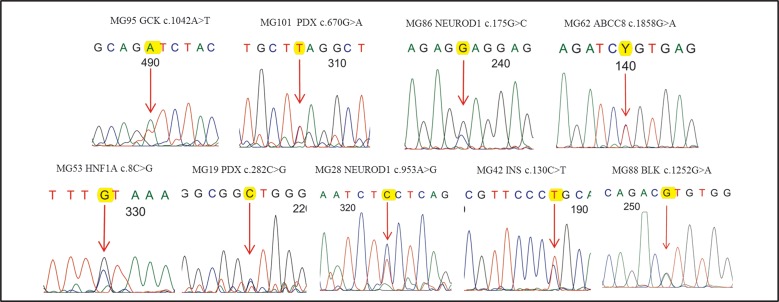
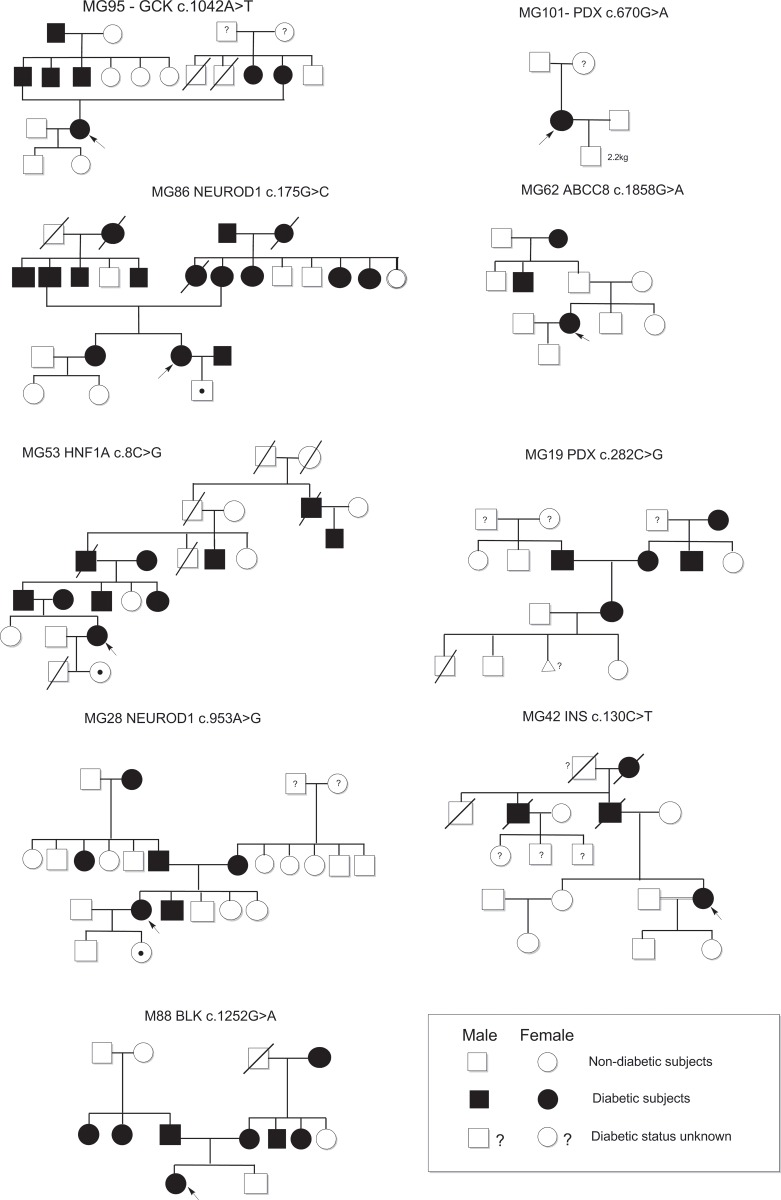
The mean NGS depth was 536X and >95% of the target was sequenced at a minimum depth of 20X covering all the coding regions. In this study, we found that 18% (9/50) of the pregnant women with diabetes carried MODY related variants. Among these subjects, five were with pre-GDM and four had overt GDM.The mean age at diagnosis of MODY positive subjects was 29.6 years. The mean BMI was 27.1 kg/m2 and only two subjects had a BMI of <25kg/m2. Overall, there was no significant difference between the subjects (MODY positive and negative) with regards to their age at diagnosis, BMI and birth weight of the offspring. The details of the identified variants along with the Sanger confirmation are summarized in Table 2 & Fig 2 and the pedigree charts in Fig 3. Three of the nine MODY related variants identified in this study were novel and were not present in 110 control subjects and the 1000 genome and the EXAC database. The reported and the novel variants identified have been classified as per ACMG 2015 guidlines based on in-silico analysis[19] and family studies (in five cases).
Table 2. Mutations identified by targeted next-generation sequencing in pregnant women with diabetes.
| MEPN | AGE | AOD | DOD | GAOD | TYPE | BMI | TP | GENE | Mutation | AA change | FS | SIFT | PP2 | MT | COMMENT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MG95 | 23 | 23 | NA | 21 wks | OVERT GDM | 24 | DIET | GCK | c.1042T>A | Ile348Phe | F | DAM | PD | DC | Likely pathogenic |
| MG101 | 26 | 10 | 16 yrs | NA | PREGDM | 23 | INS | PDX | c.670 G>A | Glu224Lys | M | DAM | PD | DC | Pathogenic |
| MG86 | 36 | 36 | NA | 24 wks | OVERT GDM | 30 | MET+ SU | NEUROD1 | c.175 G>C | Glu59Gln | M, Sis, S | Tol | B | DC | |
| Uncertain significance | |||||||||||||||
| MG 62 | 30 | 24 | 6 yrs | NA | PREGDM | MET | ABCC8 | c.1858G>A | Arg620Cys | DAM | Tol | DC | |||
| Uncertain significance | |||||||||||||||
| MG53 | 25 | 22 | 3 yrs | NA | PREGDM | 30 | INS | HNF1A | c.8C>G | Ser3Cys* | F, D | DAM | PD | DC | Likely pathogenic |
| MG19 | 35 | 35 | NA | 12 wks | OVERT GDM | 27 | MET | PDX | c.282C>G | His94Gln* | Tol | B | DC | Uncertain significance | |
| MG28 | 29 | 27 | 2 yrs | NA | PREGDM | 30 | MET+ SU | NEUROD1 | c.953A>G | Phe318Ser* | F, D | DAM | B | DC | Likely pathogenic |
| MG42 | 35 | 35 | NA | 28 wks | OVERT GDM | 27 | DIET | INS | c.130C>T | Gly44Arg | DAM | PD | DC | ||
| Uncertain significance | |||||||||||||||
| MG 88 | 23 | 23 | 4 wks | NA | PRE GDM | 30 | MET+ INS | BLK | c.1252G>A | Val418Met | DAM | PD | DC | Likely pathogenic |
MEPN–Molecular Endocrinology Pin Number, AOD: Age of diagnosis, DOD: Duration of diabetes, GAOD: Gestational age of Diagnosis, BMI: Body mass index, AA: Amino acid, FS: Family members screened, F:Father, M:Mother, Sis: Sister, S: Son, D: Daughter, *Novel variant, SIFT: Sorting intolerant from tolerant; PolyPhen 2:Polymorphism Phenotyping v2; MT: Mutation taster, DAM: Damaging, Tol: Tolerated, PD: Probably damaging, B: Benign, DC: Disease causing, HNF4A:Hepatocyte Nuclear factor 4 alpha; GCK: Glucokinase; HNF1A:Hepatocyte nuclear factor 1 alpha; PDX1: Pancreatic and duodenal homeobox 1; HNF1B:Hepatocyte nuclear factor 1 beta; NEUROD1, Neurogenic differentiation factor 1; INS, Insulin gene, MET: Metformin, SU: Sulfonylurea.
Fig 2. Sanger confirmation of MODY variants identified in the study.
Fig 3. Detailed pedigree charts of MODY positive pregnant women with diabetes.
Interestingly, in this study, we have identified mutations/rare variants only in patients with pre GDM and overt GDM. In addition, eight (29.6%) of the 27 subjects with an autosomal dominant family history, were positive for MODY related variants, whereas only one (4.3%) of the 23 subjects without a 2/3 generation family history of diabetes carried a MODY variant. Even in our previous study on MODY revealed likely pathogenic MODY variants only in clinically suspected MODY patients and not in young diabetes subjects who do fit into clinical criteria of MODY. Therefore, we believe that few of these variants described in this study even with uncertain significance (variable penetrance or weaker effect) require further evaluation and functional studies to dissect the role of these variants in diabetes.
Genotype Phenotype Correlation
In the present study, a patient with pre-GDM MG53 was positive for a novel p.S3C HNF1A variant. This variant with a paternal inheritance was further transmitted to her child. Her father, who was diagnosed with diabetes at the age of 32 years is currently managed with SU therapy. MG53 during her first pregnancy, had uncontrolled hyperglycemia and delivered a male neonate with transposition of great vessels (TGV) with pulmonary atresia, who expired at the age of 1 month.
We identified MODY4 PDX1 variants in two subjects, MG19 with overt diabetes and MG101 with pre-gestational diabetes. M19 was positive for a novel H94Q PDX1 variant and following delivery, requires SU therapy for glycemic control. Her previous pregnancies were complicated by miscarriage and a low birth weight neonate with an early postnatal death. MG101 was positive for a reported E224K PDX1 pathogenic variant [24] and was diagnosed with diabetes at the age of 10 years with ketoacidosis at onset, negative for autoantibodies and has been on Insulin therapy. She delivered a preterm baby with low birth weight (2240 grams) who was negative for the PDX1 pathogenic variant. Interestingly, her mother who is a carrier of this PDX1 E224K has impaired glucose tolerance.
MG86 with overt GDM and MG28 with pre-GDM were found to positive for a reported E59Q variant and a novel F318S NEUROD1 (MODY-6) variant respectively. MG86 with a BMI of 30kg/m2 was diagnosed to have diabetes at 12 weeks postpartum and is currently on metformin with glipizide. Her child is also a carrier of this variant. She has inherited this variant from her mother who was diagnosed to have diabetes at the age of 40 years and had delivered a macrosomic baby with a birth weight of 4000 grams in 1974 at the age of 21 years. MG28, with a novel F318S NEUROD1 likely pathogenic variant is obese with a BMI of 30kg/m2and has a paternal inheritance and her first offspring is a carrier with low birth weight.
MG42 with overt GDM was positive for a novel G44R INS gene variant. Postpartum she has impaired glucose tolerance and has been managed with dietary modification alone. Both her parents, her grandfather and paternal uncle who had diabetes, have expired. Her children are negative for this variant.
MG88 had pre GDM and was identified to have a novel BLK likely pathogenic variant. Postpartum she had diabetes and is managed with metformin. She is obese with a BMI of 30kg/m2, and similar reports of patients with BLK mutation and obesity have been published by Kim et.al [25]
MG95 was positive for a reported GCK Ile348Phe mutation and was diagnosed with overt diabetes when screened at 21 weeks of gestation with a HbA1c of 7%. She had GDM in her previous pregnancy and was managed with Insulin. She presented with recurrent gestational diabetes with postpartum normal blood glucose. She had inherited this variant from her father who was diagnosed at the age of 40 years and is currently being managed with SU therapy. The clinical phenotype in carriers of the GCK variant in the present family has been heterogeneous without the characteristic fasting hyperglycemia as seen in other patients with GCK gene mutation[26].
MG62, a 30 year old woman with pre-GDM was positive for Arg620Cys ABCC8 variant[27] which was previously reported in congenital hyperinsulinism. She was diagnosed with diabetes six years prior to conception and has been managed with metformin and SU therapy. This subject with an ABCC8 variant has early onset of diabetes without a family history of diabetes or hypoglycemia.
Discussion
Utilizing the NGS based 2GDMODY protocol[12] we have screened a comprehensive panel of thirteen MODY genes in 50 pregnant women with diabetes and identified MODY related variants in 9(18%) subjects. These variants were detected in subjects with overt GDM and pre-GDM (9/32) and the women with GDM were negative for novel or reported pathogenic variants (0/18). The word “overt GDM” has been used for the diagnosis of “overt diabetes in pregnancy” based on the guidelines released by the Association of Diabetes and Pregnancy Study Groups (IADPSG) consensus panel in 2010[16] and The Endocrine Society guidelines on diabetes and pregnancy in 2013[28]. The reason for earlier detection of the disorder during pregnancy at our centre is a result of robust screening done at the first visit since the IADPSG guidelines are followed. These women would have been labelled as GDM if screening had been carried out based on the previous criteria at 24–28 weeks[16]. As previously defined, GDM also included a subgroup with more severe hyperglycemia that presented with special issues concerning management during pregnancy and postpartum follow-up.
Women who meet the criteria for diabetes are now classified as a separate category named “overt diabetes in pregnancy” by the IADPSG criteria and represent the "highest at risk" GDM cohort, who have an increased risk of congenital abnormalities and diabetes related complications. The future risk of these women for developing diabetes appears unclear at present[29]. A retrospective study of 254 women meeting the criteria for overt diabetes has demonstrated that 41% had a normal glucose tolerance at 6–8 weeks postpartum and hence all women classified as “overt diabetes in pregnancy” cannot be considered as having pre-existing diabetes prior to conception[17]. It would indeed be useful to look at this group carefully in future studies on hyperglycemic disorders in pregnancy. A further prospective follow-up comparing women meeting both sets of the IADPSG criteria would therefore be useful in further refining the risk in these patients.
In GDM patients, MODY related variants were present seven times higher (29.6% compared to 4.3%, OR = 9.2, 95% CI 1.06–80.93, P = 0.027) in those with a three generation family history of diabetes. This is similar to earlier reports, wherein the selective screening for pathogenic variants in subjects with the typical phenotype with the presence of a family history of diabetes has shown a prevalence which ranges from 12–80% [10]. Recent study by Flannick et.al., showed that among the general population only 32% of of the subject’s positive for HGMD MODY variants and 31% with likely pathogenic variants had IFG or diabetes[30]. However, when these individuals preselected based on the phenotype, carried an excess low frequency nonsynonymous variants (4.7% compared to 1.5%, OR = 3.2, P = 0.011) and an apparent excess of likely pathogenic or HGMD MODY variants in obese old subjects with diabetes when compared to young lean diabetes subjects[30]. These results suggest that the classification of the identified MODY variants needs to based on the phenotype and require further family screening before filtering these variants based on Exac data sets which does contain individual with disease.
NEUROD1 variants (MODY 6) have been reported from only 10 families [12,31–35]. However, in contrast to this study, the Norwegian study [31] did not find NEUROD1 as a candidate gene in GDM. Although the clinical phenotype of some PDX1 and NEUROD1 variants remains unclear, increased body weight has been reported in earlier studies in subjects with NEUROD1 mutation [32–34]. Indeed, the mean BMI of the subjects who were NEUROD1 positive (30kg/m2) in our study was higher than those with other MODY related variants (26.1kg/m2), which signifies the additional role of diabetogenic factors in MODY. Twelve weeks postpartum, in these subjects with NEUROD1 variants and obesity, we performed an oral glucose tolerance test; both showed a significant reduction in insulin release at 1 and 2 hours when compared with T2D and control subjects (S1 Fig).
MG101 with pre-GDM was positive for a reported E224K PDX1 mutation which was shown to result in reduced tranactivation and was also found to co-segregating with early-onset diabetes or impaired glucose tolerance in an Indo-Trinidadian family[24]. Further, the E224K, was shown to disrupt the ability of nuclear factor PCIF1 (PDX C-terminus Interacting Factor-1) to inhibit PDX1 transactivation, suggesting that the interaction between PDX1 and PCIF1are required for normal glucose homeostasis[36]. However, with 190 allele carreiers in 1000 genome and Exac database[18] could mislead the interpretation of this variant as benign. But with the evidence from functional studies[24] and with recent findings of digenic mutations NEUROD1 –PDX1[12] included in annotated digenic diseases database (DIDA) identifiers: dd220[37,38] suggest that this variant affects the beta cell functioning and may requires additional genetic or environmental factors to express the disease phenotype.
Subject MG19 with overt-GDM who was positive for a novel H94Q PDX1 variant had persistently elevated glucose levels in the diabetic range post-partum and is currently being managed with metformin and SU therapy. Further, PDX1 variant carriers MG101 & MG19 had delivered babies with low birth weight. Gragnoli C et.al., in their study found that four out of five pregnant women who were carriers of the PDX1 pathogenic variant(P33T)[39], had their pregnancies complicated by reduced birth weights, miscarriages, or early postnatal deaths, similar to that noticed in our subjects MG19 and MG101. These findings suggest that the PDX1 pathogenic variants may provide an increased susceptibility to the occurrence of low birth weight in the offspring of pregnant women with MODY 4 [39]. Therefore, it is important to note the PDX1 variants in pregnant women with diabetes which may predispose these subjects and play a role during gestation and clinical outcomes.
Insulin gene (INS) mutations (MODY 10) cause permanent neonatal diabetes (PNDM) and are a very occasional cause of diabetes diagnosed in childhood or adulthood. In the present study, MG42 was positive for a novel G44R INS gene variant. She was diagnosed to have overt diabetes in her second pregnancy. In a study by Boesgaard et.al., a novel heterozygous insulin gene variant c.17G>A, R6H was identified in a 27 year old woman with GDM and up to a 30% reduction in beta-cell function measured by an insulinogenic index was noted in the family members(37). The family members with diabetes in this study [40] with insulin gene variants c.17G>A, R6H were not insulin-dependent, as was observed in MG42 in this study.
MG88, a 23 year old woman with a novel BLK variant (MODY-11) was obese with a BMI of 30kg/m2 similar to earlier reports by Kim et.al., who had noted that there was a higher prevalence of obesity in individuals with diabetes linked to the BLK gene than in diabetic individuals with MODY linked to other loci[25]. Borowiecet et.al., also suggested that the diabetogenic environment conferred by an increased body weight might be necessary for translation of the beta-cell abnormalities caused by the BLK gene variant into diabetes[41].
Further, we have performed Sanger sequencing for all the available first degree relatives of six of the nine women who were positive for MODY related variants and who provided consent for family screening. All the parents and siblings who screened negative for the identified pathogenic variants did not have diabetes. All parents who were positive for the identified variants had diabetes, except the mother of MG101 with a E224K PDX1 pathogenic variant had impaired glucose tolerance. Further, three of the seven (45%) neonates who were screened were found to carry the identified variant (GCK,HNF1A,NEUROD1). However, the offspring would be labelled as carriers at present and would need long-term follow up. Postpartum, three of the four women with pre-GDM responded to sulphonylureas (SU) whereas one continued on insulin(PDX1); additionally, of the five MODY subjects with overt diabetes in pregnancy who underwent a follow-up oral glucose tolerance test, two of these subjects with PDX1, NEUROD1 progressed to develop diabetes, two subjects with INS, PDX1 showed impaired glucose tolerance, one with a GCK gene variant at 12 weeks postpartum reverted to normal glucose tolerance.
Contemporary literature has reported on MODY pathogenic variants in GDM, suggesting that GCK pathogenic variants are the commonest, followed by HNF1A, HNF4A and PDX1 genes[10]. To date, there were no reports on NEUROD1, INS, BLK and ABCC8 variants in pregnant women with diabetes. This may be attributable either to the lower prevalence of pathogenic variants in the screened population or due to the sequential screening by the conventional Sanger sequencing methodology, which is limited by cost and scalability. Interestingly, in this study population, utilizing the NGS we have identified MODY variants in NEUROD1, PDX1 and BLK genes in comparison to the more commonly reported HNF4A, GCK and HNF1A gene variants. The variable pattern of identified variants in our population in comparison to earlier reports may be attributed to the differential prevalence of various MODY pathogenic variants across the world [12,42]. To the best of our knowledge, this is the first NGS based comprehensive screening of 13 MODY genes in pregnant women with diabetes with the first report of NEUROD1, INS, BLK and ABCC8 gene variants in pregnant women with diabetes from India.
Phenotypic heterogeneity in MODY is a well-known fact in individual members within a family who share the pathogenic variants has been documented in recent studies[12,26,43]. Furthermore, some of the variants like heterozygous E1506K pathogenic variant in the ABCC8 gene resulting in congenital hyperinsulinism in infancy, loss of insulin secretory capacity in early adulthood, and autosomal dominant diabetes in middle age in different members[44]. Therefore, parallel NGS approach will help in identifying additional variants in other genes which may contribute to the heterogeneity seen in MODY patients. The novel variants identified in this study were absent in 110 control subjects and the 1000 genome, EXAC database and were predicted to be causative by at least one of the bioinformatics tool. However the absence of these variants in a large number of exomes and variation in the conserved region do not confer pathogenicity, but suggest the need to investigate further to identify the role of these novel variants in diabetes. Indeed, in the absence of detailed familial co-segregation and functional studies, we acknowledge the difficulties and limitations in interpreting the sequencing data in categorizing the novel MODY variants. Further, with thousands of exome sequencing data from the Exac database which includes rare variants causing late onset disorders[18] poses a challenge to classify these variants in conditions such as GDM[45]. However, interpretation of data from NGS based parallel multigene testing would become clinically significant when the genomics data from patients is linked with detailed phenotype and this publication would help in such an effort in MODY and GDM.
Conclusions
Comprehensive NGS based parallelized multi-gene screening revealed MODY mutations or rare variants only in patients with pre-GDM and overt GDM with an autosomal dominant family history. Therefore, this subset of patients could benefit through MODY gene screening, confirmed diagnosis and appropriate therapy in few cases.
Utilizing the ExAC database resource in variant filtering and classification in young onset or reproductive age onset diseases such as GDM requires caution and need to based on the functional studies, phenotype, and family screening, to confirm the role of these variants in MODY.
We also believe that further family studies and long-term follow up of the offspring would provide an opportunity to understand the heterogeneity, penetrance and pathogenicity of MODY related variants and their response to sulphonylurea treatment. This may also help in deciphering potential implications for treatment during pregnancy and the role of specific pathogenic variants in the occurrence of comorbidities like macrosomia, preterm deliveries, low birth weight, pregnancy loss and neonatal complications. Further, this would enable early detection of glucose intolerance, intervention and appropriate therapy for patients and their families.
Supporting Information
(DOCX)
Acknowledgments
We thank Dr Nishanth Arulappan, Dr Shweta Nadig, Ms.Banu, Mr Kali and Miss Amulya Ruby for their contribution to various aspects of the study.
Data Availability
We have submitted the NGS data to the European Nucleotide Archive and the details are given below. Study accession number is: PRJEB10688. Study unique name is: ena-STUDY-CHRISTIAN MEDICAL COLLEGE-07-09-2015-09:00:08:518-122. Study title is: MODY GENE SCREENING IN PREGNANT WOMEN WITH DIABETES. Sample accession Secondary accession Sample unique name ERS845523 SAMEA3538374 GM53 ERS845524 SAMEA3538375 GM42 ERS845525 SAMEA3538376 GM101 ERS845526 SAMEA3538377 GM86 ERS845527 SAMEA3538378 GM95 ERS845528 SAMEA3538379 GM95b ERS845529 SAMEA3538380 GM19 ERS845530 SAMEA3538381 GM28 ERS845531 SAMEA3538382 GM62 ERS845532 SAMEA3538383 GM88 All other relevant data is included within the paper.
Funding Statement
Funded by Ministry of Science and Technology, Department of Biotechnology, Government of India. NO.BT/PR6461/MED/12/576/2012.
References
- 1.Chan JCN, Malik V, Jia W, Kadowaki T, Yajnik CS, Yoon K- H, et al. Diabetes in Asia: epidemiology, risk factors, and pathophysiology. JAMA. 2009;301: 2129–2140. 10.1001/jama.2009.726 [DOI] [PubMed] [Google Scholar]
- 2.Seshiah V, Balaji V, Balaji MS, Paneerselvam A, Kapur A. Pregnancy and diabetes scenario around the world: India. Int J Gynaecol Obstet Off Organ Int Fed Gynaecol Obstet. 2009;104 Suppl 1: S35–38. [DOI] [PubMed] [Google Scholar]
- 3.Seshiah V, Balaji V, Balaji MS, Paneerselvam A, Arthi T, Thamizharasi M, et al. Prevalence of gestational diabetes mellitus in South India (Tamil Nadu)—a community based study. J Assoc Physicians India. 2008;56: 329–333. [PubMed] [Google Scholar]
- 4.Kim C, Berger DK, Chamany S. Recurrence of gestational diabetes mellitus: a systematic review. Diabetes Care. 2007;30: 1314–1319. 10.2337/dc06-2517 [DOI] [PubMed] [Google Scholar]
- 5.Weng J, Ekelund M, Lehto M, Li H, Ekberg G, Frid A, et al. Screening for MODY mutations, GAD antibodies, and type 1 diabetes—associated HLA genotypes in women with gestational diabetes mellitus. Diabetes Care. 2002;25: 68–71. [DOI] [PubMed] [Google Scholar]
- 6.Molven A, Njølstad PR. Role of molecular genetics in transforming diagnosis of diabetes mellitus. Expert Rev Mol Diagn. 2011;11: 313–320. 10.1586/erm.10.123 [DOI] [PubMed] [Google Scholar]
- 7.Shepherd M, Shields B, Ellard S, Rubio-Cabezas O, Hattersley AT. A genetic diagnosis of HNF1A diabetes alters treatment and improves glycaemic control in the majority of insulin-treated patients. Diabet Med J Br Diabet Assoc. 2009;26: 437–441. [DOI] [PubMed] [Google Scholar]
- 8.Vaxillaire M, Froguel P. Monogenic diabetes in the young, pharmacogenetics and relevance to multifactorial forms of type 2 diabetes. Endocr Rev. 2008;29: 254–264. 10.1210/er.2007-0024 [DOI] [PubMed] [Google Scholar]
- 9.Kavvoura FK, Owen KR. Maturity onset diabetes of the young: clinical characteristics, diagnosis and management. Pediatr Endocrinol Rev PER. 2012;10: 234–242. [PubMed] [Google Scholar]
- 10.Colom C, Corcoy R. Maturity onset diabetes of the young and pregnancy. Best Pract Res Clin Endocrinol Metab. 2010;24: 605–615. 10.1016/j.beem.2010.05.008 [DOI] [PubMed] [Google Scholar]
- 11.Zhang W, Cui H, Wong L-JC. Application of next generation sequencing to molecular diagnosis of inherited diseases. Top Curr Chem. 2014;336: 19–45. 10.1007/128_2012_325 [DOI] [PubMed] [Google Scholar]
- 12.Chapla A, Mruthyunjaya MD, Asha HS, Varghese D, Varshney M, Vasan SK, et al. Maturity onset diabetes of the young in India—a distinctive mutation pattern identified through targeted next-generation sequencing. Clin Endocrinol (Oxf). 2015;82: 533–542. [DOI] [PubMed] [Google Scholar]
- 13.Colclough K, Saint-Martin C, Timsit J, Ellard S, Bellanné-Chantelot C. Clinical utility gene card for: Maturity-onset diabetes of the young. Eur J Hum Genet. 2014;22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thanabalasingham G, Owen KR. Diagnosis and management of maturity onset diabetes of the young (MODY). BMJ. 2011;343: d6044 10.1136/bmj.d6044 [DOI] [PubMed] [Google Scholar]
- 15.Owen KR. Monogenic diabetes: old and new approaches to diagnosis. Clin Med Lond Engl. 2013;13: 278–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metzger BE, Coustan DR. Summary and recommendations of the Fourth International Workshop-Conference on Gestational Diabetes Mellitus. The Organizing Committee. Diabetes Care. 1998;21 Suppl 2: B161–167. [PubMed] [Google Scholar]
- 17.Wong T, Ross GP, Jalaludin BB, Flack JR. The clinical significance of overt diabetes in pregnancy. Diabet Med J Br Diabet Assoc. 2013;30: 468–474. [DOI] [PubMed] [Google Scholar]
- 18.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536: 285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics…—PubMed—NCBI [Internet]. [cited 11 Oct 2016]. Available: https://www.ncbi.nlm.nih.gov/pubmed/25741868 [DOI] [PMC free article] [PubMed]
- 20.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7: 248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11: 863–874. 10.1101/gr.176601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7: 575–576. 10.1038/nmeth0810-575 [DOI] [PubMed] [Google Scholar]
- 23.Cooper GM, Goode DL, Ng SB, Sidow A, Bamshad MJ, Shendure J, et al. Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nat Methods. 2010;7: 250–251. 10.1038/nmeth0410-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cockburn BN, Bermano G, Boodram L-LG, Teelucksingh S, Tsuchiya T, Mahabir D, et al. Insulin promoter factor-1 mutations and diabetes in Trinidad: identification of a novel diabetes-associated mutation (E224K) in an Indo-Trinidadian family. J Clin Endocrinol Metab. 2004;89: 971–978. 10.1210/jc.2003-031282 [DOI] [PubMed] [Google Scholar]
- 25.Kim S- H, Ma X, Weremowicz S, Ercolino T, Powers C, Mlynarski W, et al. Identification of a locus for maturity-onset diabetes of the young on chromosome 8p23. Diabetes. 2004;53: 1375–1384. [DOI] [PubMed] [Google Scholar]
- 26.Cuesta-Muñoz AL, Tuomi T, Cobo-Vuilleumier N, Koskela H, Odili S, Stride A, et al. Clinical heterogeneity in monogenic diabetes caused by mutations in the glucokinase gene (GCK-MODY). Diabetes Care. 2010;33: 290–292. 10.2337/dc09-0681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James C, Kapoor RR, Ismail D, Hussain K. The genetic basis of congenital hyperinsulinism. J Med Genet. 2009;46: 289–299. 10.1136/jmg.2008.064337 [DOI] [PubMed] [Google Scholar]
- 28.Blumer I, Hadar E, Hadden DR, Jovanovič L, Mestman JH, Murad MH, et al. Diabetes and pregnancy: an endocrine society clinical practice guideline. J Clin Endocrinol Metab. 2013;98: 4227–4249. 10.1210/jc.2013-2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noctor E, Dunne FP. Type 2 diabetes after gestational diabetes: The influence of changing diagnostic criteria. World J Diabetes. 2015;6: 234–244. 10.4239/wjd.v6.i2.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flannick J, Beer NL, Bick AG, Agarwala V, Molnes J, Gupta N, et al. Assessing the phenotypic effects in the general population of rare variants in genes for a dominant Mendelian form of diabetes. Nat Genet. 2013;45: 1380–1385. 10.1038/ng.2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malecki MT, Jhala US, Antonellis A, Fields L, Doria A, Orban T, et al. Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nat Genet. 1999;23: 323–328. 10.1038/15500 [DOI] [PubMed] [Google Scholar]
- 32.Kristinsson SY, Thorolfsdottir ET, Talseth B, Steingrimsson E, Thorsson AV, Helgason T, et al. MODY in Iceland is associated with mutations in HNF-1alpha and a novel mutation in NeuroD1. Diabetologia. 2001;44: 2098–2103. 10.1007/s001250100016 [DOI] [PubMed] [Google Scholar]
- 33.Liu L, Furuta H, Minami A, Zheng T, Jia W, Nanjo K, et al. A novel mutation, Ser159Pro in the NeuroD1/BETA2 gene contributes to the development of diabetes in a Chinese potential MODY family. Mol Cell Biochem. 2007;303: 115–120. 10.1007/s11010-007-9463-0 [DOI] [PubMed] [Google Scholar]
- 34.Gonsorcíková L, Průhová S, Cinek O, Ek J, Pelikánová T, Jørgensen T, et al. Autosomal inheritance of diabetes in two families characterized by obesity and a novel H241Q mutation in NEUROD1. Pediatr Diabetes. 2008;9: 367–372. 10.1111/j.1399-5448.2008.00379.x [DOI] [PubMed] [Google Scholar]
- 35.Sagen JV, Baumann ME, Salvesen HB, Molven A, Søvik O, Njølstad PR. Diagnostic screening of NEUROD1 (MODY6) in subjects with MODY or gestational diabetes mellitus. Diabet Med J Br Diabet Assoc. 2005;22: 1012–1015. [DOI] [PubMed] [Google Scholar]
- 36.Liu A, Oliver-Krasinski J, Stoffers DA. Two conserved domains in PCIF1 mediate interaction with pancreatic transcription factor PDX-1. FEBS Lett. 2006;580: 6701–6706. 10.1016/j.febslet.2006.11.021 [DOI] [PubMed] [Google Scholar]
- 37.Gazzo AM, Daneels D, Cilia E, Bonduelle M, Abramowicz M, Van Dooren S, et al. DIDA: A curated and annotated digenic diseases database. Nucleic Acids Res. 2016;44: D900–D907. 10.1093/nar/gkv1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DETAIL | DIDA [Internet]. [cited 12 Oct 2016]. Available: http://dida.ibsquare.be/detail/?dc=dd220
- 39.Gragnoli C, Stanojevic V, Gorini A, Von Preussenthal GM, Thomas MK, Habener JF. IPF-1/MODY4 gene missense mutation in an Italian family with type 2 and gestational diabetes. Metabolism. 2005;54: 983–988. 10.1016/j.metabol.2005.01.037 [DOI] [PubMed] [Google Scholar]
- 40.Boesgaard TW, Pruhova S, Andersson EA, Cinek O, Obermannova B, Lauenborg J, et al. Further evidence that mutations in INS can be a rare cause of Maturity-Onset Diabetes of the Young (MODY). BMC Med Genet. 2010;11: 42 10.1186/1471-2350-11-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borowiec M, Liew CW, Thompson R, Boonyasrisawat W, Hu J, Mlynarski WM, et al. Mutations at the BLK locus linked to maturity onset diabetes of the young and beta-cell dysfunction. Proc Natl Acad Sci U S A. 2009;106: 14460–14465. 10.1073/pnas.0906474106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woodhouse NJ, Elshafie OT, Al-Mamari AS, Mohammed NH, Al-Riyami F, Raeburn S. Clinically-Defined Maturity Onset Diabetes of the Young in Omanis: Absence of the common Caucasian gene mutations. Sultan Qaboos Univ Med J. 2010;10: 80–83. [PMC free article] [PubMed] [Google Scholar]
- 43.Fajans SS, Bell GI. MODY: history, genetics, pathophysiology, and clinical decision making. Diabetes Care. 2011;34: 1878–1884. 10.2337/dc11-0035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huopio H, Otonkoski T, Vauhkonen I, Reimann F, Ashcroft FM, Laakso M. A new subtype of autosomal dominant diabetes attributable to a mutation in the gene for sulfonylurea receptor 1. Lancet Lond Engl. 2003;361: 301–307. [DOI] [PubMed] [Google Scholar]
- 45.Pulst SM. A lot of nexts. Neurol Genet. 2015;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
We have submitted the NGS data to the European Nucleotide Archive and the details are given below. Study accession number is: PRJEB10688. Study unique name is: ena-STUDY-CHRISTIAN MEDICAL COLLEGE-07-09-2015-09:00:08:518-122. Study title is: MODY GENE SCREENING IN PREGNANT WOMEN WITH DIABETES. Sample accession Secondary accession Sample unique name ERS845523 SAMEA3538374 GM53 ERS845524 SAMEA3538375 GM42 ERS845525 SAMEA3538376 GM101 ERS845526 SAMEA3538377 GM86 ERS845527 SAMEA3538378 GM95 ERS845528 SAMEA3538379 GM95b ERS845529 SAMEA3538380 GM19 ERS845530 SAMEA3538381 GM28 ERS845531 SAMEA3538382 GM62 ERS845532 SAMEA3538383 GM88 All other relevant data is included within the paper.