SUMMARY
Fibrin is a filamentous network made in blood to stem bleeding; it forms when fibrinogen is converted into fibrin monomers that self-associate into oligomers and then to polymers. To gather structural insights into fibrin formation and properties, we combined high-resolution atomic force microscopy of fibrin(ogen) oligomers and molecular modeling of crystal structures of fibrin(ogen) and its fragments. We provided a structural basis for the intermolecular flexibility of single-stranded fibrin(ogen) oligomers and identified a hinge region at the D:D inter-monomer junction. Following computational reconstruction of the missing portions, we recreated the full-atomic structure of double-stranded fibrin oligomers that was validated by quantitative comparison with the experimental images. We characterized previously unknown intermolecular binding contacts at the D:D and D:E:D interfaces, which drive oligomerization and reinforce the intra- and inter-strand connections in fibrin besides the known knob-hole bonds. The atomic models provide valuable insights into the submolecular mechanisms of fibrin polymerization.
Graphical abstract
INTRODUCTION
Fibrin is an end product of blood clotting and forms the scaffold of hemostatic clots and obstructive thrombi in blood vessels. The molecular mechanisms of fibrin formation, its structure, and its properties are of fundamental biological importance. These are also related to clinical medicine and life-threatening pathologies, such as bleeding and thrombosis, including ischemic stroke and heart attack. Fibrin is produced from fibrinogen, a plasma protein, made of six paired polypeptide chains (Aα Bβ γ)2 held together by S-S-bonds (Figure 1). Fibrin formation is initiated by the cleavage of fibrinopeptides A and B from the N termini of the Aα and Bβ chains of fibrinogen, respectively, to produce fibrin monomer (α β γ)2. The release of fibrinopeptide A exposes an N-terminal α chain motif GPR called knob ‘A’ that binds to exposed holes ‘a’ in the globular γ-nodules located in another fibrin molecule (Everse et al., 1998; Kostelansky et al., 2002), resulting in A-a interactions (Laudano and Doolittle, 1978; Litvinov et al., 2005). Exposure of knobs ‘A’ is necessary and sufficient to form fibrin through the interaction with holes ‘a’. The release of fibrinopeptides B exposes an N-terminal β chain motif GHR called knob ‘B’, which is complementary to holes ‘b’ located in the globular β-nodules of another fibrin molecule.
Figure 1. Structure of Monomeric Fibrin.
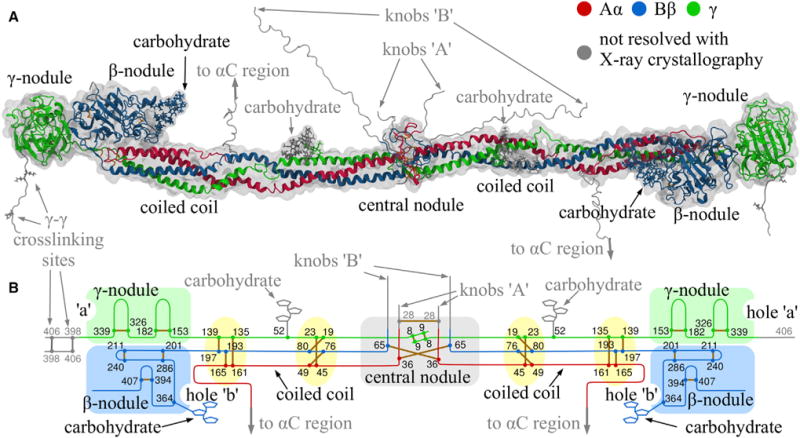
(A) Crystal structure of the resolved parts of human fibrinogen (PDB: 3GHG) within its hydrodynamic volume.
(B) Schematic of fibrin(ogen) structure reinforced by 29 disulfide bonds (yellow), with the globular parts highlighted in light green (γ-nodule), light blue (β-nodule), and light gray (central nodule).
Fibrin polymerization is initiated when two fibrin molecules interact in a half-staggered fashion via the A-a knob-hole bonds (Weisel and Litvinov, 2013). Addition of a third molecule results in an end-to-end association where the lateral D regions of two molecules form the D:D interface (Figure 2). This comprises the monomer junction in each of two strands in fibrin oligomers (Everse et al., 1998). The D:D interface, which contains residues γ275–309 (Everse et al., 1998) is weak, yielding first to a pulling force upon stretching of fibrin(ogen) oligomers (Zhmurov et al., 2011, 2012). Studies of point mutations revealed that residues γ275, γ308, and γ309 are essential for D-D interactions and elongation of fibrin strands (Hirota-Kawadobora et al., 2004; Marchi et al., 2006; Bowley and Lord, 2009). Fibrin monomers can add longitudinally to form protofibrils, two-stranded oligomers that reach a certain length to begin lateral aggregation (Hantgan et al., 1980; Fowler et al., 1981; Medved et al., 1990). Side-to-side interactions between two strands are mediated by the central E region of one fibrin molecule and two lateral D regions of two other molecules (Figures 3 and 4). The D-E-D complex is held together mainly by the A-a knob-hole bonds and, possibly, by additional interactions at the D:E and D:D interfaces (Figure 5; Kononova et al., 2013). Protofibrils self-associate to form twisted fibers of variable thickness (Medved et al., 1990) with a regular 22.5 nm repeat corresponding to half the monomer length (Weisel, 2005). Fibrin fibers branch to form a 3D fibrin network (Weisel, 2005). Therefore, formation of the end-to-end intra-strand D:D interfaces and lateral inter-strand D:E:D interfaces are critical to fibrin polymerization.
Figure 2. Structure of the Single-Stranded Fibrinogen Oligomers.
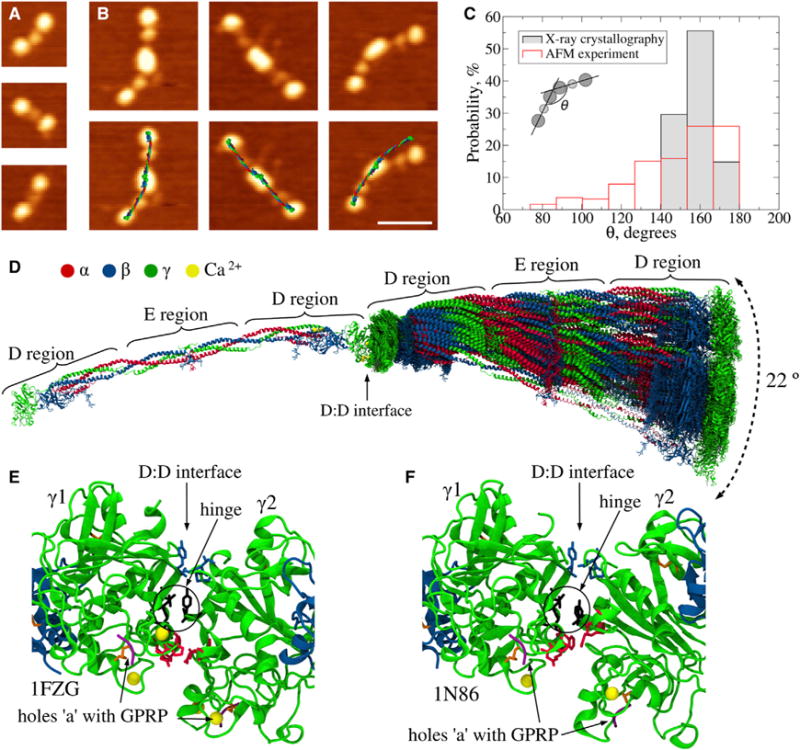
(A) AFM images of fibrinogen molecules. Three distinct globular regions are visualized; the lateral D regions are larger than the central E region.
(B) AFM images of crosslinked fibrinogen dimers Fg2 (top) and same images with computationally reconstructed dimers superimposed (bottom). The superposition of reconstructed structures over AFM images was made by computational alignment of the centers of mass of D and E regions of in silico structures, with the geometrical centers of D and E regions determined from AFM. The scale bar represents 50 nm.
(C) Histograms of the bending angles formed by adjacent fibrinogen monomers in single-stranded fibrinogen oligomers from AFM images (n = 240) and from in silico structures (n = 27; Table S1). The inset illustrates how the bending angle (θ) is defined.
(D) Distribution of positions of the second (right) monomer relative to the first (left) monomer from 27 D-D structures in Table S1 (see Data S1). There is a 22° bending angle that quantifies variations in the relative orientation of monomers.
(E and F) Detailed view of two representative D:D interfaces from the PDB structures: bent arrangement (PDB: 1FZG) and linear arrangement (PDB: 1N86). The side chains forming stable interfacial contacts are shown with sticks: residues γ1Ala279-Tyr280 and γ2Asn308-Gly309 form a hinge (black); residues γ1Ala271-Asp272 and γ2Pro299-Ser300 favor the bent conformation (red); residues γ1Ala241-Pro243 and γ2Ala279-Tyr280 form contacts stabilizing the linear arrangement (blue). See also Figure S1 for a comparison of D-D interfacial contacts found in naturally occurring D-dimers (A) with those reported as crystal contacts (B) and for the results of the normal mode analysis (D and E); Figure S5A for SDS-PAGE of single-stranded oligomers; and Data S1.
Figure 3. Step-by-Step Reconstruction and Structure of Double-Stranded Fibrin Oligomer FO2/3.
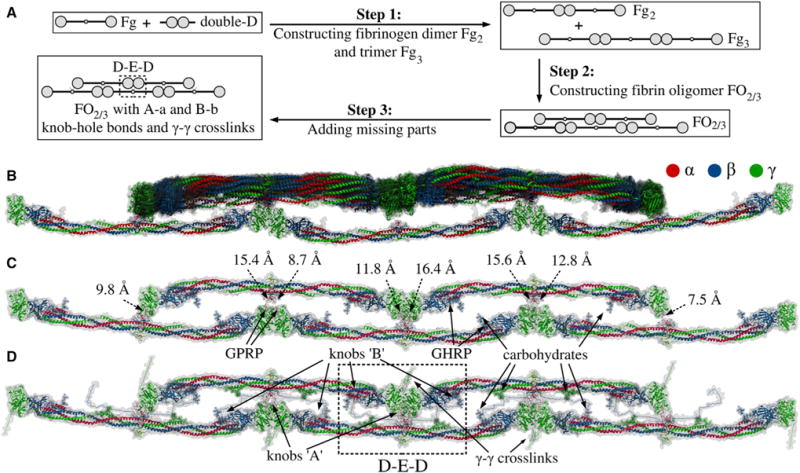
(A) Steps 1–3 summarize the procedure used to reconstruct double-stranded fibrin oligomer FO2/3 (see Experimental Procedures). As the input, we use the atomic structural models of human fibrinogen (PDB: 3GHG; Kollman et al., 2009) and double-D fragment (PDB: 1N86; Yang et al., 2002). These structures are utilized in step 1 to create fibrinogen dimer Fg2 and trimer Fg3, which are used in step 2 to construct a double-stranded fibrin oligomer FO2/3 with two/three monomers in the upper/lower strand. The initial model of FO2/3 does not contain knobs ‘A’ and ‘B’ and the γ-γ crosslinks, which are incorporated in step 3. To remove steric clashes and to minimize the potential energy of the obtained conformation of FO2/3, this step is followed by energy minimization. See also Figure S2 for geometric constraints used in Monte Carlo docking.
(B) Superposition of all 14 successfully reconstructed structures from the Monte Carlo docking (see Supplemental Information).
(C and D) Representative structures of FO2/3 before and after the incorporation of missing residues: knobs ‘A’ and ‘B’ and g-g crosslinks (see Data S2). (C) also shows the distances for all eight constraints due to A-a knob-hole bonds. See also Figure S2 and Data S2.
Figure 4. AFM Images and In Silico Structures of Two-Stranded Fibrin Oligomers.
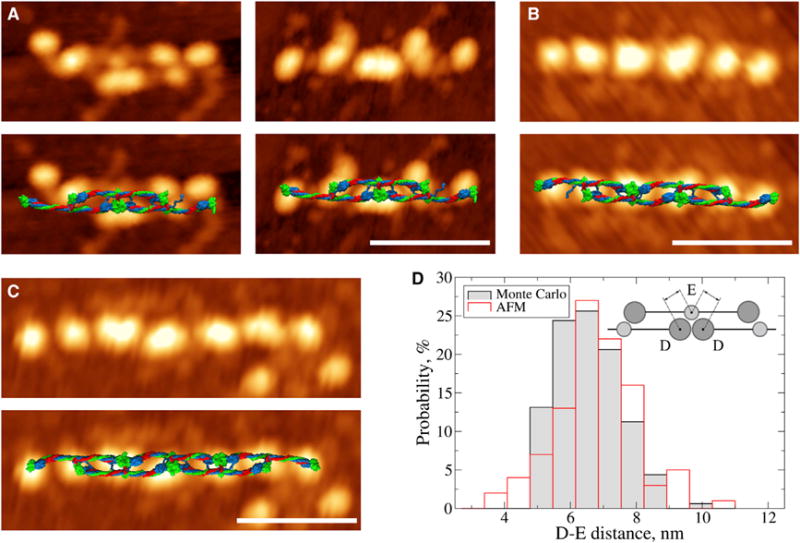
(A–C) AFM images of fibrin oligomers: trimer (A), tetramer (B), and pentamer (C). The bottom row displays AFM images with in silico structures superimposed. Due to the larger flexibility of shorter oligomers, the superposition is better for the longer structures (B and C) compared with the shorter structures (A). The scale bars represent 50 nm.
(D) Histograms of the D-E distances from AFM image analysis and in silico structures obtained in step 2 (see Figure 3). The inset in (D) shows a schematic illustrating how the distance between nodules of the D and E regions was measured. See also Figure S3 for additional AFM images.
Figure 5. Structure of the D-E-D Complex.
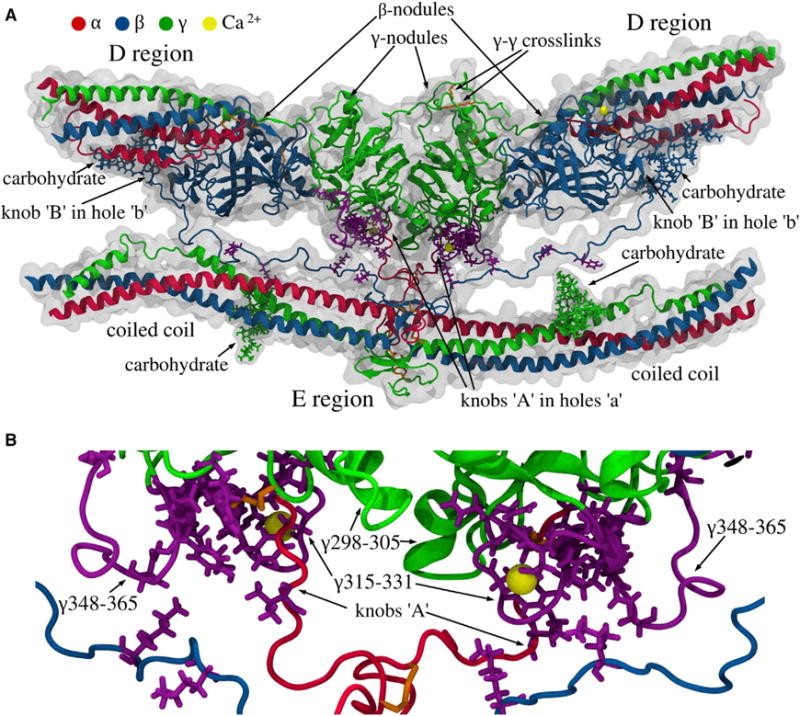
(A) The D regions of two adjacent molecules in the upper strand with the γ- and the β-nodules, and the E region of the third fibrin molecule in the lower strand with parts of the coiled coils. Also shown are the carbohydrate chains, two pairs of knobs ‘A’ and ‘B’ in the E region of the lower molecule bound, respectively, to holes ‘a’ (in the γ-nodules) and ‘b’ (in the β-nodules) of the upper molecules, the γ-γ crosslinks, and calcium ions (see Data S3).
(B) Magnified view of the D-E-D complex with the residue contacts stabilizing the D:E interfaces. The purple side chains mark the single-point mutations in fibrinogen variants (see Table S2). The loops γTyr348-Asn365 and γTrp315-Gly331 in the γ-nodules, where most of the natural mutations occur, are shown in purple. The figure is based on MD simulations with the SASA implicit solvent model (see Supplemental Information). MD simulations of the γ-E-γ fragment in explicit solvent (see Supplemental Information) show similar results, with identical residue-residue contacts between amino acids in the γ-nodules and E region. See also Figure S4 for details of the B-b knob-hole interactions; and Data S3.
Although the X-ray crystallographic studies of the fibrin(ogen) molecule and fibrin fragments provide valuable information about the structure of the D-D complex and a major part of the D-E-D complexes, this information is incomplete for the following reasons. First, the crystallographic structures do not resolve residues 395–411 at the C termini of the γ chains involved in the crosslinking initiated by factor XIIIa, the plasma transglutaminase, but these residues stabilize the D:D interface. Second, the structures do not resolve residues α17–26 and β15–57 at the N termini of the α and β chains, containing knobs ‘A’ and ‘B’, respectively, and, hence, they do not provide information about their interaction with holes ‘a’ and ‘b’. However, the A-a and B-b knob-hole interactions are important non-covalent interactions in fibrin polymerization. Third, the X-ray data present a static picture of rigid monomers, thus ignoring local conformational fluctuations due to thermal excitations in protein domains, and underestimating the role of global dynamic structural transitions in fibrin molecules due to intra- and intermolecular interactions.
To uncover the structural details of the early stages of fibrin polymerization, we performed combined atomic force microscopy (AFM) and molecular modeling studies of single- and double-stranded fibrin oligomers. Based on thorough analysis of all 27 crystal structures of human fibrin(ogen) and its fragments resolved to date, we have reconstructed atomic models of short fibrin oligomers with knob-hole bonds and γ-γ crosslinks. We correlated these models with high-resolution AFM images of fibrin oligomers and with X-ray crystallographic data on D-D and D-E interfacial junctions. These efforts have enabled us to provide a structural basis for the flexibility of fibrin oligomers due to D:D interface bending and to identify a hinge region. Furthermore, we have provided an atomic-level description of D:E:D interfaces between the central nodule and the peripheral β- and γ-nodules. We characterized the previously unknown intermolecular contacts that strengthen the A-a and B-b knob-hole bonds. The results obtained advance our knowledge of the atomic structures of fibrin oligomers populated in early stages of fibrin polymerization and provide insights into intra- and inter-strand interactions. These results define the mechanism of fibrin formation and influence the biochemical and biomechanical properties of fibrin polymers at multiple spatial scales.
RESULTS
Flexibility of End-to-End Fibrinogen Oligomers from AFM Imaging
First, we used AFM to image single-stranded end-to-end fibrinogen oligomers formed by covalent crosslinking of fibrinogen with factor XIIIa, structures normally present in fibrin polymer in clots. In a mixture of monomeric and oligomeric fibrinogen molecules adsorbed on a surface (Figure 2), the majority of single-stranded oligomers were dimers, but trimers and longer oligomers were also detected. Most of the dimers (Fg2) and trimers (Fg3) formed by crosslinked fibrinogen monomers were in the bent conformation presumably due to flexible D:D interfaces (Figures 2A and 2B). To quantify the conformational flexibility of the D:D interfaces, we measured the bending angles between two adjacent fibrinogen molecules (Figure 2C, inset). Only dimers and trimers were included in the bending angle analysis because longer oligomers had a variable shape due to surface effects, which complicated the analysis. The histogram of the D-D angles (Figure 2C) revealed that only 18% of all structures were straight with θ = 170°–180° bending angles, whereas ~79% of structures showed a θ = 90°–170° angle. Thus, the majority of single-stranded fibrinogen oligomers showed a large ~80° angle bending owing to flexible D:D interfaces forming end-to-end intermolecular contacts.
Flexibility of D:D Interfaces from X-Ray Crystallography
To determine whether the flexibility of intermolecular end-to-end connections in solution is also characteristic of the crystalline state, we used 27 crystallographically resolved D-D structures of either the D-dimer fibrin fragments or the D-D pairs of monomeric fibrinogen fragments D packed in a crystal (Table S1). We employed structure alignment to visualize possible arrangements of fibrin(ogen) molecules so that their mutual D:D interface corresponds to each of the 27 crystal structures (see Supplemental Information). For each structure, we obtained a particular end-to-end arrangement of two fibrin(ogen) molecules in which the D regions form an angle corresponding to a D-D-structure from Table S1. This allowed us to examine all possible geometries of the D-D associations in crystals (Figure 2D; Data S1). The angle formed by the monomers in the reconstructed dimers was found to fluctuate between θ = 149° and 171° (Figure 2C), further supporting our conclusion that fibrinogen oligomers are capable of bending. A comparison of the bending angles from AFM imaging and X-ray crystallography revealed that the θ-angle fluctuations in a crystalline state were within the range from AFM (Figure 2C). To verify the fibrinogen dimer model we superimposed in silico reconstructed dimers with the corresponding AFM images and obtained a very good overlap between the two types of data for relatively straight dimers (θ > 140°; Figures 2B and 2C). The agreement for bent dimers (θ < 140) was worse due to lower angle variation of the X-ray data.
Structure of the D:D Interface
Large bending angle variations observed in AFM and X-ray crystallography (Figures 2C and 2D) have enabled us to hypothesize the existence of a flexible hinge in the D:D interface. To identify the residues that comprise the pivotal point, we analyzed the interface of two γ-nodules in the double-D structures (Table S1). Owing to the asymmetry of the D-D contacts, the γ-nodules were designated γ1 and γ2 (Figures 2E and 2F). The γ1-γ2 contacts were defined as those having a Cα-Cα distance of less than the standard 8 Å cutoff distance (Mickler et al., 2007; Zhmurov et al., 2010). Analysis of the inter-residue contacts reinforcing the D-dimer’s structures enabled us to identify the amino acids that form contacts at the D:D (γ1: γ2) interface: γ1Ala241-Pro243 with γ2Ala279-Tyr280; γ1Met264 with γ2Ala279; γ1Ala271-Asp272 with γ2Pro299-Ser300; γ1Ala279-Tyr280 with γ2Thr277 and γ2Asn308-Gly309; γ1Thr277 and γ1Gly309 with γ2Phe303. Next, we profiled the root-mean-square fluctuations (RMSF) of the positions of residues in γ2 relative to the γ1-nodules and positions of residues in γ1 relative to γ2 across all crystal structures. Assuming that the strongest contacts between residues with the smallest RMSF would correspond to the “hinge region” (Figure S1C), we identified a group of four contacts formed by residues with the smallest 1.5–1.7 Å RMSF values: γ1Ala279-γ2Asn308; γ1Tyr280-γ2Asn308; γ1Ala279-γ2Gly309; and γ1Tyr280-γ2Gly309 (Figures 2E, 2F, and S1C). These contacts comprise the pivotal point in the hinge region at the D:D interface. We also performed a normal mode analysis (see Supplemental Information) for the dimer Fg2 reconstructed in silico using the PDB structure (PDB: 1FZG) for the bent D:D interface. We found several bending deformation modes for monomers around their pivotal point (Figures S1D and S1E), providing additional evidence for the hinge region at the D-D junction and revealing dynamic signatures of the D:D-interfacial flexibility.
Two Types of D:D Interfaces in Crystals
To distinguish between the inter-residue contacts forming the straight versus bent arrangements of single-stranded fibrinogen oligomers, we performed clustering analysis (see Supplemental Information) using all available D-D PDB entries (Table S1). This revealed two main structure groups, one of which clustered around the PDB structure (PDB: 1N86) for the straight connection (cluster 1; Figure S1) and the other clustered around the PDB entry (PDB: 1FZG) for the bent connection (cluster 2; Figure S1). Cluster 2 had 19 structures and cluster 1 had only five structures, implying that the bent conformation of the D-D complex was more favorable. The remaining three D-D structures neither matched any of the two groups nor formed a separate cluster, and so they were excluded from the analysis. There were common contacts in both clusters, but the contacts between residues γ1Ala241-Pro243 and γ2Ala279-Tyr280 were more frequent in the straight dimers. The bent dimers had unique contacts not detected in the straight form, namely γ1Ala271-Asp272 and γ2Pro299-Ser300. The bent and straight conformations also had very different values of solvent-accessible surface area (SD:D) for the D:D interface: SD:D ≈ 2,150 Å2 (bent) versus SD:D ≈ 1,600 Å2 (straight), indicating that the D:D interface is tighter in the bent conformation. The numbers of interfacial residue-residue contacts, i.e., 30 (bent) versus 20 (straight), indicate that the bent conformation is slightly more stable thermodynamically, also explaining why 19 out of 27 double-D crystal structures (Table S1) are bent (Figure S1).
Constructing Double-Stranded Fibrin Oligomers
To elucidate the structural basis of fibrin polymerization, we reconstructed atomic models of double-stranded fibrin oligomers formed at early steps of fibrin polymerization. Two linear strands of fibrin(ogen) molecules were associated laterally based on the spatial constraints described below. Because the end-to-end D-D interactions are the same both in fibrinogen and in fibrin, the structures of the D:D interface characterized above were used to form the linear fibrin constructs. Here, we outline the step-by-step reconstruction of the fibrin oligomer (named FO2/3), which comprises a linear strand of two fibrin molecules associated laterally in a half-staggered fashion with a linear strand of three monomers (Figure 3A). In step 1, we performed the linear alignment of the dimer and trimer (Figure 3A) using the PDB structure (PDB: 1N86) that corresponds to the D-dimer fragment from crosslinked fibrin in the straight configuration (Figures 2 and S1). In step 2, we used atomic models of single-stranded dimer Fg2 and trimer Fg3 to perform 20 Monte Carlo docking simulations (see Supplemental Information). Here, the objective was to bring the two single-stranded oligomers closer together while keeping their molecular half-staggered arrangement, so that two D-E and four D-E-D complexes could form (Figures 3 and S2). The following spatial constraints were satisfied: two fibrin strands should be within the 26.6 Å distance to allow formation of the A-a knob-hole bonds. These are formed by the nine-residue-long N-terminal peptides of the α chains bearing knobs ‘A’ and the γ-nodules with complementary holes ‘a’ (see Experimental Procedures and Figure S2A). Of 20 Monte Carlo runs, 14 were successful, i.e., all the knob-hole bond constraints were satisfied at the end of each run (there are eight A-a knob-hole bonds in FO2/3; see Figure S2B). We found that 14 final structures (Figure 3B) cluster around the average fibrin oligomer FO2/3 structure (Figure 3C) with a root-mean-square deviation <2.5 nm.
Experimental Verification of Fibrin Oligomer Models
First, we compared the molecular dimensions of pentamers FO2/3 from the virtual structure set (Figure 3B) and AFM images (Figures 4A–4C and S3A–S3D) using the D–E distance between the center of globular parts of D region in one strand and the center of the globular part of contiguous E region in the other. To better resolve the experimental D–E distance, we analyzed AFM images of fibrin oligomers from trimers FO1/2 (Figures 4A and S3A) to dodecamers FO6/6 (Figures 4B, 4C, and S3B–S3D). The average experimental D-E distance was 6.8 ± 1.2 nm, which compared well with the 6.5 ± 0.9 nm value from in silico structures of FO2/3. We also analyzed the dynamics of the D–E distance in the Monte Carlo docking simulations, which showed rapid convergence to the average structure of FO2/3 (Figure S3E). Next, we compared the histograms of experimental and theoretical D-E distances (Figure 4D), which shows the overlap of theoretical and experimental distributions. The minimal D-E distances were 4.8 versus 3.5 nm and the maximal distances were 9.7 versus 10.4 nm for the virtual models and AFM images, respectively. The excellent agreement between the D-E distances in atomic models and AFM images of fibrin oligomers validates our in silico model of FO2/3.
Incorporating Missing Residues Unresolved in X-Ray Crystallography
Two strands of fibrin oligomers are held together in the D-E-D complex consolidated mainly by the A-a and B-b knob-hole bonds (Litvinov et al., 2005, 2007). In addition, the D-E-D complex is stabilized by covalent bonds by factor XIIIa between the γ-nodules of adjacent fibrin molecules. Although the FO2/3 construct lacks the γ-γ crosslinks and A-a and B-b knob-hole bonds, it conforms to the distance constraint allowing knobs ‘A’ and ‘B’ to reach holes ‘a’ and ‘b’ (see Experimental Procedures and Figure S2). Therefore, in step 3 (Figure 3A): (1) knob ‘A’-containing N-terminal peptide (GPRV, α17–20) inserted into hole ‘a’ was tethered to the structured part of the α chain via the VERHQS motif (see Experimental Procedures) stretched between αVal20 and αAla27 (Figure S2); and (2) knob ‘B’ (GHRP, β15–18) placed into hole ‘b’ was tethered to the structured part of the β chain by the LDK KRE EAP SLR PAP PPI SGG GYR ARP AKA AAT QKK VER sequence (see Experimental Procedures and Supplemental Information) stretched between βPro18 and βLys58 (Figures 5 and S4). In step 3 we also incorporated: (3) the unresolved residues in the C-terminal parts of β and γ chains (βPro459-Gln461 and γGly395-Val411); and (4) covalent bonds linking the residues involved in the γ-γ crosslinking (γGln398, γGln399, and γLys406; see Experimental Procedures). After several rounds of extensive energy minimization (Experimental Procedures), we arrived at the complete structure of short double-stranded fibrin oligomer FO2/3 with γ-γ cross-links and A-a and B-b knob-hole bonds (Figure 3D). This is an important building block for the elongation of two-stranded fibrin oligomers and their transformation into fibrin protofibril (see Data S2).
Structure of the D:E:D Interface
In our previous study, we hypothesized that A-a knob-hole interactions are not limited to binding of knob ‘A’ to hole ‘a’, and that the D:E interfaces reinforcing the D-E-D complex structure also contain the inter-strand contacts that stabilize fibrin polymers (Litvinov et al., 2005; Kononova et al., 2013). We utilized the full atomic structure of FO2/3 to identify the binding contacts (beyond knob-hole bonds) that stabilize the interface between the E and D regions (Figure 5, Data S3). Specifically, we carried out three 50 ns Molecular Dynamics (MD) simulation runs in explicit water and three 100 ns MD simulations in implicit solvent (see Experimental Procedures and Supplemental Information) and analyzed the strong contacts that persisted at least 50% of the time. Residues αVal20-Lys29 (adjacent to the knob ‘A’ sequence αGly17-Arg19) were found to interact with the γ-nodule, especially with residues γAsp298-Phe304, γAsn319-Asn325, and γGln329 near the “entrance” to hole ‘a’. We observed contact formation between the N-terminal part of the β chain (beyond the knob ‘B’ sequence βGly15-Pro18) including residues βLys53-Pro60 and βPro70-Asp71 and residues in the γ-nodule. We also observed formation of binary contacts between residues αLeu54, αGlu57, αPhe62, αArg65, γGln33, γAsp37, and γGln49 in the coiled coil and γGln329-Asp330 in the γ-nodule. The most important residues in the D regions interacting with the E region are clustered in three loops: γTrp315-Gly331, γTyr348-Asn365, and γAsp298-Thr305 (Figure 5B). In loop γTrp315-Gly331, which is one of the main structural determinants of hole ‘a’ (Kononova et al., 2013; Pratt et al., 1997), residues γGln329 and γAsp330 are involved in A-a bond formation through interaction with residue αArg17 in knob ‘A’. Loop γTyr348-γAsn365 is also important for the D–E interactions because it forms contacts with residues in knob ‘B’ and in the coiled coils (Figure 5A).
DISCUSSION
Fibrin oligomers are intermediate structures that determine the final arrangement of the fibrin network (Weisel and Litvinov, 2013). The experimental isolation and characterization of fibrin oligomers is challenging because they form highly unstable heterogeneous structures due to rapid self-assembly. To gather structural and mechanistic information on fibrin polymerization, we performed a combined experimental and theoretical study employing experimental single-molecule imaging and computational molecular modeling of fibrin oligomers. We were able to reconstruct the atomic structural models of short double-stranded fibrin oligomers, which were correlated with high-resolution AFM images. These efforts have enabled us to provide the structural basis of the remarkable flexibility of fibrin polymers with an unprecedented degree of spatial resolution.
An important step in fibrin polymerization is the end-to-end D-D interactions in the same strands within two-stranded fibrin oligomers. To isolate these interactions from other intermolecular contacts and study them separately, we reproduced polymerization of fibrinogen catalyzed by the transglutaminase, factor XIIIa, an active form of factor XIII zymogen activated by thrombin in the presence of Ca2+. The C-terminal portion of each of fibrin(ogen)’s γ chains contains a crosslinking site at which two adjacent molecules form an intermolecular ε-(γ-glutamyl)-lysyl covalent isopeptide bond between the γLys406 in one γ chain and γGln398/399 in the other γ chain. This reaction resulting in formation of a gel without proteolytic cleavage of fibrinogen is evolutionarily older than thrombin-induced activation and polymerization of fibrin(ogen) (Lorand and Conrad, 1984; Yeh et al., 1998). Factor XIIIa-induced polymerization of fibrinogen was reproduced in earlier studies (Blomback et al., 1985; Weisel et al., 1993) in which single-stranded fibrin oligomers were synthesized in vitro. These single-stranded fibrin oligomers were visualized using transmission electron microscopy, which showed the conformational variability (Brown et al., 2007).
The high-resolution AFM imaging of crosslinked single-stranded fibrinogen oligomers revealed that the bonds formed between the lateral D regions of two adjacent monomers are relatively flexible, thus deviating from the straight configuration by a ~60° or more bending angle (Figure 2). This was confirmed by a thorough theoretical analysis of all 27 available crystal structures that comprise possible types of D-D interactions, including the natural crosslinked D-dimers from fibrin and D-D contacts between fibrinogen molecules or fibrin(ogen)-derived fragments D originating from crystal packing. Not surprisingly, the degree of flexibility of the D-D contacts in crystals was substantially smaller and varied within 22° bending angle. Because crystal packing might affect the protein structure making the protein-protein interactions potentially nonspecific and physiologically irrelevant, we divided all 27 crystal structures of the D-D complex into two groups, comprising either naturally formed or artificial D-D structures: 12 crystal structures of natural D-dimers formed in fibrin and 14 structures with D-D complexes in a crystal lattice. Figure S1 shows that the difference in the average angles for the two groups of complexes were statistically insignificant, thus confirming that the D:D interface structures most likely represent the “biological” contacts rather than “crystal” contacts. It is noteworthy that the flexibility of the D-D junction is substantially weaker than the bendability of the hinge regions in the fibrin(ogen) coiled coils, which are capable of forming kinks (Brown et al., 2000; Köhler et al., 2015).
The computational reconstruction of single-stranded fibrinogen oligomers and the structural analysis of D:D interfaces was based on resolved structures of human fibrin(ogen) and its fragments (Table S1). Using molecular structure alignment followed by Monte Carlo docking with subsequent energy minimization, we identified persistent intermolecular contacts at the D:D junction forming a pivotal point around which the angle fluctuations occur (Figures 2E and 2F). To select the functionally important interatomic contacts, we applied an 8 Å cutoff between any two Cα-atoms, which is a standard definition of “native contacts” between amino acids (Mickler et al., 2007; Zhmurov et al., 2010) from the ~7–9 Å range commonly used. We found that, depending on the structure, there are 20–30 native contacts stabilizing the interface. The number of interfacial contacts was larger for the bent configuration of D-D complex compared with the straight configuration. This finding suggests that the D:D interface has a propensity for bending, a characteristic which might be crucial for the mechanism of protofibril twisting during the formation of fibrin fibers (Weisel et al., 1987). Analysis of the D:D contacts allowed us to identify and characterize the hinge, which is localized to residues γAla279-Tyr280 in one fragment of D and residues γAsn308-Gly309 in the other. Interestingly, point mutations in these same residue positions, namely mutations in residues γAla279, γTyr280, γAsn308, and γGly309, which facilitate the D-D interactions, are also implicated in a number of dysfibrinogenemias with impaired fibrin formation (Table S2; Hanss and Biot, 2001).
To recreate complete models of two-stranded fibrin oligomers, we used crystallographic information about the fibrin(ogen) molecules, structural analysis of contacts observed in fibrin(ogen)-derived fragments and complexes, geometric constraints due to the non-covalent knob-hole bonds, and MD simulations-based molecular modeling of unresolved unstructured parts bearing highly specific binding sites, such as knobs ‘A’ and ‘B’. Following in silico reconstruction of missing portions of the fibrin molecule, we also modeled the A-a and B-b knob-hole interactions (Figures 3 and 5). The 3D arrangements revealed by reconstructed N-terminal portions of the α and β chains (knobs ‘A’ and ‘B’ and their tethers) have provided spatial constraints necessary to build the D:E:D interface, the key structural determinant of double-stranded fibrin arrangement that holds the two strands together. Although during formation of fibrin fibers B-b interactions can occur either within or between the protofibrils (Moskowitz and Budzynski, 1994; Weisel, 2007), here we satisfied all the intra-chain B-b knob-hole bonds to arrive at the most complete structure of fibrin oligomers. Irrespective of the potential significance of B-b interactions (Weisel and Litvinov, 2013), the Nβ region that contains knob ‘B’ is long enough to allow the B-b bond formation within the D-E-D complex and between fibrin strands without any steric clashes (Medved and Weisel, 2009; Figures 5 and S4). We did not add the αC region into our structures because this part of about 400 residues is highly unstructured, which makes its computational reconstruction a separate difficult task. Most importantly, the αC regions are not directly involved in the formation of short oligomers at the early stages of polymerization (Weisel and Litvinov, 2013), and so their presence or absence would not affect the structure of D:E or D:D interfaces. Nevertheless, given the functional importance of the αC regions at the later stages of fibrin polymerization, their reconstruction would be essential to study more complex fibrin structures and their properties. Importantly, all the atomic structures of double-stranded fibrin oligomers reported here were validated by high-resolution AFM imaging of fibrin oligomers (Figure S3).
The very good agreement between the experimentally determined and theoretically predicted intermolecular dimensions of double-stranded fibrin oligomers that we have demonstrated has enabled us to characterize in atomic detail the D:E interface, including the newly resolved binding sites beyond the knob ‘A’-hole ‘a’ associations. Our previous single-molecule experiments on the forced dissociation of the knob-hole bonds revealed ~6-fold stronger dissociation forces for the A-a knob-hole bonds compared with the B-b knob-hole bonds (Litvinov et al., 2005, 2007), but in our MD simulation studies these forces were found to be roughly equal (Kononova et al., 2013). We hypothesized that this disagreement was due to our modeling of knob ‘A’ using the GPR motif at the N termini of the α chains, whereas in experiments the A:a binding interface can extend beyond the GPR motif (Figure 5B). Our results fully confirm this hypothesis and show that the reconstructed N-terminal parts of fibrin’s α chains (α17–26) and β chains (β16–57) indeed form additional intermolecular binding sites besides the terminal N-terminal tripeptides (knobs) known to fit into small complementary holes.
The algorithm of reconstructing fibrin oligomers used in this work is somewhat different from the sequence of events during natural fibrin polymerization. In particular, the initial driving force of real fibrin polymerization is the A-a knob-hole interaction, while the end-to-end D-D interaction used here as the initial step to connect fibrin monomers is secondary to the half-staggered lateral association of fibrin molecules mediated by the knob-hole bonds between the D and E regions. There are at least three arguments that justify the approach used in this paper. First, the D-D bonds and the D:D interfaces are real and physiologically important, as gleaned, e.g., from the mutations in the D:D interface that affect fibrin formation and properties (Hanss and Biot, 2001). Second, the A-a knob-hole bonds are formed by the crystallographically unresolved flexible N terminus of the α chain (knob ‘A’), meaning that there is no initial structural information to model fibrin assembly starting with the A-a knob-hole interactions. Therefore, the N terminus of the α chain bearing knob ‘A’ was reconstructed computationally using the spatial constraints provided by the D-E-D complex formed based on the known crystal D-D structures and the GPRP peptide (knob ‘A’ mimetic) associated with fragments D or D-dimer. This approach can be named generally as “moving from known to unknown” while reconstructing the full-atom structures of fibrin oligomers. We would certainly have arrived at very similar results had we started with the knob-hole interactions, provided we had had the initial information necessary to model them, because the spatial limitations in the D-E-D complex are quite stringent. Third, the final structures of fibrin oligomers reconstructed in silico are in very good agreement with fibrin oligomers visualized experimentally using AFM (Figures 2, 4, and S3) or transmission electron microscopy (Fowler et al., 1981).
There is an ongoing debate about whether the factor XIIIa-mediated γ-γ crosslinking between fibrin monomers occurs longitudinally (i.e., within a strand) or transversely (between the strands of a protofibril) (Weisel, 2004a, 2004b; Mosesson, 2004a, 2004b). The topology of the D:D interface and the spatial constraints in our two-stranded models of fibrin oligomers strongly support the longitudinal γ-γ crosslinking for the following reasons. The γ-γ crosslinks connect the C-terminal parts of two γ chains, namely residues γArg398/399 and γLys406. The C-terminal part of the γ chain is unstructured downstream to residue γAsn390. There are nine residues between γAsn390 and γArg399, seven residues between γArg399 and γLys406, and nine residues between γAsn399 and γAsn390 of the other γ chain. Taking the value of 3.8 Å for a fully extended amino acid length and, hence, the value of 18 Å for the length of γArg399-γLys406 γ-γ bond, the contour length of the crosslinked γ chains connecting two γAsn390 residues is estimated to be ~113 Å. Using our two-stranded fibrin protofibril structure, we found the distances between residues γAsn390 in the two γ-nodules located in different strands to be ~230 Å, i.e., 2× farther than a fully extended polypeptide chain can span. This suggests that the transverse γ-γ crosslinking is not feasible without substantial elongation of the γ-nodules, which is potentially possible provided that elongation of the γ chain due to the C-terminal β strand pull out occurs. According to this “pull-out” hypothesis, the C-terminal β strand of the γ chain (residues γLys381-Asn390) can be drawn out of the γ-nodule without much perturbation of its native fold (Yakovlev et al., 2000). Consequently, the β strand can extend outwards, increasing the reach of the γ-γ crosslinks. We calculated the contour length of the two crosslinked g chains between γLys381 residues to be ~180 Å, while the shortest distance between two γLys381 residues in our structures is ~200 Å. This means that even if both β strands are pulled out from two crosslinked γ-nodules and the polypeptide chains forming the γ-γ crosslinks are fully extended, the γ-nodules from two different strands are still too far apart. This type of crosslinking would require additional residues (i.e., γMet379-Lys380) to be pulled out from the γ-nodule, but the removal of these residues might also cause the dissociation of A-a knob-hole bonds because these residues are close to hole ‘a’.
These results provide additional insights into the molecular mechanisms of fibrin formation. Many functionally important intermolecular contacts identified and quantitatively characterized here have never been described hitherto, while they drive fibrin oligomerization. Apart from the known A-a and B-b knob-hole bonds, the flexible interfaces described in this paper are formed due to D-D interactions as well as D:E:D contacts. Importantly, the functional significance of these intermolecular juncture sites was validated by quantitative comparison with corresponding naturally occurring genetic mutations that cause impaired fibrin formation (Table S2). Information about these contacts could never be gleaned from sequence analysis alone without the step-by-step 3D spatial reconstruction of fibrin(ogen) folded domains, and the subsequent equilibration and analysis of their tertiary and quaternary structures performed in this study.
To conclude, the atomic models reconstituted computationally in this work and validated experimentally present a significant development necessary for the structure-based understanding of unique biochemical and material properties of fibrin polymers. Atomic models of fibrin oligomers and protofibrils are available for downloading in the PDB format for atomic coordinates at http://faculty.uml.edu/vbarsegov/research/fibrin.html.
EXPERIMENTAL PROCEDURES
Formation of Crosslinked Fibrinogen Oligomers
To induce end-to-end (D-D) oligomerization of fibrinogen molecules, 10 mg/mL human fibrinogen (HYPHEN BioMed) in 20 mM Tris-HCl buffer (pH 7.4) containing 150 mM NaCl, 30 mM CaCl2, and 5 ATU/mL hirudin (Sigma) was mixed with pre-activated human factor XIIIa (Enzyme Research Laboratories) at 50 μg/mL (final concentration) at 37°C. Aliquots were taken at various time points and the crosslinking reaction was stopped by dilution and boiling the aliquots in sample buffer with 100 mM DTT, 2% SDS. The reduced samples were run on a 10% SDS-PAGE gel followed by staining with Coomassie blue R-250 (Figure S5A). The covalent crosslinking of fibrinogen molecules via the C termini of the γ chains was corroborated by the appearance of the γ-dimer band in SDS-PAGE of reduced samples. A fine γ-dimer band was visible 1 min after addition of factor XIIIa, became more pronounced at 3 min, and reached a maximum at 15 min. To obtain a solution of fibrinogen oligomers for AFM imaging, the factor XIIIa-catalyzed cross-linking was stopped with iodoacetamide (2 mM final concentration) at the beginning of gelation (~60 min) and the nascent gel was removed. The soluble fraction of monomeric and oligomeric fibrinogen that was not incorporated into the gel was diluted 2,000-fold (to about 1–3 μg/mL) with 20 mM Tris-HCl buffer (pH 7.4) containing 150 mM NaCl, 10 mM CaCl2, and 2 mM iodoacetamide.
Formation of Double-Stranded Fibrin Oligomers
Human fibrinogen (3 μg/mL) in 20 mM Tris-HCl buffer (pH 7.4) containing 150 mM NaCl and 10 mM CaCl2 was mixed with 0.05 U/mL thrombin (final concentration), incubated for 15–20 min at room temperature and immediately used for AFM imaging. Short double-stranded fibrin oligomers and longer protofibrils were predominantly obtained with this sample preparation protocol. Covalent crosslinking of these fibrin structures was ensured by formation of the γ-dimer bands in SDS-PAGE (Figure S5B).
AFM
AFM imaging of fibrinogen or fibrin oligomers was performed on the surface of modified hydrophilized graphite (Klinov et al., 2007; Protopopova et al., 2015). Three microliters of a sample solution prepared as described above was applied on the modified graphite surface and kept for 15 s at room temperature. A ~50× volume drop of fresh Milli-Q water was then carefully placed above the sample solution for 10 s and then removed with a flow of air making the surface ready for imaging. The AFM imaging was performed using a MFP-3D microscope (Asylum Research – Oxford Instruments) in AC mode with a typical scan rate of 0.8 Hz. Images were taken in air using SSS-SEIHR cantilevers (Nanosensors) with a tip radius of 3 ± 2 nm. FemtoScan Online software (http://www.femtoscanonline.com) was used to filter and analyze the AFM data.
Computational Reconstruction of Short Fibrin Oligomers
The following steps 1–3 (Figure 3A) were used to construct the atomic structures of double-stranded fibrin oligomer FO2/3 and the D-E-D construct. Steps 1–3 are described in more detail in Supplemental Information. In short, step 1 is a reconstitution of single-stranded dimer (Fg2) and trimer (Fg3) with the Kabsch rotation algorithm applied to known structures of the full-length fibrinogen molecule and the D:D interface. In step 2, the structures of Fg2 and Fg3 were used in conjunction with Monte Carlo docking to obtain fragment FO2/3. We performed 20 Monte Carlo runs both for the bent and straight conformations of the D-D junction, but none of the runs for the bent conformation converged to a structure with all eight constraints satisfied. By contrast, the Monte Carlo runs with the straight conformation showed a very good convergence, with 14 of 20 final structures having all eight constraints satisfied. In step 3, we incorporated the missing parts of fibrin molecules, followed by the energy minimization of the obtained complete structure of FO2/3. The structure of the D-E-D construct was obtained from the structure of FO2/3 (Figures 3 and 5).
Atomic Models of the D-E-D Complex
We used the energy-minimized structure of FO2/3 to extract complexes γ-E-γ and D-E-D (Figures 3D and 5). The γ-E-γ complex contains residues γLys140-Lys411 in the γ-nodules to represent adjacent D regions and residues αGly17-Phe74, βArg57-Val104, and γTyr1-Gln49 in the E region. The D-E-D complex contained residues αIle119-Gln200, βAsp154-Gln461, and γGlu91-Val411 in the D regions and residues αGly17-Lys123, βGly15-Asn158, and γTyr1-Lys95 in the E region. Two Ca2+ ions were added to γ-E-γ and D-E-D. In the γ-E-γ complex, the atoms of active sequence GPR (αGly17-Arg19) in knob ‘A’ were placed at positions of corresponding atoms in the GPRP peptide co-crystallized with fibrinogen. In the D-E-D complex, knobs ‘A’ and ‘B’ were placed at the position of the GPR and GHR residues of the corresponding synthetic peptides. The MD simulations were performed as described in Supplemental Information.
Supplementary Material
Highlights.
Atomic models for single- and double-stranded fibrin oligomer are reconstructed
Structures are validated by quantitative comparison with high-resolution AFM images
Structural basis for interfacial flexibility of fibrin oligomers is provided
Atomic structure of the D:E:D interface beyond the knob-hole bonds is characterized
Acknowledgments
We thank Dr. Leonid Medved for valuable comments and Dr. Dmitry Klinov for providing a graphite modificator. This work was supported by American Heart Association grants 15GRNT23150000 and 13GRNT16960013, NIH grants HL090774 and UO1-HL116330, NSF grant DMR 1505662, Russian Foundation for Basic Research grants 15-01-06721A and 15-37-21027, and by the Program for Competitive Growth at Kazan Federal University.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, five figures, two tables, and three 3D molecular models and can be found with this article online at http://dx.doi.org/10.1016/j.str.2016.08.009.
AUTHOR CONTRIBUTIONS
A.Z., P.Z., and V.B. performed computational reconstruction of the atomic models; A.D.P., A.R.M., R.I.L., and J.W.W. carried out the AFM experiments; A.Z. and A.D.P. statistically analyzed the data; A.Z., R.I.L., J.W.W., and V.B. designed the research and wrote the paper.
References
- Blomback B, Procyk R, Adamson L, Hessel B. FXIII induced gelation of human fibrinogen – an alternative thiol enhanced, thrombin independent pathway. Thromb Res. 1985;37:613–627. doi: 10.1016/0049-3848(85)90094-5. [DOI] [PubMed] [Google Scholar]
- Bowley SR, Lord ST. Fibrinogen variant BbD432A has normal polymerization but does not bind knob ‘B’. Blood. 2009;113:4425–4430. doi: 10.1182/blood-2008-09-178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Volkmann N, Jun G, Henschen-Edman AH, Cohen C. The crystal structure of modified bovine fibrinogen. Proc Natl Acad Sci USA. 2000;97:85–90. doi: 10.1073/pnas.97.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AEX, Litvinov RI, Discher DE, Weisel JW. Forced unfolding of coiled-coils in fibrinogen by single-molecule AFM. Biophys J. 2007;92:L39–L41. doi: 10.1529/biophysj.106.101261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everse SJ, Spraggon G, Veerapandian L, Riley M, Doolittle RF. Crystal structure of fragment double-D from human fibrin with two different bound ligands. Biochemistry. 1998;37:8637–8642. doi: 10.1021/bi9804129. [DOI] [PubMed] [Google Scholar]
- Fowler WE, Hantgan RR, Hermans J, Erickson HP. Structure of the fibrin protofibril. Proc Natl Acad Sci USA. 1981;78:4872–4876. doi: 10.1073/pnas.78.8.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanss M, Biot F. A database for human fibrinogen variants. Ann NY Acad Sci. 2001;936:89–90. doi: 10.1111/j.1749-6632.2001.tb03495.x. [DOI] [PubMed] [Google Scholar]
- Hantgan R, Fowler W, Erickson H, Hermans J. Fibrin assembly: a comparison of electron microscopic and light scattering results. Thromb Haemost. 1980;44:119–124. [PubMed] [Google Scholar]
- Hirota-Kawadobora M, Terasawa F, Suzuki T, Tozuka M, Sano K, Okumura N. Comparison of thrombin-catalyzed fibrin polymerization and factor XIIIa-catalyzed cross-linking of fibrin among three recombinant variant fibrinogens, Gamma275C, Gamma275H, and Gamma275A. J Thromb Haemost. 2004;2:1359–1367. doi: 10.1111/j.1538-7836.2004.00844.x. [DOI] [PubMed] [Google Scholar]
- Köhler S, Friederike S, Giovanni S. The internal dynamics of fibrinogen and its implications for coagulation and adsorption. PLoS Comput Biol. 2015;11:e1004346. doi: 10.1371/journal.pcbi.1004346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Pandi L, Sawaya MR, Riley M, Doolittle RF. Crystal structure of human fibrinogen. Biochemistry. 2009;48:3877–3886. doi: 10.1021/bi802205g. [DOI] [PubMed] [Google Scholar]
- Kononova O, Litvinov RI, Zhmurov A, Alekseenko A, Cheng CH, Agarwal S, Marx K, Weisel JW, Barsegov V. Molecular mechanisms, thermodynamics and dissociation kinetics of knob-hole interactions in fibrin. J Biol Chem. 2013;288:22681–22692. doi: 10.1074/jbc.M113.472365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostelansky MS, Betts L, Gorkun OV, Lord ST. 2.8Å crystal structures of recombinant fibrinogen fragment D with and without two peptide ligands: GHRP binding to the ‘b’ site disrupts its nearby calcium-binding site. Biochemistry. 2002;41:12124–12132. doi: 10.1021/bi0261894. [DOI] [PubMed] [Google Scholar]
- Klinov D, Dwir B, Kapon E, Borovok N, Molotsky T, Kotlyar A. High-resolution atomic force microscopy of duplex and triplex DNA molecules. Nanotechnology. 2007;18:225102. [Google Scholar]
- Laudano AP, Doolittle RF. Synthetic peptide derivatives that bind to fibrinogen and prevent the polymerization of fibrin monomers. Proc Natl Acad Sci USA. 1978;75:3085–3089. doi: 10.1073/pnas.75.7.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvinov RI, Gorkun OV, Owen SF, Shuman H, Weisel JW. Polymerization of fibrin: specificity, strength, and stability of knob-hole interactions studied at the single-molecule level. Blood. 2005;106:2944–2951. doi: 10.1182/blood-2005-05-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvinov RI, Gorkun OV, Galanakis DK, Yakovlev S, Medved L, Shuman H, Weisel JW. Polymerization of fibrin: direct observation and quantification of individual B:b knob-hole interactions. Blood. 2007;109:130–138. doi: 10.1182/blood-2006-07-033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorand L, Conrad SM. Transglutaminases. Mol Cell Biochem. 1984;58:9–35. doi: 10.1007/BF00240602. [DOI] [PubMed] [Google Scholar]
- Marchi RC, Carvajal Z, Boyer-Neumann C, Anglés-Cano E, Weisel JW. Functional characterization of fibrinogen Bicetre II: a gamma 308 Asn->Lys mutation located near the fibrin D: D interaction sites. Blood Coagul Fibrinolysis. 2006;17:193–201. doi: 10.1097/01.mbc.0000220241.22714.68. [DOI] [PubMed] [Google Scholar]
- Medved L, Weisel JW. Recommendations for nomenclature on fibrinogen and fibrin. J Thromb Haemost. 2009;7:355–359. doi: 10.1111/j.1538-7836.2008.03242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medved L, Ugarova T, Veklich Y, Lukinova N, Weisel JW. Electron microscope investigation of the early stages of fibrin assembly. Twisted protofibrils and fibers. J Mol Biol. 1990;216:503–509. doi: 10.1016/0022-2836(90)90376-W. [DOI] [PubMed] [Google Scholar]
- Mickler M, Dima RI, Dietz H, Hyeon C, Thirumalai D, Rief M. Revealing the bifurcation in the unfolding pathways of GFP by using single-molecule experiments and simulations. Proc Natl Acad Sci USA. 2007;104:20268–20273. doi: 10.1073/pnas.0705458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosesson MW. The fibrin cross-linking debate: cross-linked γ-chains in fibrin fibrils bridge ‘transversely’ between strands: yes. J Thromb Haemost. 2004a;2:388–393. doi: 10.1111/j.1538-7933.2004.00613.x. [DOI] [PubMed] [Google Scholar]
- Mosesson MW. Cross-linked γ-chains in a fibrin fibril are situated transversely between its strands. J Thromb Haemost. 2004b;2:1469–1471. doi: 10.1111/j.1538-7836.2004.00873.x. [DOI] [PubMed] [Google Scholar]
- Moskowitz KA, Budzynski AZ. The (DD)E complex is maintained by a composite fibrin polymerization site. Biochemistry. 1994;33:12937–12944. doi: 10.1021/bi00248a001. [DOI] [PubMed] [Google Scholar]
- Pratt KP, Cote HCF, Chung DW, Stenkamp RE, Davie EW. The primary fibrin polymerization pocket: three-dimensional structure of a 30-kDa C-terminal γ chain fragment complexed with the peptide Gly-Pro-Arg-Pro. Proc Natl Acad Sci USA. 1997;94:7176–7181. doi: 10.1073/pnas.94.14.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protopopova AD, Barinov NA, Zavyalova EG, Kopylov AM, Sergienko VI, Klinov DV. Visualization of fibrinogen αC regions and their arrangement during fibrin network formation by high-resolution AFM. J Thromb Haemost. 2015;13:570–579. doi: 10.1111/jth.12785. [DOI] [PubMed] [Google Scholar]
- Weisel JW. Cross-linked γ-chains in fibrin fibrils bridge transversely between strands: no. J Thromb Haemost. 2004a;2:394–399. doi: 10.1111/j.1538-7933.2003.00621.x. [DOI] [PubMed] [Google Scholar]
- Weisel JW. Cross-linked γ-chains in a fibrin fibril are situated transversely between its strands. J Thromb Haemost. 2004b;2:1467–1469. doi: 10.1111/j.1538-7836.2004.00873.x. [DOI] [PubMed] [Google Scholar]
- Weisel JW. Fibrinogen and fibrin. Adv Protein Chem. 2005;70:247–299. doi: 10.1016/S0065-3233(05)70008-5. [DOI] [PubMed] [Google Scholar]
- Weisel JW. Which knobs fit into which holes in fibrin polymerization? J Thromb Haemost. 2007;5:2340–2343. doi: 10.1111/j.1538-7836.2007.02794.x. [DOI] [PubMed] [Google Scholar]
- Weisel JW, Litvinov RI. Mechanisms of fibrin polymerization and clinical implications. Blood. 2013;121:1712–1719. doi: 10.1182/blood-2012-09-306639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisel JW, Nagaswami C, Makowski L. Twisting of fibrin fibers limits their radial growth. Proc Natl Acad Sci USA. 1987;84:8991–8995. doi: 10.1073/pnas.84.24.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisel JW, Francis CW, Nagaswami C, Marder VJ. Determination of the topology of factor XIIIa-induced fibrin gamma-chain cross-links by electron microscopy of ligated fragments. J Biol Chem. 1993;268:26618–26624. [PubMed] [Google Scholar]
- Yakovlev S, Litvinovich S, Loukinov D, Medved L. Role of the β-strand insert in the central domain of the fibrinogen γ-module. Biochemistry. 2000;39:15721–15729. doi: 10.1021/bi001836h. [DOI] [PubMed] [Google Scholar]
- Yang Z, Pandi L, Doolittle RF. The crystal structure of fragment double-D from cross-linked lamprey fibrin reveals isopeptide linkages across an unexpected DD interface. Biochemistry. 2002;41:15610–15617. doi: 10.1021/bi026666i. [DOI] [PubMed] [Google Scholar]
- Yeh MS, Chen YL, Tsai IH. The hemolymph clottable proteins of tiger shrimp, Penaeus monodon, and related species. Comp Biochem Phys B. 1998;121:169–176. doi: 10.1016/s0305-0491(98)10085-8. [DOI] [PubMed] [Google Scholar]
- Zhmurov A, Dima RI, Kholodov Y, Barsegov V. SOP–GPU: Accelerating biomolecular simulations in the centisecond timescale using graphics processors. Proteins. 2010;78:2984–2999. doi: 10.1002/prot.22824. [DOI] [PubMed] [Google Scholar]
- Zhmurov A, Brown AEX, Litvinov RI, Dima RI, Weisel JW, Barsegov V. Mechanism of fibrin (ogen) forced unfolding. Structure. 2011;19:1615–1624. doi: 10.1016/j.str.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhmurov A, Kononova O, Litvinov RI, Dima RI, Barsegov V, Weisel JW. Mechanical transition from α-helical coiled coils to β-sheets in fibrin(ogen) J Am Chem Soc. 2012;134:20396–20402. doi: 10.1021/ja3076428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


