Figure 2. Structure of the Single-Stranded Fibrinogen Oligomers.
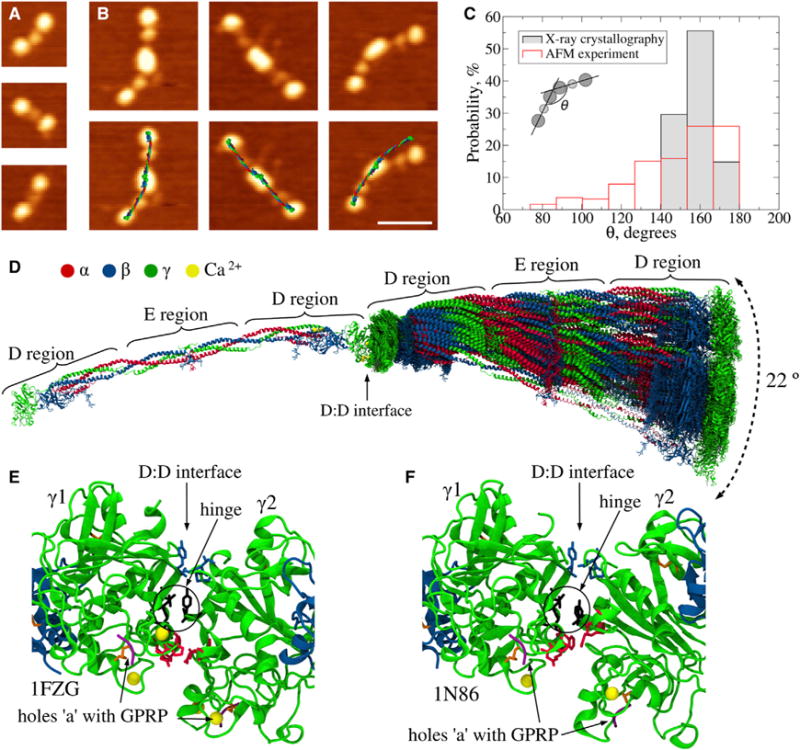
(A) AFM images of fibrinogen molecules. Three distinct globular regions are visualized; the lateral D regions are larger than the central E region.
(B) AFM images of crosslinked fibrinogen dimers Fg2 (top) and same images with computationally reconstructed dimers superimposed (bottom). The superposition of reconstructed structures over AFM images was made by computational alignment of the centers of mass of D and E regions of in silico structures, with the geometrical centers of D and E regions determined from AFM. The scale bar represents 50 nm.
(C) Histograms of the bending angles formed by adjacent fibrinogen monomers in single-stranded fibrinogen oligomers from AFM images (n = 240) and from in silico structures (n = 27; Table S1). The inset illustrates how the bending angle (θ) is defined.
(D) Distribution of positions of the second (right) monomer relative to the first (left) monomer from 27 D-D structures in Table S1 (see Data S1). There is a 22° bending angle that quantifies variations in the relative orientation of monomers.
(E and F) Detailed view of two representative D:D interfaces from the PDB structures: bent arrangement (PDB: 1FZG) and linear arrangement (PDB: 1N86). The side chains forming stable interfacial contacts are shown with sticks: residues γ1Ala279-Tyr280 and γ2Asn308-Gly309 form a hinge (black); residues γ1Ala271-Asp272 and γ2Pro299-Ser300 favor the bent conformation (red); residues γ1Ala241-Pro243 and γ2Ala279-Tyr280 form contacts stabilizing the linear arrangement (blue). See also Figure S1 for a comparison of D-D interfacial contacts found in naturally occurring D-dimers (A) with those reported as crystal contacts (B) and for the results of the normal mode analysis (D and E); Figure S5A for SDS-PAGE of single-stranded oligomers; and Data S1.
