Abstract
Prion propagation is mediated by the structural alteration of normal prion protein (PrPC) to generate pathogenic prion protein (PrPSc). To date, compounds for the inhibition of prion propagation have mainly been screened using PrPSc-infected cells. Real time–quaking-induced conversion (RT-QuIC) is one alternative screening method. In this study, we assessed the propagation inhibition effects of known anti-prion compounds using RT-QuIC and compared the results with those from a PrPSc-infected cell assay. Compounds were applied to RT-QuIC reactions at 0 h or 22 h after prion propagation to determine whether they inhibited propagation or reduced amplified aggregates. RT-QuIC reactions in presence of acridine, dextran sulfate sodium (DSS), and tannic acid inhibited seeded aggregation with sporadic Creutzfeldt-Jakob disease at 0 h. After treatment at 22 h, amplified fluorescence was decreased in wells treated with either acridine or tannic acid. Compound activities were verified by western blot of RT-QuIC products and in a dye-independent conversion assay, the Multimer Detection System. Protease K-resistant PrPSc fragments (PrPres) were reduced by DSS and tannic acid in the PrPSc-infected cell assay. Importantly, these inhibitory effects were similar despite different treatment times (0 h versus 3 days). Consequentially, RT-QuIC enabled the more specific classification of compounds according to action (i.e., inhibition of prion propagation versus reduction of amplified aggregates). RT-QuIC addresses the limitations of cell-based screening methods and can be used to further aid our understanding of the mechanisms of action of anti-prion compounds.
Introduction
Prion diseases are fatal neurodegenerative diseases that trigger the accumulation of pathogenic prion protein (PrPSc) and neuronal death in humans and animals [1]. The process of prion propagation involves the structural alteration of host-encoded cellular prion protein (PrPC) to PrPSc and the autocatalytic amplification of pathogenic protein [2]. PrPSc is largely protease-resistant, insoluble, β-sheet rich, and capable of aggregation as a hallmark of prion disease [3]. Therefore, inhibiting the conversion of PrPC to PrPSc and/or facilitating the degradation of PrPSc are primary strategies for anti-prion pharmaceutical development.
Previous studies have investigated pharmacotherapy, immunotherapy, and cell therapy approaches to prion disease [4, 5]. In recent years, most novel anti-prion compounds have been screened and validated using permanent PrPSc-infected cell models [6–8]. In this assay, cells are plated (1 × 105 cells/well), allowed to stably attach, and achieve 50% confluence within 24 h. Attached cells are then incubated with anti-prion compounds for 5–8 days. Then, within 2 days, protease K-resistant PrPSc fragments (PrPres) are detected by western blotting [9–11]. Although cell screening is mandatory to evaluate the actions of drugs in the cellular environment in vitro, this method does not differentiate between the abilities of potential anti-prion compounds to inhibit prion propagation and degrade PrPres aggregates.
Real time–quaking-induced conversion (RT-QuIC) is a recent innovation that allows the real-time detection and monitoring of the conversion of a recombinant PrP (rPrP) to synthetic PrP amyloid fibrils [12]. RT-QuIC has been successfully used to detect small amounts of PrPSc in brain tissue, cerebrospinal fluid, blood, saliva, and nasal fluid from human, cervid, ovine, bovine, and rodent species [13–18]. Amplified PrPres resultant from the conversion of rPrP to its aggregated form is shown by increased Thioflavin T (ThT) fluorescence, which directly indicates the formation of beta-sheet structures in PrPres aggregates. Here, we hypothesized that synthetic prion propagation in a RT-QuIC reaction seeded with PrPSc-infected brain homogenate would be inhibited by anti-prion compounds and easily monitored using fluorescence. Moreover, this reaction system would enable differentiation between anti-prion compound inhibition of propagation and aggregate degradation.
Acridine, dextran sulfate sodium (DSS), and tannic acid each have alleged anti-prion activity and were selected from previous reports where they were used for the treatment of other diseases or symptoms including malaria, thrombosis, and oxidation, respectively. The goal of this study was to evaluate the usefulness of RT-QuIC for screening these anti-prion compounds by comparing the results of compound activities indicated by RT-QuIC and PrPSc-infected cell screening methods.
Materials and Methods
Compounds
All compounds (acridine orange hydrochloride [318337], DSS [67578], tannic acid [403040], quinacrine dihydrochloride (Q3251) and doxycycline hyclate (D9891) were purchased from the same vendor (Sigma-Aldrich, Saint Louis, MO, USA) in order to ensure consistency in quality. Quinacrine and doxycycline were used as controls. Compounds were dissolved in distilled water or dimethyl sulfoxide (DMSO) to produce 50 mM stock solutions and stored at −80°C until use. The class, medical use, and solubility of each test compound was surveyed from manufacturer reports and the ChemSpider database (Table 1) [19–21].
Table 1. Compounds information.
Human biological samples
Brain homogenate from a confirmed case of sporadic Creutzfeldt-Jakob disease (sCJD) was obtained from the National Institute for Biological Standards and Control (NIBSC; reference code NHBX0/0001). Homogenate was serially diluted 10-fold from 10-2 to 10-7. CSF samples were obtained from Korean patients with confirmed sCJD (3 patients) and from patients with progressive dementia (24 patients) as part of routine diagnoses. A CSF sample from a patient with Alzheimer’s disease (AD) (1 patient) was provided by Dr. Kim at Seoul National University Bundang Hospital in Korea. This study was approved by the Institutional Review Board (IRB) of the Korea Centers for Disease Control and Prevention (IRB No. 2014-06EXP-04-R-A). Written informed consent was obtained from the patients or their legal guardians.
RT-QuIC reaction
Human rPrP (residues 23–231; accession no. K02234) was produced and the RT-QuIC reaction was performed as per Wilham et al. and Cramm et al. with minor modifications [12, 23]. The final composition of the reaction buffer was as follows: 162 mM phosphate buffer (pH 6.9), 170 mM NaCl, 1 mM EDTA, 10 μM ThT, and 0.1 mg rPrP. Ninety-six microliters of buffer was dispensed into each well of a clear-bottomed black 96-well microplate (Nunc, 265301). Two microliters of diluted sCJD brain homogenate was seeded into each well. Compounds were applied at different times in order to examine compound effects in the presence or absence of aggregation. Treatment at t = 0 h was a condition in which there was not considered to be any aggregation and treatment at t = 22 h was a condition in which there was considered to be a sufficient amount of aggregation. These experiments were performed in order to determine whether a given compound inhibited the conversion of PrPC to PrPres or eliminated amplified PrPres aggregates.
For treatment at 0 h, 2 μL of each compound (acridine, DSS, tannic acid, or doxycycline at 20 μM and quinacrine at 5 μM) was added to wells at t = 0 h. For treatment at 22 h, fluorescence saturation at 65,000 relative fluorescence units (rfu) was confirmed after 22 h and subsequently 2 μL of each compound was added to wells (all compounds were applied at the same concentration). DMSO treatment was used as a negative control. The plate was sealed and incubated for 90 h at 44°C with intermittent shaking using the BMG Labtech Optima Fluostar plate reader. Each reaction was performed in quadruplicate. A positive response was denoted if the average of the 2 highest readings at 72 h was greater than 2-fold of the negative control, as described in previous studies using saturation at 65,000 rfu [23, 24]. To ensure optimal RT-QuIC conditions in our laboratory, we repeatedly validated the cut-off in CSF samples from 24 progressive dementia patients, serially diluted sCJD homogenate, and sCJD or AD CSF-seeded reactions.
ScN2a cells
ScN2a cells were derived from the neuroblastoma-2a cell line obtained from the American Type Culture Collection and persistently infected with PrPSc (Rocky Mountain Laboratory [RML] strain). This cell line was generously provided by Dr. Ryu at Hanyang University in Korea. Cell culture was performed as described previously [25]. Briefly, cells were seeded in a T25 cm2 flask (TPP, Z707481) at 2 × 105 cells/flask and pre-incubated in Dulbecco’s Modified Eagle Medium (Gibco, 12430) containing 10% fetal bovine serum, 1% penicillin-streptomycin (Gibco, 15140), and 2 mM l-glutamine (Gibco, 25030) for 24 h under 5% CO2 at 37°C.
We tried to mimic different PrPres conditions in cells, similar to the RT-QuIC reactions. When compound was applied at 0 h, the cells had achieved about 50% confluence and were considered able to endure a non-toxic concentration of the compound. PrPres was not maximally produced from PrPC at this point in time; that is, minimal PrPC had been converted into PrPres. Treatment at 0 h was therefore able to test whether a given compound interfered with the conversion of PrPC to PrPres in cells. When compound was applied at 3 days, the cells had achieved about 98% confluence. Therefore, PrPres production was not acute and we assumed that all PrPC had been converted into mature PrPres. These experiments were used to confirm whether a compound inhibited the conversion of PrPC to PrPres or eliminated amplified PrPres aggregates.
For treatment at 0 h, pre-incubation media was replaced with fresh media containing test compound. After incubation for 3 additional days, the media was replaced again with fresh media containing test compound. For treatments at 3 days, pre-incubation was initially replaced with fresh media. After incubation for 3 additional days, the media was replaced with fresh media containing test compound. After total incubation for 6 days, media was removed, the cells were washed with PBS, and the remaining cells were lysed. Control experiments were treated with DMSO vehicle (0.1% v/v). Each compound was tested in duplicate in 3 independent experiments.
Cytotoxicity assay
A MTT-based cytotoxicity assay was performed as described previously [25]. Compounds applied at 5 different concentrations between 1 and 100 μM or DMSO vehicle were incubated with cells for 3 days. Cytotoxicity was expressed as the percentage of the signal observed in DMSO-treated control cells.
Western blot analysis
For the detection of PrPres in RT-QuIC products, 50 μL of solution was collected from wells treated with acridine, DSS, tannic acid, doxycycline, or DMSO vehicle upon termination of the RT-QuIC reaction. The solutions were digested with proteinase K (PK) (20 μg/mL) (Merck, 70663) for 60 min at 37°C, stopped with 2 mM Pefabloc (Roche, 11429876001), and centrifuged for 90 min at 20,000 × g at 4°C. Pellets were resuspended in SDS sample buffer (125 mM Tris-HCl, pH 6.8, 5% (vol/vol) glycerol, 6 mM EDTA, 5% (wt/vol) SDS, 0.04% (vol/vol) bromophenol blue, and 12.5% (vol/vol) β-mercaptoethanol) and separated by SDS-PAGE. Proteins were probed with the monoclonal PrP antibody, 6H4 (1:2,500) (Prionics, PRN-01-011).
Cells were rinsed once in phosphate-buffered saline (PBS) and lysed in ice-cold lysis buffer (10 mM EDTA, 10 mM Tris, pH 8.0, 100 mM NaCl, 0.5% [wt/vol] Nonidet P-40, and 0.5% [wt/vol] sodium deoxycholate). Lysates were sonicated at an amplitude of 30%. The total protein concentration was adjusted to 1 mg/mL. Subsequent PK digestion and western blotting were performed as mentioned above. Signals were visualized using ECL (Elpis Biotech, EBP-1073) and protein bands were scanned using an Image Scanner III (GE). Another blot of the cell lysates without PK digestion was probed using a β-actin antibody (1:5000) (Cell Signaling, 4970). The relative band densities are shown as the volume intensity/mm2 relative to the β-actin band density.
MDS assay
The MDS assay is a dye-independent conversion assay that has been used previously to screen anti-prion compounds [25]. The MDS assay kit was supplied by People Bio Inc. and performed according to manufacturer specifications with minor modifications [26]. Briefly, test compounds (0.05, 0.2, 1, 5, and 20 μM) were mixed with the reaction buffer containing 50 ng of recombinant PrP, 1% Triton X-100, 10% Blockace, and Tris-buffered saline containing 0.1% (vol/vol) Tween 20 (TBST) in 2 mL screw cap tubes. Doxycycline (0.05, 0.2, 1, 5, and 20 μM), quinacrine (1 and 5 μM), and DMSO (0.1% vol/vol) were used as negative, positive, and vehicle controls, respectively. The mixture was incubated with continuous shaking for 3 h at 37°C. 3E7 PrP antibody (2 μg) conjugated to magnetic beads and HRP-conjugated PrP T2 antibody (8 μg) were added to the pre-incubated mixture. After additional incubation for 1 h under the same conditions, the beads were separated and washed 3 times with TBST using a magnetic particle concentrator (Invitrogen, 120.20D). Luminescence signal was developed by adding Supersignal ELISA pico chemiluminescence substrate (Pierce, 37070) and quantified using a VICTOR3 microplate reader (Perkin Elmer, 1420–032).
Statistical analysis
Each experiment was repeated 3 times. A 1-way analysis of variance with Tukey-Kramer post hoc tests was conducted using Microsoft Excel. Differences were considered to be statistically significant when P < 0.05.
Results
Determination of optimal RT-QuIC condition
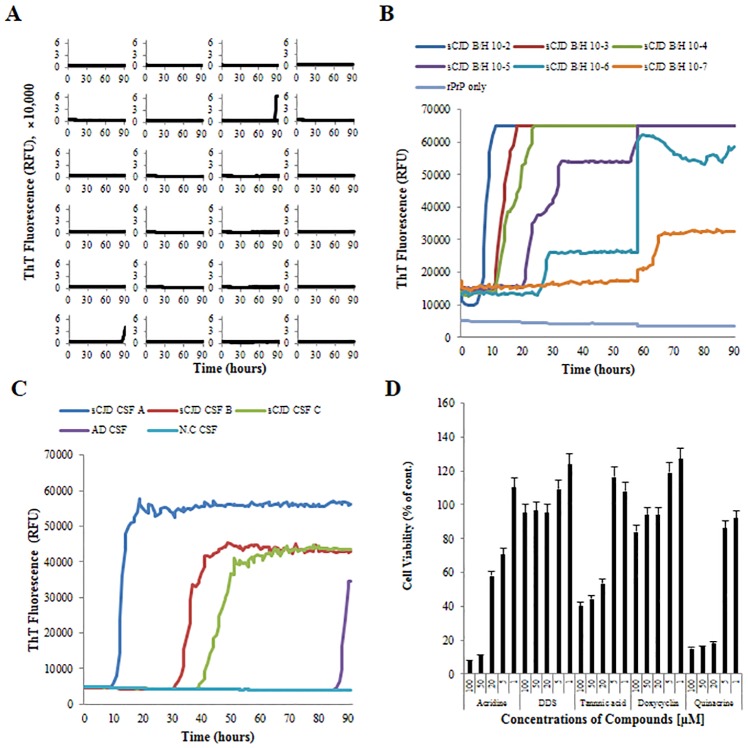
We optimized the RT-QuIC conditions prior to test compound screening. For a negative control, CSF samples from progressive dementia patients without sCJD (n = 24) were tested. Nonspecific self-aggregation was identified in 2 of 24 samples at more than 85 h (Fig 1A). For a positive control for brain seed, sCJD homogenates were serially diluted 10-fold from 10-2 to 10-7 and tested. ThT fluorescence was acutely increased in a concentration-dependent manner (Fig 1B). For a positive control for CSF, samples from sCJD and AD patients were tested. Fluorescence was increased in all sCJD CSF samples, and fluorescence in the AD CSF sample was increased after 85 h (Fig 1C). Accordingly, a 72 h time point was deemed to be reasonable and adopted for further study.
Fig 1. Optimization of RT-QuIC and determination of compound cytotoxicity.
Time point optimization was performed using CSF samples from 24 cases of progressive dementia (A) and validation was performed using serially diluted sCJD brain homogenate (B) and CSF from a sCJD or AD patient (C). Cell viability was assessed in the presence of 5 different compounds at 5 different concentrations between 1 μM and 100 μM (D). Values represent the mean of 3 independent experiments and the standard deviation is shown as error bars, P < 0.05.
Determination of compound concentrations for treatment
A MTT cytotoxicity assay was used to determine appropriate treatment concentrations for RT-QuIC and cell screening. Acridine was highly toxic at > 50 μM, tannic acid was non-toxic at < 20 μM, DSS and doxycycline permitted cell viability in excess of 90% at 100 μM. Quinacrine showed consistent toxicity (high toxicity at > 5 μM) as per a previous study [25]. Final concentrations between 1 and 20 μM were found to be optimal for treatment, and therefore, concentrations of 5 and 20 μM were selected for further study (Fig 1D).
RT-QuIC screening
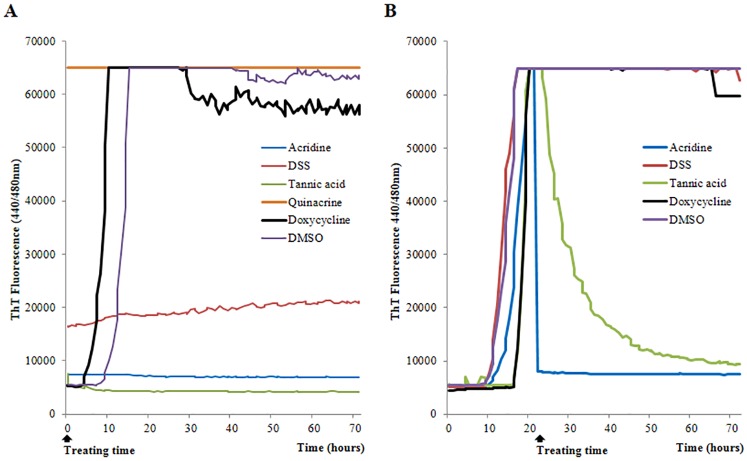
Acridine, DSS, tannic acid, and doxycycline were applied to RT-QuIC reactions at the specified times. Test wells that were treated with quinacrine were saturated at 65,000 rfu at the first reading and thus, were unable to show further amplification. Moreover, a visible yellow fluorescent color similar to that of ThT was observed during the preparation of stock solutions. Therefore, quinacrine was excluded from the following studies. Inhibition of prion propagation was observed at the initial reaction after treatment at 0 h. Increases in fluorescence were inhibited in wells that were treated with acridine or tannic acid compared to those treated with doxycycline or DMSO over the 72-h period. DSS treatment produced a slight fluorescence elevation that was less than 2-fold between the first and last readings (which was our criterion for a positive response) over the 72-h period. Alternatively, fluorescence was elevated to saturation in wells without any compound between 5 and 20 h. Thus, we confirmed that the generation of synthetic PrP aggregates was inhibited by acridine, DSS, and tannic acid (Fig 2A). Additionally, the elimination of fully aggregated PrP amyloid was observed after treatment at 22 h. Fluorescence reached saturation in untreated wells between 12 and 20 h. Thereafter, acridine, DSS, and tannic acid were added to the wells. Acridine and tannic acid reduced fluorescence to baseline levels during the total reaction. In particular, acridine took less time than tannic acid to reduce fluorescence to baseline level. DSS did not produce an observable fluorescence change and was comparable to the negative control. Thus, we confirmed that PrP aggregates were reduced by acridine and tannic acid treatments (Fig 2B).
Fig 2. Inhibitory effects in RT-QuIC screening.
Compounds were applied to RT-QuIC reactions at 0 h (A) and 22 h (B). Each sample was performed in quadruplicate. The average ThT fluorescence value of the 2 highest readings at 72 h per well was plotted against time. DMSO vehicle was used as a negative control.
Confirmation of PrPres in RT-QuIC products
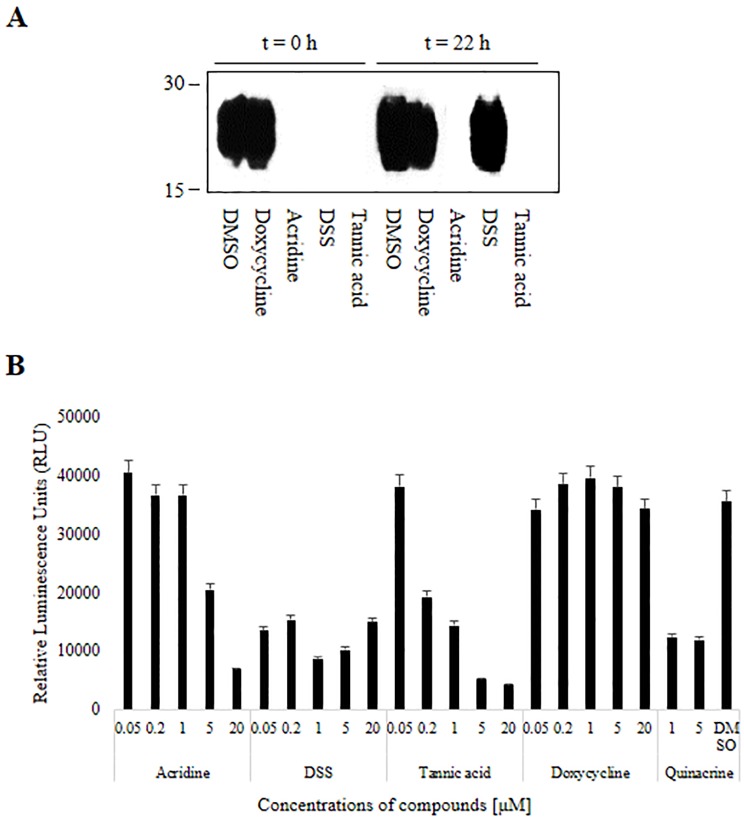
To exclude the possibility that compounds were affecting the ThT fluorescence dye rather than substrate PrPC or PrP aggregates, RT-QuIC products were collected from wells after the last reaction and PrPres was detected using western blotting. PrPres bands were not detected in RT-QuIC products from reactions that included acridine, DSS, or tannic acid at 0 h (Fig 3A, t = 0 h). PrPres was not detected in products from reactions that included acridine, DSS, or tannic acid, whereas PrPres was generated in wells with DMSO and doxycycline. Thus, we concluded that acridine, DSS, and tannic acid inhibited prion propagation in the RT-QuIC reaction and prevented the generation of PrPres. Additionally, PrPres bands were not detected in RT-QuIC products from reactions that included acridine and tannic acid at 22 h, whereas these bands were detected in products from reactions that included DSS at 22 h (Fig 3A, t = 22 h). DMSO and doxycycline controls showed consistent amounts of PrPres. Western blotting results were similar to RT-QuIC results. We therefore confirmed that flat or reduced fluorescence in our RT-QuIC results was related to effects of the compounds on PrPC or PrPres.
Fig 3. PrPres in RT-QuIC products shown by western blot and MDS.
PrPres was detected in reaction products after application of compounds to RT-QuIC reactions at 0 h or 22 h (A). Luminescence as a marker of PrPres formation is shown for each compound and concentration (B). Each compound was tested in duplicate in 3 independent experiments. Each value represents the mean and standard deviation; P < 0.05 and representative blots are shown.
Dye-independent conversion screening
To exclude the possibility that compounds were affecting the ThT fluorescence (i.e., ThT quenching) rather than substrate PrPC or PrPres aggregates, we performed a MDS enzyme-linked immunosorbent assay to quantify PrPres formation in a dye-independent conversion assay. PrPres was reduced by acridine and tannic acid in concentration-dependent manners from 0.05 to 20 μM. DSS also reduced PrPres at all concentrations tested. The IC50 values for acridine, tannic acid, and DSS were 5 μM, 0.2 μM, and 0.05 μM, respectively. Quinacrine produced a positive result and doxycycline produced a negative result in this assay (Fig 3B). Accordingly, acridine, DSS, tannic acid, doxycycline, and DMSO produced consistent results in RT-QuIC (t = 0 h) and in the present assay. Therefore, direct ThT quenching by the test compounds was unlikely in this study.
PrPSc-infected cell screening
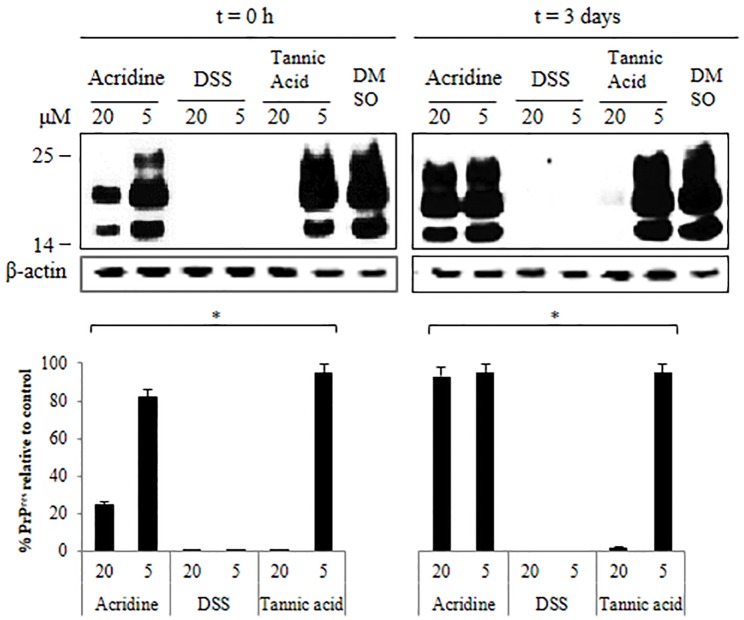
Acridine, DSS, and tannic acid were applied to PrPSc-infected cells at different concentrations (5 and 20 μM) at t = 0 h or t = 3 days. Relative amounts of PrPres produced in cells treated with compounds were very similar between the 2 time intervals. Large PrPres inhibitory effects were observed in response to 5 and 20 μM of DSS, while only 20 μM tannic acid produced a significant inhibitory effect in both experiments. Acridine only showed a partial inhibitory effect at 20 μM in the 0 h experiment (Fig 4, t = 0 h and t = 3 days). The inhibitory effects of acridine, dextran, and tannic acid in RT-QuIC and in cell-based screening are summarized in Table 2.
Fig 4. Western blots of PrPres in cell screening.
PrPres after cell screening with test compounds and subsequent cell lysis for western blotting. Each compound was tested in duplicate in 3 independent experiments. The relative band densities are shown as the volume intensity/mm2 relative to the β-actin band density. Each value represents the mean and standard deviation; P < 0.05 and representative blots are shown.
Table 2. Summary for anti-prion activities in RT-QuIC and cell-based screening.
| Compounds | RT-QuIC | Cell-based assay | ||
|---|---|---|---|---|
| Treatment t = 0 h | Treatment t = 22 h | Treatment t = 0 h | Treatment t = 3 days | |
| Acridine | +++ | +++ | +a | - |
| DSS | +++ | - | +++ | +++ |
| Tannic acid | +++ | +++ | ++b | ++b |
aPartial activity at high treatment only
bFull activity at high treatment only
Discussion
A majority of anti-prion compounds have been confirmed in vitro and to some extent in vivo [6, 8]. However, compounds can affect a variety of factors and components in cells, including DNA, protein, and metal/non-metal ions. Therefore, some compounds may not directly inhibit the conversion of PrPC or reduce the amplification of PrPres, but may have indirect effects on other cellular factors. Alternatively, the RT-QuIC assay has a relatively simple reaction environment. In the present study, possible environmental influences (especially ThT quenching by compounds) were in part excluded by performing follow-up western blotting of RT-QuIC reaction products and MDS. However, it is still unclear whether acridine inhibited PrPres and quenched ThT at the same time. It has been reported that, in dye-dependent seeded aggregation assays, low molecular weight substances can interfere with the fluorescence of dye compounds bound to amyloid fibrils [27]. Indeed, some compounds have been posited to compete with ThT for the same binding site on fibrils rather than directly inhibit the fibrilization process. Other studies have similarly described the interference of small molecules with ThT fluorescence during fibril formation [28, 29]. To address this issue, we performed a dye-independent conversion assay in order to determine whether the test compounds indeed inhibited fibrilization. We concluded that the compounds in this study were unlikely to produce ThT quenching.
The present study thus demonstrates that RT-QuIC allows anti-prion compounds to be tested easily, specifically, and reliably: first, RT-QuIC can test the ability of compounds to inhibit the conversion of PrPC to PrPres (i.e., RT-QuIC screening after compound application at 0 h). Acridine, DSS, and tannic acid clearly inhibited the conversion of PrPC to PrPres. Whether these compounds induce the inactivation of hot spot regions of PrPC related to structural conversion or inactivate small amounts of PrPSc in brain seed should be confirmed in future studies. Second, RT-QuIC can determine the ability of compounds to eliminate or reduce PrPres (i.e., RT-QuIC screening after compound application at 22 h/post-saturation). Acridine and tannic acid dramatically reduced PrPres aggregates in this paradigm; however, whether these compounds target the structural unfolding of PrPres or render pre-formed aggregates more PK-sensitive is an important topic for further study. Schmitz and colleagues suggested that their target compound bound seed particles in the RT-QuIC process and thus inhibited the transformation and aggregation steps [30]. Such a mechanism may be considered in the context of our findings.
It is important to note that, in our cell-based assays, the amount of PrPres was not determined at baseline or other pre-treatment time points, such that we could not guarantee a zero-level of PrPres in our t = 0 experiments. Treatment prior to the 24 h pre-incubation was not an option because immature cells are especially sensitive to non-toxic levels of test compounds and DMSO. Conversely, in cases of infection with PrPSc after compound treatment, a sufficient concentration of PrPres could not be ensured with short passage periods. All of these issues can be addressed using RT-QuIC; the absence or presence of PrPres can be confirmed by monitoring fluorescence during each reaction stage.
Given that amounts of PrPres could not be monitored in cells, we tried to mimic different amounts of PrPres in our RT-QuIC reactions by manipulating treatment times. We hypothesized that treatment at t = 0 h might permit compounds to affect the conversion of PrPC to PrPres easily in our reactions. Contrary to our expectation, relative amounts of PrPres were similar between the 2 time conditions (t = 0 and 3 days) in DSS and tannic acid. However, the activity of acridine was higher when applied at t = 0 h than 3 days at 20 μM. It can be considered that acridine far reduced PrPres by inhibiting the conversion of PrPC. However, there is no other existing evidence to support this activity given the past difficulty of monitoring PrPres in cells. Alternatively, our observation may have been due to a longer incubation period in the 0 h treatment condition (total incubation period = 6 days) than in the 3 days treatment condition (total incubation period = 3 days). Notably, several laboratories have employed an incubation period of 8 days or more in cell screening assays [9, 10]. Therefore, some anti-prion compounds may show activity when applied for longer incubation periods than reported.
While the inhibitory effect of acridine was weak in the cell screening assay, it had the greatest effect in the RT-QuIC screening assay. If only cell screening was used for drug development, acridine would be excluded from subsequent studies, whereas its activity in the RT-QuIC assay would qualify it for future screening and indicate the potential for multiple mechanisms of action. Tannic acid showed a delayed effect on the degradation of PrPres relative to acridine in RT-QuIC screening. This result suggests that the ability of acridine to degrade PrPres was much greater than that of tannic acid. DSS showed powerful anti-prion activity and minimal cytotoxicity in cell screening; however, it only showed inhibitory activity in RT-QuIC screening at t = 0 h. The observed differences for DSS may be evidence that DSS has a great inhibitory effect and preferentially block the conversion of PrPC. Therefore, the mechanism of action of DSS is likely related to its effects on the generation of PrPres.
We can also infer possible mechanisms of action for acridine, DSS, and tannic acid with reference to other studies. A previous study used Fourier transform infrared spectroscopy and hydrogen-deuterium exchange to demonstrate structural differences between PrPSc derived from different strains and PrPres generated by in vitro shaking, although the PK-resistant bands on western blotting were similar [31, 32]. The ScN2a cell line used in this study was infected with the RML strain, and RT-QuIC was based on in vitro shaking. Hence, it can be speculated that acridine was more effective at inhibiting PrPres structures derived from the RT-QuIC assay than PrPSc structures from the strain in ScN2a cells. Additionally, it has been reported that unknown cofactors used for the conversion of PrPC to PrPSc may be reduced during the PMCA reaction [33]. Another study showed that phosphatidylethanolamine was required as a cofactor for the amplification of PrPSc by a protein misfolding cyclic amplification [34]. One possible interpretation of our study is therefore that the drug performance of acridine may be interrupted by the presence of cofactors in cell screening. Alternatively, the activity of tannic acid was independent of the origin of PrPSc structures and cofactors, and consistent across both screening methods. Hence, tannic acid may have multiple mechanisms of action and be a candidate for advancement in future drug development studies.
Ideally, results from both RT-QuIC and cell screening should be consistent; however, we do not believe that it is necessary for these results to agree. Each screening method considers different mechanisms of action that are helpful for the further validation and understanding of anti-prion compounds. Of note, results from cell screening assays have not always been consistent with in vivo results. Many anti-prion compounds have only been studied in vivo as therapeutic agents given that (1) it is difficult to distinguish prophylactic versus therapeutic activity in vitro, (2) changes in PrPres concentration are not easily monitored in vitro, and (3) it is difficult to determine whether a given cell incubation stage is suitable for prophylactic or therapeutic study. We suggest that RT-QuIC can be a preferable tool for the discovery of anti-prion compounds because of its versatility (for determining prophylactic versus therapeutic potential), reliability, and consistency. Thus, this study not only provides insights into prion biology, but also offers information for future studies and for prion disease therapeutic development.
Acknowledgments
We would like to thank Dr. Chongsuk Ryou at Hanyang University in Korea for providing the ScN2a cell line and Dr. Sang Yun Kim at Seoul University Bundang Hospital in Korea for providing AD patient CSF.
Data Availability
Supporting data have been uploaded to figshare (https://figshare.com/s/6fae340e8e3d60830b9b).
Funding Statement
This work was supported by the Korea Centers for Disease Control and Prevention (4800-4838-303-210-13 and 2014-NG52003-00). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216(4542):136–44. Epub 1982/04/09. [DOI] [PubMed] [Google Scholar]
- 2.Bieschke J, Weber P, Sarafoff N, Beekes M, Giese A, Kretzschmar H. Autocatalytic self-propagation of misfolded prion protein. Proc Natl Acad Sci U S A. 2004;101(33):12207–11. Epub 2004/08/07. 10.1073/pnas.0404650101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manson JC, Jamieson E, Baybutt H, Tuzi NL, Barron R, McConnell I, et al. A single amino acid alteration (101L) introduced into murine PrP dramatically alters incubation time of transmissible spongiform encephalopathy. EMBO J. 1999;18(23):6855–64. Epub 1999/12/03. 10.1093/emboj/18.23.6855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song CH, Honmou O, Ohsawa N, Nakamura K, Hamada H, Furuoka H, et al. Effect of transplantation of bone marrow-derived mesenchymal stem cells on mice infected with prions. J Virol. 2009;83(11):5918–27. Epub 2009/03/20. 10.1128/JVI.00165-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Relano-Gines A, Lehmann S, Bencsik A, Herva ME, Torres JM, Crozet CA. Stem cell therapy extends incubation and survival time in prion-infected mice in a time window-dependant manner. J Infect Dis. 2011;204(7):1038–45. Epub 2011/09/02. 10.1093/infdis/jir484 [DOI] [PubMed] [Google Scholar]
- 6.Geissen M, Leidel F, Eiden M, Hirschberger T, Fast C, Bertsch U, et al. From high-throughput cell culture screening to mouse model: identification of new inhibitor classes against prion disease. ChemMedChem. 2011;6(10):1928–37. Epub 2011/07/15. 10.1002/cmdc.201100119 [DOI] [PubMed] [Google Scholar]
- 7.Bach S, Talarek N, Andrieu T, Vierfond JM, Mettey Y, Galons H, et al. Isolation of drugs active against mammalian prions using a yeast-based screening assay. Nat Biotechnol. 2003;21(9):1075–81. Epub 2003/08/12. 10.1038/nbt855 [DOI] [PubMed] [Google Scholar]
- 8.Solassol J, Crozet C, Lehmann S. Prion propagation in cultured cells. Br Med Bull. 2003;66:87–97. Epub 2003/10/03. [DOI] [PubMed] [Google Scholar]
- 9.Bate C, Langeveld J, Williams A. Manipulation of PrPres production in scrapie-infected neuroblastoma cells. J Neurosci Methods. 2004;138(1–2):217–23. Epub 2004/08/25. 10.1016/j.jneumeth.2004.04.001 [DOI] [PubMed] [Google Scholar]
- 10.Stanton JB, Schneider DA, Dinkel KD, Balmer BF, Baszler TV, Mathison BA, et al. Discovery of a novel, monocationic, small-molecule inhibitor of scrapie prion accumulation in cultured sheep microglia and Rov cells. PLoS One. 2012;7(11):e51173 Epub 2012/12/12. 10.1371/journal.pone.0051173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon D, Herva ME, Benitez MJ, Garrido JJ, Rojo AI, Cuadrado A, et al. Dysfunction of the PI3K-Akt-GSK-3 pathway is a common feature in cell culture and in vivo models of prion disease. Neuropathol Appl Neurobiol. 2014;40(3):311–26. Epub 2013/06/08. 10.1111/nan.12066 [DOI] [PubMed] [Google Scholar]
- 12.Wilham JM, Orru CD, Bessen RA, Atarashi R, Sano K, Race B, et al. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 2010;6(12):e1001217 Epub 2010/12/15. 10.1371/journal.ppat.1001217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atarashi R, Satoh K, Sano K, Fuse T, Yamaguchi N, Ishibashi D, et al. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nat Med. 2011;17(2):175–8. Epub 2011/02/01. 10.1038/nm.2294 [DOI] [PubMed] [Google Scholar]
- 14.Orru CD, Bongianni M, Tonoli G, Ferrari S, Hughson AG, Groveman BR, et al. A test for Creutzfeldt-Jakob disease using nasal brushings. N Engl J Med. 2014;371(6):519–29. Epub 2014/08/08. 10.1056/NEJMoa1315200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vascellari S, Orru CD, Hughson AG, King D, Barron R, Wilham JM, et al. Prion seeding activities of mouse scrapie strains with divergent PrPSc protease sensitivities and amyloid plaque content using RT-QuIC and eQuIC. PLoS One. 2012;7(11):e48969 Epub 2012/11/10. 10.1371/journal.pone.0048969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orru CD, Wilham JM, Raymond LD, Kuhn F, Schroeder B, Raeber AJ, et al. Prion disease blood test using immunoprecipitation and improved quaking-induced conversion. MBio. 2011;2(3):e00078–11. Epub 2011/05/12. 10.1128/mBio.00078-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson DM, Manca M, Haley NJ, Denkers ND, Nalls AV, Mathiason CK, et al. Rapid antemortem detection of CWD prions in deer saliva. PLoS One. 2013;8(9):e74377 Epub 2013/09/17. 10.1371/journal.pone.0074377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elder AM, Henderson DM, Nalls AV, Wilham JM, Caughey BW, Hoover EA, et al. In vitro detection of prionemia in TSE-infected cervids and hamsters. PLoS One. 2013;8(11):e80203 Epub 2013/11/14. 10.1371/journal.pone.0080203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villa V, Tonelli M, Thellung S, Corsaro A, Tasso B, Novelli F, et al. Efficacy of novel acridine derivatives in the inhibition of hPrP90-231 prion protein fragment toxicity. Neurotox Res. 2011;19(4):556–74. Epub 2010/04/21. 10.1007/s12640-010-9189-8 [DOI] [PubMed] [Google Scholar]
- 20.Adjou KT, Simoneau S, Sales N, Lamoury F, Dormont D, Papy-Garcia D, et al. A novel generation of heparan sulfate mimetics for the treatment of prion diseases. J Gen Virol. 2003;84(Pt 9):2595–603. Epub 2003/08/15. 10.1099/vir.0.19073-0 [DOI] [PubMed] [Google Scholar]
- 21.Kocisko DA, Baron GS, Rubenstein R, Chen J, Kuizon S, Caughey B. New inhibitors of scrapie-associated prion protein formation in a library of 2000 drugs and natural products. J Virol. 2003;77(19):10288–94. Epub 2003/09/13. 10.1128/JVI.77.19.10288-10294.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Getz TM, Manne BK, Buitrago L, Mao Y, Kunapuli SP. Dextran sulphate induces fibrinogen receptor activation through a novel Syk-independent PI-3 kinase-mediated tyrosine kinase pathway in platelets. Thromb Haemost. 2013;109(6):1131–40. Epub 2013/04/11. 10.1160/TH12-09-0645 [DOI] [PubMed] [Google Scholar]
- 23.Cramm M, Schmitz M, Karch A, Zafar S, Varges D, Mitrova E, et al. Characteristic CSF prion seeding efficiency in humans with prion diseases. Mol Neurobiol. 2015;51(1):396–405. Epub 2014/05/09. 10.1007/s12035-014-8709-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGuire LI, Peden AH, Orru CD, Wilham JM, Appleford NE, Mallinson G, et al. Real time quaking-induced conversion analysis of cerebrospinal fluid in sporadic Creutzfeldt-Jakob disease. Ann Neurol. 2012;72(2):278–85. Epub 2012/08/29. 10.1002/ana.23589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyeon JW, Choi J, Kim SY, Govindaraj RG, Jam Hwang K, Lee YS, et al. Discovery of Novel Anti-prion Compounds Using In Silico and In Vitro Approaches. Sci Rep. 2015;5:14944 Epub 2015/10/10. 10.1038/srep14944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.An SS, Lim KT, Oh HJ, Lee BS, Zukic E, Ju YR, et al. Differentiating blood samples from scrapie infected and non-infected hamsters by detecting disease-associated prion proteins using Multimer Detection System. Biochem Biophys Res Commun. 2010;392(4):505–9. 10.1016/j.bbrc.2010.01.053 [DOI] [PubMed] [Google Scholar]
- 27.Noormagi A, Primar K, Tougu V, Palumaa P. Interference of low-molecular substances with the thioflavin-T fluorescence assay of amyloid fibrils. J Pept Sci. 2012;18(1):59–64. 10.1002/psc.1416 [DOI] [PubMed] [Google Scholar]
- 28.Hudson SA, Ecroyd H, Kee TW, Carver JA. The thioflavin T fluorescence assay for amyloid fibril detection can be biased by the presence of exogenous compounds. FEBS J. 2009;276(20):5960–72. 10.1111/j.1742-4658.2009.07307.x [DOI] [PubMed] [Google Scholar]
- 29.Jameson LP, Smith NW, Dzyuba SV. Dye-binding assays for evaluation of the effects of small molecule inhibitors on amyloid (abeta) self-assembly. ACS Chem Neurosci. 2012;3(11):807–19. 10.1021/cn300076x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitz M, Cramm M, Llorens F, Candelise N, Muller-Cramm D, Varges D, et al. Application of an in vitro-amplification assay as a novel pre-screening test for compounds inhibiting the aggregation of prion protein scrapie. Sci Rep. 2016;6:28711 10.1038/srep28711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smirnovas V, Baron GS, Offerdahl DK, Raymond GJ, Caughey B, Surewicz WK. Structural organization of brain-derived mammalian prions examined by hydrogen-deuterium exchange. Nat Struct Mol Biol. 2011;18(4):504–6. Epub 2011/03/29. 10.1038/nsmb.2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sano K, Atarashi R, Nishida N. Structural conservation of prion strain specificities in recombinant prion protein fibrils in real-time quaking-induced conversion. Prion. 2015;9(4):237–43. Epub 2015/08/19. 10.1080/19336896.2015.1062201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim JI, Cali I, Surewicz K, Kong Q, Raymond GJ, Atarashi R, et al. Mammalian prions generated from bacterially expressed prion protein in the absence of any mammalian cofactors. J Biol Chem. 2010;285(19):14083–7. Epub 2010/03/23. 10.1074/jbc.C110.113464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deleault NR, Piro JR, Walsh DJ, Wang F, Ma J, Geoghegan JC, et al. Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc Natl Acad Sci U S A. 2012;109(22):8546–51. Epub 2012/05/16. 10.1073/pnas.1204498109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supporting data have been uploaded to figshare (https://figshare.com/s/6fae340e8e3d60830b9b).