Abstract
Spontaneous neural differentiation of embryonic stem cells is induced by Noggin-mediated inhibition of bone morphogenetic protein 4 (BMP4) signaling. RhoA is a guanosine triphosphatase (GTPase) that regulates cytoskeletal dynamics and gene expression, both of which control stem cell fate. We found that disruption of Syx, a gene encoding a RhoA-specific guanine nucleotide exchange factor, accelerated retinoic acid–induced neural differentiation in murine embryonic stem cells aggregated into embryoid bodies. Cells from Syx+/+ and Syx−/− embryoid bodies had different abundances of proteins implicated in stem cell pluripotency. The differentiation-promoting proteins Noggin and RARγ (a retinoic acid receptor) were more abundant in cells of Syx−/− embryoid bodies, whereas the differentiation-suppressing proteins SIRT1 (a protein deacetylase) and the phosphorylated form of SMAD1 (the active form of this transcription factor) were more abundant in cells of Syx+/+ embryoid bodies. These differences were blocked by the overexpression of constitutively active RhoA, indicating that the abundance of these proteins was maintained, at least in part, by RhoA activity. The peripheral stress fibers in cells from Syx−/− embryoid bodies were thinner than those in Syx+/+ cells. Furthermore, less Noggin and fewer vesicles containing Rab3d, a GTPase that mediates Noggin trafficking, were detected in cells from Syx−/− embryoid bodies, which could result from increased Noggin exocytosis. These results suggested that, in addition to inhibiting Noggin transcription, RhoA activity in wild-type murine embryonic stem cells also prevented neural differentiation by limiting Noggin secretion.
INTRODUCTION
Embryonic stem cells (ESCs) are pluripotent, maintaining the potential to differentiate into various somatic cell types. Understanding the molecular mechanisms that control ESC differentiation is relevant to both basic research and clinical applications. This is pertinent particularly for neuronal differentiation, the understanding of which is key to inducing nerve regeneration. Both intrinsic and extrinsic factors control ESC fate. The intrinsic factors Oct4, Nanog, and SOX2 are the core transcription factors that confer ESC self-renewal and pluripotency (1–4). Extrinsically, several cytokines, including bone morphogenetic protein (BMP) (5) and Wnt-β-catenin (6–10), play important roles in mouse ESC (mESC) renewal and differentiation. Among the extrinsic signals, BMPs are crucial for directing both self-renewal and differentiation of ESCs (11–14). BMPs are members of the transforming growth factor–β (TGFβ) superfamily that modulates gene expression through the small mothers against decapentaplegic (SMAD) transcription factors. Through the receptor-regulated SMADs SMAD1 and SMAD5 and the common-mediator SMAD SMAD4, BMP4 attenuates extracellular signal–regulated kinase (ERK) activity by stimulating dual specificity protein phosphatase 9 (DUSP9) (15). This reinforces ESC self-renewal in response to leukemia inhibitory factor (LIF) (15, 16). When deprived of LIF, murine ESCs (mESCs) automatically undergo early neural differentiation caused by a reduction in the abundance of signal transducer and activator of transcription 3 (STAT3) and the concomitant conversion of BMP4 from supporting self-renewal to promoting lineage commitment (15, 17). BMP4 signaling can be augmented by several extrinsic factors, such as valproic acid (VPA), a chemical inhibitor of glycogen synthase kinase 3β that increases the expression of Bmp4 mRNA and the abundance of BMP4 protein (18). BMP4 signaling is inhibited by the antagonistic factor Noggin, which interferes with its binding to the BMP receptor (BMPR) (19). The inhibition of BMP4 signaling by Noggin induces neural differentiation by activating the phosphatidylinositol 3-kinase/Akt signaling pathway (20) and by increasing Pax6 expression (21).
Retinoic acid (RA) is a biologically active form of vitamin A that plays an important role in neural differentiation (22–24). High concentration of RA promotes neural gene expression and represses mesodermal gene expression during embryoid body (EB) formation (23, 25). Moreover, RA can promote the degradation of phosphorylated (active) SMAD1 and antagonize BMP and SMAD signaling (26). The effects of RA are mediated by specific nuclear RA receptors (RARs) that heterodimerize with retinoid X receptors to induce transcription of target genes. RA signaling is modulated by SIRT1, a nuclear nicotinamide adenine dinucleotide (NAD+)–dependent protein deacetylase (27) that deacetylates cellular RA-binding protein II (CRABPII) (28). CRABPII is required for RA translocation into the nucleus to facilitate RA binding to RARs (29–31).
Rho guanosine triphosphatases (GTPases) are key intracellular signal mediators that transduce extracellular stimuli to the cytoskeleton. They control intercellular adhesion, cell polarity, and migration, as well as gene expression (32, 33). RhoA is required in early embryogenesis for the maintenance of intercellular junctions in mESCs (34) and for BMP2-induced osteogenesis (35). Moreover, previous studies implicated RhoA in the inhibition of neural differentiation (36, 37). Rho family proteins are activated by guanine nucleotide exchange factors (GEFs) (38). We have previously shown that the gene encoding the RhoA-specific GEF Syx (39–41) (also called PLEKHG5 or Tech), which is expressed in human ESCs (42), is required for vascular development in the mouse and the zebrafish (43). We therefore sought to determine the role of Syx in mESC differentiation.
We compared differentiation of Syx+/+ and Syx−/− mESCs in EBs, a common approach for modeling early embryonic development (44). In the absence of Syx, mESCs underwent accelerated neural differentiation. This was partially prevented by rescuing Syx−/− cells with Syx or a constitutively active form of RhoA (CA-RhoA). We determined that the balance of pluripotency-conserving versus differentiation-inducing factors favors neural differentiation of Syx−/− cells. In addition, we identified a relationship between RhoA activity and Noggin’s effects on differentiation by comparing the filamentous actin (F-actin) patterns in Syx+/+ and Syx−/− cells. We found that the peripheral stress fibers were thinner in Syx−/− cells, whereas these cells’ contents of Noggin and of Rab3d, the GTPase that mediates the trafficking of Noggin-containing vesicles, were lower than in Syx+/+ cells. On the basis of these findings, we propose that RhoA prevents the differentiation of stem cells, at least in part, by maintaining a physical barrier that interferes with the secretion of Noggin-loaded vesicles at the cell membrane.
RESULTS
Loss of Syx accelerates RA-dependent neural differentiation of mESCs
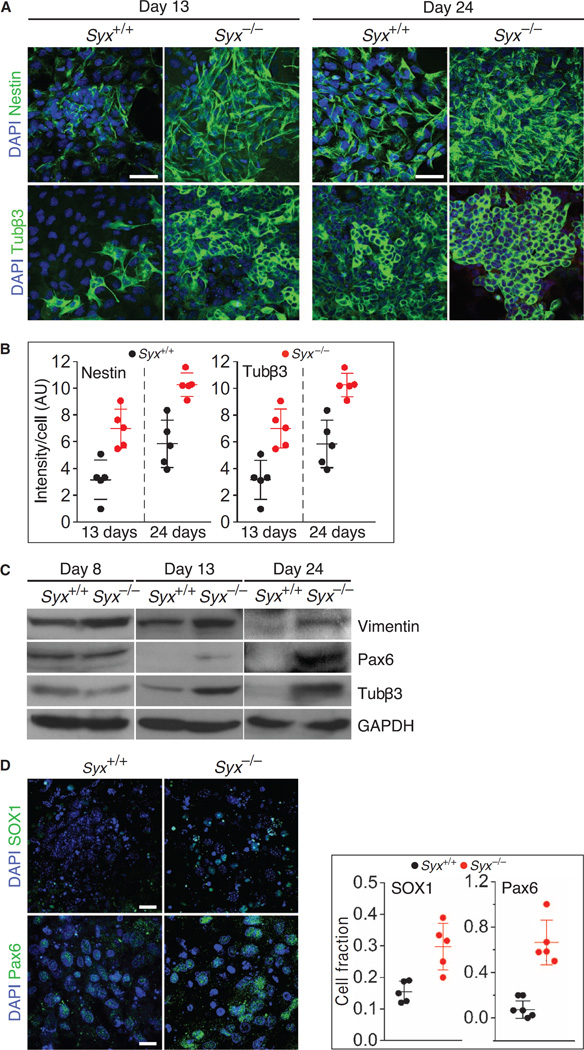
We compared the pluripotency of Syx+/+ and Syx−/− mESCs by immunofluorescence (fig. S1A; full-length immunoblots are shown in fig. S4) and immunoblotting (fig. S1B) of the core transcription factors that confer stemness— Oct4, Nanog, and SOX2. The abundance of each protein was similar in both assays. The ablation of Syx is expected to reduce cellular RhoA activity, rather than abolish it, because mESCs likely express other genes encoding RhoA-specific GEFs. We confirmed that RhoA activity was reduced in Syx−/− cells (fig. S1C).
To compare the spontaneous differentiation of RA-naïve Syx+/+ and Syx−/− mESCs, we analyzed the development of EBs of each genotype into the three germ layers using markers for the ectodermal, mesodermal, and endodermal lineages. The expression of ectodermal and endodermal genes was up to 15 or 14 times higher, respectively, in Syx−/− EBs, whereas that of mesodermal markers was reduced by more than half (fig. S1D). Because neural progenitor cells originate from the ectoderm (45), we compared the abundances of the neural differentiation markers tubulin b3 (Tubβ3) (46) and vimentin (47) in Syx+/+ and Syx−/− EBs. Both Tubβ3 and vimentin were present in spontaneously differentiating Syx−/− EBs, but were not detectable in Syx+/+ EBs (fig. S1E), in agreement with the increased expression of ectodermal genes in Syx−/− mESCs. We compared the neural differentiation of Syx+/+ and Syx−/− EBs by testing their responses to RA, which drives this process (20, 21), using Tubβ3 and the neural differentiation marker nestin (48). We noted an increase in the abundance of nestin and Tubβ3 in Syx−/− cells compared to Syx+/+ cells at days 13 and 24 after EB aggregation (Fig. 1A). The abundance of these proteins was significantly higher in Syx−/− cells at both time points (Fig. 1B). We selected vimentin and the neural progenitor cell marker Pax6 (44, 49, 50) to follow the progress of neural differentiation in EBs by immunoblotting. The differences between these neural differentiation markers in cells from Syx−/− and Syx+/+ EBs were similar to the immunofluorescence results. At day 8, the abundance of the markers was similar in Syx+/+ and Syx−/− EBs, but at days 13 and 24, the abundance of both markers was substantially higher in Syx−/− EBs (Fig. 1C).
Fig. 1. Neural differentiation in cells from Syx+/+ and Syx−/− EBs.
(A) Immunofluorescence images of nestin- or Tubβ3-labeled cells that were dissociated from RA-treated Syx+/+ and Syx−/− EBs at the indicated times (scale bars, 50 µm). DAPI, 4′,6-diamidino-2-phenylindole. (B) Histograms quantifying nestin and Tubβ3 immunofluorescence intensities shown in (A) [two-way analysis of variance (ANOVA), n = 5 fields, each containing >90 Syx+/+ cells or >130 Syx−/− cells, acquired in two independent experiments; mean ± SD, P < 0.001 for all differences]. AU, arbitrary units. (C) Immunoblot of neural differentiation markers vimentin, Pax6, and Tubβ3 in one of two independent experiments with cells dissociated from RA-treated Syx+/+ and Syx−/− EBs at the indicated time points. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as a loading control. (D) Immunofluorescence images of the neural progenitor cell markers SOX1 and Pax6 in cells from RA-treated Syx+/+ and Syx−/− EBs 12 days after EB aggregation (scale bars, 50 µm). Quantification of the immunofluorescence of each marker is shown in the adjoining histograms [n = 5 fields containing 138 (Pax6, Syx+/+), 82 (Pax6, Syx−/−), 122 (SOX1, Syx+/+), and 93 (SOX1, Syx−/−) cells from two independent experiments; Pax6: 26.2 higher odds for the presence of Pax6 in Syx−/− rather than in Syx+/+ cells (95% confidence interval, 10.6 to 64.8; P < 0.001); SOX1: 2.4 higher odds for the presence of SOX1 in Syx−/− rather than in Syx+/+ cells (95% confidence interval, 1.6 to 3.4; P < 0.001)].
Because neural stem cells progress to progenitor cells before fully differentiating to neurons, we quantified the fractions of cells of Syx+/+ and Syx−/− EBs that were neural progenitors using Pax6 and SOX1 (21), and the fractions that were neurons with the nuclear neuronal marker FOX3 (51). The fraction of cells from Syx−/− EBs that were progenitor cells was at least twice that of cells from Syx+/+ EBs (Fig. 1D). As expected, at this relatively early stage of differentiation, no neurons were present in cells from either Syx+/+ or Syx−/− EBs because we did not detect FOX3 in any cell nuclei (fig. S1F).
Syx and RhoA inhibit neural differentiation
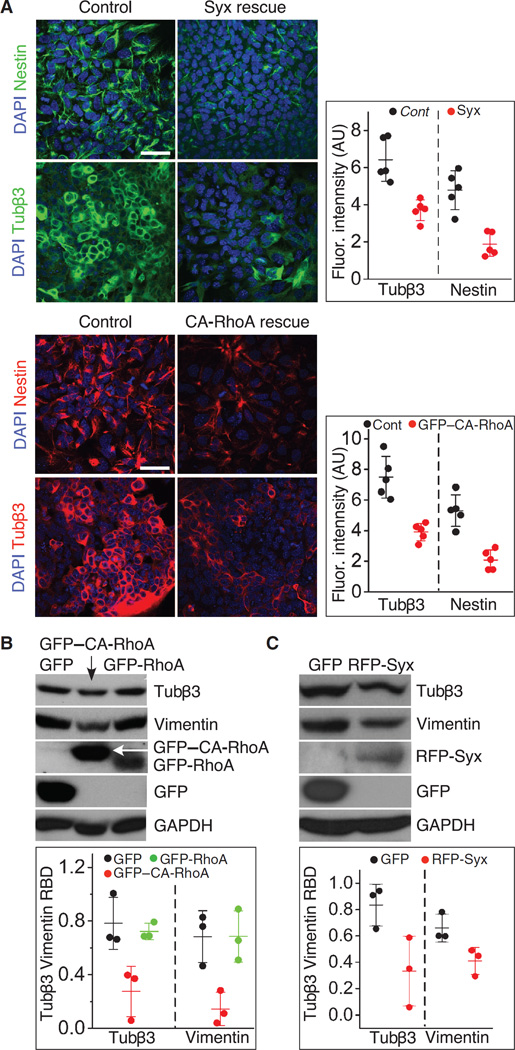
To verify that the accelerated neural differentiation was caused specifically by the deletion of Syx, we rescued cells that were dissociated from RA-treated Syx−/− EBs by transfecting Syx (fig. S2A). The reintroduction of Syx into Syx−/− mESCs reduced the abundance of nestin and Tubβ3 compared to the negative control cells that were transfected with an empty plasmid. The amounts of both proteins were significantly lower in the rescued cells (Fig. 2A). Because Syx activates RhoA, we rescued cells from RA-treated Syx−/− EBs by transfection with CA-RhoA (RhoA-Q63L) (fig. S2B) (52). Similar to cells rescued by Syx, CA-RhoA–transfected cells produced less nestin and Tubβ3 than the cells transfected with an empty vector control (Fig. 2A). The difference was evident also in the immunoblots of green fluorescent protein (GFP)–CA-RhoA– or GFP-RhoA–expressing cells and negative control cells (Fig. 2B). Rescue with red fluorescent protein (RFP)–Syx had a similar effect on Tubβ3 and nestin abundance (Fig. 2C). This effect could result from Syx activities other than guanine exchange. Collectively, these results suggest that inactivation of RhoA enhances neural differentiation.
Fig. 2. Expression of Syx or CA-RhoA reduced neural differentiation in cells from Syx−/− EBs.
(A) Immunofluorescence images of cells dissociated from RA-treated Syx−/− EBs, transfected with RFP-Syx or GFP–CA-RhoA, and immunolabeled as indicated. GFP was used as a negative control. The adjoining histograms show the quantification of Tubβ3 and nestin immunofluorescence in images of cells from each group (two-sample t test, equal variance, n = 5 fields from three independent experiments, each containing >130 cells, mean ± SD; Syx rescue: P = 0.003 for Tubβ3, P < 0.001 for nestin; CA-RhoA rescue: P = 0.012 for Tubβ3, P < 0.001 for nestin). (B) Immunoblot of the neural differentiation markers Tubβ3 and nestin in cells from RA-treated Syx+/+ EBs transfected with GFP, GFP-RhoA, or GFP–CA-RhoA. The histogram shows the quantification of Tubβ3 and vimentin band densities normalized to GAPDH band density in cells transfected with either Syx or CA-RhoA (one-way ANOVA, n = 3 independent experiments, mean ± SD; Tubβ3: P = 0.015 for overall difference between the three groups, P = 0.008 for GFP–CA-RhoA to GFP, P = 0.0013 for GFP– CA-RhoA to GFP-RhoA; vimentin: P = 0.013 for overall difference between the three groups, P = 0.009 for GFP–CA-RhoA to GFP, P = 0.009 for GFP– CA-RhoA to GFP-RhoA). RBD, relative band density. (C) As in (B), in Syx−/− cells rescued by GFP or by RFP-Syx (two-sample t test, equal variance, n = 3 independent experiments, mean ± SD; P = 0.048 for Tubβ3, P = 0.043 for vimentin). GAPDH is a loading control.
Loss of Syx antagonizes BMP4 signaling by reducing SMAD1 phosphorylation
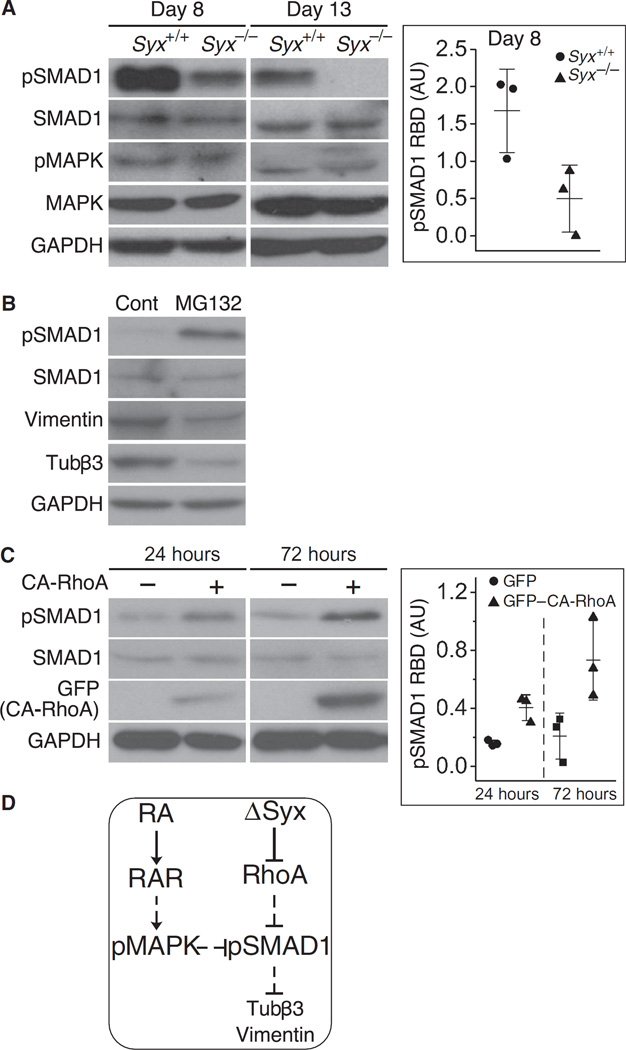
Phosphorylation of SMAD1 by BMPRs (53) is critical for the transcriptional response to BMP4, which mediates neural differentiation (26, 54). To test whether Syx is involved in BMP4 signaling, we compared the phosphorylation of SMAD1 in cells dissociated from Syx+/+ or Syx−/− EBs at several time points. The phosphorylation of SMAD1 decreased in Syx−/− cells to below the detection threshold by day 13, but not in Syx+/+ cells (Fig. 3A). Furthermore, the phosphorylation of mitogen-activated protein kinase (MAPK) on day 13 was higher in Syx−/− cells than in Syx+/+ cells (Fig. 3A). The phosphorylation and subsequent activation of MAPK, mediated through RA-induced increase in the production of Gadd45, which binds to and activates the upstream MAPK kinase kinase (55), are required for RA-promoted degradation of phosphorylated SMAD (26). To examine the impact of phosphorylated SMAD1 in Syx−/− EBs on neural differentiation, we treated EB-dissociated cells with MG132, a pharmacological inhibitor of proteasome-mediated degradation (26, 56). Immunoblotting showed that treatment by MG132 partially restored the phosphorylation of SMAD1 and delayed the neural differentiation of Syx−/− cells (Fig. 3B).
Fig. 3. SMAD1 phosphorylation negatively correlated with neural differentiation.
(A) Immunoblots showing the phosphorylation of SMAD1 (pSMAD1) and MAPK (pMAPK) in cells dissociated from RA-treated Syx+/+ or Syx−/− EBs at the indicated times (two-sample t test, equal variance, n = 3 independent experiments, mean ± SD, P = 0.049). GAPDH is a loading control. (B) Cells dissociated from RA-treated Syx+/+ or Syx−/− EBs 4 days after aggregation were treated with MG132 to inhibit proteasomal degradation or with dimethyl sulfoxide as a negative control (Cont) followed by immunoblotting with the indicated antibodies (one of two independent experiments). (C) Immunoblots of cells from RA-treated Syx−/− EBs transfected with GFP–CA-RhoA and probed at the indicated times to detect pSMAD1 (two-sample t test, equal variance, n = 3 independent experiments, mean ± SD, P = 0.0095 for 24 hours, P = 0.046 for 72 hours). (D) Signaling scheme representing the data shown in (A) to (C). Solid lines represent direct regulatory events, whereas dashed lines represent events that either are indirect or have not been shown to be direct.
To determine whether the difference in SMAD1 phosphorylation between Syx+/+ and Syx−/− cells was caused by inactivation of RhoA, we rescued cells dissociated from Syx−/− EBs by GFP–CA-RhoA. The phosphorylation of SMAD1 in these cells increased as differentiation progressed (Fig. 3C), suggesting that disruption of Syx decreased SMAD1 phosphorylation, thus impairing BMP4 signaling in RA-treated cells. These results suggest that the ablation of Syx contributes to RA-induced acceleration of neural differentiation by reducing the abundance of phosphorylated SMAD1 (Fig. 3D).
RAR abundance is increased in Syx−/− EBs
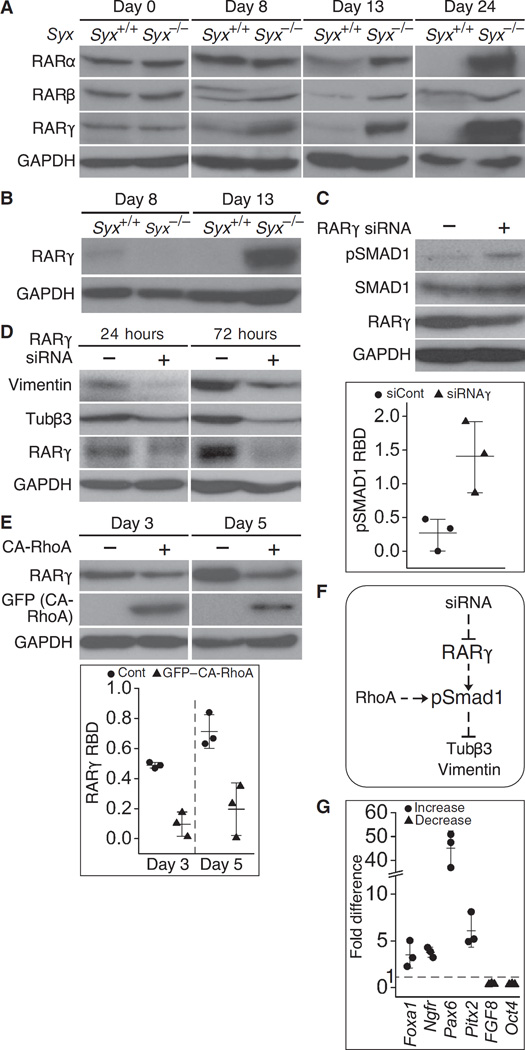
RA modulates developmental processes that are controlled by growth factors, including neurogenesis (57). Although part of the RA absorbed by a cell is oxidized and degraded, the remaining RA can serve as a ligand for nuclear RARs (RARα, RARβ, and RARγ), which bind to DNA and promote transcription when liganded (30, 58). We compared the abundances of RARα, RARβ, and RARγ in cells dissociated from Syx+/+ or Syx−/− EBs at several time points after aggregation. On day 0 (before aggregation and RA treatment) and on day 8 (after aggregation and RA treatment), the amount of the RAR isoforms did not differ substantially between Syx+/+ and Syx−/− cells (Fig. 4A). On days 13 and 24 (after aggregation and RA treatment), however, the amount of each RAR isoform was considerably higher in Syx−/− cells compared to Syx+/+ cells (Fig. 4A), indicating that Syx−/− EBs are more responsive to RA. Even in the absence of RA, RARγ was abundant in Syx−/− EBs at day 13, arguing that the loss of Syx alone was sufficient to increase RARγ production (Fig. 4B). Silencing RARγ in cells dissociated from RA-treated Syx−/− EBs resulted in higher SMAD1 phosphorylation (Fig. 4C), in agreement with previous studies (26).
Fig. 4. RAR production was increased in cells from Syx−/− EBs.
(A) Immunoblot of RARs at the indicated times in RA-treated Syx+/+ and Syx−/− EBs (one of two independent experiments). GAPDH is a loading control. (B) Immunoblot showing RARγ production at day 13 in Syx−/− EBs in the absence of RA (one of two independent experiments). (C) Immunoblot showing phosphorylated SMAD1 (pSMAD1) after RARγ knockdown in cells dissociated from RA-treated Syx−/− EBs. Quantification of normalized pSMAD1 band density is shown in the histogram (two-sample t test, unequal variance, n = 3 independent experiments, mean ± SD, P = 0.047). (D). Immunoblot showing markers of neural differentiation vimentin and Tubβ3 after RARγ knockdown in cells dissociated from RA-treated Syx−/− EBs (one of two independent experiments). (E) Immunoblot showing RARγ in cells dissociated from Syx−/− EBs at the indicated times and transfected with GFP–CA-RhoA. Quantification is shown in the histogram (two-way ANOVA, n = 3 independent experiments, mean ± SD, P < 0.001 for both times). (F) Signaling scheme representing the data shown in (A) to (E). Solid lines represent direct regulatory events, whereas dashed lines represent events that either are indirect or have not been shown to be direct. (G) Quantitative real-time polymerase chain reaction (qRT-PCR) measurements of mRNA abundances of the indicated genes in Syx−/− relative to Syx+/+ cells from 13-day-old RA-naïve EBs (n = 3 replicates).
To test whether RARs affect neural differentiation, we knocked down Rarγ by small interfering RNA (siRNA). Neural differentiation was delayed, as indicated by the reduced abundances of Tubβ3 and vimentin (Fig. 4D). To determine the effect of RhoA activity on RARγ production, we rescued cells from dissociated Syx−/− EBs by transfection of GFP–CA-RhoA. RARγ abundance was lower than in GFP-transfected control cells, indicating that active RhoA reduced RARγ production (Fig. 4E). These results suggest that, in the absence of RA signaling, neural differentiation is slowed down as a result of increase in the abundance of phosphorylated SMAD1 (Fig. 4F).
The increased responsiveness to RA suggested that the loss of Syx may have deregulated RA signaling in Syx−/− EBs. To test this premise, we compared the expression of known RA target genes in Syx+/+ and Syx−/− EBs that were not treated with RA. We have already shown that the protein encoded by one of these genes, Pax6 (21), was substantially more abundant in RA-treated Syx−/− EBs (Fig. 1, C and D). Similarly, qRT-PCR results yielded a 45-fold higher expression of Pax6 in RA-naïve Syx−/− EBs (fig. S1D). We selected additional RA target genes for which expression was shown to either increase [Ngfr (59), Foxa1 (60), and Pitx2 (61)] or decrease [Pou5f1/Oct4 (62) and Fgf8 (63)] in response to RA. The expression of these genes in RA-naïve Syx−/− relative to Syx+/+ EBs had similar increase or decrease patterns (Fig. 4G), supporting the notion that the loss of Syx sensitizes mESCs to RA.
Neural differentiation in EBs is inhibited by BMP4 and augmented by Noggin
The balance between BMP4 and its antagonist Noggin determines mESC fate (64). We compared BMP4 and Noggin abundances in cells dissociated from Syx+/+ and Syx−/− EBs. BMP4 abundance was similar in cells from Syx+/+ and Syx−/− EBs during neural differentiation (fig. S3A). Likewise, the expression of the genes encoding the BMP4 receptors did not differ significantly between the two genotypes (fig. S3B). In contrast, Noggin was not detected in cells from Syx+/+ EBs, whereas it was abundant in cells from Syx−/− EBs 8 and 13 days after aggregation (fig. S3A).
To probe the functions of BMP4 and Noggin in the neural differentiation of Syx−/− mESCs, we applied two methods for changing the balance of their production. First, we treated EBs with VPA, which increases BMP4 production by activating the Wnt–β-catenin pathway (18). Second, we knocked down the expression of endogenous Noggin by siRNA. The abundance of nestin and Tubβ3 decreased in Syx−/− mESCs after VPA treatment (fig. S3C), whereas bmp4 expression increased by 80% as measured by qRT-PCR (fig. S3D), as expected. Similarly, Noggin knockdown reduced the abundance of the neural markers vimentin and Tubβ3 (fig. S3E). Conversely, overexpression of Noggin in Syx+/+ cells increased vimentin and Tubβ3 abundances (fig. S3F).
RhoA reduces Noggin expression, and RARγ binds Syx and rhophilin-2
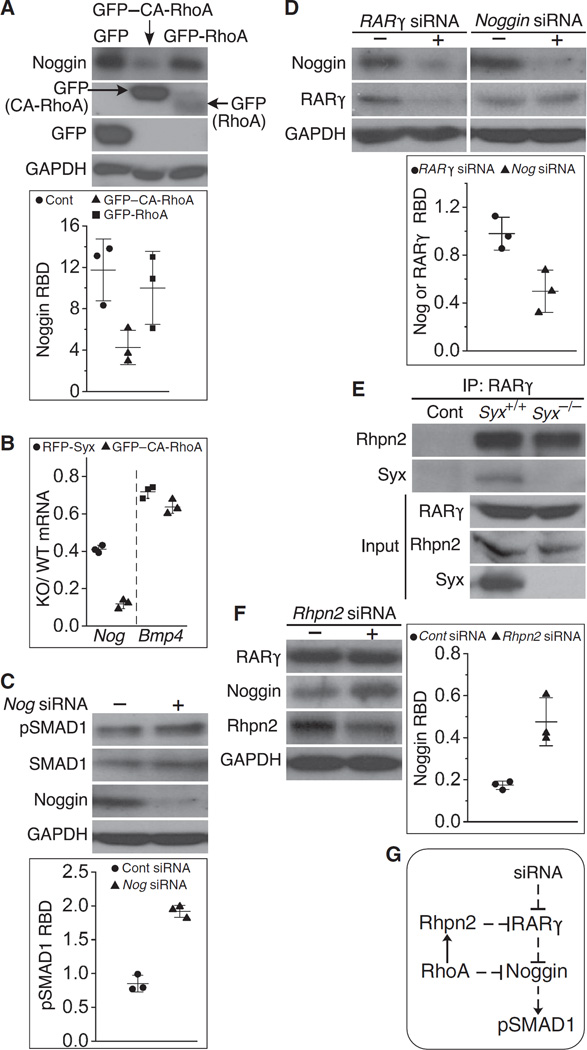
To determine whether RhoA is involved in regulating Noggin expression, we rescued Syx−/− cells by transfection of GFP–CA-RhoA or GFP-RhoA. GFP–CA-RhoA transfection decreased Noggin production markedly, whereas GFP-RhoA and GFP transfections did not (Fig. 5A), indicating that RhoA activity affects Noggin expression. To confirm this result, we measured Noggin transcription in GFP–CA-RhoA–transfected cells and in RFP-Syx–transfected cells by qRT-PCR. Both GFP–CA-RhoA and RFP-Syx transfections decreased Noggin transcription (Fig. 5B). Notably, transfection of RFP–CA-RhoA decreased Noggin transcription more than transfection with RFP-Syx, in agreement with the role of RhoA downstream of Syx (Fig. 5B). Additionally, we found that Noggin knockdown increased SMAD1 phosphorylation, which could potentially delay neural differentiation (Fig. 5C). Collectively, these results suggest that Noggin is essential for neural differentiation in Syx−/− ESCs. Because both Noggin and Rarγ promote neural differentiation, we compared the expression of these two genes. Whereas Rarγ knockdown reduced Noggin abundance, Noggin knockdown had no effect on RARγ abundance (Fig. 5D), in agreement with the upstream function of RARγ in RA-induced Noggin expression.
Fig. 5. RhoA activity reduced Noggin production, potentially through recruitment of RARγ to RhoA through Rhpn2.
(A) Transfection of cells from RA-treated Syx−/− EBs with GFP–CA-RhoA, but not GFP-RhoA, reduced Noggin production (one-way ANOVA, n = 3 independent experiments, mean ± SD, P = 0.018 for GFP to CA-RhoA). GAPDH is a loading control. (B) qRT-PCR results showing the effects of Syx or CA-RhoA transfection on the relative abundances of Noggin (Nog) and Bmp4 in cells from RA-treated Syx+/+ and Syx−/− EBs (two-sample t test, equal variance, mean ± SD, n = 3 independent experiments, P < 0.001 for Nog, P = 0.045 for Bmp4). (C) Immunoblot showing that Noggin knockdown increased SMAD1 phosphorylation (pSMAD1) in cells from RA-treated EBs (two-sample t test, unequal variance, mean ± SD, n = 3 independent experiments, P < 0.001). (D) Immunoblot showing that Rarγ silencing in cells from RA-treated EBs reduced Noggin production, but Nog silencing did not reduce RARγ production (two-sample t test, unequal variance, mean ± SD, n = 3 independent experiments, P = 0.047). (E) RARγ coimmunoprecipitated with RHPN2 and Syx in extracts from mESCs (one of two independent experiments). (F) Immunoblot showing RARγ, Noggin, and RHPN2 after transfection of cells from RA-treated EBs with siRNA targeting RHPN2 or a control siRNA (two-sample t test, unequal variance, n = 3 three independent experiments, mean ± SD, P = 0.041). (G) Signaling scheme representing the data shown in (A), (C), and (D). Solid lines represent direct regulatory events, whereas dashed lines represent events that either are indirect or have not been shown to be direct.
RARγ binds to the RhoA effector rhophilin-2 (RHPN2) (65, 66), an interaction that could potentially target RhoA to RARγ. We confirmed that RARγ coimmunoprecipitated with Rhpn2 and Syx in extracts from differentiating mESCs (Fig. 5E). To gauge the effect of Rhpn2 binding on RARγ-dependent Noggin production, we knocked down Rhpn2 in differentiating mESCs and compared Noggin abundance in the silenced cells to cells transfected by a control nontargeting siRNA. Partial Rhpn2 knockdown was accompanied by a significant increase in Noggin abundance (Fig. 5F), suggesting that Rhpn2 inhibits RARγ signaling. The results shown in Fig. 5 suggest that RhoA reduces Noggin’s abundance, in part, through Rhpn2 (Fig. 5G).
SIRT1 is required for the induction of SMAD1 phosphorylation
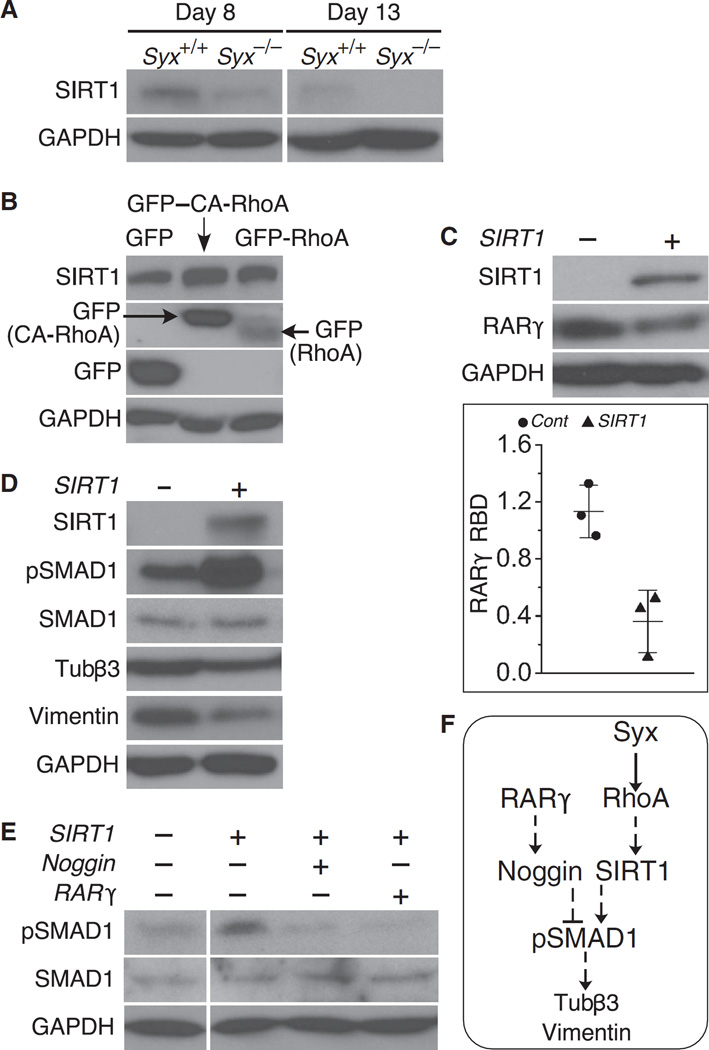
SIRT1 is a nuclear NAD+-dependent deacetylase that inhibits cellular RA signaling by deacetylating CRABPII (28). CRABPII is a cellular RA carrier that translocates from the cytosol into the nucleus upon RA binding, where it activates RARs (29, 31). Loss of SIRT1 increases the nuclear accumulation of CRABPII and thus enhances RA signaling (28). To test the role of SIRT1 in neural differentiation, we compared SIRT1 abundance in Syx+/+ and Syx−/− EBs. SIRT1 was less abundant in Syx−/− EBs (Fig. 6A). To confirm that the reduced abundance of SIRT1 was caused by the inactivation of RhoA, we transfected GFP–CA-RhoA into cells from dissociated RA-treated Syx−/− EBs. This manipulation increased SIRT1 production relative to GFP-RhoA or negative control GFP-producing cells (Fig. 6B).
Fig. 6. SIRT1 production was lower in Syx−/− EBs and was partially inhibited by RhoA.
(A) Immunoblot showing SIRT1 abundance in dissociated Syx+/+ or Syx−/− EBs at the indicated time points (one of two independent experiments). GAPDH is a loading control. (B) Immunoblot of cells from dissociated RA-treated Syx−/− EBs after transfection by GFP–CA-RhoA and GFP–wild-type RhoA (one of two independent experiments). (C) Immunoblot showing the effect of SIRT1 transfection on RARγ production in cells dissociated from RA-treated EBs (two-sample t test, equal variance, mean ± SD, n = 3 independent experiments, P = 0.01). (D) Immunoblot showing that transfection of cells dissociated from RA-treated Syx+/+ EBs with SIRT1 increased SMAD1 phosphorylation (pSMAD1) and reduced the abundance of the neural differentiation markers Tubβ3 and vimentin (one of two independent experiments). (E) Effect of SIRT1 coexpression with Noggin or Rarγ on SMAD1 phosphorylation in 8-day EBs transfected by the indicated plasmids (one of two independent experiments; the left lanes were immunoblotted on a separate membrane). (F) Signaling scheme representing the data shown in (A) to (E). Solid lines represent direct regulatory events, whereas dashed lines represent events that either are indirect or have not been shown to be direct.
To further explore the role of SIRT1 during neural differentiation, we overexpressed SIRT1 in cells that were dissociated from RA-treated Syx−/− EBs. RARγ production was reduced in SIRT1-transfected mESCs (Fig. 6C), whereas SMAD1 phosphorylation increased (Fig. 6D), indicating that neural differentiation was suppressed in the SIRT1-transfected cells. Coexpression of either Noggin or Rarγ in SIRT1-transfected Syx−/− cells prevented SMAD1 phosphorylation and restored neural differentiation (Fig. 6E). These results suggest that RA signaling, mediated by Noggin, reduces pSMAD1 abundance, whereas Syx and RhoA sustain SMAD1 phosphorylation through SIRT1 (Fig. 6F).
Peripheral stress fibers are thinner, and Rab3d and Noggin are reduced in Syx−/− cells
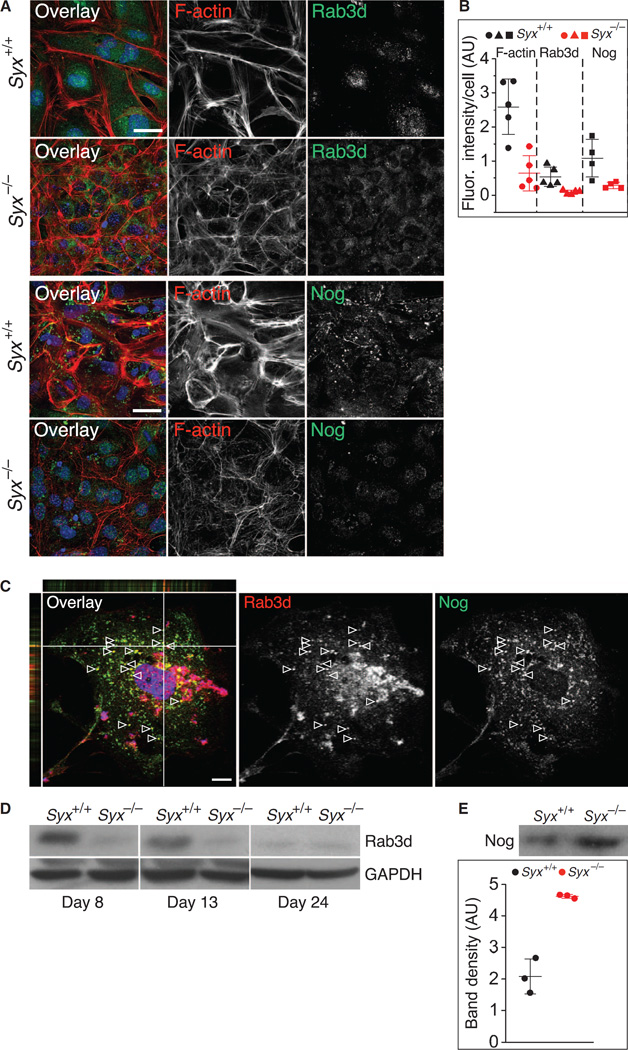
Loss of Syx only partially reduced RhoA activity (fig. S1D). Given that Syx-dependent RhoA activation occurs at the cell periphery (41), we expected to see differences between the peripheral stress fibers in Syx+/+ and Syx−/− cells. Peripheral stress fibers, visualized by staining for F-actin, were thinner in Syx−/− cells compared to Syx+/+ cells (Fig. 7A). Quantification of F-actin immunofluorescence in cells of each genotype estimated the difference at about fivefold (Fig. 7, A and B). Because Rab3d facilitates Noggin secretion (67) and because these two proteins colocalized in Syx+/+ cells (Fig. 7C), we compared the abundance of Rab3d-associated vesicles in Syx+/+ and Syx−/− cells. Syx+/+ cells retained more vesicular Rab3d (Fig. 7B). Immunoblotting for Rab3d confirmed this result (Fig. 7D). Similarly, the abundance of Noggin in Syx−/− cells was significantly lower than that in Syx+/+ cells (Fig. 7B). Although the lower Noggin abundance detected by imaging Syx−/− cells appears to contradict other immunoblotting results (fig. S3A), it should be noted that immunoblotting detects Noggin in the cytoplasm and on the cell surface, whereas imaging detects only cytoplasmic Noggin. Secreted Noggin is likely to associate with the cell surface because it binds tightly to heparan sulfate proteoglycans (68). The physiologically relevant Noggin is the secreted extracellular pool that inhibits BMP4 signaling by interfering with its binding to BMPRs. We tested the abundance of extracellular Noggin by immuno-blotting equal volumes of medium from Syx+/+ and Syx−/− cells. Noggin abundance was more than twofold greater in the medium of Syx−/− cells (Fig. 7E).
Fig. 7. Stress fibers, Rab3d, and Noggin in cells dissociated from Syx+/+ and Syx−/− EBs.
(A) Immunofluorescence of F-actin and the vesicular marker Rab3d or Noggin (Nog) in cells dissociated from RA-treated EBs (scale bars, 25 µm). (B) Quantification of F-actin, Rab3d, and Noggin immunofluorescence intensity per cell in cells from RA-treated Syx+/+ or Syx−/− EBs (two-sample t test, unequal variance, mean ± SD, n = 5 fields from two independent experiments, 35 to 50 cells per field; P = 0.008 for F-actin, P = 0.007 for Rab3d, P = 0.017 for Noggin). (C) Immunofluorescence image of a Syx+/+ mESC showing colocalization of FLAG-tagged Rab3d with endogenous Noggin in cytoplasmic punctae in the focal plane (arrowheads) and in optical sections along the white lines (one of two independent experiments). (D) Immunoblot of Rab3d in cells from RA-treated Syx+/+ and Syx−/− EBs at the indicted time points (one of two independent experiments). GAPDH is a loading control. (E) Immunoblot of Noggin in the medium of cells from RA-treated Syx+/+ and Syx−/− EBs (two-sample t test, unequal variance, mean ± SD, n = 3 independent experiments, P = 0.015).
DISCUSSION
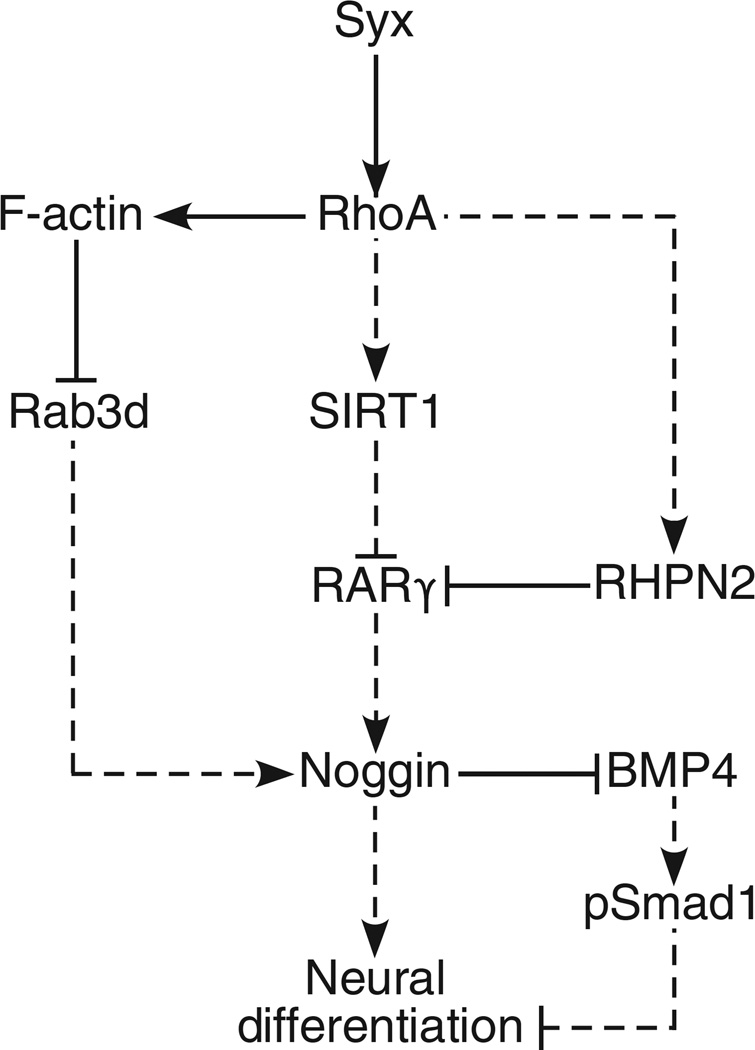
We propose two pathways for the inhibition of neural differentiation by RhoA (Fig. 8). The first is transcriptional, consisting of connections between RhoA and downstream target genes implicated in neural differentiation: SIRT1 (28), RARγ (29, 31), and, further downstream, SMAD1 (15, 16) and Noggin (69). The inhibition of RARγ suppresses neural differentiation because of the concomitant decrease in Noggin expression, thus allowing BMP4 to bind to BMPRs (19). The mechanism by which RARγ increases Noggin production is unknown. Published data suggest that RARγ could increase Noggin expression by associating with a large transcription factor complex (70) that includes Myc, for which there are binding sites in the 5′ promoter region of Noggin (71).
Fig. 8. Schematic representation of a dual signaling pathway downstream of Syx and RhoA that suppresses neural differentiation.
Solid lines represent direct regulatory events, whereas dashed lines represent events that either are indirect or have not been shown to be direct.
We propose a second mechanism for the inhibition of neural differentiation by RhoA that is mediated by the actin cytoskeleton. We observed that, compared to Syx−/− cells, the cytoplasm of Syx+/+ cells contained more vesicular Noggin and Rab3d, a GTPase that supports the trafficking of Noggin-containing secretory vesicles and the development of the nervous system (67). These results imply that Syx−/− cells either made less Noggin than Syx+/+ cells or secreted more Noggin than Syx+/+ cells. The latter explanation is supported by our observation that Noggin abundance was increased in the medium of Syx−/− cells compared to Syx+/+ cells. The peripheral stress fiber bundles in Syx+/+ cells, which are thicker than the bundles around Syx−/− cells, may impede the secretion of Noggin-containing vesicles. Similar to previous observations in neutrophils (72), stress fibers can act as a physical barrier that obstructs the translocation of Noggin-carrying vesicles from the cytoplasm to the plasma membrane. Consequently, Noggin exocytosis would be lower, thus slowing neural differentiation. Human ESC survival and growth also require the maintenance of robust circumferential stress fibers (42).
The substantially increased expression of ectodermal markers (fig. S1A) and the proteins they encode (fig. S1B) suggests that the accelerated neural differentiation of Syx−/− EBs reflects faster acquisition of neural fate by cells of the ectodermal germ layer. Alternatively, neural differentiation could conceivably be expedited by transdifferentiation of mesodermal cells into ectodermal cells, afforded by the loss of Syx and the concomitant deregulation of downstream transcription factors.
The antagonistic relationship between RA and RhoA appears to be bidirectional. The RhoA effector Rhpn2 binds to RARγ (65) and may inhibit RARγ activity (Fig. 5E), but Rhpn2 may also inhibit RhoA by sequestration (66). RhoA is present in the nucleus (73), thus enabling this putative association with RARγ through Rhpn2. Notably, binding to RARγ inhibits the transcriptional activity of serum response factor (74), a major mediator of RhoA-dependent transcriptional regulation (75). Together with our study, these findings reveal previously unappreciated crosstalk between RhoA and RA signaling in transcriptional regulation.
MATERIALS AND METHODS
mESC harvesting and maintenance
mESCs (C57BL/6) were purchased from Life Technologies. Syx−/− mESCs were isolated from blastocysts collected from pregnant Syx−/− mice (43) at embryonic day 3.5. The inner cell mass was isolated as described (76, 77) and cultured on a layer of mitomycin C–treated primary mouse embryonic fibroblast (MEF) feeder cells (EMD Millipore). mESCs were grown in complete Iscove’s modified Dulbecco’s medium (IMDM) containing 15% fetal bovine serum (FBS) and LIF (1000 U/ml), 0.1 µM nonessential amino acids, 55 µM 2-mercaptoethanol, penicillin (100 U/ml) and streptomycin (100 µg/ml), gentamicin reagent solution (200 µg/ml; Life Technologies), and 0.2% MycoZap Plus-PR (Lonza). Before differentiation, the cell mixture was transferred to a newly prepared petri dish and incubated for 40 min to remove differentiated ESCs and MEF cells. ESCs were cultured in gelatin-coated dish without feeder cells for two passages at 37°C, 5% CO2.
EB generation and induction of differentiation
mESCs were detached and dissociated by 0.25% trypsin-EDTA (Life Technologies), suspended in fresh IMDM without LIF, counted, and diluted to 5 × 105/ml in IMDM supplemented with 0.5 µM RA (Sigma). A 20-µl drop was placed on a petri dish, which was then inverted and incubated for 4 days at 37°C, 5% CO2, until EBs were formed. EBs were collected from the hanging drops, placed in a petri dish in RA-free IMDM, and grown for 4 days. Only the EBs used for the immunoblots shown in Fig. 4B and fig. S1 (B and C), for the immunofluorescence images in fig. S1A, and for the qRT-PCR in fig. S1D were not treated with RA. EBs were dissociated with 0.25% collagenase II (Worthington) for 1 hour and washed three times in phosphate-buffered saline (PBS) before placement on fibronectin or gelatin-coated coverslips for analysis.
RNA isolation and qRT-PCR amplification
RNA was isolated from mESCs or EBs by TRIzol (Ambion) and reverse-transcribed to complementary DNA (Takara). qRT-PCR was performed using SYBR Green (Takara). To quantify relative mRNA transcription, the data were normalized relative to the GAPDH mRNA reading. The ΔΔCT method was used to quantify mRNA (78). The qRT-PCR primers are listed in table S1.
Plasmid transfection into mESCs
pCDNA3.1, pcDNA3-EGFP-RhoA-Q63L (Addgene), pEGFP-SIRT1, pEGFP-Noggin, pCMV-3×Flag-Noggin or Rab3d, and pcDNA3-mRFP-Syx plasmids were transfected by Effectene (Qiagen), according to the manufacturers’ instructions. mRNA for qRT-PCR analysis was extracted after 48 hours of transfection. Cells were lysed for immunoblotting 72 to 96 hours after transfection. Cells were imaged by immunofluorescence 72 hours after transfection.
RNA interference in mESCs
siRNA targeting Noggin, Rarγ, and Rhpn2 and nontargeting siRNA (Santa Cruz Biotechnology) were transfected by Effectene. RNA for qRT-PCR analysis was extracted 48 hours after transfection.
Detection of secreted Noggin
EBs generated by equal numbers of either Syx+/+ or Syx−/− ESCs were grown and differentiated for equal durations, as described above, in equal volumes of medium in wells of a 24-well plate. The medium was replaced 8 days after cell aggregation. Five hundred microliters of medium was collected from each well on day 13 and concentrated by centrifugation through 10-kD filters (Microcon, EMD Millipore) to a final volume of 25 µl.
Coimmunoprecipitation assays
mESCs were cultured without LIF for 2 days and then lysed on ice (Pierce IP Lysis Buffer, Thermo Fisher Scientific) with protease inhibitors (cOmplete Protease Inhibitor Cocktail, Roche). Cell lysates were incubated with the antibodies specified in Results and conjugated to immobilized protein G (Dynabeads, Thermo Fisher Scientific) at 4°C overnight. Immune complexes were pelleted and washed three times with cold IP buffer. Proteins were eluted by glycine-HCl (pH 3.0). Samples were analyzed by immunoblotting.
Immunofluorescence staining of mESCs and EBs
Cells were plated on fibronectin-coated coverslips, washed three times in PBS, fixed with 4% paraformaldehyde for 30 min, washed again three times in PBS, and permeabilized with 1% Triton X-100 for 20 min. After blocking for 1 hour with 5% bovine serum albumin, samples were probed with primary antibodies recognizing Nanog (Bethyl Laboratories), FLAG, FOX3, GFP, SOX1 (Cell Signaling), Rab3d (Proteintech), nestin, Oct4, SOX2, Tubβ3 (Santa Cruz Biotechnology), or Noggin (Thermo Fisher Scientific) or with phalloidin–Alexa Flour 568 (Thermo Fisher Scientific) according to the manufacturers’ instructions. The GFP antibody was also used to detect RFP because it cross-reacts with it. Secondary antibodies [Alexa Fluor 488 or Alexa Fluor 568 Goat anti-Mouse IgG (H+L), Thermo Fisher Scientific] were applied according to the manufacturer’s instructions. The coverslips were mounted with ProLong Gold (Thermo Fisher Scientific). EBs were plated on coverslips for 4 days after growing in hanging drops for 4 days and processed like mESCs. Images were acquired on TCS SP (Leica) or LSM 510 (Zeiss) confocal microscopes. The immunofluorescence intensities in the images shown in Figs. 1A, 2A, and 7A were quantified by integrating the above-background pixel intensities of the relevant channels with ImageJ (79). The images shown in Fig. 1D were quantified by counting the number of nuclei in which either the SOX1 or Pax6 signal was present in each field.
Immunoblotting
Cell extracts were separated by SDS–polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and probed with the following antibodies according to the manufacturer’s instructions: Noggin (Abcam); BMP4 (Sigma); GAPDH (Sigma); Tubβ3 (Santa Cruz Biotechnology); Rab3d, Rhpn2, and Syx (Plekhg5) (Proteintech); RARα, RARγ, vimentin, Pax6, GFP, SMAD1, and pSMAD1 (Cell Signaling); MAPK and pMAPK (EMD Millipore); and RARβ (Sigma). Primary antibodies were detected with horseradish peroxidase–conjugated secondary antibodies (Cell Signaling) followed by enhanced chemiluminescence (Denville Scientific). The proteasome inhibitor MG132 was purchased from Sigma-Aldrich. Immunoblots were quantified by band densitometry with Image Studio Lite (LI-COR).
Rho activity assay
mESCs were serum-starved for 24 hours, treated by lysophosphatidic acid (1 µg/ml; Tocris) for 3 min, and lysed on ice. Guanosine triphosphate (GTP)–RhoA was pulled down by immobilized rhotekin RhoA binding domain (Cytoskeleton) for 1 hour at 4°C. RhoA was detected by immunoblotting with anti-RhoA (Cytoskeleton).
Development of definitive endoderm for lineage analysis
mESCs were grown in FBS-free medium containing activin A (100 ng/ml) and Wnt3a (25 ng/ml) (R&D Systems) for 1 day. The medium was supplemented with 0.2% FBS on the second day and by additional 1.8% on the next 2 days, after which mRNA was isolated for qRT-PCR amplification as above.
Statistical analysis
All experiments were run at least in duplicate. Data were averaged for n ≥ 3 samples and shown with SDs. Differences were considered statistically significant for P ≤ 0.05. Two-sample t test was used for two-group comparisons if there was no evidence of violation of the normal distribution assumption. The assumption of equal or unequal variance for the two groups was used as appropriate to the nature of the data. The number of positive cells among the total counted cells in each field of either Pax6- or SOX1-immunolableled Syx+/+ and Syx−/− cells in Fig. 1D was analyzed by repeated-measures logistic regression, adjusting for correlation between cells from the same field using the generalized estimating equation approach (80). Relative band density measurements in three sample groups (Fig. 2B) were analyzed by one-way ANOVA. Data of two classification categories (genotype and time after EB aggregation; Figs. 1B and 4E) were analyzed by two-way ANOVA only for the effect of genotype, blocking the effect of time. The reported differences represent only the effect of genotype. Residuals in ANOVA analysis were examined for consistency with the assumption of normal distribution. No multiple testing adjustments were made because of the relatively small sample sizes. Data analysis was performed by R software package (81).
Supplementary Material
Acknowledgments
We thank P. Tesar (Case Western Reserve Medical School) for advice. Funding: This study was supported by NIH grant R01 HL119984 to A.H. L.J. is supported by William K. Bowes Jr. Foundation, the Swedish Research Council (K201299X-22005-01-3), the Swedish Cancer Society (110604), the Cardiovascular Program, and the Strategic Research Program in Neuroscience at Karolinska Institute, Jeanssons Stiftelser, and Magnus Bergvalls Stiftelse.
Footnotes
www.sciencesignaling.org/cgi/content/full/9/438/ra76/DC1
Fig. S1. Comparisons of gene expression patterns and GTP-RhoA abundance in Syx−/− EBs or mESCs, respectively, versus their Syx+/+ counterparts.
Fig. S2. Transfection of RFP-Syx or GFP–CA-RhoA into Syx−/− cells.
Fig. S3. Noggin production increased during neural differentiation, and VPA inhibited neural differentiation.
Fig. S4. Full-length images of the immunoblots shown in Figs. 1 to 7 and figs. S1 and S3.
Table S1. Primer sequences for qRT-PCRs.
Author contributions: J.Y. designed the project, performed experiments, and wrote the manuscript; C.W. and I.S. performed experiments; L.J. provided technical expertise and wrote the manuscript; I.C. performed statistical analysis of the results and wrote the manuscript; and A.H designed the project and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh Y-H, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung W-K, Clarke ND, Wei C-L, Ng H-H. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 2.Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 3.Loh Y-H, Wu Q, Chew J-L, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong K-Y, Sung KW, Lee CWH, Zhao X-D, Chiu K-P, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei C-L, Ruan Y, Lim B, Ng H-H. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 4.Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- 5.Fei T, Xia K, Li Z, Zhou B, Zhu S, Chen H, Zhang J, Chen Z, Xiao H, Han J-DJ, Chen Y-G. Genome-wide mapping of SMAD target genes reveals the role of BMP signaling in embryonic stem cell fate determination. Genome Res. 2010;20:36–44. doi: 10.1101/gr.092114.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dravid G, Ye Z, Hammond H, Chen G, Pyle A, Donovan P, Yu X, Cheng L. Defining the role of Wnt/β-catenin signaling in the survival, proliferation, and self-renewal of human embryonic stem cells. Stem Cells. 2005;23:1489–1501. doi: 10.1634/stemcells.2005-0034. [DOI] [PubMed] [Google Scholar]
- 7.Hao J, Li T-G, Qi X, Zhao D-F, Zhao G-Q. WNT/β-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Dev. Biol. 2006;290:81–91. doi: 10.1016/j.ydbio.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Kelly KF, Ng DY, Jayakumaran G, Wood GA, Koide H, Doble BW. β-Catenin enhances Oct-4 activity and reinforces pluripotency through a TCF-independent mechanism. Cell Stem Cell. 2011;8:214–227. doi: 10.1016/j.stem.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyabayashi T, Teo J-L, Yamamoto M, McMillan M, Nguyen C, Kahn M. Wnt/β-catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5668–5673. doi: 10.1073/pnas.0701331104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yun M-S, Kim S-E, Jeon SH, Lee J-S, Choi K-Y. Both ERK and Wnt/β-catenin pathways are involved in Wnt3a–induced proliferation. J. Cell Sci. 2005;118:313–322. doi: 10.1242/jcs.01601. [DOI] [PubMed] [Google Scholar]
- 11.Di-Gregorio A, Sancho M, Stuckey DW, Crompton LA, Godwin J, Mishina Y, Rodriguez TA. BMP signalling inhibits premature neural differentiation in the mouse embryo. Development. 2007;134:3359–3369. doi: 10.1242/dev.005967. [DOI] [PubMed] [Google Scholar]
- 12.James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFβ/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 13.Michon F, Forest L, Collomb E, Demongeot J, Dhouailly D. BMP2 and BMP7 play antagonistic roles in feather induction. Development. 2008;135:2797–2805. doi: 10.1242/dev.018341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pera MF, Andrade J, Houssami S, Reubinoff B, Trounson A, Stanley EG, Ward-van Oostwaard D, Mummery C. Regulation of human embryonic stem cell differentiation by BMP-2 and its antagonist noggin. J. Cell Sci. 2004;117:1269–1280. doi: 10.1242/jcs.00970. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Fei T, Zhang J, Zhu G, Wang L, Lu D, Chi X, Teng Y, Hou N, Yang X, Zhang H, Han J-DJ, Chen Y-G. BMP4 signaling acts via dual-specificity phosphatase 9 to control ERK activity in mouse embryonic stem cells. Cell Stem Cell. 2012;10:171–182. doi: 10.1016/j.stem.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 16.Qi X, Li T-G, Hao J, Hu J, Wang J, Simmons H, Miura S, Mishina Y, Zhao G-Q. BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6027–6032. doi: 10.1073/pnas.0401367101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ying Q-L, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 18.Jung G-A, Yoon J-Y, Moon B-S, Yang D-H, Kim H-Y, Lee S-H, Bryja V, Arenas E, Choi K-Y. Valproic acid induces differentiation and inhibition of proliferation in neural progenitor cells via the beta-catenin-Ras-ERK-p21Cip/WAF1 pathway. BMC Cell Biol. 2008;9:66. doi: 10.1186/1471-2121-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirsinger E, Duprez D, Jouve C, Malapert P, Cooke J, Pourquie O. Noggin acts downstream of Wnt and Sonic Hedgehog to antagonize BMP4 in avian somite patterning. Development. 1997;124:4605–4614. doi: 10.1242/dev.124.22.4605. [DOI] [PubMed] [Google Scholar]
- 20.López-Carballo G, Moreno L, Masiá S, Pérez P, Barettino D. Activation of the phosphatidylinositol 3-kinase/Akt signaling pathway by retinoic acid is required for neural differentiation of SH-SY5Y human neuroblastoma cells. J. Biol. Chem. 2002;277:25297–25304. doi: 10.1074/jbc.M201869200. [DOI] [PubMed] [Google Scholar]
- 21.Gajović S, St-Onge L, Yokota Y, Gruss P. Retinoic acid mediates Pax6 expression during in vitro differentiation of embryonic stem cells. Differentiation. 1997;62:187–192. doi: 10.1046/j.1432-0436.1998.6240187.x. [DOI] [PubMed] [Google Scholar]
- 22.Blumberg B. An essential role for retinoid signaling in anteroposterior neural specification and neuronal differentiation. Semin. Cell Dev. Biol. 1997;8:417–428. doi: 10.1006/scdb.1997.0165. [DOI] [PubMed] [Google Scholar]
- 23.Okada Y, Shimazaki T, Sobue G, Okano H. Retinoic-acid-concentration-dependent acquisition of neural cell identity during in vitro differentiation of mouse embryonic stem cells. Dev. Biol. 2004;275:124–142. doi: 10.1016/j.ydbio.2004.07.038. [DOI] [PubMed] [Google Scholar]
- 24.Ross SA, McCaffery PJ, Drager UC, De Luca LM. Retinoids in embryonal development. Physiol. Rev. 2000;80:1021–1054. doi: 10.1152/physrev.2000.80.3.1021. [DOI] [PubMed] [Google Scholar]
- 25.Bain G, Ray WJ, Yao M, Gottlieb DI. Retinoic acid promotes neural and represses mesodermal gene expression in mouse embryonic stem cells in culture. Biochem. Biophys. Res. Commun. 1996;223:691–694. doi: 10.1006/bbrc.1996.0957. [DOI] [PubMed] [Google Scholar]
- 26.Sheng N, Xie Z, Wang C, Bai G, Zhang K, Zhu Q, Song J, Guillemot F, Chen Y-G, Lin A, Jing N. Retinoic acid regulates bone morphogenic protein signal duration by promoting the degradation of phosphorylated Smad1. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18886–18891. doi: 10.1073/pnas.1009244107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, Yamada KA, Imai S-i. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013;18:416–430. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang S, Huang G, Fan W, Chen Y, Ward JM, Xu X, Xu Q, Kang A, McBurney MW, Fargo DC, Hu G, Baumgart-Vogt E, Zhao Y, Li X. SIRT1-mediated deacetylation of CRABPII regulates cellular retinoic acid signaling and modulates embryonic stem cell differentiation. Mol. Cell. 2014;55:843–855. doi: 10.1016/j.molcel.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delva L, Bastie J-N, Rochette-Egly C, Kraiba R, Balitrand N, Despouy G, Chambon P, Chomienne C. Physical and functional interactions between cellular retinoic acid binding protein II and the retinoic acid-dependent nuclear complex. Mol. Cell. Biol. 1999;19:7158–7167. doi: 10.1128/mcb.19.10.7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong D, Ruuska SE, Levinthal DJ, Noy N. Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid. J. Biol. Chem. 1999;274:23695–23698. doi: 10.1074/jbc.274.34.23695. [DOI] [PubMed] [Google Scholar]
- 31.Sessler RJ, Noy N. A ligand-activated nuclear localization signal in cellular retinoic acid binding protein-II. Mol. Cell. 2005;18:343–353. doi: 10.1016/j.molcel.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 32.Hall A. Rho family GTPases. Biochem. Soc. Trans. 2012;40:1378–1382. doi: 10.1042/BST20120103. [DOI] [PubMed] [Google Scholar]
- 33.Stankiewicz TR, Linseman DA. Rho family GTPases: Key players in neuronal development, neuronal survival, and neurodegeneration. Front. Cell. Neurosci. 2014;8:314. doi: 10.3389/fncel.2014.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harb N, Archer TK, Sato N. The Rho-Rock-Myosin signaling axis determines cell-cell integrity of self-renewing pluripotent stem cells. PLOS One. 2008;3:e3001. doi: 10.1371/journal.pone.0003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y-K, Yu X, Cohen DM, Wozniak MA, Yang MT, Gao L, Eyckmans J, Chen CS. Bone morphogenetic protein-2-induced signaling and osteogenesis is regulated by cell shape, RhoA/ROCK, and cytoskeletal tension. Stem Cells Dev. 2012;21:1176–1186. doi: 10.1089/scd.2011.0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu H, Yu SP, Gutekunst CA, Gross RE, Wei L. Inhibition of the Rho signaling pathway improves neurite outgrowth and neuronal differentiation of mouse neural stem cells. Int. J. Physiol. Pathophysiol. Pharmacol. 2013;5:11–20. [PMC free article] [PubMed] [Google Scholar]
- 37.Lu W, Wang J, Wen T. Downregulation of Rho-GDI g promotes differentiation of neural stem cells. Mol. Cell. Biochem. 2008;311:233–240. doi: 10.1007/s11010-008-9713-9. [DOI] [PubMed] [Google Scholar]
- 38.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 39.De Toledo M, Coulon V, Schmidt S, Fort P, Blangy A. The gene for a new brain specific RhoA exchange factor maps to the highly unstable chromosomal region 1p36.2-1p36.3. Oncogene. 2001;20:7307–7317. doi: 10.1038/sj.onc.1204921. [DOI] [PubMed] [Google Scholar]
- 40.Marx R, Henderson J, Wang J, Baraban JM. Tech: A RhoA GEF selectively expressed in hippocampal and cortical neurons. J. Neurochem. 2005;92:850–858. doi: 10.1111/j.1471-4159.2004.02930.x. [DOI] [PubMed] [Google Scholar]
- 41.Ngok SP, Geyer R, Liu M, Kourtidis A, Agrawal S, Wu C, Seerapu HR, Lewis-Tuffin LJ, Moodie KL, Huveldt D, Marx R, Baraban JM, Storz P, Horowitz A, Anastasiadis PZ. VEGF and Angiopoietin-1 exert opposing effects on cell junctions by regulating the Rho GEF Syx. J. Cell Biol. 2012;199:1103–1115. doi: 10.1083/jcb.201207009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohgushi M, Minaguchi M, Sasai Y. Rho-signaling-directed YAP/TAZ activity underlies the long-term survival and expansion of human embryonic stem cells. Cell Stem Cell. 2015;17:448–461. doi: 10.1016/j.stem.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 43.Garnaas MK, Moodie KL, Liu M-L, Samant GV, Li K, Marx R, Baraban JM, Horowitz A, Ramchandran R. Syx, a RhoA guanine exchange factor, is essential for angiogenesis in vivo. Circ. Res. 2008;103:710–716. doi: 10.1161/CIRCRESAHA.108.181388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang S-C, Wernig M, Duncan ID, Brüstle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 45.Beddington RS. Induction of a second neural axis by the mouse node. Development. 1994;120:613–620. doi: 10.1242/dev.120.3.613. [DOI] [PubMed] [Google Scholar]
- 46.Lee MK, Tuttle JB, Rebhun LI, Cleveland DW, Frankfurter A. The expression and posttranslational modification of a neuron-specific β-tubulin isotype during chick embryogenesis. Cell Motil. Cytoskeleton. 1990;17:118–132. doi: 10.1002/cm.970170207. [DOI] [PubMed] [Google Scholar]
- 47.Schnitzer J, Franke WW, Schachner M. Immunocytochemical demonstration of vimentin in astrocytes and ependymal cells of developing and adult mouse nervous system. J. Cell Biol. 1981;90:435–447. doi: 10.1083/jcb.90.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lendahl U, Zimmerman LB, McKay RDG. CNS stem cells express a new class of intermediate filament protein. Cell. 1990;60:585–595. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- 49.Pankratz MT, Li XJ, Lavaute TM, Lyons EA, Chen X, Zhang SC. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells. 2007;25:1511–1520. doi: 10.1634/stemcells.2006-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reubinoff BE, Itsykson P, Turetsky T, Pera MF, Reinhartz E, Itzik A, Ben-Hur T. Neural progenitors from human embryonic stem cells. Nat. Biotechnol. 2001;19:1134–1140. doi: 10.1038/nbt1201-1134. [DOI] [PubMed] [Google Scholar]
- 51.Mullen RJ, Buck CR, Smith AM, Neu N. a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- 52.Subauste MC, Von Herrath M, Benard V, Chamberlain CE, Chuang T-H, Chu K, Bokoch GM, Hahn KM. Rho family proteins modulate rapid apoptosis induced by cytotoxic T lymphocytes and Fas. J. Biol. Chem. 2000;275:9725–9733. doi: 10.1074/jbc.275.13.9725. [DOI] [PubMed] [Google Scholar]
- 53.Kretzschmar M, Liu F, Hata A, Doody J, Massagué J. The TGF-β family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev. 1997;11:984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- 54.Eivers E, Fuentealba LC, De Robertis EM. Integrating positional information at the level of Smad1/5/8. Curr. Opin. Genet. Dev. 2008;18:304–310. doi: 10.1016/j.gde.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takekawa M, Saito H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell. 1998;95:521–530. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 56.Wu WKK, Sung JJY, To KF, Yu L, Li HT, Li ZJ, Chu KM, Yu J, Cho CH. The host defense peptide LL-37 activates the tumor-suppressing bone morphogenetic protein signaling via inhibition of proteasome in gastric cancer cells. J. Cell. Physiol. 2010;223:178–186. doi: 10.1002/jcp.22026. [DOI] [PubMed] [Google Scholar]
- 57.Maden M. Retinoid signalling in the development of the central nervous system. Nat. Rev. Neurosci. 2002;3:843–853. doi: 10.1038/nrn963. [DOI] [PubMed] [Google Scholar]
- 58.Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 59.Scheibe RJ, Wagner JA. Retinoic acid regulates both expression of the nerve growth factor receptor and sensitivity to nerve growth factor. J. Biol. Chem. 1992;267:17611–17616. [PubMed] [Google Scholar]
- 60.Jacob A, Budhiraja S, Qian X, Clevidence D, Costa RH, Reichel RR. Retinoic acid-mediated activation of HNF-3α during EC stem cell differentiation. Nucleic Acids Res. 1994;22:2126–2133. doi: 10.1093/nar/22.11.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar S, Duester G. Retinoic acid signaling in perioptic mesenchyme represses Wnt signaling via induction of Pitx2 and Dkk2. Dev. Biol. 2010;340:67–74. doi: 10.1016/j.ydbio.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schoorlemmer J, van Puijenbroek A, van Den Eijnden M, Jonk L, Pals C, Kruijer W. Characterization of a negative retinoic acid response element in the murine Oct4 promoter. Mol. Cell. Biol. 1994;14:1122–1136. doi: 10.1128/mcb.14.2.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumar S, Duester G. Retinoic acid controls body axis extension by directly repressing Fgf8 transcription. Development. 2014;141:2972–2977. doi: 10.1242/dev.112367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sela-Donenfeld D, Kalcheim C. Regulation of the onset of neural crest migration by coordinated activity of BMP4 and Noggin in the dorsal neural tube. Development. 1999;126:4749–4762. doi: 10.1242/dev.126.21.4749. [DOI] [PubMed] [Google Scholar]
- 65.Peck JW, Oberst M, Bouker KB, Bowden E, Burbelo PD. The RhoA-binding protein, rhophilin-2, regulates actin cytoskeleton organization. J. Biol. Chem. 2002;277:43924–43932. doi: 10.1074/jbc.M203569200. [DOI] [PubMed] [Google Scholar]
- 66.Wang J, Huo K, Ma L, Tang L, Li D, Huang X, Yuan Y, Li C, Wang W, Guan W, Chen H, Jin C, Wei J, Zhang W, Yang Y, Liu Q, Zhou Y, Zhang C, Wu Z, Xu W, Zhang Y, Liu T, Yu D, Zhang Y, Chen L, Zhu D, Zhong X, Kang L, Gan X, Yu X, Ma Q, Yan J, Zhou L, Liu Z, Zhu Y, Zhou T, He F, Yang X. Toward an understanding of the protein interaction network of the human liver. Mol. Syst. Biol. 2011;7:536. doi: 10.1038/msb.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim H, Han JK. Rab3d is required for Xenopus anterior neurulation by regulating Noggin secretion. Dev. Dyn. 2011;240:1430–1439. doi: 10.1002/dvdy.22643. [DOI] [PubMed] [Google Scholar]
- 68.Paine-Saunders S, Viviano BL, Economides AN, Saunders S. Heparan sulfate proteoglycans retain Noggin at the cell surface: A potential mechanism for shaping bone morphogenetic protein gradients. J. Biol. Chem. 2002;277:2089–2096. doi: 10.1074/jbc.M109151200. [DOI] [PubMed] [Google Scholar]
- 69.Lamb TM, Knecht AK, Smith WC, Stachel SE, Economides AN, Stahl N, Yancopolous GD, Harland RM. Neural induction by the secreted polypeptide noggin. Science. 1993;262:713–718. doi: 10.1126/science.8235591. [DOI] [PubMed] [Google Scholar]
- 70.Liu Z, Merkurjev D, Yang F, Li W, Oh S, Friedman MJ, Song X, Zhang F, Ma Q, Ohgi KA, Krones A, Rosenfeld MG. Enhancer activation requires trans-recruitment of a mega transcription factor complex. Cell. 2014;159:358–373. doi: 10.1016/j.cell.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.EpiTect ChIP qPCR Primers. www.sabiosciences.com/chipqpcrsearch.php?app=TFBS.
- 72.Johnson JL, Monfregola J, Napolitano G, Kiosses WB, Catz SD. Vesicular trafficking through cortical actin during exocytosis is regulated by the Rab27a effector JFC1/Slp1 and the RhoA-GTPase-activating protein Gem-interacting protein. Mol. Biol. Cell. 2012;23:1902–1916. doi: 10.1091/mbc.E11-12-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rajakyla EK, Vartiainen MK. Rho, nuclear actin, and actin-binding proteins in the regulation of transcription and gene expression. Small GTPases. 2014;5:e27539. doi: 10.4161/sgtp.27539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim S-W, Kim H-J, Jung D-J, Lee S-K, Kim Y-S, Kim JH, Kim TS, Lee JW. Retinoid-dependent antagonism of serum response factor transactivation mediated by transcriptional coactivator proteins. Oncogene. 2001;20:6638–6642. doi: 10.1038/sj.onc.1204695. [DOI] [PubMed] [Google Scholar]
- 75.Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 76.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 77.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 78.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 79.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.L Zeger S, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–130. [PubMed] [Google Scholar]
- 81.R Core Team. 2R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2015. www.r-project.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.