Abstract
Background
Radiation-induced salivary hypofunction is a common side-effect of treatment for head and neck cancers. Patients suffer significant morbidity and there is no suitable conventional therapy. We are conducting a Phase I clinical trial, using a first-generation serotype 5 adenoviral (Ad5) vector encoding human aquaporin-1 (AdhAQP1) to treat such patients. One week after the administration of AdhAQP1 to an enrolled, generally healthy patient, E1-containing adenovirus was detected in parotid saliva.
Methods
The real-time quantitative polymerase chain reactuion (PCR) was used to measure the Ad5 E1 gene and AdhAQP1 in saliva and serum. PCR and sequencing were used to characterize viral/vector DNA extracted from saliva. The presence of infectious adenovirus was assessed by the inoculation of A549 cells with aliquots of saliva. Serum Ad5 neutralizing antibodies were measured by the inhibition of 293-cell transduction with an Ad5 vector encoding luciferase. Multiple clinical evaluations were performed.
Results
On day 7 after AdhAQP1 delivery, low levels of the Ad5 E1 gene were detected in parotid saliva (82 copies/μl). In addition, significant levels of AdhAQP1 were also detected (1.5 × 103 copies/μl). The patient was asymptomatic and subsequent analysis of parotid saliva samples prior to day 7 and after day 7 until day 42 was negative for both virus and vector. No virus or vector was detected in serum at any time. Detailed PCR analyses of DNA extracted from the day 7 parotid saliva sample suggested the absence of a recombination event, and no infectious virus was found.
Conclusions
The patient most likely had a latent Ad5 infection in the targeted parotid gland that was activated after gene transfer and was without clinical consequence. Published in 2009 by John Wiley & Sons, Ltd.
Keywords: adenoviral vector, clinical trial, E1 gene, parotid gland, saliva
Introduction
Radiation-induced salivary hypofunction is a common, significant and persistent sequela of the treatment of head and neck cancers [1]. Patients with this condition lack acinar cells, which are the only cells in salivary glands able to secrete fluid [2,3], and suffer considerable morbidity, including dysphagia, frequent oral infections, reduced healing of oral mucosal tissues and general oral discomfort [4]. At present, there is no effective treatment available. We previously showed that the delivery of a serotype 5, adenoviral (Ad5) vector, encoding human aquaporin-1 (hAQP1 [5], AdhAQP1) to rat and miniature pig salivary glands after irradiation led to near normal levels of salivary flow [6,7]. Subsequently, we submitted an application for a Phase I, single-dose escalation clinical trial that has received all the required approvals (http://www.clinicaltrials.gov/ct/show/NCT00372320?order=1). All patients are seen at the NIH Clinical Center in Bethesda, MD, USA. In the present study, we describe the detection and investigation of E1-containing adenovirus in a single parotid saliva sample from a patient (#25) enrolled in the present study.
Materials and methods
Quantitative polymerase chain reaction (QPCR) assay for the Ad5 E1 gene
The E1 gene of wild-type (WT) Ad5 encodes the E1a [from nucleotides (nt) 468–1632 of WT Ad5 (access number: AY339865)] and E1b [from nt 1672–3509 of WT Ad5 (access number: AY339865)] proteins, which are required for the replication of WT Ad5. The E1 gene is deleted in AdhAQP1. To detect whether the Ad5 E1 gene was present in patient serum and saliva samples, as a surrogate assay for the presence of WT Ad5, we designed a set of TaqMan primers and a probe using the Primer Express Primer Design software (PE Applied Biosystems, Foster City, CA, USA). The sequences of the primers and probe chosen were: E1q1 (5′-TGTGCCCCATTAAACCAGTTG-3′) [locates from nt 1421–1441 of WT Ad5 (access number: AY339865)], E1q2 (5′-TCCTCGATACATTCCACAGCC-3′) [locates from nt 1467–1487 of WT Ad5 (access number: AY339865)] and E1probe1 (5′-/56-FAM/CGTGAGAGTTGGTGGGCGTCGC/36-TSMTSp/-3′) [locates from nt 1443–1464 of WT Ad5 (access number: AY339865)]. The quantitative (Q) PCR assay results in the amplification of a 66-bp fragment from the E1a region of WT Ad5.
The DNA standard used for this QPCR assay was a 648-bp PCR product (termed PCR 1; Figure 1) that originates from the E1 region of WT Ad5 (nt 1387–2035). It was amplified using a genomic DNA sample from 293 cells with primers E1F58 (5′-AACACACCTCCTGAGATACACCCG-3′) [locates from nt 1387–1410 of WT Ad5 (access number: AY339865)] and E1B98 (5′-GGGTTTCTTCGCTCCATTTATCC-3′) [locates from nt 2013–2035 of WT Ad5 (access number: AY339865)].
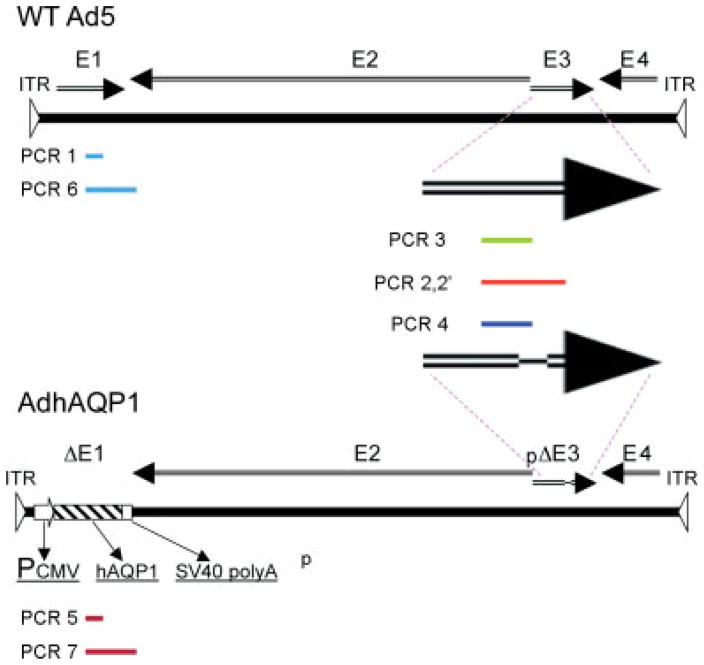
Figure 1.
Schematic representation of the PCR products studied. Linear representations of the WT serotype 5 adenovirus (WT Ad5) genome and the AdhAQP1 genome are shown. Arrows indicate different genomic regions. The PCR products studied, and their locations, are shown as bars in different colors. The two large arrows show the regions amplified in the Ad5 E3 region. For details of the primers used and PCR reaction conditions, see Materials and methods. Additional information on the sequence of the PCR products is provided in Table 2 and Figure 3
Genomic DNA for the QPCR assay was extracted from saliva and serum samples with the QIAamp DNA Blood Mini kit (Qiagen, Valencia, CA, USA). One, 5 or 10 μl of extracted DNA sample were used for the QPCR reactions. WT Ad5 was used as positive control. The negative control was genomic DNA obtained from a normal human serum sample. In the assay, there were no template controls. Each reaction contained 25 μl of primers (10 pmol of each primer), probe (15 pmol of probe), the sample to be tested, plus 25 μl of TaqMan Universal PCR Master Mix (Applied Biosystems). The assays were performed in an ABI Prism 7700 Sequence Detector (Applied Biosystems). The conditions used for the QPCR assays were: 95 °C for 2 min, 95 °C for 8 min, 95 °C for 15 s and 60 °C for 1 min for 40 cycles.
PCR assays and QPCR assay for AdhAQP1
Genomic DNA for all PCR reactions, and for the QPCR assay to detect AdhAQP1, was also extracted from saliva and serum samples with the QIAamp DNA Blood Mini kit (Qiagen). Five microliters of the extracted DNA sample were used for the PCR reactions and 1, 5 or 10 μl of extracted DNA sample were used for the AdhAQP1 QPCR assay.
For the QPCR assay to determine the presence of the AdhAQP1 vector in extracted DNA samples, the primers and probe used to amplify an 83-bp fragment from cytomegalovirus (CMV) promoter and human AQP1 cDNA junction were: CMV-AQP1primer1 (5′-CGTGTACGGTGGGAGGTCTATATAA-3′), CMV-AQP1primer2 (5′-GCTGGTACCGAGCTCGAATT-3′) and CMV-AQP1-probe1 (5′-/56-FAM/CTCGTTTAGTGAACCGTCAGATCCGGTC/36-TSMTSp/-3′). For this assay, the plasmid pACCMV-hAQP1 [6] was used as a standard.
To characterize the viral/vector DNA present in the day 7 Ad5 E1-positive parotid saliva sample from patient #25, we used PCR to amplify multiple sequences in the Ad5 E1 and E3 regions. Figure 1 depicts the eight PCR products that we attempted to generate from the DNA extracted, along with their locations in WT Ad5 and AdhAQP1. The products are named as indicated below and in Figure 1. We postulated that this strategy would allow us to obtain a ‘fingerprint’ of the genetic information present in the DNA extracted from the day 7 right parotid saliva sample of interest. The primers used to generate these PCR products for sequencing were: (i) E1F58 and E1B98 (above) to generate a 648-bp fragment [termed PCR 1] of the Ad5 E1 gene (only found in WT Ad5); (ii) E3F122 (5′-AGAGATGACCAACACAACCAACG-3′); a sequence that locates from nt 29481–29503 of WT Ad5 (access number: AY339865) or from nt 30686–30708 of AdhAQP1) and E3B114 (5′-ATGGATACACGGGGTTGAAGG-3′); a sequence that locates from nt 31065–31085 of WT Ad5 (access number: AY339865) or from nt 32189–32169 of AdhAQP1). These will generate either a 1604-bp fragment from E3 region of WT Ad5 [termed PCR 2] or a 1503-bp fragment from AdhAQP1 [termed PCR 2′]; (iii) E3F122 and Ad5WTE3B117 (5′-GCGTTCCAGCCAATGTCAAG-3′) to generate from PCR 2 an 817-bp fragment from E3 region of WT Ad5 [termed PCR 3]; (iv) E3F122 and Ad5AQPE3B110 (5′-CTGTAAAGCACAACTCCCACCTG-3′) to generate from PCR 2′ a 937-bp fragment from the E3 region of AdhAQP1 [termed PCR 4]; (v) AQPE2F12 (5′-TGTCTTCATCAGCATCGGTTCTG-3′) and AQPE2B6 (5′-ATCCCACAGCCAGTGTAGTCAATAG-3′) to generate a 500-bp fragment from the human AQP1 cDNA [termed PCR 5]; (vi) E1F58 and AQPE2B15 (5′-CTCACAATGCTTCCATCAAACGAG-3′) to generate a 2256-bp fragment from the E1/E2 junctional region of WT Ad5 [termed PCR 6]; and AQPE2F21 (5′-TACTGCCTGACCTTGGAATCGTCC-3′) and AQPE2B15 to generate a 2239-bp fragment from the AQP1/E2 region of AdhAQP1 [termed PCR 7].
The sequences for all forward and reverse primers, and probes for the QPCR assays, were selected using Primer Express Primer Design software (PE Applied Biosystems) and the assays performed in an ABI Prism 7700 Sequence Detector. The conditions used for the QPCR assays were: 95 °C for 2 min, 95 °C for 8 min, 95 °C for 15 s and 60 °C for 1 min for 40 cycles. The sequences for all forward and reverse primers for the PCR reactions were selected using MacVector software (MacVector, Inc, Cary, NC, USA). All PCR reactions were carried out in DNA Thermal Cycler (Perkin Elmer, Waltham, MA, USA). The conditions used for the PCR reactions were: 94 °C for 10 min, then a temperature cycle of 94 °C for 1 min, 55 °C or 60 °C for 1 min and 72 °C for 3 min, for 30 cycles.
Infectious assay methods
A549 cells were plated in two wells of a six-well plate and allowed to reach confluence. Thereafter, 7 μl of the day 7 right parotid saliva sample was added to one well and 3 μl of this saliva sample was added to the other well. Both saliva aliquots were from a fresh thaw of the sample. Thereafter, 1 ml of RPMI-1640 medium (with 10% fetal calf serum; double antibiotics; all obtained from Invitrogen, Carlsbad, CA, USA) was added to each well, followed by an additional 2 ml of complete growth medium after 2 h. Growth medium was changed every 2 days and the cells monitored for the occurrence of cytopathic effects for 24 days. Two wells of control cells, in a separate six-well plate, were treated similarly, but without added aliquots of saliva, and served as a negative control. As a positive control, we used WT Ad5 (102–104 particles) to infect A549 cells at three multiplicities of infection (MOIs) over a 10-day period. The assay was capable of detecting the presence of WT Ad5 at a MOI of 0.025 (104 particles/4 × 105 cells) in 5 days and at a MOI of 0.0025 (103 particles) in 7 days.
Assay for neutralizing antibodies
Anti-Ad5 neutralizing antibodies in serum samples were measured by the ability to prevent transduction of 293 cells by a recombinant Ad5 vector [8,9]. Briefly, 293 cells were plated at 105 cells/well in 96-well plates, approximately 24 h prior to exposure to an Ad5 vector (AdCMVLuc; approximately 106 particles/well) encoding the luciferase reporter gene. For 30 min prior to adding vector to cells, the vector was incubated in serum dilutions (total volume 50 μl). Thereafter, the vector and the diluted serum samples were added to each well for 1 h, followed by the further addition of 150 μl of fresh culture media. After subsequent incubation for 24 h, cells in each well were lysed and luciferase activity determined in a luminometer. Titers are reported as the dilution of serum that resulted in 50% inhibition of the transduction of 293 cells with AdCMVLuc.
Saliva collection
To collect parotid saliva from participants, a Teflon device, called a Carlson-Crittenden cup, was placed over the orifice of Stensen’s duct [10]. Saliva, if present, flows from the duct orifice, into the cup, and then through a plastic tube into the collection vial (a pre-weighed Eppendorf tube). After completion of a timed, stimulated (with 2% citrate swabbing of the dorsum of the tongue) parotid saliva collection, there typically is some saliva still present in the Teflon cup, as well as in the plastic tube that drains into the collection vial. This extra parotid saliva is retained in a second pre-weighed Eppendorf tube with a notation as ‘extra’ saliva. Although this extra sample does not constitute the timed primary saliva sample for determining salivary flow rates per study protocol, it is available for analyses if needed (see below).
Results
General clinical course
Patient #25 is a 58-year-old Caucasian male, with a history of irradiation-induced salivary hypofunction subsequent to treatment for a poorly differentiated squamous cell carcinoma in the right tonsil that was metastatic to a single node (stage 4 A). He was enrolled in NIH study 06-D-0206 under IND 13 102 after meeting all eligibility criteria. He was the second participant to be treated with the first dose level of AdhAQP1 (4.8 × 107 vector particles). Prior to this treatment, all of his clinical laboratory tests (hematology, serum chemistry, urinalysis) were within normal limits, except for slightly lower values for white blood cells and serum amylase (Table 1). Vector was administered to the patient’s right parotid gland on day 1 (Figure 2).
Table 1.
Selected clinical laboratory parameters after AdhAQP1 gene transfer to patient #25
| Day 1 (pre-vector) | +6 h | +12 h | +24 h | +48 h | Day 7 | Day 14 | Day 28 | Day 30 | Day 42 | |
|---|---|---|---|---|---|---|---|---|---|---|
| WBC | SL ↓ | SL ↑ | WNL | WNL | WNL | WNL | WNL | WNL | ND | WNL |
| ESR | WNL | WNL | WNL | WNL | WNL | WNL | WNL | WNL | ND | WNL |
| CRP | WNL | WNL | WNL | WNL | WNL | WNL | WNL | WNL | ND | WNL |
| AMY | SL ↓ | SL ↓ | WNL | WNL | WNL | WNL | WNL | WNL | ND | WNL |
| IgG | WNL | WNL | WNL | WNL | WNL | WNL | WNL | WNL | ND | WNL |
| LDH | WNL | WNL | WNL | WNL | WNL | WNL | WNL | WNL | WNL | WNL |
| ALT | WNL | WNL | WNL | WNL | WNL | WNL | WNL | 42 ↑ | 44 ↑ | WNL |
| AST | WNL | WNL | WNL | WNL | WNL | WNL | WNL | 75 ↑ | 45 ↑ | WNL |
| CK | WNL | WNL | WNL | WNL | WNL | WNL | WNL | 3370 ↑ | 812 ↑ | WNL |
WNL, within normal limits; SL, slight; ↑, decrease; ↓, increase; WBC, white blood cells; ESR, erythrocyte sedimentation rate; CRP, C-reactive protein; AMY, serum amylase; IgG, serum immunoglobulin G; LDH, lactate dehydrogenase; ALT, alanine aminotransferase (U/l); AST, aspartate aminotransferase (U/l); CK, creatine kinase (U/l).
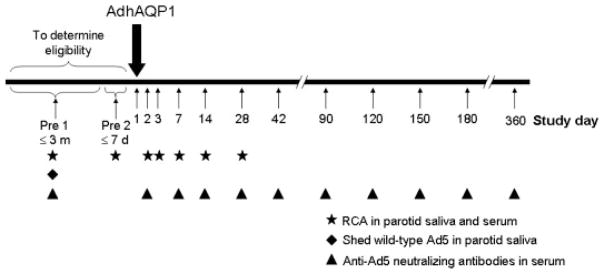
Figure 2.
Timeline depicting the general design of NIH clinical protocol 06-D-0206. Patients are seen for two screening visits to determine their eligilility. The first visit (Pre 1) must occur within 3 months of vector delivery, whereas the second (Pre 2) must occur within 1 week of vector delivery. Patients meeting all inclusion and exclusion criteria are administered AdhAQP1 on day 1 (large downward arrow). After vector administration, patients are seen on each of the study days indicated (small upward arrows). Saliva and serum samples are assayed for the WT serotype 5 adenovirus (Ad5) E1 gene (RCA; surrogate QPCR assay for replication competent adenovirus), shed, infectious WT Ad5, and anti-Ad5 neutralizing antibodies, on the study days indicated by specific symbols. At each visit, patients receive a physical examination, medical history review, oral/head and neck examination, saliva collection, and multiple clinical laboratory analyses (e.g. hematology, serum chemistry). Additionally, multiple imaging procedures are performed throughout the study
After vector administration, the patient was seen for regularly scheduled evaluations and testing per protocol (Figure 2). At each time point, collection of parotid saliva was made and blood samples were obtained. Throughout the thus far 180-day post-AdhAQP1 delivery period, the patient has reported no untoward symptoms, and has had essentially normal system reviews, physical examinations, and detailed head and neck examinations. Additionally, his clinical laboratory test results (Table 1) were generally within normal limits. On day 28, significant elevations in aspartate aminotransferase (serum glutamic oxaloacetic transaminase), alanine aminotransferase (serum glutamic pyruvic transaminase) and creatine kinase levels were documented (Table 1). These elevations were recorded as adverse events unrelated to the delivery of study drug or protocol procedures, and were considered to be related to recent strenuous exercise reported by the participant. By day 30, after refraining from vigorous physical activity, these three serum values were approaching normal levels and, at day 42, all three were within normal limits.
In addition, all special imaging studies performed on this patient have revealed no unexpected or untoward findings. These included a single pre-treatment contrast X-ray (sialogram) of the targeted right parotid gland, a single pre-treatment computed tomogram of the mandible and neck, multiple magnetic resonance images of the face/sinuses/neck from pre-treatment to day 42, multiple technetium scintiscans of the salivary glands from pre-treatment to day 42, two gallium scans of the parotid glands (pre-treatment and day 2), and a single pre-treatment computed tomogram of the chest.
Initial finding of adenoviral E1 gene in saliva specimen
At the day 7 visit, the patient had no general or oral symptoms and had an unremarkable head and neck examination, unremarkable review of systems and normal physical examination. Saliva samples were collected and, per protocol, tested for the presence of replication competent adenovirus using the surrogate QPCR assay for detecting the WT Ad5 E1 gene described above. The initial assay of DNA extracted (1 μl) from a collected sample of right parotid saliva on day 7 was positive at low levels for the E1 gene. Subsequently, multiple extracted DNA samples from the day 7 right parotid saliva sample were tested using three different amounts of extracted DNA per assay (1 μl, n = 10; 5 μl, n = 5; 10 μl, n = 6). A total of two 1-μl samples, one 5-μl sample, and all six 10-μl samples were positive. On the basis of the values from the six 10-μl samples, a calculated value of 82 E1 gene copies/μl parotid saliva was obtained. This value represents a total number of 7872 E1 gene copies in the 96-μl sample of right parotid saliva collected during the protocol’s timed collection period (1 min). In this same saliva sample, we also detected a total of 7.2 × 105 copies of the AdhAQP1 vector (1.5 × 103 copies/μl saliva; assayed by QPCR; see above). Importantly, all serum samples tested from this patient visit were negative for the presence of the Ad5 E1 gene and AdhAQP1. On the basis of these aggregate results, further study enrollment was suspended on day 9 pending a detailed understanding and resolution of this event.
Given the finding of the Ad5 E1 gene in the primary saliva sample, we tested the additional extra saliva from the same time point for the Ad5 E1 gene. When multiple samples of extracted DNA obtained from this extra right parotid saliva (1 μl, n = 9; 10 μl, n = 3) were tested for the presence of the Ad5 E1 gene by QPCR, all were negative. All other collected saliva samples, and all serum samples, from patient #25 (i.e. those obtained before day 7 and subsequently on days 14, 28 and 42), tested negative for the presence of the Ad5 E1 gene and the AdhAQP1 vector.
We also measured the Ad5 neutralizing antibody titer present in the day 14-serum sample collected. Although this time point was relatively early after vector administration to allow a classic antibody response, given that the participant’s neutralizing antibody titer at his pre-vector administration visit was ≤1:1024, we considered that, if there was a significant level of replicating Ad5 vector in his gland, we would expect to see a robust antibody response at day 14. There was none, however (i.e. his serum neutralizing antibody titer at day 14 was ≤ 1:512).
It is noteworthy that the other two patients enrolled in this study, who received the same vector dose as patient #25, tolerated the procedures and treatment well. These patients, #40 and #19, are Caucasian males, and were aged 66 and 56 years, respectively, at the time of AdhAQP1 administration. Prior to vector delivery, their anti-Ad5 neutralizing antibody levels were ≤1:4 and ≤1:256, respectively. Routine testing (Figure 2) of their parotid saliva and serum specimens, from prior to vector delivery until the day 28 visit, gave negative results for the Ad5 E1 gene with the QPCR assay.
Hypothesis and characterization of adenoviral DNA in parotid saliva sample
On the basis of the above findings with patient #25, we hypothesized two possible explanations regarding how the Ad5 E1 gene came to be present in the patient’s day 7 right parotid saliva sample. The first possibility was that the detected E1 gene was the result of contamination with WT Ad5 that occurred either during collection, when making aliquots or during the assay. The likelihood of a contamination of the primary timed saliva sample (versus the extra saliva sample or serum samples) is unlikely in view of the presence of AdhAQP1 in the primary saliva sample, although it is impossible to unequivocally rule out. Second, we considered that it was possible that patient #25 had a latent Ad5 infection (i.e. asymptomatic) in his right parotid gland and the AdhAQP1 vector transduced some of the cells containing WT Ad5. Adenoviruses can infect parotid glands, although frank clinical manifestations (parotitis) are only reported for immunocompromised patients [11]. The presence of an asymptomatic Ad5 infection in this patient might then permit either: (i) a recombination event to occur between the WT Ad5 and AdhAQP1 or (ii) reactivation of the WT Ad5 and subsequent replication of both the WT Ad5 and the AdhAQP1 vector, independently, in the absence of a recombination event.
With either scenario (i) or (ii), virus/vector replication could have lysed the cells in which it was occurring. Shed virus/vector would then have entered the duct lumen and remained, because this participant makes no resting (unstimulated) saliva (i.e. until a stimulus to secretion occurred, in this case during our timed collection). This sudden burst of saliva secretion would in effect wash out the duct of virus/vector. This circumstance would also provide an explanation for the finding that only the primary timed right parotid saliva sample was Ad5 E1 gene positive, but not the extra sample. A variation on this hypothesis is also possible (i.e. even if the cells that contained virus/vector were intact and not lysed, the virus/vector would likely be in an apical location in these polarized epithelial cells and would be released on the initial secretory stimulus). With a small number of cells affected, the saliva collected subsequently would be negative. Scenarios (i) and (ii) also are consistent with the finding that no WT Ad5 (or AdhAQP1) was found in serum at any time (i.e. no release of virus/vector occurred across the basolateral pole of the cell).
Initially, we generated the PCR products PCR 2 (1604 bp) and PCR 2′ (1503 bp) for the E3 targeted sequences in WT Ad5 and AdhAQP1, respectively, with extracted DNA from the positive day 7 right parotid saliva sample. For both products the same 5′-primer was used because the corresponding sequence occurs in both pJM17 and WT Ad5. However, PCR 2 would only be generated from WT Ad5 (i.e. it targets a sequence deleted in pJM17). Reciprocally, PCR 2′ would only be present in the AdhAQP1 vector (i.e. it is derived from the pJM17 mutated E3 sequence). Four rounds of PCR led to good bands, at the expected sizes, in duplicate samples for each primer pair (not shown). These reaction products then served as the template for generating PCR products 3 and 4 (Figure 1). These two products were generated, and the PCR product bands were extracted and then sequenced (Figure 3). Distinct results were seen for each product in the duplicate products generated: a sequence matching the WT Ad5 E3 region and a sequence matching the pJM17 E3 sequence. All of the other PCR products were generated, except for PCR 7, and had sequences identical to those expected (Table 2).
Figure 3.
Sequences of PCR products 3 and 4. For the location of these two PCR products in the WT serotype 5 adenovirus and AdhAQP1 genomes, see Figure 1. The underlined font in PCR 4 represents the mutation in the Ad5 E3 region found in pJM17. For details on the primers used and PCR reaction conditions, see the Materials and methods
Table 2.
Summary of PCR product sequencing results
| PCR product | Sequence result |
|---|---|
| PCR 1 | Identical with WT Ad5 |
| PCR 2 | Generated only |
| PCR 2′ | Generated only |
| PCR 3 | Identical with WT Ad5 |
| PCR 4 | Identical with AdhAQP1 |
| PCR 5 | Identical with AdhAQP1 |
| PCR 6 | Identical with WT Ad5 |
| PCR 7 | Unable to generate |
All generated products, except PCR 2 and PCR 2′, were isolated from agarose gels and subjected to conventional sequencing. PCR 2 and PCR 2′ were, in effect, the templates for the PCR reactions used to generate PCR 3 and PCR 4.
We also tested the ability of two aliquots of a fresh thaw of the day 7 parotid saliva sample of patient #25 to infect A549 cells. Neither of the two wells of A549 cells inoculated with the saliva sample showed any evidence for cytopathic effects after 24 days of observation.
Discussion
The aggregate results from all of the experiments performed with the parotid saliva and serum samples from patient #25 are consistent with the notion that he had a sub-clinical, low level, latent, WT Ad5 infection of his right parotid gland at the time of gene transfer. Administration of AdhAQP1 to that gland led to WT Ad5 reactivation and AdhAQP1 replication. Thereafter, after cell lysis and virus/vector release into the gland lumen, the small amount of virus/vector was washed out of the gland by stimulating secretion of parotid saliva. The PCR findings are consistent with the presence of separate WT Ad5 and AdhAQP1 in the day 7 right parotid saliva sample, and thus favor the absence of any recombination event having occurred in the patient’s gland. Nonetheless, the PCR results cannot completely exclude the possibility (and potential for) a recombination. The patient has remained in good health up to his day 180 visit, and showed no evidence of WT Ad5/AdhAQP1 vector in all other saliva samples, and all serum samples up to day 42 visit. These findings support a view that the occurrence of the Ad5 E1 gene in the patient’s day 7 parotid saliva sample was an idiosyncratic episode with no discernable clinical consequences.
Several factors may have led to the detection of the Ad5 E1 gene in the day 7 saliva sample of patient #25. First, adenoviral infections are quite common across widely diverse populations [12–15], with latent infections present in many organs and in lymphoid tissue [16]. Second, the reactivation of latent adenovirus can occur in immunosuppressed patients, such as in post-transplant or HIV-AIDS patients [11,16,17]. Furthermore, in transplant patients, this reactivation appears to be facilitated during antiviral (CMV) prophylaxis with either ganciclovir or valganciclovir [16]. Finally, although the precise mechanism(s) involved in such reactivations are not entirely clear, reactivation may also involve exposure to inflammatory cytokines [18,19] and/or interactions with other viral proteins that may be present in the same tissues [20,21].
In the context of the above general factors, there are two specific considerations regarding the clinical status of patient #25 that may have contributed to the reactivation of the latent WT Ad5. First, the patient previously was treated for a right tonsillar carcinoma and experienced considerable anatomical deformity after both his original surgery, as well as from subsequent surgery needed to correct radiation-induced osteonecrosis. As a result, it is reasonable to suggest that normal lymphatic drainage in this anatomical area of the patient would be compromised. Second, the patient uses, on a daily basis, valacyclovir for prophylactic treatment of Herpes infections. Although the effects of valacyclovir were not studied in the solid organ transplant studies of latent virus reactivation [16], it is a similar anti-viral drug to those studied [22]. These considerations, along with the administration of AdhAQP1, may have provided the circumstances to allow reactivation of the latent WT Ad5 present in his right parotid gland.
As indicated above, we consider this event was an idiosyncratic episode that could occur after Ad5 vector-mediated gene transfer to any tissue. The approved clinical protocol originally included two screening assays to assess the previous exposure of potential patients to Ad5: (i) determination of Ad5 E1 gene presence in parotid saliva and serum and (ii) measurement of serum neutralizing antibody levels. None of the three patients treated with 4.8 × 107 particles of AdhAQP1 showed evidence for Ad5 E1 gene presence in samples obtained prior to vector administration. All three, however, had neutralizing antibodies present in their serum. Interestingly, patient #25 exhibited the highest serum neutralizing antibody level of the three patients. Typically, 35–50% of a study population displays neutralizing antibodies to Ad5 in their serum [12–15]. The titer found for patient #25 prior to AdhAQP1 delivery was well above the average level observed in these studies. As a further safety precaution with this clinical trial in the future, with any patient for whom a stimulated saliva sample can be collected from the target parotid gland during the screening visit, an aliquot of that parotid saliva sample will be used to inoculate A549 cells to test for the presence of shedding infectious virus (i.e. an assay comparable to that used in the present study; Figure 2). If infectious virus emerges during a subsequent 10-day follow-up period, then the patient will no longer be eligible for treatment and will be withdrawn from the study. However, if no cytopathic effects are observed, the patient may be treated provided that all other eligibility criteria are met. This additional screening step, although potentially helpful, may still be inadequate to identify patients with a latent parotid gland Ad5 infection. Indeed, the day 7 parotid saliva sample from patient #25 was negative in this infectious assay. Although that finding may be a result of inadequate assay sensitivity, it more likely reflects the high level and widely diverse nature of anti-viral activities present in saliva [23–26] (i.e. contact of any shed virus/vector with saliva may simply render the virus/vector non-infectious). Therefore, it appears to be most prudent to view that a positive QPCR assay for the Ad5 E1 gene in a saliva sample at screening may indicate the presence of a latent Ad5 infection in the targeted gland. As a result, and as is currently carried out, the patient should be excluded from enrollment. However, and as noted earlier, this criterion would not have led to the exclusion of patient #25 from the present study. Given the addition of the screening assay for shedding infectious virus, the medical status of the patients being studied in this trial, and the findings from the current PCR and other investigations with saliva samples from patient #25, no further steps appear to be necessary with respect to preventing an event such as this from re-occurring.
Acknowledgments
The Division of Intramural Research, National Institute of Dental and Craniofacial Research, NIH, provided all the support for this research. We thank Ms Jennifer Bacik and Dr Robert Lindblad for their comments on portions of this manuscript.
Footnotes
This article is a U.S. Government work and is in the public domain in the U.S.A.
References
- 1.Cox JD, Steitz J, Pajak TF. Toxicity criteria of the Radiation Therapy Oncology Group (RTOG) and the European Organization for research and treatment of cancer. Int J Radiat Oncol Biol Phys. 1995;31:1341–1346. doi: 10.1016/0360-3016(95)00060-C. [DOI] [PubMed] [Google Scholar]
- 2.Baum BJ. Principles of saliva secretion. Ann NY Acad Sci. 1993;694:17–23. doi: 10.1111/j.1749-6632.1993.tb18338.x. [DOI] [PubMed] [Google Scholar]
- 3.Vitolo JM, Baum BJ. The use of gene transfer for the protection and repair of salivary glands. Oral Dis. 2002;8:183–191. doi: 10.1034/j.1601-0825.2002.02865.x. [DOI] [PubMed] [Google Scholar]
- 4.Vissink A, Jansma J, Spijkervet FK, et al. Oral sequelae of head and neck radiotherapy. Crit Rev Oral Biol Med. 2002;14:199–212. doi: 10.1177/154411130301400305. [DOI] [PubMed] [Google Scholar]
- 5.Preston GM, Agre P. Isolation of the cDNA for erythrocyte integral membrane protein of 28 kDa: member of an ancient channel family. Proc Natl Acad Sci USA. 1991;88:11110–11114. doi: 10.1073/pnas.88.24.11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delporte C, O’Connell BC, He X, et al. Increased fluid secretion after adenoviral-mediated transfer of the aquaporin-1 cDNA to irradiated rat salivary glands. Proc Natl Acad Sci USA. 1997;94:3268–3273. doi: 10.1073/pnas.94.7.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shan Z, Li J, Zheng C, et al. Increased fluid secretion after adenoviral-mediated transfer of the human aquaporin-1 cDNA to irradiated miniature pig parotid glands. Mol Ther. 2005;11:444–451. doi: 10.1016/j.ymthe.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Sprangers MC, Lakhal W, Koudstaal W, et al. Quantifying adenovirus-neutralizing antibodies by luciferase transgene detection: addressing pre-existing immunity to vaccine and gene therapy vectors. J Clin Microbiol. 2003;41:5046–5052. doi: 10.1128/JCM.41.11.5046-5052.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng C, Vitolo JM, Zhang W, et al. Extended transgene expression from a nonintegrating adenoviral vector containing retroviral elements. Mol Ther. 2008;16:1089–1097. doi: 10.1038/mt.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ship JA, Fox PC, Baum BJ. How much saliva is enough?. Normal function defined. J Am Dent Assoc. 1991;122:63–69. doi: 10.14219/jada.archive.1991.0098. [DOI] [PubMed] [Google Scholar]
- 11.Gelfand MS, Cleveland KO, Lancaster D, et al. Adenovirus parotitis in patients with AIDS. Clin Infect Dis. 1994;19:1045–1048. doi: 10.1093/clinids/19.6.1045. [DOI] [PubMed] [Google Scholar]
- 12.D’Ambrosio E, Del Grosso N, Chicca A, Midulla M. Neutralizing antibodies against 33 human adenoviruses in normal children in Rome. J Hygiene. 1982;89:155–161. doi: 10.1017/s0022172400070650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chirmule N, Propert KJ, Magosin SA, et al. Immune responses to adénovirus and adeno-associated virus in humans. Gene Ther. 1999:1574–1583. doi: 10.1038/sj.gt.3300994. [DOI] [PubMed] [Google Scholar]
- 14.Nwanegbo E, Vardas E, Gao W, et al. Prevalence of neutralizing antibodies to adenoviral serotypes 5 and 35 in the adult populations of The Gambia, South Africa and the United States. Clin Diag Lab Immunol. 2002;11:351–357. doi: 10.1128/CDLI.11.2.351-357.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang Z, Li Y, Cun A, et al. Chimpanzee adénovirus antibodies in humans, sub-Saharan Africa. Emerg Infect Dis. 2006:1596–1599. doi: 10.3201/eid1210.060078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humar A. Reactivation of viruses in solid organ transplant patients receiving cytomegalovirus prophylaxis. Transplantation. 2006;82:S9–S14. doi: 10.1097/01.tp.0000230432.39447.8b. [DOI] [PubMed] [Google Scholar]
- 17.Durepaire N, Ranger-Rogez S, Gandji JA, et al. Enteric prevalence of adenovirus in human immunodeficiency patients. J Med Virol. 1995;45:56–60. doi: 10.1002/jmv.1890450111. [DOI] [PubMed] [Google Scholar]
- 18.Higashimoto Y, Elliott WM, Behzad AR, et al. Inflammatory mediator mRNA expression by adenovirus E1A-transfected bronchial epithelial cells. Am J Respir Crit Care Med. 2002;166:200–207. doi: 10.1164/rccm.2111032. [DOI] [PubMed] [Google Scholar]
- 19.Leen A, Ratnayake M, Foster A, et al. Contact-activated monocyt: efficient antigen presenting cells for the stimulation of antigen-specific T cells. J Immunother. 2007;30:96–107. doi: 10.1097/01.cji.0000211325.30525.84. [DOI] [PubMed] [Google Scholar]
- 20.Metcalf JP, Monick MM, Stinski MF, et al. Adenovirus E1A 13S gene product up-regulates the cytomegalovirus major intermediate early promoter. Am J Respir Cell Mol Biol. 1994;10:448–452. doi: 10.1165/ajrcmb.10.4.8136160. [DOI] [PubMed] [Google Scholar]
- 21.Westphal E-M, Mauser A, Swenson J, et al. Induction of lytic Epstein–Barr virus (EBV) infection in EBV-associated malignancies using adenovirus vectors in vitro and in vivo. Cancer Res. 1999;59:1485–1491. [PubMed] [Google Scholar]
- 22.Reischig T, Opatrny K, Jr, Treska V, et al. Prospective comparison of valacyclovir aand oral ganciclovir for the prevention of cytomegalovirus disease in high-risk renal transplant patients. Kidney Blood Press Res. 2005;28:218–225. doi: 10.1159/000087129. [DOI] [PubMed] [Google Scholar]
- 23.Yeh C-K, Handelman B, Fox PC, Baum BJ. Further studies of salivary inhibition of HIV-1 infectivity. J Acquir Immune Defic Syndr. 1992;5:898–903. [PubMed] [Google Scholar]
- 24.Bergey EJ, Gu M, Collins AR, et al. Modulation of herpes simplex virus type 1 replication by human salivary secretions. Oral Microbiol Immunol. 1993;8:89–93. doi: 10.1111/j.1399-302x.1993.tb00551.x. [DOI] [PubMed] [Google Scholar]
- 25.Drobni P, Naslund J, Evander M. Lactoferrin inhibits human papillomavirus binding and uptake in vitro. Antiviral Res. 2004;64:63–68. doi: 10.1016/j.antiviral.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 26.White MR, Helmerhorst EJ, Ligtenberg, et al. Multiple components contribute to ability of saliva to inhibit influenza viruses. Oral Microbiol Immunol. 2009;24:18–24. doi: 10.1111/j.1399-302X.2008.00468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]