Abstract
Use of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) is associated with dramatic, durable, and tolerable responses and side effect profiles when applied for palliation of advanced EGFR-mutated non-small-cell lung cancers (NSCLCs). Expert guidelines recommend that EGFR mutation testing results should be available within 10 working days of receipt of tumor specimen by the testing laboratory; in circumstances where the tumor specimen needs to be sent to an external laboratory for testing, the sample should be sent within 3 working days of receiving the request for testing. We report here 2 cases, out of 109 EGFR-mutated (exon 19 deletion or L858R) NSCLCs seen at our institution, experiencing rapid clinical deterioration and death within the window of time prescribed by consensus testing guidelines. We hypothesize that a faster turn-around time may have changed the clinical outcome. Improving rapid turnaround times for tumor genotyping may afford more optimal palliation vis-à-vis early initiation of oral targeted therapy in patients with advanced EGFR-mutated NSCLC.
Keywords: EGFR, TKI, NSCLC, gefitinib, erlotinib, afatinib
Introduction
In 2013, the International Association for the Study of Lung Cancer (IASLC), Association for Molecular Pathology (AMP), and College of American Pathologists (CAP) published guidelines for molecular testing of epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK) gene rearrangements in advanced lung adenocarcinomas (LACs). Expert consensus calls for EGFR mutation (exon 19 deletions and L858R genotype) and ALK rearrangement results to be available within 10 working days of receipt of tumor specimen by the testing laboratory; if send out testing is required, the specimen should be submitted to the outside vendor within 3 working days of receiving the request for testing1. It is unknown if a shorter turn-around time would impact outcomes of patients with advanced LAC.
Here, we report two cases of rapidly fatal LACs harboring sensitizing EGFR mutations. Patients seen at Beth Israel Deaconess Medical Center whose tumors were genotyped were identified through an ongoing Institutional Review Board-approved protocol2; 109 cases with EGFR exon 19 deletions (n=65) or L858R (n=44) were identified. Median overall survival was 37.6 months (95%CI 29.9-45.3) for advanced LACs (n =89). Of these, 2 (2.2%) patients died within 28 days of tissue diagnosis.
Discussion
Case Presentation #1
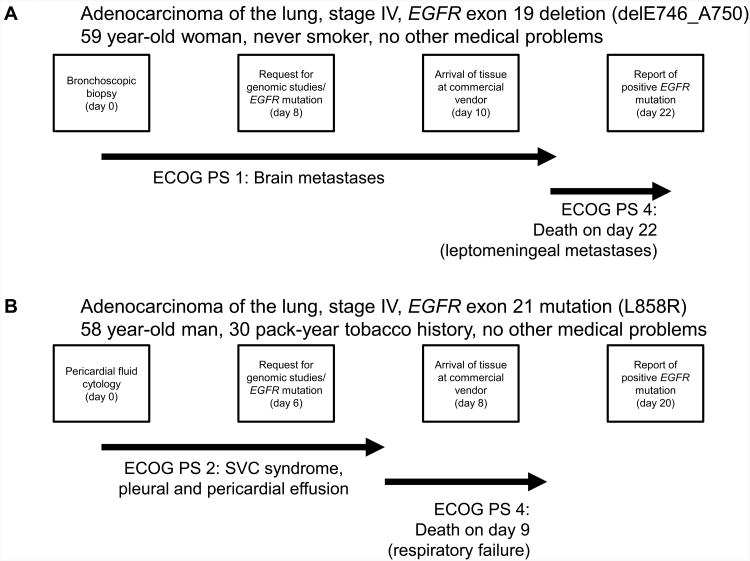
A previously healthy 59-year-old white female and non-smoker presented with 3 months of cough and dyspnea, subsequently developing acute headaches and intermittent confusion over one week. Imaging studies showed a left upper lung mass and multiple brain metastases. Eastern Cooperative Oncology Group (ECOG) performance status (PS) at presentation was 1. Transbronchial biopsy was performed (day 0), and final pathologic report showed LAC (day 4). A request for tumor genotyping (EGFR sequencing and ALK FISH) was placed (day 8), and tissue was submitted to an outside vendor (received on day 10). The patient completed palliative whole brain radiotherapy (3000cGy) during this interim. PS was noted to have declined to 3, precluding cytotoxic chemotherapy. On day 8, she suffered further abrupt clinical deterioration (PS of 4) due to encephalopathy. Cerebrospinal fluid analysis confirmed malignant leptomeningeal involvement. Refractory encephalopathy led to withdrawal of care on day 16, and the patient died on day 22. A finalized report disclosing the EGFR genotype was received on the same day as death (day 22), showing a sensitizing exon 19 deletion (E746_A750, Figure 1A).
Figure 1.
Tumor genotype procedures and timeframe. A.) Initial tissue sampling, genomic study request, arrival to vendor, issue of final report, and concise clinical course for case 1. B.) Initial tissue sampling, genomic study request, arrival to vendor, issue of final report, and concise clinical course for case number 2. Date of tissue acquisition in both cases = Day 0.
Case Presentation #2
A 58 year-old gentleman with a 30 pack-year tobacco history presented with 2 months of progressive dyspnea and PS of 2. Imaging studies demonstrated a large right-sided mediastinal mass causing superior vena cava (SVC) obstruction, pleural, and pericardial effusions. Pericardial fluid cytology (day 0) revealed LAC (final report on day 5). A request for genomic testing was made (day 6), and the sample was received by the offsite vendor (day 8). Due to unstable clinical status, cytotoxic chemotherapy could not be initiated. Pericardiocentesis, thoracentesis, SVC stenting, and tracheal stenting with intra-bronchial tumor debridement were performed, but the patient suffered respiratory failure with further decline in PS to 4 by day 5. He required ventilator support and died on day 9. A finalized report disclosing the EGFR genotype was received on day 20, showing a sensitizing exon 21 L858R mutation (Figure 1B).
Conclusions
Cases of rapidly fatal (within the first month following a diagnostic procedure) EGFR-mutated LAC were rare (<2%) in our cohort. In most circumstances, testing turnaround times (TATs) within the window currently endorsed by expert groups will allow for timely and efficacious care. We also note that testing paradigms may differ in various cancer centers/care settings, i.e. internal testing (which may offer more rapid results as compared to send-out testing). In those cases where patients are progressing rapidly and cannot wait for molecular testing results (but are otherwise appropriate candidates for cytotoxic chemotherapy), systemic therapy should not be withheld; patients can be transitioned to targeted therapies either once molecular data is known or upon completion of the planned initial course of chemotherapy3.
However, the experiences reported here highlight instances in which the current approach may hinder care in a small subset of patients with advanced disease, i.e. those (as in the patients presented here) in whom use of cytotoxic chemotherapies is precluded by inadequate clinical/performance status. Here, rapid knowledge of sensitizing oncogenic driver mutations may afford treatment with a relatively nontoxic, highly efficacious therapy within a narrow therapeutic window—and use of such agents in those with poor performance status (PS 3-4) is supported by evidence-based guidelines3 Despite tumor submission, testing, and reporting periods that fall within the timelines endorsed by expert groups1,4, the current strategy may provide results too late for therapeutic intervention in these circumstances.
Availability of point-of-care (POC) genomic testing may allow for rapid initiation of EGFR TKIs (gefitinib, erlotinib, afatinib), which are associated with initial rapid tumor regression in almost all cases of EGFR exon 19 deletion or L858R-mutated LACs5,6. It is therefore speculative to consider whether the patients presented here may have had more favorable outcomes if EGFR-targeted therapy had commenced soon after diagnosis. We propose POC testing with TATs in the 5-7 day range as an actionable target—the technology to accomplish this is being explored, but will take time to adequately develop for widespread or commercial use. Noninvasive genotyping methods using peripheral blood to assess either cell-free DNA (cfDNA) and/or circulating tumor cells (CTCs) are a rapidly evolving domain for improving the ease and rapidity of testing. In a recently published study, peripheral blood cfDNA testing with digital droplet PCR afforded rapid TATs (median 3 days) and with moderate sensitivity (69-80%)7; however, such technologies remain investigational for the time being. As such platforms evolve, maintaining precision while maximizing speed and accessibility will remain important priorities in the effort to optimally match patients with therapies.
Acknowledgments
Funding/Grant Support: This work was funded in part through a Lung Cancer Foundation of America-International Association for the Study of Lung Cancer grant (to DBC), an American Cancer Society grant RSG 11-186 (to DBC), and National Cancer Institute grants CA090578 (to DBC).
Footnotes
Conflict of interest: Daniel B. Costa has received consulting fees and honoraria from Pfizer (unrelated to the current work), and honoraria from Boehringer Ingelheim (unrelated to the current work). EF has served as scientific consultant for Boston Scientific (unrelated to the current work), education consultant for Olympus (unrelated to the current work) and is the principal investigator for an ongoing trial in navigation bronchoscopy funded by Medtronic (unrelated to the current work). No other conflict of interest is stated.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2013 Jul;8(7):823–859. doi: 10.1097/JTO.0b013e318290868f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vanderlaan PA, Yamaguchi N, Folch E, et al. Success and failure rates of tumor genotyping techniques in routine pathological samples with non-small-cell lung cancer. Lung cancer. 2014 Apr;84(1):39–44. doi: 10.1016/j.lungcan.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ettinger DS, Wood DE, Akerley W, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 4.2016. Journal of the National Comprehensive Cancer Network: JNCCN. 2016 Mar;14(3):255–264. doi: 10.6004/jnccn.2016.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leighl NB, Rekhtman N, Biermann WA, et al. Molecular testing for selection of patients with lung cancer for epidermal growth factor receptor and anaplastic lymphoma kinase tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the College of American Pathologists/International Association for the study of lung cancer/association for molecular pathology guideline. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014 Nov 10;32(32):3673–3679. doi: 10.1200/JCO.2014.57.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jorge SE, Kobayashi SS, Costa DB. Epidermal growth factor receptor (EGFR) mutations in lung cancer: preclinical and clinical data. Brazilian journal of medical and biological research = Revista brasileira depesquisas medicas e biologicas / Sociedade Brasileira de Biofisica … [et al] 2014 Nov;47(11):929–939. doi: 10.1590/1414-431X20144099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. The Lancet Oncology. 2012 Mar;13(3):239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 7.Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA oncology. 2016 Apr 7; doi: 10.1001/jamaoncol.2016.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]