SUMMARY
System-wide quantitative analysis of ubiquitylomes has proven to be a valuable tool for elucidating targets and mechanisms of the ubiquitin-driven signaling systems, as well as gaining insights into neurodegenerative diseases and cancer. Current mass spectrometry methods for ubiquitylome detection require large amounts of starting material and rely on stochastic data collection to increase replicate analyses. We describe a method compatible with cell line and tissue samples for large-scale quantification of 5,000–9,000 ubiquitylation forms across ten samples simultaneously. Using this method, we reveal site-specific ubiquitylation in mammalian brain and liver tissues, as well as in cancer cells undergoing proteasome inhibition. To demonstrate the power of the approach for signal-dependent ubiquitylation, we examined protein and ubiquitylation dynamics for mitochondria undergoing PARKIN- and PINK1-dependent mitophagy. This analysis revealed the largest collection of PARKIN- and PINK1-dependent ubiquitylation targets to date in a single experiment, and it also revealed a subset of proteins recruited to the mitochondria during mitophagy.
Graphical Abstract
INTRODUCTION
Ubiquitylation, the process through which ubiquitin (UB) is covalently linked via an E1-E2-E3 cascade to lysine residues in cellular proteins, constitutes a central pathway through which cellular decisions are made (Finley, 2009; Komander and Rape, 2012). This reflects roles for ubiquitylation in both the rapid removal of proteins from the cell via UB-dependent proteasomal degradation and rapid changes in the architecture of signaling complexes reliant on UB assemblies as scaffolds. Ubiquitylation is also a highly versatile post-translational modification, and it can involve assemblies of eight distinct chain-linkage types (M1, K6, K11, K27, K29, K33, K48, and K63), wherein K11 and K48 contribute preferentially to proteasomal turnover. In keeping with such central roles in biology, components of the UB system are frequently mutated in disease (Ciechanover and Kwon, 2015; Morrow et al., 2015).
A central feature of the UB system is its dynamicity. It is now clear that cellular signals such as phosphorylation can promote the following: (1) intrinsic activation of the UB conjugation machinery itself; (2) recognition of substrates by the conjugation apparatus; and (3) removal of UB from proteins via deubiquitylating enzymes, among other forms of regulation. Despite these well-known avenues of cellular control, we rarely have a quantitative understanding of how the ubiquitylation state of particular proteins and particular ubiquitylation sites in proteins are altered in a signal-dependent manner and how modification may be altered in the context of genetic defects in the signaling pathway. Moreover, very little is known concerning how the ubiquitylomes of tissues vary. These limitations are due in part to the absence of methods that allow facile quantitative profiling of ubiquitylation events. Quantitative analysis of ubiquitylated peptides to elucidate UB signaling events has traditionally employed label-free- (Yu et al., 2015) or metabolic label- (Kim et al., 2011; Udeshi et al., 2013a) based mass spectrometry (MS) approaches in combination with enrichment using di-glycine remnant antibodies, which capture ubiquitylated peptides from proteins post-trypsinization (Ordureau et al., 2015b). However, these methods measure biological replicates in multiple nano liquid chromatography tandem mass spectrometry (nLC-MS/MS) experiments, limiting the number of peptides quantified across all samples, and they typically require large amounts of starting material (5–40 mg/sample) (Kim et al., 2011; Na et al., 2012; Udeshi et al., 2013b). Further complicating systems-based approaches for understanding UB signaling is the potential stochastic sampling afforded when using the di-glycine capture approach, as evidenced by incomplete overlap of di-glycine-modified peptides identified in biological replicate experiments.
Isobaric labels offer many advantages for quantifying post-translational modifications (PTMs) by MS, such as phosphorylation (Erickson et al., 2015). Isobaric labels facilitate a decrease in starting material used for enrichment, because peptides from multiple samples are mixed before analysis and appear as one peak in a mass spectrum. Additionally, isobaric labels minimize missing values by enabling measurements across all replicates with a single MSn event. However, chemical tags label the primary amine of the di-glycine remnant, thereby inhibiting enrichment via immunoprecipitation (IP) with anti-di-glycine antibodies after samples are labeled and mixed.
The perceived variability of di-glycine enrichment has dissuaded separate enrichment of ubiquitylated samples and, by extension, the use of isobaric labels. If the di-glycine remnant IP variability was sufficiently low (e.g., coefficient of variation [CV] < 10%), it would be possible to utilize isobaric labeling to enable deep, multiplexed quantitation of ubiquitylomes from cell culture, tissue, and clinical samples. In this report, we describe the use of isobaric tagging coupled with di-glycine enrichment to profile quantitative changes in the ubiquitylomes of cells and tissues, largely overcoming current limitations with label-free and metabolic-labeling approaches. Moreover, we demonstrate the utility of this approach for systematically elucidating signal-dependent ubiquitylation on a global scale in the context of the PARKIN-PINK1 pathway responsible for depolarization-dependent mitochondrial ubiquitylation promoting mitophagy.
RESULTS AND DISCUSSION
Isobaric Tagging Allows Multiplexed Quantitative Di-glycine Remnant Profiling
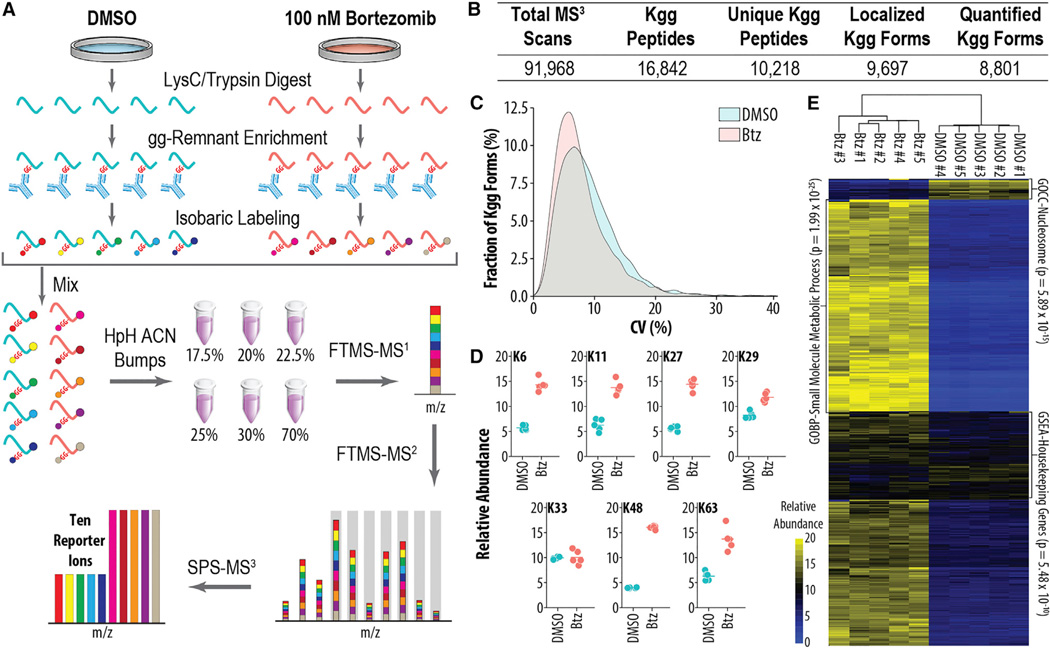
The ability to perform quantitative di-glycine profiling in a multiplexed manner would greatly facilitate the analysis of dynamic ubiquitylation processes. To assess the technical reproducibility of the di-glycine remnant antibody enrichment and subsequent isobaric labeling, HTC116 cells were treated for 16 hr with either DMSO or the proteasome inhibitor bortezomib (Btz) (Figure 1). Proteasome inhibition greatly increases the abundance of ubiquitylated proteins and provides an effective setting for examining reproducibility. Peptides were generated by trypsin digestion, cleaving UB from modified lysine residues and producing the di-glycine remnant (Figure 1A) (Peng et al., 2003). To isolate the variance induced by the antibody enrichment and isobaric labeling, the resulting peptide pool was split into five technical replicates, each containing 1 mg of material. The peptide mixtures were then enriched using a di-glycine remnant antibody, labeled with ten-plex tandem mass tags (TMTs) (McAlister et al., 2012; Werner et al., 2012), combined, separated into six fractions using a high-pH reversed-phase spin cartridge, and analyzed in 3-hr nLC-MS/MS experiments using an synchronous precursor selection MS3 (SPS-MS3) method on a tribrid mass spectrometer (Orbitrap Fusion; Figure 1A) (McAlister et al., 2014; Ting et al., 2011). Preliminary experiments revealed that isobaric tags increased gas phase charge of ubiquitylated peptides more drastically than non-ubiquitylated peptides due to an additional tag on the di-glycine residue (Figure S1). Excluding +2 precursors from selection during MS analysis effectively ignored ~40% of all non-ubiquitylated peptides, thereby substantially increasing the amount of instrument time dedicated to ubiquitylated peptides (Figure S1B).
Figure 1. Isobaric Labeling Method for Reproducible, Multiplexed Quantitation of Ubiquitylomes.
(A) Sample preparation and instrumental workflow. Proteins from samples are isolated and digested yielding peptides that are enriched, labeled with isobaric tags, and mixed. Peptides are separated by offline high pH reversed phase (HpH-RP) before analysis by MS using the SPS-MS3 method.
(B) Summary statistics of experiment depicted in (A) are shown.
(C) Coefficient of variation (CV) (n = 5) for either DMSO- or Btz-treated cells is shown.
(D) Quantitative values for ubiquitylated lysine residues on UB are shown.
(E) Global changes in ubiquitylation upon Btz treatment are shown.
In total, we identified 10,218 unique modified peptides and quantified 8,801 sites of ubiquitylation across all ten 1 mg samples in just 18 hr of analysis time (1.8 hr/ubiquitylome) (Figure 1B). Encouragingly, the median CV among five technical replicates was 8.13% and 6.88% for the DMSO- and Btz-treated samples, respectively (Figure 1C). Measurements of the ubiquitylated lysine residues on UB itself enabled quantification of sites involved in various UB biologies. For example, K6 was significantly altered by 2.51 ± 0.31-fold, while K48 exhibited the largest change of 4.04 ± 0.12-fold (Figure 1D). Hierarchical clustering revealed that technical replicates clustered tightly, and the vast majority of ubiquitylation sites increased in abundance when the proteasome was inhibited (Figure 1E), as expected based on previous studies (Kim et al., 2011). Note that it is important to consider the ratio of antibody to peptide input when performing di-glycine remnant IP. For a 16-hr Btz treatment, accumulation of UB K48 peptide prohibited large amounts of starting material; for example, beginning with 10 mg/sample yielded reproducible quantification of 18,471 ubiquitylation forms (Figure S2A and S2B), but quantitation of UB chains differed from previous studies (Kim et al., 2011). This was addressed by lowering starting amount to 1 mg/sample, but also could be addressed by increasing the amount of antibody used for the IP.
Multiplexed Di-glycine Remnant Profiling Allows Quantitative UB Profiling in Tissues
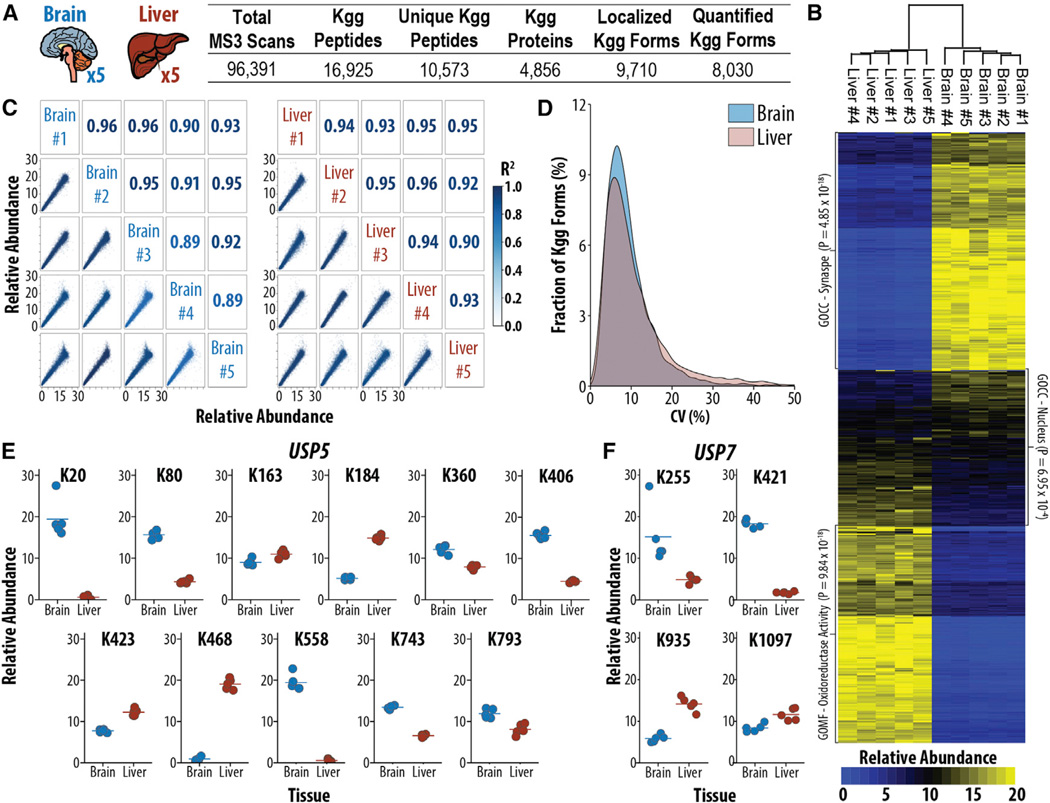
Quantitative analysis of proteins and PTMs from mouse tissues provides fundamental insights for analyzing mouse models of human diseases (Humphrey et al., 2015; Morris et al., 2015). Label-free approaches have become a popular alternative to metabolic labeling of mice (Robles et al., 2014), but they typically start with large amounts of material (e.g., 40 mg/sample) (Na et al., 2012) and rely on multiple experiments to obtain replicate measurements. Thus, we sought to explore the compatibility of isobaric label-based ubiquitylome analysis with mammalian tissue samples. Using the method outlined above, we enriched 7 mg peptide input from five biological replicates of liver or brain. In total, using only 18 hr of analysis time (1.8 hr/ubiquitylome), we identified 10,573 ubiquitylated peptides of which 8,030 were localized and quantified across all ten samples (Figure 2A). Quantification of ubiquitylomes from biological replicates of tissues was highly correlated, with R2 values ≥ 0.89 for all comparisons of biological replicates (Figure 2B). Encouragingly, the CV of the five biological replicates was 8.08% and 8.49% for brain and liver, respectively, demonstrating that biological variation did not prohibit system-wide ubiquitylome analyses of tissue samples (Figure 2C). Encouragingly, this amount of variability enabled statistically significant (p < 0.05) measurements of fold changes as low as 20%, with a statistical power >0.9.
Figure 2. Multiplexed Ubiquitylome Analysis Reveals Tissue-Specific Ubiquitylation Events on Key Proteins in the UB-Proteasome System.
(A) Experimental design and summary statistics for the analysis of ubiquitylation in biological replicates of mouse brain and liver (n = 5) are shown.
(B) Global differences between brain and liver for protein normalized ubiquitylation abundances are shown.
(C) Correlation of biological replicates in each tissue. R2 values are displayed with color corresponding to the degree of correlation.
(D) CV (n = 5) for either brain or liver biological replicates is shown.
(E and F) Protein normalized ubiquitylation quantification for USP5 (E) and USP7 (F) reveals tissue-specific ubiquitylation events.
To account for variations in protein expression, we adjusted ubiquitylation measurements to protein levels resulting in 6,382 normalized ubiquitylation forms (Figure 2C). Curiously, a subset of proteins exhibited varying tissue specificity of multiple ubiquitylation sites. Two proteins involved in the ubiquitin proteasome system (UPS), USP5 and USP7, exhibited at least one site specifically ubiquitylated in either brain or liver tissue (Figures 2E and 2F). USP5, a deubiquitinating enzyme involved in the processing of UB precursors (Grou et al., 2015) and cell growth (via the p53-dependent apoptotic pathway) (Potu et al., 2014), exhibited four significantly increased (fold change > 2, p < 0.01) ubiquitylation sites in the brain (K163, K184, K423, and K468) and two significantly increased sites in the liver (K20 and K558; Figure 2E). Likewise, USP7, a deubiquitinating enzyme involved in tumor suppression, DNA repair, and immune responses (Kessler et al., 2007; Sowa et al., 2009), had two sites significantly increased in the liver (K935 and K1097) and one site significantly increased in the brain (K421; Figure 2F). These results demonstrate that a deeper analysis of tissue-specific ubiquitylation afforded by isobaric labels has the potential to reveal and quantify a large number of tissue-specific ubiquitylation events in a highly reproducible manner.
Temporal Analysis of Protein Ubiquitylation via Di-glycine Remnant Multiplexing
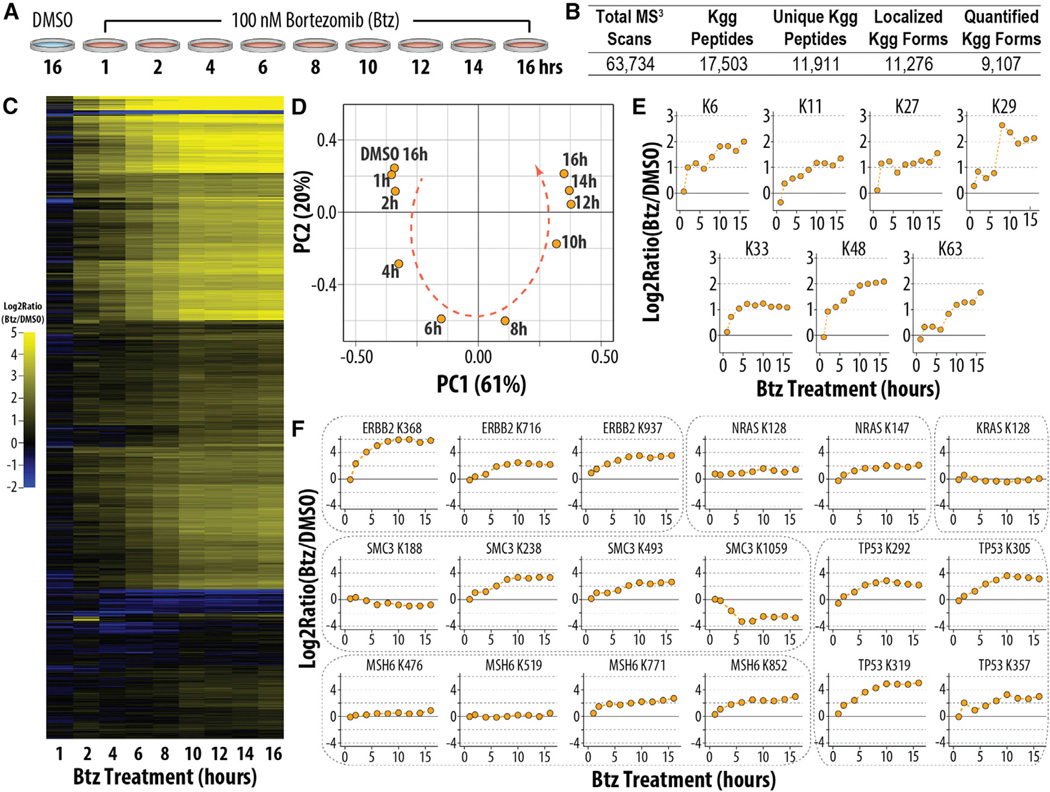
Unlike metabolic-labeling approaches, isobaric labeling enables the simultaneous analysis of up to ten biological conditions, permitting the analysis of multi-point time course data in one experiment (Murphy et al., 2015). We treated cells with Btz and harvested them at 1, 2, 4, 6, 8, 10, 12, 14, and 16 hr post-treatment; additionally, we treated cells for 16 hr with DMSO as a control (Figure 3A). Starting from 1 mg/sample, we identified 11,911 unique ubiquitylated peptides of which 9,036 were localized and quantified in 18 hr of analysis time (1.8 hr/ubiquitylome) (Figure 3B). As expected, nearly all ubiquitylation sites increased in response to Btz treatment (Figure 3C). Principal-component analysis demonstrated that the DMSO-treated control was most similar to the 1-hr Btz-treated cells and depicted a clear progression from earlier to later time points (Figure 3D). Ubiquitylation of lysine residues on UB increased after Btz treatment, and nearly all measurements corroborated those made in the initial experiment (Figure 1D). For example, K48 was measured as changing 4.0- and 4.2-fold in the initial and time course experiments, respectively (Figure 3E). A subset of these residues was ubiquitylated more than 2-fold after only 1 hr of Btz treatment (e.g., K6, K27, and K48), while others increased more gradually (e.g., K11 or K63) (Dammer et al., 2011).
Figure 3. Multiplexed Ubiquitylome Analysis Monitors Cellular Response to Btz over Nine Time Points.
(A) Experimental design of Btz treatment time course is shown.
(B) Summary statistics for the experiment analyzing the samples in (A) are shown.
(C) Global ubiquitylome changes in response to treatment with Btz are shown.
(D) Principal-component analysis of changes in ubiquitylation upon Btz treatment is shown.
(E) Time-dependent changes in ubiquitylation of ubiquitinated lysine residues on UB are shown.
(F) Ubiquitylation changes of proteins involved in cancer pathways upon treatment with Btz are shown.
HTC116 cells are a colon-derived cancer cell line permitting the quantification of ubiquitylation dynamics of proteins involved in cancer pathways (Figure 3F). As expected, the majority of ubiquitylation sites increased after Btz treatment; for example, ubiquitylation of K368 on the tyrosine kinase ERBB2 increased from ~4-fold after 2 hr to ~64-fold after 8 hr of Btz treatment (Figure 3F). Four sites of ubiquitylation were quantified on MSH6, a protein involved in DNA damage repair. Two sites (K771 and K852) increased over the duration of the time course, while the others (K476 and K519) did not change substantially. These results demonstrated that not all sites are regulated equally upon proteasomal inhibition, and they suggested that particular sites were either selectively ubiquitylated or protected from ubiquitylation.
Multiplexed Proteomics Reveal Dynamic Alterations in Mitochondrial Ubiquitylation and Protein Recruitment during PARKIN-Dependent Mitophagy
The high level of multiplexing offered by isobaric labels enables complex experimental designs that include multiple replicates, time points, and cellular conditions. To demonstrate the power of multiplexing for ubiquitylome analysis, we used isobaric labels to characterize proteome and ubiquitylome dynamics during mitophagy. In this process, mitochondrial depolarization leads to the accumulation of the PINK1 protein kinase on the mitochondrial outer membrane (MOM), which then promotes activation of MOM protein ubiquitylation by the PARKIN UB ligase (Ordureau et al., 2015b; Pickrell and Youle, 2015; Yamano et al., 2016). PINK1 stimulates this process through a feedforward mechanism involving both phosphorylation of S65 in PARKIN’s N-terminal UB-like (UBL) domain, which activates its UB chain assembly activity, and through phosphorylation of S65 on UB chains assembled on mitochondria (Ordureau et al., 2014, 2015a). pS65-UB serves to bind and further activate PARKIN. Together these events lead to the depolarization-induced ubiquitylation of dozens of primary sites in multiple MOM proteins (Sarraf et al., 2013; Bingol et al., 2014), as well as the assembly of K6, K11, K48, and K63 UB chains, which are phosphorylated on S65 with a stoichiometry of ~0.2 via PINK1 (Heo et al., 2015; Ordureau et al., 2014, 2015a).
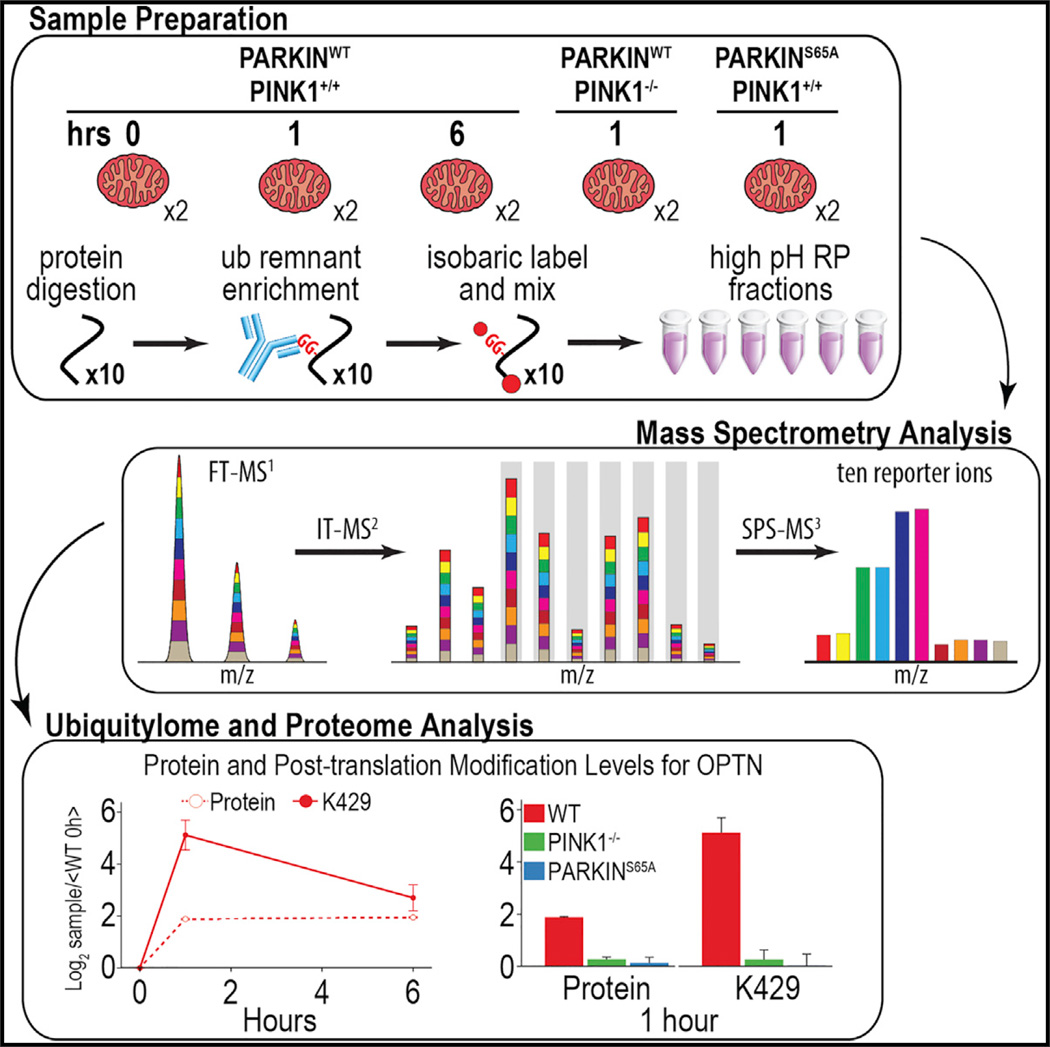
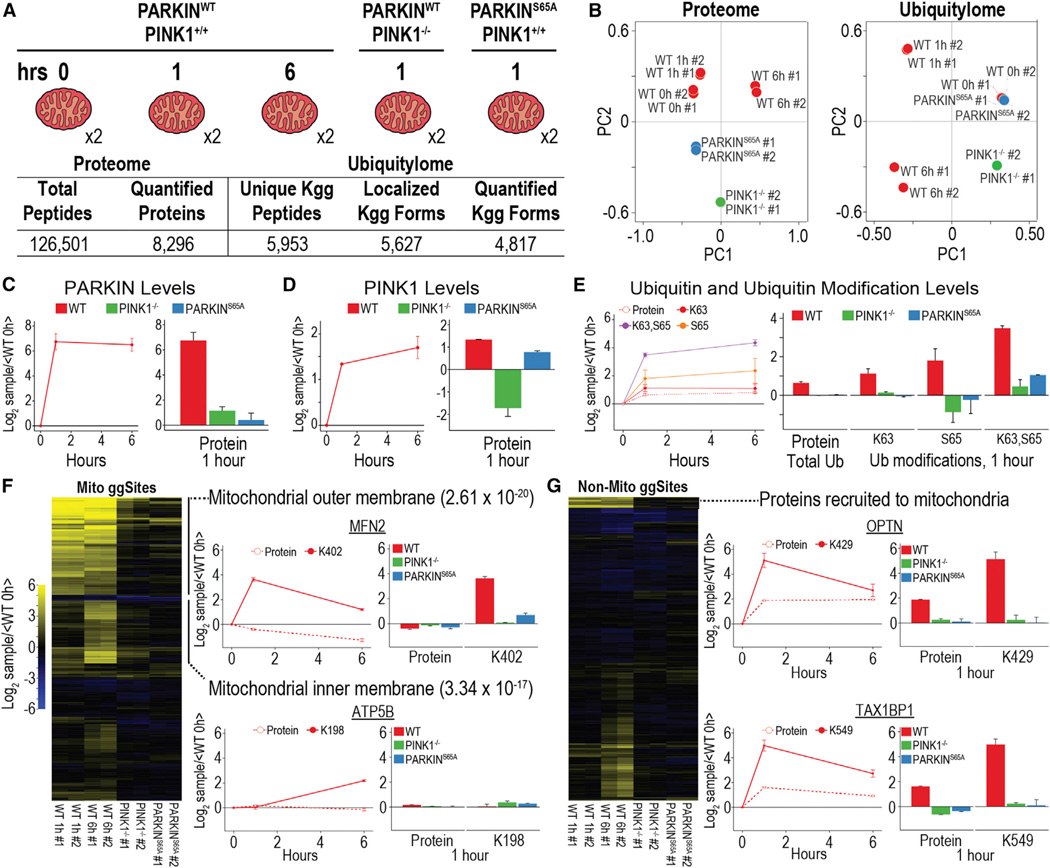
Prior di-glycine capture studies in this system were performed using primarily binary comparisons and did not directly assess the requirement for PINK1 activity and PARKIN S65 phosphorylation in MOM ubiquitylation (Sarraf et al., 2013; Bingol et al., 2014), making comparisons across experiments and conditions challenging. In addition, previous studies have indicated that autophagy adaptors and other regulatory proteins, in addition to PARKIN, are recruited to mitochondria in response to depolarization (Heo et al.,2015; Lazarou et al.,2015; Richter et al., 2016); but, the full repertoire of proteins recruited to depolarized mitochondria is unknown. To capture a complete and dynamic view of the mitophagy induction and the role of PINK1 activity and PARKIN phosphorylation, we measured total protein and ubiquitylation dynamics in enriched mitochondrial samples from HeLa cells conditionally expressing PARKINWT at 0, 1, and 6 hr after the induction of mitophagy by depolarization. Additionally, we measured the effect of a 1-hr treatment on PARKINWT-expressing cells lacking PINK1 (PINK1−/−) or cells expressing non-phosphorylatable PARKINS65A (Figure 4A). All five conditions were simultaneously measured in biological duplicate in a single ten-plex quantitative MS experiment. In total, 8,296 proteins and 4,817 di-glycine-containing peptides from 2,015 proteins were quantified across all ten samples in only 36 and 18 hr, respectively (Figure 4A). Principal-component analysis demonstrated superb reproducibility of measurements among biological replicates and that PINK1−/− and PARKINS65A ubiquitylomes closely resemble PARKINWT cells without depolarization (0 hr) (Figure 4B).
Figure 4. Proteome and Ubiquitylome Dynamics of Enriched Mitochondria Undergoing Mitophagy.
(A) Experimental design and summary statistics of proteome and ubiquitylome analysis are shown.
(B) Principal-component analysis for proteome and ubiquitylome data is shown.
(C and D) PARKIN (C) and PINK1 (D) protein expression during mitophagy for three time points in WT cells and one time point in PINK1−/− and PARKINS65A cells is shown.
(E) Expression levels for total levels of UB protein and modifications of UB, including K63 ubiquitylation, S65 phosphorylation, and a form of UB containing both modifications, are shown.
(F) Hierarchical clustering for ubiquitylation forms belonging to mitochondrial proteins. Protein and ubiquitylation expression for mitochondrial outer (MFN2) and inner membrane (ATP5B) proteins is shown.
(G) Hierarchical clustering for ubiquitylation forms belonging to non-mitochondrial proteins. Protein and ubiquitylation expression for proteins recruited to the mitochondria during mitophagy (OPTN and TAX1BP1) is shown.
All points and bars represent the mean ratio for biological replicates (± 1 SD).
Crude enrichment of mitochondria resulted in the quantification of 1,001 of 1,158 MitoCarta 2.0 proteins, and it enabled the clear identification of cytosolic proteins recruited to the mitochondria, using either proteome- or ubiquitylome-level quantification. As expected, PINK1 and PARKIN abundance on mitochondria rapidly increased upon depolarization. Protein measurements confirmed this, as PINK1 and PARKIN protein levels increased in wild-type (WT) cells after depolarization, and, as expected based on prior studies (Ordureau et al., 2014, 2015a), PARKIN was not efficiently recruited in cells lacking PINK1 or in the context of the PARKINS65A mutant (Figures 4C and 4D). A form of UB-containing phosphorylation of S65 (singly and in combination with K63 di-glycine modification) can be used as a direct measure of PINK1-PARKIN activity in response to depolarization (Ordureau et al., 2014). This phosphorylated form of UB increased dramatically in PARKINWT cells, while exhibiting a muted response in the absence of PINK1 or in the context of PARKINS65A, which isn’t efficiently recruited to mitochondria and therefore doesn’t efficiently build UB chains on mitochondria (Heo et al., 2015; Ordureau et al., 2014, 2015a) (Figure 4E). Total UB protein level as well as the ubiquitylation of K6, K11, K48, and K63 increased in PARKINWT cells, as previously shown (Cunningham et al., 2015; Ordureau et al., 2014), while remaining virtually unchanged in both PINK1−/− and PARKINS65A cells (Figure 4E; Table S5).
Hierarchical clustering of the PARKIN-dependent ubiquitylome revealed distinct trends for mitochondrial and non-mitochondrial ubiquitylation forms (Figures 4F and 4G). MOM proteins, which constitute the primary substrate for ubiquitylation events during mitophagy, demonstrated dramatic increases in ubiquitylation at the 1-hr time point in WT cells, and they exhibited little ubiquitylation in either the PINK1−/− or PARKINS65A cells. For example, Mitofusin-2 (MFN2) is a MOM protein that mediates mitochondrial fusion and is widely recognized as a target of PARKIN in response to depolarization (Pickrell and Youle, 2015). Time course measurements demonstrated an ~50% decrease in MFN2 protein expression in PARKINWT cells, consistent with previous studies indicating a role for the proteasome and p97 in PARKIN-dependent MFN2 turnover (Tanaka et al., 2010). In contrast, ubiquitylation of the previously identified primary ubiquitylation site (K402; Sarraf et al., 2013) demonstrated a dramatic increase at the 1-hr time point, an event that was dependent upon PINK1 and S65 in PARKIN (Figure 4F). In total, 130 proteins known to be localized to mitochondria had di-glycine-containing peptides whose abundance increased by at least 1.8-fold at 1 and/or 6 hr post-depolarization in a manner that required PINK1 activity or PARKIN phosphorylation on Ser 65 (Table S6). This includes 99 ubiquitylation sites in 42 proteins identified in prior di-glycine enrichment studies (Sarraf et al., 2013) (Figure S3A).
Importantly, isobaric tagging-based analysis provided deeper coverage of ubiquitylation sites than previous isotope labeling-based studies (Sarraf et al., 2013). For example, as compared to Sarraf et al. (2013), isobaric labeling quantified 16 additional ubiquitylation sites on TOMM70A (Figure S3A), nine of which were detected previously in a non-quantitative analysis of PARKIN ubiquitylation targets (Bingol et al., 2014). Likewise, additional ubiquitylation sites were found in several other MOM proteins, including CPT1A, CYB5R1, and VDAC1/VDAC2 (Figure S3A). Previous studies indicate PARKIN-dependent membrane rupture after extended periods of mitochondrial depolarization (Yoshii et al., 2011). Hierarchical clustering identified a second group of 30 inner mitochondrial membrane/matrix proteins (Table S7) that also are ubiquitylated preferentially at 6 hr of depolarization in a PINK1-dependent manner, with little or no ubiquitylation occurring at 1 hr. This group included ubiquitylation of K198 in ATP5B, a component of ATP synthase, as well as K96 and K252 in VDAC1 (Figure 4F; Figure S3B). Previous studies also identified ubiquitylation of ATP5B at long time points (Sarraf et al., 2013). Taken together, these results suggest that, during depolarization, PARKIN gains access to a cohort of mitochondrial inner membrane proteins possibly as a result of MOM rupture, although we cannot formally rule out stalled translocation of inner membrane proteins with concomitant ubiquitylation by PARKIN (Sarraf et al., 2013), which is known to associate with the translocon via PINK1 (Lazarou et al., 2012).
The multiplexing approach when combined with an enrichment step also can provide a means by which to examine signal-dependent recruitment of proteins to a particular cellular organelle. Indeed, we quantified recruitment of all the previously reported UB-binding mitophagy adaptors (OPTN, TAX1BP1, SQSTM1 [p62], CALCOCO2 [NDP52], and NBR1) known to be recruited to mitochondria to promote mitophagy (Heo et al., 2015; Lazarou et al., 2015; Richter et al., 2016). For example, protein levels for OPTN and TAX1BP1 were found to be dynamically enriched on mitochondria at 1 hr post-depolarization, and this correlated with ubiquitylation of K429 and K549 for OPTN and TAX1BP1, respectively (Figure 4G). As expected, this recruitment was dependent on PINK1 and PARKIN activity. Moreover, our systematic data suggest that PINK1- and PARKIN-dependent recruitment of proteins to mitochondria during mitophagy is perhaps much more widespread than previously appreciated. Indeed, hierarchical clustering revealed a group of 137 ubiquitylated peptides derived from 85 proteins classified in Uniprot as cytoplasmic that exhibit ubiquitylation dynamics, and in many cases enrichment on mitochondria, consistent with mitochondrial recruitment from the cytosol.
Conclusions
Here we report a novel method for system-wide, multiplexed ubiquitylation analysis of both cell culture- and tissue-derived samples. We demonstrate that independent IP of ubiquitylated peptides from parallel samples followed by isobaric labeling is highly reproducible, producing CVs of ~8% for both technical and biological replicates. Furthermore, isobaric labeling enables deep quantitation of ubiquitylomes (8,000–9,000 quantified sites) starting with only 1 and 7 mg peptide per sample for cell culture or tissue samples, respectively. Simultaneous measurement with isobaric labels allows quantification of ten ubiquitylomes in only 18 hr of analysis (1.8 hr/ubiquitylome), representing a drastic decrease in instrument time compared to current methods. For example, initial quantitative analysis of the PARKIN system (Sarraf et al., 2013) involved 72 independent binary IP-MS experiments, including 16 experiments employing HeLa cells as performed here, in order to overcome stoichastic sampling and analysis of multiple time points. The use of isobaric tagging greatly simplifies such analyses, providing a large number of known and new candidate PARKIN targets in a single experiment that has the flexibility to incorporate genetic controls for pathway activation.
UB-driven signaling is central to many cellular processes and also is disrupted in disease, including cancer, neurodegeneration, and immunity (Popovic et al., 2014; Weathington and Mallampalli, 2014). The methods described here will greatly facilitate large-scale quantitative analysis of ubiquitylation dynamics, an understanding of global protein homeostasis, and a system-wide level. Moreover, this method has the potential to aid in the elucidation of how ubiquitylation pathways are quantitatively altered in disease through applications that involve tissue analysis in the context of animal models, an experimental target that is not possible with current technology.
STAR*METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
EXPERIMENTAL MODEL AND SUBJECT DETAILS
- METHOD DETAILS
- Mammalian Culture Cell Lysate
- Mice Tissues Lysate
- Mammalian Culture Cell and Mice Tissues Lysate Digestion
- Mitochondrial Isolation and Digestion
- Immunoprecipitation of diGly-Containing Peptides
- TMT Labeling and Sample Fractionation
- Orbitrap Fusion and Lumos Parameters
- Data Processing and Spectra Assignment
QUANTIFICATION AND STATISTICAL ANALYSIS
DATA AND SOFTWARE AVAILABILITY
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PTMScan Ubiquitin Remnant Motif (K-ε-GG) Kit | Cell Signaling Technology | Cat#5562 |
| Protein A Plus Ultralink resin | ThermoFisher Scientific | Cat#53142 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Bortezomib | ApexBio Technology | Cat#A2614 |
| Antimycin A | Sigma-Aldrich | Cat#A8674 |
| Oligomycin A | Sigma-Aldrich | Cat#75351 |
| Tandem Mass Tags | ThermoFisher Scientific | Cat#90406 |
| cOmplete, Mini Protease Inhibitor Cocktail | Sigma-Aldrich | Cat#11836153001 |
| PhosSTOP, Phosphatase Inhibitor | Sigma-Aldrich | Cat#4906837001 |
| EPPS | Sigma-Aldrich | Cat#E9502 |
| Rapigest SF Surfactant | Glixx Laboratories | Cat#GLXC-07089 |
| 2-Chloroacetamide | Sigma-Aldrich | Cat#C0267 |
| Aprotinin | Roche | Cat#10981532001 |
| Leupeptin | Roche | Cat#11034626001 |
| AEBSF Protease Inhibitor | Gold Biotechnology | Cat#A-540 |
| TCEP | Sigma-Aldrich | Cat#646547 |
| NEM | Sigma-Aldrich | Cat#E3876 |
| PR-619 | Selleck Chem | Cat# S7130 |
| Critical Commercial Assays | ||
| BCA Protein Assay Kit | ThermoFisher Scientific | Cat#23225 |
| Quantitative Colorimetric Peptide Assay | Thermo Fisher Scientific | Cat#23275 |
| High pH Reversed-Phase Peptide Fractionation Kit | ThermoFisher Scientific | Cat#84868 |
| Deposited Data | ||
| Raw Mass Spectrometry Data Files | This paper; Proteome Xchange | PXD004628 |
| Experimental Models: Cell Lines | ||
| HCT-116, Human colorectal carcinoma | ATCC | Cat#CCL-247 |
| HeLa Flp-In T-REx (HFT) PARKINWT | Ordureau et al., 2014 | N/A |
| HeLa Flp-In T-REx (HFT) PARKIN S65A | Ordureau et al., 2014 | N/A |
| HeLa Flp-In T-REx (HFT) PINK1−/− | Ordureau et al., 2014 | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6 | Jackson Laboratories | Stock#000664 |
| Software and Algorithms | ||
| In-house mass spectrometry data analysis software | Huttlin et al., 2010 | N/A |
| SEQUEST | Eng et al., 1994 | N/A |
| Perseus | Tyanova et al., 2016 | http://www.coxdocs.org/doku.php?id=perseus:start |
| R | https://www.r-project.org/ | N/A |
| Other | ||
| Orbitrap Fusion Mass Spectrometer | ThermoFisher Scientific | Cat#IQLAAEGAAPFADBMBCX |
| Orbitrap Fusion Lumos Mass Spectrometer | ThermoFisher Scientific | Cat#IQLAAEGAAPFADBMBHQ |
| Easy-nLC 1000 | ThermoFisher Scientific | Cat#LC120 |
STAR*METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
For inquiries about reagent and resource sharing please contact Steven Gygi (steven_gygi@hms.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human colorectal carcinoma cells (HCT116) were used for method development and time course analysis. Inducible HeLa Flp-In T-REx (HFT) cells with or without expression of PARKINWT or PARKINS65A proteins, or lacking PINK1 (Ordureau et al., 2014) were used for mitophagy studies.
All mice were lean male mice (8 weeks of age) obtained from The Jackson Laboratory (C57BL/6) and used to compare ubiquitylomes of brain and liver. Mice were housed at 23°C under a 12-hr light/dark cycle with free access to food and water. All animal experiments were performed according to procedures approved by the Institutional Animal Care and Use Committee (IACUC) of the Beth Israel Deaconess Medical Center.
METHOD DETAILS
Mammalian Culture Cell Lysate
Colorectal carcinoma cells (HCT116) were grown in DMEM supplemented with 10% FBS (heat treated, Seradigm), 2 mM glutamine and 100 units/ml penicillin/streptomycin (GIBCO). Cells were plated in 150 mm culture dishes (Corning) and allowed to grow 24 hr before treating them with bortezomib (Btz) or DMSO (ApexBio Technology) at the indicated times. All cells were harvested at a 70%–80% confluency. Cells were washed on ice twice with ice-cold PBS, scraped into 50 ml conical tubes and centrifuged at 215x g for 5 min at 4°C. Cells were re-suspended in 2 ml of lysis buffer [8 M urea, 50 mM HEPES (pH 7.2), 75 mM NaCl, 10 mM N-Ethylmaleimide (NEM), 1x protease inhibitor cocktail (Roche) and 1x PhosSTOP phosphatase inhibitor Cocktail (Roche)] and passed through a 21G needle 10 times.
Mice Tissues Lysate
Five mice were sacrificed individually by cervical dislocation and their brains and livers were harvested, washed twice with PBS, and snap frozen in liquid nitrogen. Brain/liver tissues were mechanically lysed with an Omni mixer homogenizer in SDS lysis buffer [2% SDS w/v, 50 mM HEPES (pH 7.2), 150 mM NaCl, 10 mM NEM, 1x protease inhibitor cocktail (Roche) and 1x PhosSTOP phosphatase inhibitor Cocktail (Roche)].
Mammalian Culture Cell and Mice Tissues Lysate Digestion
Suspensions were centrifuged at maximum speed for 15 min at 4°C and lysates were transferred to a clean 50 ml tubes. Lysates were reduced for 1 hr at room temperature with 10 mM DTT, and cysteine residues were then alkylated with 20 mM NEM (room temperature, 30 min). Protein content was extracted twice by methanol-chloroform precipitation and subsequent ice-cold acetone washes. Protein pellets were resuspended in 8 M urea, 50 mM HEPES (pH 8.2) buffer and protein concentrations were measured by BCA assay (Thermo Fisher Scientific). Samples were then diluted to 4 M urea with 50 mM HEPES (pH 8.2) and digested at 30°C for 6 hr with endoproteinase Lys-C (Wako, Japan) at a 1/100 enzyme/protein ratio. The mixtures were then diluted to 1 M urea with 50 mM HEPES (pH 8.2) and trypsin was added at a 1/50 enzyme/protein ratio. The reaction was incubated overnight at 37°C with gentle end-over-end rotation and stopped by acidification with formic acid (FA) 0.5% (v/v) (pH ~2). Peptides were subjected to tC18 SepPak solid-phase extraction cartridges (SPE) (Waters) and lyophilized. Peptide concentrations were determined using the microBCA assay (Thermo Fisher Scientific).
Mitochondrial Isolation and Digestion
Inducible HeLa Flp-In T-REx (HFT) cells with or without expression of PARKINWT or PARKINS65A proteins, or lacking PINK1 were previously described (Ordureau et al., 2014). To induce lower protein expression for each protein of interest, cells were treated with 0.5 µM doxycycline (DOX) for 16 hr to induce low levels of expression of the proteins of interest. Cells were either left untreated or depolarized with a mixture of Antimycin A (10 µM) and Oligomycin A (5 µM) (Sigma Chemical Company) (referred to as AO) for the indicated time period. At the indicated times, cells were washed twice with ice cold PBS containing 200 mM chloroacetamide and mitochondria were purified as described previously (Ordureau et al., 2014). Crude mitochondrial prep were lysed in 50 mM Tris/HCl pH 7.5,1 mM EDTA, 1 mM EGTA, 50 mM NaF, 5 mM sodium pyrophosphate, 10 mM sodium 2-glycerol 1-phosphate, 1 mM sodium orthovanadate, 1% (v/v) NP-40,1 mg/ml aprotinin, 1 µg/ml leupeptin, 1 mM benzamidine, 1 mM AEBSF, 10 µM PR-619,100 mM chloroacatemide and 1x PhosSTOP phosphatase inhibitor Cocktail (Roche). Crude mitochondrial extracts were sonicated, clarified by centrifugation (16000xg for 10 min at 4°C) followed by filtration through a 0.45 µM filter and protein concentrations determined by the Bradford assay.
Samples were subjected to reduction (5 mMTCEP – 15 min) and alkylation (30 mM chloroacetamide – 15 min) followed by cholorform/methanol precipitation. Samples were digested overnight at 25°C with Lys-C (1:100) [in 200 mM EPPS pH8.0, 0.1% Rapigest (Waters Corporation)] and trypsin (1:100) was added the next morning and samples incubated for a further 6 hr at 37°C. Digests were acidified with an equal volume of 2% (vol/vol) formic acid (FA) to a pH of ~2 for 30 min. Peptides were then subjected to tC18 SepPak solid-phase extraction cartridges (SPE) (Waters) and lyophilized.
Immunoprecipitation of diGly-Containing Peptides
The diGly monoclonal antibody (Cell Signaling Technology) (32 µg/IP) was coupled to Protein A Plus Ultralink resin (40 µl slurry/IP) (Thermo Fisher Scientific) overnight at 4°C prior to its chemical cross-linking reaction (Udeshi et al., 2013a). Dried peptides (1 mg for mammalian samples treated with Btz; 7 mg for mice tissues) were resuspended in 1.5 ml of ice-cold IAP buffer [50 mM MOPS (pH 7.2), 10 mM sodium phosphate and 50 mM NaCl] and centrifuged at maximum speed for 5 min at 4°C to remove any insoluble material. Supernatants (pH ~7.2) were incubated with the antibody beads for 2 hr at 4°C with gentle end-over-end rotation. After centrifugation at 215x g for 2 min, beads were washed three times with ice-cold IAP buffer and twice with ice-cold PBS. The diGly peptides were eluted twice with 0.15% TFA, desalted using homemade StageTips (Rappsilber et al., 2003) and dried via vacuum centrifugation. Each lysate was immunoprecipitated twice.
TMT Labeling and Sample Fractionation
Enriched diGly containing peptides were resuspended in 18 µl of 200 mM HEPES (pH 8.2). Isobaric labeling of the peptides was performed using the 10-plex tandem mass tag (TMT) reagents (Thermo Fisher Scientific). TMT reagents (0.8 mg) were dissolved in 40 µl anhydrous acetonitrile (ACN) of which 3 µl were added to the peptides along with 4 µl of ACN (30% (v/v) ACN final concentration). After 1 hr at at room temperature, the reaction was quenched with 2 µl of 5% hydroxylamine. Labeled peptides were combined, acidified with FA (pH ~2) and dried via vacuum centrifugation.
For ubiquitylated peptides the high pH reversed-phase peptide fractionation kit (Thermo Fisher Scientific) to separate peptides into six fractions (17.5% ACN, 20% ACN, 22.5% ACN, 25% ACN, 30% ACN and 70% ACN) according to manufacturer’s instructions; for mitochondrial enriched samples the 70% ACN fraction was replaced by a 27.5% ACN fraction due to low numbers of identifications in 70% ACN fractions. For whole proteome analysis peptides were separated by basic-phase reversed-phase chromatography, as previously described (Isasa et al., 2015). Lyophilized peptides were desalted using homemade StageTips and resuspended with 5 µl of 5% FA, 4% ACN prior to mass spectrometry analysis.
Orbitrap Fusion and Lumos Parameters
All spectra were acquired on an Oribtrap Fusion or Lumos mass spectrometer (Thermo Fisher Scientific) coupled to an Easy-nLC 1000 (Thermo Fisher Scientific) ultra-high pressure liquid chromatography (UHPLC) pump. Peptides were separated on an in-house packed 100 µM inner diameter column containing 0.5 cm of Magic C4 resin (5 mm, 100 Å, Michrom Bioresources) followed by 25 cm of Sepax Technologies GP-C18 resin (1.8 µm, 120 Å) with a gradient consisting of 8%-28% (ACN, 0.1% FA) over 180 min at ~450nL/min. The scan sequence began with FTMS1 spectra (resolution of 120,000; mass range 400–1250 m/z; automatic gain control (AGC) target 5 × 105, max injection time of 100 ms). The ten most intense MS1 ions were selected for MS2 analysis. Precursors were filtered according to charge state ≥ 2 or ≥ 3 for proteome and ubiquitylome analysis, respectively. Monoisotopic peak assignment was used and previously interrogated precursors were excluded using a dynamic window (60 s ± 10 or 7.5 ppm for ubiquitylome and proteome analysis, respectively).
For ubiquitylome analysis the MS2 precursors were isolated using the quadrupole (0.5 Th window) and analyzed in the Orbitrap (FTMS2) at 15,000 resolution, with an AGC target of 1 × 105 and a max injection time of 300 ms. Precursors were fragmented by CID at a normalized collision energy (NCE) of 35%. Following acquisition of each MS2 spectrum, a synchronous-precursor-selection (SPS) MS3 scan was collected on the top 10 most intense ions in the MS2 spectrum. SPS-MS3 precursors were fragmented by high energy collision-induced dissociation (HCD) and analyzed using the Orbitrap (NCE = 55%, AGC =1 × 105, maximum injection time = 500 ms, and resolution = 60K).
For proteome analysis the MS2 precursors were isolated using the quadrupole (0.5 Th window), with an AGC target of 2.5 × 104 and a max injection time of 150 ms. Precursors were fragmented by CID at a normalized collision energy (NCE) of 35% and analyzed in the ion trap. Following acquisition of each MS2 spectrum, a synchronous-precursor-selection (SPS) MS3 scan was collected on the top 10 most intense ions in the MS2 spectrum. SPS-MS3 precursors were fragmented by high energy collision-induced dissociation (HCD) and analyzed using the Orbitrap (NCE = 55%, AGC = 2.2 × 105, maximum injection time = 150 ms, and resolution = 60K).
Data Processing and Spectra Assignment
A compilation of in-house software was used to convert mass spectrometric data (Thermo “.raw” files) to mzXML format, as well as to correct monoisotopic m/z measurements and erroneous peptide charge state assignments (Huttlin et al., 2010). Assignment of MS/ MS spectra was performed using the SEQUEST algorithm (Eng et al., 1994). The Btz-treated cells utilized the Homo sapiens UniProt database (downloaded on 2014). The mice tissues analysis utilized the Mouse UniProt database (downloaded on 2014). Each database was concatenated with a database composed of all protein sequences in the reversed order as well as known contaminants (human keratins). Searches were performed using a 20 ppm precursor ion tolerance for total protein level analysis. The product ion tolerance was set to 0.03 Da for ubiquitylome analysis and 0.9 Da for proteome analysis. Peptide’s N/C terminus was required to have lysC/trypsin specificity [1 1 KR] and allowing up to 4 missed cleavages. TMT tags on peptide N termini/lysine residues (+229.16293 Da) and cysteines NEM-alkylation (+125.04767 Da) were set as static modifications; while methionine oxidation (+15.99492 Da) and lysine ubiquitylation (+ 114.04293 Da) were set as dynamic modifications. For mitochondrial enriched samples chloroacetamide was used in place of NEM for cysteine alkylation and the corresponding static mass shift (+57.02146 Da) was used for database searching. Additionally, mitochondrial enriched samples were searched with utilizing a variable mod of phosphorylation (+79.96633 Da) to identify phosphorylation sites on ubiquitin.
Peptide-spectrum matches (PSMs) were adjusted to the 1% false discovery rate (FDR) (Elias and Gygi, 2007). PSM filtering was performed using an in-house linear discrimination analysis algorithm (Huttlin et al., 2010), considering the following parameters: XCorr, ΔCn-difference score, peptide ion mass accuracy, charge state, missed cleavages and precursor mass accuracy. Linear discrimination scores were used to assign probabilities to each MS2 spectrum. These probabilities were then used to filter the dataset to a final protein-level FDR of 1%. Additionally, principles of parsimony were used to assign redundant peptides to proteins.
To quantify the confidence of each ubiquitylation site, we used a modified version of Ascore (Erickson et al., 2015). To confidently localize ubiquitylation sites only the ones with Ascore values > 13 (p ≤ 0.05) were considered.
QUANTIFICATION AND STATISTICAL ANALYSIS
For quantification, reporter ion counts were summed across all matching PSMs using an in-house software (Huttlin et al., 2010). Briefly, a 0.003 Th window around the theoretical m/z value of each reporter ion was scanned for ions, and the maximum intensity nearest the theoretical m/z was used. Reporter ion intensities were adjusted to correct for the isotopic impurities of the different TMT reagents according to manufacturer specifications. Lastly, for each protein signal to noise measurements of the peptides were summed and then normalized to 100 across the 10 samples yielding a “relative abundance” measurement.
MS3 spectra with nine missing TMT reporter ion channels, MS3 spectra with TMT reporter summed signal to noise ratio < 200, or isolation purities < 50% were excluded from quantitation (McAlister et al., 2012). t tests with Welch’s correction for unequal variances were used for all 5 × 5 replicates (DMSO versus Btz treatment, mouse brain versus mouse liver). Multiple test correction was performed by adjusting the calculated p values according to Benjamini-Hochberg. All data analysis was performed using R (http://www.R-project.org). Heat maps and associated gene ontology enrichment were generated with Perseus (http://www.coxdocs.org/doku.php?id=perseus:start) (Tyanova et al., 2016).
DATA AND SOFTWARE AVAILABILITY
The accession number for the mass spectrometry data reported in this paper is available through the PRIDE Archive, a member of the ProteomeXchange (PX) consortium and can be accessed with the following identifier Proteome Xchange: PXD004628.
Supplementary Material
In Brief.
Large-scale simultaneous ubiquitylome analysis of ten cell line or tissue samples results in the quantitation of 5,000–9,000 ubiquitylation forms, and it reveals PINK1- and PARKIN-dependent ubiquitylation events during early and late mitophagy.
Highlights.
Reproducible, accurate method for quantitative measurement of ubiquitylation sites
Analysis of 5,000–9,000 ubiquitylation forms across ten tissue or cell culture samples
Site-specific ubiquitylation of cancer-related proteins upon bortezomib treatment
PINK1- and PARKIN-dependent ubiquitylation events during early and late mitophagy
Acknowledgments
This work was supported by NIH grants GM67945 and CA185137 to S.P.G.; R37NS083524, GM095567, and AG011085 to J.W.H.; and R01-GM043601 to D.J.F. and by a postdoctoral fellowship from the Edward R. and Anne G. Lefler Center (A.O.). M.I. was supported by an EMBO Long Term Fellowship (EMBO LTF ALTF 315–2012).
Footnotes
Supplemental Information includes three figures and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.cels.2016.08.009.
AUTHOR CONTRIBUTIONS
C.M.R., M.I., A.O., M.A.P., J.W.H., and S.P.G. designed the experiments. D.J.F., J.W.H., and S.P.G. oversaw experiments. C.M.R., M.I., A.O., M.A.P., and M.P.J. prepared samples. S.A.B advised on sample preparation procedures. C.M.R. and M.I. performed MS experiments. C.M.R., M.I., A.O., J.W.H., and S.P.G. analyzed the data and wrote the manuscript. All authors edited the manuscript.
REFERENCES
- Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS, Sheng M. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;19:370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Kwon YT. Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp. Mol. Med. 2015;47:e147. doi: 10.1038/emm.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CN, Baughman JM, Phu L, Tea JS, Yu C, Coons M, Kirkpatrick DS, Bingol B, Corn JE. USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nat. Cell Biol. 2015;17:160–169. doi: 10.1038/ncb3097. [DOI] [PubMed] [Google Scholar]
- Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D, Gearing M, Rees H, Lah JJ, Levey AI, et al. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J. Biol. Chem. 2011;286:10457–10465. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Erickson BK, Jedrychowski MP, McAlister GC, Everley RA, Kunz R, Gygi SP. Evaluating multiplexed quantitative phosphopeptide analysis on a hybrid quadrupole mass filter/linear ion trap/orbitrap mass spectrometer. Anal. Chem. 2015;87:1241–1249. doi: 10.1021/ac503934f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grou CP, Pinto MP, Mendes AV, Domingues P, Azevedo JE. The de novo synthesis of ubiquitin: identification of deubiquitinases acting on ubiquitin precursors. Sci. Rep. 2015;5:12836. doi: 10.1038/srep12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J-M, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol. Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey SJ, Azimifar SB, Mann M. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 2015;33:990–995. doi: 10.1038/nbt.3327. [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isasa M, Rose CM, Elsasser S, Navarrete-Perea J, Paulo JA, Finley DJ, Gygi SP. Multiplexed, proteome-wide protein expression profiling: yeast deubiquitylating enzyme knockout strains. J. Proteome Res. 2015;14:5306–5317. doi: 10.1021/acs.jproteome.5b00802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler BM, Fortunati E, Melis M, Pals CEGM, Clevers H, Maurice MM. Proteome changes induced by knock-down of the deubiquitylating enzyme HAUSP/USP7. J. Proteome Res. 2007;6:4163–4172. doi: 10.1021/pr0702161. [DOI] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Lazarou M, Jin SM, Kane LA, Youle RJ. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell. 2012;22:320–333. doi: 10.1016/j.devcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, Kuhn K, Pike I, Grothe RA, Blethrow JD, Gygi SP. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal. Chem. 2012;84:7469–7478. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister GC, Nusinow DP, Jedrychowski MP, Wühr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M, Knudsen GM, Maeda S, Trinidad JC, Ioanoviciu A, Burlingame AL, Mucke L. Tau post-translational modifications in wild-type and human amyloid precursor protein transgenic mice. Nat. Neurosci. 2015;18:1183–1189. doi: 10.1038/nn.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow JK, Lin H-K, Sun S-C, Zhang S. Targeting ubiquitination for cancer therapies. Future Med. Chem. 2015;7:2333–2350. doi: 10.4155/fmc.15.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JP, Stepanova E, Everley RA, Paulo JA, Gygi SP. Comprehensive temporal protein dynamics during the diauxic shift in Saccharomyces cerevisiae. Mol. Cell. Proteomics. 2015;14:2454–2465. doi: 10.1074/mcp.M114.045849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na CH, Jones DR, Yang Y, Wang X, Xu Y, Peng J. Synaptic protein ubiquitination in rat brain revealed by antibody-based ubiquitome analysis. J. Proteome Res. 2012;11:4722–4732. doi: 10.1021/pr300536k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau A, Sarraf SA, Duda DM, Heo J-M, Jedrychowski MP, Sviderskiy VO, Olszewski JL, Koerber JT, Xie T, Beausoleil SA, et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell. 2014;56:360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau A, Heo J-M, Duda DM, Paulo JA, Olszewski JL, Yanishevski D, Rinehart J, Schulman BA, Harper JW. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1in mitochondrial quality control using a ubiquitin replacement strategy. Proc. Natl. Acad. Sci. USA. 2015a;112:6637–6642. doi: 10.1073/pnas.1506593112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau A, Münch C, Harper JW. Quantifying ubiquitin signaling. Mol. Cell. 2015b;58:660–676. doi: 10.1016/j.molcel.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014;20:1242–1253. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- Potu H, Peterson LF, Pal A, Verhaegen M, Cao J, Talpaz M, Donato NJ. Usp5 links suppression of p53 and FAS levels in melanoma to the BRAF pathway. Oncotarget. 2014;5:5559–5569. doi: 10.18632/oncotarget.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, Zaffagnini G, Wild P, Martens S, Wagner SA, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. USA. 2016;113:4039–4044. doi: 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles MS, Cox J, Mann M. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014;10:e1004047. doi: 10.1371/journal.pgen.1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;18:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen D-F, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods. 2011;8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Udeshi ND, Mertins P, Svinkina T, Carr SA. Large-scale identification of ubiquitination sites by mass spectrometry. Nat. Protoc. 2013a;8:1950–1960. doi: 10.1038/nprot.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udeshi ND, Svinkina T, Mertins P, Kuhn E, Mani DR, Qiao JW, Carr SA. Refined preparation and use of anti-diglycine remnant (K-ε-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell. Proteomics. 2013b;12:825–831. doi: 10.1074/mcp.O112.027094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weathington NM, Mallampalli RK. Emerging therapies targeting the ubiquitin proteasome system in cancer. J. Clin. Invest. 2014;124:6–12. doi: 10.1172/JCI71602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Becher I, Sweetman G, Doce C, Savitski MM, Bantscheff M. High-resolution enabled TMT 8-plexing. Anal. Chem. 2012;84:7188–7194. doi: 10.1021/ac301553x. [DOI] [PubMed] [Google Scholar]
- Yamano K, Matsuda N, Tanaka K. The ubiquitin signal and autophagy: an orchestrated dance leading to mitochondrial degradation. EMBO Rep. 2016;17:300–316. doi: 10.15252/embr.201541486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii SR, Kishi C, Ishihara N, Mizushima N. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J. Biol. Chem. 2011;286:19630–19640. doi: 10.1074/jbc.M110.209338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Phu L, Varfolomeev E, Bustos D, Vucic D, Kirkpatrick DS. Immunoaffinity enrichment coupled to quantitative mass spectrometry reveals ubiquitin-mediated signaling events. J. Mol. Biol. 2015;427:2121–2134. doi: 10.1016/j.jmb.2015.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.