Abstract
Background
The equilibrium of oral microbiome may be altered by environmental factors, including cigarette smoking. Several recent studies also suggest that oral pathogens causing periodontal disease, such as Fusobacterium nucleatum, are involved in pathogenesis of colorectal cancer.
Methods
For this study oral rinse DNA samples from 190 participants in a population-based case-control study for colorectal cancer were used to amplify a V3-V4 region of bacterial 16S rRNA gene. The amplicons were sequenced using Illumina MiSeq paired end chemistry on two runs, yielding approximately 35 million filtered reads which were assigned to bacterial phyla.
Results
No association was found between Fusobacterium abundance or presence and colorectal cancer. However, adjusted for age and experimental batch, colorectal cancer history was associated with increased presence of genus Lactobacillus and increased relative abundance of Rothia by 28% and current smoking was associated with a 33% decrease in relative counts of Betaproteobacteria (primarily Neisseria) and 23% increase in relative abundance of Veillonellaceae family. We also found that smoking had significant effects on the 2nd component scores and 2nd coordinate distances in principal component and coordinate analyses.
Conclusions
It remains to be elucidated whether the observed differences can be translated into biochemical changes in oral environment, thus potentially affecting oral health.
Keywords: Oral microbiome, Cigarette smoking, Colorectal cancer
1. Introduction
The microbiota, and the genes comprising its microbiome play important roles in human health.[1, 2] Advances in metagenomics have revealed that microhabitats exist throughout the human body and that each microhabitat maintains a unique ecosystem with distinct atmospheric and nutritional compositions that foster symbiotic interactions.[3] However, this dynamic equilibrium can be altered by environmental factors and external interferences, such as the use of antibiotics.[2] These alterations frequently lead to microbial imbalances, a phenomenon called dysbiosis, which has been linked to various pathological conditions, including cancer.[3, 4]
Oral microbiome has been relatively understudied compared with the gut microbiome. The human oral microbiome comprises more than 2,000 bacterial taxa[5, 6] , including a large number of opportunistic pathogens that are involved in periodontal, respiratory and cardiovascular and other systemic diseases[5, 6] and is considered to be the second most diverse community following stool.[6, 7] Several recent studies also suggest that oral pathogens causing periodontal disease, such as Porphyromonas (P) gingivalis and Fusobacterium (F) nucleatum, may be involved in pathogenesis of not only oral cancer[8–10] but also cancer in the digestive organs (e.g., colorectum and pancreas),[11–13] which may be exposed to oral microbiome through saliva or by the blood stream. Consistent observations support that F. nucleatum and Fusobacteria family are enriched in cancer tissue or stool samples from colorectal cancer patients,[11, 14–16] while acceleration of tumorigenesis has been observed in a mouse model exposed to F. nucleatum.[17] Interestingly, cancers that showed a potential link to the oral pathogens are all smoking related[18, 19] and smoking is a major environmental factor that influences orodental pathophysiology.[20, 21] Smokers are known to harbor a more pathogenic anaerobic subgingival microbe population than non-smokers,[22] but earlier studies have generated vastly mixed results concerning relative abundance of specific bacterial groups between smokers and non-smokers, except positive correlation of smoking with F. nucleatum or Fusobacteria class.[20–23] Gut microbiome is acquired through the mouth and thus oral microbiome possibly serves as a reservoir for the microbiome in the distal gastrointestinal tract. Besides, sampling oral microbiome through mouthwash or saliva is more amendable for epidemiologic research than collecting fecal samples and less invasive than obtaining mucosal biopsy through endoscopy. Therefore, in the present study we aimed to interrogate the associations of history of colorectal cancer and cigarette smoking with oral microbiome composition among population-based samples using high throughput 16S rRNA DNA sequencing.
2. Materials and Methods
2.1 Study design
This study was designed as secondary analyses of biological specimens and epidemiologic data collected for the published studies described elsewhere.[24] The study was approved by the WSU Human Investigation Committee. Eligible study subjects were residents in the Metropolitan Detroit Tri-County (Wayne, Oakland and Macomb) area, between 45 and 80 years of age at time of ascertainment, with a working telephone and no prior history of any invasive cancer, insitu colorectal cancer or colectomy. Eligible colorectal cancer cases were histologically diagnosed between January 1, 2003 and September 30, 2005, and were identified through a rapid case-reporting system implemented in the Metropolitan Detroit Cancer Surveillance System (MDCSS), a founding member of the National Cancer Institute’s Surveillance, Epidemiology, and End Results-cancer registries, which allowed access to patients within 3–4 months from their diagnosis. A total of 3,746 potentially eligible cases were identified. Following receipt of case reports, the physicians on the record were contacted for their clearance and then the patents were contacted by mail. Among these cases, physician consent was not obtained for 385. Of the remaining, 110 subjects were further excluded because no subject contact information was available or other administrative reasons. An additional 47 cases were found to be ineligible before enrollment, leaving a total of 3,204 subjects. Among these 1,335 consented to the study (41.7%). Population controls were identified though random digit dialing (RDD). These controls were frequency matched to the cases by 5-year age group, gender, county of residence and race (African American [AA] vs. non-AA), which were projected based on the data from preceding years. The RDD telephone numbers were generated by Survey Sampling (Fairfield, CT) with pre-screening for business and non-working numbers. RDD interviewers were instructed to call each selected telephone number up to 9 times at different times of the day and week including evenings and weekends, in order to obtain a household census to identify potentially eligible study subjects. A total of 36,936 unique RDD telephone numbers were surveyed. 36% of the numbers were excluded, 33% were screened for household census information, and 31% were not able to be screened. The excluded numbers were business, public or disconnected numbers, out of study area, dataline (modem, fax etc.), or other miscellaneous reasons. The numbers not screened were due to no answers throughout 9 attempts and refusals to give any household information and communication problems in either language or hearing ability. Among the remaining numbers screened for household census, 47% were ineligible primarily due to ages of household members. Of the eligible households (53%), 75% agreed to receive a study invitation letter and the rest refused to provide a mailing address. To balance the age distribution of cases and controls, we selected 2,831 for enrollment and 1 682 completed the study (59.4%). Of the total of 3,017 participants, 135 controls were considered to be ineligible according to their age and medical history in the questionnaires, and 130 cases were also considered ineligible because the final MD-CSS reports did not confirm the eligibility or because of previous cancer or colectomy reported in the completed questionnaires. The subjects were interviewed over the telephone using structured questionnaires regarding their usual diet and other risk factors for colorectal cancer for the time-period preceding cancer diagnosis (approximately 2 years prior to the interview). At the same time or shortly after the interview, the study participants provided one of the following types of biospecimens; (1) peripheral blood through home phlebotomy service, (2) buccal cells collected by oral rinse with commercial mouthwash liquid and (3) archived (grossly normal) tissue blocks. The oral rinse samples were collected by 30 second swishing with a commercial mouthwash liquid containing 15% alcohol. The participants were instructed not to brush teeth, rinse mouth, eat or drink for at least 1 hour before collection. DNA was isolated with the Gentra Autopure system and then stored at −80°C until analysis at the Wayne State University Applied Genomics Technology Center. Among the controls, 71% provided blood samples and 29% gave mouthwash samples. Among the colorectal cancer cases, phlebotomy samples were collected from 66%, followed by mouthwash 27% and tissue blocks 7%. From among the participants who provided oral rinse samples, we randomly selected 192 samples for this exploratory study, oversampling smokers, after excluding those with insufficient residual volume. To minimize potential effects of immunosuppression from the disease and treatment, colorectal cancer cases were further limited to those who did not have distant metastasis or receive chemotherapy. As a result, colorectal cancer cases included in this study (n = 68) consisted of 74% local and 26% regional stages and 75% colon and 25% rectal cancer. Approximately 90% of cancer were well to moderately differentiated among those with histological grade information (68%). All but one patient had surgery while one radiation. Median time from diagnosis to biospecimen acquisition was 9.8 months.
2.2 Sequencing
Amplification and sequencing of the 16S ribosomal RNA gene was achieved following the Illumina 16S Metagenomic Sequencing Library Preparation protocol. The primer pair, the S-D-Bact-0341-b-S-17/S-D-Bact-0785-a-A-21, was used to amplify 464 bp of the V3 and V4 regions of the 16S gene. The correct size of the product was confirmed on an Agilent Tape Station. The samples were cleaned using AMPure XP beads to remove excess primers and primer dimers and indexed using the 96 sample Nextera XT Index Kit, giving each sample a unique identifier with dual 8 base adapters. The indexed product was cleaned with AMPure XP beads and the size checked with the Tape station to confirm indexing success. After fluorometric sample quantification using Invitrogen’s Qubit 2.0, samples were diluted to 4 nM and pooled. The library pool was then denatured, diluted to 20 pM and a PhiX control was added. The pool was sequenced on an Illumina MiSeq using paired 300 bp reads with MiSeq V3 reagents and the data were processed using the MiSeq Reporter software.
2.3 Bioinformatic analysis
The online portal for Illumina data analysis (http://www.illumina.com/) was used to access Basespace at https://basespace.illumina.com/home/index, and the 16S metagenomics Basespace application was applied to the data. 16S metagenomics analysis uses DNA from amplicon sequencing of prokaryotic 16S small subunit rRNA genes with the high performance version of RDP Naïve Bayes algorithm.[25] FASTQ sequences were uploaded to Basespace and the 16S metagenomics application was executed. After assembling, full length sequences from paired ends were referenced against the Illumina curated version of Greengenes database (May 2013) at 97% identity level. We also used the Quantitative Insights for Molecular Ecology (Qiime) suite of programs[26] to compute several α and β diversity indices including Chao1, Shannon and Simpson index values were generated after rarefying samples at 10,851 reads and to perform principal coordinate analysis (PCoA) based on weighted Unifrac distance matrices.[27] Xming and Emperor Plot[28] were used to visualize PCoA plots.
2.4 Statistical analysis
Basespace summary text files reporting number of reads for each identified taxonomic unit were aggregated for all samples from 2 separate runs. We removed 2 samples that failed to generate 20,000 classifiable reads, leaving 190 analytical samples. Analyses were restricted to bacterial groups that represented, on average at least 1% of total reads classifiable at each taxonomic level and two a priori selected species, F. nucleatum and P. gingivalis. The associations of relative abundance of each taxonomic unit with sample/subject’s characteristics were analyzed by negative binomial model as an extension of Poisson regression for count data, to deal with overdispersion,[29] with adjustment of selected covariates. Zero-inflated models were also employed when the count was 0 for more than 5% of the samples, which produced the parameter estimates for both counts and zero proportion. To ease interpretation for the latter estimate, we presented its reverse term (powered-1) as an estimate for presence (non-zero). In both analyses the total number of reads classifiable at each taxonomic level was used as an offset variable. These regression models have been recently adopted to analyze microbial count data taking into other covariates into account.[30, 31] In addition, we performed principal component analyses at class, family and species levels where relative abundance of each bacterial group was square root transformed as it performed better than other transformations, such as natural log or arcsine. Analysis of variance was used to compare principal component scores as well as UniFrac distances of principal coordinates between subject groups with adjustment for selected covariates where first coordinate distances were natural log-transformed to improve deviation from normal distribution. Most subjects’ characteristics were dichotomized. “Current smokers” were defined as ever smokers who smoked at least one cigarette per day for six months or longer and were smoking within 2 year prior to interview. “Habitual alcohol drinkers” were defined as subjects reporting 7 or more alcoholic drinks per week. “Advanced age” was defined as 65 years or older (approximate median of the whole study population). All statistical analyses were performed using SAS version 9.
3. Results
Table 1 presents characteristics of selected study samples. Colorectal cancer cases were 68 (36%), while the rest were controls. In both groups approximately 14% were African Americans and the rest (86%) other races (primarily Caucasians). While 24% of the colorectal cancer cases and 30% of the controls were considered to be current smokers, 16% of the colorectal cancer cases and 11% of the controls were habitual alcohol drinkers. The number of total classifiable reads was substantially higher in run 1 than in run 2 (19215417 vs. 15457823). Due to these differences in performance, run was a significant confounder in most analyses, and was therefore always included in statistical models.
Table 1.
Characteristics of study subjects
| Disease history | |||||
|---|---|---|---|---|---|
| Characteristics | Categories | Colorectal cancer (N = 68) |
Controls (N = 122) |
||
| No. | % | No. | % | ||
| Gender | Female | 36 | 52.9% | 81 | 66.4% |
| Male | 32 | 47.1% | 41 | 33.6% | |
| Race | African American |
10 | 14.7% | 17 | 13.9% |
| Other | 58 | 85.3% | 105 | 86.1% | |
| Age | 45–54 | 12 | 17.7% | 32 | 26.2% |
| 55–64 | 19 | 27.9% | 40 | 32.8% | |
| 65+ | 37 | 54.4% | 50 | 41.0% | |
| Smoking | Never | 26 | 38.2% | 49 | 40.2% |
| Ex | 26 | 38.2% | 37 | 30.3% | |
| Current | 16 | 23.5% | 36 | 29.5% | |
| Alcohol | < 1 | 38 | 55.9% | 74 | 64.9% |
| 1–6/week | 19 | 27.9% | 27 | 23.7% | |
| 7+/week | 11 | 16.2% | 13 | 11.4% | |
| Body mass index (kg/m2) | < 25 | 18 | 26.9% | 55 | 45.8% |
| 25–30 | 35 | 52.2% | 40 | 33.3% | |
| 30+ | 14 | 20.9% | 25 | 20.8% | |
| Non-steroidal anti- | No | 54 | 79.4% | 83 | 68.0% |
| inflammatory drug use | Yes | 14 | 20.6% | 39 | 32.0% |
| Experimental batch | Run 1 | 46 | 67.6% | 48 | 39.3% |
| Run 2 | 22 | 32.4% | 74 | 60.7% | |
As shown in Table 2, Firmicutes were a highly dominant phylum, and the majority of these belonged to Streptococcaceae at the family level. Bacteroidetes and Actinobacteria both contributed 13% at the phylum level, and their dominant families were Prevotellaceae and Micrococcaceae, respectively. Phylum Proteobacteria (8.6%) were equally divided into Beta and Gamma at the class level, and Neisseriaceae and Pasteurellaceae were respective dominant families. Fusobacteria represented a relatively small portion (3.7%) of phyla.
Table 2.
Relative abundance (%) of bacterial groups representing at least 1% of classifiable reads (in parentheses) at each taxonomic level
| Phyla (34508276) |
% | Classes (34325631) | % | Orders (34271401) |
% | Families (33775084) | % | Genera (33507332) |
% | Species (26368975) | % |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Firmicutes | 61.3 | Bacilli | 47.8 | Lactobacillales | 44.0 | Streptococcaceae | 40.5 | Streptococcus | 40.7 | S_tigurinus | 12.4 |
| S_vestibularis | 9.7 | ||||||||||
| S_parasanguinis | 4.0 | ||||||||||
| S_oralis | 3.4 | ||||||||||
| S_pseudopneumoniae | 3.3 | ||||||||||
| S_bovis | 1.8 | ||||||||||
| S_infantis | 1.3 | ||||||||||
| S_gordonii | 1.0 | ||||||||||
| Carnobacteriaceae | 1.9 | Granulicatella | 1.9 | G_adiacens | 2.1 | ||||||
| Lactobacillaceae | 1.3 | Lactobacillus | 1.3 | ||||||||
| Gemellales | 3.6 | Gemellaceae | 3.7 | Gemella | 3.7 | G_cunicula | 1.5 | ||||
| G_sanguinis | 1.3 | ||||||||||
| Clostridia | 13.4 | Clostridiales | 12.6 | Veillonellaceae | 10.9 | Veillonella | 10.2 | V_atypica | 6.7 | ||
| V_dispar | 2.0 | ||||||||||
| Lachnospiraceae | 1.2 | ||||||||||
| Bacteroidetes | 13.2 | Bacteroidia | 12.3 | Bacteroidales | 12.3 | Prevotellaceae | 9.4 | Prevotella | 10.2 | P_melaninogenica | 5.2 |
| P_histicola | 2.0 | ||||||||||
| Porphyromonadaceae | 1.6 | Porphyromonas | 1.5 | ||||||||
| Actinobacteria | 12.7 | Actinobacteria | 12.8 | Actinomycetales | 12.3 | Micrococcaceae | 7.7 | Rothia | 7.6 | R_mucilaginosa | 4.1 |
| Actinomycetaceae | 3.2 | Actinomyces | 2.2 | ||||||||
| Proteobacteria | 8.6 | Betaproteobacteria | 4.0 | Neisseriales | 3.7 | Neisseriaceae | 3.8 | Neisseria | 3.7 | N_mucosa | 2.9 |
| Gammaproteobacteria | 3.9 | Pasteurellales | 3.8 | Pasteurellaceae | 3.9 | Haemophilus | 1.9 | H_parainfluenzae | 1.9 | ||
| Mannheimia | 1.2 | M_caviae | 1.6 | ||||||||
| Fusobacteria | 3.7 | Fusobacteria | 3.7 | Fusobacteriales | 3.7 | Fusobacteriaceae | 1.9 | Fusobacterium | 1.9 | ||
| Leptotrichiaceae | 1.9 | Leptotrichia | 1.8 | ||||||||
| Other | 0.5 | Other | 2.0 | Other | 3.9 | Other | 7.2 | Other | 10.2 | Other | 31.9 |
Next we analyzed the effect of subject’s characteristics on the relative abundance of bacterial groups at each taxonomic level (see Table ??). Advanced age (> 65 years) was associated with changes in microbiome contents, showing increased relative abundance of class Bacilli, order Lactobacillales and genus Streptococcus by 7%-8%. Although P. gingivalis represented only 0.3% of the classified species, relative abundance among subjects harboring this bacterium was 2.6 fold (95% CI 1.32–5.23) increased with advanced age. On the other hand, the relative abundance of Micrococcaceae family decreased by approximately 20% (0.81, 95% CI 0.66–1.00) with advanced age. Neither gender nor African American race shows associations with these bacterial relative abundances (data not shown). Due to the several observed associations with advanced age, we included advanced age as a binary covariate in subsequent analyses.
Adjusted for advanced age and experimental batch, colorectal cancer history was associated with increased presence of genus Lactobacillus by almost 2 times (95% CI 1.08–4.00) and increased relative abundance of Rothia by 28% (1.28; 95% CI 1.02–1.59). Adjusted for advanced age and experimental batch, current smoking was associated with a decrease in relative counts of Betaproteobacteria (primarily Neisseria) by 33% (0.66; 95 CI 0.47–0.93) but a 23% increase in relative abundance of Veillonellaceae family (1.23: 95% CI 1.00–1.51). Although simultaneous adjustment of colorectal cancer status did not reduce the strength of these associations (data not shown), changes in relative abundance by smoking status were generally more pronounced in the cases with colorectal cancer history than controls. For example, the estimates for Betaproteobacteria and Veillonellaceae were 0.29 (0.14–0.59) and 1.53 (0.93–2.52), respectively, in the cases, whereas they were 0.77 (0.51–1.14) and 1.13 (0.94–1.36), respectively, in the controls. But the interaction was modestly significant only for Betaproteobacteria (p = .04).
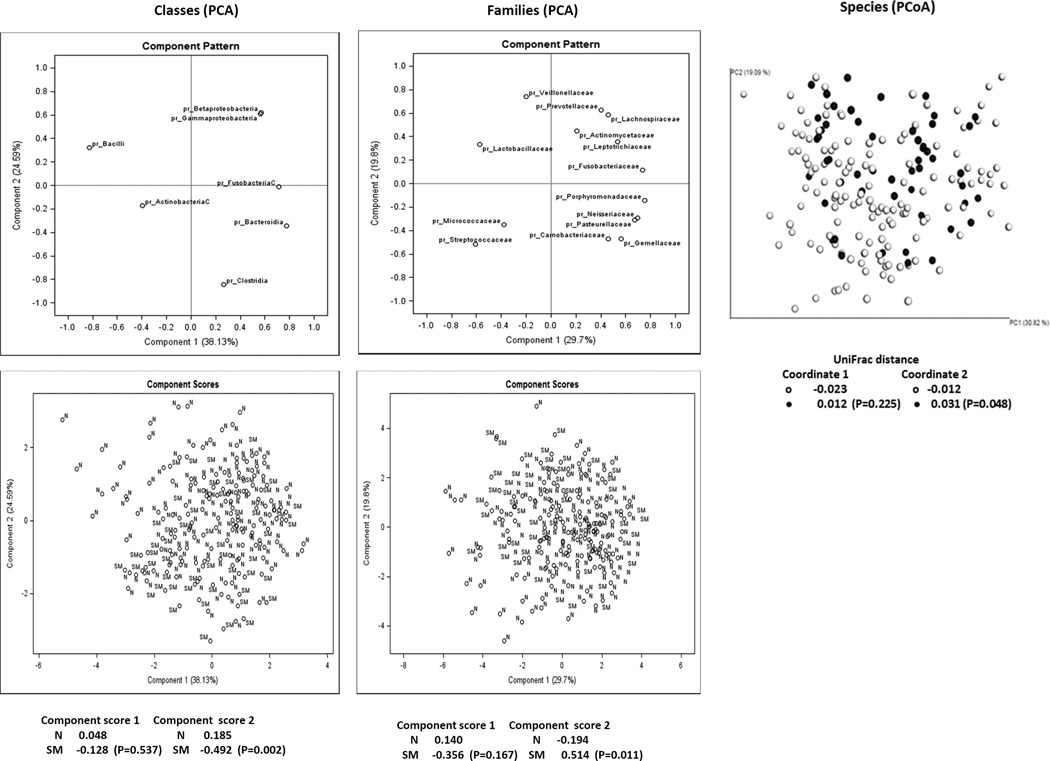
None of the bacterial groups from phylum to genus levels were associated with habitual alcohol consumption, but relative counts of P. gingivalis increased by 3 times (2.97: 95% CI 1.16–7.61) with habitual alcohol intake among the subjects carrying this bacterium (data not shown). None of the diversity or richness indexes estimated by Qiime showed associations with smoking, colorectal cancer history or advanced age (data not shown). The main results of principal component analysis (PCA) from Basespace data and of principal coordinate analysis (PCoA) based on Qiime analysis are illustrated in Figure 1, along with the 1st and 2nd principal component scores adjusted for advanced age and experimental batch and 1st an 2nd principal coordinate Unifrac distances adjusted for experimental batch for current smokers and non-smokers. PCA plots for the class and family levels show that some family members within a class were clustered (e.g., Proteobacteria), but not for others (i.e., Firmicutes, Bacteroidetes and Actinobacteria), suggesting that relative abundances of major family members within a class were not necessarily correlated each other. The 2nd principal component scores at the class and family levels from PCA and the 2nd coordinate distances from PCoA were statistically significantly different between current smokers and the others (P = .002–.048). When we tested the scores/distances of these components and coordinate stratified by colorectal cancer status, the differences between current smokers and non-smokers were generally more apparent in the cases with colorectal cancer history than the controls. At the class level the 2nd component scores in current-smokers vs. non-smokers were −1.097 vs. 0.065 in the cases and −0.246 vs. 0.268 in the controls and the corresponding scores at the family levels were 1.071 vs. −0.125 in the cases and 0.271 vs. −0.237 in the controls, whereas the 2nd coordinate distances at the species level for current smokers and non-smokers were 0.127 vs. −0.015 in the cases and −0.009 vs. −0.011 in the controls, respectively.
Figure 1.
Principal component analyses (PCA) at the class (leftmost) and family (middle) levels for taxa with at least 1% average relative abundance and principal coordinate analysis (PCoA) at the species (rightmost) level. For PCA, upper panels show taxa distribution and lower panels sample distribution, while for PCoA the upper panel illustrates sample distribution. SM or closed circles indicate current smokers and N or open circles non-smokers. Covariate adjusted mean component scores and coordinate distances for the 1st and 2nd components/coordinates were calculated and tested between current smokers and non-smokers and presented underneath each panel
4. Discussion
This exploratory study of the oral microbiome revealed several intriguing findings. Specifically, we observed associations of smoking and colorectal cancer history with relative abundances of specific bacterial groups; and changes in microbiome composition associated with smoking, not altering diversity or richness. While the associations with specific bacterial groups were rather modest and may represent chance findings, the changes in microbial composition were reproducible by two different approaches. We discuss our results here in comparison with other published microbiome papers, noticing differences and limitations of the present study in this rapidly expanding area of research.
The results presented here are likely influenced by the methods employed to collect and measure oral microbes., The microbiome of each organ is distinct and substantial variability occurs even within oral cavity, partly due to spatial variations in the availability of oxygen.[7] The oral rinse samples used in this study are likely to over-represent microbes present in the surface of the oral cavity as well as saliva, and less likely to include microbes from dental plaques or gingival crevicular fluid, which may explain the dominance of Firmicutes, as noted in buccal mucosa.[7] Salivary microbiome has been reported to have higher richness and diversity compared with gingival plaque microbiome, and to be less susceptible to changes in periodontal conditions.[32] Second, taxonomic classification in most metagenomic analyses is based on hypervariable sequences (V1-V9) of bacterial 16S rRNA gene. While several different primer sets are available, it has been well documented that the choice of the V regions to be amplified significantly influences the results.[33, 34] We used primers for combined regions including V4, which has been reported to be capable of assigning sequences down to genus level with good accuracy.[35]
Despite distinct microbial community by anatomic site, correlations exist between stool and oral microbiome,[36, 37] which corroborates earlier reports pointing to associations of colorectal cancer with oral pathogens. Nonetheless, the present study did not confirm the association between colorectal cancer and F. nucleatum. Although this epidemiologic association has been rather consistent[11, 14–16] and although proinflammatory properties of this bacterium were reported in an animal model,[17] causality of this association has been unclear. On the other hand, changes in mucosal barriers and metabolic and morphologic characteristic of established cancer cells may have facilitated colonization by certain bacteria. In the present study, we found that Lactobacillus and Rothia were associated with colorectal cancer history. Contrary to the well-known probiotic role in the gut,[38] Lactobacilli are oral pathogens strongly associated with dental caries.[39] While it is unlikely Lactobacilli possess enteric oncogenicity, it may serve as a surrogate marker for poor oral hygiene or poor oral health. In fact, epidemiologic studies have found connections of periodontal disease with risk of dying from,[13] and developing colorectal cancer[40] as well as with positive results to colorectal cancer screening.[41] The genus Rothia is usually considered to be a benign oral commensal. However, Rothia has recently been recognized as an opportunistic pathogen causing a number of diseases in immunocompromised hosts[42] and has been associated with periodontitis in AIDS patients.[43] Although our cases did not undergo chemotherapy, individuals who develop or developed cancer may suffer underlying suboptimal immune function.
Instead, we found smokers had decreased relative abundance of Neisseria and increased relative abundance of Veillonellaceae families. While some Neisseria species are well known human pathogens, causing meningitis and gonorrhea,[48] most Nesseria are indeed commensal and are common in upper aerodigestive tract.[48, 49] Importantly, Neisseria has been shown to be a bacterial group that colonizes on the surface of natural teeth in much higher density than on denture teeth.[50] Smokers are known to be susceptible to periodontal diseases,[21–23] leading to tooth loss, which possibly accounts for the observed association in this study. Corroboratively, lower abundance of Neisseria in smokers has been reported by other studies.[20, 23] Veillonellaceae are also considered to be benign commensals in the oral cavity, abundantly present in saliva and on dorsal and lateral surfaces of the tongue,[49] but prior studies have found conflicting (positive, negative or no) associations with cigarette smoking in periodontitis patients.[20, 23, 51] An interesting trait of this bacterium is its specific metabolic activity. Because up to 25% of ingested nitrate is actively taken up by the salivary grand, concentrated up to 20 fold, saliva contains high concentrations of nitrate and nitrite.[52] The conversion of nitrate to nitrite is catalyzed by nitrate reductase of oral bacteria.[52] Veillonella (V) species, including V. atypica, are one of the major bacterial groups with this metabolic activity.[53–55] These compounds are potentially toxic, because nitrite can be further converted to carcinogenic nitrosamines[56] and pro-inflammatory nitric oxide.[52] Tobacco not only provides nitrate but also alkaloids to form nitrosamines,[56] which can be detected in saliva of tobacco users.[57] Therefore, these bacterial metabolic activities have pathophysiological consequences on oral and systemic health.
In addition to specific bacterial abundance, we found moderate but statistically significant differences in overall composition of microbiome by smoking status in both PCA and PCoA. These relatively small differences along with the lack of differences in diversity indexes may be attributable to the fact that the beta (between subjects) diversity of salivary microbiome is one of the lowest, while the alpha (within subjects) diversity is one of the highest,[7] among various anatomic sites. Yet, differences in salivary,[58] mucosal, and subgingival[22] microbiome structure according to smoking status have been reported using either PCA or PCoA by others. One of these authors also confirmed no effect of alcohol drinking.[59] We also realize that classification of smokers differs by studies, which may affect the interpretation of the data. Our questionnaire asked about exposure up to 2 years prior to the interview in order to minimize effects from the disease, and accordingly our current smokers included past smokers who stopped smoking within 2 years. The rest of past smokers were combined with never smokers because we assumed that direct exposure to cigarette smoke rather than oral conditions caused by smoking should produce the main impact. Initial screening analyses based on three strata also suggested the data of past smokers were in fact closer to never smokers than current smokers. Nevertheless, misclassification of smoking status is a concern due to lack of objective measures (tobacco metabolites) of smoking in urine or breath and because misclassification of past smokers as current smokes are likely to be differential (more common in the cases). Although the misclassification may have diluted the overall observed differences, the differential bias itself may have limited effect.
Considering several limitations in this exploratory study, we note first that although the sample size of 190 may be viewed as relatively large for metagenomic studies, it is small for traditional epidemiologic studies to detect modest sizes of differences which are typical in homogenous human populations. This may be especially the case, given the known low beta diversity of oral microbiome[7] and for observational studies where investigators have little control for known and unknown confounders. Second, the retrospective nature of the case-control comparisons limits the ability to infer causal relationships for observed associations. Specifically, temporal relations between exposures and outcomes cannot be ascertained in this study. Third, because the parent study was designed to address medical questions, information was not collected about oral hygiene practices, nor about oral and dental disease histories, which are likely to be critical determinants of oral microbiome, contributing to variations in the exposure. Accordingly, the data from this study should be interpreted with caution. In addition there may be unmeasured confounding factors as the effects of smoking on bacterial composition were more pronounced in the cases than in the controls, although this may alternatively indicate possible smoking-microbial interactions in smoking-associated cancer. However, given the nature of a posteriori analyses as well as the limited number of current smokers included in the cases, we should not over-interpret the results of these stratified analyses. Finally, even with a newer generation of sequencing technology, 96 multiplexed runs did not yield sufficient sequence depth to detect some of the pathogens that have been reported by culture and DNA hybridization techniques in periodontitis patients,[49–51] probably due to low frequencies in saliva. On the other hand, strengths include a relatively diverse study population (non-clinic based) and greater coverage of microbiome from global oral locations, adjustment for important confounders, such as experimental variability, and quantitative analyses beyond graphic presentation typically used in metagenomic studies.
5. Conclusions
In summary, the results of the present study failed to support earlier observations that an oral pathogen, F. nucleatum, was associated with colorectal cancer. On the other hand, we identified smoking-associated changes in oral microbiome composition. Further multi-disciplinary studies are warranted to elucidate whether the observed differences can be translated into biochemical changes in the oral environment, thus potentially affecting oral health, including the risk of developing oral and gastrointestinal cancer.
Table 3.
The effects** and 95% confidence intervals (in parenthesis) of age, cancer history and current smoking on relative bacterial count at each taxonomic level based on negative binomial models
| Phyla | Classes | Orders | Families | Genera | Species | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Old (≥ 65 yrs) vs. young | |||||||||||
| Firmicutes | 1.05 (1.00–1.11) | Bacilli | 1.07 (1.00–1.15) | Lactobacillales | 1.08 (1.00–1.17) | Streptococcaceae | 1.08 (0.99–1.17) | Streptococcus | 1.08 (0.99–1.17) | S_gordonii | 1.49 (1.06–1.93) |
| 1.61 (0.81–3.19) | |||||||||||
| Bacteroidetes | 0.98 (0.77–1.24) | Bacteroidia | 0.96 (0.75–1.24) | Bacteroidales | 0.96 (0.75–1.24) | Porphyromonadaceae* | 0.93 (0.69–1.25) | Porphyromonas* | 0.96 (0.71–1.29) | P_gingivalis* | 2.63 (1.32–5.23) |
| 0.83 (0.43–1.59) | 0.81 (0.44–1.52) | 1.22 (0.60–2.49) | |||||||||
| Actinobacteria | 0.85 (0.74–0.99) | Actinobacteria | 0.85 (0.74–0.98) | Actinomycetales | 0.84 (0.73–0.98) | Micrococcaceae | 0.81 (0.66–1.00) | Rothia | 0.81 (0.66–1.00) | R_mucilaginosa | 0.79 (0.62–1.00) |
| Proteobacteria | 0.93 (0.69–1.25) | ||||||||||
| Fusobacteria* | 0.95 (0.77–1.18) | ||||||||||
| 0.81 (0.29–4.27) | |||||||||||
| Colorectal cancer vs. controls | |||||||||||
| Firmicutes | 1.02 (0.96–1.07) | Bacilli | 1.01 (0.94–1.09) | Lactobacillales | 1.01 (0.93–1.10) | Lactobacillaceae* | 1.55 (0.77–3.08) | Lactobacillus* | 1.54 (0.77–3.07) | - | |
| 2.08 (1.08–4.00) | 2.08 (1.08–4.00) | ||||||||||
| Bacteroidetes | 0.89 (0.69–1.15) | ||||||||||
| Actinobacteria | 1.22 (1.04–1.42) | Actinobacteria | 1.22 (1.04–1.42) | Actinomycetales | 1.20 (1.02–1.41) | Micrococcaceae | 1.27 (1.02–1.59) | Rothia | 1.28 (1.02–1.59) | R_mucilaginosa | 1.26 (0.98–1.62) |
| Proteobacteria | 0.87 (0.63–1.21) | ||||||||||
| Fusobacteria* | 0.92 (0.73–1.11) | ||||||||||
| 0.58 (0.19–1.75) | |||||||||||
| Current smoking vs . not smoking | |||||||||||
| Firmicutes | 1.05 (0.99–1.12) | Clostridia | 1.20 (0.98–1.47) | Clostridiales | 1.20 (0.98–1.47) | Veillonellaceae | 1.23 (1.00–1.51) | Veillonella | 1.23 (1.00–1.52) | V_atypica | 1.33 (1.06–1.66) |
| Bacteroidetes | 1.03 (0.78–1.37) | ||||||||||
| Actinobacteria | 1.02 (0.86–1.21) | ||||||||||
| Proteobacteria | 0.66 (0.47–0.92) | Betaproteobacteria* | 0.66 (0.47–0.93) | Neisseriales* | 0.67 (0.48–0.95) | Neisseriaceae* | 0.67 (0.48–0.95) | Neisseria* | 0.68 (0.48–0.96) | N_mucosa* | 0.71 (0.49–1.02) |
| 0.42 (0.20–0.89) | 0.40 (0.19–0.84) | 0.40 (0.19–0.84) | 0.42 (0.20–0.87) | 0.42 (0.20–0.87) | |||||||
| Fusobacteria* | 0.94 (0.74–1.19) | ||||||||||
| 1.43 (0.36–5.65) | |||||||||||
Effect: 1.00: No effect, > 1: Times increase in the relative count; < 1: times decrease in the relative count by respective factor.
Adjusted for experimental batch for age, and age and experimental batch for smoking and colorectal cancer status
Based on a zero-inflated model: upper cell indicates effect on the relative counts beyond 0: lower indicates effect on the prevalence (non- zero) of a given bacterial gro
Acknowledgments
This research was supported by a grant from National Institutes of Health R01-CA93817 (IK) and P30CA022453 (Cancer Center Support Grant). The authors thank the study participants for their generosity in donating time and biospecimens, the Metropolitan Detroit Cancer Surveillance System for rapid case-ascertainment and Ms. Barbara Rusin, Dr. Maria Samerson and Ms. Ann Bankowski for their excellent technical assistance.
Footnotes
Conflicts of Interest Disclosure
The authors declare no conflicts of interest.
References
- 1.Sharon G, Garg N, Debelius J, et al. Specialized metabolites from the microbiome in health and disease. Cell metabolism. 2014;20:719–730. doi: 10.1016/j.cmet.2014.10.016. PMid:25440054 http://dx.doi.org/10.1016/j.cmet.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martín R, Miquel S, Langella P, et al. The role of metagenomics in understanding the human microbiome in health and disease. Virulence. 2014;5:413–423. doi: 10.4161/viru.27864. PMid:24429972 http://dx.doi.org/10.4161/viru.27864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zarco MF, Vess TJ, Ginsburg GS. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Diseases. 2012;18:109–120. doi: 10.1111/j.1601-0825.2011.01851.x. http://dx.doi.org/10.1111/j.1601-0825.2011.01851.x. [DOI] [PubMed] [Google Scholar]
- 4.Schwabe RF, Jobin C. The microbiome and cancer. Nature reviews Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. PMid:24132111 http://dx.doi.org/10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warinner C, Rodrigues JFM, Vyas R, et al. Pathogens and host immunity in the ancient human oral cavity. Nat Genet. 2014;46:336–344. doi: 10.1038/ng.2906. http://dx.doi.org/10.1038/ng.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wade WG. The oral microbiome in health and disease. Pharmacological Research. 2013;69:137–143. doi: 10.1016/j.phrs.2012.11.006. PMid:23201354 http://dx.doi.org/10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Human_Microbiome_Project_Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. http://dx.doi.org/10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitmore SE, Lamont RJ. Oral Bacteria and Cancer. PLoS Pathog. 2014;10:e1003933. doi: 10.1371/journal.ppat.1003933. PMid:24676390 http://dx.doi.org/10.1371/journal.ppat.1003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy KN, Sonkodi I, Szöke I, et al. The microflora associated with human oral carcinomas. Oral Oncology. 1998;34:304–308. http://dx.doi.org/10.1016/S1368-8375(98)80012-2. [PubMed] [Google Scholar]
- 10.Katz J, Onate MD, Pauley KM, et al. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. Int J Oral Sci. 2011;3:209–215. doi: 10.4248/IJOS11075. http://dx.doi.org/10.4248/IJOS11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zackular JP, Rogers MAM, Ruffin MT, et al. The Human Gut Microbiome as a Screening Tool for Colorectal Cancer. Cancer Prevention Research. 2014;7:1112–1121. doi: 10.1158/1940-6207.CAPR-14-0129. PMid:25104642 http://dx.doi.org/10.1158/1940-6207.CAPR-14-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michaud DS, Izard J, Wilhelm-Benartzi CS, et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut. 2013;62:1764–1770. doi: 10.1136/gutjnl-2012-303006. PMid:22990306 http://dx.doi.org/10.1136/gutjnl-2012-303006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn J, Segers S, Hayes RB. Periodontal disease, Porphyromonas gingivalis serum antibody levels and orodigestive cancer mortality. Carcinogenesis. 2012;33:1055–1058. doi: 10.1093/carcin/bgs112. PMid:22367402 http://dx.doi.org/10.1093/carcin/bgs112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Molecular Systems Biology. 2014;10 doi: 10.15252/msb.20145645. PMid:25432777 http://dx.doi.org/10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahn J, Sinha R, Pei Z, et al. Human Gut Microbiome and Risk for Colorectal Cancer. Journal of the National Cancer Institute. 2013;105:1907–1911. doi: 10.1093/jnci/djt300. PMid:24316595 http://dx.doi.org/10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu N, Yang X, Zhang R, et al. Dysbiosis Signature of Fecal Microbiota in Colorectal Cancer Patients. Microb Ecol. 2013;66:462–470. doi: 10.1007/s00248-013-0245-9. http://dx.doi.org/10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 17.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor immune microenvironment. Cell host & microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. PMid:23954159 http://dx.doi.org/10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.IARC. Tobacco Smoke and Involuntary Smoking. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Vol 83. Lyon France: International Agency for Research on Cancer; 2004. [PMC free article] [PubMed] [Google Scholar]
- 19.Secretan B, Straif K, Baan R, et al. A review of human carcinogens– Part E: tobacco, areca nut, alcohol, coal smoke, and salted fish. The Lancet Oncology. 2009;10:1033–1034. doi: 10.1016/s1470-2045(09)70326-2. http://dx.doi.org/10.1016/S1470-2045(09)70326-2. [DOI] [PubMed] [Google Scholar]
- 20.Moon JH, Lee JH, Lee JY. Subgingival microbiome in smokers and non-smokers in Korean chronic periodontitis patients. Molecular Oral Microbiology. 2014 doi: 10.1111/omi.12086. n/a-n/a. http://dx.doi.org/10.1111/omi.12086. [DOI] [PubMed] [Google Scholar]
- 21.Bizzarro S, Loos BG, Laine ML, et al. Subgingival microbiome in smokers and non-smokers in periodontitis: an exploratory study using traditional targeted techniques and a next-generation sequencing. Journal of Clinical Periodontology. 2013;40:483–492. doi: 10.1111/jcpe.12087. http://dx.doi.org/10.1111/jcpe.12087. [DOI] [PubMed] [Google Scholar]
- 22.Mason MR, Preshaw PM, Nagaraja HN, et al. The subgingival microbiome of clinically healthy current and never smokers. ISME J. 2014 doi: 10.1038/ismej.2014.114. http://dx.doi.org/10.1038/ismej.2014.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shchipkova AY, Nagaraja HN, Kumar PS. Subgingival Microbial Profiles of Smokers with Periodontitis. Journal of Dental Research. 2010;89:1247–1253. doi: 10.1177/0022034510377203. PMid:20739702 http://dx.doi.org/10.1177/0022034510377203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato I, Land S, Majumdar AP, et al. Functional polymorphisms to modulate luminal lipid exposure and risk of colorectal cancer. Cancer epidemiology. 2010;34:291–297. doi: 10.1016/j.canep.2010.02.010. PMid:20308031 http://dx.doi.org/10.1016/j.canep.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Illumina. 16S Metagenomics App 15055860 A ed2014. Available from: https://support.illumina.com/content/dam/illumina-support/documents/documentation/software_documentation/basespace/16s-metagenomics-user-guide-15055860-a.pdf. [Google Scholar]
- 26.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. PMid:20383131 http://dx.doi.org/10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Applied and environmental microbiology. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. PMid:16332807 http://dx.doi.org/10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vazquez-Baeza Y, Pirrung M, Gonzalez A, et al. EMPeror: a tool for visualizing high-throughput microbial community data. GigaScience. 2013;2:16. doi: 10.1186/2047-217X-2-16. PMid:24280061 http://dx.doi.org/10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnold T. SAS/EST 9.22 User’s Guide. Cary, NC: SAS Institute; 2010. [Google Scholar]
- 30.Gonzales-Barron U, Cadavez V, Butler F. Conducting inferential statistics for low microbial counts in foods using the Poisson-gamma regression. Food Control. 2014;37:385–394. http://dx.doi.org/10.1016/j.foodcont.2013.09.032. [Google Scholar]
- 31.Park S, Navratil S, Gregory A, et al. Multifactorial Effects of Ambient Temperature, Precipitation, Farm Management, and Environmental Factors Determine the Level of Generic Escherichia coli Contamination on Preharvested Spinach. Applied and Environmental Microbiology. 2015;81:2635–2650. doi: 10.1128/AEM.03793-14. PMid:25636850 http://dx.doi.org/10.1128/AEM.03793-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamanaka W, Takeshita T, Shibata Y, et al. Compositional Stability of a Salivary Bacterial Population against Supragingival Microbiota Shift following Periodontal Therapy. PLoS ONE. 2012;7:e42806. doi: 10.1371/journal.pone.0042806. PMid:22916162 http://dx.doi.org/10.1371/journal.pone.0042806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Garrity GM, Tiedje JM, et al. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Applied and Environmental Microbiology. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. PMid:17586664 http://dx.doi.org/10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Youssef N, Sheik CS, Krumholz LR, et al. Comparison of Species Richness Estimates Obtained Using Nearly Complete Fragments and Simulated Pyrosequencing-Generated Fragments in 16S rRNA Gene-Based Environmental Surveys. Applied and Environmental Microbiology. 2009;75:5227–5236. doi: 10.1128/AEM.00592-09. PMid:19561178 http://dx.doi.org/10.1128/AEM.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claesson MJ, O’Sullivan O, Wang Q, et al. Comparative Analysis of Pyrosequencing and a Phylogenetic Microarray for Exploring Microbial Community Structures in the Human Distal Intestine. PLoS ONE. 2009;4:e6669. doi: 10.1371/journal.pone.0006669. PMid:19693277 http://dx.doi.org/10.1371/journal.pone.0006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–360. doi: 10.1038/nature13178. http://dx.doi.org/10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Zhang D, Jia H, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21:895–905. doi: 10.1038/nm.3914. http://dx.doi.org/10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 38.Saxelin M. Probiotic Formulations and Applications, the Current Probiotics Market, and Changes in the Marketplace: A European Perspective. Clinical Infectious Diseases. 2008;46:S76–S79. doi: 10.1086/523337. PMid:18181728 http://dx.doi.org/10.1086/523337. [DOI] [PubMed] [Google Scholar]
- 39.Badet C, Thebaud NB. Ecology of Lactobacilli in the Oral Cavity: A Review of Literature. The Open Microbiology Journal. 2008;2:38–48. doi: 10.2174/1874285800802010038. PMid:19088910 http://dx.doi.org/10.2174/1874285800802010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arora M, Weuve J, Fall K, et al. An Exploration of Shared Genetic Risk Factors Between Periodontal Disease and Cancers: A Prospective Co-Twin Study. American Journal of Epidemiology. 2010;171:253–259. doi: 10.1093/aje/kwp340. PMid:19969528 http://dx.doi.org/10.1093/aje/kwp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yen AMF, Lai H, Fann JCY, et al. Relationship between Community Periodontal Index and Fecal Hemoglobin Concentration, an Indicator for Colorectal Neoplasm. Journal of Dental Research. 2014;93:760–766. doi: 10.1177/0022034514539976. PMid:24938273 http://dx.doi.org/10.1177/0022034514539976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan ST, Ahamed M, Musarrat J, et al. Anti-biofilm and antibacterial activities of zinc oxide nanoparticles against the oral opportunistic pathogens Rothia dentocariosa and Rothia mucilaginosa. European Journal of Oral Sciences. 2014;122:397–403. doi: 10.1111/eos.12152. http://dx.doi.org/10.1111/eos.12152. [DOI] [PubMed] [Google Scholar]
- 43.Zhang F, He S, Jin J, et al. Exploring salivary microbiota in AIDS patients with different periodontal statuses using 454 GS-FLX Titanium pyrosequencing. Frontiers in Cellular and Infection Microbiology. 2015;5:55. doi: 10.3389/fcimb.2015.00055. PMid:26191508 http://dx.doi.org/10.3389/fcimb.2015.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brook I, Gober AE. Recovery of potential pathogens and interfering bacteria in the nasopharynx of smokers and nonsmokers. Chest. 2005;127:2072–2075. doi: 10.1378/chest.127.6.2072. PMid:15947322 http://dx.doi.org/10.1378/chest.127.6.2072. [DOI] [PubMed] [Google Scholar]
- 45.Brook I, Gober AE. Effect of smoking cessation on the microbial flora. Archives of otolaryngology–head & neck surgery. 2007;133:135–138. doi: 10.1001/archotol.133.2.135. PMid:17309981 http://dx.doi.org/10.1001/archotol.133.2.135. [DOI] [PubMed] [Google Scholar]
- 46.Biedermann L, Zeitz J, Mwinyi J, et al. Smoking Cessation Induces Profound Changes in the Composition of the Intestinal Microbiota in Humans. PLoS ONE. 2013;8:e59260. doi: 10.1371/journal.pone.0059260. PMid:23516617 http://dx.doi.org/10.1371/journal.pone.0059260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kato I, Nechvatal JM, Dzinic S, et al. Smoking and other personal characteristics as potential predictors for fecal bacteria populations in humans. Medical science monitor: International Medical Journal of Experimental and Clinical Research. 2010;16:Cr1–Cr7. PMid:20037488. [PMC free article] [PubMed] [Google Scholar]
- 48.Feder HM, Jr, Garibaldi RA. The Significance of Nongonococcal, Nonmeningococcal Neisseria Isolates from Blood Cultures. Reviews of Infectious Diseases. 1984;6:181–188. doi: 10.1093/clinids/6.2.181. PMid:6374834 http://dx.doi.org/10.1093/clinids/6.2.181. [DOI] [PubMed] [Google Scholar]
- 49.Mager DL, Ximenez-Fyvie LA, Haffajee AD, et al. Distribution of selected bacterial species on intraoral surfaces. Journal of Clinical Periodontology. 2003;30:644–654. doi: 10.1034/j.1600-051x.2003.00376.x. http://dx.doi.org/10.1034/j.1600-051X.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- 50.Teles FR, Teles RP, Sachdeo A, et al. Comparison of microbial changes in early re-developing biofilms on natural teeth and dentures. Journal of periodontology. 2012;83:1139–1148. doi: 10.1902/jop.2012.110506. PMid:22443543 http://dx.doi.org/10.1902/jop.2012.110506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamma JJ, Nakou M. Subgingival Microflora in Smokers with Early Onset Periodontitis. Anaerobe. 1997;3:153–157. doi: 10.1006/anae.1997.0095. PMid:16887581 http://dx.doi.org/10.1006/anae.1997.0095. [DOI] [PubMed] [Google Scholar]
- 52.Lundberg JO, Govoni M. Inorganic nitrate is a possible source for systemic generation of nitric oxide. Free Radical Biology and Medicine. 2004;37:395–400. doi: 10.1016/j.freeradbiomed.2004.04.027. PMid:15223073 http://dx.doi.org/10.1016/j.freeradbiomed.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 53.Hyde ER, Andrade F, Vaksman Z, et al. Metagenomic Analysis of Nitrate-Reducing Bacteria in the Oral Cavity: Implications for Nitric Oxide Homeostasis. PLoS ONE. 2014;9:e88645. doi: 10.1371/journal.pone.0088645. PMid:24670812 http://dx.doi.org/10.1371/journal.pone.0088645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doel JJ, Benjamin N, Hector MP, et al. Evaluation of bacterial nitrate reduction in the human oral cavity. European Journal of Oral Sciences. 2005;113:14–19. doi: 10.1111/j.1600-0722.2004.00184.x. http://dx.doi.org/10.1111/j.1600-0722.2004.00184.x. [DOI] [PubMed] [Google Scholar]
- 55.Kanady JA, Aruni AW, Ninnis JR, et al. Nitrate reductase activity of bacteria in saliva of term and preterm infants. Nitric Oxide. 2012;27:193–200. doi: 10.1016/j.niox.2012.07.004. PMid:22842223 http://dx.doi.org/10.1016/j.niox.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stepanov I, Jensen J, Hatsukami D, et al. New and traditional smokeless tobacco: comparison of toxicant and carcinogen levels. Nicotine & tobacco research: Official Journal of the Society for Research on Nicotine and Tobacco. 2008;10:1773–1782. doi: 10.1080/14622200802443544. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2892835/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sipahimalani AT, Chadha MS, Bhide SV, et al. Detection of Nnitrosamines in the saliva of habitual chewers of tobacco. Food and chemical toxicology: an international journal published for the British Industrial Biological Research Association. 1984;22:261–264. doi: 10.1016/0278-6915(84)90003-6. http://dx.doi.org/10.1016/0278-6915(84)90003-6. [DOI] [PubMed] [Google Scholar]
- 58.Belstrøm D, Holmstrup P, Nielsen CH, et al. Bacterial profiles of saliva in relation to diet, lifestyle factors, and socioeconomic status. Journal of Oral Microbiology. 2014;6 doi: 10.3402/jom.v6.23609. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3974179/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bebek G, Bennett KL, Funchain P, et al. Microbiomic subprofiles and MDR1 promoter methylation in head and neck squamous cell carcinoma. Human Molecular Genetics. 2012;21:1557–1565. doi: 10.1093/hmg/ddr593. PMid:22180460 http://dx.doi.org/10.1093/hmg/ddr593. [DOI] [PMC free article] [PubMed] [Google Scholar]