Abstract
Defining treatment susceptible or resistant populations of cancer patients through the use of genetically defined biomarkers has revolutionized cancer care in recent years for some disease/patient groups. Research continues to show that histologically defined diseases are diverse in their expression of unique mutations or other genetic alterations, however, which presents both opportunities for the development of personalized cancer treatments, but increased difficulty in testing these therapies because potential patient populations are divided into ever-smaller numbers. To address some of the growing challenges in biomarker development and clinical trial design, NCCN assembled a group of experts across specialties and solid tumor disease types to begin to define the problems and to consider alternate ways of designing clinical trials in the era of multiple biomarkers and targeted therapies. Results from that discussion are presented, focusing on issues of clinical trial design from the perspective of statisticians, clinical researchers, regulators, pathologists and information developers.
Introduction
Clinical trials in oncology are more likely to fail than those in any other disease area1. Many fail in Phase III, after great expense and many years of effort2,3. Assuming that most decisions to proceed to phase III are based on reasonable data sets, it is critical to understand why the phase III trial results are so often not as expected.
The heterogeneity of cancer across and within a disease is likely one of the reasons. We now know that cancer is primarily a disease driven by genetic and epigenetic changes that affect key cellular processes associated with the malignant phenotype4. The identification of the large array of different genetic alterations in both common and rare cancers has led to a reclassification of these diseases. Cancers that have historically been grouped together according to histologic attributes can now be categorized into ever smaller subgroups, often with unique prognoses and responses to both standard and targeted treatments. The heterogeneity is not just amongst cancers in different patients but also exists between and within tumor sites in the same person5. Therefore, specific therapeutic interventions may apply a selection pressure, which in this heterogeneous background can lead to a Darwinian evolution of the tumor’s characteristics6.
The recognition of this heterogeneity has led us away from a ‘one size fits all’ model of cancer therapeutics and fueled many of the recent major advances in cancer care. For example, testing for epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer (NSCLC) to preferentially direct the 10–15% of Western patients who test positive for the mutation toward an EGFR tyrosine kinase inhibitor for first-line treatment of advanced disease is now standard. This treatment has been shown to be superior to chemotherapy in this population in terms of response rate and progression free survival (PFS)7,8. Similarly, ERBB2(HER2) expression/amplification in breast cancer informs both advanced and early stage treatment decision-making. Testing for HER2 and use of HER2 targeted therapy in positive patients has become the standard of care9–13. Because genomic aberrations often do not segregate by organ of origin, reclassification of cancer based on genomic alteration across tumor types may yield larger subgroups.
Although the use of predictive biomarkers to individualize treatment has revolutionized care in some cancer subtypes, the paradigm of one companion diagnostic, one drug and one clinical trial may not represent the most efficient or effective use of resources in the current medical, scientific and economic environment.
In order to further this conversation, NCCN convened a panel of experts in the areas of clinical research in multiple solid tumor types, clinical trial design, statistics, pathology, and regulatory affairs to discuss recent results, lessons learned, current challenges, future strategies, and concerns that are relevant across specialties. This article presents the results and recommendations of the NCCN Working Group on Clinical Trial Design in the Era of Multiple Biomarkers and Targeted Therapies.
Novel Trial Design
Novel clinical trial designs are being explored in an attempt to expedite the development and approval of new drugs. I-SPY2 (Investigation of Serial Studies to Predict Your Therapeutic Response With Imaging and Molecular Analysis 2) is a phase II screening study that uses an adaptive randomization strategy to test multiple drugs and biomarkers in the early treatment of breast cancer. The real-time assessments of each therapy enables more rapid decisions to be made regarding the likelihood of success, with the expectation that less-hopeful drugs will be abandoned while promising agents graduate to smaller targeted phase III trials. A Phase II/III strategy, Lung-MAP (Lung Cancer Master Protocol) study, has recently begun to accrue patients. In this study, next generation sequencing (NGS) is being used to assign patients with squamous NSCLC to 1 of 5 substudies, each based upon a single biomarker and matched drug. The bucket or basket navigation trial assigns patients to a treatment group based upon biomarker profile without consideration of histology or tissue of origin. This has been used in one case to direct patients to appropriate phase I clinical trials (PREDICT), and other such studies are in the planning stages (MATCH).
Lessons Learned From an Adaptive Phase II Screening Trial: The I-SPY2 Trial in Breast Cancer
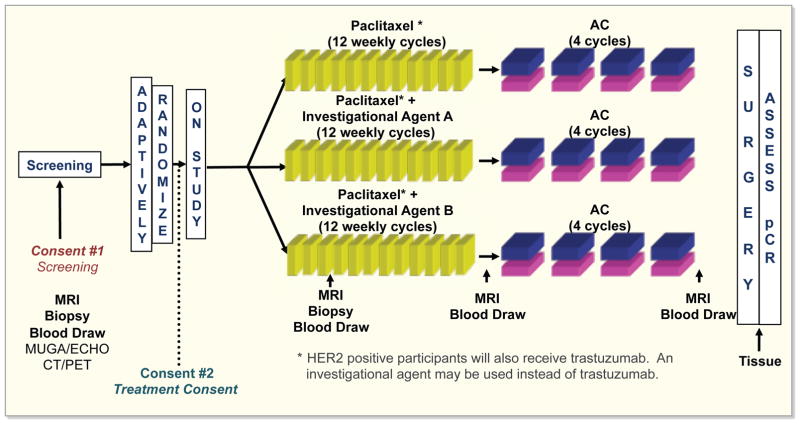
I-SPY2 is a phase II trial in breast cancer targeting women with large primary tumors (>3 cm) in the neoadjuvant setting. All candidates are screened for biomarker status, including estrogen receptor, ESR1(ER) and progesterone receptor, PGR(PR) expression (+/−), HER2 expression/amplification (+/−) and MammaPrint® 70 gene expression prognostic signature (MP high, MP low)14. The focus of the study is on disease at an early, potentially curable state. Patients with MP low, Her2−, ER/PGR+ tumors are excluded from participation, because neoadjuvant chemotherapy is not an effective therapeutic strategy for most of these women15. The study includes 10 of the most prevalent biomarker signatures, divided among 8 or more experimental treatment arms. The design of the study involves addition of experimental drugs to chemotherapy with paclitaxel for 12 weeks, or paclitaxel plus HER2 inhibitors for those subsets with HER2 positive tumors (Figure 1). Then patients are treated with doxorubicin and cyclophosphamide chemotherapy, followed by surgery. Biopsy samples are collected at 4 points during the treatment cycle. The endpoint of the study is defined as pathologic complete response (pCR), based upon tumor tissue collected at surgery14. The FDA has recently released a draft guidance outlining the terms under which pCR is an appropriate endpoint for accelerated approval of antineoplastic agents for breast cancer when used in the neoadjuvant setting16.
Figure 1. I-SPY2 trial design.
Abbreviations: AC, anthracycline; ECHO, echocardiogram; ERBB2(HER2), human epidermal growth factor receptor 2; I-SPY2, Investigation of Serial Studies to Predict Your Therapeutic Response With Imaging and Molecular Analysis 2; MUGA, multi gated acquisition scan; pCR, pathologic complete response.
aHER2+ participants will also receive trastuzumab. An investigational agent may be used instead of trastuzumab.
The trial uses a Bayesian adaptive design, meaning that the probabilities that the current patient’s tumor will benefit from each of the available experimental therapies are calculated and the patients therapy is assigned proportional to these probabilities. This predictive probability is then used to select incoming patients for the experimental treatment arms that are most likely to succeed, based upon the patients’ biomarker signatures. Drugs from several companies are used in the trial, and regimens are paired with biomarkers in a continual screening process. The trial encompasses 20 centers and more thano 650 patients have been enrolled to date.
Two of the first 7 drugs tested have met the predetermined experimental threshold for graduation from the trial, defined as a calculated 85% Bayesian predictive probability of success if that combination of agents were tested against a control in a randomized phase III clinical trial of 300 patients, using a primary endpoint of pCR. Veliparib, a poly(ADP-ribose) polymerase inhibitor17, is one of the first “graduates”. It graduated with a signature of triple-negative breast cancer (HER2−, ER−, PR−). Preliminary results were presented at the San Antonio Breast Cancer (SABC) meeting in December 2013, based upon 72 patients overall compared with 44 control patients. The study showed that among triple-negative patients veliparib plus carboplatin has a probability of 0.99 of being superior to control treatment and a predicted probability of success of 0.90 in a 300 patient phase II trial18–20. Neratinib, an EGFR, HER2, HER4 inhibitor,21 is the other graduate. In February 2014 investigators presented the results of 115 patients who received neratinib compared with 78 control patients at the American Association for Cancer Research annual meeting. The drug’s graduating signature was HER2+/ER−PR−.22
Whether these drugs will in fact show the efficacy that is expected and will be superior to other drugs already in use remains to be seen. However, an “extended adjuvant” trial was announced in July 2014 showing a statistically significant 33% reduction in the risk of disease events for patients with HER2+ tumors treated with neratinib after trastuzumab.23
Benefits to adaptive platform design illustrated by I-SPY2
Adaptive trial designs as illustrated by I-SPY2 allow investigators to formulate an estimate of the likelihood of success when matching drugs with biomarker profiles, thus fast-tracking therapies to be more easily tested in smaller, targeted phase III trials. Interim analyses and selection of a validated surrogate endpoint (pCR) also allow the trial to reach initial conclusions more rapidly. Other efficiencies are accomplished by the multidrug trial using a single control arm for multiple experimental therapies. The fact that it is more likely for a patient to be assigned to a therapy, particularly a therapy that is statistically more likely to work with the individual biology of that patient’s tumor could also increase patient satisfaction and lead to increased trial participation14,24–28.
Lessons Learned: Designing the Lung Cancer Master Protocol
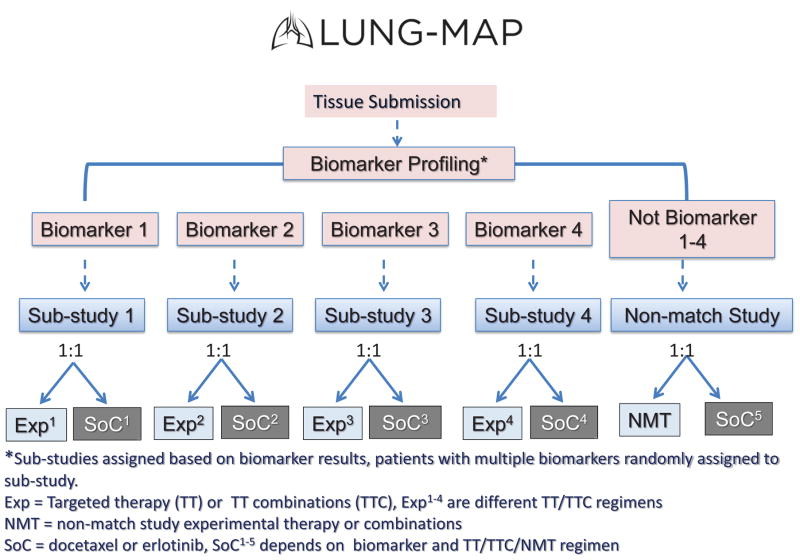
Another study that uses a novel design to test multiple therapies simultaneously is the Lung-MAP study29–32. This study recently began accruing patients to second-line therapies for squamous cell lung cancer. The initial version of the study includes 5 parallel, independently conducted substudies testing investigational drugs or regimens against a randomized control arm of standard licensed second-line therapies in NSCLC (mostly docetaxel) in groups with the same biomarker profile. Investigational drugs are selected based upon the availability of an analytically valid biomarker, and include a cyclin-dependant kinase inhibitor, a PI3 kinase inhibitor, an anti-hepatocyte growth factor (HGF) monoclonal antibody, and a fibroblast growth factor receptor (FGFR) tyrosine kinase inhibitor. NGS, provided by Foundation Medicine and immunohistochemistry (IHC) will be used to establish biomarker profiles. Biomarker status determines patient assignment to substudy, and sufficient flexibility exists to allow for additional markers and agents to roll into the trial (Figure 2).
Figure 2. Lung Master Protocol (Lung MAP) trial design.
Exp 1–4 are different TT/TTC regimens.
SoC 1–5 depends on biomarker and TT/TTC/NMT regimen.
Abbreviations: Exp, experimental therapy (TT or TTC); NMT, non-match study experimental therapy or combinations; SoC, standard of care (docetaxel or erlotinib); TT, targeted therapy; TTC, targeted therapy combination.
aSubstudies assigned based on biomarker results; patients with multiple biomarkers randomly assigned to substudy.
Patients will be assigned to a substudy based upon biomarker profiling by NGS of 250 target genes, or in the case of the HGF inhibitor, by IHC for MET expression. Patients who do not have any of the qualifying biomarkers will be assigned to the “non-match” sub-study, and randomized to treatment with MedImmune’s PDL1 inhibitor or docetaxel, thus giving all patients entering the trial a possibility of receiving a new therapeutic agent.
The study is a multi-substudy phase II/III trial, with PFS as the primary endpoint of the phase II portion of the trial, assessed as an interim analysis after 55 progression events. If futility is established, that arm of the trial is closed to further accrual. The phase III portion of the trial, which can seamlessly follow the phase II interim analysis, uses PFS and overall survival (OS) as co-primary endpoints. Approximately 500 to 1000 patients are targeted to be enrolled per year, with new agents and markers rolling into the trial as new substudies. Final analysis will begin when 256 OS and 290 PFS events have occurred. Unfortunately, given the natural history of this disease in these patients (median OS of 8 to 9n months and PFS of approximately 3 months), the results of this trial will be known quickly.
This master protocol is groundbreaking for a variety of reasons: the creation of a framework able to simultaneously test therapies from a variety of companies using a single (or limited) type of eligibility testing—in this case NGS—is a key step. Using a single platform for biomarker screening, rather than an individual companion diagnostic paired with each experimental therapy, should improve efficiency within this trial, and may provide an example that can be extended to other diseases. The combined phase II/III design allows for interim analyses to stop a study early, especially because the investigators have established a high bar for drugs to continue past the phase II portion of the trial. MASTER will be followed closely and the lessons learned will inform studies in other diseases.
Lessons Learned From the MD Anderson Cancer Center (MDACC) Phase I PREDICT Basket Navigation Trial
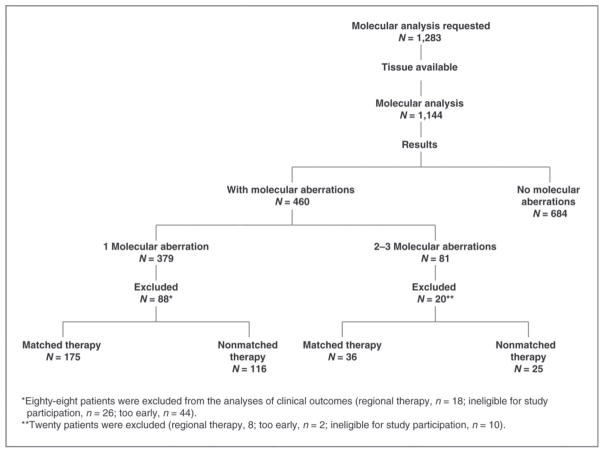
A third type of clinical trial that uses biomarker testing in a novel application is the disease site of origin agnostic ‘basket’ navigation type of trial, whereby patients are screened for mutations or other biomarkers, and then assigned to treatment groups or clinical trials based upon biomarker screening, regardless of the site of the tumor. In other words, the molecular profile is used to make a “diagnosis” that leads to therapy. One example of this type of trial implemented at MDACC in the form of a phase I clinical trial program was the PREDICT (Profile-Related Evidence Determining Individualized Cancer Therapy) program. Patients with advanced or metastatic cancers refractory to therapy, who had experienced relapse after standard therapy, or who had a tumor for which there was no standard therapy were referred to the phase I program. As one of the criteria for entry into the clinical trials program, patients were assessed by PCR based sequencing for several molecular aberrations, including mutations in PIK3CA, BRAF, KRAS, NRAS, EGFR, KIT, GNAQ, TP53, MET, and RET33. Based upon their mutation/aberration profile, they were then matched with targeted therapies that were available either on prescription or within existing trials within the institution. Therapies were considered to be matched if the drug in the trial was known to inhibit the target in question at low nmol concentrations however each biomarker was not necessarily already proven to be an effective predictive biomarker, nor the drug an effective inhibitor of that pathway in the clinic. 1283 molecular analyses were requested, 1144 were able to be performed, and of these, 60% showed no aberrations in the genes tested, 33% had single gene aberrations, 6.4% had two gene aberrations and 0.7% (8 patients) had three or more of the aberrations tested (Figure 3). Of the patients with single gene aberrations referred to phase I clinical trials, 175 received matched therapy, and 116 received therapy without matching. In the group that received matched therapy, 27% obtained a complete (2%) or partial (25%) response, compared with 5% (all partial responses) of those receiving non-matched therapies (Figure 3).
Figure 3. CONSORT diagram, PREDICT program.
aRegional therapy, 18; ineligible for study participation, 26; too early, 44.
bRegional therapy, 8; ineligible for participation, 10; too early, 2.
From Tsimberidou AM, Iskander NG, Hong DS, et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res 2012;18:6373–6383; with permission.
Results of this trial were sufficiently positive in the opinion of some investigators that further randomized studies are planned, although the success will be dependent upon the quality of both predictive biomarkers and the drugs in question. In addition, a National Clinical Trials Network (NCTN) sponsored phase II trial, MATCH, is currently in the planning phase.32 This trial will use NGS of approximately 200 genes supplemented by fluorescence in situ hybridization (FISH) and IHC. Genes selected for sequencing have all been matched with targeted therapies, either FDA approved for treatment in some type of cancer, or with evidence for activity against one of the selected targets. Patients enrolled in the study will be followed for response and PFS. When their tumors progress, patients will be removed from the study. An estimated 3000 patients will need to be screened to enroll 1000 with targetable aberrations, and the study is expected to take 3 to 4 years to complete. It is anticipated that follow-up phase II trials will be needed to provide further clinical evidence of efficacy for the drugs shown to have activity in one of the biomarker defined populations.
One obstacle to the more general implementation of this strategy of clinical trial design is the definition of an “actionable” mutation. In terms of proven efficacy, most patients screened did not show mutation profiles with an actionable genetic alteration. However, in the years since this program began more patients are found to have alterations that could be deemed “actionable”. The expense incurred by screening all patients to target treatment to a small minority is a significant barrier, and funding sources need to be determined. However, as a method by which patients can be triaged into appropriate clinical trials, this expense is justified. The percentage of treatable patients will potentially increase assuming more clinically active targeted agents are identified, and as the cost for comprehensive mutation screening continues to fall, comprehensive molecular genetic profiling will be more practical in the general cancer population as a part of routine cancer care.
In summary, novel strategies will be needed to address the increasing complexity of cancer. Trials will need to be designed with earlier endpoints and intermediate analyses, allowing for sequential therapies as treatments fail or resistance emerges.
Validation of Technology and Tissue Sampling
Another area that must be considered when developing a clinical trial design is the testing technology itself, and the allocation and use of necessary tissue samples for performing the test. At the most basic level, the quantity and quality of the tissue sample is critical for adequate and accurate testing. The quality of the sample may be affected by several preanalytic variables including collection methods, handling, fixation and storage. Standard operating procedures must be generated for techniques of tissue sampling, handling and storage to meet the requirement of the test. Guidelines for tissue suitability must be established, including acceptable limits of nonneoplastic tissue in the biopsy samples, specific processing of bone lesions to preserve the integrity of nucleic acid, and specific limitations on tissue quantity for adequate performance of the assay.
Samples from metastatic sites in particular can be problematic, and may be the focus of clinical trial testing in oncology. Lesions metastatic to bone, for example, would require specific handling and avoidance of routine decalcification methods to preserve suitability of the tissue for molecular studies. Depending on the sensitivity of the assay chosen, lesions metastatic to lymph nodes may prove difficult to test because of the presence of lymphoid tissue, which can rapidly dilute the tumor content, whereas local changes surrounding metastatic loci may contain fibrosis and inflammation, causing a similar diluting effect. Furthermore, a large biopsy does not necessarily translate into a large amount of suitable tumor tissue. In some cases, aspirates have been shown to be equally suitable and often preferable to biopsies, owing to a larger percentage of tumor cells and less fragmented DNA. Consultations with institutional pathologists, surgeons and interventional radiologists to determine the best procedures to obtain high-quality samples are vital to establishing parameters for biomarker testing within clinical trials.
Validating an assay and its associated technology is also extremely important. Performance characteristics for assays that derive from the newest technologies can be difficult to prove, because of a paucity of historical data and a lack of determined gold standards for comparison. Other risks are associated with introducing new technologies into clinical practice and clinical trials. Technologies may not be mature and the sources of technical bias may not yet be known. On the other hand, now that the genomic drivers of cancer are better understood, performing clinical trials without molecular selection is likely to continue to yield many trial failures. Further, technological advancements in genomic sequencing have occurred at a startling pace, and fully understanding the implications of these technologies requires their rapid incorporation into clinical trials that permit learning throughout the trial life cycle.
As an example of the challenges, consider the situation where a trial required validated testing for specific BRAF mutations across a variety of tumor types. Methods available to accomplish this included: direct sequencing, locked nucleic acid-peptide nucleic acid PCR clamp-based testing, matrix-assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry, NGS methods, or IHC with mutation-specific antibodies. Tissue samples for this study could be variable, and come from several different neoplastic disease processes, including hairy cell leukemia, plasma cell neoplasms, thyroid carcinoma, colon cancer, NSCLC and melanoma34–36. The initial approach taken was to attempt to validate IHC as a screening method. IHC has the advantage of being readily available in most pathology labs, is able to be automated, and has minimal requirements for tissue. However, further attempts to validate the IHC assay showed that this methodology also has some distinct disadvantages. Poorly fixed or old archival tissue leads to suboptimal antibody performance, and standard techniques for decalcification (mainly with HCL and PCN samples)also lead to poor performance. A high variability of staining was seen depending on the tissue source, and interpretation varied also with the type of malignancy.
DNA based methods, such as MALDI-TOF and Sanger sequencing, are highly reproducible and have low requirements for DNA, but are not sufficiently sensitive for use with samples with a low tumor content, a problem anticipated with hairy cell leukemia and plasma cell neoplasia samples, and small biopsies and fluid samples with low numbers of neoplastic cells. NGS methods were also considered, using a custom panel and Amplicon capture methodology that was available in-house within the institution. However, this had a higher requirement for DNA, a longer turnaround time and was insufficiently sensitive in many cases. Thus, for a single study requiring analysis of a single biomarker, it was necessary to validate several platforms and establish multiple workflows in order to accommodate differences and quality of sample types.
Considering all of the potential issues with using variable in-house methodology for gene mutation analysis, multisite site studies should standardize the techniques used and consider a central provider of these services whenever possible. Because of the additional variability when multiple single-gene assays are used, plus the inefficient use of tissue and expense with this approach, multiplexed assays are becoming increasingly popular. Although both single gene and multiplex assays can be conducted in an academic setting, commercial vendors do exist. However, some factors must be considered when using centralized commercial laboratory services. First, it is plausible that if this approach becomes widespread, restricting molecular diagnostics to one or only a small set of vendors will increase the costs significantly because of lack of competition, with the end result that fewer patients will have molecular profiling performed. In some commercial cases, results are delivered without details on the underlying data or of the bioinformatics tools used to perform the analyses—essentially results are presented in a black box. Bioinformatic programs may be platform specific or unavailable for outside analysis, with limited data for direct comparison of performance among the available programs, limiting the ability of researchers to independently validate results of the analyses. Separate from the issues with commercial providers, NGS platforms, wherever performed, can have other technical issues associated with data interpretation, including difficulties in sequencing GC rich regions, sequencing errors that appear at the ends of reads, and homopolymers being susceptible to insertion-deletion errors. Thus, thresholds to call variants can vary across platforms, programs and users.
In light of these many sources of variation, standards and guidelines for every step of the process from tissue acquisition to data analysis are absolutely required to avoid site-to-site variability in a multi-site trial. Centralized testing may be one solution, but does come with a price in terms of turnaround time and potentially also in transparency if a commercial vendor is used. Perhaps the most important takeaway is that close communication between the assay designers and pathologists is necessary to ensure that complexities of biopsy, tumor, and disease processes and assays are taken into consideration, so that variants are accurately assessed and patients are appropriately assigned to trial categories.
Challenge: An Education Gap
Implementing many of these concepts into clinical practice has been challenging at best. Complexity of testing and molecular data generation has led to a formidable education gap. The recognition that aberrations in signal transduction pathways, DNA repair pathways, apoptosis, chromosomal instability and other central cellular processes may be the direct cause of malignant transformation does not immediately translate into clinical utility. Oncologists in practice will be hard-pressed to stay abreast of the burgeoning field and to counsel patients on the ramifications of aberrations found in their tumors. A need exists for cancer genetic information that can be used by physicians to elucidate this morass to their patients.
One such resource available on the Vanderbilt University website is the My Cancer Genome web based information portal37. This site began by covering mutations in NSCLC and melanoma and has since expanded to include 21 disease sites, 56 genes, and 416 disease-gene-variant relationships, and will continue to include more disease entities and genetic aberrations38. The site is designed to be used by physicians at the point of care, so that if a patient’s mutation results from a multigene panel were available, the oncologist could then use the site to find data underlying the association of a particular mutation with the disease and could then learn whether targeted treatments were available or in development. Data displayed include details on the particular mutation, its frequency in the disease and implications for targeted therapies, and links to ongoing clinical trials. Dates of most recent information update and direct links to relevant references are also included.
This information has also been linked directly to the electronic health record system in use at the Vanderbilt University hospital39,40 so that when multigene mutation results are included in a patient’s record, selecting the mutation on the electronic record will automatically access the relevant mutation page. Because of the direct link to mutation-based clinical trials, this can also facilitate greater trial referral and enrollment. Participation in clinical trials hovers at 3 to 4% nationally, and if treatments are to be developed for all of the genetic variants that are currently being discovered, this number will have to increase dramatically at both community sites and academic centers. A thoughtfully built and integrated information resource like My Cancer Genome can help facilitate this process. Future plans for the site include a patient centric view.
Issues In Specific Tumor Types
BRAF Mutations in Melanoma
Melanoma is among the cancers with increasing incidence in the United States, with at least 76,000 anticipated diagnoses and an estimated 10,000 deaths occurring in 201441. Although 84% of patients are diagnosed at stage 1, with 5 year survival rates greater than 90%, this plummets to a median survival of only 6–9 months for those diagnosed with metastatic disease, with 16% surviving at 5 years. The BRAF gene, which encodes a serine/threonine protein kinase in the MAP kinase (MAPK) pathway downstream of RAS, is mutated in approximately 40% of human melanomas42. Most of these mutations are the result of valine (V) to glutamate (E) or lysine (K) substitutions at amino acid position 600, resulting in a constitutively active form of the kinase43. Vemurafenib and dabrafenib are selective kinase inhibitors that bind to the mutated form of BRAF in the ATP binding site, thereby blocking further downstream signaling. These agents have been FDA-approved for the treatment of patients with unresectable or metastatic melanoma who harbor a BRAF V600E mutation43–46. Importantly, both inhibitors have been shown to improve OS and PFS, with medians of approximately 13 to 15 months and 6 to 7 months, respectively. In parallel with the FDA approvals, development of a paired companion diagnostic test for each agent is also noteworthy.
Understanding the relevant signal-transduction pathways is critical to further drug development. When a patient harbors an activating mutation in BRAF, the downstream kinase MEK is also activated. This recognition led to the development of trametinib, a MEK inhibitor45,47,48. Trametinib is indicated as a single agent and in combination with dabrafenib in patients with unresectable or metastatic melanoma that harbor a BRAF V600E or V600K mutation. It is important to understand that MEK inhibition in BRAF mutation-positive patients appears to have a lower response rate and shorter PFS and OS when compared to the BRAF inhibitors, and that MEK inhibition is not effective in patients who have progressed on a prior BRAF inhibitor. However, the combination of BRAF and MEK inhibition in the appropriate patient population does improve response rate and PFS, although somewhat more modestly than one would hope.45,49 Collectively, these targeted therapies represent a substantial step forward in the treatment of patients with BRAF-mutated melanoma.
Nonetheless, despite recognition of a driver mutation and the availability of targeted therapies, responses to single agent MEK or BRAF inhibitors, or the combination of both MEK and BRAF inhibition, are only approximately 25%, 50% and 70%, respectively. Furthermore, as indicated previously, responses tend not to be durable, with most patients found to have progressive disease in 6 to 7 months. Understanding mechanisms of resistance and the development of other effective therapies is paramount for further advances in the treatment of melanoma.
Several resistance mechanisms have been proposed. For example, MAPK pathway activation, such as CRAF activation or upregulation of COT/Tp12 has been implicated in resistance to BRAF targeted therapy.50 Interestingly, this paradoxical activation has been proposed as an etiologic factor in the development of squamous cell carcinomas of the skin, which may occur in patients treated with BRAF inhibitors.51,52
Resistance is also thought to be mediated by reactivation of the MAPK pathway through upstream (NRAS) or downstream (MEK) mechanisms. Furthermore, resistance not mediated by the MAPK pathway has been shown to arise through upregulation of other signal transduction pathways such as the PI3 kinase/AKT pathway. Therefore, approaches utilizing inhibition of two parallel pathways are also being explored.50
It is worthy to note that sensitivity to BRAF inhibitors is not only dependent on presence of the activating mutation, but also of the tumor type under study42,46,50,53,54. For example, although the BRAF inhibitors have led to impressive advances in the treatment of melanoma, and some success in patients with BRAF-mutated NSCLC55, they seem to have a relative lack of efficacy in colorectal cancer, even in patients who have a BRAF V600E mutation56,57.
The Development of ALK Inhibitors in NSCLC
Molecular analyses have proven value in non-small lung cancer with a variety of driver mutations identified and active targeted therapies either licensed or being explored in clinical trials.58 Although the exploitation of activating EGFR mutations as the major determinants of responsiveness to the EGFR TKIs erlotinib, afatinib, or gefitinib largely occurred retrospectively after the initial development of these drugs, newer examples of personalized therapy in lung cancer have occurred prospectively59–61. Perhaps the best example is represented by activating rearrangements of the anaplastic lymphoma kinase (ALK) gene, which have been found in 4 to 7% of NSCLC patients. ALK inhibitors have demonstrated significant clinical activity in patients with tumors harboring these rearrangements 62. Lessons learned through the development of biomarker tests and associated targeted therapies directed against ALK can potentially be applied to clinical trial design in other malignancies63.
The initial phase I trial of crizotinib, a potent ALK and MET inhibitor, was designed to enroll a series of expanded cohorts at the recommended phase II dose based not on histology per se, but on tumors with proven evidence of ALK or MET activation64. Following the discovery of ALK in lung cancer that occurred after the phase I study had begun accrual, the protocol was amended to allow an ALK+ NSCLC cohort to be added. One lesson from this approach was that the strong signal that eventually led to the rapid approval of crizotinib in ALK+ NSCLC based on its activity in several hundred patients was apparent very early on. Waterfall plots of the first 19 patients and those from over 100 patients had almost identical shapes65,66 (overall response rate, 53% vs 61%). When the efficacy signals are so strong it is difficult to know how to determine the necessary numbers of patients for a drug approval. This issue becomes all the more pressing when the specific subtype of cancer being targeted is rare, because large randomized studies comparing the treatment with standard therapies may not be feasible. Increasingly, regulatory agencies have shown flexibility, with accelerated and conditional approvals based on dramatic activity even in small numbers of patients. Most recently, the advent of FDA “breakthrough status” for promising drugs/development approaches allows sponsors much closer contact with senior members of the agency to try and maximize the chances of successfully developing effective agents.
The second lesson learned is that there needs to be balance between the perceived heterogeneity of local biomarker testing for clinical trial eligibility and the perception that testing in a large central laboratory will necessarily be superior in all cases. During start-up of a new testing technology, central laboratories will have their own learning curve for implementation and for detailed and transparent recordkeeping. For the crizotinib trials, ALK-FISH analysis was initially performed locally, and then confirmed (or not) by a centralized testing laboratory. Later, the phase II/III crizotinib studies required testing in a central laboratory, even in cases deemed positive in local laboratory testing, and tests that were positive in the first instance and negative in the second were initially recorded as “negative” and not “discordant”. This initially resulted in some confusion regarding the activity of crizotinib in ALK− tumors. Although some ALK− tumors do respond to crizotinib potentially because of the presence of one of the other known drivers of sensitivity to crizotinib, such as ROS1 and MET, some of the responses seen initially in patients with presumed ALK− may have been the result of central testing recording a “negative” for a tumor that had been documented as being positive first on local testing. Consequently, it is vitally necessary that the basis for a discordant test be documented in detail, including image capture where appropriate and the cases noted as “discordant” rather than “negative”. If the chances of a false positive are considered lower than for a false negative, it may make sense to allow enrollment of positive tumors based upon local testing, and reserve central testing for retrospective confirmation. In addition, even if not discordant, sequential testing before entry to confirm a local result centrally also increases the delay in initiating treatment.
A third observation regarding biomarker subsets in NSCLC was that molecular selection may alter the reaction to therapy for more than just the experimental targeted drug. For example, pemetrexed and docetaxel, when given as single agents as second-line treatment in NSCLC67 resulted in nearly identical treatment response rates, and OS and PFS. However, when these 2 drugs were compared with crizotinib in a randomized trial of patients with ALK+ NSCLC (N=343, for crizotinib n=172, pemetrexed n=99 and docetaxel n=72), the pemetrexed treated patients had double the median PFS (4.2 months vs. 2.6 months for docetaxel) and more than 3 times the response rate (29% vs 7%) compared with docetaxel.68 Consequently, when considering comparing a standard chemotherapy to a targeted therapy in a molecularly preselected population, knowing the degree of benefit in the same molecular population, as opposed to extrapolating directly from an unselected population, would help to more accurately size these randomized studies. For this reason, the design of the Lung-MAP is appealing becasue it assesses efficacy differences between the experimental and control (e.g. docetaxel) treatments in each biomarker preselected population separately.
The crizotinib trials also highlighted a fourth lesson to be learned: that grouping central nervous system (CNS) progression together with extra-CNS progression in the determination of PFS may not be the best approach in the future. Even if the disease can be controlled in the body, the variable rates of penetrance of drugs into CNS tissues mean that the brain can be a significant site for progression, through pharmacologic rather than biological means.63 For example, one study showed that for 46% of patients on crizotinib, the CNS represents the site of first progression, and that the CNS was the sole site of progression in 85% of these patients.69 It was also shown, at least in one case report, that less than 0.3% of the plasma concentration of crizotinib enters the cerebrospinal fluid, indicating that crizotinib may not present at therapeutic concentrations in the brain.70 The activity of a drug in the brain, especially in diseases with a high rate of brain metastases, such as NSCLC, may be better addressed in future trials through defining separate cohorts of patients with specific oncogenic driver and CNS disease, and adding those cohorts to the earliest phase I studies of the relevant molecularly targeted therapies. In addition, in the future, trials may well introduce distinct formal end points, such as CNS and extra-CNS time to progression or CNS and extra-CNS response rates to allow the degree of benefit to be tracked separately in the CNS from disease elsewhere in the body.71
The final observation from the ALK trials in NSCLC was that treatment beyond progression can be beneficial. Specifically, at the time of progression many drug sensitive clones may still be suppressed and can be documented to flare up when the TKI is stopped.72 Consequently, although traditionally progression has prompted discontinuation of the drug, if isolated areas in the CNS or even outside the CNS progress and most of the rest of the disease is still under control with the targeted therapy, local treatment, such as with radiation of the progressing disease and continuation of the TKI may be associated with many months of benefit before a new progressive site develops73.
Colorectal Cancer
Stage II colon cancer provides a prime example of a circumstance when treatment selection should be informed by molecular analyses of tumors. Patients with node negative stage II disease will be cured by surgery alone approximately 80% of the time, but identifying high-risk patients has been elusive. Therefore, generic use of adjuvant chemotherapy after surgery has been shown to confer only a slight benefit. For example, in the QUASAR clinical trial, patients with stage II colon cancer were randomized either to receive adjuvant chemotherapy with 5-FU, or to observation only, with chemotherapy given on recurrence. Results of this trial showed an approximately 2 to 3% improvement in OS for patients treated with chemotherapy, indicating that chemotherapy provided no benefit for 97% of patients with stage II disease.74–76 Therefore, the ability to selectively target the highest-risk patients for therapy is clearly needed.
Attempts have been made to use molecular signatures and mutation profiles to define subsets of patients with colon cancer to identify those most in need of post-surgical adjuvant chemotherapy. Thus far, disruptions in 2 major molecular pathways have been shown to be associated with colon cancer. By far the most common, occurring in roughly 85% of cases, are molecular changes that can be traced to chromosomal instability. Chromosomal instability and inactivation of the adenomatous polyposis coli gene lead to the genetic and epigenetic events such as the mutations, global hypomethylation and allelic deletions that have been identified in colon cancer. The most commonly occurring mutations in colon cancer are KRAS, PIK3CA, BRAF, and NRAS, with mutation frequencies of 38%, 20%, 14% and 3%, respectively.77 Reported mutation frequencies can vary somewhat, depending upon detection methods. KRAS and NRAS mutations are known to be predictive for lack of response to EGFR-targeting antibody therapy for patients with advanced disease.78,79 BRAF is downstream of RAS on the RAS/RAF/MAPK signaling pathway, and PIK3CA mutations have been implicated in the genesis of several kinds of cancer.80
Elements of the DNA mismatch repair (MMR) pathway, disrupted in the remaining 15% of colon cancer patients include protein products of the MLH1, MSH2, and PMS2 genes. These proteins are components of the enzymatic system responsible for recognizing and repairing insertion, deletion, and substitution errors that can arise during DNA replication and recombination and for repairing some forms of DNA damage.
MLH1, MSH2, MSH6, and PMS2 proteins can be altered through germline mutations or epigenetic inactivation of MLH1 via promoter methylation. Germline mutation in a mismatch repair enzyme, most often MLH1 or MSH2, leads to the development of hereditary non-polyposis colon cancer, also known as Lynch Syndrome, which is responsible for 2 to 3% of colon cancers. Sporadic cases are most often associated with loss of expression of an MMR gene (MLH1) caused by promoter hypermethylation.
When analyzed for ability to predict response to oxaliplatin, mutations in KRAS, NRAS, PIK3CA, and BRAF and MMR status were found not associated with resistance or response.77 BRAF V600E mutation status and MMR proficiency were both prognostic factors associated with shorter overall survival times.77,81 Other studies using multi-gene expression tests have not been able to show predictive value. Thus, identifying the molecular characteristics of colon tumors most likely to benefit from chemotherapy and the development of targeted agents remain high priorities for clinical trial design.
The ASSIGN trial
A molecularly directed clinical trial is being contemplated for metastatic colorectal cancer. A working group has been formed to explore the possibilities of such a trial in collaboration with the NCI, FDA, industry and advocacy groups. All would agree that the overarching goal of choosing colon cancer therapies based on molecular tumor characteristics is laudatory. Furthermore, sufficient advances in technology have made large scale sequencing feasible, and targeted therapies directed at some key driver pathways are now becoming available.
The study has a name – ASSIGN – but no other details are yet determined. Study designs with similarity to the lung master protocol are currently being debated. Platform technologies for screening are anticipated to be next generation sequencing or the equivalent to identify aberrations in 200 to 400 genes. The platform will be able to measure amplifications, and possibly translocations and should enable the accurate definitions of even complex pathways, such as the PI3 kinase pathway, which is also a prime target in rectal cancer.
For all of the enthusiasm this effort is generating, however, one minor detail remains: identifying agents that can inhibit targets as monotherapy or finding a means to combine targeted therapies. To date, colorectal cancer has defied single agent targeted therapies in contrast to lung or breast cancer, for example. Preliminary data are needed before launching such an ambitious effort.
Small Cell Lung Cancer
Small cell lung cancer (SCLC) represents approximately 15% of all lung cancers and is almost exclusively smoking related. It is an exceptionally aggressive disease, characterized by early metastatic potential and a high proliferative index, which accounts for it being the sixth leading cause of cancer deaths in the United States. Surgery is only appropriate for the less than 5% of patients diagnosed with very early-stage disease. At least 70% of patients have extensive stage disease at presentation. Most chemotherapy regimens show high initial overall response rates in both extensive and limited-stage disease, but almost all patients rapidly relapse, leading to overall poor survival rates. In looking to future strategies for treating SCLC, traditional antiproliferative chemotherapy alone seems unlikely to improve outcomes. Identifying the molecular targets that are driving cell survival, proliferation and metastasis is critical, as is identification of lung cancer progenitor cells (cancer stem cells) which are presumably responsible for the high frequency of recurrence following the initial response to anti-proliferative therapy.
A wide variety of molecularly targeted agents have failed to demonstrate clinical activity against SCLC in clinical trials despite being supported by solid preclinical rationale. These include matrix metalloprotease inhibitors (marimastat, BAY-12-9566); angiogenesis inhibitors (thalidomide, vandetanib, sunitinib, sorafenib, cedarinib, aflibercet, bevacizumab); growth factor pathway inhibitors (imatinib, 2A11 (an anti-GRP MoAb), tipifarnib, gefitinib, dasatinib, everolimus, temsirolimus, ATRA); and apoptotic pathway stimulators (oblimersen, obatoclax, AT-101); the HDAC inhibitor romidepsin, immuno-targeted vaccines (BEC2) and immunotoxins (NCAM-ricin).82
In addition to the limited efficacy of targeted therapy in SCLC, obtaining adequate tissue for appropriate biomarker analysis has also been challenging because of the high frequency of diagnosis from bronchoscopic biopsy samples. Obtaining serial biopsies for clinical trials is even more challenging83 further confounding attempts to perform pharmacodynamic studies or to characterize the molecular nature of these tumors as the disease progresses.
Recent studies have attempted to identify common, rational targets in SCLC tumor samples and cell lines. Peifer, et al. performed SNP array analysis, exome sequencing, transcriptome analysis and whole genome sequencing in subsets of 63 SCLC tumor samples.84 They reported that SCLC has a very high mutation rate, on the order of that observed in melanoma and squamous cell lung cancer, which is presumably because of the prevalence of this tumor among heavy smokers who have high exposure to mutagenic carcinogens in tobacco smoke. Aberrations in TP53 and RB1 were nearly universal, and derangements of histone modifiers, MYC amplification, PTEN deletion, SLIT2 mutation and FGFR1 amplification were found in 5–20% of tumors.
Another study, using SCLC tumors and cell lines found similarly high rates of mutations, with frequent mutations in TP53, RB1, PIK3CA, CDKN2A and PTEN, along with aberrations in chromatin modifiers and DNA-repair and checkpoint pathway components.85 Interestingly, SOX2 amplification was found in 27% of samples. SOX proteins are involved in stem cell renewal, as are the NOTCH and Hedgehog pathways, which were also frequently dysregulated. Because residual cancer stem cells seem to be responsible for the high rate of relapse in SCLC these pathways may be rational targets for further study.
Another recent study attempted to define molecular predictors for chemotherapy response by testing 267 potential therapeutic compounds against 44 SCLC molecularly characterized cell lines 86. MYC gene amplification was identified in 44% of cell lines, and these cell lines showed a high degree of sensitivity to aurora kinase B (AURKB) inhibitors. Because AURKB inhibitors are currently in clinical development, this finding provides a sound rationale for testing these compounds in molecularly selected patients with SCLC. Other genetic alterations that were found to be associated with sensitivity to specific agents included FGFR1 amplification with FGFR inhibitors, and PTEN loss with HSP inhibitors, but, interestingly, not PI3K inhibitors.
Attempts at targeting KIT provide the best example of how sound preclinical rationale can lead to disappointing clinical findings. Expression of KIT and its ligand, stem cell factor (SCF), are found in 30 to 100% of SCLCs,87–92 though mutations such as those that drive gastrointestinal stromal tumors are generally not present. Numerous preclinical studies showed that KIT inhibitors, including imatinib, could inhibit both kinase activity and cellular proliferation in SCLC. However, several phase II trials of imatinib in SCLC failed to demonstrate any clinical activity, despite specific selection of patients whose tumors expressed KIT.93–95 A common theme is that the benefit of targeted agents is largely limited to patients with tumors that harbor activating mutations in the target gene. These studies further highlight the point that mere expression does not define the biological relevance of a potential therapeutic target.
In summary, although the clinical need for effective treatment for SCLC is great, experience with targeted therapy in this disease illustrates many of the most pressing problems in applying molecular medicine to clinical trials in solid tumors. The extreme aggressiveness of SCLC and the general debility of many patients when they present with the disease creates substantial difficulties in attaining adequate accrual to clinical trials, particularly those with strict eligibility criteria and the need for molecular characterization. In addition, adequate tumor samples are difficult to obtain, further hindering molecular diagnostic studies. The high mutation frequency complicates the identification of driver mutations and leads to the rapid evolution of resistant clones, making it highly unlikely that single-agent targeted therapy will lead to substantial clinical gains. More likely, advances will only be made through the evaluation of combinations of agents targeting disparate pathways, a strategy that creates further hurdles regarding toxicity and the need for greater inter-industry cooperation.
Renal Cell Carcinoma
Renal cell carcinoma (RCC) is diagnosed in approximately 65,000 patients annually, and leads to approximately 13,000 deaths per year, representing approximately 4% of cancers diagnosed. SEER data indicate that the 5 year survival rate is approximately 12% for those with distant metastases.96 One reason for the high mortality associated with RCC is the lack of treatments that can induce durable long-term remissions of metastatic disease and the absence of effective adjuvant therapies for high-risk localized disease. The lack of clinically useful biomarkers to define treatment sensitive or resistant populations is one barrier to the development of effective treatments in RCC. However, several significant barriers exist to discovering better molecular markers. RCC has been shown to be a significantly genomically heterogeneous tumor type5, which may hinder molecular characterization and development of personalized therapies. It has been estimated that nearly 70% of somatic mutations present in RCC tissues are not detectable across all tumor sites. Another barrier to biomarker development is the inconsistency and variability in tissue preparation and biomarker evaluation,97 which will affect accuracy of the biomarker data that are collected and interpreted. Efforts to define prognostic or predictive factors have largely shown that clinical features, including tumor grade, local extent of tumor, regional lymph node metastases and evidence of metastases at presentation remain the most consistent features associated with survival.
Clear cell carcinomas constitute 85% of RCCs, whereas the remaining 15%, though called non-clear cell RCC, contain many different histologies including type I and type 2 papillary and chromophobe tumors. Among the clear cell subset, 2 groups of molecularly targeted therapies have emerged for the treatment of metastatic disease; 5 agents target the VEGF pathway, and 2 agents are mTOR inhibitors. All of these therapies have been shown to prolong time to disease progression in the metastatic population, and together represent the first new therapeutic classes for RCC in many years. However in most cases, clinical benefit remains transitory and patients eventually experience disease progression.98,99
Another new area pertains to the recent development of agents targeting the PD-1/PD-L1 axis. PD-1 and one of its ligands, PD-L1, are normally expressed on cells of the immune system, with PD-1 appearing on activated T cells and PD-L1 appearing on activated antigen presenting and other cells.100 RCC tumors with high PD-L1 expression have been shown to be more aggressive.101 Antibodies that bind PD-1 and interfere with the interaction of PD-1 and PD-L1 are in clinical development for a variety of tumor types. A phase I trial involving several solid tumor histologies showed that tumors that do not express PD-L1 are not responsive to anti-PD-1 immunotherapy, whereas roughly 36% of the tumors positive for PD-L1 expression showed a response to anti-PD-1 antibody therapy. Of the 42 specimens tested for PD-L1 expression, 5 were from RCC.102 Nevertheless, more recent data103,104 in RCC are showing that patients with PD-L1 negative tumors can still respond to agents that target the PD-1/PD-L1 axis. Further refinement of this data and standardization of PD-L1 staining are warranted.
Ovarian Cancer
Ovarian cancer is another malignancy that presents a variety of challenges for genomic and personalized medicine development. It is the fifth most common cause of cancer death in US women, with 22,240 cases diagnosed in 2013, and approximately 14,000 deaths from the disease per year. A total of 70% of patients present with advanced disease (stage III and IV) at diagnosis. Long term survival data show that within the patient population with advanced stage III disease, the 5 year PFS rate is 15 to 25% with 5 year OS rates of 50 to 60% in patients receiving appropriate aggressive care.105
Ovarian cancer has 4 main histotypes. All of these were previously thought to arise from ovarian surface epithelial cells; however, more recent evidence suggests that the most common high-grade ovarian cancer subtype, serous carcinoma, arises from cells originating from the fimbriated end of the Fallopian tube.106 Other primary histotypes include: endometrioid, mucinous, and clear cell. These types differ in molecular makeup and response to therapy 107. Molecular characterization of ovarian tumors has resulted in a division into 2 groups. Type I tumors principally have mutations in PTEN, PI3K, KRAS, BRAF, or β-catenin, have genomically stable tumors and rare TP53 mutations. These are often found as low grade epithelial or endometrioid tumors, and borderline, mucinous and clear cell. Type 2 tumors are characterized by TP53 mutations and generalized genomic instability, and are most often high grade endometrioid or serous tumors. Tumors that are BRCA1 or BRCA2+ are also found in this subgroup.108
An early biomarker, MUC16(CA-125), a carbohydrate cell surface antigen shed into the serum in a variety malignant and benign conditions, was discovered by using ovarian cancer cells as an immunogen for the production of monoclonal antibodies. The antibody generated recognized a molecule highly expressed on ovarian tumor cells, but not on other cells isolated from the same patient. It has been extensively studied for a variety of uses as a biomarker in ovarian cancer, including monitoring response to therapy and detecting persistent disease.109,110 Attempts have been made to use serum CA-125 levels as an early detection or screening test for asymptomatic women, but variability in baseline levels and changing expression in a variety of benign conditions have made this strategy not optimally useful on a population wide basis.109 CA-125 is a highly glycosylated single transmembrane cell surface protein. Phosphorylation of the intracellular domain leads to cleavage of the extracellular domain and release from the membrane. One function of CA-125 is to bind mesothelin (MSLN), a molecule that mediates heterotypic cell adhesion. This interaction may be important for mediating adhesion between tumor cells migrating into the peritoneum and the mesothelial epithelium.111
The utility of using CA-125 levels as a marker of early recurrence in ovarian cancer thus enabling earlier treatment, is based on the observation that CA-125 levels may rise several months before disease recurrence becomes symptomatic. This finding was used to study whether or not earlier detection of recurrence followed by earlier chemotherapy treatment would lead to better outcomes. Women with ovarian cancer in complete remission after first-line platinum-based chemotherapy with a normal CA-125 serum level were enrolled in a prospective clinical trial and followed by clinical examination and measurement of CA-125 levels every 3 months. If levels increased more than twice the upper limit of normal, patients were randomized to receive either early chemotherapy or continued monitoring until other clinical symptoms of relapse appeared. In all, 529 patients were randomized to treatment groups with 265 assigned to early treatment and 264 to delayed treatment. With a median follow-up of 56.9 months and 186 deaths in the early treatment group and 184 deaths in the delayed treatment group, analysis of OS data showed no survival difference between those groups.112 These results suggest that routine monitoring of CA-125 for disease recurrence may have little role, because early treatment in asymptomatic women did not result in a survival benefit. However, in this study a variety of standard chemotherapy regimens were used. It is possible that the best chemotherapy or targeted agent for this population has not been identified and that this biomarker may be more appropriately used to better identify patient populations suitable for inclusion in clinical trials.
Other issues in ovarian cancer clinical trial design
Because close to 80% of patients experience initial response to platinum-based chemotherapies, these regimens will continue to be important for the treatment of ovarian cancer. Thus, new or targeted agents will need to be administered in combination with cytotoxic agents. A recent trial using bevacizumab in combination with cytotoxic chemotherapy provided only a small improvement in PFS when used in the up-front treatment setting. Unfortunately no predictive biomarkers have yet been identified that allow a determination of which patient subsets may benefit from the addition of bevacizumab to chemotherapy. Patients with ovarian cancer may remain platinum-sensitive through several rounds of therapy, though all do eventually become resistant. Focusing on the platinum resistant patients is an important area for development, because fewer options are available at recurrence.
Breast Cancer
Because of the availability of tissue biopsy material, large numbers of patients and an active research and advocate community, breast cancer has been a leader in the use of clinical biomarkers. Biomarkers have been used in breast cancer to determine risk, for screening purposes, to determine and refine diagnoses, in the determination of prognosis, including the prediction of relapse, disease progression and survival, and as markers to predict response to therapy, including hormone therapies, chemotherapy and other novel therapy. Biomarkers are also used to monitor the course of disease. ESR1(ER) expression is a weak prognostic marker and its presence is strongly predictive for response to endocrine manipulations. More recently, expression or amplification of HER2 (HER2 positivity) has been recognized as an indicator of a population of breast cancer patients that had an inferior prognosis to those with HER2-negative tumors. More importantly, HER2-positivity is a strong predictive marker for those patients who were able to respond to trastuzumab or other agents targeting HER2 or its associated kinase activity.
Multiparameter gene expression profiling has been used to produce biomarker signatures that are predictive of benefits from chemotherapy in certain patient populations113, and have provided the means to describe a variety of unique breast cancer subtypes. Gene expression profiling has been used to divide breast cancer into phenotypically distinct subtypes114, which correlate with therapeutic response and overall prognosis. Roughly 80% of so-called triple negative breast tumors (those that test negative for HER2, ER, and PGR(PR)) are contained within the basal epithelial-like expression cluster, whereas HER2+ tumors cluster separately. Another cluster contains tumors with a normal breast-like gene expression pattern. Hormone receptor positive tumors are found within the luminal subtype, which can be further divided into a luminal A subtype, which is marked by expression of ER and PGR, is HER2 negative, and has a low risk of recurrence. Luminal B subtype tumors are HER2 negative and ER positive, and carry a higher recurrence risk. Another luminal B subtype is characterized by expression of HER2. The luminal A subtype, owing to its sensitivity to hormonal therapy also shows the longest relapse free and OS among all groups.
It has since been shown that measurement of a smaller set of clinical markers can duplicate the division of breast cancer into these subtypes115, thus allowing better definition for treatment and identification of groups to target in clinical trials without needing expression analysis to be performed on every patient. Future clinical trials in breast cancer can thus be conducted in a subtype specific manner.
Hormone Receptor-Positive Disease
New therapies in breast cancer are often tested first in the metastatic setting, and only when efficacy is demonstrated in this population are studies undertaken in earlier disease in the adjuvant setting by measuring disease free and OS. However, because of past successes and longer survival times, especially in hormone receptor positive subsets, it has become difficult to use this model because of the long periods of follow up required. Several assays are commercially available to help differentiate women whose tumors are likely hormone sensitive and therefore may not need chemotherapy, from those for whom chemotherapy should be added. The assays, such as Oncotype DX®113, Prosigna™116 and EndoPredict®117, can also be used to stratify patients at high risk for enrollment in clinical trials investigating the use of chemotherapy or new targeted therapies.
The excellent outcomes experienced by most women with hormone receptor-positive disease have also led to the inclusion of newer therapies in the pre-surgical neoadjuvant setting as described previously in reference to the I-SPY2 clinical trial. Studies have shown that there is no difference in long term outcome whether chemotherapy is delivered before or after surgery and that pre-operative chemotherapy enhances breast conservation.118 In these situations, an alternative endpoint can be used, because all patients will have disease when the study is initiated. A wide ranging meta-analysis119 has shown that patients who experience a pCR after neoadjuvant therapy have improved survival, especially in aggressive subsets (triple negative; high grade hormone receptor positive, HER2-negative; and hormone receptor negative, HER2 positive), but an increase in pCR did not predict improved OS or event free survival.
Another marker, MKI67(Ki-67), which is an indicator of proliferative potential, may be useful in predicting the efficacy of neo-adjuvant therapies for hormone-receptor positive subsets. With recommendations suggesting 5 to 10 years of endocrine therapies, most women will become long term survivors, and therefore showed that improvement in adjuvant therapy within this group is quite difficult. The neoadjuvant model can also be used to evaluate combination of novel therapies with standard hormonal agents. For example, the randomized phase III study Alternate Approaches for Clinical Stage II or III Estrogen Receptor Positive Breast Cancer NeoAdjuvant Treatment (ALTERNATE) is being conducted with the goal of developing a biomarker strategy based upon Ki-67 expression level in the neoadjuvant setting, and to use response and modulation in the marker to predict long term outcomes for hormone receptor-positive in postmenopausal women with breast cancer.120 Treatment arms include fulvestrant and anastrozole alone and in combination. The ALTERNATE trial will establish a comparison of Ki-67 levels at baseline, 4 weeks, and 24 months, and use these to predict long term outcomes.
HER2+ Disease
HER2-targeted therapies have been successful in the treatment of HER2-positive disease, and have revolutionized the prognosis of this subtype. Nonetheless, opportunities still exist for improvement and benefits to further understanding the heterogeneity of this population. In some large clinical studies, where HER2 testing was first completed locally and shown to be positive, but then central testing was negative, benefits were still seen from HER2 targeted therapy.121 The underlying molecular basis for this is not known, and ongoing studies are addressing this important question. Studies have also shown that dual targeting of HER2, using either lapatinib or pertuzumab is associated with an increased benefit compared to trastuzumab alone. HER2+ disease can also be subdivided based upon hormone receptor expression. HER2+, hormone receptor-positive tumors show a lower rate of pCR compared with HER2+ hormone receptor-negative tumors. This difference in response might indicate a need to assess the populations separately in future studies. Further molecular characterization of these tumors may also lead to targeted treatments, and obviate the need for chemotherapy, or lead to a better understanding of mechanisms of resistance.
Triple Negative Breast Cancer
Approximately 20% of newly diagnosed breast cancers are triple negative (ER−, PR− and HER2−). Most are characterized by a high proliferative index, TP53 mutation and genomic instability, and are common in BRCA1 mutation carriers. Most women with triple negative tumors will respond to chemotherapy to some extent, but many will experience recurrence or metastatic disease.122 Responses in this subset are heterogeneous, and further stratification and development of new targeted therapies are needed.
Recently, Vanderbilt investigators were able to use gene expression profiling of 587 triple negative breast cancer tumors to further divide this group into roughly 6 new subsets. Of these 47% were basal-like in expression pattern, with 2 clusters of basal-like tumors defined, characterized by high expression of genes involved in proliferation and DNA damage response123. Another subtype was enriched in immunomodulatory genes. Two further subsets were mesenchymal-like, with enrichment in genes involved in epithelial mesenchymal transition and cell motility pathways. The final subset contained cells with a luminal androgen receptor positive expression pattern. This type of subdivision showed numerous opportunities for new targeted drug development, but also illustrated the problem of small numbers of patients and the difficulty in designing a drug development and clinical trial strategy for these types of populations.
For effective future development of therapies for breast cancer, new studies will need to be designed to address specific questions in well-defined subtypes. Another point to consider is the rational removal of unneeded therapies from multi-agent regimens. How to design studies that can provide data about the possibility of eliminating components of multi-component regimens without compromising efficacy and outcomes remains a significant challenge.
Conclusions and Recommendations
The examples provided of solid tumor studies illustrate that we are far from understanding the molecular complexity of any tumor type. For some of the tumors addressed here, such as SCLC, no effective molecularly targeted therapies exist. For others, treatments for some subsets have been defined, but even within targeted groups, not all tumors respond, and responses are often transient, as illustrated by ALK-rearranged NSCLC and BRAF-mutated melanoma. Even in tumors with relatively effective treatments and good long term outcomes, such as hormone receptor positive breast cancer, relapse eventually occurs in most patients. The molecular basis of resistance to targeted therapies is an area of ongoing investigation, and these resistance mechanisms vary widely among different tumor types. In EGFR mutation positive NSCLC, resistance usually develops because of secondary mutations in the target itself, whereas in other tumor types, such as BRAF-mutant melanoma, resistance usually arises through upregulation of other compensatory signal transduction pathways. All of these issues indicate the need for more comprehensive, multiplexed molecular data for more patients at all stages of disease across tumor types. Sequential testing for individual genetic aberrations quickly exhausts valuable specimens and does not contribute as much to the overall understanding of carcinogenesis, treatment response, and the development of resistance. As illustrated by ALK+ lung cancer, responses to treatments other than the targeted agent itself (such as to some standard cytotoxic therapies) can also be affected by the tumor’s molecular milieu.
The NCI recently announced that it is providing $12 million to fund tissue repositories for clinical trial samples at 3 institutions.124,125 More such tissue and data banks along with alternative sources of funding will be needed if the scope of personalized cancer therapy is to continue to expand. Building data resources will also require the development of guidelines for technology and tissue handling to ensure that the data that is captured today will continue to be useful in the future.
The clinical trial process itself must be streamlined to allow the evaluation of multiple promising targeted strategies at once, as in the Lung MAP and the I-SPY2 trial, and to broaden the testing of novel strategies across tumor types that share predictive biomarkers, as is being developed in the MATCH protocol. Regulatory agencies have been open to new approaches and further advances will require continued flexibility.
Another critical point is the need to enlist greater participation in clinical trials. The current level of 3 to 4% participation in the U.S. will not sustain the trials needed to validate the ever increasing number of promising agents that target smaller and smaller molecularly selected patient subsets. Improvement in trial participation is tightly linked with efforts to close the education gap. All oncologists, both in the community and at academic centers, will need to understand the complexities and limitations of the molecular characterization of tumors, and be able to communicate relevant information to their patients in an understandable and useful manner to allow shared clinical decision-making. Avenues for communication between clinical researchers and molecular pathologists need to be developed to allow for appropriate prioritization of resources and seamless screening and enrollment of patients onto clinical trials requiring biomarker assessment. Reliable online information resources that can be accessed everywhere and linked to electronic health record systems will be vitally important to reduce the complexity of molecular results and simplify the link between test results and treatment or trial participation.
Currently, far too many targets and associated agents are in the pipeline to allow for traditional clinical trial assessment of every strategy, particularly considering the ever decreasing, less common subsets of patients who are most likely to benefit from these treatments. As multiplexed assays and genome/exome wide sequencing become more common in oncologic practice, greater numbers of patients will be receiving rational, target-driven therapy in the off-label, non-clinical trial setting. The clinical outcomes of these patients will represent a great potential resource to expand the understanding of the efficacy of targeted therapies in complex molecular environments. The development of large, national databases to capture this off-trial data across tumor types would be an invaluable asset in the efforts to unlock the true promise of personalized therapy in cancer care.
Npvel collaborations will be needed to adequately assess new therapies and biomarkers in future clinical trials. Institutions, the pharmaceutical and diagnostic industries, foundations, NCI, FDA, insurance carriers, patients and advocates can all make important contributions in these efforts. Recently, Lung MAP and MATCH trials have been able to build networks that include the restructured National Clinical Trials Network as one component, and I-SPY, PREDICT and BATTLE are notable for success outside the NCI. Effective, efficient partnerships that can move rapidly to take advantage of the quickly evolving technology and drug development can be built and will be essential for success.
References
- 1.Arrowsmith J, Miller P. Trial watch: phase II and phase III attrition rates 2011–2012. Nat Rev Drug Discov. 2013;12:569. doi: 10.1038/nrd4090. [DOI] [PubMed] [Google Scholar]
- 2.Amiri-Kordestani L, Fojo T. Why do phase III clinical trials in oncology fail so often? J Natl Cancer Inst. 2012;104:568–569. doi: 10.1093/jnci/djs180. [DOI] [PubMed] [Google Scholar]
- 3.Gan HK, You B, Pond GR, Chen EX. Assumptions of expected benefits in randomized phase III trials evaluating systemic treatments for cancer. J Natl Cancer Inst. 2012;104:590–598. doi: 10.1093/jnci/djs141. [DOI] [PubMed] [Google Scholar]
- 4.Munoz J, Swanton C, Kurzrock R. Molecular profiling and the reclassification of cancer: divide and conquer. Am Soc Clin Oncol Educ Book. 2013;2013:127–134. doi: 10.14694/EdBook_AM.2013.33.127. [DOI] [PubMed] [Google Scholar]
- 5.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gridelli C, Rossi A. EURTAC first-line phase III randomized study in advanced non-small cell lung cancer: Erlotinib works also in European population. J Thorac Dis. 2012;4:219–220. doi: 10.3978/j.issn.2072-1439.2012.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31:3327–3334. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
- 9.Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 10.Slamon D. Herceptin: increasing survival in metastatic breast cancer. Eur J Oncol Nurs. 2000;4:24–29. doi: 10.1054/ejon.2000.0070. [DOI] [PubMed] [Google Scholar]
- 11.Slamon D, Eiermann W, Robert N, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365:1273–1283. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 13.The NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Breast Cancer (Version 3.2014). © 2014 National Comprehensive Cancer Network, Inc. Available at: NCCN.org. Accessed [July 16, 2014]. To view the most recent and complete version of the NCCN Guidelines®, go online to NCCN.org. Available at: http://www.nccn.org/professionals/physician_gls/pdf/breast.pdf. Accessed July 16.
- 14.Barker AD, Sigman CC, Kelloff GJ, et al. I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin Pharmacol Ther. 2009;86:97–100. doi: 10.1038/clpt.2009.68. [DOI] [PubMed] [Google Scholar]
- 15.Albain KS, Barlow WE, Ravdin PM, et al. Adjuvant chemotherapy and timing of tamoxifen in postmenopausal patients with endocrine-responsive, node-positive breast cancer: a phase 3, open-label, randomised controlled trial. Lancet. 2009;374:2055–2063. doi: 10.1016/S0140-6736(09)61523-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guidance for Industry: Pathologic Complete Response in Neoadjuvant Treatment of High-Risk Early-Stage Breast Cancer: Use as an Endpoint to Support Accelerated Approval DRAFT GUIDANCE. [Accessed August 8];2012 Available at: http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM305501.pdf.
- 17.Antolin AA, Mestres J. Linking off-target kinase pharmacology to the differential cellular effects observed among PARP inhibitors. Oncotarget. 2014;5:3023–3028. doi: 10.18632/oncotarget.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. [Accessed August 8];I-SPY 2: Novel Study Design. 2013 Available at: http://www.sabcs.org/UserPortal/Documents/SABCS_2013_Issue4.pdf.
- 19. [Accessed August 8, 2014];New presurgery combination therapy for triple-negative breast cancer. 2013 See more at: http://www.centerwatch.com/news-online/article/5737/new-presurgery-combination-therapy-for-triple-negative-breast-cancer#sthash.lxxDiwJK.1WpOa3bu.dpuf. Available at: http://www.centerwatch.com/news-online/article/5737/new-presurgery-combination-therapy-for-triple-negative-breast-cancer#sthash.lxxDiwJK.1WpOa3bu.dpbs.
- 20.Printz C. I-SPY2 trial yields first results on combination therapy for triple-negative breast cancer. Cancer. 2014;120:773. doi: 10.1002/cncr.28627. [DOI] [PubMed] [Google Scholar]
- 21.Bose P, Ozer H. Neratinib: an oral, irreversible dual EGFR/HER2 inhibitor for breast and non-small cell lung cancer. Expert Opin Investig Drugs. 2009;18:1735–1751. doi: 10.1517/13543780903305428. [DOI] [PubMed] [Google Scholar]
- 22.Neratinib Graduates to I-SPY 3. Cancer Discov. 2014;4:624. doi: 10.1158/2159-8290.CD-NB2014-055. [DOI] [PubMed] [Google Scholar]
- 23. [Accessed Sept. 8];Puma Biotechnology Announces Positive Top Line Results from Phase III PB272 Trial in Adjuvant Breast Cancer (ExteNET Trial) 2014 Available at: http://www.businesswire.com/news/home/20140722005059/en/Puma-Biotechnology-Announces-Positive-Top-Line-Results#.VA3vma90zkh.
- 24.Jones D. Adaptive trials receive boost. Nat Rev Drug Discov. 2010;9:345–348. doi: 10.1038/nrd3174. [DOI] [PubMed] [Google Scholar]
- 25.Berry DA. Bayesian clinical trials. Nat Rev Drug Discov. 2006;5:27–36. doi: 10.1038/nrd1927. [DOI] [PubMed] [Google Scholar]
- 26.Berry DA. Next generation clinical trials. Clin Adv Hematol Oncol. 2011;9:601–603. [PubMed] [Google Scholar]
- 27.Berry DA. Adaptive clinical trials: the promise and the caution. J Clin Oncol. 2011;29:606–609. doi: 10.1200/JCO.2010.32.2685. [DOI] [PubMed] [Google Scholar]
- 28.Berry DA. Adaptive clinical trials in oncology. Nat Rev Clin Oncol. 2012;9:199–207. doi: 10.1038/nrclinonc.2011.165. [DOI] [PubMed] [Google Scholar]
- 29. [Accessed August 8];Lung Cancer Master Protocol Activation Announcement. 2013 Available at: http://www.focr.org/events/2013-friends-brookings-conference-clinical-cancer-research.
- 30.Trial offers new model for drug development. Cancer Discov. 2014;4:266–267. doi: 10.1158/2159-8290.CD-ND2014-003. [DOI] [PubMed] [Google Scholar]
- 31.Fox JL. Master Protocol for squamous cell lung cancer readies for launch. Nat Biotechnol. 2014;32:116–118. doi: 10.1038/nbt0214-116b. [DOI] [PubMed] [Google Scholar]
- 32.Abrams J, Conley B, Mooney M, et al. National Cancer Institute’s Precision Medicine Initiatives for the New National Clinical Trials Network. Am Soc Clin Oncol Educ Book. 2014;34:71–76. doi: 10.14694/EdBook_AM.2014.34.71. [DOI] [PubMed] [Google Scholar]
- 33.Tsimberidou AM, Iskander NG, Hong DS, et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res. 2012;18:6373–6383. doi: 10.1158/1078-0432.CCR-12-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arcila M, Lau C, Nafa K, Ladanyi M. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn. 2011;13:64–73. doi: 10.1016/j.jmoldx.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabiawu-Ajise OEF JT, Soslow R, Levine D, Ladanyi M, Arcila ME. 2060 Assessment of BRAF V600E Mutation Status by Immunohistochemistry in Lung and Ovarian Carcinomas. Mod Pathol. 2013:26. [Google Scholar]
- 36.Liu YCJ, Nafa K, Teruya-Feldstein J, Arcila ME. Detection of BRAF V600E Mutation in Hairy Cell Leukemia: Comparison of High-Sensitivity Locked Nucleic Acid-PCR Sequencing and IHC with the VE1 Mutation-Specific Antibody. Journal of Molecular Diagnostics. 2013;15:843–945. [Google Scholar]
- 37.Levy MA, Lovly CM, Horn L, et al. My Cancer Genome: Web-based clinical decision support for genome-directed lung cancer treatment. J Clin Oncol. 2011:29. [Google Scholar]
- 38.Van Allen EM, Wagle N, Levy MA. Clinical analysis and interpretation of cancer genome data. J Clin Oncol. 2013;31:1825–1833. doi: 10.1200/JCO.2013.48.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carney PH. Information Technology and Precision Medicine. Seminars in Oncology Nursing. 30:124–129. doi: 10.1016/j.soncn.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 40.Levy MA, Lovly CM, Pao W. Translating genomic information into clinical medicine: lung cancer as a paradigm. Genome Res. 2012;22:2101–2108. doi: 10.1101/gr.131128.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. [Accessed August 8];SEER Stat Fact Sheets: Melanoma of the Skin. Available at: http://seer.cancer.gov/statfacts/html/melan.html.
- 42.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 43.Hall RD, Kudchadkar RR. BRAF Mutations: Signaling, Epidemiology, and Clinical Experience in Multiple Malignancies. Cancer Control. 2014;21:221–230. doi: 10.1177/107327481402100307. [DOI] [PubMed] [Google Scholar]
- 44.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flaherty KT, Infante JR, Daud A, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turajlic S, Furney SJ, Stamp G, et al. Whole-genome sequencing reveals complex mechanisms of intrinsic resistance to BRAF inhibition. Ann Oncol. 2014;25:959–967. doi: 10.1093/annonc/mdu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Falchook GS, Lewis KD, Infante JR, et al. Activity of the oral MEK inhibitor trametinib in patients with advanced melanoma: a phase 1 dose-escalation trial. Lancet Oncol. 2012;13:782–789. doi: 10.1016/S1470-2045(12)70269-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Falchook GS, Long GV, Kurzrock R, et al. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: a phase 1 dose-escalation trial. Lancet. 2012;379:1893–1901. doi: 10.1016/S0140-6736(12)60398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sosman JA, Daud A, Weber JS, et al. BRAF inhibitor (BRAFi) dabrafenib in combination with the MEK1/2 inhibitor (MEKi) trametinib in BRAFi-naive and BRAFi-resistant patients (pts) with BRAF mutation-positive metastatic melanoma (MM) ASCO Meeting Abstracts. 2013;31:9005. [Google Scholar]
- 50.Haarberg HE, Smalley KS. Resistance to Raf inhibition in cancer. Drug Discov Today Technol. 2014;11:27–32. doi: 10.1016/j.ddtec.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang V, Hepper D, Anadkat M, Cornelius L. Cutaneous toxic effects associated with vemurafenib and inhibition of the BRAF pathway. Arch Dermatol. 2012;148:628–633. doi: 10.1001/archdermatol.2012.125. [DOI] [PubMed] [Google Scholar]
- 52.Sloot S, Fedorenko IV, Smalley KS, Gibney GT. Long-term effects of BRAF inhibitors in melanoma treatment: friend or foe? Expert Opin Pharmacother. 2014;15:589–592. doi: 10.1517/14656566.2014.881471. [DOI] [PubMed] [Google Scholar]
- 53.Perez-Lorenzo R, Zheng B. Targeted inhibition of BRAF kinase: opportunities and challenges for therapeutics in melanoma. Biosci Rep. 2012;32:25–33. doi: 10.1042/BSR20110068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer. 2014;14:455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Planchard D, Mazieres J, Riely GJ, et al. Interim results of phase II study BRF113928 of dabrafenib in BRAF V600E mutation-positive non-small cell lung cancer (NSCLC) patients. ASCO Meeting Abstracts. 2013;31:8009. [Google Scholar]
- 56.Kopetz S, Desai J, Chan E, et al. PLX4032 in metastatic colorectal cancer patients with mutant BRAF tumors. ASCO Meeting Abstracts. 2010;28:3534. [Google Scholar]
- 57.Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 58.Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 60.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 61.Yang JC, Shih JY, Su WC, et al. Afatinib for patients with lung adenocarcinoma and epidermal growth factor receptor mutations (LUX-Lung 2): a phase 2 trial. Lancet Oncol. 2012;13:539–548. doi: 10.1016/S1470-2045(12)70086-4. [DOI] [PubMed] [Google Scholar]
- 62.Shaw AT, Engelman JA. ALK in lung cancer: past, present, and future. J Clin Oncol. 2013;31:1105–1111. doi: 10.1200/JCO.2012.44.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol. 2014 doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- 64.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Camidge DR, Bang YJ, Kwak EL, et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol. 2012;13:1011–1019. doi: 10.1016/S1470-2045(12)70344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kwak EL, Camidge DR, Clark J, et al. Clinical activity observed in a phase I dose escalation trial of an oral c-met and ALK inhibitor, PF-02341066. ASCO Meeting Abstracts. 2009;27:3509. [Google Scholar]
- 67.Hanna N, Shepherd FA, Fossella FV, et al. Randomized phase III trial of pemetrexed versus docetaxel in patients with non-small-cell lung cancer previously treated with chemotherapy. J Clin Oncol. 2004;22:1589–1597. doi: 10.1200/JCO.2004.08.163. [DOI] [PubMed] [Google Scholar]
- 68.Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 69.Weickhardt AJ, Scheier B, Burke JM, et al. Local ablative therapy of oligoprogressive disease prolongs disease control by tyrosine kinase inhibitors in oncogene-addicted non-small-cell lung cancer. J Thorac Oncol. 2012;7:1807–1814. doi: 10.1097/JTO.0b013e3182745948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Costa DB, Kobayashi S, Pandya SS, et al. CSF concentration of the anaplastic lymphoma kinase inhibitor crizotinib. J Clin Oncol. 2011;29:e443–445. doi: 10.1200/JCO.2010.34.1313. [DOI] [PubMed] [Google Scholar]
- 71.Lin NU, Lee EQ, Aoyama H, et al. Challenges relating to solid tumour brain metastases in clinical trials, part 1: patient population, response, and progression. A report from the RANO group. Lancet Oncol. 2013;14:e396–406. doi: 10.1016/S1470-2045(13)70311-5. [DOI] [PubMed] [Google Scholar]
- 72.Riely GJ, Kris MG, Zhao B, et al. Prospective assessment of discontinuation and reinitiation of erlotinib or gefitinib in patients with acquired resistance to erlotinib or gefitinib followed by the addition of everolimus. Clin Cancer Res. 2007;13:5150–5155. doi: 10.1158/1078-0432.CCR-07-0560. [DOI] [PubMed] [Google Scholar]
- 73.Weickhardt A, Doebele R, Oton A, et al. A phase I/II study of erlotinib in combination with the anti-insulin-like growth factor-1 receptor monoclonal antibody IMC-A12 (cixutumumab) in patients with advanced non-small cell lung cancer. J Thorac Oncol. 2012;7:419–426. doi: 10.1097/JTO.0b013e31823c5b11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hutchins G, Southward K, Handley K, et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29:1261–1270. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 75.Marshall JL. Risk assessment in Stage II colorectal cancer. Oncology (Williston Park) 2010;24:9–13. [PubMed] [Google Scholar]
- 76.Sanz-Pamplona R, Berenguer A, Cordero D, et al. Clinical value of prognosis gene expression signatures in colorectal cancer: a systematic review. PLoS One. 2012;7:e48877. doi: 10.1371/journal.pone.0048877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gavin PG, Colangelo LH, Fumagalli D, et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: an assessment of their prognostic and oxaliplatin predictive value. Clin Cancer Res. 2012;18:6531–6541. doi: 10.1158/1078-0432.CCR-12-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Douillard JY, Rong A, Sidhu R. RAS mutations in colorectal cancer. N Engl J Med. 2013;369:2159–2160. doi: 10.1056/NEJMc1312697. [DOI] [PubMed] [Google Scholar]
- 79.Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 80. [Accessed August 8];COSMIC: Catalogue of Somatic Mutations in Cancer. Available at: http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/
- 81.Lochhead P, Kuchiba A, Imamura Y, et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105:1151–1156. doi: 10.1093/jnci/djt173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hann CL, Rudin CM. Fast, hungry and unstable: finding the Achilles’ heel of small-cell lung cancer. Trends Mol Med. 2007;13:150–157. doi: 10.1016/j.molmed.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kalemkerian GP, Jiroutek M, Ettinger DS, et al. A phase II study of all-trans-retinoic acid plus cisplatin and etoposide in patients with extensive stage small cell lung carcinoma: an Eastern Cooperative Oncology Group Study. Cancer. 1998;83:1102–1108. doi: 10.1002/(sici)1097-0142(19980915)83:6<1102::aid-cncr8>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 84.Peifer M, Fernandez-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–1116. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sos ML, Dietlein F, Peifer M, et al. A framework for identification of actionable cancer genome dependencies in small cell lung cancer. Proc Natl Acad Sci U S A. 2012;109:17034–17039. doi: 10.1073/pnas.1207310109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boldrini L, Ursino S, Gisfredi S, et al. Expression and mutational status of c-kit in small-cell lung cancer: prognostic relevance. Clin Cancer Res. 2004;10:4101–4108. doi: 10.1158/1078-0432.CCR-03-0664. [DOI] [PubMed] [Google Scholar]
- 88.LaPoint RJ, Bourne PA, Wang HL, Xu H. Coexpression of c-kit and bcl-2 in small cell carcinoma and large cell neuroendocrine carcinoma of the lung. Appl Immunohistochem Mol Morphol. 2007;15:401–406. doi: 10.1097/01.pai.0000213153.41440.7d. [DOI] [PubMed] [Google Scholar]
- 89.Lopez-Martin A, Ballestin C, Garcia-Carbonero R, et al. Prognostic value of KIT expression in small cell lung cancer. Lung Cancer. 2007;56:405–413. doi: 10.1016/j.lungcan.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 90.Naeem M, Dahiya M, Clark JI, et al. Analysis of c-kit protein expression in small-cell lung carcinoma and its implication for prognosis. Hum Pathol. 2002;33:1182–1187. doi: 10.1053/hupa.2002.129199. [DOI] [PubMed] [Google Scholar]
- 91.Rossi G, Cavazza A, Marchioni A, et al. Kit expression in small cell carcinomas of the lung: effects of chemotherapy. Mod Pathol. 2003;16:1041–1047. doi: 10.1097/01.MP.0000089780.30006.DE. [DOI] [PubMed] [Google Scholar]
- 92.Sihto H, Sarlomo-Rikala M, Tynninen O, et al. KIT and platelet-derived growth factor receptor alpha tyrosine kinase gene mutations and KIT amplifications in human solid tumors. J Clin Oncol. 2005;23:49–57. doi: 10.1200/JCO.2005.02.093. [DOI] [PubMed] [Google Scholar]
- 93.Dy GK, Miller AA, Mandrekar SJ, et al. A phase II trial of imatinib (ST1571) in patients with c-kit expressing relapsed small-cell lung cancer: a CALGB and NCCTG study. Ann Oncol. 2005;16:1811–1816. doi: 10.1093/annonc/mdi365. [DOI] [PubMed] [Google Scholar]
- 94.Krug LM, Crapanzano JP, Azzoli CG, et al. Imatinib mesylate lacks activity in small cell lung carcinoma expressing c-kit protein: a phase II clinical trial. Cancer. 2005;103:2128–2131. doi: 10.1002/cncr.21000. [DOI] [PubMed] [Google Scholar]
- 95.Schneider BJ, Kalemkerian GP, Ramnath N, et al. Phase II trial of imatinib maintenance therapy after irinotecan and cisplatin in patients with c-Kit-positive, extensive-stage small-cell lung cancer. Clin Lung Cancer. 2010;11:223–227. doi: 10.3816/CLC.2010.n.028. [DOI] [PubMed] [Google Scholar]
- 96. [Accessed August 8];SEER Stat Fact Sheets: Kidney and Renal Pelvis Cancer. Available at: http://seer.cancer.gov/statfacts/html/kidrp.html.
- 97.Di Napoli A, Signoretti S. Tissue biomarkers in renal cell carcinoma: issues and solutions. Cancer. 2009;115:2290–2297. doi: 10.1002/cncr.24233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Choueiri TK. Clinical treatment decisions for advanced renal cell cancer. J Natl Compr Canc Netw. 2013;11:694–697. doi: 10.6004/jnccn.2013.0204. [DOI] [PubMed] [Google Scholar]
- 99.Hudes GR, Carducci MA, Choueiri TK, et al. NCCN Task Force report: optimizing treatment of advanced renal cell carcinoma with molecular targeted therapy. J Natl Compr Canc Netw. 2011;9(Suppl 1):S1–29. doi: 10.6004/jnccn.2011.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Topalian SL, Drake CG, Pardoll DM. Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor immunity. Curr Opin Immunol. 2012;24:207–212. doi: 10.1016/j.coi.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thompson RH, Dong H, Kwon ED. Implications of B7-H1 expression in clear cell carcinoma of the kidney for prognostication and therapy. Clin Cancer Res. 2007;13:709s–715s. doi: 10.1158/1078-0432.CCR-06-1868. [DOI] [PubMed] [Google Scholar]
- 102.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cho DC, Sosman JA, Sznol M, et al. Clinical activity, safety, and biomarkers of MPDL3280A, an engineered PD-L1 antibody in patients with metastatic renal cell carcinoma (mRCC) ASCO Meeting Abstracts. 2013;31:4505. [Google Scholar]
- 104.Choueiri TK, Fishman MN, Escudier BJ, et al. Immunomodulatory activity of nivolumab in previously treated and untreated metastatic renal cell carcinoma (mRCC): Biomarker-based results from a randomized clinical trial. ASCO Meeting Abstracts. 2014;32:5012. [Google Scholar]
- 105.Armstrong DK, Bundy B, Wenzel L, et al. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N Engl J Med. 2006;354:34–43. doi: 10.1056/NEJMoa052985. [DOI] [PubMed] [Google Scholar]
- 106.McCluggage WG. Morphological subtypes of ovarian carcinoma: a review with emphasis on new developments and pathogenesis. Pathology. 2011;43:420–432. doi: 10.1097/PAT.0b013e328348a6e7. [DOI] [PubMed] [Google Scholar]
- 107.Bookman MA. Molecular wanderings through the DNA damage response and risk for ovarian cancer. J Natl Cancer Inst. 2014;106:djt350. doi: 10.1093/jnci/djt350. [DOI] [PubMed] [Google Scholar]
- 108.Bast RC, Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9:415–428. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bast RC., Jr CA 125 and the detection of recurrent ovarian cancer: a reasonably accurate biomarker for a difficult disease. Cancer. 2010;116:2850–2853. doi: 10.1002/cncr.25203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bast RC, Jr, Feeney M, Lazarus H, et al. Reactivity of a monoclonal antibody with human ovarian carcinoma. J Clin Invest. 1981;68:1331–1337. doi: 10.1172/JCI110380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kaneko O, Gong L, Zhang J, et al. A binding domain on mesothelin for CA125/MUC16. J Biol Chem. 2009;284:3739–3749. doi: 10.1074/jbc.M806776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rustin GJ, van der Burg ME, Griffin CL, et al. Early versus delayed treatment of relapsed ovarian cancer (MRC OV05/EORTC 55955): a randomised trial. Lancet. 2010;376:1155–1163. doi: 10.1016/S0140-6736(10)61268-8. [DOI] [PubMed] [Google Scholar]
- 113.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 114.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goldhirsch A, Winer EP, Coates AS, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nielsen T, Wallden B, Schaper C, et al. Analytical validation of the PAM50-based Prosigna Breast Cancer Prognostic Gene Signature Assay and nCounter Analysis System using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer. 2014;14:177. doi: 10.1186/1471-2407-14-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Martin M, Brase JC, Calvo L, et al. Clinical validation of the EndoPredict test in node-positive, chemotherapy-treated ER+/HER2− breast cancer patients: results from the GEICAM 9906 trial. Breast Cancer Res. 2014;16:R38. doi: 10.1186/bcr3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Connolly RM, Stearns V. Current approaches for neoadjuvant chemotherapy in breast cancer. Eur J Pharmacol. 2013;717:58–66. doi: 10.1016/j.ejphar.2013.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014 doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
- 120.Goncalves R, Ma C, Luo J, et al. Use of neoadjuvant data to design adjuvant endocrine therapy trials for breast cancer. Nat Rev Clin Oncol. 2012;9:223–229. doi: 10.1038/nrclinonc.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Paik S, Kim C, Wolmark N. HER2 status and benefit from adjuvant trastuzumab in breast cancer. N Engl J Med. 2008;358:1409–1411. doi: 10.1056/NEJMc0801440. [DOI] [PubMed] [Google Scholar]
- 122.Carey LA, Dees EC, Sawyer L, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 123.Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. [Accessed August 8];Limited Competition: Biospecimen Banks to Support NCI-Clinical Trials Network (NCTN) (U24) See more at: http://grants.nih.gov/grants/guide/rfa-files/RFA-CA-14-501.html#_Part_2._Full. Available at: http://grants.nih.gov/grants/guide/rfa-files/RFA-CA-14-501.html#_Part_2._Full.
- 125. [Accessed August 6];NCI to Provide $12M for Biobanks to Support Clinical Trials Network. Available at: http://www.genomeweb.com/nci-provide-12m-biobanks-support-clinical-trials-network.