Abstract
Neocortical excitatory and inhibitory neurons derive from distinct progenitor domains during embryonic development, and migrate to their final positions, where they assemble into functional circuits. This process appears to be influenced by lineage relationships among locally born excitatory neurons, raising the intriguing possibility that this might be true for cortical interneurons. Two recent articles by the Fishell laboratory and our own used retrovirus-encoded DNA barcodes as unambiguous lineage tracing tools to address this question, finding that clonally related inhibitory interneurons dispersed widely across the forebrain (Harwell et al., 2015; Mayer et al., 2015). In the accompanying report, Sultan et al. reanalyze the datasets from both studies and propose a new interpretation, whereby clonally related interneurons would be considered clustered according to specific spatial constraints. After studying the report from Sultan et al. and carefully revisiting previously published studies, we find no evidence of lineage-dependent MGE/PoA-derived interneuron clustering in the forebrain.
eTOC blurb
Turrero García et al. revise previous studies in response to Sultan et al., confirming that present experimental evidence supports the idea that clonally related MGE/PoA-derived interneurons do not form spatially isolated clusters, but rather disperse within and across forebrain regions.
Introduction
The way that neurons in our brain are assembled into functional circuits remains largely unknown. During embryonic development, the different neuronal subtypes that will form cortical circuits are born from spatially distinct areas. Progenitors that give rise to excitatory neurons divide locally within the prospective neocortex. Most inhibitory neurons, or interneurons, destined for the cortex are born from progenitors located in ventral structures of the embryonic brain known as the medial ganglionic eminence (MGE) and preoptic area (PoA), and must then migrate great distances to integrate into developing circuits (Bartolini et al., 2013; Kepecs and Fishell, 2014; Marín and Müller, 2014; Wonders and Anderson, 2006). In recent years, there has been tremendous interest in the role that clonal lineage may play in these processes. One of the first examples of this came from the work of the Shi laboratory examining the role of lineage in preferential synapse relationships between lineage-related cells in ontogenic columns of cortical excitatory projection neurons (Yu et al., 2009, 2012). More recently several groups have been interested in whether clonal lineage relationships are also responsible for regulating the organization and assembly of inhibitory circuitry in the cerebral cortex (Brown et al., 2011; Ciceri et al., 2013; Harwell et al., 2015; Mayer et al., 2015; Sultan et al., 2014). Since inhibitory interneurons migrate great distances tangentially from their birthplace in order to reach their final position in the cortex, keeping track of clonal lineage relationships among these neurons has been a challenge.
Two recent studies attempted to track the organization of lineage-related interneurons by labeling progenitors in the MGE/PoA with either one or two fluorescent protein markers, encoded by retroviruses that were used to infect embryonic brains “at clonal density” (Brown et al., 2011; Ciceri et al., 2013). Using the spatial relationships among labeled cells in the cortex to assign clonal relationships (i.e., considering that cells located closely in space were derived from the same MGE/PoA progenitors), Brown et al. concluded that presumptive interneuron clones were vertically and horizontally aligned in a manner reminiscent of excitatory projection neurons. This raised the intriguing possibility that lineage relationships could guide either the formation of connections between clonally related interneurons or their integration into functionally related circuits. Ciceri et al. used an unsupervised, unbiased method to detect potential clusters of fluorescent protein-labeled interneurons. The vast majority of the clusters detected in the mature brain were composed of cells located either within a single neocortical layer (46.85 ± 4.06 %) or in two adjacent ones (39.45 ± 2.14 %), rather than vertically aligned. When MGE/PoA progenitors were labeled using a combination of two retroviruses encoding different fluorescent proteins, most clusters (66.83 ± 1.4 %) contained a mixture of cells expressing both fluorophores. From this, the authors concluded that clonal lineage could not be the sole responsible factor driving interneuron clustering: “Although the mechanisms underlying the clustering of interneurons in the cortex remain unclear, our experiments suggest that this process is not univocally linked to their shared clonal origin” (Ciceri et al., 2013). These findings have important implications for understanding the rules that guide the assembly and function of brain circuits; however, new technical approaches were necessary to investigate the lineage relationships among cortical interneurons. Our group, concurrently with the Fishell laboratory, recently addressed this issue (Harwell et al., 2015; Mayer et al., 2015), taking advantage of a replication-incompetent Moloney Murine Leukemia Virus (M-MLV)-based retrovirus library, consisting of 105 unique 24-bp sequences or barcodes. This tool allowed us to track unambiguous clonal relationships between MGE-derived interneurons in the mature cortex. Both studies found that neurons belonging to the same clonal lineage were widely dispersed throughout the cortex and other forebrain structures.
The conclusions of previous studies were based on the a priori assumption that the non-random (i.e., clustered) distribution of virally labeled cortical interneurons was due to the clonal relationships between them (Brown et al., 2011). This was impossible to assess without appropriate tools to definitively determine cell lineages. Indeed, when such tools were applied, unequivocal interneuron clones were not clustered, but rather widely dispersed, both within and across different brain structures (Harwell et al., 2015; Mayer et al., 2015). In the current issue of Neuron, Sultan et al. conduct an extensive reanalysis of our data and those of Mayer et al. They suggest that those studies could be interpreted as support for a model such as the one proposed by Brown et al., whereby cortical interneurons would be functionally organized into spatially isolated local clusters according to their lineage. The authors attempt to find clustered sister cells within the datasets provided by the Fishell laboratory and our own, and are only able to do so at distances that are far too large for any functional connections to be established between clonally related interneurons. We believe that the Sultan et al. report has largely arisen from misunderstanding and/or misinterpreting our data and conclusions, and aim to address here some of the issues the authors bring up. The accompanying reply by Mayer et al. highlights and further clarifies other points worth discussing.
Definition of important terms
We believe that throughout the accompanying report from the Shi laboratory there are a series of terms that are not clearly and unequivocally defined. We believe that this might lead to ambiguous interpretations and confusion. For the sake of clarity, we would like to point them out below, along with our considerations about their use.
Random / wide / non-specific distribution of interneurons. Sultan et al. use these terms indistinctly, despite their different meanings; neither our previous report (Harwell et al., 2015) nor the one from the Fishell laboratory (Mayer et al., 2015) claim that MGE/PoA derived interneurons are randomly allocated to different brain structures, but rather claim that they are widely distributed across them, meaning that clonally related interneurons can be found in anatomically and functionally distinct areas. Likewise, the term non-specific was never used to describe interneuron dispersion in any of the reports discussed here, and we believe it to be too imprecise to be used in this context.
Clonal density / clonal labeling / clonally related / clonal clusters. Infection of MGE/PoA progenitors with low-titer viruses encoding fluorescent reporters allows the study of ontogenetic radial units during embryonic development (Brown et al., 2011; Ciceri et al., 2013; Harwell et al., 2015; Mayer et al., 2015). However, this labeling at clonal density is not equivalent to clonal labeling, since it is impossible to unequivocally identify interneurons generated from the same ontogenetic radial unit (i.e., clonally related) after their migration into the neocortex, based solely on their expression of a reporter protein. It is thus incorrect to make the conceptual leap from observing GFP-positive interneurons in spatial proximity to assuming that they are clonal clusters.
Considerations about the use of barcoded retrovirus
Despite the fact that the technical advantages and caveats of using a DNA-barcoded retroviral library were already discussed in our original report, as well as in that from the Fishell laboratory (Harwell et al., 2015; Mayer et al., 2015), Sultan et al. bring up some technical concerns that we feel should be discussed here. The first such concern is the role that viral genome silencing could play in the analysis of barcode-labeled clones, since it might lead to an underestimation of the total size of the clones. This is a valid point, which is by no means exclusive to M-MLV barcode libraries but common to all retroviruses, including those used in Brown et al., 2011, and Ciceri et al., 2013 (Katz et al., 2007), and it was addressed in the original reports (Harwell et al., 2015; Mayer et al., 2015). Barcode recovery from GFP-negative tissue confirmed that clonally related cells, irrespective of silencing, were widely dispersed throughout the brain (Mayer et al., 2015).
With regards to the potential overrepresentation of certain barcodes in the retroviral library we used, we would like to point out that the only barcode that was detected more than once across the infected brains in our original study, which could thus have been deemed as overrepresented in the library, was excluded from our analyses (note the absence of “Clone 6” in Fig. 4D of Harwell et al., 2015). The claim from Sultan et al. that “~6% (1 out of 16; the barcode of clone 6) of recovered barcodes were found in more than one brain” is clearly misconstrued: while it does represent that percentage of the number of clones that were mapped and further analyzed within one brain, it only represents ~1.8 % of all recovered barcodes within said brain (n = 136 barcoded cells, including 16 clones comprising a total of 41 cells [Harwell et al., 2015]).
We cannot agree with Sultan et al. when they state “it is necessary to exclude the single barcoded cells with no real siblings from quantitative clonal analysis”. Single barcoded cells are irrelevant to the conclusions we obtained from cells with shared barcodes (namely, that they can be found spread through and across brain structures and that they do not form spatially isolated clusters). Using the spatial information of any virally labeled cells, irrespective of their barcode or clonal lineage, provides valuable information for performing spatial analyses, as is clear from the work of other groups (Brown et al., 2011; Ciceri et al., 2013), much of which would be rendered invalid if only confirmed clonal relationships could be used to perform distance and clustering analyses, as Sultan et al. suggest. We do not believe this to be the case, and stand by our use of virally labeled cells, with or without barcode recovery, as the best proxy to analyze MGE/PoA-derived interneuron distribution within and across different forebrain areas in the context of the experimental datasets discussed here. As Sultan et al. themselves put it, elsewhere in their report, “these single barcoded, non-clonally related cells were born at a similar time as clonally related cells in the datasets, thereby serving as a good experimental control”.
All in all, we consider the use of a barcode-containing viral library as a significant technical advance over previous tools, since it can unequivocally assign clonal relationships among infected cells, regardless of the caveats that the Fishell laboratory and us already discussed in the original reports (Harwell et al., 2015; Mayer et al., 2015).
Non-random distribution of interneurons
Sultan et al. state that clonally related interneurons do not randomly disperse in the forebrain. We completely agree with this. We think it is important to highlight that nowhere in our previous study do we claim that the production or organization of forebrain interneurons are random events. We concluded that interneurons derived from the same clonal lineages were widely dispersed, occupying distant positions throughout the cortex, and could even be distributed through different forebrain structures (Harwell et al., 2015; Mayer et al., 2015), but this does in no way mean that the allocation of interneurons to those structures would be randomized, as implied in the analyses of Sultan et al.
Sultan et al. use the idea of the ‘random walk’ behavior of some migrating interneurons described by other authors (Ang et al., 2003; Tanaka et al. 2009) to suggest that cortical interneurons might migrate towards their final positions through “random diffusion”, making this a focal point of their discussion. This idea was also the basis for the null hypothesis in Brown et al., 2011. However, the reports cited by them refer only to the behavior of a subset of cortical interneurons (those entering the cortex through the marginal zone migratory route) during their migration, and not to their final allocation within the brain, which is far from randomized (Marín, 2013; Guo and Anton, 2014).
MGE/PoA-derived interneurons are allocated to brain structures in different proportions
One of the two main questions posed by Sultan et al. is “(1) Do a majority of interneuron clones disperse across different anatomic divisions or brain structures?” Both the Mayer et al. and Harwell et al. articles already answered this question, reporting that the majority of multi-cell clones were located in a single brain structure (71 % and 57.9 %, respectively; n = 3 brains [Mayer et al., 2015; Harwell et al., 2015 – note that Figure 3 in the Sultan et al. report is based on a single brain from the Harwell et al. dataset, not on the actual total numbers reported in that study, which were collected from a total of 3 brains]). However, Sultan et al. perform some analyses that we believe merit additional consideration.
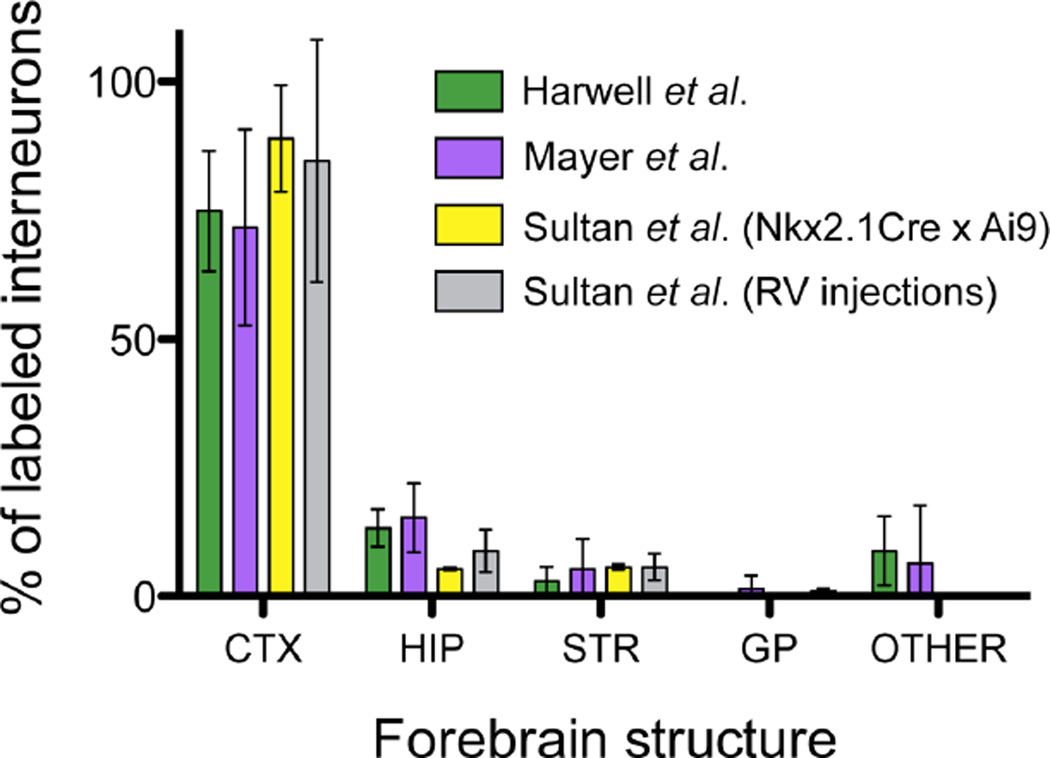
Sultan et al. use two different labeling methods to examine the proportion of MGE/PoA-derived interneurons that are allocated to different forebrain structures, and determine that the vast majority of these neurons (85–89 %) are located in the cortex, with smaller proportions in hippocampus (5–9 %), striatum (5–6 %) and globus pallidus (1.6 %). They then go on to perform a series of mathematical simulations based on the combination of these proportions with the location of the 32 multi-cell clones detected by Mayer et al., finding statistically significant differences when comparing the “expected” allocation of their simulated multi-cell clones with the experimental ones reported by Mayer et al. We believe that these simulations, which are not really randomized (since they are in fact constrained by different sets of experimental data), do not add any biologically relevant information to this discussion, but rather serve as an artificial source of statistical significance. This is particularly clear in Fig. S3, where the authors find statistically significant differences between the interneuron distribution data obtained by Mayer et al. and a simulation of itself. We believe that the proportions obtained by Sultan et al. are much more useful when taken at face value. We find similar proportions of retrovirus-labeled interneurons in each brain structure in our own dataset, considering all labeled cells irrespective of barcode recovery (74.9 ± 11.7 % cortex, 13.2 ± 3.6 % hippocampus, 2.9 ± 2.8 % striatum, 0.2 ± 0.3 % globus pallidus; n = 3 brains). The results from Mayer et al. follow the same trend (71.7 ± 19.0 % cortex, 15.1 ± 6.7 % hippocampus, 5.3 ± 5.8 % striatum, 1.4 ± 2.5 % globus pallidus; n = 3 brains). Neither study obtained interneuron allocation proportions significantly different from those reported by Sultan et al. (Figure 1). We believe that the mathematical simulations performed by Sultan et al. are an overly complicated way to test a hypothesis (i.e., that multi-cell interneuron clones are randomly distributed throughout the brain) that was never in dispute.
FIG. 1. The allocation of interneurons in the forebrain is not random.
Graph displaying the proportions (in %) of MGE/PoA-derived interneurons across different brain structures (CTX: cortex; HIP: hippocampus; STR: striatum; GP: globus pallidus), as obtained from four different experimental datasets: Harwell et al., 2015 (green, n = 3 brains, 504 cells; note: an additional brain from the same experimental dataset as those described in Harwell et al., 2015 was analyzed for this figure); Mayer et al., 2015 (purple, n = 3 brains, 84 cells across 32 multi-cell clones); and the two different approaches followed by Sultan et al. (yellow, n = 3 brains; and gray, n = 7 brains). Interneurons were labeled either by intraventricular injection of a library of replication-incompetent Moloney Murine Leukemia retrovirus encoding DNA barcode and GFP in E12 or E10 mouse embryos (Harwell et al. and Mayer et al., respectively), by analysis of a fluorescent reporter in MGE/PoA-derived cells (Nkx2.1CrexAi9 mice; Sultan et al., yellow bars), or injection of GFP-encoding retrovirus in E12 mouse embryos (Sultan et al., gray bars). Data are presented as average ± S.D.; no significant differences were found between studies for any of the brain regions (one-way ANOVA).
Defining Neuronal Clusters
As correctly pointed out by Sultan et al., in our previous article we found that MGE/PoA-derived cortical interneurons (although not “clonally labeled”, since clonal identities were not considered for this particular analysis) were significantly closer than expected by randomized simulation, “suggesting that the overall population distribution is not random, consistent with previous studies” (Harwell et al., 2015). We believe that there is enough scientific evidence to consider that these interneurons are distributed in clusters throughout the neocortex (Brown et al., 2011; Ciceri et al., 2013; Harwell et al., 2015; Mayer et al., 2015). However, we fully stand by our conclusion that any local clustering of interneurons in the brain appears to occur largely independent of clonal lineage relationships. Since the exact definition of interneuron clusters (or lack thereof) seems to be the primary source of the reinterpretation of our data proposed by Sultan et al., we will try to address this issue. Clusters should be defined as groups of cells positioned closely together in space, with all other considerations (such as relative distances, cell density within any given dataset, etc.) being secondary to this simple concept.
In their report, Sultan et al. claim that distance measurements are not an adequate resource to analyze clustering of interneurons (“Mayer et al. analyzed the clustering solely based on distance measurement, which can be inaccurate for the following reasons. (…)”). This statement is surprising not only given previous studies by the same group, where conclusions were based exclusively on distance measurements (Brown et al., 2011), but also in light of the spatial analyses performed throughout the report from Sultan et al. in order to dispute the conclusions of Mayer et al. and our own. These analyses invariably rely on the Euclidean distances between data points, either in absolute terms or indirectly by analyzing the lowest hierarchical branches within dendrograms, i.e., the pairs of cells located closest to each other (see more detailed discussion about dendrogram analysis in the accompanying reply by Mayer et al.). It is hard for us to understand how spatial clustering, which is a measure of relative distance between points in space, can be detached from this type of measurements. According to Sultan et al., “clustering in spatial distribution is relative to a wide and non-specific dispersion, but does not necessarily correspond to the absolute shortest distance between the data points per se”. In the context discussed here, we fail to imagine any biologically relevant scenarios in which clusters should not be defined by the closest distances between cells, except for cases where such a “cluster” would encompass different anatomical structures.
Spatial dimensions of interneuron clusters
In their report, Sultan et al. choose to define interneuron “local clusters” as the lowest hierarchical branches in a dendrogram that deliberately excludes all labeled cells that do not belong to multi-cell clones, rather than performing unbiased, unsupervised clustering analysis based on proximity relationships among all labeled cells (Ciceri et al., 2013). This heavily skews their results, eliminating the requirement for groups of cells to be spatially isolated in order to be considered as clusters (Brown et al., 2011) and making their subsequent analyses difficult to interpret within any biologically meaningful context. Within these already very limited datasets, Sultan et al. then calculate the distance between cells belonging to the same multi-cell clone, regardless of their location within the brain, and compare it to the distance between non-related cells, obtaining average values consistently greater than 1500 µm (greater than 2000 µm in the case of the Harwell et al. dataset). With these analyses, sister cells are considered “locally clustered” despite being further than 1 mm apart, and regardless of the possible presence of other, non-related cells in their spatial proximity. It is worth noting that a previous study from the same group considered that sparsely labeled interneurons “did not randomly disperse but formed spatially isolated clusters that were often more than 500 µm apart” (Brown et al., 2011). If 500 µm are deemed as a sufficient distance to consider two clusters of interneurons “spatially isolated”, it stands to reason that any pair of interneurons located further than 1500 µm apart should not be considered to form a local cluster.
Use of nearest neighbor distance as a measure of clustering
Sultan et al. dispute the validity of our conclusions based on nearest neighbor distances (NND; Harwell et al., 2015, Figure 4F), since such measurements depend on the number of data points. This is indeed true, since NNDs that are considered ‘close’ with few data points could be considered ‘far away’ with more points (i.e., the overall distances between cells are scaled down by the number of data points). However, the simulations of complete spatial randomness that were used as our statistical standard for judging ‘clustering’, ‘randomness’ or ‘uniformity’ take into account the total number of data points (cell density) of any given brain. Additionally, Sultan et al. refer to the distance plotted in Harwell et al. Fig. 4E as that of “clonally labeled interneurons”, while this graph does not distinguish between clonally related or unrelated interneurons, and rather points out the fact that all interneurons seem to be located closer together than expected by random distribution. Mayer et al., in this same issue, clarify this point further by providing additional considerations, as well as a detailed, density-independent analysis of the average intra- and inter-clonal distances of their dataset.
The dimensions of excitatory neuron clusters are insufficient to encompass clonally related interneurons
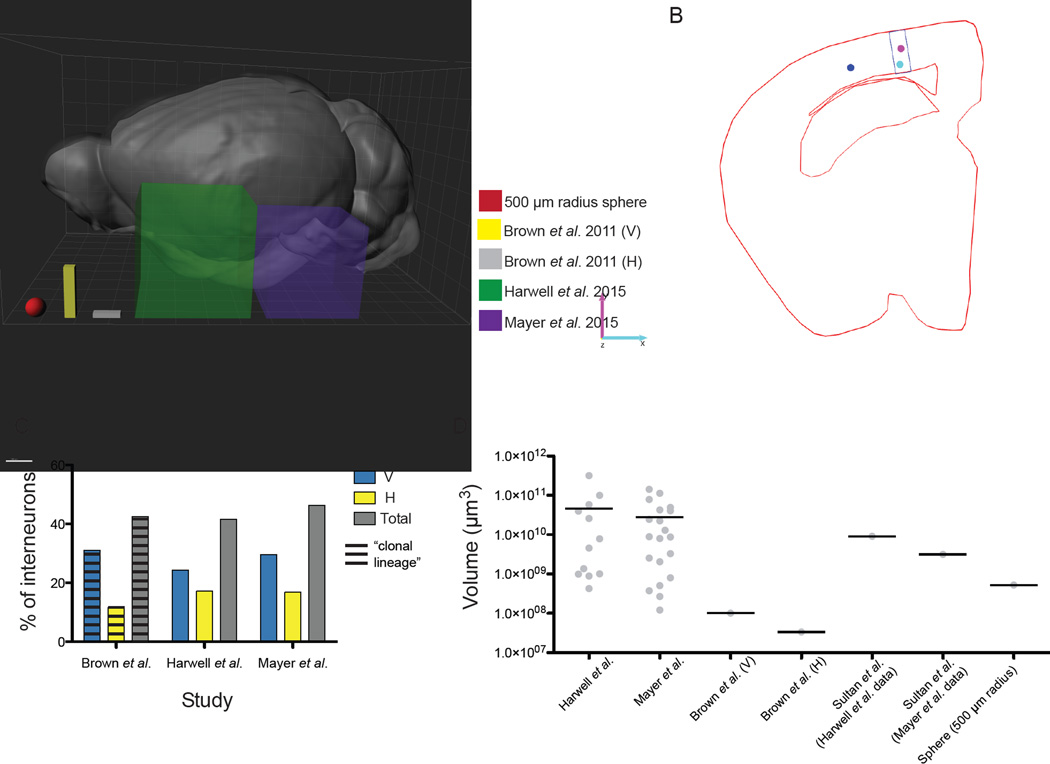
The definition of a cluster of cells by Sultan et al. is based on specific spatial constraints, which seem to vary drastically with respect to the data analysis performed in Brown et al., where the authors concluded that ‘Individual clonal clusters consisting of interneurons expressing the same or distinct neurochemical markers exhibited clear vertical or horizontal organization’. In that study, the authors detected interneuron clusters using the spatial dispersion parameters they calculated from vertical and horizontal clusters of excitatory neurons in the neocortex. Although the exact algorithm they applied is unclear, they used these values to define the dimensions of two different cuboidal matrices to detect vertical and horizontal clusters (300 × 1400–1600 × 210 µm for vertical clusters, and 800 × 200 × 210 µm for horizontal ones). When the cortices within their dataset were “scanned”, any group of interneurons fitting into either matrix was considered a cluster of the corresponding type (Figure 2A, yellow and gray cuboids). We decided to use the same approach with our own dataset and that of Mayer et al., where lineage relationships could be unequivocally assigned. We first tested the ability of cuboidal matrices of the dimensions and orientations defined by Brown et al. (in fact, since the thickness of the brain sections in Harwell et al. was 25 µm, the cuboidal matrices we defined were slightly bigger than those of Brown et al., 2011 [225 µm instead of 210 in the z-dimension]) to detect clusters in our dataset, as well as that of Mayer et al. Given that the “very low density” at which Brown et al. labeled the brains in their dataset yielded greatly variable numbers of cells (average ± S.D. of 199 ± 150 interneurons per brain, n = 13 P21–30 brains; data kindly provided by S.-H. Shi), we assumed that those matrices would be robust enough to be applied to the cell densities obtained in our own dataset and that of Mayer et al. We were unable to detect any pairs of clonally related interneurons that could be comprised within any cuboids of the dimensions and orientations proposed by Brown et al., in either of the datasets (Figure 2C). When we were able to find several cells of known clonal lineage that would fit within a matrix of those dimensions (and thus could be considered clustered according to the criteria proposed by Brown et al.), those interneurons were invariably unrelated (Figure 2B). Additionally, when we performed these analyses only on multi-cell clones, irrespective of the clonal relationships among clustered cells, we observed proportions of clustered interneurons similar to those reported by Brown et al. (Figure 2C), further reinforcing the possible existence of clonal lineage-independent clustering of cortical interneurons. Sultan et al. did not to perform these analyses on our dataset or that of Mayer et al., choosing instead to show a graph (Fig. S8) in which the maximum spatial dimensions of the cuboids described in Brown et al. are plotted against the distances between pairs of clonally related cells in said datasets. We believe that this is not a correct reanalysis, since it eliminates the geometrical considerations (i.e., the orientation of cells into distinct vertical and horizontal clusters of virally labeled interneurons) that were central to the conclusions of Brown et al. (Brown et al., 2011).
FIG. 2. Cortical interneurons are not clustered according to their lineage.
(A) Side view of a 3-D rendering of a P28 mouse brain (created from data from the Allen Brain Institute), with cuboids of the dimensions used to detect interneuron vertical (V, yellow) and horizontal (H, gray) clusters (i.e., “clones”) in Brown et al. 2011, compared to cuboids of the average dimensions necessary to detect pairs of clonally related interneurons in the Harwell et al., 2015 (green) and Mayer et al., 2015 (purple) datasets. A sphere with a radius of 500 µm (red), as a distance relevant to the size of local circuits, is shown for comparison (see text for details). Grid lines: 1 mm. (B) Representative Neurolucida tracing (red) from the Harwell et al. 2015 dataset, showing a group of interneurons (colored dots) fitting a “vertical cluster” cuboidal matrix as proposed by Brown et al. (black); the clonal identity of each cell within the cluster is indicated by their color. Note that each neuron in this plot belongs to a different clone. (C) Graph depicting the proportion of interneurons grouped into either vertical (V, blue) or horizontal (H, yellow) clusters, as well as the total proportion of clustered interneurons (gray), in the indicated studies. Clusters were detected as in Brown et al., 2011. Horizontal lines depict the proportion of cells assumed to belong to the same clone (Brown et al.); note that no such relationship could be detected experimentally (Harwell et al., Mayer et al.). (D) Plot of the volume of minimum-dimension cuboids necessary to encompass unequivocally clonally related interneurons in the datasets of Harwell et al., 2015, Mayer et al., 2015, and Brown et al., 2011 (vertical “(V)” and horizontal “(H)”). For comparison, the volume of a 500 µm-radius sphere (relevant to the size of local circuits) and the volumes of spheres with diameters equivalent to the average intra-clonal distances calculated by Sultan et al. for the Harwell et al. and Mayer et al. datasets are also plotted (see text for details). All data points are plotted where possible; horizontal lines mark average values (n = 1 brain, 16 clones [Harwell et al.]; 3 brains, 20 clones [Mayer et al.]). Notice the logarithmic scale of the y-axis.
We then took the reverse approach, taking into consideration the clonal identities we had obtained in order to calculate the minimum dimensions of cuboidal matrices that could comprise pairs of cells belonging to unequivocal cortical clones from our dataset and that of Mayer et al (Figure 2A). We found that any such matrix would be substantially larger than those used for detection of radial and horizontal clones in Brown et al. (Figure 2A, D). We plotted the minimum volumes necessary to encompass the average intra-clonal distances calculated by Sultan et al. for our dataset and that of Mayer et al., and found them to be at least one order of magnitude greater than the volumes used to detect clones in Brown et al. (Figure 2D).
Meaning of clustering: functional unit size
We would like to consider now the possible biological meaning of interneuron clustering, even when defined based on the spatial measurements reported by Sultan et al. As outlined above, the average distances between clonally related neurons in both the Mayer et al. and our datasets calculated by Sultan et al. are consistently greater than 1500 µm. To put that distance in context, cortical MGE/PoA-derived interneurons barely display any connectivity to pyramidal cells beyond 500 µm of intersomatic distance (Packer et al., 2013) (Figure 2A,D). The intra-clonal distances calculated by Sultan et al. are also beyond the boundaries of functional units based on the connectivity of cortical pyramidal cells, as noted by Mayer et al.: “This analysis shows that sibling interneurons reside in a volume that exceeds functional cortical units, such as whisker barrels of the somatosensory cortex (400 µm (…))” (Mayer et al., 2015). It is thus hard to reconcile these distances with the notion of local circuits or any other function-based spatial units that could potentially drive the clustering of clonally related interneurons, as proposed by Brown et al.: “the predictable spatial organization of clonally related sister inhibitory interneurons raises the possibility of a lineage-dependent functional organization of inhibitory interneurons in the mammalian neocortex” (Brown et al., 2011). In line with these considerations, the unsupervised agglomerative hierarchical clustering analysis performed by Ciceri et al. was able to detect clones with an average threshold value (i.e., maximum distance at which cells can be considered to form part of a cluster) of 389 ± 18 µm, which notably falls within the size range of functional units mentioned above.
Conclusions
While we do appreciate the thorough efforts of Sultan et al. towards reanalyzing our data, as well as the useful corrections they make regarding our unfortunate alignment mistakes, we believe that their adjustments do not fundamentally alter the conclusions we reached in Harwell et al. 2015. In summary:
Barcode-containing retroviruses represent a major technical advance with respect to previous studies and are at present, to the best of our knowledge, the only available tool that permits the study of unequivocal clonal relationships within the experimental framework discussed here.
We completely agree that allocation of MGE/PoA-derived interneurons to different structures within the forebrain is not random, which was never a point of contention in our original study.
We cannot agree that clonally related cells located farther than 1.5 mm apart, often across anatomical boundaries, can be considered to form a local cluster.
We remain convinced that the results presented by our group and the Fishell laboratory (Harwell et al., 2015; Mayer et al., 2015) provide compelling evidence for the dispersion of clonally related MGE/PoA-derived interneurons, both within and across different forebrain structures. The relationship between cell lineage and circuit assembly remains a potentially fundamental aspect of brain development that needs further investigation. Unambiguous lineage markers, as well as detailed and careful analyses of experimental data obtained through them, will be the key to address this extremely interesting topic. We hope we have properly dispelled any concerns that the report from Sultan et al. might have raised, and instead of discussing the finer points of their report, we invite them and any other scientists interested in this topic to experimentally confirm or dispute our previous study, utilizing the freely available tools described therein. In fact, we believe that our conclusions are further reinforced after the thorough revisiting of the datasets initiated by Sultan et al. and addressed here both by Mayer et al. and us, and would thus like to express our gratitude to all authors involved for this chance to clarify and confirm our conclusions.
Highlights.
Barcoded retrovirus libraries allow unequivocal assignment of lineage relationships
Interneurons are not randomly distributed throughout different forebrain regions
Interneuron clones disperse widely within and across forebrain structures
Clonal lineage does not determine spatially isolated local interneuron clustering
Acknowledgments
We are grateful to Hunter Elliott at the Harvard Medical School Image and Data Analysis Core for help with data visualization, as well as to the Enhanced Neuroimaging Core at the Harvard Neurodiscovery Center for access to their equipment. We would like to thank A. R. Kriegstein (UCSF), in whose lab the Harwell et al. 2015 study was initiated, and S. R. Datta (HMS) for comments on this manuscript. M.T.G. is partially supported by The Ellen R. and Melvin J. Gordon Center for the Cure and Treatment of Paralysis. Research in the Harwell laboratory is supported by the NIH (K01NS089720).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
All authors analyzed data. M.T.G. and C.C.H. wrote the paper with input from E.M.
References
- Ang ES, Jr, Haydar TF, Gluncic V, Rakic P. Four-dimensional migratory coordinates of GABAergic interneurons in the developing mouse cortex. J Neurosci. 2003;23(13):5805–5815. doi: 10.1523/JNEUROSCI.23-13-05805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini G, Ciceri G, Marín O. Integration of GABAergic interneurons into cortical cell assemblies: lessons from embryos and adults. Neuron. 2013;79(5):849–864. doi: 10.1016/j.neuron.2013.08.014. [DOI] [PubMed] [Google Scholar]
- Brown KN, Chen S, Han Z, Lu CH, Tan X, Zhang XJ, Ding L, Lopez-Cruz A, Saur D, Anderson SA, Huang K, Shi SH. Clonal production and organization of inhibitory interneurons in the neocortex. Science. 2011;334(6055):480–486. doi: 10.1126/science.1208884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciceri G, Dehorter N, Sols I, Huang ZJ, Maravall M, Marín O. Lineage-specific laminar organization of cortical GABAergic interneurons. Nat Neurosci. 2013;16(9):1199–1210. doi: 10.1038/nn.3485. [DOI] [PubMed] [Google Scholar]
- Guo J, Anton ES. Decision making during interneuron migration in the developing cerebral cortex. Trends Cell Biol. 2014;24(6):342–351. doi: 10.1016/j.tcb.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwell CC, Fuentealba LC, Gonzalez-Cerrillo A, Parker PR, Gertz CC, Mazzola E, Garcia MT, Alvarez-Buylla A, Cepko CL, Kriegstein AR. Wide Dispersion and Diversity of Clonally Related Inhibitory Interneurons. Neuron. 2015;87(5):999–1007. doi: 10.1016/j.neuron.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz RA, Jack-Scott E, Narezkina A, Palagin I, Boimel P, Kulkosky J, Nicolas E, Greger JG, Skalka AM. High-frequency epigenetic repression and silencing of retroviruses can be antagonized by histone deacetylase inhibitors and transcriptional activators, but uniform reactivation in cell clones is restricted by additional mechanisms. J Virol. 2007;81(6):2592–2604. doi: 10.1128/JVI.01643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepecs A, Fishell G. Interneuron cell types are fit to function. Nature. 2014;505(7483):318–326. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín O. Cellular and molecular mechanisms controlling the migration of neocortical interneurons. Eur J Neurosci. 2013;38(1):2019–2029. doi: 10.1111/ejn.12225. [DOI] [PubMed] [Google Scholar]
- Marín O, Müller U. Lineage origins of GABAergic versus glutamatergic neurons in the neocortex. Curr Opin Neurobiol. 26:132–141. doi: 10.1016/j.conb.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Jaglin XH, Cobbs LV, Bandler RC, Streicher C, Cepko CL, Hippenmeyer S, Fishell G. Clonally Related Forebrain Interneurons Disperse Broadly across Both Functional Areas and Structural Boundaries. Neuron. 2015;87(5):989–998. doi: 10.1016/j.neuron.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, McConnell DJ, Fino E, Yuste R. Axo-dendritic overlap and laminar projection can explain interneuron connectivity to pyramidal cells. Cereb Cortex. 2013;23(12):2790–2802. doi: 10.1093/cercor/bhs210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan KT, Shi W, Shi SH. Clonal origins of neocortical interneurons. Curr Opin Neurobiol. 2014;26:125–131. doi: 10.1016/j.conb.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka DH, Yanagida M, Zhu Y, Mikami S, Nagasawa T, Miyazaki J, Yanagawa Y, Obata K, Murakami F. Random walk behavior of migrating cortical interneurons in the marginal zone: time-lapse analysis in flat-mount cortex. J Neurosci. 2009;29(5):1300–1311. doi: 10.1523/JNEUROSCI.5446-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7(9):687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Yu YC, Bultje RS, Wang X, Shi SH. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature. 2009;458(7237):501–504. doi: 10.1038/nature07722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YC, He S, Chen S, Fu Y, Brown KN, Yao XH, Ma J, Gao KP, Sosinsky GE, Huang K, Shi SH. Preferential electrical coupling regulates neocortical lineage-dependent microcircuit assembly. Nature. 2012;486(7401):113–117. doi: 10.1038/nature10958. [DOI] [PMC free article] [PubMed] [Google Scholar]