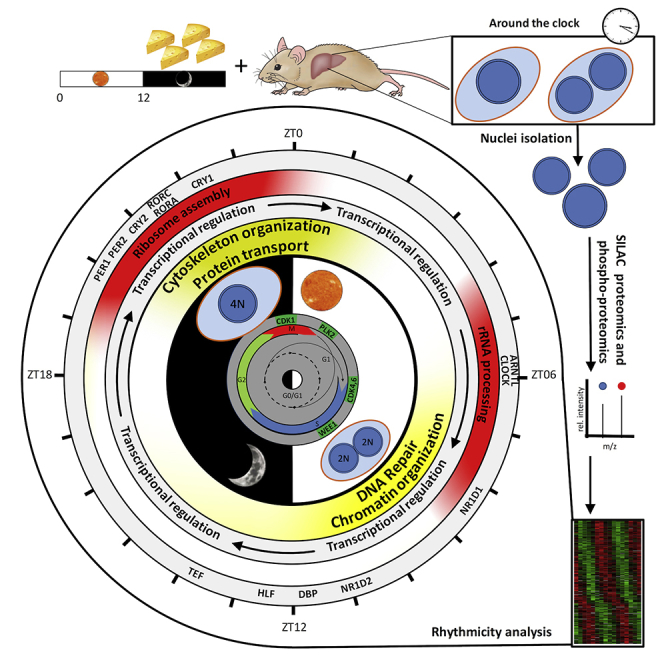
Summary
Diurnal oscillations of gene expression controlled by the circadian clock and its connected feeding rhythm enable organisms to coordinate their physiologies with daily environmental cycles. While available techniques yielded crucial insights into regulation at the transcriptional level, much less is known about temporally controlled functions within the nucleus and their regulation at the protein level. Here, we quantified the temporal nuclear accumulation of proteins and phosphoproteins from mouse liver by SILAC proteomics. We identified around 5,000 nuclear proteins, over 500 of which showed a diurnal accumulation. Parallel analysis of the nuclear phosphoproteome enabled the inference of the temporal activity of kinases accounting for rhythmic phosphorylation. Many identified rhythmic proteins were parts of nuclear complexes involved in transcriptional regulation, ribosome biogenesis, DNA repair, and the cell cycle and its potentially associated diurnal rhythm of hepatocyte polyploidy. Taken together, these findings provide unprecedented insights into the diurnal regulatory landscape of the mouse liver nucleus.
Graphical Abstract
Highlights
-
•
SILAC nuclear proteomics uncovered new diurnal regulatory landscape of mouse liver
-
•
Regulation of the diurnal nuclear proteome is mostly post-translational
-
•
Diurnal proteins regulate transcription, ribosome biogenesis, DNA repair, and cell cycle
-
•
Hepatocyte polyploidy and size oscillate diurnally
Wang et al. quantify the temporal nuclear accumulation of proteins and phosphoproteins in the mouse liver and reveal that 13% of nuclear proteins exhibit a diurnal rhythm regulated at the post-translational level through nuclear transport of protein complexes involved in transcription, DNA repair, ribosome biogenesis, cell cycle, and polyploidy.
Introduction
While the human and mouse genomes have been available for over a decade, progress in measuring the expression of gene products has been made mainly on the level of mRNA abundance (Melé et al., 2015) and, to a lesser extent, proteins (Geiger et al., 2013, Kim et al., 2014), with little information on their cellular localization and dynamic regulation. Indeed, cellular functions in eukaryotes often rely on membrane-enclosed organelles with specialized and compartmentalized functions, interacting dynamically with each other. The cell nucleus can sense signals from biochemical or mechanical origins and translate these into molecular response, notably through control of gene expression. Proteomic studies have therefore characterized the protein composition of different nuclear compartments, notably the nuclear membrane (Schirmer et al., 2003), the nuclear pore (Cronshaw et al., 2002), the nucleolus (Andersen et al., 2005), the centrosome (Andersen et al., 2003), or interchromatin granules (Saitoh et al., 2004). However, apart from analyses of the brain and neurons (Dammer et al., 2013, Ren et al., 2015), heart (Franklin et al., 2011), liver (Zhang et al., 2013), or multiple tissues in parallel (Foster et al., 2006, Kislinger et al., 2006), only very few comprehensive total nuclear proteomes are available in mammalian cells and tissues. In all these cases, the obtained coverage was still fairly low compared to the predicted mammalian nuclear proteome (Bauer et al., 2011, Fink et al., 2008). Moreover, quantitative proteomics techniques such as SILAC (stable isotope labeling with amino acids in culture) have rarely been employed, and no studies addressed dynamic aspects or genotype dependency of nuclear proteomes. In addition, while the whole-liver phosphoproteome has been previously described at a very high coverage (Humphrey et al., 2015), or as part of multiple tissue experiments (Huttlin et al., 2010, Lundby et al., 2012), no specific nuclear phosphoproteome has been analyzed experimentally on mammalian healthy tissue, though organelle-specific phosphoproteomes have been predicted computationally based on whole-cell studies (Chen et al., 2015).
Here, we focus on nuclear functions measured temporally in the mouse liver, as animals are exposed to diurnal and feeding-fasting cycles. In those conditions, the liver is spectacularly dynamic, changing not only its entire gene expression landscape (Doherty and Kay, 2010), but also its morphology in response to feeding and hormonal cues (Gerber et al., 2013, Uchiyama and Asari, 1984). These changes are thought to allow the separation of incompatible metabolic processes occurring at different times of the day (Gachon et al., 2004), and are regulated through transcriptional, post-transcriptional, translational, and post-translational regulations (Asher and Sassone-Corsi, 2015). The circadian clock consists of an endogenous and autonomous oscillator with a period of nearly 24 hr, which coordinates most aspects of physiology and behavior in mammals, including humans (Gerhart-Hines and Lazar, 2015). This oscillatory clockwork is organized hierarchically, with a master clock in the suprachiasmatic nuclei of the hypothalamus that communicates timing signals to enslave oscillators in peripheral organs. Rhythms in gene products are generated by molecular feedback loops, in which multiple layers of control, including temporal post-transcriptional and post-translational regulation, contribute (Crane and Young, 2014). While transcriptional regulation orchestrated by the circadian clock has been well studied (Koike et al., 2012, Le Martelot et al., 2012, Vollmers et al., 2012), regulations at the proteome and phosphoproteome levels are largely unexplored, despite recent description of rhythmic protein levels in whole-tissue extracts (Mauvoisin et al., 2014, Robles et al., 2014).
Here, we report a quantitative and high-resolution analysis of the diurnal nuclear proteome and phosphoproteome in mouse liver, highlighting the deep impact of diurnal rhythms on liver function. In addition to transcriptional regulation, we found that crucial cellular functions like DNA repair, ribosome biogenesis, cell cycle, and polyploidy are also subject to diurnal regulation, mostly at the post-translational level. In this context, organelle-specific time-resolved quantitative proteomics provides an outstanding tool to systemically reveal regulated cellular functions, which would be inaccessible with genomic or whole-cell proteomic approaches.
Results
High-Coverage Nuclear Proteome Quantified by SILAC-Based Mass Spectrometry in Mouse Liver
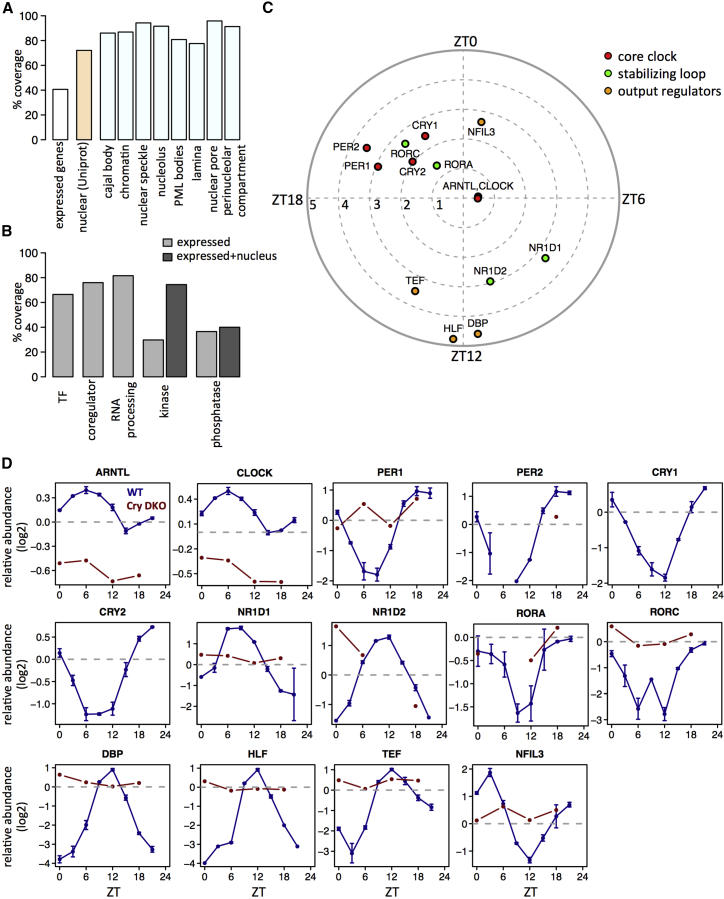
To measure the diurnal accumulation of proteins in the nucleus of mouse liver, we designed a quantitative SILAC mass spectrometry (MS) experiment in which nuclear protein extracts were harvested from mice liver every 3 hr for 2 days, yielding two biological replicates at each of eight time points. Relative protein abundance in those 16 samples was quantified against a common reference sample obtained by in vivo total stable isotope labeling of mouse tissues (SILAC) as described before (Figure S1A, available online; related to Figure 1) (Mauvoisin et al., 2014). This SILAC-based analysis identified a total of 4,820 distinct proteins, of which 84% (4,035) yielded relative measurements in at least 8 out of the 16 samples (Figure S1B; Table S1; related to Figure 1). We globally obtained a high correlation between biological replicates (70% correlation on average), with the exception of zeitgeber time (ZT; ZT0, lights on; ZT12, lights off) 21 due to a potential contamination of one sample during the nuclei preparation (Figure S1C). Among the detected proteins with known subcellular localization in Uniprot (UniProt Consortium, 2015), around 75% were known to localize in the nucleus or to shuttle between the nucleus and cytoplasm. Moreover, 93% of the obtained MS raw signal was from nuclear/shuttling proteins (Figures S1D and S1E), and more than 80% of all identified proteins were nuclear according to the COMPARTMENTS database (Binder et al., 2014) (Figure S1F). Compared with existing compendia of nuclear proteomes, for instance, the experimentally determined 824 proteins in Kislinger et al. (2006) (Figure S1G), the computationally defined proteins (∼3,500) in Bauer et al. (2011) and Fink et al. (2008), and the approximately 4,220 proteins annotated as nuclear in Uniprot, our liver data achieved higher coverage. In fact, we covered nearly 70% of all known proteins expressed in the liver nucleus (Figure 1A) and close to 90% for proteins annotated as parts of nuclear compartments in mouse liver (Figures 1A and S1H). The coverage for proteins involved in important nuclear functions, such as transcription factors (TFs), transcriptional co-regulators, and RNA processing proteins, was above 60% (Figure 1B).
Figure 1.
High-Coverage Nuclear Proteome by SILAC-Based MS Identifies Robust Diurnal Rhythms in Core Clock and Clock-Regulated Proteins in Mouse Liver
(A) Coverage of liver-expressed genes (assessed by RNA-seq, requiring mean reads per kilobase of transcript per million mapped reads [RPKM] > 0.5 in wild-type data from; Atger et al., 2015; white bar), annotated as nuclear (Uniprot, beige) and annotated as belonging to nuclear sub-compartments (light blue).
(B) Coverage of liver-expressed genes with specific functions. For kinases and phosphatases, the coverage, taking into account nuclear annotation, is also shown.
(C) Core clock and clock-regulated proteins quantified in the nuclear proteome. Phases and amplitudes are indicated by the angles and distances to the center, respectively.
(D) Temporal nuclear accumulations of individual proteins for wild-type (blue line) and Cry1/2 DKO (red line). The error bars in wild-type represent SEM between two biological replicates.
To study temporal regulations, we first analyzed the nuclear accumulation of protein involved in circadian rhythms. We could detect all the known components of the core clock, as well as clock-controlled TFs (e.g., members of PAR bZip family; Gachon, 2007; and E4BP4/NFIL3 in Figures 1C and 1D). All these components rhythmically accumulated in the nucleus with expected phases and amplitudes. Such rhythms were disrupted in clock-deficient Cry1/2 double-knockout (DKO) mice (van der Horst et al., 1999), in which no CRY proteins could be detected (Figure S1I).
Extensive Rhythms of Nuclear Protein Abundance Are Mainly Regulated at the Post-transcriptional Level
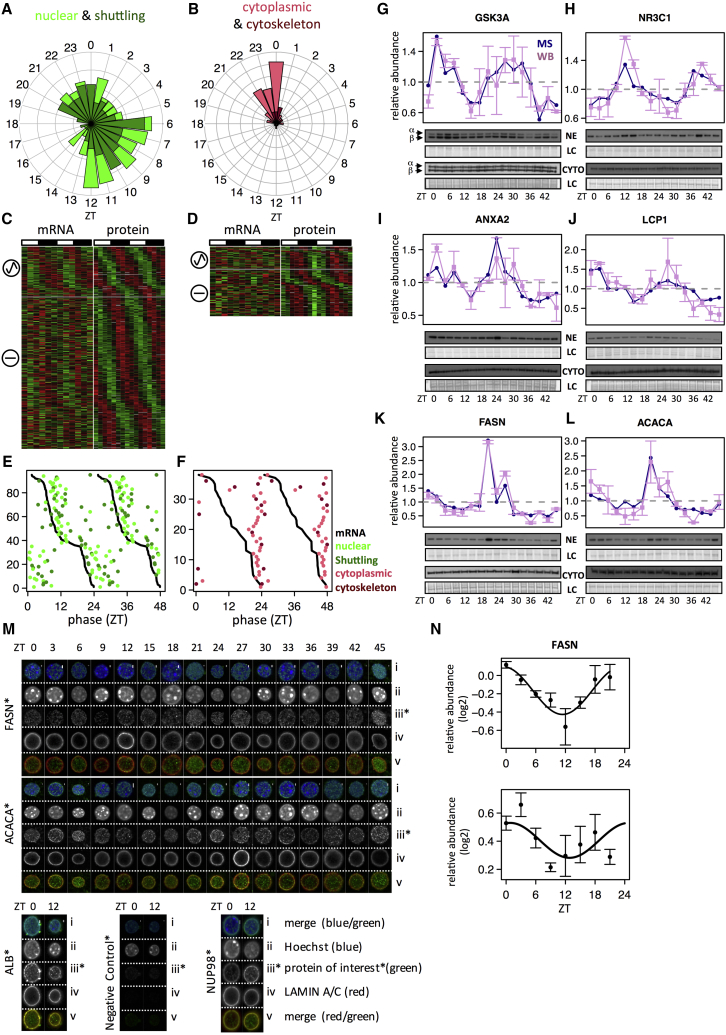
We identified 522 (13%, false discovery rate [FDR] = 5%) proteins that rhythmically accumulated in the nucleus, or 1,835 using a less stringent criterion (45%, FDR = 0.25) (Figure S2A; Table S2; related to Figure 2). Our previous characterization of the whole-cell rhythmic proteome using the exact same MS and analysis (Mauvoisin et al., 2014) identified only 195 rhythmic proteins (∼5%, FDR = 0.25), indicating that the nuclear proteome is subject to extensively more diurnally rhythmic regulation. The 522 rhythmic nuclear proteins showed a bimodal peak time distribution similar to that in the total proteome (Mauvoisin et al., 2014), with peaks at the end of the light and dark periods (Figure S2B). In addition to an increased number of rhythmic proteins, nuclear proteins also displayed increased peak-to-trough amplitudes compared to total protein extracts, with maxima above 30-fold (Figure S2C). The potential contamination at ZT21 (sample 1) by cytoplasmic proteins had only a minor effect on the global analysis of rhythmicity (Figure S2D). The majority (400 out of 522) of the rhythmic proteins are known to localize in the nucleus or shuttle from cytoplasm to the nucleus (Figure 2A). These proteins showed a bimodal distribution of peak times, with maxima toward the end of day and night periods. Unexpectedly, a fraction (122 out of 522, 23%) of proteins annotated to be mostly cytoplasmic as well as constituents of the cytoskeleton showed rhythmic accumulation in the nucleus with a sharp phase distribution around ZT0, albeit with lower amplitudes compared to nuclear and shuttling proteins (Figures 2B and S2E).
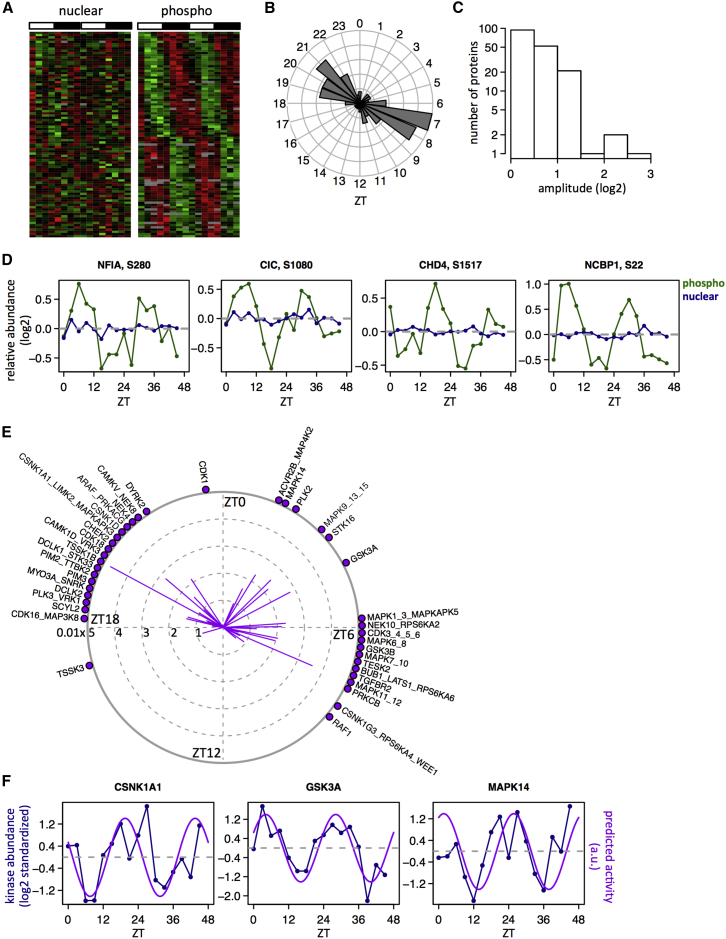
Figure 2.
Rhythm of Nuclear Proteins Is Mainly Regulated at the Post-transcriptional Level
A total of 522 proteins were identified as rhythmic in nuclear extracts (FDR < 0.05). They were divided into two classes according to annotations of cellular localization: C1 for nuclear (n = 232) and shuttling proteins (n = 168); C2 for cytoplasmic (n = 94) and cytoskeleton proteins (n = 28).
(A) Peak time distributions in ZT for proteins in C1 (nuclear, light green; shuttling, dark green).
(B) Same as (A) for C2 (cytoplasm, light red; cytoskeleton, dark red).
(C and D) Heatmap representation of proteins in C1 (C) and C2 (D) and their corresponding mRNAs measured by total RNA-seq in the same experimental conditions (LD and night-restricted feeding) (Atger et al., 2015). Data were standardized by rows, and gray blocks indicate missing data. Sinusoidal icon means rhythmic mRNA, and straight line constant mRNA.
(E and F) For rhythmic proteins encoded by rhythmic mRNA (FDR < 0.05), phase correlations between proteins and mRNAs for C1 (E) (statistically significant with p < 10−15, circular correlation test) and C2 (F) (not significant with p = 0.21).
(G–L) Examples of rhythmic proteins in C1 (G, GSK3α [quantified] and GSK3β; H, NR3C1) and C2 (I, ANXA2; J, LCP1; K, FASN; and L, ACACA) confirmed by western blot (WB) analysis in nuclear extracts (NE; upper blots) and cytoplasmic extracts (CYTO; lower blots). The two biological replicates are shown as ZT0–ZT21 (replica 1) and ZT24–ZT45 (replica 2). MS and WB data are normalized to the temporal mean, and the 16 time points show the mean and SEM from two independent biological samples. Naphtol blue-black staining of the membranes was used as a loading control (LC) and serves as a reference for normalization of the quantified values.
(M) Examples of rhythmic proteins in C2 (FASN, ACACA, and ALB) detected in the nucleus by confocal microscopy. Secondary antibodies alone were used as negative control and the nuclear pore complex NUP98 as positive control. Nuclei were stained and confocal z stack images were acquired at 0.20 μm intervals (see z stacks in Figure S2K). Vertical white scale bars represent 1 μm.
(N) Densitometry analyses for proteins FASN (peak time = 23.4 hr, rhythmicity test with p < 10−4) and ACACA (peak = 0.8 hr, p = 0.11) with confocal signal (iii) normalized by LAMIN A/C signal (iv) from (M). Data show the mean and SEM from at least three nuclei.
We next denote nuclear/shuttling proteins as the C1 group and cytoplasmic/cytoskeleton proteins as C2. Strikingly, only a fraction of rhythmic nuclear proteins was encoded by rhythmic mRNAs, quantified by RNA sequencing (RNA-seq) in the same light-dark (LD) and feeding conditions for both C1 (23%) and C2 (33%) (Figures 2C and 2D), highlighting the importance of post-transcriptional regulation in generating these rhythms in nuclear proteins. Among the fraction of proteins with corresponding rhythmic mRNAs, the phases of protein in C1 were highly correlated with the phases of their cognate mRNAs, with an average delay of 3 hr (Figures 2E and S2F). This delay is shorter than what we found for total proteins (Mauvoisin et al., 2014) and probably reflects short protein half-lives, which is supported by the observation that these proteins were enriched in TFs (Figure S2G). In contrast, the peak times of C2 proteins in the nucleus did not correlate with mRNA peaks (Figure 2F). Furthermore, only a small fraction of the rhythmic nuclear proteins also showed rhythms in total extracts for both C1 (6%) and C2 (17%) (Figure S2H), suggesting that the protein translocation from the cytoplasm into the nucleus is an important regulatory mechanism. The proteins with rhythms both in the total and nuclear extracts coincided with food-driven rhythmically secreted proteins (Mauvoisin et al., 2014) and showed a clear phase preference at ZT19 in the total and ZT22 in the nuclear extracts (Figure S2I).
To further assess the diurnal nuclear accumulation of proteins annotated as cytoplasmic, we independently quantified protein abundance in nuclear and cytoplasmic extracts using western blots (WBs) for proteins in both C1 (GSK3A and NR3C1 in Figures 2G and 2H) and C2 (ANXA2, LCP1, FASN, ACACA, and ALB in Figures 2I–2L and S2J). This confirmed the rhythms and peak accumulation times of these proteins in the nuclear extracts, while rhythms in the cytoplasmic extracts were absent. In addition, we performed immunofluorescence experiments on purified nuclei on the cytoplasmic proteins FASN, ACACA, and ALB, all showing rhythmic nuclear accumulation. Confocal microscopy indeed confirmed that these proteins rhythmically accumulate in the nucleus with the same phases as in the MS, whereas structural components of nuclei, LAMIN A/C, and NUP98 did not (Figures 2M, 2N, S2K, and S2L).
Rhythmic Accumulation of Nuclear Protein Complexes
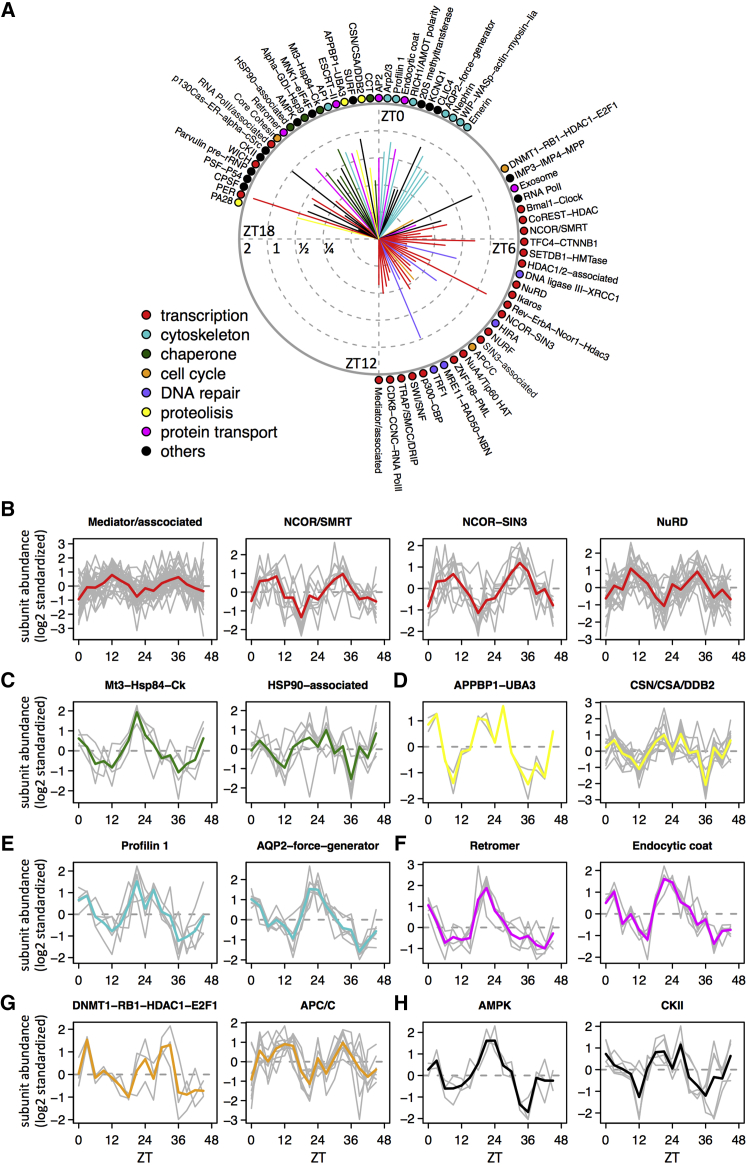
Many quantified nuclear proteins were subunits of well-characterized nuclear protein complexes. In fact, the subunit coverage of known complexes was very high, even among ones with numerous subunits (Figure S3A; Table S3; related to Figure 3). Often, proteins belonging to the same complex showed highly similar diurnal profiles (Figures S3B and S3C). Since not all subunits of annotated complexes might follow the same temporal patterns, we identified nuclear protein complexes with synchronized rhythmic subunits using singular value decomposition (SVD). We retained complexes in which the fraction of variances captured by the first singular component was significant (p < 0.05), and the rhythm of the complex was estimated from that of the first component (Experimental Procedures). Unexpectedly, such analysis yielded 360 complexes with synchronized subunits out of the 993 detected (at least two distinct subunits detected) (Figure S3D), of which 185 showed diurnal accumulation representing diverse functions, peak times, and amplitudes (Figure 3A). Some of these complexes showed low average amplitudes, potentially reflecting that some subunits are shared by multiple complexes not necessarily expressed in the same phase, or that amplitudes among subunits are heterogeneous (Figure S3E). Most of the complexes peaking during the day were involved in transcriptional regulation and DNA repair, whereas ones peaking at night are more enriched in cytoskeleton organization, protein transport, proteolysis, and chaperoning of proteins (Figures 3A–3G). Of note, the multimeric kinases AMPK and CKII, involved in circadian clock regulation (Lamia et al., 2009, Maier et al., 2009, Tamaru et al., 2009), both show rhythmic nuclear accumulations with a maximum at the end of the dark period (Figure 3H). Hence, our results strongly suggest temporal compartmentalization of fundamental nuclear processes in the liver.
Figure 3.
Diurnal Accumulation of Nuclear Protein Complexes
(A) Nuclear protein complexes with rhythmic accumulation were identified by the singular value decomposition (SVD). Peak times (identified from the first eigengene in the SVD) and mean peak-to-trough amplitudes (mean of amplitudes of rhythmic subunits within the same complex) are indicated by the angles (reference ZT times are indicated) and length of associated solid lines (the tick labels show log2 amplitudes). Specific functions of these rhythmic complexes are color coded.
(B–H) Temporal profiles of subunits of some rhythmic nuclear protein complexes are shown, e.g., mediator/associated, NCOR/SMRT, NCOR-SIN3, and NuRD complexes for transcription (B); Mt3-Hsp84-Ck and HSP90-associated complexes for chaperone (C); APPBP1-UBA3 and CSN/CSA/DDB2 complexes for proteolysis (D); Profilin 1 and AQP2-force-generator complexes for cytoskeleton (E); retromer and endocytic coat complexes for protein transport (F); DNMT1-RB1-HDAC1-E2F1 and APC/C complexes for cell cycle (G); and AMPK and CKII complexes for kinases (H).
Temporal Organization of Ribosome Biogenesis and DNA Repair
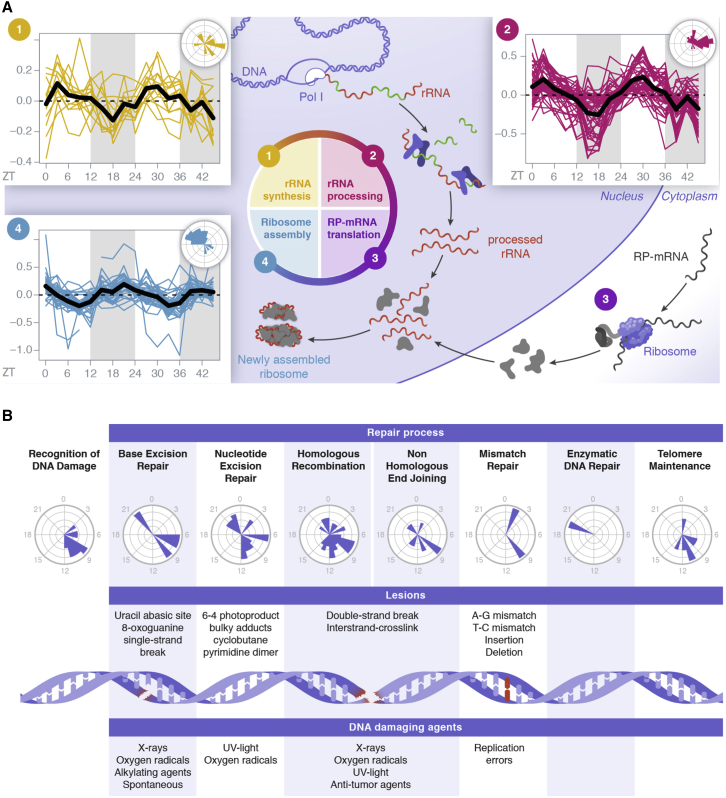
The functions of rhythmic nuclear proteins and complexes pointed toward temporal organization of ribosome biogenesis and DNA repair. Ribosome biogenesis was represented by 99 proteins showing different phases of nuclear accumulation, corresponding to distinct steps (de la Cruz et al., 2015) (Figure 4A; Table S4; related to Figure 4). First, proteins involved in rRNA transcription, including RNA polymerase I subunits, showed a maximum nuclear accumulation around ZT6, consistent with the transcription of the 45S rRNA (Jouffe et al., 2013) (Figure S4A; related to Figure 4). Proteins and complexes involved in rRNA processing and pre-ribosome assembly also peaked at ZT6, including the small-subunit processome (SSU) (Phipps et al., 2011), the PeBoW complex (Lapik et al., 2004), and the exosome (Lykke-Andersen et al., 2009) (Figures 4A and S4A). Second, rRNA synthesis and maturation during the day were followed by ribosomal protein synthesis, shown to take place in the cytoplasm around ZT18 (Atger et al., 2015, Jouffe et al., 2013). Lastly, the final assembly of the ribosomes in the nucleolus involves the rhythmic nuclear accumulation of the pre-60S ribosome (Nissan et al., 2002) and the large-subunit processome (LSU) (McCann et al., 2015) with peak phases near ZT22 (Figure S4A). Altogether, ribosome biogenesis, one of the most energy-consuming cellular processes (Warner, 1999), appeared diurnally and sequentially orchestrated, possibly to occur in sync with sufficient nutrient availability.
Figure 4.
Temporal Organization of Ribosome Biogenesis and DNA Repair
(A) Temporal accumulations and phase distributions of rhythmic nuclear proteins involved in sequential steps of ribosome biogenesis, namely rRNA synthesis (1) and processing (2), and ribosome assembly (4). Temporal profiles of several complexes involved are found in Figure S4B.
(B) Peak time distributions of rhythmic nuclear proteins in the different mechanisms involved in DNA repair. Temporal profiles of several complexes involved are found in Figure S4C.
DNA repair, a crucial process for the maintenance of genome integrity (Ciccia and Elledge, 2010) (Figure 4B; Table S4), was represented by 96 rhythmic nuclear proteins. The majority of rhythmic proteins involved in DNA repair peak between ZT7 and ZT12 (Figure S4B). Enzymatic DNA repair (Martineau-Pivoteau et al., 1996) and telomere maintenance (Chen et al., 2014) were shown to have maximum activity during the night, whereas nucleotide excision repair (NER) peaked at the end of the light period (Kang et al., 2009), like other processes involved in ionizing radiation-induced DNA damage (Palombo et al., 2015). The circadian clock has been involved in the process (Fu et al., 2002, Kang et al., 2010). Here, we found that the proteins involved in all DNA repair mechanisms show a rhythmic nuclear accumulation (Figures 4B and S4C). In addition, we observed enrichment in proteins involved in DNA replication-associated DNA repair around ZT9 (Mjelle et al., 2015), corresponding to the time of maximal diurnal DNA synthesis in mouse liver (Barnum et al., 1958, Echave Llanos et al., 1970). This observation suggests that increased DNA repair activity may be associated with increased DNA replication around ZT9.
Diurnal Nuclear Phosphoproteome and Predicted Rhythmic Kinase Activities
Phosphorylation is involved in the regulation of both core clock and clock outputs (Reischl and Kramer, 2011). We found two kinase complexes, AMPK and CKII, showing diurnal accumulation in the nucleus (Figure 3H). To further study the diurnal phosphoproteome and its associated kinome activity, we used the same nuclear extracts and performed SILAC MS after enriching for phosphopeptides. Among the 4,689 phosphosites identified, 1,448 could be quantified in at least 8 out of 16 samples and mainly comprised phospho-serine (Figure S5A; related to Figure 5). The limited ratio of quantified over identified phosphopeptides came from a limitation of the SILAC technique, for which only lysine-containing phosphopeptides can be quantified. In total, 154 of these quantified phosphosites (11%), distributed within 113 canonical proteins, showed rhythmic nuclear accumulation (FDR < 5%) (Table S5; related to Figure 5), with a bimodal distribution of peak times located in the middle of the day (ZT7) and night (ZT21), and amplitudes up to 60-fold (Figures S5B and S5C). We could compare the rhythmic phosphorylation levels with the levels of the corresponding proteins for ∼90% of identified phosphoproteins (Figure S5D). To identify putative rhythmic phosphorylation activities, we distinguished two classes of rhythmic nuclear phosphosites, those with or without corresponding rhythmic nuclear protein accumulation (FDR < 0.05). The first class of phosphosites (n = 52, 36%) with rhythmic nuclear proteins showed biphasic phase distribution and the same high amplitudes as the corresponding proteins (Figures S5E–S5G). Also, the rhythms of phosphosites were highly correlated to the rhythms of nuclear proteins (Figures S5H and S5I), indicating that rhythmic phosphorylation passively reflects the rhythmic protein accumulation. Rhythmic phosphosites in the second class (64%) corresponded to non-rhythmic proteins (Figure 5A) and showed a similar biphasic peak time distribution (Figure 5B), but had more moderate amplitudes compared to the first class (Figure 5C). Such rhythms (examples in Figure 5D) suggested active regulation of phosphorylation by either kinases or phosphatases. We exploited known kinase specificities (Hu et al., 2014) to identify putative kinases with rhythmic activities. Using two methods, a linear model and phase enrichment analysis (Experimental Procedures), we predicted 39 kinase motifs with rhythmic activities (Figure 5E; Table S5). Three of these predicted kinases were detected in the nuclear proteome and showed rhythmic accumulation with the same phases as the predicted corresponding motif activities (Figures 5F and S5J), suggesting that in those cases the rhythmic phosphorylation is regulated by the cyclic nuclear accumulation of the kinase. Among these kinases, several were known as regulators of the circadian clock. For example, GSK3α and GSK3β, which are known to phosphorylate and regulate BMAL1, CLOCK, and REV-ERBα (Reischl and Kramer, 2011), presented a maximum activity and/or nuclear accumulation during the day, whereas CKIα and CKIδ, known as regulators of PER proteins, showed a maximum activity and/or nuclear accumulation during the night. In addition, we predicted several kinases involved in the regulation of the cell cycle as having diurnal activities. Notably, the cell-cycle-related kinases CDK4 and CDK6 showed a maximum activity around ZT6, whereas CDK1 had a maximum activity around ZT23, in opposite phase to its inhibitory kinase WEE1 (Vermeulen et al., 2003).
Figure 5.
Rhythmic Nuclear Phosphorylation and Inferred Kinase Activities
(A) Heatmap representation of rhythmically phosphorylated peptides (n = 92, right panel) with associated non-rhythmic nuclear proteins (left panel).
(B and C) Phase (B) and peak-to-trough (C) amplitude (log2) distributions for rhythmic nuclear phosphoproteins in (A).
(D) Individual examples of rhythmic phosphorylated sites (blue line, nuclear proteins; green line, nuclear phosphosites).
(E) Kinase motifs displaying rhythmic activities inferred from non-rhythmic nuclear proteins containing rhythmic phosphorylation sites through a linear model with elastic-net regularization. Direction of lines indicates peak activity times, and distances to the center of solid lines indicate activity amplitudes of each motif.
(F) Inferred rhythmic activities of CSNK1A1, GSK3A, and MAPK14 compared with their respective nuclear protein accumulations.
Comprehensive Diurnal Transcriptional Landscape in Mouse Liver
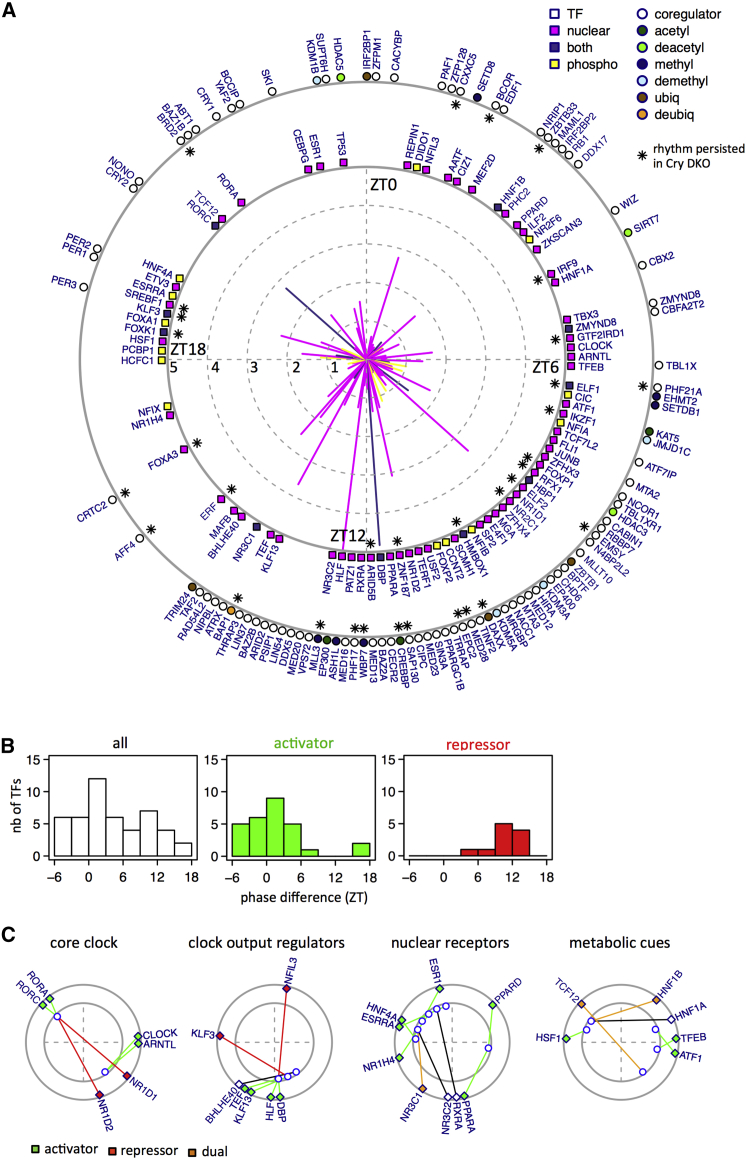
Proteins involved in transcription regulation were highly represented among rhythmic nuclear proteins and phosphoproteins (Table S6; related to Figure 6). Indeed, we identified 80 TFs and 99 transcription co-regulators showing robust diurnal nuclear accumulation (FDR < 0.05; Figures 6A and S6A–S6D; related to Figure 6). Of those, the rhythms of 16 TFs and 17 co-regulators persisted in Cry1/2 DKO mice (Figures 6A, S6B, and S6D), indicating that these are most likely driven by feeding rhythms. Among the rhythmic TFs identified in wild-type, we found core clock components, known clock output regulators (e.g., DBP, HLF, TEF, and NFIL3), and factors previously shown to be involved in the coupling between the clock and metabolism—for example, HSF1 (Reinke et al., 2008), FOXA family members (Rouyer et al., 1997), the sterol-regulated SREBP1 (Gilardi et al., 2014), and ETS family members, recently predicted to be implicated in diurnal transcriptional activity in mouse liver (Fang et al., 2014). Among other identified rhythmic TFs, many are related to hormone and metabolic regulations, involving several nuclear receptors, notably GR (NR3C1), MR (NR3C2), NR1H4, PPARα, and PPARδ. We also noted the starvation/feeding-dependent regulator of autophagy TFEB and ZKSCAN3 (Chauhan et al., 2013, Settembre et al., 2013), possibly linked with rhythmic autophagy in mouse liver (Ma et al., 2011). Another intriguing observation is the opposite phase between the antagonistic regulators of liver zonation TCF4 (encoded by the Tcf7l2 gene) and HNF4α (Gougelet et al., 2014).
Figure 6.
Eighty TFs and ∼100 Transcriptional Co-regulators with Clear Diurnal Oscillations Identified in the Nuclear Proteome and Phosphoproteome
(A) Rhythmic TFs are represented by the squares around the inner circle and co-regulators by the empty dots around the outer circle. Each line represents one TF; direction of lines indicates peak time; lengths of solid lines represent peak-to-trough amplitudes (log2). Colors of the lines and squares encode if TFs are rhythmic only in the nuclear proteome (pink), the phosphoproteome (yellow), or both datasets (blue). For the co-regulators, dot colors indicate their enzymatic activities. Persistence of the rhythmicity in Cry1/2 DKO mice is indicated by a black asterisk.
(B) Time differences between TF accumulations and corresponding motif activities for all rhythmic TFs identified (white), or for TFs already characterized as activators (green) and repressors (red).
(C) Peak times of TF nuclear accumulations (outer circle) compared with activity phases of corresponding DNA binding motif (inner circle) for core clock regulators, clock output regulators, nuclear receptors, and TFs regulated by metabolic cues. Color code indicated the commonly accepted transcriptional function of each TF, namely activator (green), repressor (red), or dual regulation (orange).
We next assessed whether these diurnally accumulating TFs in the nucleus induced rhythmic transcriptional activities. Specifically, we used RNA-seq data under the same LD and night-restricted feeding conditions (Atger et al., 2015) and considered pre-mRNA accumulation as a proxy for transcription (Gaidatzis et al., 2015). We then modeled transcription output as a linear combination of transcriptional activities associated with known DNA binding motifs of TFs (Balwierz et al., 2014, Rey et al., 2011) located within DNase 1 hypersensitive regions (from mouse ENCODE) and near gene promoters in mouse liver (Figure S6E; Table S6). The availability of nuclear protein expression patterns allowed us to make more specific hypotheses regarding the proteins responsible for time-specific transcriptional activities. In particular, the delays between TF accumulations and maximal predicted activities of cognate motifs showed a bimodal distribution around 0 and 12 hr (Figure 6B). Strikingly, TFs annotated as transcriptional activators tended to peak in phase with their motif activities, while repressors were in opposite phase with motif activities. For example, the RORE element was predicted as maximally active near ZT21, which coincided with maximal expression of the activators RORA/RORC, while the repressors NR1D1/NR1D2 (REV-ERBα and REV-ERBβ) peaked in opposition (Figure 6C). Further examples included the E-box, with ARNTL (BMAL1) and CLOCK (activators); the D-box and DBP, HLF, TEF (activators), and NFIL3 (repressors); as well as nuclear receptors and regulators of hepatic metabolism, including the recently proposed ETS family members (Fang et al., 2014) (Figures 6C and S6F).
Many of the transcriptional co-regulators with acetylase and deacetylase, or methylase and demethylase activities, quantified in our dataset showed a rhythmic nuclear accumulation, with peak phases between ZT6 and ZT12 (Figures S6G and S6H). We tested for correlations between the rhythm of these enzymes and global changes in histone acetylation and methylation by WB (Figure S6I). Despite a general decrease in histone modification near ZT10, we did not detect clear rhythm of these modifications, consistent with genome-wide chromatin immunoprecipitation (ChIP) studies on those histone marks (Le Martelot et al., 2012, Vollmers et al., 2012). In sum, the measured accumulation rhythms of many transcription regulators significantly help understanding the origins of diurnal rhythms in transcription.
Rhythmic Orchestration of Polyploidy in Mouse Liver
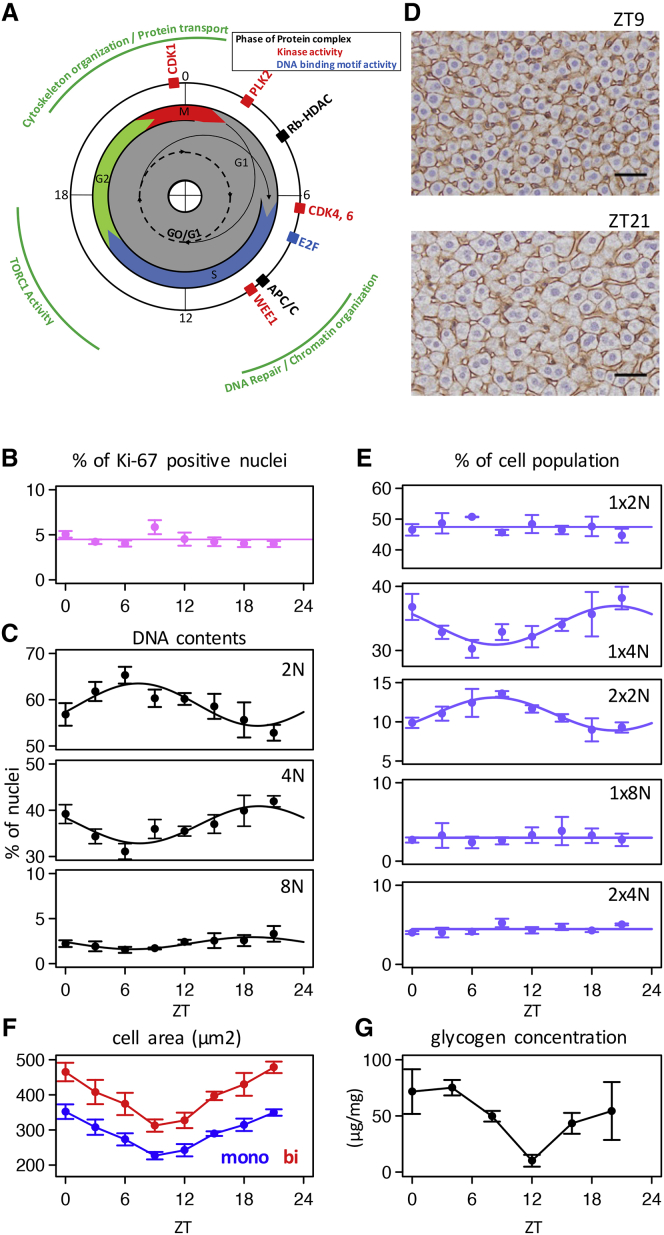
Numerous rhythmic nuclear proteins, phosphosites, and transcriptional activities are involved in cell-cycle regulations, which occurred at specific times during the day in a consistent temporal ordering (Figure 7A). Indeed, PLK2 kinase activity is involved in centriole duplication at the G1/S transition (Warnke et al., 2004) and peaked near ZT2 (Figure 5E). G1/S is also characterized by the inhibition of the E2F TF by the RB-HDAC complex, which reached its maximum of nuclear accumulation around ZT4 (Figure 3G) (Magnaghi-Jaulin et al., 1998). These findings thus place the G1 phase between ZT0 and ZT6, synchronized with the increase of nuclear MCM proteins (Figure S7D) known to assemble at replication origins during late G1 and to be active during the S phase (Costa et al., 2013). S phase entry is controlled by RB phosphorylation in part by the cyclin D-CDK4/6 complex, thus activating the E2F TF. Interestingly, CDK4/6 kinase reached its maximum activity around ZT6 (Figure 5E), just a few hours before the maximum activity of the E2F motif (Figure S6E). RB accumulation and phosphorylation measured by WB in the nucleus and the cytoplasm showed decreased nuclear RB levels synchronized with E2F motif activity, potentially due to the nuclear exclusion of phosphorylated RB (Jiao et al., 2006), preceding its subsequent degradation in the cytosol (Uchida et al., 2005) (Figures S7A and S7B; related to Figure 7). Taken together, these findings suggested that S phase occurs between ZT6 and ZT15, concomitant with the observed maximum of DNA repair (Figure 4B) and chromatin organization (Atger et al., 2015). Also, the maximum nuclear accumulation of the APC/C complex, which, when associated with CDH1, prevents premature S phase entry (Li and Zhang, 2009), and the maximum of WEE1 activity, a kinase that prevents premature mitosis by inhibiting CDK1 but also plays a role in chromatin synthesis in S phase (Mahajan and Mahajan, 2013), further support this hypothesis. Following S phase, G2 phase is characterized by cell growth. This step is in part under the control of the TORC1 pathway, which coordinates cell growth and cell-cycle progression according to nutrient availability, in part through the control of ribosomes biogenesis (Thapa et al., 2013). Since TORC1 activity is rhythmic, with a maximum near ZT16, it is likely that this phase overlaps with the G2 phase (Atger et al., 2015, Jouffe et al., 2013). Finally, M phase entry is controlled by the CDC25-dependent activation of cyclin B-CDK1 complex and characterized by the increased phosphorylation of histone H3 on Ser10 (Figure S7C) (Hendzel et al., 1997) and important modifications of the cellular cytoskeleton (Heng and Koh, 2010). Considering the maximum CDK1 activity found at the end of the night period (Figure 5E) synchronized with maximum nuclear localization of complexes involved in cytoskeleton organization (Figure 3), it is plausible that M phase occurs near the night-day transition, which is temporally consistent with the mitotic activity observed following partial hepatectomy (Matsuo et al., 2003).
Figure 7.
Diurnal Orchestration of Polyploidy in Mouse Liver
(A) Summary of cell-cycle markers identified by the analysis of nuclear proteome and phosphoproteome (outer circle) and inferred temporal windows of cell-cycle phases around the clock (inner disk).
(B) Proportion of Ki-67-positive nuclei around the clock. Each of these eight time points shows the mean and SEM of four independent biological samples.
(C) Proportions of nuclei with different DNA contents (2N, 4N, and 8N) measured by FACS analysis show diurnal variations. Each of these eight time points shows the mean and SEM of four independent biological samples. Cosine-fit curves are also shown.
(D) Representative IHC images of mouse liver sections harvested at ZT9 and ZT21 (4 μm thick liver slices). Plasma membrane is stained using β-catenin, and Mayer’s hematoxylin is used for nuclear staining. Horizontal black scale bars represent 40 μm.
(E) Proportions of mono- (1 × 2N) and bi-nucleated diploid (2 × 2N), mono- (1 × 4N) and bi-nucleated tetraploid (2 × 4N), and mono-nucleated octaploid (1 × 8N) hepatocytes around the clock extracted from IHC image analysis. Each time point shows the mean and SEM of four independent biological samples.
(F) Diurnal oscillations of cell areas for mono- and bi-nucleated hepatocytes extracted from IHC image analysis. Each time point shows the mean and SEM of four independent biological samples.
(G) Temporal liver glycogen concentration. Each time point shows the mean and SEM of three independent biological samples.
Thus, all evidence pointed toward aligned diurnal and cell cycles (Bieler et al., 2014, Feillet et al., 2014), which was surprising given that normal livers of 8- to 12-week-old mice are not actively proliferating. We estimated the fraction of cycling cells by staining for Ki-67 and found ∼5% of positive nuclei throughout the day (Figure 7B). In the absence of clear readouts concerning rhythmic cell division, we investigated whether the detected cell-cycle activities might be linked with ploidy, since polyploidy in hepatocytes of adult mouse is widespread and increases with age (Gentric and Desdouets, 2014). We thus analyzed DNA content of liver nuclei by flow cytometry. Surprisingly, this revealed antiphasic proportions of diploid nuclei (with two copies of chromosomes, 2N) compared to tetraploid nuclei (4N) (Figure 7C), suggesting a daily pattern of polyploidy. We then performed histological slices of mouse liver, which, in addition to DNA content, also allowed us to monitor the number of nuclei per cell, as well as the cell sizes (Figures 7D, 7E, and S7E–S7G). The fraction of bi-nucleated cells showed a diurnal pattern with maximum at ZT9 (Figure S7E). Using nuclear size to estimate DNA content (Figures S7F and S7G) (Martin et al., 2002), we could distinguish a number of hepatocyte subtypes. This revealed that the observed rhythm of DNA content (Figure 7C) came from two populations of cells, namely bi-nucleated diploid (2 × 2N) and mono-nucleated tetraploid (1 × 4N) cells, oscillating in opposite phases, while other populations showed no rhythm (Figure 7E). Moreover, we also observed a dramatic diurnal rhythm (60% increase at the peak) in hepatocytes size, both for mono- and bi-nucleated cells, with a maximum size at the night-day transition (Figure 7F), in agreement with previous reports (Echave Llanos et al., 1971, Gerber et al., 2013). This rhythm was in sync with the glycogen content (Figure 7G), consistent with reports that insulin-dependent cell swelling might positively regulate glycogen synthesis (Lang et al., 1998). Thus, although the liver constitutes a largely quiescent organ, we uncovered a diurnal rhythm in polyploidy.
Discussion
Though recent whole-cell diurnal proteomics studies could detect more than 5,000 proteins, the coverage of lowly expressed regulatory proteins such as TFs was poor (Chiang et al., 2014, Mauvoisin et al., 2014, Robles et al., 2014). To reduce sample complexity, we here performed a proteomics and phosphoproteomics analyses of liver nuclei, while a similar strategy recently analyzed diurnal rhythms in the mitochondria (Neufeld-Cohen et al., 2016). Combining the accuracy of the SILAC technology with time-series sampling in the liver allowed us to reach above 70% coverage of the nuclear proteome. The obtained signals allowed quantifications of all core-clock components and many clock-controlled output TFs, in wild-type mice as well as in clock-disrupted Cry1/2 DKO mice, with temporal patterns that showed comparable quality as ones obtained in transcriptome studies (Figure 1). These time series data allowed us to show that the nuclear proteome is subjected to very significant diurnal regulation (Figure 2A) orchestrated mainly at the post-transcriptional level. As the diurnal regulation of translation efficiency was recently shown to concern only a few classes of genes (Atger et al., 2015), our findings suggest that the diurnal regulation of protein stability and nuclear transport of complexes are likely the most important causes of rhythms in nuclear protein abundance (Figure 2). In particular, our quantitative and temporal approach allowed us to dynamically monitor well-studied nuclear protein complexes (Figure 3). This dynamic complexome identified complexes peaking during the day, which were involved in transcriptional regulation and also DNA repair. Of interest, the proteins involved in DNA repair (Figure 4) constitute 18% of the rhythmic proteins identified and are synchronized with the predicted time of DNA replication. On the other hand, complexes peaking during the night were enriched in cytoskeleton organization, protein transport, proteolysis, and chaperoning of proteins, suggesting a temporal compartmentalization of generic biological functions of the liver nucleus. Also, the translation of components of ribosomes and the translation machinery occurred during the night, when nutrients are available. In fact, ribosome biogenesis, representing 18% of the identified rhythmic nuclear proteome, was rhythmically orchestrated in accordance with our previous work (Atger et al., 2015, Jouffe et al., 2013) (Figure 4).
We then performed a temporal nuclear SILAC phosphoproteome quantification, which identified hundreds of rhythmic phosphorylation sites and allowed us to infer rhythmic kinase activities (Figure 5). Although we cannot exclude the contribution of rhythmic phosphatase activities (Reischl and Kramer, 2011), some of the predicted kinase activities most probably originate from the synchronized accumulation of the kinases in the nucleus, as seen for CSNK1A1, GSK3A, and MAPK14 (Figure 5F). Phosphorylation events can also regulate transcriptional activity. In addition, for the 80 TFs and 99 co-regulators showing diurnal rhythms in our nuclear proteomic screen, our phosphoproteome approach also identified numerous TFs and co-regulators with rhythmic phosphorylation sites (Figure 6). We also identified rhythmic TFs whose rhythms persisted in clock-disrupted mice, which allows us to distinguish effects of the circadian clock versus metabolic cues and feeding/fasting-driven regulation of diurnal transcription. Comprehensive and temporal measurements on the abundance of transcription regulators in nuclei are clearly powerful to explain rhythms in mRNA transcription, since most previous efforts were limited to indirect computational inference using DNA motifs (Bozek et al., 2009, Fang et al., 2014, Rey et al., 2011).
In addition to the diurnal regulation of transcription, DNA repair, and ribosome biogenesis, one striking observation consisted in the diurnal orchestration of cell-cycle activities and the modifications of cell morphology and size (Figure 7). Modification of hepatocyte size, and even liver size, was previously observed (Echave Llanos et al., 1970, Echave Llanos et al., 1971, Gerber et al., 2013, Leveille and Chakrabarty, 1967), but the mechanisms are not fully explained. Maximal hepatocyte size occurred during the hepatocyte growth phase (Figure 7F), in phase with maximal glycogen synthesis (Figure 7G), but also ribosome biogenesis (Figure 4A), which is a proxy for protein synthesis. Meanwhile, the shrinking phase corresponded to glycogen breakdown and liver protein secretion (Mauvoisin et al., 2014). However, the typical fraction of liver mass represented by glycogen is about 5%, which is small compared to the observed 60% increase in cell size. The regulation of cell volume was shown to play an important role in hepatocyte metabolism, including glycogen (Baquet et al., 1990) and protein synthesis (Stoll et al., 1992), both of which are stimulated by cell swelling and inhibited by cell shrinkage, in the absence of other stimuli (Lang et al., 1998), implicating that cell size fluctuations could even be the cause of these phenomena. Among the several factors reported to regulate cell size, insulin is an important regulator of cell swelling, which is counteracted by the opposite action of glucagon (Schliess and Häussinger, 2003). In parallel, insulin can also induce actin polymerization (Theodoropoulos et al., 1992), a phenomenon linked to hepatocyte swelling (Gerber et al., 2013). Thus, insulin signaling may partly exert its influence on rhythmic liver metabolism via the regulation of cell size and hepatocyte structure. Diurnal hepatocyte swelling may also be associated with the observed nuclear accumulation of cytoplasmic proteins when cells reached their largest size (Figures 2B and 7F). Indeed, while this might also reflect the presence of nucleoplasmic structures (Malhas et al., 2011), recent studies in cancer cells showed that cellular and nuclear deformation, following cell migration, caused nuclear envelope rupture, followed by entry of cytoplasmic proteins (Denais et al., 2016, Raab et al., 2016). Such migration involves the same cytoskeleton complexes that we found to accumulate rhythmically at the night-day transition (Figure 3A) (Etienne-Manneville, 2013).
Finally, we observed diurnal fluctuation of hepatocyte polyploidy. Namely, the fraction of bi-nucleated diploid hepatocytes peaked during the day, and mono-nucleated tetraploid hepatocytes peaked in opposite phase during the night, similar to observations in rat liver (Barbărasă, 1976, Bucher and Suppan, 1967). In liver, polyploidy increases with age and stress and is thought to confer resistance to xenobiotic or nutritional injuries (Gentric and Desdouets, 2014). However, depending on the type of ploidy, hepatocytes do not have the same capacity to divide after partial hepatectomy. Namely, while both mono- and bi-nucleated cells entered the cell cycle, only a few of them fully completed cell division. Indeed, only bi-nucleated cell number decreases during regeneration, showing their important contribution to the hepatocyte repopulation by giving rise to two daughter cells (Miyaoka et al., 2012). Interestingly, liver regeneration after partial hepatectomy (Barbason, 1970, Matsuo et al., 2003, Souto and Llanos, 1985) follows a diurnal rhythm with more rapid regeneration when the liver damage occurred around ZT8, when bi-nucleated diploid cells reach their maximum level (Figure S7E). It is thus conceivable that the diurnal proportion of these bi-nucleated diploid cells might be responsible for the observed diurnal liver regeneration.
In conclusion, our in vivo measured quantitative nuclear temporal proteomes and phosphoproteomes contribute an important step toward the identification of a new diurnal biological function orchestrated by the circadian clock and/or feeding rhythms.
Experimental Procedures
Animals
Animal studies conformed to the regulations of the veterinary office of the Canton of Vaud. Cry1/2 DKO mice (van der Horst et al., 1999) in the C57BL/6J genetic background are described in Bur et al. (2009). Ten-week-old mice had free access to food and water in 12 hr light/12 hr dark cycles under standard animal housing conditions. For all experiments, animals were fed only at night starting 4 days before the experiment to control for genotype-dependent feeding rhythms. SILAC mice were prepared as previously described (Krüger et al., 2008, Mauvoisin et al., 2014).
Preparation of Whole-Cell Protein Extracts
Whole-cell extracts (TEs) were prepared as described in the Supplemental Experimental Procedures.
Preparation of Nuclei and Cytoplasmic and Nuclear Protein Extracts
Nuclei were purified and extracts prepared as described in the Supplemental Experimental Procedures.
SILAC-Based MS Analysis of Nuclear Proteomics and Phosphoproteomics
Tandem MS (MS/MS)-based SILAC proteomic analysis is mostly done as previously described (Mauvoisin et al., 2014) (details in Supplemental Experimental Procedures). Raw MS data and search engine outputs were deposited in the ProteomeXchange Consortium (proteomexchange.org) via the PRIDE partner repository with the identifiers ProteomeXchange: PXD003818 for nuclear proteomics and ProteomeXchange: PXD004191 for nuclear phosphoproteomics.
Annotation of Protein Localization
We used Uniprot to annotate protein localization, i.e., nuclear, shuttling, cytoplasmic, or cytoskeleton, for nuclear and total extracts. In addition, localization of 522 rhythmic proteins in nuclear extract (Table S2) was manually corrected or added according to the literature if protein localization in Uniprot was missing.
Rhythmicity Analysis for Nuclear Proteins and Phosphoproteins
Rhythmicity in temporal nuclear accumulation of proteins and phosphoproteins used harmonic regression, as described previously (Mauvoisin et al., 2014). To test whether rhythm of nuclear proteins in wild-type persists in Cry1/2 DKO mice, we applied linear regressions combined with model selection, similar to Atger et al. (2015) (details in the Supplemental Experimental Procedures).
Identification of Rhythmic Protein Complexes with SVD
To identify protein complexes showing diurnal accumulation in the nucleus, we applied SVD to the matrix , in which each row represents the standardized temporal profile of each subunit belonging to the same protein complex (details in the Supplemental Experimental Procedures).
Inference of Rhythmic Motif Activity of TFs with DNase Hypersensitive Sites and Pre-mRNAs
We defined a non-redundant motif library and inferred rhythmic motif activity of TFs (details in the Supplemental Experimental Procedures).
Inference of Kinases with Rhythmic Activities
We used phase enrichment analysis and linear model with regularization to infer kinases with rhythmic activities (details in the Supplemental Experimental Procedures).
Immunofluorescence and Confocal Microscopy
Purified nuclei were fixed using PFA 2% in PBS and seeded onto glass coverslips coated with poly-l-ornithine. Fixed nuclei were washed with PBS and permeabilized with Eth-OH / Met-OH for 5 min. After washing with PBS, nuclei were incubated with LAMIN A/C and ACACA, FASN, NUP98 or ALB (antibodies in PBS 1% Horse Serum (HS) for 1 hr at RT. Nuclei were rinsed with PBS followed by incubation with Alexa Fluor 488 and 555 coupled secondary antibodies in PBS 1% HS containing Hoechst (Life technologies) for 1 hr at RT. They were imaged using Leica TCS SP8 confocal microscope after a final PBS rinsing with images collected at 63x magnification. References for the antibodies are given in Table S7, related to Experimental Procedures.
FACS Analysis
Fluorescence-activated cell sorting (FACS) analysis of purified nuclei is detailed in the Supplemental Experimental Procedures.
Immunohistochemistry Experiment
Immunohistochemistry (IHC) analysis of mouse liver is detailed in the Supplemental Experimental Procedures.
IHC Image Segmentation and Estimation of Different Polyploid Cell Populations
Whole IHC images were automatically segmented and nuclear areas, as proxies of DNA contents, were estimated. It allowed us to estimate fractions of all different polyploid cell populations (details in the Supplemental Experimental Procedures).
Author Contributions
Conceptualization, J.W., D.M., F.N., and F.G.; Formal Analysis, J.W. and F.N.; Investigation, D.M., E.M., F.A., A.N.G., L.D., F.S., A.P., M.K., P.W., and M.Q.; Visualization, J.W. and D.M.; Writing – Original Draft, J.W., D.M., V.D., F.N., and F.G.; Writing – Review & Editing, all authors; Supervision, F.N. and F.G.; Funding Acquisition, F.N. and F.G.
Acknowledgments
This research was supported by the Swiss National Science Foundation (through individual research grant 31003A-153340 to F.N.), the Ecole Polytechnique Fédérale de Lausanne (EPFL), the European Research Council (through individual starting grants ERC-2010-StG-260988 to F.G. and ERC-2010-StG-260667 to F.N.), and the Leenaards Foundation (to F.G. and F.N.). We thank Laura Symul for her help on figure artwork. We thank Jessica Sordet-Dessimoz at the Histology Core Facility, EPFL, for the assistance in IHC. D.M., E.M., F.A., A.N.G., L.D., F.S., A.P., M.K., and F.G. are employees of Nestlé Institute of Health Sciences S.A.
Published: November 3, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.cmet.2016.10.003.
Contributor Information
Felix Naef, Email: felix.naef@epfl.ch.
Frédéric Gachon, Email: frederic.gachon@rd.nestle.com.
Accession Numbers
Raw MS data and search engine outputs have been deposited in the ProteomeXchange Consortium under ID codes ProteomeXchange: PXD003818 (nuclear proteomics) and PXD004191 (nuclear phosphoproteomics).
Supplemental Information
Related to Figure 1. Sheet1: Relative abundance (normalized L/H ratios from the MaxQuant analysis) for all distinct 4,820 proteins identified by SILAC MS in the nucleus of mouse liver at different time points and in all replicates under LD conditions. Sheet2: Raw data with peptide counts from MaxQuant analysis at different time points in all replicates. Sheet3: Description of the column headers for Sheet1. Sheet4: Description of the column headers for Sheet2.
Related to Figure 2. Sheet 1. Biochemical functions and cellular localization of these rhythmic nuclear proteins. Sheet 2. Relative abundance of rhythmic nuclear proteins and their corresponding phases and amplitudes in wild-type mice and their correlation in Cry1/2 DKO. Sheet 3. Corresponding mRNA expression measured by total RNA-seq. Sheet 4. Corresponding previously published proteomic data in whole-liver extracts extracts (Mauvoisin et al., 2014). Sheet 5. Description of the column headers for Sheets1–4.
Related to Figure 3. Sheet 1. Phase, amplitudes, and related functions of the identified rhythmic complexes. Sheet 2. All known complexes with at least two detected subunits in nuclear proteome and corresponding SVD analysis. Sheet 3. Description of the column headers for Sheets1 and 2.
Related to Figure 4. Sheet 1. Rhythmic nuclear proteins with identified phases and amplitudes involved in different steps of ribosome biogenesis. Sheet 2. Rhythmic nuclear proteins with identified phases and amplitudes involved in different steps of DNA repair process. Sheet 3. Description of the column headers for Sheets1 and 2.
Related to Figure 5. Sheet 1. Complete diurnal phosphoproteome dataset and corresponding rhythmicity analysis. Sheet 2. Kinase motifs with rhythmic activities inferred using linear model with regularization. Sheet 3. Description of the column headers for Sheets1 and 2.
Related to Figure 6. Sheet 1. Rhythmic TF quantified in nuclear proteome and/or nuclear phosphoproteome. Sheet 2. Rhythmic co-regulators quantified in nuclear proteome. Sheet 3. TF motifs with rhythmic activities inferred from DNase hypersensitive sites (DHSs) and pre-mRNA datasets. Sheet 4. Comparison between rhythmic TFs and their corresponding motif activity. Sheet 5. Description of the column headers for Sheets1–4.
Related to Experimental Procedures
References
- Andersen J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Andersen J.S., Lam Y.W., Leung A.K.L., Ong S.-E., Lyon C.E., Lamond A.I., Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Asher G., Sassone-Corsi P. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161:84–92. doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- Atger F., Gobet C., Marquis J., Martin E., Wang J., Weger B., Lefebvre G., Descombes P., Naef F., Gachon F. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc. Natl. Acad. Sci. USA. 2015;112:E6579–E6588. doi: 10.1073/pnas.1515308112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balwierz P.J., Pachkov M., Arnold P., Gruber A.J., Zavolan M., van Nimwegen E. ISMARA: automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res. 2014;24:869–884. doi: 10.1101/gr.169508.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquet A., Hue L., Meijer A.J., van Woerkom G.M., Plomp P.J. Swelling of rat hepatocytes stimulates glycogen synthesis. J. Biol. Chem. 1990;265:955–959. [PubMed] [Google Scholar]
- Barbărasă C. Rhythmic circadian activity of the rat hepatic lobule reflected at cellular level. Morphol. Embryol. (Bucur.) 1976;22:33–39. [PubMed] [Google Scholar]
- Barbason H.R. [Influence of the rhythm of circadian activity on mitotic index during hepatic regeneration] C. R. Acad. Sci. Hebd. Seances Acad. Sci. D. 1970;270:3295–3298. [PubMed] [Google Scholar]
- Barnum C.P., Jardetzky C.D., Halberg F. Time relations among metabolic and morphologic 24-hour changes in mouse liver. Am. J. Physiol. 1958;195:301–310. doi: 10.1152/ajplegacy.1958.195.2.301. [DOI] [PubMed] [Google Scholar]
- Bauer D.C., Willadsen K., Buske F.A., Lê Cao K.-A., Bailey T.L., Dellaire G., Bodén M. Sorting the nuclear proteome. Bioinformatics. 2011;27:i7–i14. doi: 10.1093/bioinformatics/btr217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieler J., Cannavo R., Gustafson K., Gobet C., Gatfield D., Naef F. Robust synchronization of coupled circadian and cell cycle oscillators in single mammalian cells. Mol. Syst. Biol. 2014;10:739. doi: 10.15252/msb.20145218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder J.X., Pletscher-Frankild S., Tsafou K., Stolte C., O’Donoghue S.I., Schneider R., Jensen L.J. COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database (Oxford) 2014;2014:bau012. doi: 10.1093/database/bau012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozek K., Relógio A., Kielbasa S.M., Heine M., Dame C., Kramer A., Herzel H. Regulation of clock-controlled genes in mammals. PLoS ONE. 2009;4:e4882. doi: 10.1371/journal.pone.0004882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher O., Suppan P. Amitose et fusion nucléaires au cours du rythme circadien. In: von Mayersbach H., editor. The Cellular Aspects of Biorhythms. Springer Berlin Heidelberg; 1967. pp. 124–132. [Google Scholar]
- Bur I.M., Cohen-Solal A.M., Carmignac D., Abecassis P.-Y., Chauvet N., Martin A.O., van der Horst G.T.J., Robinson I.C.A.F., Maurel P., Mollard P., Bonnefont X. The circadian clock components CRY1 and CRY2 are necessary to sustain sex dimorphism in mouse liver metabolism. J. Biol. Chem. 2009;284:9066–9073. doi: 10.1074/jbc.M808360200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S., Goodwin J.G., Chauhan S., Manyam G., Wang J., Kamat A.M., Boyd D.D. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol. Cell. 2013;50:16–28. doi: 10.1016/j.molcel.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.-D., Wen M.-S., Shie S.-S., Lo Y.-L., Wo H.-T., Wang C.-C., Hsieh I.C., Lee T.-H., Wang C.-Y. The circadian rhythm controls telomeres and telomerase activity. Biochem. Biophys. Res. Commun. 2014;451:408–414. doi: 10.1016/j.bbrc.2014.07.138. [DOI] [PubMed] [Google Scholar]
- Chen X., Shi S.-P., Suo S.-B., Xu H.-D., Qiu J.-D. Proteomic analysis and prediction of human phosphorylation sites in subcellular level reveal subcellular specificity. Bioinformatics. 2015;31:194–200. doi: 10.1093/bioinformatics/btu598. [DOI] [PubMed] [Google Scholar]
- Chiang C.-K., Mehta N., Patel A., Zhang P., Ning Z., Mayne J., Sun W.Y.L., Cheng H.-Y.M., Figeys D. The proteomic landscape of the suprachiasmatic nucleus clock reveals large-scale coordination of key biological processes. PLoS Genet. 2014;10:e1004695. doi: 10.1371/journal.pgen.1004695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., Elledge S.J. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A., Hood I.V., Berger J.M. Mechanisms for initiating cellular DNA replication. Annu. Rev. Biochem. 2013;82:25–54. doi: 10.1146/annurev-biochem-052610-094414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane B.R., Young M.W. Interactive features of proteins composing eukaryotic circadian clocks. Annu. Rev. Biochem. 2014;83:191–219. doi: 10.1146/annurev-biochem-060713-035644. [DOI] [PubMed] [Google Scholar]
- Cronshaw J.M., Krutchinsky A.N., Zhang W., Chait B.T., Matunis M.J. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammer E.B., Duong D.M., Diner I., Gearing M., Feng Y., Lah J.J., Levey A.I., Seyfried N.T. Neuron enriched nuclear proteome isolated from human brain. J. Proteome Res. 2013;12:3193–3206. doi: 10.1021/pr400246t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Karbstein K., Woolford J.L., Jr. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu. Rev. Biochem. 2015;84:93–129. doi: 10.1146/annurev-biochem-060614-033917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais C.M., Gilbert R.M., Isermann P., McGregor A.L., te Lindert M., Weigelin B., Davidson P.M., Friedl P., Wolf K., Lammerding J. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty C.J., Kay S.A. Circadian control of global gene expression patterns. Annu. Rev. Genet. 2010;44:419–444. doi: 10.1146/annurev-genet-102209-163432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echave Llanos J.M., de Vaccaro M.E.E., Surur J.M. 24-hour variations in DNA of the liver in young and adult male mice. J. Interdiscipl. Cycle Res. 1970;1:161–171. [Google Scholar]
- Echave Llanos J.M., Aloisso M.D., Souto M., Balduzzi R., Surur J.M. Circadian variations of DNA synthesis, mitotic activity, and cell size of hepatocyte population in young immature male mouse growing liver. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 1971;8:309–317. doi: 10.1007/BF02893540. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. Microtubules in cell migration. Annu. Rev. Cell Dev. Biol. 2013;29:471–499. doi: 10.1146/annurev-cellbio-101011-155711. [DOI] [PubMed] [Google Scholar]
- Fang B., Everett L.J., Jager J., Briggs E., Armour S.M., Feng D., Roy A., Gerhart-Hines Z., Sun Z., Lazar M.A. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 2014;159:1140–1152. doi: 10.1016/j.cell.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feillet C., Krusche P., Tamanini F., Janssens R.C., Downey M.J., Martin P., Teboul M., Saito S., Lévi F.A., Bretschneider T. Phase locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle. Proc. Natl. Acad. Sci. USA. 2014;111:9828–9833. doi: 10.1073/pnas.1320474111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink J.L., Karunaratne S., Mittal A., Gardiner D.M., Hamilton N., Mahony D., Kai C., Suzuki H., Hayashizaki Y., Teasdale R.D. Towards defining the nuclear proteome. Genome Biol. 2008;9:R15. doi: 10.1186/gb-2008-9-1-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster L.J., de Hoog C.L., Zhang Y., Zhang Y., Xie X., Mootha V.K., Mann M. A mammalian organelle map by protein correlation profiling. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Franklin S., Zhang M.J., Chen H., Paulsson A.K., Mitchell-Jordan S.A., Li Y., Ping P., Vondriska T.M. Specialized compartments of cardiac nuclei exhibit distinct proteomic anatomy. Mol. Cell. Proteomics. 2011;10:000703. doi: 10.1074/mcp.M110.000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Pelicano H., Liu J., Huang P., Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- Gachon F. Physiological function of PARbZip circadian clock-controlled transcription factors. Ann. Med. 2007;39:562–571. doi: 10.1080/07853890701491034. [DOI] [PubMed] [Google Scholar]
- Gachon F., Nagoshi E., Brown S.A., Ripperger J., Schibler U. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004;113:103–112. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- Gaidatzis D., Burger L., Stadler M.B. Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat. Biotechnol. 2015;33:722–729. doi: 10.1038/nbt.3269. [DOI] [PubMed] [Google Scholar]
- Geiger T., Velic A., Macek B., Lundberg E., Kampf C., Nagaraj N., Uhlen M., Cox J., Mann M. Initial quantitative proteomic map of 28 mouse tissues using the SILAC mouse. Mol. Cell. Proteomics. 2013;12:1709–1722. doi: 10.1074/mcp.M112.024919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentric G., Desdouets C. Polyploidization in liver tissue. Am. J. Pathol. 2014;184:322–331. doi: 10.1016/j.ajpath.2013.06.035. [DOI] [PubMed] [Google Scholar]
- Gerber A., Esnault C., Aubert G., Treisman R., Pralong F., Schibler U. Blood-borne circadian signal stimulates daily oscillations in actin dynamics and SRF activity. Cell. 2013;152:492–503. doi: 10.1016/j.cell.2012.12.027. [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z., Lazar M.A. Circadian metabolism in the light of evolution. Endocr. Rev. 2015;36:289–304. doi: 10.1210/er.2015-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilardi F., Migliavacca E., Naldi A., Baruchet M., Canella D., Le Martelot G., Guex N., Desvergne B., CycliX Consortium Genome-wide analysis of SREBP1 activity around the clock reveals its combined dependency on nutrient and circadian signals. PLoS Genet. 2014;10:e1004155. doi: 10.1371/journal.pgen.1004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gougelet A., Torre C., Veber P., Sartor C., Bachelot L., Denechaud P.-D., Godard C., Moldes M., Burnol A.-F., Dubuquoy C. T-cell factor 4 and β-catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology. 2014;59:2344–2357. doi: 10.1002/hep.26924. [DOI] [PubMed] [Google Scholar]
- Hendzel M.J., Wei Y., Mancini M.A., Van Hooser A., Ranalli T., Brinkley B.R., Bazett-Jones D.P., Allis C.D. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Heng Y.-W., Koh C.-G. Actin cytoskeleton dynamics and the cell division cycle. Int. J. Biochem. Cell Biol. 2010;42:1622–1633. doi: 10.1016/j.biocel.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Hu J., Rho H.-S., Newman R.H., Zhang J., Zhu H., Qian J. PhosphoNetworks: a database for human phosphorylation networks. Bioinformatics. 2014;30:141–142. doi: 10.1093/bioinformatics/btt627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey S.J., Azimifar S.B., Mann M. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 2015;33:990–995. doi: 10.1038/nbt.3327. [DOI] [PubMed] [Google Scholar]
- Huttlin E.L., Jedrychowski M.P., Elias J.E., Goswami T., Rad R., Beausoleil S.A., Villén J., Haas W., Sowa M.E., Gygi S.P. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao W., Datta J., Lin H.-M., Dundr M., Rane S.G. Nucleocytoplasmic shuttling of the retinoblastoma tumor suppressor protein via Cdk phosphorylation-dependent nuclear export. J. Biol. Chem. 2006;281:38098–38108. doi: 10.1074/jbc.M605271200. [DOI] [PubMed] [Google Scholar]
- Jouffe C., Cretenet G., Symul L., Martin E., Atger F., Naef F., Gachon F. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 2013;11:e1001455. doi: 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T.-H., Reardon J.T., Kemp M., Sancar A. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc. Natl. Acad. Sci. USA. 2009;106:2864–2867. doi: 10.1073/pnas.0812638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T.-H., Lindsey-Boltz L.A., Reardon J.T., Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc. Natl. Acad. Sci. USA. 2010;107:4890–4895. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.-S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislinger T., Cox B., Kannan A., Chung C., Hu P., Ignatchenko A., Scott M.S., Gramolini A.O., Morris Q., Hallett M.T. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- Koike N., Yoo S.-H., Huang H.-C., Kumar V., Lee C., Kim T.-K., Takahashi J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger M., Moser M., Ussar S., Thievessen I., Luber C.A., Forner F., Schmidt S., Zanivan S., Fässler R., Mann M. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- Lamia K.A., Sachdeva U.M., DiTacchio L., Williams E.C., Alvarez J.G., Egan D.F., Vasquez D.S., Juguilon H., Panda S., Shaw R.J. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang F., Busch G.L., Ritter M., Völkl H., Waldegger S., Gulbins E., Häussinger D. Functional significance of cell volume regulatory mechanisms. Physiol. Rev. 1998;78:247–306. doi: 10.1152/physrev.1998.78.1.247. [DOI] [PubMed] [Google Scholar]
- Lapik Y.R., Fernandes C.J., Lau L.F., Pestov D.G. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol. Cell. 2004;15:17–29. doi: 10.1016/j.molcel.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Le Martelot G., Canella D., Symul L., Migliavacca E., Gilardi F., Liechti R., Martin O., Harshman K., Delorenzi M., Desvergne B., CycliX Consortium Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 2012;10:e1001442. doi: 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveille G.A., Chakrabarty K. Diurnal variations in tissue glycogen and liver weight of meal-fed rats. J. Nutr. 1967;93:546–554. doi: 10.1093/jn/93.4.546. [DOI] [PubMed] [Google Scholar]
- Li M., Zhang P. The function of APC/CCdh1 in cell cycle and beyond. Cell Div. 2009;4:2. doi: 10.1186/1747-1028-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundby A., Secher A., Lage K., Nordsborg N.B., Dmytriyev A., Lundby C., Olsen J.V. Quantitative maps of protein phosphorylation sites across 14 different rat organs and tissues. Nat. Commun. 2012;3:876. doi: 10.1038/ncomms1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S., Brodersen D.E., Jensen T.H. Origins and activities of the eukaryotic exosome. J. Cell Sci. 2009;122:1487–1494. doi: 10.1242/jcs.047399. [DOI] [PubMed] [Google Scholar]
- Ma D., Panda S., Lin J.D. Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO J. 2011;30:4642–4651. doi: 10.1038/emboj.2011.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L., Groisman R., Naguibneva I., Robin P., Lorain S., Le Villain J.P., Troalen F., Trouche D., Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- Mahajan K., Mahajan N.P. WEE1 tyrosine kinase, a novel epigenetic modifier. Trends Genet. 2013;29:394–402. doi: 10.1016/j.tig.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B., Wendt S., Vanselow J.T., Wallach T., Reischl S., Oehmke S., Schlosser A., Kramer A. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 2009;23:708–718. doi: 10.1101/gad.512209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhas A., Goulbourne C., Vaux D.J. The nucleoplasmic reticulum: form and function. Trends Cell Biol. 2011;21:362–373. doi: 10.1016/j.tcb.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Martin N.C., McCullough C.T., Bush P.G., Sharp L., Hall A.C., Harrison D.J. Functional analysis of mouse hepatocytes differing in DNA content: volume, receptor expression, and effect of IFNgamma. J. Cell. Physiol. 2002;191:138–144. doi: 10.1002/jcp.10057. [DOI] [PubMed] [Google Scholar]
- Martineau-Pivoteau N., Cussac-Buchdahl C., Chollet P., Rolhion C., Debiton E., Rapp M., Kwiatkowski F., Madelmont J.C., Levi F. Circadian variation in O6-methylguanine-DNA methyltransferase activity in mouse liver. Anticancer Drugs. 1996;7:703–709. doi: 10.1097/00001813-199608000-00012. [DOI] [PubMed] [Google Scholar]
- Matsuo T., Yamaguchi S., Mitsui S., Emi A., Shimoda F., Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- Mauvoisin D., Wang J., Jouffe C., Martin E., Atger F., Waridel P., Quadroni M., Gachon F., Naef F. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc. Natl. Acad. Sci. USA. 2014;111:167–172. doi: 10.1073/pnas.1314066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann K.L., Charette J.M., Vincent N.G., Baserga S.J. A protein interaction map of the LSU processome. Genes Dev. 2015;29:862–875. doi: 10.1101/gad.256370.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melé M., Ferreira P.G., Reverter F., DeLuca D.S., Monlong J., Sammeth M., Young T.R., Goldmann J.M., Pervouchine D.D., Sullivan T.J., GTEx Consortium Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y., Ebato K., Kato H., Arakawa S., Shimizu S., Miyajima A. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr. Biol. 2012;22:1166–1175. doi: 10.1016/j.cub.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Mjelle R., Hegre S.A., Aas P.A., Slupphaug G., Drabløs F., Saetrom P., Krokan H.E. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair (Amst.) 2015;30:53–67. doi: 10.1016/j.dnarep.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Neufeld-Cohen A., Robles M.S., Aviram R., Manella G., Adamovich Y., Ladeuix B., Nir D., Rousso-Noori L., Kuperman Y., Golik M. Circadian control of oscillations in mitochondrial rate-limiting enzymes and nutrient utilization by PERIOD proteins. Proc. Natl. Acad. Sci. USA. 2016;113:E1673–E1682. doi: 10.1073/pnas.1519650113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan T.A., Bassler J., Petfalski E., Tollervey D., Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombo P., Moreno-Villanueva M., Mangerich A. Day and night variations in the repair of ionizing-radiation-induced DNA damage in mouse splenocytes. DNA Repair (Amst.) 2015;28:37–47. doi: 10.1016/j.dnarep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Phipps K.R., Charette J., Baserga S.J. The small subunit processome in ribosome biogenesis—progress and prospects. Wiley Interdiscip. Rev. RNA. 2011;2:1–21. doi: 10.1002/wrna.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M., Gentili M., de Belly H., Thiam H.R., Vargas P., Jimenez A.J., Lautenschlaeger F., Voituriez R., Lennon-Duménil A.M., Manel N., Piel M. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 2016;352:359–362. doi: 10.1126/science.aad7611. [DOI] [PubMed] [Google Scholar]
- Reinke H., Saini C., Fleury-Olela F., Dibner C., Benjamin I.J., Schibler U. Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev. 2008;22:331–345. doi: 10.1101/gad.453808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischl S., Kramer A. Kinases and phosphatases in the mammalian circadian clock. FEBS Lett. 2011;585:1393–1399. doi: 10.1016/j.febslet.2011.02.038. [DOI] [PubMed] [Google Scholar]
- Ren X., Bai X., Zhang X., Li Z., Tang L., Zhao X., Li Z., Ren Y., Wei S., Wang Q. Quantitative nuclear proteomics identifies that miR-137-mediated EZH2 reduction regulates resveratrol-induced apoptosis of neuroblastoma cells. Mol. Cell. Proteomics. 2015;14:316–328. doi: 10.1074/mcp.M114.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey G., Cesbron F., Rougemont J., Reinke H., Brunner M., Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles M.S., Cox J., Mann M. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 2014;10:e1004047. doi: 10.1371/journal.pgen.1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouyer F., Rachidi M., Pikielny C., Rosbash M. A new gene encoding a putative transcription factor regulated by the Drosophila circadian clock. EMBO J. 1997;16:3944–3954. doi: 10.1093/emboj/16.13.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N., Spahr C.S., Patterson S.D., Bubulya P., Neuwald A.F., Spector D.L. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell. 2004;15:3876–3890. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer E.C., Florens L., Guan T., Yates J.R., 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- Schliess F., Häussinger D. Call volume and insulin signaling. Int. Rev. Cytol. 2003;225:187–228. doi: 10.1016/s0074-7696(05)25005-2. [DOI] [PubMed] [Google Scholar]
- Settembre C., De Cegli R., Mansueto G., Saha P.K., Vetrini F., Visvikis O., Huynh T., Carissimo A., Palmer D., Klisch T.J. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souto M., Llanos J.M. The circadian optimal time for hepatectomy in the study of liver regeneration. Chronobiol. Int. 1985;2:169–175. doi: 10.3109/07420528509055556. [DOI] [PubMed] [Google Scholar]
- Stoll B., Gerok W., Lang F., Häussinger D. Liver cell volume and protein synthesis. Biochem. J. 1992;287:217–222. doi: 10.1042/bj2870217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru T., Hirayama J., Isojima Y., Nagai K., Norioka S., Takamatsu K., Sassone-Corsi P. CK2α phosphorylates BMAL1 to regulate the mammalian clock. Nat. Struct. Mol. Biol. 2009;16:446–448. doi: 10.1038/nsmb.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa M., Bommakanti A., Shamsuzzaman M., Gregory B., Samsel L., Zengel J.M., Lindahl L. Repressed synthesis of ribosomal proteins generates protein-specific cell cycle and morphological phenotypes. Mol. Biol. Cell. 2013;24:3620–3633. doi: 10.1091/mbc.E13-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoropoulos P.A., Stournaras C., Stoll B., Markogiannakis E., Lang F., Gravanis A., Häussinger D. Hepatocyte swelling leads to rapid decrease of the G-/total actin ratio and increases actin mRNA levels. FEBS Lett. 1992;311:241–245. doi: 10.1016/0014-5793(92)81111-x. [DOI] [PubMed] [Google Scholar]
- Uchida C., Miwa S., Kitagawa K., Hattori T., Isobe T., Otani S., Oda T., Sugimura H., Kamijo T., Ookawa K. Enhanced Mdm2 activity inhibits pRB function via ubiquitin-dependent degradation. EMBO J. 2005;24:160–169. doi: 10.1038/sj.emboj.7600486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama Y., Asari A. A morphometric study of the variations in subcellular structures of rat hepatocytes during 24 hours. Cell Tissue Res. 1984;236:305–315. doi: 10.1007/BF00214231. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst G.T.J., Muijtjens M., Kobayashi K., Takano R., Kanno S., Takao M., de Wit J., Verkerk A., Eker A.P.M., van Leenen D. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Vermeulen K., Van Bockstaele D.R., Berneman Z.N. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003;36:131–149. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers C., Schmitz R.J., Nathanson J., Yeo G., Ecker J.R., Panda S. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 2012;16:833–845. doi: 10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Warnke S., Kemmler S., Hames R.S., Tsai H.-L., Hoffmann-Rohrer U., Fry A.M., Hoffmann I. Polo-like kinase-2 is required for centriole duplication in mammalian cells. Curr. Biol. 2004;14:1200–1207. doi: 10.1016/j.cub.2004.06.059. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Fang C., Bao H., Fan H., Shen H., Yang P. Nuclear proteome profile of C57BL/6J mouse liver. Sci. China Life Sci. 2013;56:513–523. doi: 10.1007/s11427-013-4488-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figure 1. Sheet1: Relative abundance (normalized L/H ratios from the MaxQuant analysis) for all distinct 4,820 proteins identified by SILAC MS in the nucleus of mouse liver at different time points and in all replicates under LD conditions. Sheet2: Raw data with peptide counts from MaxQuant analysis at different time points in all replicates. Sheet3: Description of the column headers for Sheet1. Sheet4: Description of the column headers for Sheet2.
Related to Figure 2. Sheet 1. Biochemical functions and cellular localization of these rhythmic nuclear proteins. Sheet 2. Relative abundance of rhythmic nuclear proteins and their corresponding phases and amplitudes in wild-type mice and their correlation in Cry1/2 DKO. Sheet 3. Corresponding mRNA expression measured by total RNA-seq. Sheet 4. Corresponding previously published proteomic data in whole-liver extracts extracts (Mauvoisin et al., 2014). Sheet 5. Description of the column headers for Sheets1–4.
Related to Figure 3. Sheet 1. Phase, amplitudes, and related functions of the identified rhythmic complexes. Sheet 2. All known complexes with at least two detected subunits in nuclear proteome and corresponding SVD analysis. Sheet 3. Description of the column headers for Sheets1 and 2.
Related to Figure 4. Sheet 1. Rhythmic nuclear proteins with identified phases and amplitudes involved in different steps of ribosome biogenesis. Sheet 2. Rhythmic nuclear proteins with identified phases and amplitudes involved in different steps of DNA repair process. Sheet 3. Description of the column headers for Sheets1 and 2.
Related to Figure 5. Sheet 1. Complete diurnal phosphoproteome dataset and corresponding rhythmicity analysis. Sheet 2. Kinase motifs with rhythmic activities inferred using linear model with regularization. Sheet 3. Description of the column headers for Sheets1 and 2.
Related to Figure 6. Sheet 1. Rhythmic TF quantified in nuclear proteome and/or nuclear phosphoproteome. Sheet 2. Rhythmic co-regulators quantified in nuclear proteome. Sheet 3. TF motifs with rhythmic activities inferred from DNase hypersensitive sites (DHSs) and pre-mRNA datasets. Sheet 4. Comparison between rhythmic TFs and their corresponding motif activity. Sheet 5. Description of the column headers for Sheets1–4.
Related to Experimental Procedures