Abstract
Spinocerebellar ataxia type 6 (SCA6) is a dominantly inherited neurodegenerative disease characterized by slowly progressive ataxia and Purkinje cell degeneration. SCA6 is caused by a polyglutamine repeat expansion within a second CACNA1A gene product, α1ACT. α1ACT expression is under the control of an internal ribosomal entry site (IRES) present within the CACNA1A coding region. Whereas SCA6 allele knock-in mice show indistinguishable phenotypes from wild-type littermates, expression of SCA6-associated α1ACT (α1ACTSCA6) driven by a Purkinje cell–specific promoter in mice produces slowly progressive ataxia and cerebellar atrophy. We developed an early-onset SCA6 mouse model using an adeno-associated virus (AAV)–based gene delivery system to ectopically express CACNA1A IRES–driven α1ACTSCA6 to test the potential of CACNA1A IRES–targeting therapies. Mice expressing AAV9-mediated CACNA1A IRES–driven α1ACTSCA6 exhibited early-onset ataxia, motor deficits, and Purkinje cell degeneration. We identified miR-3191-5p as a microRNA (miRNA) that targeted CACNA1A IRES and preferentially inhibited the CACNA1A IRES–driven translation of α1ACT in an Argonaute 4 (Ago4)–dependent manner. We found that eukaryotic initiation factors (eIFs), eIF4AII and eIF4GII, interacted with the CACNA1A IRES to enhance α1ACT translation. Ago4-bound miR-3191-5p blocked the interaction of eIF4AII and eIF4GII with the CACNA1A IRES, attenuating IRES-driven α1ACT translation. Furthermore, AAV9-mediated delivery of miR-3191-5p protected mice from the ataxia, motor deficits, and Purkinje cell degeneration caused by CACNA1A IRES–driven α1ACTSCA6. We have established proof of principle that viral delivery of an miRNA can rescue a disease phenotype through modulation of cellular IRES activity in a mouse model.
INTRODUCTION
Spinocerebellar ataxias (SCAs) are a genetically heterogeneous group of dominantly inherited neurodegenerative diseases characterized by progressive ataxia and Purkinje cell degeneration (1–3). To date, more than 30 SCAs have been characterized, each being associated with distinct genes and mutations and therefore requiring individual therapeutic approaches (2, 3). The lack of efficacious therapeutics and the large number of genetic events that result in SCAs have highlighted the need for effective preclinical models to identify and test “druggable” targets.
SCA type 6 (SCA6) is one of the most common forms of autosomal dominant SCAs, representing 10 to 20% of patients with dominantly inherited ataxia. It has an incidence of about 5/100,000 persons (2–8). Patients with SCA6 develop slowly progressive cerebellar ataxia with extensive selective Purkinje cell degeneration, usually beginning at 40 to 50 years of age (2–8). SCA6 is caused by an expanded CAG repeat in the CACNA1A gene, which results in an expanded polyglutamine (polyQ) tract. Previous studies unexpectedly found that the expanded polyQ tract does not affect the function or kinetics of the α1A (Cav2.1, P/Q-type) voltage-gated Ca2+ channel subunit, a gene product of the full-length CACNA1A gene (9, 10). Additionally, SCA6 allele knock-in mice were indistinguishable from wild-type littermates, even in old age (9, 10).
We recently discovered that the CACNA1A gene is bicistronic, that is, it encodes both the full-length α1A subunit and a newly recognized transcription factor, α1ACT, consisting of 547 amino acids of the C terminus encoded within a separate open reading frame (ORF) of the same mRNA. The second cistron is translated from a newly identified internal ribosomal entry site (IRES) upstream of the second ORF. We have also characterized the cellular physiological and pathological properties of both wild-type α1ACT and an expanded polyQ tract containing α1ACT in vivo, showing that the expanded polyQ tract in α1ACT results in the SCA6 phenotype. Additionally, we showed that elimination of the portion of the IRES sequence from the human mRNA encoding SCA6-associated α1A selectively eliminated the expression of the SCA6-associated α1ACT (α1ACTSCA6) fragment and was protective in a cell culture model of α1ACTSCA6 toxicity (11). Because the complete silencing of CACNA1A gene expression would be lethal (11, 12), a more suitable therapeutic approach for SCA6 would be to selectively eliminate expression of α1ACT while sparing α1A expression. We reasoned that modulating the expression of microRNAs (miRNAs) targeting the CACNA1A IRES could offer a new therapeutic approach for treating SCA6 through regulation of IRES-dependent α1ACTSCA6 translation. On the basis of this hypothesis, we developed a mouse model in which α1ACTSCA6 was expressed in a CACNA1A IRES–dependent manner.
RESULTS
Somatic gene transfer of CACNA1A IRES–driven α1ACTSCA6 causes Purkinje cell degeneration in mice
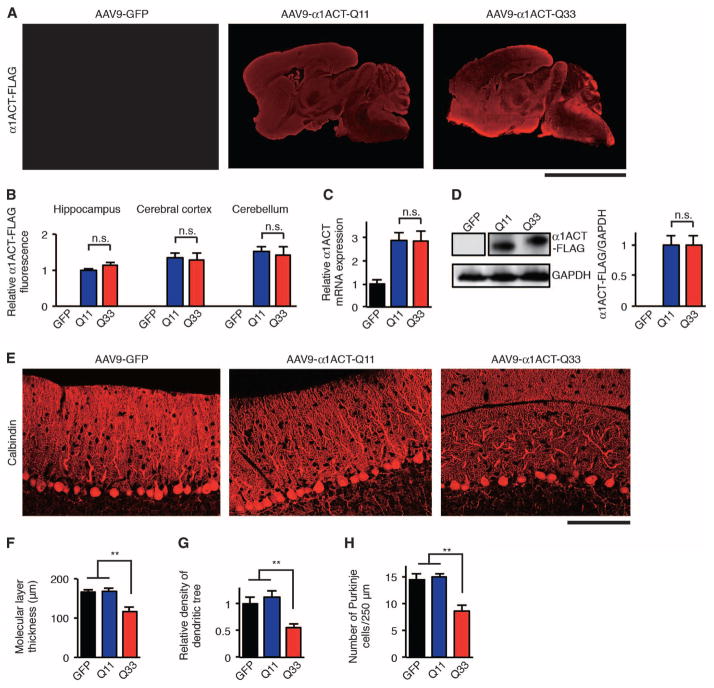
We established an in vivo adeno-associated virus type 9 (AAV9) delivery system using the constructs of α1ACT transgenes tagged with a C-terminal FLAG epitope, expressed under the control of CACNA1A IRES (IRES-α1ACT-Q11 and IRES-α1ACT-Q33, fig. S1A; AAV9-α1ACT-Q11 and AAV9-α1ACT-Q33, fig. S1B). We also prepared a control AAV9 vector expressing green fluorescent protein (GFP) (AAV9-GFP). A viral load of 1010 vector genomes (vg) of each of these constructs was directly injected into the right lateral ventricle of neonatal wild-type C57/BL6J mice at postnatal day 1. In vivo delivery of AAV9 by intraventricular injection resulted in more than 75% of Purkinje cells being transduced with the AAV9 vector (fig. S1, C and D). Four weeks after AAV9 vector injection, we sacrificed the mice and found widespread expression of α1ACT-Q11-FLAG or α1ACT-Q33-FLAG that was predominantly observed in the cerebral cortex and cerebellum of wild-type mice injected with AAV9-α1ACT-Q11 (AAV9–α1ACT-Q11 mice) or AAV9-α1ACT-Q33 (AAV9-α1ACT-Q33 mice) (Fig. 1, A and B). Quantitative real-time polymerase chain reaction (qRT-PCR) of total RNA from the cerebellum of AAV9-injected mice revealed that α1ACT mRNA in the cerebellum of AAV9-α1ACT-Q11 and AAV9-α1ACT-Q33 mice showed a greater than 2.5-fold up-regulation compared to mouse endogenous CACNA1A mRNA in the cerebellum of wild-type mice injected with AAV9-GFP (AAV9-GFP mice). Expression of IRES-α1ACT-Q11 mRNA in AAV9-α1ACT-Q11 mice was comparable to that of IRES-α1ACT-Q33 mRNA in AAV9-α1ACT-Q33 mice (Fig. 1C). CACNA1A IRES–driven α1ACT protein expression in the cerebellum was also comparable between AAV9-α1ACT-Q11 and AAV9-α1ACT-Q33 mice (Fig. 1D).
Fig. 1. Somatic gene transfer of CACNA1A IRES–driven α1ACTSCA6 causes Purkinje cell degeneration in mice.
(A) Representative immunofluorescence images of AAV9-injected mouse brain and cerebellum from 4-week-old mice stained with FLAG-specific antibodies and fluorescent secondary antibody. Scale bar, 5 mm. (B) Relative α1ACT-FLAG immunofluorescence intensities in AAV9-injected mouse hippocampus, cerebral cortex, and cerebellum (n = 6). Relative α1ACT-FLAG immunofluorescence is expressed as signal intensities per unit area quantified by the National Institutes of Health (NIH) ImageJ software. n.s., not significant. (C) Relative α1ACT mRNA expression in the cerebellum of AAV9-injected mice at 4 weeks of age (n = 4). (D) Western blot and densitometric analyses showing α1ACT-Q11-FLAG and α1ACT-Q33-FLAG expression in the cerebellum of AAV9-injected mice at 4 weeks of age (n = 4). (E) Representative immunofluorescence images of AAV9-injected mouse cerebellum from 4-week-old mice stained with calbindin-specific antibodies and fluorescent secondary antibody. Scale bar, 100 μm. (F to H) Molecular layer thickness (F), density of dendritic tree (G), and Purkinje cell count of AAV9-injected mouse cerebellum from 4-week-old mice (n = 6) (H). GFP, AAV9-GFP mice; Q11, AAV9-α1ACT-Q11 mice; Q33, AAV9-α1ACT-Q33 mice. All data represent means ± SEM. **P < 0.01. Student’s t test in (B) to (D). One-way analysis of variance (ANOVA) in (F) to (H).
We found that, compared with AAV9-α1ACT-Q11 mice, AAV9-α1ACT-Q33 mice showed a significant decrease in the molecular layer thickness (P < 0.01) and the density of the Purkinje cell dendritic tree (P < 0.01) (Fig. 1, E to G). We also found that AAV9-α1ACT-Q33 caused a 50% loss of Purkinje cells in the cerebellum (Fig. 1H) but no obvious pathological changes in the cerebral cortex and hippocampus (fig. S2, A and B). These pathological features in the cerebellum of AAV9-α1ACT-Q33 mice resembled those of Purkinje cell degeneration in patients with SCA6 (13–15).
CACNA1A IRES–driven α1ACTSCA6 causes an early-onset ataxia and motor deficits in mice
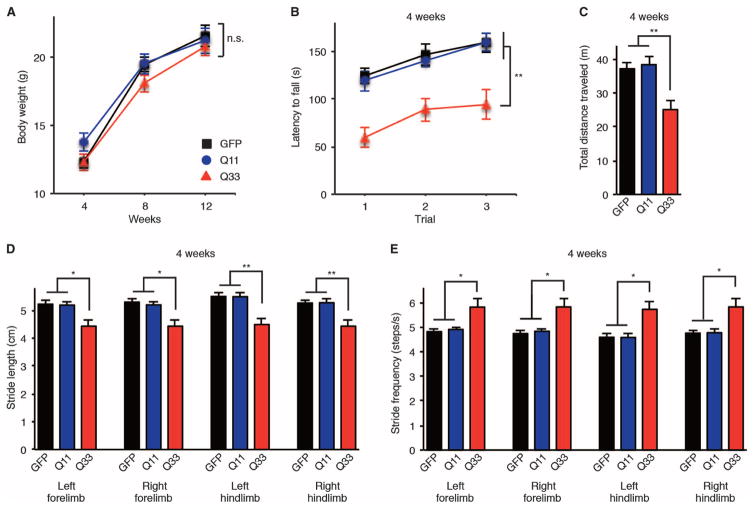
To assess the behavioral phenotype of AAV9-α1ACT-Q33 mice, we used a rotarod performance test, an open-field assay, and a gait stability assessment using a video-assisted computerized treadmill (DigiGait) for gait analysis. AAV9-α1ACT-Q33 mice grew at roughly the same rate and had a comparable body weight to AAV9-α1ACT-Q11 and AAV9-GFP mice (Fig. 2A). However, AAV9-α1ACT-Q33 mice showed defects in motor functions as assessed by the rotarod test (Fig. 2B), activity in the open-field assay (Fig. 2C), and gait stability by the gait analysis assay (Fig. 2, D and E). AAV9-α1ACT-Q33 caused impaired performance on the accelerating rotarod test in mice beginning at 4 weeks of age (P < 0.01) (Fig. 2B). Ambulatory distance in the open-field test was significantly shorter in AAV9-α1ACT-Q33 mice than in AAV9-α1ACT-Q11 and AAV9-GFP mice (P < 0.01) (Fig. 2C). DigiGait analyses revealed that AAV9-α1ACT-Q33 mice exhibited shorter stride length (P < 0.05) (Fig. 2D) and greater stride frequencies (P < 0.05) (Fig. 2E) in all four limbs from 4 weeks of age, indicating early-onset instability during walking. AAV9-α1ACT-Q33 mice often showed weaving from side to side on a treadmill, whereas AAV9-α1ACT-Q11 and AAV9-GFP mice typically walked straight (movies S1 to S3). Furthermore, the deficit in behavioral phenotypes of AAV9–α1ACT-Q33 mice and continuous CACNA1A IRES–driven α1ACT expression in the cerebellum were observed over a long-term follow-up of 30 weeks (fig. S3, A to G). These findings indicate that AAV9-mediated somatic gene transfer of α1ACTSCA6 under the control of CACNA1A IRES caused severe ataxia in mice at much earlier ages and more reproducibly than the Purkinje cell–specific promoter-driven transgenic SCA6 mouse model we previously developed (11).
Fig. 2. CACNA1A IRES–driven α1ACTSCA6 causes an early-onset ataxia and motor deficits in mice.
(A to E) Clinical features of AAV9-injected mice. Body weight (A), rotarod test (B), and open-field assay (C) at 4 weeks of age. Stride length (D) and stride frequencies (E) assessed by DigiGait analysis at 4 weeks of age (n = 12, 6 male and 6 female mice per group). GFP, AAV9-GFP mice; Q11, AAV9-α1ACT-Q11 mice; Q33, AAV9-α1ACT-Q33 mice. All data represent means ± SEM. *P < 0.05, **P < 0.01. Two-way ANOVA in (A) and (B). One-way ANOVA in (C) to (E).
miR-3191-5p inhibits the CACNA1A IRES–driven translation of α1ACT while sparing α1A and CACNA1A mRNA expression
To develop a CACNA1A IRES–directed therapeutic approach for SCA6, we used an miRNA-mediated approach. miRNAs have been increasingly recognized to play a role in the regulation of gene expression, in many cases by both translational repression and mRNA destabilization (16, 17). To date, miRNAs have not been used to preferentially regulate the translation of disease genes driven by a cellular IRES. Although prevailing evidence has pointed to the role of natural miRNAs in gene silencing and translational repression through binding to 3′ untranslated regions (UTRs), or rarely 5′UTR, of targeted mRNAs (16–21), predicted miRNA binding sites are found throughout the genome including in both coding and noncoding regions (22, 23).
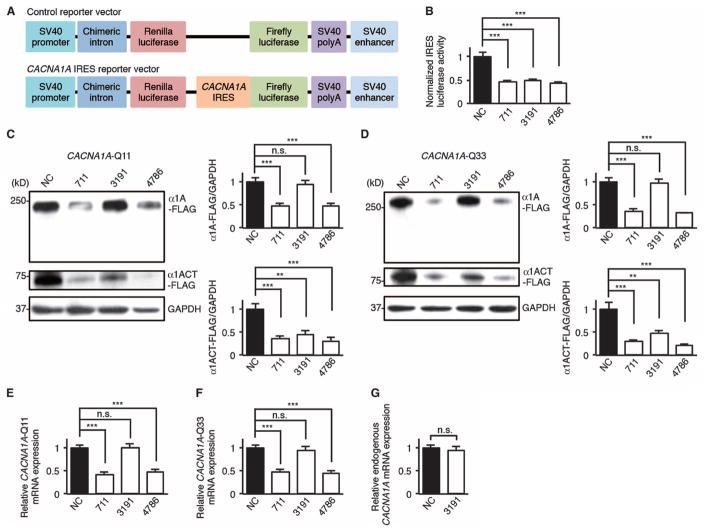
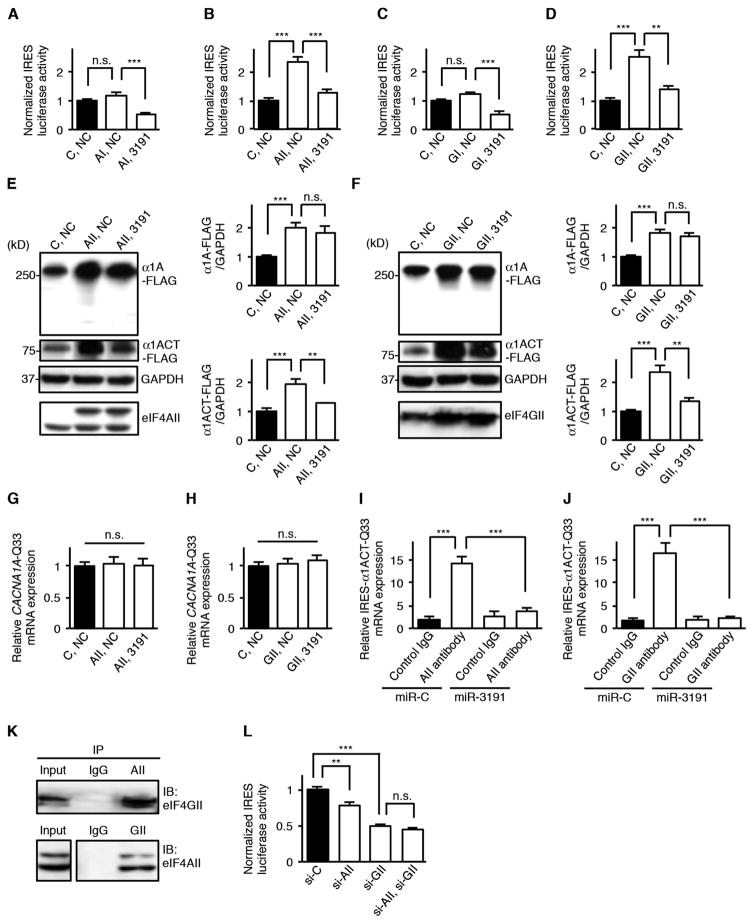
We used the miRNA_Targets program (22) to predict that miRNAs—miR-711 (accession number, MIMAT0012734), miR-3191-5p (accession number, MIMAT0022732), and miR-4786-3p (accession number, MIMAT0019955)—bind to sequences within the stem-loop structure of CACNA1A IRES (fig. S4, A to D) (24). To examine the effects of these miRNAs on CACNA1A IRES–driven translation, we cotransfected these miRNAs with either a bicistronic reporter vector bearing CACNA1A IRES or a control vector into human embryonic kidney (HEK) 293 cells (Fig. 3A). Dual-luciferase assays revealed that miR-711, miR-3191-5p, and miR-4786-3p down-regulated CACNA1A IRES–driven luciferase activities compared to a negative control miRNA (Fig. 3B). We tested the effects of these miRNAs on CACNA1A-encoded C-terminal FLAG–tagged peptides (α1A-FLAG and α1ACT-FLAG) with the normal (Q11) or pathological (Q33) polyQ tract in transfected HEK293 cells. Although miR-711 and miR-4786-3p decreased full-length α1A-FLAG and α1ACT-FLAG expression, miR-3191-5p down-regulated α1ACT-FLAG expression but spared α1A-FLAG expression (Fig. 3, C and D). This was the case with α1A-FLAG and α1ACT-FLAG in mice with the normal (Q11) or pathological (Q33) polyQ tract. qRT-PCR studies of total RNA showed that miR-711 and miR-4786-3p decreased CACNA1A mRNA expression relative to a negative control miRNA, but miR-3191-5p did not affect either CACNA1A-Q11 or CACNA1A-Q33 mRNA expression (Fig. 3, E and F). We also found that miR-3191-5p did not affect endogenous CACNA1A mRNA expression in HEK293 cells (Fig. 3G).
Fig. 3. miR-3191-5p inhibits the CACNA1A IRES–driven translation of α1ACT while sparing α1A and CACNA1A mRNA expression in HEK293 cells.
(A) Schematic representation of a bicistronic control reporter vector and a bicistronic CACNA1A IRES reporter vector. SV40, simian virus 40; polyA, polyadenylation signal sequence. (B) Relative dual-luciferase CACNA1A IRES reporter activities in HEK293 cells treated with miR-711, miR-3191-5p, miR-4786-3p, or an miRNA negative control (NC) (n = 6). The ratio of firefly luciferase to Renilla luciferase activities of a bicistronic CACNA1A IRES reporter vector was normalized to that of a bicistronic control reporter vector. (C and D) Western blot and densitometric analyses showing α1A-FLAG and α1ACT-FLAG expression in HEK293 cells treated with three miRNAs or NC (n = 6). (C) CACNA1A-Q11. (D) CACNA1A-Q33. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (E and F) Relative CACNA1A-Q11 (E) and CACNA1A-Q33 (F) mRNA expression in HEK293 cells treated with three miRNAs or NC (n = 6). (G) Relative endogenous CACNA1A mRNA expression in HEK293 cells treated with miR-3191-5p or NC (n = 6). All data represent means ± SEM. **P < 0.01, ***P < 0.001. Student’s t test in (B) to (G).
To determine whether miR-3191-5p interacts with regions within the CACNA1A mRNA other than the CACNA1A IRES, we prepared mutated CACNA1A IRES templates, resistant to the binding of miR-3191-5p (CACNA1A IRESmut) (fig. S5, A to C). CACNA1A IRESmut functioned normally both in the CACNA1A IRESmut dual-luciferase assays and in the α1A-FLAG– and α1ACT-FLAG–expressing HEK293 cells, but inhibition of expression by miR-3191-5p was prevented (fig. S5, D and E). These results suggest that miR-3191-5p interacts with CACNA1A IRES only through its predicted binding site and inhibits the CACNA1A IRES–driven translation of α1ACT while sparing α1A expression and CACNA1A mRNA expression.
Ago4 is required for miR-3191-5p–mediated inhibition of α1ACT translation
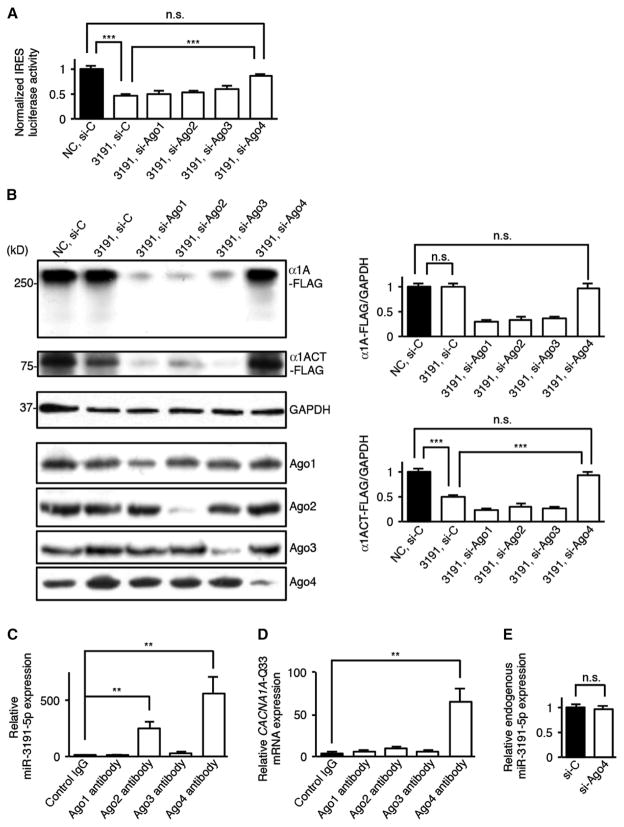
miRNAs generally act on targeted mRNAs in collaboration with the miRNA-induced silencing complex (miRISC)–guided Argonaute (Ago) proteins (25–29). To examine the role of Ago proteins on the inhibitory effects of miR-3191-5p, we cotransfected HEK293 cells with miR-3191-5p and small interfering RNAs (siRNAs) targeting Ago1 to Ago4 carried on either a bicistronic reporter vector bearing the CACNA1A IRES or a control vector. Dual-luciferase assays revealed that knockdown of Ago4, but not Ago1 to Ago3, reversed the silencing effects of miR-3191-5p on the CACNA1A IRES–driven luciferase activities (Fig. 4A). Western blot analyses showed that silencing of Ago4, but not Ago1 to Ago3, also prevented the down-regulation of α1ACT-FLAG expression by miR-3191-5p, suggesting that Ago4 is required for miR-3191-5p–mediated inhibition of α1ACT translation (Fig. 4B).
Fig. 4. Ago4 is required for miR-3191-5p–mediated inhibition of CACNA1A IRES–driven α1ACT translation.
(A) Relative dual-luciferase CACNA1A IRES reporter activities in HEK293 cells treated with si-Ago1, si-Ago2, si-Ago3, si-Ago4, or a scrambled control siRNA (si-C) in the presence of miR-3191-5p or an miRNA NC (n = 6). The ratio of firefly luciferase to Renilla luciferase activities of a bicistronic CACNA1A IRES reporter vector was normalized to that of a bicistronic control reporter vector. (B) Western blot and densitometric analyses showing α1A-Q33-FLAG and α1ACT-Q33-FLAG expression in HEK293 cells treated with four siRNAs or si-C in the presence of miR-3191-5p or NC (n = 6). (C and D) Amount of miR-3191-5p (C) and CACNA1A-Q33 (D) mRNA binding to Ago1 to Ago4 that was immunoprecipitated using antibodies against Ago1 to Ago4 or a control immunoglobulin G (IgG) (n = 6). (E) Relative amount of endogenous miR-3191-5p in HEK293 cells treated with si-Ago4 or si-C (n = 6). All data represent means ± SEM. **P < 0.01, ***P < 0.001. Student’s t test in (A) to (E).
To determine whether Ago4 directly associates with both miR-3191-5p and CACNA1A mRNA or whether Ago4 indirectly mediates the silencing effects of miR-3191-5p, we performed RNA immunoprecipitation–coupled qRT-PCR studies using antibodies specific to Ago1 to Ago4. We found that Ago2- and Ago4-specific antibodies preferentially precipitated miR-3191-5p to a greater extent than did control IgG. Only Ago4-specific antibodies specifically precipitated CACNA1A mRNA, indicating that Ago4 selectively bound to both miR-3191-5p and CACNA1A mRNA (Fig. 4, C and D). Ago1- and Ago3-specific antibodies had no effect. We also confirmed that knockdown of Ago4 did not affect miR-3191-5p expression in HEK293 cells (Fig. 4E). These results suggest that miR-3191-5p bound to Ago4 directly interacts with the CACNA1A IRES and selectively inhibits the CACNA1A IRES–driven α1ACT translation.
miR-3191-5p inhibits the translational initiation of CACNA1A IRES–driven α1ACT by eIF4AII and eIF4GII
Because both eukaryotic initiation factors (eIFs) eIF4A and eIF4G have been previously shown to be involved in IRES-dependent translation in several types of viral IRESs (17), we examined the effects of these on CACNA1A IRES–driven α1ACT translation. We found that overexpression of either eIF4AII or eIF4GII, but not eIF4AI or eIF4GI, increased the CACNA1A IRES–driven luciferase activities and that the effects of eIF4AII and eIF4GII were blocked by cotransfection with miR-3191-5p (Fig. 5, A to D). Western blot analyses showed that overexpression of either eIF4AII or eIF4GII increased both α1A-FLAG and α1ACT-FLAG expression and that miR-3191-5preversedthe up-regulating effects of eIF4AII and eIF4GII on α1ACT-FLAG expression without affecting eIF4AII and eIF4GII expression (Fig. 5, E and F). We also found that overexpression of either eIF4AII or eIF4GII in the presence or absence of miR-3191-5p did not affect CACNA1A mRNA expression (Fig. 5, G and H). When we used CACNA1A IRESmut templates, the miR-3191-5p–mediated inhibition of the up-regulating effects of eIF4AII and eIF4GII on both CACNA1A IRESmut dual-luciferase activities and α1ACT-FLAG expression were prevented, indicating that the effects of miR-3191-5p depended on its interactions with CACNA1A IRES (fig. S6, A to D).
Fig. 5. miR-3191-5p inhibits the translational initiation of CACNA1A IRES–driven α1ACT by eIF4AII and eIF4GII.
(A to D) Relative dual-luciferase CACNA1A IRES reporter activities in HEK293 cells treated by overexpression of either eIF4AI (AI) (A), eIF4AII (AII) (B), eIF4GI (GI) (C), eIF4GII (GII) (D), or a control vector (C) in the presence of miR-3191-5p or an miRNA NC (n = 6). The ratio of firefly luciferase to Renilla luciferase activities of a bicistronic CACNA1A IRES reporter vector was normalized to that of a bicistronic control reporter vector. (E and F) Western blot and densitometric analyses showing α1A-Q33-FLAG and α1ACT-Q33-FLAG expression in HEK293 cells treated by overexpression of either AII (E), GII (F), or C in the presence of miR-3191-5p or NC (n = 6). (G and H) Relative CACNA1A-Q33 mRNA expression in HEK293 cells treated by overexpression of either AII (G), GII (H), or C in the presence of miR-3191-5p or NC (n = 6). (I and J) Amount of IRES-α1ACT-Q33 mRNA binding to AII (I), GII (J), or a control IgG in the presence of vectors expressing miR-3191-5p (miR-3191) or an miRNA control (miR-C) that was immunoprecipitated using antibodies against AII, GII, or a control IgG (n = 6). (K) Immunoprecipitation (IP) from HEK293 cells transfected with AII- and GII-expressing vectors using an antibody against AII, GII, or a control IgG. Recovered proteins were analyzed using Western blot. Input, 10%. (L) Relative dual-luciferase CACNA1A IRES reporter activities in HEK293 cells treated with si-AII, si-GII, or a scrambled si-C (n = 6). The ratio of firefly luciferase to Renilla luciferase activities of a bicistronic CACNA1A IRES reporter vector was normalized to that of a bicistronic control reporter vector. All data represent means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. Student’s t test in (A) to (F), (I), (J), and (L). One-way ANOVA in (G) and (H).
Because both eIF4AII and eIF4GII affected not only CACNA1A IRES–driven translation of α1ACT but also cap-dependent translation of full-length α1A (Fig. 5, E and F), we used IRES-α1ACT vectors (fig. S1A) to test the binding affinities of eIF4AII and eIF4GII to CACNA1A IRES. RNA immunoprecipitation–coupled qRT-PCR studies revealed that both eIF4AII- and eIF4GII-specific antibodies precipitated IRES-α1ACT mRNAs to a greater extent than did control IgG, and that binding affinities between IRES-α1ACT mRNAs and both eIF4AII and eIF4GII were decreased by treatment with miR-3191-5p (Fig. 5, I and J). Coimmunoprecipitation studies revealed that eIF4AII associated with eIF4GII (Fig. 5K). We also found that knockdown of eIF4GII had a greater effect on the down-regulation of CACNA1A IRES–driven α1ACT translation than did knockdown of eIF4AII (Fig. 5L). On the basis of these findings, we concluded that the complex of eIF4AII and eIF4GII directly acts on the CACNA1A IRES to enhance IRES-driven α1ACT translation (fig. S6E). miR-3191-5p bound to Ago4 selectively inhibits the translational initiation of CACNA1A IRES–driven α1ACT by eIF4AII and eIF4GII (fig. S6F).
miR-3191-5p prevents Purkinje cell degeneration in mice caused by CACNA1A IRES–driven α1ACTSCA6
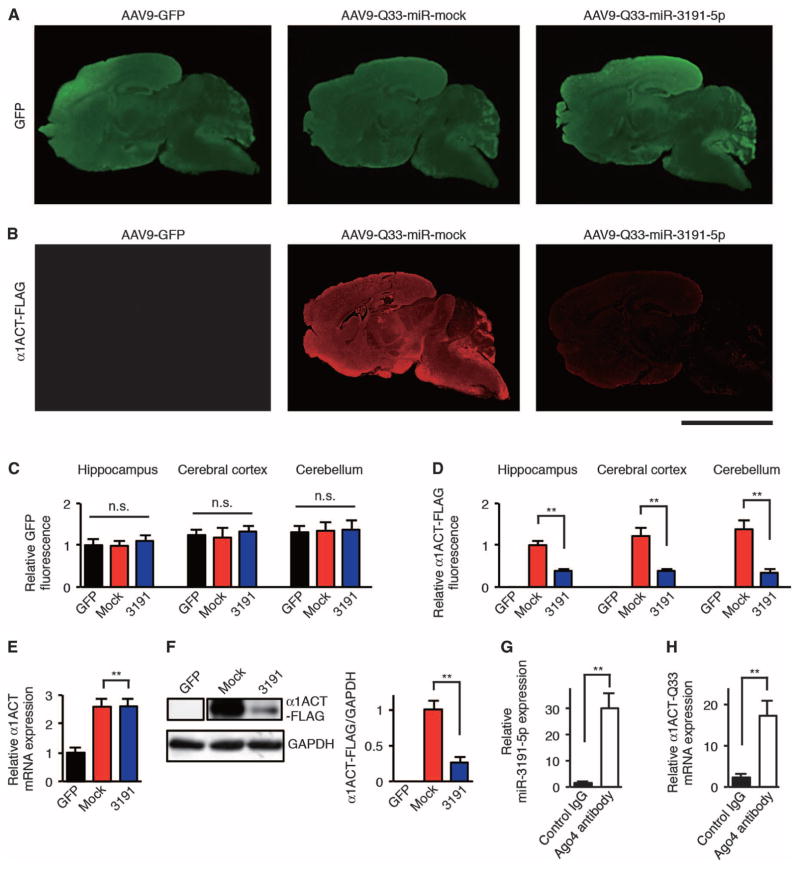
To examine the therapeutic effects of miR-3191-5p–mediated reduction of CACNA1A IRES–driven α1ACTSCA6 translation on the disease phenotype of SCA6 mice, we constructed an AAV9 vector that allowed for the simultaneous expression of GFP and either miR-3191-5p (AAV9–miR-3191-5p) or a nonspecific mock miRNA (AAV9–miR-mock) (fig. S7A). We then co-injected AAV9-α1ACT-Q33 with either AAV9–miR-3191-5p (AAV9-Q33–miR-3191-5p) or AAV9–miR-mock (AAV9–Q33–miR-mock) into the right lateral ventricle of neonatal wild-type C57/BL6J mice at postnatal day 1. Four weeks after AAV9 vector injection, we sacrificed AAV9-injected mice and found widespread transduction of miR-mock and miR-3191-5p throughout the brain and cerebellum (Fig. 6, A and C). We also found a high efficiency (>80%) of co-transduction of AAV9-α1ACT-Q33 and either AAV9–miR-mock or AAV9–miR-3191-5p in the cerebellum of AAV9-injected mice (fig. S7, B and C). There was a decrease in CACNA1A IRES–driven α1ACT-Q33-FLAG protein expression in the brain and cerebellum of AAV9–Q33–miR-3191-5p mice as compared to AAV9–Q33–miR-mock mice (Fig. 6, B and D). Whereas IRES-α1ACT-Q33 mRNA expression in the cerebellum of AAV9–Q33–miR-3191-5p mice was comparable to that for AAV9–Q33–miR-mock mice (Fig. 6E), Western blot analyses supported the silencing effect of miR-3191-5p on CACNA1A IRES–driven α1ACT-Q33-FLAG protein expression in the cerebellum of AAV9–Q33–miR-3191-5p mice compared to AAV9–Q33–miR-mock mice (Fig. 6F). Using RNA immunoprecipitation–coupled qRT-PCR studies, we also confirmed that Ago4 bound to both miR-3191-5p and IRES-α1ACT-Q33 mRNA in the cerebellum of AAV9–Q33–miR-3191-5pmice(Fig. 6, G and H).
Fig. 6. AAV9-mediated therapeutic delivery of miR-3191-5p blocks CACNA1A IRES–driven α1ACT translation in mice.
(A and B) Representative immunofluorescence images of AAV9-injected mouse brain and cerebellum from 4-week-old mice stained with GFP-specific (A) and FLAG-specific (B) antibodies and fluorescent secondary antibody. Scale bar, 5 mm. (C and D) Relative GFP (C) and α1ACT-FLAG (D) immunofluorescence intensities in AAV9-injected mouse hippocampus, cerebral cortex, and cerebellum (n = 6). Relative GFP and relative FLAG immunofluorescence are expressed as signal intensities per unit area quantified by NIH ImageJ software. (E) Relative α1ACT mRNA expression in the cerebellum of AAV9-injected mice at 4 weeks of age (n = 4). (F) Western blot and densitometric analyses showing α1ACT-Q33-FLAG expression in the cerebellum of AAV9-injected mice at 4 weeks of age (n = 4). (G and H) Amount of miR-3191-5p (G) and α1ACT-Q33 (H) mRNA binding to Ago4 that was immunoprecipitated using an antibody against Ago4 or a control IgG in the cerebellum of AAV9–Q33–miR-3191-5p mice at 4 weeks of age (n = 6). GFP, AAV9-GFP mice; mock, AAV9–Q33–miR-mock mice; 3191, AAV9–Q33–miR-3191-5p mice. All data represent means ± SEM. **P < 0.01. One-way ANOVA in (C). Student’s t test in (D) to (H).
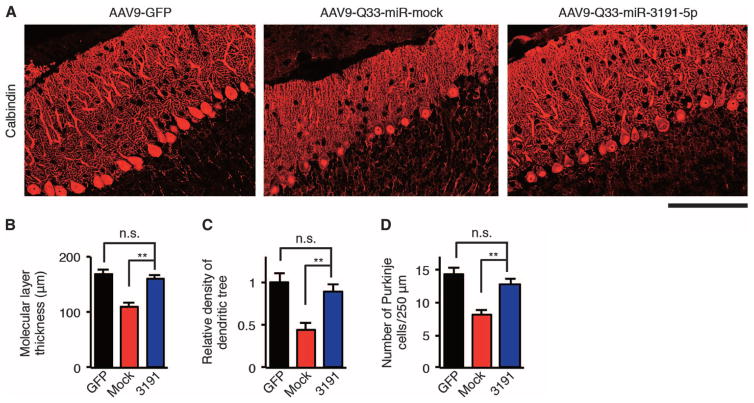
Immunohistochemical examination of AAV9–Q33–miR-3191-5p mouse cerebellum showed that the treatment with AAV9–miR-3191-5p protected Purkinje cells from degenerative changes (Fig. 7, A to D). As compared to AAV9–Q33–miR-mock mice, AAV9–miR-3191-5p mice showed protection from thinning of the molecular layer of the cerebellum (P < 0.01) (Fig. 7B), decreased density of the dendritic tree (P < 0.01) (Fig. 7C), and decreased number of Purkinje cells (P < 0.01) (Fig. 7D) caused by AAV9-delivered IRES-driven α1ACT-Q33. These results indicate that AAV9-delivered miR-3191-5p inhibited Purkinje cell degeneration caused by CACNA1A IRES–driven α1ACTSCA6 in mice.
Fig. 7. miR-3191-5p prevents Purkinje cell degeneration caused by CACNA1A IRES–driven α1ACTSCA6 in mice.
(A) Representative immunofluorescence images of AAV9-injected mouse cerebellum from 4-week-old mice stained with calbindin-specific antibodies and fluorescent secondary antibody. Scale bar, 100 μm. (B to D) Molecular layer thickness (B), density of dendritic tree (C), and Purkinje cell counts (D) in AAV9-injected mouse cerebellum from 4-week-old mice (n = 6). GFP, AAV9-GFP mice; mock, AAV9–Q33–miR-mock mice; 3191, AAV9–Q33–miR-3191-5p mice. All data represent means ± SEM. **P < 0.01. Student’s t test in (B) to (D).
miR-3191-5p prevents the ataxia and motor deficits caused by CACNA1A IRES–driven α1ACTSCA6 in mice
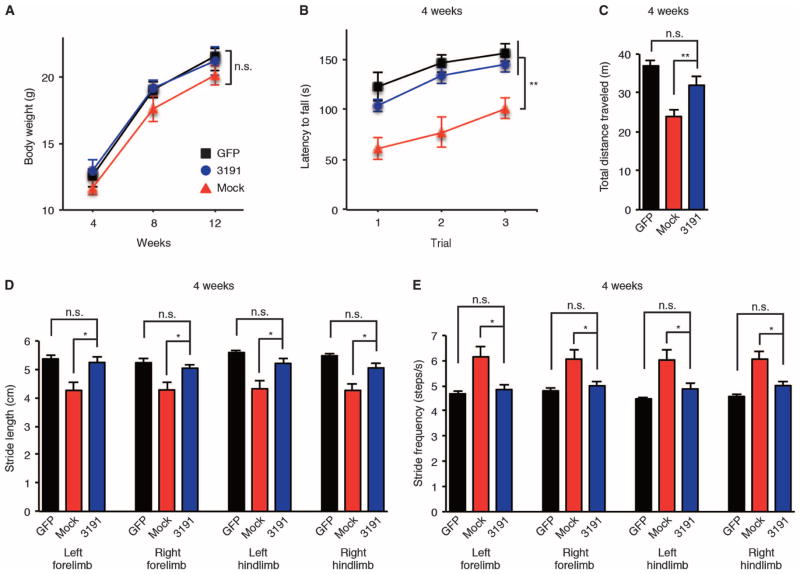
We further examined the therapeutic effect of AAV9–miR-3191-5p on mouse behavioral phenotypes caused by CACNA1A IRES–driven α1ACTSCA6. Although the treatment with AAV9–miR-3191-5p did not affect the body weight of AAV9–Q33–miR-3191-5p mice compared to AAV9–Q33–miR-mock mice (Fig. 8A), we found that the CACNA1A IRES–driven α1ACTSCA6-associated disease phenotypes in AAV9–Q33–miR-3191-5p mice were prevented, as assessed by the rotarod test (Fig. 8B), open-field assay (Fig. 8C), and DigiGait for gait analysis (Fig. 8, D and E). AAV9–Q33–miR-3191-5p mice performed significantly better on the accelerating rotarod test (P < 0.01) (Fig. 8B) and ambulated a greater distance in the open-field test than did AAV9–Q33–miR-mock mice (P < 0.01) (Fig. 8C) at 4 weeks of age. As compared to AAV9–Q33–miR-mock mice, AAV9–Q33–miR-3191-5p mice exhibited improvement of gait instability in all four limbs caused by AAV9-delivered IRES-driven α1ACT-Q33 (P < 0.05) (Fig. 8, D and E). We also found that treatment with AAV9–miR-3191-5p prevented the weaving steps of AAV9–Q33–miR-3191-5p mice (movies S4 and S5). Furthermore, the therapeutic effect of AAV9–miR-3191-5p on mouse behavioral phenotypes and CACNA1A IRES–driven α1ACT-Q33 expression in the cerebellum of AAV9–Q33–miR-3191-5p mice persisted during long-term follow-up of 30 weeks (fig. S8, A to G).
Fig. 8. AAV9–miR-3191-5p prevents the ataxia and motor deficits caused by CACNA1A IRES–driven α1ACTSCA6 in mice.
(A to E) Clinical features of AAV9-injected mice. Body weight (A), rotarod test (B), and open-field assay (C) at 4 weeks of age. Stride length (D) and stride frequencies (E) assessed by DigiGait analysis at 4 weeks of age (n = 12, 6 male and 6 female mice per group). GFP, AAV9-GFP mice; mock, AAV9–Q33–miR-mock mice; 3191, AAV9–Q33–miR-3191-5p mice. All data represent means ± SEM. *P < 0.05, **P < 0.01. Two-way ANOVA in (A) and (B). Student’s t test in (C) to (E).
Although both mature and stem-loop sequences of hsa–miR-3191-5p used in our study are identified in the human genome (miRBase: www.mirbase.org/), those of mmu-miR-3191-5p have not yet been identified in the mouse genome. To identify other potential candidates of hsa–miR-3191-5p–targeting mRNAs in mouse, we first used TargetScanHuman 7.0 (www.targetscan.org/vert_70/) to find the top 100 ranked hsa–miR-3191-5p–targeting human mRNAs. We selected 13 genes with high reliability (cumulative weighted context++ score <−1 and total context++ score < −1) (fig. S9A). Among these, seven genes (PRX, ZNF781, C22orf46, ZNF23, ZNF286A, ERBB4, and PTBP1) have hsa–miR-3191-5p binding sites within the 3′UTR of their conserved mouse orthologs. We examined the mRNA levels of these seven genes in the cerebellum of wild-type mice injected with AAV9–miR-3191-5p compared to those injected with AAV9–miR-mock. We found that AAV9–miR-3191-5p down-regulated the mRNA expression of mouse orthologs of ZNF781, C22orf46, ZNF23, and ERBB4 by about 50%, whereas those of the other three genes were unchanged, indicating that not all of the predicted mRNA targets were affected by the treatment with AAV9–miR-3191-5p in mice (fig. S9, B to H).
To determine whether an overall change in whole-gene expression in the mouse brain and cerebellum after AAV9–miR-3191-5p injections leads to significant adverse and toxic effects, we monitored wild-type mice injected with AAV9–miR-3191-5p for 12 months and found no abnormal phenotypes. Also, we found no histopathological abnormalities in the brain, cerebellum, heart, lung, liver, and kidney of wild-type mice injected with AAV9–miR-3191-5p (fig. S10, A to F). On the basis of these findings, we conclude that AAV9-mediated delivery of miR-3191-5p is a well-tolerated and successful therapeutic approach for treating the disease phenotype in our SCA6 mouse model.
DISCUSSION
To explore possible therapeutic strategies for treating SCA6, we developed a robust SCA6 mouse model using AAV9-mediated somatic gene transfer of the CACNA1A second cistron to express α1ACTSCA6 under the control of CACNA1A IRES. We found prominent α1ACTSCA6 expression that was predominantly in cerebral cortex and cerebellum of AAV9-injected mice. Mice injected with AAV9 expressing α1ACTSCA6 showed unambiguous disease phenotypes associated with the pathological and clinical features of patients with SCA6 (2–8) much earlier and more reproducibly than the Purkinje cell–specific promoter-driven transgenic SCA6 mouse model we previously developed (11) or the knock-in SCA6 mouse model with polyQ expansion size observed in patients with SCA6 (9, 10). By immunofluorescence analysis, we found that administration of a total of 1010 vg of AAV9 expressing CACNA1A IRES–driven α1ACTSCA6 into the lateral ventricle caused an ~50% decrease in the number of Purkinje cells that was sufficient to lead to early-onset ataxia and motor deficits from an early age. These findings indicate that AAV9-mediated CACNA1A IRES–driven α1ACTSCA6 expression is stronger and therefore more toxic in mouse cerebellum than α1ACTSCA6 expression in previously developed SCA6 mouse models (9–11). These early and severe ataxic phenotypes are also observed in Pumilio1-deficient mice, mimicking SCA1, another type of autosomal dominant SCA (30).
Although we have previously shown that α1ACT functions as a transcription factor essential for cerebellar development (11), the detailed mechanism by which α1ACTSCA6 causes Purkinje cell degeneration in adulthood remains unknown. Because most individuals with SCA6 are heterozygous for the SCA6-causing expansion, that is, they still have one copy of the normal CACNA1A second cistron, they express some levels of the normal α1ACT protein. On the other hand, SCA6 homozygotes that express two copies of mutant α1ACT protein and no normal α1ACT protein develop normally and have nearly the same age of onset as SCA6 heterozygotes (7, 8, 31–33). This confirms that the mutant α1ACT protein fulfills some of the normal α1ACT protein functions in development and that SCA6 arises by a dominant “gain-of-function” mechanism.
In cell culture experiments, we have discovered that miR-3191-5p directly acts on the CACNA1A IRES to regulate the translation of α1ACT in an Ago4-dependent manner. Previous studies have shown that miRNAs, by forming miRISC with Ago1 or Ago2, block the assembly of translation initiation factors to repress either cap- or IRES-dependent translation through their binding to the 3′UTR of targeted mRNA (18–20). There are four members of the RNA-induced silencing complex–related Ago protein family in human Ago1, Ago2, Ago3, and Ago4 (27). Very little is known about Ago4, although the absence of a catalytic domain suggests that it might have a distinct role in gene silencing compared with Ago2 (34–39). Our data showing that Ago4 interacts with miR-3191-5p and CACNA1A IRES to repress translation of the second cistron may suggest that it has a specific role for the mRNA-sparing translational repression via miRISC, possibly even selectively for the translational repression of IRESs.
We have also demonstrated that miR-3191-5p bound to Ago4 selectively inhibits the translational initiation of α1ACT by eIFs, eIF4AII and eIF4GII, which enhance α1ACT expression. Although the list of cellular mRNAs that are thought to contain IRESs is growing (17, 40–42), cellular IRESs show little structural relationship to each other, and their underlying mechanism remains largely unknown but is thought to follow the picornavirus paradigm of binding to the eIF4A-eIF4G complex (17). Identification of eIF4AII, which may provide helicase activity, and eIF4GII, which may serve as a scaffold (17, 43–45), in the mechanism of CACNA1A IRES–mediated translation supports recent findings that show the effect of eIFs on cellular IRESs in human disease (40–42). Presumably, in addition to these eIFs, other IRES transacting factors and small RNAs may play a part in the regulation of CACNA1A IRES–mediated translation (17, 40, 42). The recently recognized abundance of IRES-mediated translational mechanisms in the human genome raises the potential for their role in human disease (46).
We have developed an miRNA-mediated therapy for SCA6 using selective translational blockade of the CACNA1A second cistron in mice. Our findings indicate that continuous inhibition of α1ACTSCA6 expression by the AAV9 vector–mediated delivery of miR-3191-5p has a substantial therapeutic effect on the SCA6 disease phenotype in mice. Considering that an ~50% decrease in the number of Purkinje cells is sufficient to cause ataxia and motor deficits in our mouse model, miRNA-mediated α1ACTSCA6 silencing by blockade of CACNA1A IRES–driven translation in greater than half of Purkinje cells would have great potential benefit in presymptomatic or symptomatic SCA6 patients (13–15).
Although AAV9-mediated intervention with miR-3191-5p did not regulate all of the predicted mRNA targets in our mouse model, we showed that miR-3191-5p has a selective silencing effect on the CACNA1A IRES–driven translation of α1ACTSCA6 in mice. Targeting CACNA1A IRES in SCA6 patients through miRNAs or small molecules may down-regulate both normal and mutant α1ACT. However, AAV9-mediated administration of miR-3191-5p into the lateral ventricle did not lead to any toxic effects in mice during long-term observations over 1 year. Other studies in our group also showed that the effect of α1ACT on establishing normal cerebellar development appeared to be irreversible (47).
There are several limitations to our study. Although patients with SCA6 develop ataxic phenotypes in adulthood (2–8), the targeted expression of α1ACTSCA6 in late adult mouse cerebellum has not been evaluated. Also, the off-target effects due to viral transduction of an miRNA may depend on viral titer and differ among species. A series of reports have suggested that miRNAs may have therapeutic promise for treating cancer, metabolic disease, and inflammation (48–50). However, before miRNAs can have success as therapeutic agents, many challenges remain to be overcome, including their efficient delivery and off-target effects (51, 52). Further studies are needed to validate more efficient and specific targeting strategies to develop an miRNA-based therapy for SCA6 patients in the future.
Although therapy targeting IRES activity has been a mainstay of antiviral therapy, it has not been used for genetic diseases (53–55). IRES-dependent translation also plays a role in the expression of some oncogenes, such as c-myc (56), L-myc (57), and epidermal growth factor receptor (58), in tumors. Our study opens the door to the development of therapies using a new strategy for the selective suppression of IRES-driven pathogenic gene products based on delivery of disease-specific miRNAs.
MATERIALS AND METHODS
Study design
Our study was based on our previous findings that SCA6 is attributable to a polyQ repeat expansion within a second CACNA1A gene product, α1ACT, and that α1ACT expression is under the control of an IRES present within the CACNA1A coding region. The objectives of the current study were threefold. The first objective was to develop an early-onset ataxia model using an AAV9-based gene delivery system to express CACNA1A IRES–driven α1ACTSCA6. The second objective was to identify miRNAs that target CACNA1A IRES and preferentially block the CACNA1A IRES–driven translation of α1ACT. Finally, we investigated the therapeutic potential of ectopic miRNA expression for the treatment of SCA6 by AAV9-mediated transduction in our previously developed mouse model of CACNA1A IRES–driven SCA6. On the basis of our previous studies (11, 59), four to six biological replicates were used for each in vitro or in vivo biochemical and histological analysis, whereas a sample size of 12 mice (6 male and 6 female mice) per group was used for behavioral testing. For AAV9 injections, neonatal mice were randomly assigned to treatment conditions with equivalent numbers in each group. All behavioral analyses were performed by experimenters who were blind to the identity of treatment conditions. Data collection and the biochemical and histological analysis for mouse samples were performed with the investigators unaware of the sample identities until statistical analyses. All source data are in the Supplementary Materials (table S1).
Construction of DNA plasmids
We used pcDNA3 vectors expressing CACNA1A-encoded FLAG-tagged peptides (α1A-FLAG and α1ACT-FLAG), a bicistronic CACNA1A IRES reporter vector, and a control reporter vector as described previously (11). We constructed the truncated transgenes of CACNA1A second cistron that lacked the sequence of 5′ upstream from CACNA1A IRES (pcDNA3-IRES-α1ACT vectors; fig. S1A) and inserted them into AAV9 genomes to prepare AAV9-α1ACT-Q11 and AAV9-α1ACT-Q33 (fig. S1B). Briefly, the sequence of nucleotides 4962 to 7757 of full-length CACNA1A complementary DNA (cDNA) (NM_001127222.1), corresponding to the sequence of CACNA1A IRES and α1ACT ORF, was amplified using IRES-α1ACT primers [5′-ATCAGGATCCGCCCTCAACACCATCGTGC-3′ (forward) and 5′-GAATCTAGATTACTTGTCATCG-3′ (reverse)] from pcDNA3 vectors expressing CACNA1A-encoded FLAG-tagged peptides. The PCR products were inserted into the Bam HI and Xba I sites of pcDNA3 vectors. We also modified the sequence of nucleotides 5013 to 5015 from “TAT” to “TAG” of stop codon (fig. S1A).
To obtain mutated CACNA1A IRES transcripts that are resistant to the binding of miR-3191-5p (CACNA1A IRESmut vectors; fig. S5, B and C), we performed C→G and G→C substitutions within the predicted miR-3191-5p binding site. We also performed C→G and G→C substitutions within the complementary binding site of the sequence targeted by miR-3191-5p to maintain the stem-loop structure of CACNA1A IRESmut as same as that of the original. Briefly, we amplified the predicted binding site of miR-3191-5p using miR mut primers [5′-ATGTCTCCGCCCCTGGGTCTccccAAcAAcTcTggccCCAGAGTGGCTTACAAGCG-3′ (forward) and 5′-CGCTTGTAAGCCACTCTGGggccAgAgTTgTTggggAGACCCAGGGGCGGAGACAT-3′ (reverse)] and the complementary binding site of the sequence targeted by miR-3191-5p using anti–miR mut primers [5′-ACTGAGCA-CAATAACTTggccAggTTgTTggAGGCCCTCATGCTTCTC-3′ (forward) and 5′-GAGAAGCATGAGGGCCTccAAcAAccTggccAAGTTATTGTGCTCAGT-3′ (reverse)]. Subsequently, we annealed PCR products using primers anti–miR mut forward and miR mut reverse and inserted the annealed PCR products into the BIp I sites of pcDNA3 vectors expressing CACNA1A-encoded FLAG-tagged peptides and into the Eco RI and Nco I sites of a bicistronic CACNA1A IRES reporter vector.
pDEST-GFP-Ago1 (Addgene, plasmid #21534) and pDEST-GFP-Ago4 (Addgene, plasmid #21536) were gifts from E. Chan (60). pEGFP-hAgo2 (Addgene, plasmid #21981) was a gift from P. Sharp (61). pGEX-GST-AGO3 (Addgene, plasmid #24318) was a gift from C. Novina (34). To create pEGFP-AGO3, we restriction-digested pGEX-GST-AGO3 with Eco RI and Bam HI and inserted it into the plasmid backbone of the pEGFP-hAgo2 after Eco RI and Bam HI digestion.
pcDNA3-HA–eIF4GI and pcDNA3-HA–eIF4GII were gifts from N. Sonenberg (McGill University, Canada) (62). M. Bushell (Medical Research Council, UK) provided us with the plasmids expressing human eIF4AI and eIF4AII (18). pcDNA3.1-HisXpress vector was a gift from T. Cooper (Baylor College of Medicine). From the plasmids expressing human eIF4AI and eIF4AII, we amplified human eIF4AI and eIF4AII cDNAs using eIF4AI-PCR primers [5′-ATCAGGATCCATGTCTGCGAGCCAGGAT-3′ (forward) and 5′-ATCAGGGCCCAGGTCAGCAACATTGAGG-3′ (reverse)] and eIF4AII-PCR primers [5′-ATCAGGATCCATGTCTGGTGGCTCCGCG-3′ (forward) and 5′-ATCAGGGCCCTTAAATAAGGTCAGCCAC-3′ (reverse)], respectively. Subsequently, we inserted the PCR products into Bam HI– and Apa I–digested pcDNA3.1-HisXpress vectors to obtain pcDNA3.1-HisXpress–eIF4AI and pcDNA3.1-HisXpress–eIF4AII. We used the original, either pcDNA3.1-HisC or pcDNA3-HA, vectors lacking the insertions in the multiple cloning sites as control vectors.
Cotransfection of DNA plasmids with either synthetic miRNA or siRNA into cultured cells
HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. All miRNAs and siRNAs were purchased from Life Technologies. ID numbers are shown as follows: miR-711 (MC15715), miR-3191-5p (MC23769), miR-4786-3p (MC21295), a negative control miRNA (#4464059), Ago1 (s25500), Ago2 (s25931), Ago3 (s46947), Ago4 (s46949), eIF4AII (s4570), eIF4GII (s16519), and an siRNA negative control (#12935-300).
We plated HEK293 cells onto six-well dishes and cotransfected each dish with 1.0 μg of the vector expressing the following: bicistronic CACNA1A IRES reporter, bicistronic CACNA1A IRESmut reporter, control reporter, and α1A-FLAG and α1ACT-FLAG; 0.5 μg of the vector expressing the following: eIF4AI, eIF4AII, eIF4GI, and eIF4GII; and either 20 nM synthetic miRNA or 20 nM siRNA molecules. We used Lipofectamine 2000 (Life Technologies) as a transfection reagent in all cases. Neither the miRNA negative control nor the siRNA negative control matched any human mRNA. The transfected cells were cultured for 48 hours before being processed for RNA and protein analysis.
Luciferase assay
HEK293 cells were plated onto six-well dishes and cotransfected with vectors as shown above. Forty-eight hours after cotransfection, the activities of firefly and Renilla luciferase in lysates prepared from transfected cells using the Dual-Luciferase Assay System (Promega) were measured by using a Wallac 1420 VICTOR3 V luminometer with a 1-s integration time (PerkinElmer). The ratio of firefly luciferase to Renilla luciferase activities of a bicistronic CACNA1A IRES reporter vector was normalized to that of a bicistronic control reporter vector.
Protein expression analysis
We did Western blot analysis as previously described (11, 63). We used the following primary antibodies: FLAG-specific antibody (1:5000, A8592 and 1:5000, F1804; Sigma-Aldrich); Ago1-specific antibody (1:2000, 9388S; Cell Signaling Technology); Ago2-specific antibody (1:2000, SAB4200085; Sigma-Aldrich); Ago3-specific antibody (1:2000, 5054S; Cell Signaling Technology); Ago4-specific antibody (1:2000, 6913S; Cell Signaling Technology); eIF4AII-specific antibody (1:2000, ab31218; Abcam); eIF4GII-specific antibody (1:1000, sc-100732; Santa Cruz Biotechnology); GFP-specific antibody (1:2000, M048-3; MBL); and GAPDH-specific antibody (1:5000, AM4300; Life Technologies). We used NIH ImageJ software to quantify the specific expression of individual proteins and demonstrated the relative signal intensities of individual proteins normalized to those of GAPDH in each sample.
Quantitative real-time polymerase chain reaction
The total RNA was extracted from HEK293 cells and the cerebellum of mice using the miRNeasy Mini Kit (Qiagen) and reverse-transcribed using the SuperScript VILO (Life Technologies) for mRNA and NCode VILO (Life Technologies) for miRNA. The cDNAs were then used for real-time PCR using the iQ SYBR Green Supermix (Bio-Rad Laboratories). We did the amplification, detection, and data analysis using a Bio-Rad iCycler system (Bio-Rad Laboratories). The crossing threshold values for the mRNAs of the individual genes were normalized to β-actin. The crossing threshold values for miR-3191-5p were normalized to U6 small nuclear RNA. Changes in the expression of mRNA and miRNA were expressed as a fold change relative to the control. We used the following primers. The sequences of the hsa-CACNA1A primers were 5′-GTCTGGGGAAGAAGTGTCCG-3′ (forward) and 5′-GCTCCTCCCTTGGCAATCTT-3′ (reverse). These hsa-CACNA1A primers discriminated between the human CACNA1A mRNA and the mouse CACNA1A mRNA. The sequences of the hsa-IRES-α1ACT primers were 5′-GTACCTCACCCGAGACTCCT-3′ (forward) and 5′-CGGACACTTCTTCCCCAGAC-3′ (reverse). These primers can also detect the mouse endogenous CACNA1A mRNA. The sequences of the hsa–β-actin primers were 5′-GCGGGAAATCGTGCGTGACATT-3′ (forward) and 5′-GATGGAGTTGAAGGTAGTTTCGTG-3′ (reverse). The sequences of the mmu-Prx primers were 5′-TTGGTGGAGATTATCGTGGAG-3′ (forward) and 5′-TCTTGCAAGCTGAGGCTCTTA-3′ (reverse). The sequences of the mmu-Zfp781 primers were 5′-CCTATGAGGATGTGCATGTGA-3′ (forward) and 5′-TGGGGTCCAGAGTGACAGATA-3′ (reverse). The sequences of the mmu-4930407I10Rik primers were 5′-GAATTCCCAAGGGCTAAAGTG-3′ (forward) and 5′-TTTCACACCATCTCCACTTCC-3′ (reverse). The sequences of the mmu-Zfp612 primers were 5′-AAGGCAGCCCTCAAGTTAATC-3′ (forward) and 5′-AAGTCTCTGATGCCAGACGAA-3′ (reverse). The sequences of the mmu-Zfp286 primers were 5′-CATGGAAACCAGACCTGAGAG-3′ (forward) and 5′-ACGCTCACATTCAAGAGCAGT-3′ (reverse). The sequences of the mmu-Erbb4 primers were 5′-CGCTAGAACTCCACTGATTGC-3′ (forward) and 5′-TACCAGCTCTGTCTCCAGGAA-3′ (reverse). The sequences of the mmu-Ptbp1 primers were 5′-TCACCAAGAACAACCAGTTCC-3′ (forward) and 5′-GTGAGCTTGGAGAAGTCGATG-3′ (reverse). The sequences of the mmu–β-actin primers were 5′-GCTACAGCTTCACCACCACA-3′ (forward) and 5′-TCTCCAGGGAGGAAGAGGAT-3′ (reverse). The sequences of the miR-3191-5p primers were 5′-GCTCTCTGGCCGTCTAC-3′ (forward) and 5′-GTCCAGTTTTTTTTTTTTTTTGGAAG-3′ (reverse). The sequences of the U6 small nuclear RNA primers were 5′-CTTCGGCAGCACATATACTAAA-3′ (forward) and 5′-AAAATATGGAACGCTTCACG-3′ (reverse). We designed the miR-3191-5p primers using a bioinformatics program (64).
Coimmunoprecipitation
We plated HEK293 cells onto 100-mm dishes and cotransfected each dish with 3.0 μg of the vectors expressing eIF4AII and eIF4GII. Forty-eight hours after transfection, we harvested HEK293 cells for immunoprecipitation using eIF4AII-specific antibody (5 μg per sample, ab31218; Abcam), eIF4GII-specific antibody (5 μg per sample, sc-100732; Santa Cruz Biotechnology), and Dynabeads Protein G Immunoprecipitation Kit (Life Technologies) according to the manufacturer’s suggested protocols. A rabbit IgG (5 μg per sample, PP64B; Millipore) and mouse IgG (5 μg per sample, CS200621; Millipore) were used as controls.
Immunoprecipitation-coupled qRT-PCR
We plated HEK293 cells onto 100-mm dishes and cotransfected each dish with 2.0 μg of the vector expressing the following: full-length CACNA1A-Q33–encoded FLAG-tagged peptides, IRES-α1ACT-Q33, miR-3191-5p (SC401396; OriGene), Ago1, Ago2, Ago3, Ago4, eIF4AII, and eIF4GII. Forty-eight hours after transfection, we harvested HEK293 cells for coimmunoprecipitation using Ago1-specific antibody (5 μg per sample, 9388S; Cell Signaling Technology), Ago2-specific antibody (5 μg per sample, 2897S; Cell Signaling Technology), Ago3-specific antibody (5 μg per sample, 5054S; Cell Signaling Technology), Ago4-specific antibody (5 μg per sample, 6913S; Cell Signaling Technology), eIF4AII-specific antibody (5 μg per sample, ab31218; Abcam), eIF4GII-specific antibody (5 μg per sample, sc-100732; Santa Cruz Biotechnology), and Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s suggested protocols. A rabbit IgG (5 μg per sample, PP64B; Millipore) and mouse IgG (5 μg per sample, CS200621; Millipore) supplied by the manufacturer were used as controls. The immunoprecipitated RNA was reverse-transcribed using SuperScript VILO (Life Technologies) for mRNA and NCode VILO (Life Technologies) for miRNA and analyzed by qRT-PCR for the differential expression of CACNA1A-Q33 mRNA and IRES-α1ACT-Q33 mRNA using the following primers: 5′-GTCTGGGGAAGAAGTGTCCG-3′ (forward) and 5′-GCTCCTCCCTTGGCAATCTT-3′ (reverse), and miR-3191-5p, 5′-GCTCTCTGGCCGTCTAC-3′ (forward) and 5′-GTCCAGTTTTTTTTTTTTTTTGGAAG-3′ (reverse). We also extracted RNA and protein complex from the cerebellum of AAV9-injected mice and harvested them for coimmunoprecipitation as same as shown above.
Development of the AAV9 vectors
The AAV9 vector plasmids contained an expression cassette, consisting of a human cytomegalovirus immediate-early promoter followed by cDNA encoding gene of our interest as shown below, woodchuck hepatitis virus posttranscriptional regulatory element (WPRE), and a simian virus 40 polyadenylation signal sequence between the inverted terminal repeats of the AAV3 genome. The AAV9 vectors expressing α1ACT with either normal CAG repeat size (AAV9-α1ACT-Q11) or mutant CAG repeat size (AAV9-α1ACT-Q33) contained the truncated transgenes of CACNA1A corresponding to the sequence of CACNA1A IRES and α1ACT ORF (IRES-α1ACT; fig. S1, A and B). AAV9-GFP contained cDNA encoding GFP sequence. AAV9–miR-3191-5p contained cDNA encoding GFP and miR-3191-5p sequence (fig. S7A). AAV9–miR-mock contained cDNA encoding GFP and the miR-mock sequence (fig. S7A). The sequences of miR-3191-5p and miR-mock are 5′-GGGGTCACCTCTCTGGCCGTCTACCTTCCACACTGACAAGGGCCGTGGGGACGTAGCTGGCCAGACAGGTGACCCC-3′ (miR-3191-5p) and 5′-GTATTGCGTCTGTACACTCACCGTTTTGGCCACTGACTGACGGTGAGTGCAGACGCAATA-3′ (miR-mock).
We synthesized the AAV9 vp cDNA as previously described with the substitution of thymidine for adenine 1337, which introduced an amino acid change from tyrosine to phenylalanine at position 446 (65). Recombinant AAV9 vectors were produced by transient transfection into HEK293 cells using the vector plasmid, an AAV3 rep and AAV9 vp expression plasmid, and the adenoviral helper plasmid pHelper (Agilent Technologies). We purified the recombinant viruses by isolation from two sequential continuous CsCl gradients, and the viral titers were determined by qRT-PCR as follows: 40 cycles of 95°C/15 s, 60°C/30 s, 72°C/90 s, and 75°C/15 s with WPRE forward primer (5′-ATTGCTTCCCGTATGGCTTTCA-3′) and WPRE reverse primer (5′-TCAGCAAACACAGTGCACACCA-3′) to amplify the sequence of nucleotides 1319 to 1201 of woodchuck hepatitis virus 2.
Injection of AAV9 into the ventricle of neonatal wild-type mice
The C57/BL6J mice were purchased from Jackson Laboratory and maintained in our breeding colony. At postnatal day 1, neonatal C57/BL6J mice were individually anesthetized on ice, and a total of 1010 vg in 2 to 4 μl of AAV9 solution were injected into the right lateral ventricle of neonatal C57/BL6J mice with a 10-μl Hamilton syringe attached to a 32-gauge needle (Hamilton Company). The viral solution contained 0.04% trypan blue (Sigma-Aldrich) to help determine whether the ventricles were indeed injected. Only those neonatal C57/BL6J mice in which the lateral ventricles were filled with viral solution were analyzed. Six male and six female mice were enrolled into each group: two groups of AAV9-GFP mice, AAV9-α1ACT-Q11 mice, AAV9-α1ACT-Q33 mice, AAV9–Q33–miR-mock mice, and AAV9–Q33–miR-3191-5p mice. All animal experiments were approved and carried out in accordance with the regulations and guidelines for the care and use of experimental animals at the Institutional Animal Care and Use Committee of the University of Chicago.
The behavioral assessments of AAV9-injected mice
We examined the behavioral assessments of AAV9-injected mice at 4, 8, 12, and 30 weeks of age. The investigators who carried out the behavioral assessments were blinded to the treatment conditions.
Rotarod
We analyzed rotarod test of mice using Economex Rotarod (Columbus Instruments) with accelerating mode (4 to 40 rpm, acceleration with 0.1 rpm per 0.8 s). We performed three consecutive trials with 10-min intervals between each trial.
Open-field assay
We examined open-field assay using Mouse Open Field Arena and 48 Channel IR Controller for Open Field Activity (ENV-510 and ENV-520, Med Associates Inc.). Briefly, mice were placed in the center of the open-field area, and their movements were monitored through the side-mounted photobeams for 30 min. We analyzed multiple parameters using Activity Monitor software (Med Associates Inc.) and adopted total distance traveled to assess the activity of each mouse.
DigiGait analysis
We also examined a video-assisted computerized treadmill for mouse gait analysis using a DigiGait with DigiGait software (Mouse Specifics). All mice were tested at the speed of 25 cm/s.
Immunohistochemistry, immunofluorescence, and histopathology
Immunofluorescence was performed as previously described (11, 59) except as modified below. We cut paraffin-embedded sagittally oriented 5-μm sections of mouse brains and cerebellums at 20-μm intervals. We used comparable sections from vermis, medial hemisphere, and lateral hemisphere for staining and histopathological assessments. Paraffin-embedded sections of perfused brains and cerebellums were dewaxed, rehydrated, and then steamed for 20 min in antigen retrieval solution (Reveal, Biocare Medical). Sections were blocked and exposed to primary antibodies for 12 hours at 4°C. After washing, fluorescent secondary antibodies in phosphate-buffered saline and 0.05% Tween 20 were added for 1 hour at room temperature. Confocal fluorescence images were captured with a Leica TCS SP2 laser scanning confocal microscope (Leica Microsystems Inc.).
We used NIH ImageJ software to quantify the FLAG and GFP expression in each section and demonstrated the relative signal intensities of the FLAG and GFP fluorescence per unit area of the mouse hippocampus, cerebral cortex, and cerebellum in each section. The molecular layer thickness and density of Purkinje dendritic trees were calculated as previously described (11, 66). We selected Purkinje cells well stained with GFP- and FLAG-specific antibodies, indicating well transduced with AAV9, and analyzed Purkinje cells (100 to 250 cells per sample) in the entire area of each section to calculate the mean of the density of Purkinje dendritic trees. The dendritic trees of the captured Purkinje cell image and the area enclosed were outlined and measured using NIH ImageJ software. We also calculated the number of Purkinje cells (100 to 250 cells per sample) in the entire area of each section and expressed the results as the number per 250 μm.
We used the following primary antibodies: FLAG-specific antibody (1:200, F1804 and 1:200, F7425; Sigma-Aldrich), GFP-specific antibody (1:200, M048-3; MBL), and calbindin-specific antibody (1:200, CB38a; Swant). Goat Alexa Fluor–conjugated anti-mouse and goat Alexa Fluor–conjugated anti-rabbit IgG antibodies (Life Technologies) were used for secondary fluorescence detection. For the tissue sections stained with hematoxylin and eosin, digital image files were created with a 3D Histech Pannoramic Scan whole slide scanner (PerkinElmer) with a Stingray F146C color camera (Allied Vision Technologies). Individual images were analyzed using the 3D Histech Pannoramic Viewer software (PerkinElmer).
Statistical analysis
Statistical analysis was performed using ANOVA and Student’s t test, unless specified, with the IBM SPSS Statistics 23.0. Two-tailed unpaired t test was used to compare two conditions. One-way ANOVA was used for comparison among multiple experimental conditions. Bonferroni post hoc test was used when comparing among each condition. For the analysis of mouse body weight and rotarod performance, two-way ANOVA was used for comparison among groups. All data represent means ± SEM. Statistical significance in figures: *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary Material
Fig. S1. Schematic representation of AAV9 vectors and AAV9 transduction efficiency into Purkinje cells.
Fig. S2. Histopathological examination of the AAV9-injected mouse cerebral cortex and hippocampus.
Fig. S3. The long-term follow-up of AAV9-injected mouse behavioral phenotypes and CACNA1A IRES–driven α1ACT expression in mice.
Fig. S4. Stem-loop structure of CACNA1A IRES and the predicted binding sites of miR-711, miR-3191-5p, and miR-4786-3p.
Fig. S5. Effects of miR-3191-5p on mutated CACNA1A IRES templates.
Fig. S6. Effects of eIF4AII and eIF4GII on mutated CACNA1A IRES templates in the presence or absence of miR-3191-5p.
Fig. S7. Cotransduction of AAV9-α1ACT-Q33 with either AAV9–miR-mock or AAV9–miR-3191-5p.
Fig. S8. The long-term follow-up of therapeutic effect of miR-3191-5p on mouse behavioral phenotypes and CACNA1A IRES–driven α1ACTSCA6 expression in mice.
Fig. S9. Potential mouse mRNAs targeted by human miR-3191-5p.
Fig. S10. Histopathological examination of the brain, cerebellum, heart, lung, liver, and kidney of wild-type mice injected with AAV9–miR-3191-5p.
Table S1. Source data.
Movie S1. DigiGait video of an 8-week-old AAV9-GFP mouse on treadmill.
Movie S2. DigiGait video of an 8-week-old AAV9-α1ACT-Q11 mouse on treadmill.
Movie S3. DigiGait video of an 8-week-old AAV9-α1ACT-Q33 mouse on treadmill.
Movie S4. DigiGait video of an 8-week-old AAV9–Q33–miR-mock mouse on treadmill.
Movie S5. DigiGait video of an 8-week-old AAV9–Q33–miR-3191-5p mouse on treadmill.
Acknowledgments
We thank M. Ito and N. Takino (Jichi Medical University, Japan) for their help with the production of the AAV vectors, Z. Huang (University of Occupational and Environmental Health, Japan) for expert technical support, K. Sahashi (Nagoya University, Japan) for the instruction on AAV injections, C. Labno (University of Chicago) for her special contributions to imaging, C. S. Jimenez (University of Chicago) for his help with cell culture experiments, and K. Robinson and C. Wei (University of Chicago) for their help with animal experiments. We also thank the Floyd family for their support of SCA6 research.
Funding: This work was supported by grants from the NIH (R01NS082788 and R01NS094665), National Ataxia Foundation Post-Doctoral Fellowship Award, JSPS KAKENHI grant no. 26293213, Japan Agency for Medical Research and Development, Lilly Scientific Fellowship Program Award, Kanae Foundation for the Promotion of Medical Science, and Uehara Memorial Foundation.
Footnotes
Author contributions: Project planning was performed by Y.M., X.D., S.-i.M., and C.G.; plasmid construction was performed by Y.M. and X.D.; cell culture experiments were performed by Y.M. and X.D.; AAV vector production was performed by S.-i.M.; AAV injection was performed by Y.M.; the biochemical and histological experiments for mouse samples were performed by Y.M. and X.D.; data analysis was performed by Y.M., X.D., S.-i.M., and C.G.; the first draft of the manuscript was written by Y.M., X.D., S.-i.M., and C.G.
Competing interests: S.-i.M. owns equity in a gene therapy company (Gene Therapy Research Institution) that commercializes the use of AAV vectors for gene therapy applications.
Data and materials availability: The source data for this study are in the Supplementary Materials. All materials generated are available for distribution.
REFERENCES AND NOTES
- 1.La Spada AR, Taylor JP. Repeat expansion disease: Progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paulson HL. The spinocerebellar ataxias. J Neuroophthalmol. 2009;29:227–237. doi: 10.1097/WNO0b013e3181b416de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durr A. Autosomal dominant cerebellar ataxias: Polyglutamine expansions and beyond. Lancet Neurol. 2010;9:885–894. doi: 10.1016/S1474-4422(10)70183-6. [DOI] [PubMed] [Google Scholar]
- 4.Zhuchenko O, Bailey J, Bonnen P, Ashizawa T, Stockton DW, Amos C, Dobyns WB, Subramony SH, Zoghbi HY, Lee CC. Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the α1A-voltage-dependent calcium channel. Nat Genet. 1997;15:62–69. doi: 10.1038/ng0197-62. [DOI] [PubMed] [Google Scholar]
- 5.Craig K, Keers SM, Archibald K, Curtis A, Chinnery PF. Molecular epidemiology of spinocerebellar ataxia type 6. Ann Neurol. 2004;55:752–755. doi: 10.1002/ana.20110. [DOI] [PubMed] [Google Scholar]
- 6.Gomez CM, Thompson RM, Gammack JT, Perlman SL, Dobyns WB, Truwit CL, Zee DS, Clark HB, Anderson JH. Spinocerebellar ataxia type 6: Gaze-evoked and vertical nystagmus, Purkinje cell degeneration and variable age of onset. Ann Neurol. 1997;42:933–950. doi: 10.1002/ana.410420616. [DOI] [PubMed] [Google Scholar]
- 7.Ikeuchi T, Takano H, Koide R, Igarashi S, Tanaka H, Tsuji S, Takahashi H, Horikawa Y, Honma Y, Onishi Y, Nakao N, Sahashi K, Tsukagoshi H, Inoue K. Spinocerebellar ataxia type 6: CAG repeat expansion in α1a voltage-dependent calcium channel gene and clinical variations in Japanese population. Ann Neurol. 1997;42:879–884. doi: 10.1002/ana.410420609. [DOI] [PubMed] [Google Scholar]
- 8.Matsuyama Z, Kawakami H, Maruyama H, Izumi Y, Komure O, Udaka F, Kameyama M, Nishio T, Kuroda Y, Nishimura M, Nakamura S. Molecular features of the CAG repeats of spinocerebellar ataxia 6 (SCA6) Hum Mol Genet. 1997;6:1283–1287. doi: 10.1093/hmg/6.8.1283. [DOI] [PubMed] [Google Scholar]
- 9.Watase K, Barrett CF, Miyazaki T, Ishiguro T, Ishikawa K, Hu Y, Unno T, Sun Y, Kasai S, Watanabe M, Gomez CM, Mizusawa H, Tsien RW, Zoghbi HY. Spinocerebellar ataxia type 6 knockin mice develop a progressive neuronal dysfunction with age-dependent accumulation of mutant CaV2.1 channels. Proc Natl Acad Sci USA. 2008;105:11987–11992. doi: 10.1073/pnas.0804350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saegusa H, Wakamori M, Matsuda Y, Wang J, Mori Y, Zong S, Tanabe T. Properties of human Cav2.1 channel with a spinocerebellar ataxia type 6 mutation expressed in Purkinje cells. Mol Cell Neurosci. 2007;34:261–270. doi: 10.1016/j.mcn.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Du X, Wang J, Zhu H, Rinaldo L, Lamar K-M, Palmenberg AC, Hansel C, Gomez CM. Second cistron in CACNA1A gene encodes a transcription factor mediating cerebellar development and SCA6. Cell. 2013;154:118–133. doi: 10.1016/j.cell.2013.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jun K, Piedras-Rentería ES, Smith SM, Wheeler DB, Lee SB, Lee TG, Chin H, Adams ME, Scheller RH, Tsien RW, Shin H-S. Ablation of P/Q-type Ca2+ channel currents, altered synaptic transmission, and progressive ataxia in mice lacking the α1A-subunit. Proc Natl Acad Sci USA. 1999;96:15245–15250. doi: 10.1073/pnas.96.26.15245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seidel K, Siswanto S, Brunt ERP, den Dunnen W, Korf H-W, Rüb U. Brain pathology of spinocerebellar ataxias. Acta Neuropathol. 2012;124:1–21. doi: 10.1007/s00401-012-1000-x. [DOI] [PubMed] [Google Scholar]
- 14.Rüb U, Schöls L, Paulson H, Auburger G, Kermer P, Jen JC, Seidel K, Korf H-W, Deller T. Clinical features, neurogenetics and neuropathology of the polyglutamine spinocerebellar ataxias type 1, 2, 3, 6 and 7. Prog Neurobiol. 2013;104:38–66. doi: 10.1016/j.pneurobio.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Ishiguro T, Ishikawa K, Takahashi M, Obayashi M, Amino T, Sato N, Sakamoto M, Fujigasaki H, Tsuruta F, Dolmetsch R, Arai T, Sasaki H, Nagashima K, Kato T, Yamada M, Takahashi H, Hashizume Y, Mizusawa H. The carboxy-terminal fragment of α1A calcium channel preferentially aggregates in the cytoplasm of human spinocerebellar ataxia type 6 Purkinje cells. Acta Neuropathol. 2010;119:447–464. doi: 10.1007/s00401-009-0630-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 17.Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340:82–85. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- 19.Fukaya T, Iwakawa H-o, Tomari Y. MicroRNAs block assembly of eIF4F translation initiation complex in. Drosophila Mol Cell. 2014;56:67–78. doi: 10.1016/j.molcel.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Fukao A, Mishima Y, Takizawa N, Oka S, Imataka H, Pelletier J, Sonenberg N, Thoma C, Fujiwara T. MicroRNAs trigger dissociation of eIF4AI and eIF4AII from target mRNAs in humans. Mol Cell. 2014;56:79–89. doi: 10.1016/j.molcel.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar A, Wong AK-L, Tizard ML, Moore RJ, Lefèvre C. miRNA_Targets: A database for miRNA target predictions in coding and non-coding regions of mRNAs. Genomics. 2012;100:352–356. doi: 10.1016/j.ygeno.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Shin C, Nam J-W, Farh KK-H, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: Functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato K, Hamada M, Asai K, Mituyama T. CENTROIDFOLD: A web server for RNA secondary structure prediction. Nucleic Acids Res. 2009;37:W277–W280. doi: 10.1093/nar/gkp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davidson BL, McCray PB., Jr Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kole R, Krainer AR, Altman S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters L, Meister G. Argonaute proteins: Mediators of RNA silencing. Mol Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song J-J, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 29.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Gennarino VA, Singh RK, White JJ, De Maio A, Han K, Kim J-Y, Jafar-Nejad P, di Ronza A, Kang H, Sayegh LS, Cooper TA, Orr HT, Sillitoe RV, Zoghbi HY. Pumilio1 haplo-insufficiency leads to SCA1-like neurodegeneration by increasing wild-type Ataxin1 levels. Cell. 2015;160:1087–1098. doi: 10.1016/j.cell.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geschwind DH, Perlman S, Figueroa KP, Karrim J, Baloh RW, Pulst SM. Spinocerebellar ataxia type 6. Frequency of the mutation and genotype-phenotype correlations. Neurology. 1997;49:1247–1251. doi: 10.1212/wnl.49.5.1247. [DOI] [PubMed] [Google Scholar]
- 32.Matsumura R, Futamura N, Fujimoto Y, Yanagimoto S, Horikawa H, Suzumura A, Takayanagi T. Spinocerebellar ataxia type 6. Molecular and clinical features of 35 Japanese patients including one homozygous for the CAG repeat expansion. Neurology. 1997;49:1238–1243. doi: 10.1212/wnl.49.5.1238. [DOI] [PubMed] [Google Scholar]
- 33.Takiyama Y, Sakoe K, Namekawa M, Soutome M, Esumi E, Ogawa T, Ishikawa K-y, Mizusawa H, Nakano I, Nishizawa M. A Japanese family with spinocerebellar ataxia type 6 which includes three individuals homozygous for an expanded CAG repeat in the SCA6/CACNL1A4 gene. J Neurol Sci. 1998;158:141–147. doi: 10.1016/s0022-510x(98)00108-7. [DOI] [PubMed] [Google Scholar]
- 34.Wang B, Li S, Qi HH, Chowdhury D, Shi Y, Novina CD. Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat Struct Mol Biol. 2009;16:1259–1266. doi: 10.1038/nsmb.1712. [DOI] [PubMed] [Google Scholar]
- 35.Juvvuna PK, Khandelia P, Lee LM, Makeyev EV. Argonaute identity defines the length of mature mammalian microRNAs. Nucleic Acids Res. 2012;40:6808–6820. doi: 10.1093/nar/gks293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei N, Zhang L, Huang H, Chen Y, Zheng J, Zhou X, Yi F, Du Q, Liang Z. siRNA has greatly elevated mismatch tolerance at 3′-UTR sites. PLOS One. 2012;7:e49309. doi: 10.1371/journal.pone.0049309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valdmanis PN, Gu S, Schüermann N, Sethupathy P, Grimm D, Kay MA. Expression determinants of mammalian argonaute proteins in mediating gene silencing. Nucleic Acids Res. 2012;40:3704–3713. doi: 10.1093/nar/gkr1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu S, Jin L, Huang Y, Zhang F, Kay MA. Slicing-independent RISC activation requires the argonaute PAZ domain. Curr Biol. 2012;22:1536–1542. doi: 10.1016/j.cub.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hauptmann J, Kater L, Löffler P, Merkl R, Meister G. Generation of catalytic human Ago4 identifies structural elements important for RNA cleavage. RNA. 2014;20:1532–1538. doi: 10.1261/rna.045203.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai BP, Jimenez J, Lim S, Fitzgerald KD, Zhang M, Chuah CTH, Axelrod H, Wilson L, Ong ST, Semler BL, Waterman ML. A novel Bcr-Abl–mTOR–eIF4A axis regulates IRES-mediated translation of LEF-1. Open Biol. 2014;4:140180. doi: 10.1098/rsob.140180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weingarten-Gabbay S, Khan D, Liberman N, Yoffe Y, Bialik S, Das S, Oren M, Kimchi A. The translation initiation factor DAP5 promotes IRES-driven translation of p53 mRNA. Oncogene. 2014;33:611–618. doi: 10.1038/onc.2012.626. [DOI] [PubMed] [Google Scholar]
- 42.Khosrow A. Translational control mechanisms in metabolic regulation: Critical role of RNA binding proteins, microRNAs, and cytoplasmic RNA granules. Am J Physiol Endocrinol Metab. 2011;301:E1051–E1064. doi: 10.1152/ajpendo.00399.2011. [DOI] [PubMed] [Google Scholar]
- 43.Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pestova TV, Shatsky IN, Hellen CU. Functional dissection of eukaryotic initiation factor 4F: The 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CUT. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci USA. 2009;106:9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science. 2016;351:aad4939. doi: 10.1126/science.aad4939. [DOI] [PubMed] [Google Scholar]
- 47.Gomez CM. Two Proteins Encoded by the CACNA1A Gene and Their Role in Cerebellar Development and Disease. Cerebellum Gordon Research Conference; Lewiston, ME. August 12, 2015. [Google Scholar]
- 48.Ibrahim AF, Weirauch U, Thomas M, Grünweller A, Hartmann RK, Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011;71:5214–5224. doi: 10.1158/0008-5472.CAN-10-4645. [DOI] [PubMed] [Google Scholar]
- 49.van Rooij E, Purcell AL, Levin AA. Developing microRNA therapeutics. Circ Res. 2012;110:496–507. doi: 10.1161/CIRCRESAHA.111.247916. [DOI] [PubMed] [Google Scholar]
- 50.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Näär AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burnett JC, Rossi JJ. RNA-based therapeutics: Current progress and future prospects. Chem Biol. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X, Li Y, Chen YE, Chen J, Ma PX. Cell-free 3D scaffold with two-stage delivery of miRNA-26a to regenerate critical-sized bone defects. Nat Commun. 2016;7:10376. doi: 10.1038/ncomms10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dasgupta A, Das S, Izumi R, Venkatesan A, Barat B. Targeting internal ribosome entry site (IRES)-mediated translation to block hepatitis C and other RNA viruses. FEMS Microbiol Lett. 2004;234:189–199. doi: 10.1016/j.femsle.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 54.Gasparian AV, Neznanov N, Jha S, Galkin O, Moran JJ, Gudkov AV, Gurova KV, Komar AA. Inhibition of encephalomyocarditis virus and poliovirus replication by quinacrine: Implications for the design and discovery of novel antiviral drugs. J Virol. 2010;84:9390–9397. doi: 10.1128/JVI.02569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis DR, Seth PP. Therapeutic targeting of HCV internal ribosomal entry site RNA. Antivir Chem Chemother. 2011;21:117–128. doi: 10.3851/IMP1693. [DOI] [PubMed] [Google Scholar]
- 56.Stoneley M, Subkhankulova T, Le Quesne JPC, Coldwell MJ, Jopling CL, Belsham GJ, Willis AE. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 2000;28:687–694. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jopling CL, Spriggs KA, Mitchell SA, Stoneley M, Willis AE. L-Myc protein synthesis is initiated by internal ribosome entry. RNA. 2004;10:287–298. doi: 10.1261/rna.5138804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Webb TE, Hughes A, Smalley DS, Spriggs KA. An internal ribosome entry site in the 5′ untranslated region of epidermal growth factor receptor allows hypoxic expression. Oncogenesis. 2015;4:e134. doi: 10.1038/oncsis.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kordasiewicz HB, Thompson RM, Clark HB, Gomez CM. C-termini of P/Q-type Ca2+ channel α1A subunits translocate to nuclei and promote polyglutamine-mediated toxicity. Hum Mol Genet. 2006;15:1587–1599. doi: 10.1093/hmg/ddl080. [DOI] [PubMed] [Google Scholar]
- 60.Lian SL, Li S, Abadal GX, Pauley BA, Fritzler MJ, Chan EKL. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA. 2009;15:804–813. doi: 10.1261/rna.1229409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leung AKL, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gradi A, Imataka H, Svitkin YV, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miyazaki Y, Adachi H, Katsuno M, Minamiyama M, Jiang Y-M, Huang Z, Doi H, Matsumoto S, Kondo N, Iida M, Tohnai G, Tanaka F, Muramatsu S-i, Sobue G. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat Med. 2012;18:1136–1141. doi: 10.1038/nm.2791. [DOI] [PubMed] [Google Scholar]
- 64.Busk PK. A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinformatics. 2014;15:29. doi: 10.1186/1471-2105-15-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iida A, Takino N, Miyauchi H, Shimazaki K, Muramatsu S-i. Systemic delivery of tyrosine-mutant AAV vectors results in robust transduction of neurons in adult mice. Biomed Res Int. 2013;2013:974819. doi: 10.1155/2013/974819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miyazaki T, Watanabe M. Development of an anatomical technique for visualizing the mode of climbing fiber innervation in Purkinje cells and its application to mutant mice lacking GluRδ2 and Cav2.1. Anat Sci Int. 2011;86:10–18. doi: 10.1007/s12565-010-0095-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Schematic representation of AAV9 vectors and AAV9 transduction efficiency into Purkinje cells.
Fig. S2. Histopathological examination of the AAV9-injected mouse cerebral cortex and hippocampus.
Fig. S3. The long-term follow-up of AAV9-injected mouse behavioral phenotypes and CACNA1A IRES–driven α1ACT expression in mice.
Fig. S4. Stem-loop structure of CACNA1A IRES and the predicted binding sites of miR-711, miR-3191-5p, and miR-4786-3p.
Fig. S5. Effects of miR-3191-5p on mutated CACNA1A IRES templates.
Fig. S6. Effects of eIF4AII and eIF4GII on mutated CACNA1A IRES templates in the presence or absence of miR-3191-5p.
Fig. S7. Cotransduction of AAV9-α1ACT-Q33 with either AAV9–miR-mock or AAV9–miR-3191-5p.
Fig. S8. The long-term follow-up of therapeutic effect of miR-3191-5p on mouse behavioral phenotypes and CACNA1A IRES–driven α1ACTSCA6 expression in mice.
Fig. S9. Potential mouse mRNAs targeted by human miR-3191-5p.
Fig. S10. Histopathological examination of the brain, cerebellum, heart, lung, liver, and kidney of wild-type mice injected with AAV9–miR-3191-5p.
Table S1. Source data.
Movie S1. DigiGait video of an 8-week-old AAV9-GFP mouse on treadmill.
Movie S2. DigiGait video of an 8-week-old AAV9-α1ACT-Q11 mouse on treadmill.
Movie S3. DigiGait video of an 8-week-old AAV9-α1ACT-Q33 mouse on treadmill.
Movie S4. DigiGait video of an 8-week-old AAV9–Q33–miR-mock mouse on treadmill.
Movie S5. DigiGait video of an 8-week-old AAV9–Q33–miR-3191-5p mouse on treadmill.