Abstract
Next-generation sequencing (NGS) technology has demonstrated that the cancer genomes are peppered with mutations. Although most somatic tumour mutations are unlikely to have any role in the cancer process per se, the spectra of DNA sequence changes in tumour mutation catalogues have the potential to identify the mutagens, and to reveal the mutagenic processes responsible for human cancer. Very recently, a novel approach for data mining of the vast compilations of tumour NGS data succeeded in separating and precisely defining at least 30 distinct patterns of sequence change hidden in mutation databases. At least half of these mutational signatures can be readily assigned to known human carcinogenic exposures or endogenous mechanisms of mutagenesis. A quantum leap in our knowledge of mutagenesis in human cancers has resulted, stimulating a flurry of research activity. We trace here the major findings leading first to the hypothesis that carcinogenic insults leave characteristic imprints on the DNA sequence of tumours, and culminating in empirical evidence from NGS data that well-defined carcinogen mutational signatures are indeed present in tumour genomic DNA from a variety of cancer types. The notion that tumour DNAs can divulge environmental sources of mutation is now a well-accepted fact. This approach to cancer aetiology has also incriminated various endogenous, enzyme-driven processes that increase the somatic mutation load in sporadic cancers. The tasks now confronting the field of molecular epidemiology are to assign mutagenic processes to orphan and newly discovered tumour mutation patterns, and to determine whether avoidable cancer risk factors influence signatures produced by endogenous enzymatic mechanisms. Innovative research with experimental models and exploitation of the geographical heterogeneity in cancer incidence can address these challenges.
Introduction
Cancer is a genetic disease, and mutations in genes that drive cancer constitute the overriding molecular events leading to malignant growth. During the first decade of the next-generation sequencing (NGS) revolution, the focus of whole-genome and whole-exome cancer sequencing projects was to describe the genome landscape of major human cancers, that is, to identify groups of genes (driver genes) that contribute to the growth of different types of tumours when mutated.1, 2 This massive effort has made it clear that the set of mutated driver genes in cancer genomes typically consists of fewer than 10 in any given tumour. Driver genes provide a blueprint of the malignant process, and offer targets for specific therapies.3, 4, 5 In less than a decade, NGS identified most genes in the genome that can provide a growth advantage to a cell if mutated. Fewer than 1% of all human genes appear to have this potential to drive neoplastic development. A characteristic subset of driver genes harbouring deleterious mutations has been identified for each major cancer type, corroborating the notion that cancer is many diseases, each type following an underlying developmental path. Although there is considerable heterogeneity in the genome landscapes of different cancers, it appears that all driver gene products affect a common set of biological pathways.5, 6
Although the first goal of tumour NGS data analysis was to identify driver gene mutations buried amongst a plethora of accumulated sequence changes, it became apparent that the frequency and types of common base substitutions differed substantially across cancer types. Furthermore, mutation pattern heterogeneity could arise in distinct sets of tumours of the same type.3, 7 Although it is generally accepted that some of this diversity stems from differences in patient exposure history, cursory perusal of mutation profiles did not lead significantly further in identifying the sources of mutations beyond what had been achieved previously through mutation spectra analysis of single cancer genes. Once methods were applied to parse enigmatic mutation catalogues into specific mutational signatures, however, the picture changed entirely. Computational mining of information previously locked in mutation databases allowed tight associations to be made between specific cancer risk factors and unique patterns of sequence changes in tumours.
Essential observations from sequencing single cancer genes in tumours: implications for cancer aetiology
Mutation patterns amongst different cancer types are different
Mutation analysis of individual cancer genes, which preceded scrutiny of NGS mutational catalogues, provided the first evidence that carcinogenic insults leave mutational ‘fingerprints' on tumour DNA. In the decades leading up to tumour NGS studies, catalogues of DNA sequence changes in frequently mutated genes such as the TP53 tumour suppressor gene or the K-ras, and B-raf oncogenes offered a first glimpse of the mutational pathways operating in human cancers. Analysis of skin and lung tumours provided convincing demonstration of an environmental impact on tumour mutation patterns.8, 9, 10 Numerous reports contributed to the understanding that exposure to ultraviolet (UV) light, the primary cause of skin cancer, is responsible for the uniquely characteristic C to T transitions at dipyrimidines in skin tumours, and that tobacco smoking causes G to T transversions, the predominant sequence changes present in lung tumours.11 (Note: When possible, we describe a mutation by naming the base proposed to carry the pre-mutagenic lesion rather than by using the COSMIC system (Catalogue of Somatic Mutations in Cancer; cancer.sanger.ac.uk), which uniformly names the pyrimidine of the Watson-Crick base pair. When the pre-mutagenic lesion is currently unknown, we employ the COSMIC system.) Despite the limited scope of single-gene sequencing, these and other valuable insights from such projects continue to emerge, particularly from the analysis of TP53. One reason why sequencing TP53 is particularly informative in revealing sources of mutagenic insult is that any one of numerous single base changes along the coding sequence is sufficient to disrupt its proper function.12, 13 Such diversity of potential mutations and sequence contexts can reveal discrete mutation profiles. Each TP53 mutation in a set of tumours of a specific type is classified according to the type of base change, strand orientation, and sequence location, and the frequencies of specific alterations are then analysed. The tumour-specific patterns that emerge (such as the TP53 G to T transversions on the non-transcribed strand in smokers' lung tumours clustering at hotspots in codons 157, 158 and 273) represent rudimentary ‘signatures' produced by the action of mutagenic processes.11, 14 As fundamental DNA-damaging properties of human carcinogenic agents such as UV light and tobacco carcinogens had been well-characterized in the laboratory,15, 16, 17 the effects of these agents on DNA were promptly recognized in skin and lung tumour TP53 mutation spectra, a major step forward at the time.
Within this single-gene framework, however, the mutation spectrum is small in scale and the approach is fraught with limitations. First, as each patient analysis typically contributes just one mutation, fingerprints only begin to emerge as data from many individuals are pooled. Second, as driver gene mutations are selected during cancer development, the types of tumour mutations likely to be detected are generally limited to the specific changes and gene locations capable of unleashing oncogenic potential are not necessarily characteristic of the genome's mutation load as a whole. The B-raf mutation spectrum in melanomas illustrates the limitations in single-gene analysis in revealing sources of a somatic mutation burden. In the B-raf driver gene, the mutagenic risk factor fails to leave its identifying fingerprint.8 Almost all B-raf mutations in melanoma are T to A transversions, yet the primary risk factor is UV light, powerful mutagen that produces C to T and CC to TT base changes at pyrimidine dinucleotides. An explanation for this anomaly is that most oncogenic B-raf mutations occur at a hotspot, the second nucleotide of B-raf codon 600. The sequence context (ACA GTG AAA) cannot capture the hallmark dinucleotide target of UV radiation. In contrast, melanoma mutations in TP53 are dispersed across the locus and do indeed display the UV-characteristic C to T transitions at dipyrimidines and CC to TT tandem mutations. (Of general note, not all mutations that a carcinogen induces will be typical of its action on DNA. Thus, T to A mutations in the B-raf gene of melanoma, although uncharacteristic of UV exposure, may well have arisen from exposure to UV, even though a T to A substitution is not the most likely molecular change that sunlight generates.)
Within a cancer type, mutation patterns in a single gene can diverge widely when groups of patients with different exposure histories are examined
Whilst it was highly plausible that risk factors are responsible for some of the mutation pattern diversity amongst different types of cancer, demonstration of a specific risk factor mutation pattern present in tumours from exposed patients, but absent in non-exposed patients with the same type of cancer, strengthens the argument considerably. Extensive supporting evidence has come from TP53 analysis of lung, urothelial and liver cancers. The G to T mutation fingerprint discovered in lung cancers and linked to tobacco smoking is not evident in lung cancer patients who are never-smokers, and the greater the tobacco smoke exposure, the more pronounced is the G to T mutation load in sentinel driver genes such as TP53.10 In urothelial cancers from patients exposed to the plant carcinogen aristolochic acid (AA), there is a striking preponderance of TP53 A to T mutations on the non-transcribed strand of DNA, the primary type of mutation induced in laboratory mutagenesis experiments with AA.18, 19 The signature does not appear in patients with no history of AA exposure. Finally, a unique liver cancer TP53 mutation pattern, characterized by strand-biased G to T substitutions predominantly at codon 249, is present in hepatocellular carcinomas (HCC) from geographical regions (for example, parts of China and sub-Saharan Africa) where there is chronic, high-level exposure to aflatoxin B1 (AFB1), and hepatitis B virus infection is prevalent.17, 20, 21 In populations where other risk factors prevail and exposure to AFB1 is minimal or absent, TP53 mutations in HCC are diverse in type and location.22 A variety of laboratory test systems demonstrated that AFB1 induces primarily G to T mutations. The codon 249 G to T hotspot mutation has shown its use as a powerful molecular biomarker of HCC risk and disease burden in regions where exposure to AFB1 is high, but it would be of little value as a biomarker in cohorts with no exposure to this carcinogen.
Overall, DNA sequencing of TP53 continues to generate evidence supporting the prediction that two cohorts with the same cancer type but exposed to different environmental risk factors can have different characteristic mutations in their tumours. Mutation spectra in oncogenes and tumour suppressor genes have also indicated that the multiplicity of distinct risk-associated TP53 mutation patterns in human tumours presaged the diversity in mutation patterns now emerging from tumour NGS data.
The game changer: genome-wide sequencing data and computational analysis
Mutation research has been witness to three seminal advances, each of which prompted a flurry of activity in laboratories around the world. First, in the 1970s, development of the rapid Salmonella/microsome assay for testing mutagenicity of chemicals, and subsequently the report on test results with 300 chemicals, established the fact that the majority of known and suspected human carcinogens are mutagenic.23 More than a decade later, Vogelstein and colleagues discovered that colorectal cancers harbour a variety of inactivating point mutations in TP53.24 This finding prompted a deluge of reports describing TP53 mutations in a variety of human tumours. The fact that the mutations were found in target sequences large and complex enough to reveal different mutation patterns in various tumour types was a turning point because tumour TP53 mutations provided the first comprehensive evidence in clinical samples that exposure to mutagenic carcinogens leave fingerprints on tumour DNA.25 With the advent of NGS technologies, the third quantum leap in mutation research on cancer aetiology is now upon us. NGS-derived mutation data constitutes a blurred mixture of fingerprints from different mutagenic processes, however, necessitating de-confounding computational procedures to identify discrete mutational signatures in simple mathematical terms.26 The somatic mutations found in cancer genomes are approximated as a linear mixture of multiple mutational signatures, each contributing a different number of mutations to different genomes:
In principle, the known set of mutations in cancer genomes is used to find the optimal set of signatures and respective exposures that best describe the original catalogues of somatic mutations. This problem can be considered as a specific case of a blind source separation problem, and the challenge is to unscramble not-observed latent variables (that is, mutational signatures and their exposures) from a set of mixtures (that is, somatic mutations in cancer genomes). To ‘unmix' and reconstruct the original sources from the records, a blind source separation algorithm is needed for best possible extraction of original signals from mixtures. The unmixing and reconstruction of the original signals is based on constrained and/or regularized optimization procedure minimizing an objective cost function together with a few imposed constraints, such as maximum variability, statistical independence, non-negativity, smoothness, sparsity, simplicity, and so on. The choice of optimization constraints is based on prior knowledge about the processed data, and hence the constraints could be different for every particular case. The non-negative nature of somatic mutations requires at the very least applying a non-negative constraint for solving the cancer genomics blind source separation problem. Alexandrov et al.26 used a widely applied approach designated non-negative matrix factorization (NMF; Figure 1) to provide an effective solution.27 NMF does not seek statistical independence or constrain any other statistical property of the mixed signals, and thus allows the estimated sources to be partially or entirely correlated. When tumour mutational catalogues are analysed with mathematical procedures such as NMF, numerous carcinogenic fingerprints hidden in a vast set of human NGS-analysed tumours can be separated and identified with unprecedented clarity, fast-forwarding our understanding of mutation origins during the evolution of cancer.26, 28
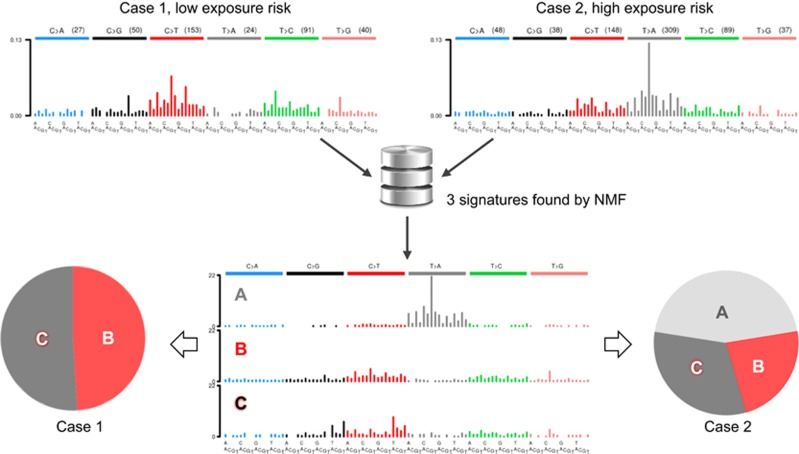
Figure 1.
When patients with the same cancer type have different exposure histories, the mutation patterns in their tumours can be strikingly different. Two representative cases of upper urinary tract urothelial tumours from regions of either low or high risk of exposure to the carcinogen aristolochic acid97 were analysed using whole-exome sequencing. The single-base substitution distribution spectra are shown on top. Performing NMF on the studied case series identified three distinct mutational signatures (A, B and C; middle panel). The pie charts show the proportionate contribution of individual signatures to the mutational load in each tumour. The absence of signature A in case 1 argues that the two tumours have distinct aetiologies.
Despite the apparent neutrality of bystander mutations in the cancer process, their sheer numbers promise to provide a far more powerful way than individual onco-mutation analysis to observe signatures of mutagenic activity. Understanding the mutagenic processes corresponding to NGS mutational signatures, however, continues to rely on finding matches with experimentally induced signatures or other laboratory data.
Diverse mutational processes are responsible for the heterogeneity in tumour NGS mutation spectra amongst different cancer types
The first NMF-based pan-analysis of NGS data from a broad assortment of different cancers demonstrated unequivocally that tumour types differ in their genome-wide mutation profiles, and presented compelling argument that distinct risk factors associated with each cancer type are likely to explain much of the heterogeneity in mutation spectra across tumour types.28 Twenty-one distinct mutational signatures were extracted from mutation data on 30 types of cancer from 7042 patients in this unprecedented study, and a known cancer risk factor or endogenous molecular process was putatively assigned to many of the signatures. The number of distinct mutational signatures is now at 30 (source: COSMIC) and may soon approach 50 as the results of pan-cancer analyses become validated, and as patients from geographic areas not previously tested become examined. Table 1 describes five signatures assigned to specific human carcinogenic exposures. (Note: In the following discussion, different signatures are referred to according to their unique identifying number. See http://cancer.sanger.ac.uk/cosmic/signatures)
Table 1. Mutational signatures assigned to IARC Group 1 carcinogen exposures.
| Name | Exposure | Group 1 carcinogen | Chemical class | Characteristic pre-mutagenic DNA lesion | Signature hallmarks | Prominent trinucleotide target in the signature |
|---|---|---|---|---|---|---|
| Signature 4 | Tobacco smoke | Benzo[a]pyrene | PAH | (+)benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide-dG adduct | G to T GG to TT tandem mutations Transcriptional strand bias | GGGGGA |
| Signature 7 | Sunlight | Ultraviolet light | NA | Py-Py photodimers | C to T at dipyrimidines CC to TT tandem mutations Transcriptional strand bias | TCC |
| Signature 11 | Chemotherapy | Temozolomide | Alkylating agent | O6-methylguanine | G to A Transcriptional strand bias | GGGGGA |
| Signature 22 | Dietary contaminant (grain); herbal medicine | Aristolochic acid | Plant alkaloid | 7-(deoxyadenosin-N(6)-yl) aristolactam I adduct | A to T Transcriptional strand bias | CAG |
| Signature 24 | Dietary contaminant (groundnuts) | Aflatoxins | Fungal toxin | 8,9-dihydro-8-(N7-guanyl)-9-hydroxyaflatoxin B1 adduct | G to T Transcriptional strand bias | GGC |
| Signature 29 | Tobacco chewing | Unspecified | Unspecified | Unspecified | G to T Transcriptional strand bias | TGCGGCTGT |
Abbreviations: IARC, International Agency for Research on Cancer; NA, not applicable; PAH, polycyclic aromatic hydrocarbon; Py, pyrimidine.
Group 1 refers to the IARC classification of substances for which there is sufficient evidence of carcinogenicity to humans. The third column lists sources of exposure with documented links to cancer risk. Bases and context sequence are shown to reflect the base targeted by the mutagen (for example, as 5′-GGA-3′ for B[a]P, but as 5′-TCC-3′ for UV). The targeted base in its preferred sequence context is underlined in the last column. The targeted trinucleotides in the last column are extracted from signature analysis of human tumours; analysis of experimental models with single, controlled exposures recapitulates major features from the human data (Figure 2).
The diversity in mutation patterns amongst cancer types can be illustrated by a comparison of signatures in small cell lung cancer, acute myeloid leukaemia and cutaneous melanoma.28 In each of these cancer types one signature (but not the same one) contributed >85% of the total mutational burden. The tobacco smoking-associated signature 4, characterized by G to T transversions with transcriptional strand bias, dominated in small cell lung cancers, whereas acute myeloid leukaemia mutations were overwhelmingly C to T transitions at CpG dinucleotides (signature 1), presumably attributable to spontaneous deamination of 5-methylcytosine, and clearly distinguishable from the UV signature C to T transitions at dipyrimidines (signature 7) in melanoma.
The mutation spectrum derived from NGS of a tumour is composed of superimposed signatures left by various mutagenic insults
In most cancer types, parsing of NGS mutational catalogues demonstrated the presence of several distinct mutational signatures, in keeping with cancer aetiologies where multiple exposures are thought to significantly contribute to risk. The fact that in NGS analysis, each tumour provides an entire spectrum of mutations (rather than a set of tumours required for single gene-based analysis) has offered unprecedented opportunity to explore the multi-factor aspect of human cancer. Despite caveats mentioned below regarding signatures in branch mutations accumulating during clonal evolution, genome-wide mutations in a tumour can be displayed as a weighted composite of distinct mutational signatures, allowing a first approximation of the relative contribution of each risk-associated signature to the total mutation burden in the tumour. With NMF or similar mathematical approaches,27, 28, 29, 30 a rough estimate of the relative impact of multiple risk factors on the total mutation load can be obtained, a goal that was out of reach in the single-gene mutational analysis era. In the initial study applying NMF to NGS data from 30 different tumour types, liver cancer displayed the greatest number of distinct mutational signatures, presumably reflecting the multi-factorial aetiology of cancer at this site discernible from the data archives used. Seven signatures were identified, amongst them signature 16, apparently unique to liver cancers, which was detected in 90% of the tumours sequenced, and contributed anywhere from a few percentages to over half of all the somatic mutations recorded in a given sample. The cause of signature 16 mutations, characterized by strand-biased A to G transitions at NpApT sites, is unclear. This observation is intriguing because HCC is one of the few cancers with several known major risk factors, notably infection by hepatitis B or C viruses, alcohol consumption and exposure to AFB1. A recent study uncovered signature 24, one of the signatures characterized by frequent strand-biased G to T transversions, in six hepatitis B virus-infected HCC patients originating from subtropical Africa.31 Extended cohort-specific as well as experimental studies are warranted to strengthen the proposed link between this signature and aflatoxin B1 exposure.
At present, of the first 30 distinct signatures defined, 60% have been provisionally assigned to known carcinogens or mutational processes. The remaining orphan signatures highlight the dearth of experimental mutation research, sending out a priority research call.
Specific endogenous mutational processes have a major impact on the mutation burden in human populations
The risk of sporadic adult cancer increases exponentially with age.32 Deamination of 5-methylcytosine, a well-studied endogenous spontaneous mutagenic process known to erode DNA sequence integrity, presents as C to T transitions at CpG dinucleotides, the ubiquitous age-associated signature labelled signature 1.33 Tumour mutation catalogues from almost all 30 types of cancers in the seminal study of Alexandrov et al.28 had at least some trace of this signature, and in some cancers signature 1 predominated.
The accumulation of this and other classes of mutational events, such as those stemming from spontaneous base hydrolysis or the inherent infidelity of DNA replication and repair,34 is to a certain extent essentially inevitable, as are some cancers. A recent study suggested that 10–30% of cancers can be primarily attributed to intrinsic factors,35 although some argument persists regarding the proportion of human cancers that presumably cannot be avoided by changes in lifestyle or environment.36 However, much remains to be understood with regard to the effect of external exposures on endogenous pro-mutagenic processes mentioned. On the basis of geographical disparities in cancer incidence within cancer types,37, 38 current estimates suggest ~90% of the global cancer burden could in principle be avoided, a large fraction of which may harbour mutational signatures that could be linked to patient exposure history. In contrast, two signatures of endogenous mutational processes discernible in practically all cancer types, signature 1 (C to T at CpG) mentioned above, and signature 5 (a diffuse pattern produced by unknown underlying molecular mechanism(s)), have been linked to age, the most inevitable and ubiquitous cancer risk factor. These two mutation patterns, attributed to ‘clock-like' cellular processes, are the only signatures described thus far for which a correlation was found between the number of such mutations and the chronological age of patients at diagnosis.33 Although it is unclear to what extent genetic background or external factors can accelerate this internal clock in normal cells, the tumours in which these signatures predominate are more likely to be those that contribute to the baseline incidence of cancer in humans.35
Surprisingly, of the first 30 signatures revealed by NMF, almost half correspond to patterns generated by enzymatic processes affecting DNA homoeostasis.39 For example, signatures 9 and 10 are similar to mutation patterns left in the wake of DNA repair polymerases eta and epsilon, respectively, and signatures 6, 15 and 20 imply defective DNA mismatch repair. Further, signature 3 has been found in the majority of samples harbouring pathogenic BRCA1/2 mutations indicating that this signature reflects failure of DNA double strand repair by homologous recombination.40 It has been long recognized that cancer patients with inherited deleterious mutations in DNA repair enzymes have tumours with a hypermutator phenotype.41 However, inherited cancer syndromes of this class are relatively rare, so the demonstration that enzymatic DNA maintenance mechanisms appear to contribute to diverse types of sporadic cancers raises the question as to whether avoidable, known cancer risk factors can influence the impact from these pathways on the human mutation burden. In particular, the extent to which cancer risk factors that do not act through a direct mutational mechanism exert an influence on genome-altering cellular processes is one of the most enticing areas of cancer research, offering rich opportunities for laboratory science and epidemiology.
It is worth remembering that the human tumours subjected to NGS in the first phase of studies were not selected to address hypotheses about aetiology. Patients were not necessarily representative of the patient population for a given type of cancer, being typically recruited from a small number of high-income countries, and little epidemiological data were available or collected on the exposure history of the subjects. It is thus premature to draw conclusions about the number or prevalence of distinct mutational signatures occurring for a given cancer worldwide.
Modulation of the activity of the APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) family of deaminases
Remarkably, the first signature analysis of NGS data28 revealed that 16 of the 30 cancer types displayed signatures that matched the mutator activities of APOBEC deaminases (signatures 2 and 13). In connection with its eponymous function, this large family of enzymes has several biological tasks, including viral restriction and suppression of retrotransposition.42 The collateral damage these enzymes inflict on single-stranded genomic DNA has been characterized extensively in experimental model systems, facilitating recognition of their mutational impact on human tumour DNA.43, 44, 45, 46 On the basis of this characterization, a role for APOBEC3A and/or APOBEC3B in human cancer is more likely than for other members of the family. The putative contribution of the APOBEC3 enzyme activity to the total tumour mutation load reported in several independent NGS studies of breast tumours47, 48 is an important clue in elucidating the incompletely understood aetiology of sporadic breast cancer. With respect to APOBEC3 dysregulation in this cancer type, alterations at the gene locus itself (coding sequence or promoter mutation, gene copy-number polymorphism) and induction of enzymatic activity by factors in the cellular environment may be responsible. 49, 50, 51
In-depth exploration of APOBEC expression modulation by cancer risk factors is needed in the wake of these recent surprising discoveries on the putative impact of APOBEC on the human mutation burden. Interestingly, significant numbers of signature 2 mutations are present in cervical cancer and in head and neck tumours,52 two types of cancer with human papillomavirus (HPV) involvement.53 Elevated APOBEC3 activity in HPV-infected cells would be a further manifestation of the APOBEC gene family responses to viral infection.54, 55 In a recent study, mutations related to the APOBEC signatures 2 and 13 found in HPV-positive head and neck cancers were reported enriched relative to the HPV-negative counterparts.56 In most cancer types exhibiting APOBEC dysregulation, however, the underlying causes remain enigmatic, with the exception of the small numbers of tumours found to harbour gene copy polymorphisms or deleterious mutations involving the APOBEC locus.
Physicochemical mutational processes, ‘amorphous' risk factors and co-mutagenic agents: SEVERAL elephants in the room?
In general, the mutagenic impact of reactive chemicals in the internal environment of the cell, and external influences on the mutagenic potential of endogenous enzymes are difficult to assess. Furthermore, it is not known how or to what extent established but ‘amorphous' risk factors with no assigned genome-wide mutational signature, such as obesity, chronic inflammation, physical inactivity, and reproductive history, modulate mutation patterns. The chemical properties of reactive oxygen species, nitrogen radicals and lipid peroxidation products associated with oxidative stress and chronic inflammation link them directly to DNA damage and these molecules are considered an important source of tumour mutations.57, 58 Nevertheless, information on the relative contribution from such sources to tumour mutation load is imprecise, and the specific patterns in base substitution distribution they might produce are ill-defined. Attack on DNA by endogenous cellular chemicals has been shown in numerous studies to elicit specific classes of base substitution, however. A recent study reported that DNA exposed to hydrochlorous acid, a chemical secreted by neutrophils in inflamed tissues, acquired 5-chlorocytosine residues, a modification that caused transitions to T, a common mutation type overall in human cancers even when the particular subclass CpG to TpG, attributable to deamination of 5-methylcytosine (signature 1), is not considered.59
Mathematical analysis of data from fit-for-purpose NGS studies, for example by comparing mutations in distinct risk cohorts, should bring more clarity to this prickly topic.60 ‘Amorphous' risk factors present no small challenge; whereas many chemical carcinogens produce unique DNA adducts that serve as traces of exposure, episodic exposures from endogenous chemical flux, or exposure to a non-mutagenic agent acting on endogenous mutational processes from a distance, are difficult to pinpoint. Finally, some risk factors may impact risk primarily by modulating a different trajectory of cancer development such as immune surveillance of cancerous cells, and not by increasing the mutation load.
Episodic exposures in cancer evolution
Two recent reports on the multi-clonal evolution of lung cancer and the role of APOBEC3B activity, in which truncal (early) mutations were compared against branch (more recent) mutations illustrate how temporal shifts in mutation patterns feed into the mutational landscape of a full-blown cancer.61, 62 The two studies, which traced lung cancer development by sampling tumours at multiple locations, concluded that APOBEC3B dysfunction typically exerts effects later in the evolution of the primary clone. The enzyme's signature was evident amongst branch mutations but not in truncal mutations. Thus, whilst parsing of a mutational catalogue can estimate the relative importance of multiple signatures, and hence exposures in the natural history of the cancer, the percentage of the total mutation burden in the late stages of cancer that are attributable to a given signature/exposure may not necessarily indicate the relative importance of multiple environmental exposures in initiating a cancer. Obtaining multiple biopsies of an exposed organ or cancer to assess tissue burden of mutant cells, or to retrace the evolution of the mutational load and the timing of distinct mutational insults, is a strategy that gains power from NGS and mutational signature analysis.61, 63, 64, 65, 66, 67, 68 In principle, one could revisit the migrant studies or time-trend studies of descriptive epidemiology but with genome-wide mutational analysis. For example, changes in mutation pattern following changes in risk factor exposure over a lifetime could be tracked in cancers from migrant populations, particularly when the difference in exposure patterns and cancer incidence between the patients' country of origin and the subsequent place of residence is extreme. An example would be migrants from Africa to Europe where exposure to the dietary carcinogen AFB1 is markedly different. The International Agency for Research on Cancer World Cancer Report 2014 contains numerous examples of widely differing exposure patterns and more than 10-fold geographical discrepancy in incidence for a number of common cancers.38 Alternatively, one could examine cancer types that have seen rapid changes in incidence over time, an example being the increases in countries undergoing rapid development, offering opportunities to compare spectra for the same tumour in the same population but in the face of different environmental and lifestyle exposures.
Examples of mutation spectra heterogeneity within a cancer type, attributable to differences in risk factor exposures
Evidence for a direct role of external risk factors in shaping human tumour mutation spectra is now accumulating from NGS projects specifically designed to capture this information by comparison of groups of patients with the same type of cancer, but differing in exposure to a known cancer-causing agent. Investigations along these lines show that mutation patterns can indeed be heterogeneous within a cancer type and that differences in risk factor exposures explain this variation. Three prominent examples are discussed here that parallel observations from earlier single-gene studies.
Mutation patterns attributable to tobacco smoking
The great majority of lung cancers worldwide arise in patients who smoke or have smoked tobacco. The outstanding features of the lung cancer NGS mutation spectrum, corroborated by several projects involving hundreds of lung cancer patients, are (i) the presence of a distinct strand-biased G to T transversion signature in smokers but, crucially, absent in never-smokers, and (ii) the high numbers of somatic mutations per tumour.7, 28, 69, 70 Computational methods have defined signatures provisionally attributable to tobacco smoke exposure although preferred sequence contexts where presumptive tobacco-associated transversions accumulate in respiratory tract cancers are not fully established, perhaps reflecting the chemical complexity of tobacco smoke. It is unlikely that NMF of mutational catalogues will differentiate between fingerprints of two distinct carcinogens that both induce strand-biased G to T substitutions should the preferred sequence contexts of the two chemicals overlap significantly. Tobacco smoke (along with alcohol consumption and HPV infection) is a principal risk factor for head and neck cancers as well as lung cancer. As expected, NGS analysis of 74 head and neck cancers, 89% of which were from patients with a history of tobacco use, identified a prominent strand-biased G to T mutation pattern similar to findings in smokers' lung tumours.71 The highest prevalence of the transversions occurred in tumours with the highest mutation burden overall, suggesting that G to T mutations could serve as a readout of tobacco smoke exposure. The mutagenic impact of tobacco carcinogens across various tissues is not uniform, however; bladder cancers of smokers do not have the same mutation profile as smokers' lung tumours.72 Differences in tissue distribution and metabolism of carcinogens in tobacco smoke are two of the many factors potentially responsible for multiple tumour type-specific mutation patterns produced by a given exposure. With respect to head and neck cancers, the tissue-specific effect of tobacco smoke is a particularly complex issue when tumours of many different cell types and subsites are grouped together for analysis. A recent NGS study that addressed this problem revealed that mutations in tongue squamous cell carcinomas do not exhibit a pattern corresponding to the spectrum found in smokers' lung cancers, whereas mutations in tumours of the larynx do.73
The AA fingerprint
Epidemiological and experimental evidence have long conspired to incriminate AA in the aetiology of upper urinary tract urothelial carcinoma (UTUC).74 AA is a potent plant mutagen that contaminates grain in some regions, such as rural areas along the lower Danube River, and is present in Aristolochia-containing herbal medicines popular in a number of countries. In two recent groundbreaking NGS studies in which the specified objective was to examine genome-wide mutation patterns in AA-associated UTUC,75, 76 the causal link between AA exposure and cancer could be established beyond reasonable doubt because of the convergence of several findings. First, the AA mutational signature was confined primarily to patients with documented exposure to AA (measurements of AA-derived adducts on adenine and/or patient exposure history). Second, the AA signature was reproduced in cells experimentally, and third, AA signature mutations (A to T transversions on the non-transcribed DNA strand at CpApG trinucleotides) were detected in somatically mutated driver genes. Clear cell and chromophobe renal cancers of patients from some regions of Eastern Europe also display this remarkably distinctive mutation pattern.77, 78 From mutational signature analysis it is now suspected that AA exposure may also be a contributing factor in causing hepatobiliary and bladder cancers.76, 79, 80
The mutational signature of a chemotherapeutic alkylating agent
Temozolomide (TMZ) is a human carcinogen, and a strong DNA alkylating agent used in the treatment of brain cancer and melanoma. Given its mutagenic properties, it came as no surprise to find that recurrent tumours of glioma patients treated with the compound displayed a heavy burden of G to A transitions, a base substitution induced by this class of chemicals when the alkylated deoxyguanine (O6-methyldeoxyguanine) mispairs with thymine. The naturally occurring ‘control group', patients not offered TMZ therapy, also have C:G to T:A transitions in their recurrent tumours, but these occur primarily at the CpG sequence contexts (attributable to spontaneous deamination of methylated cytosine), unlike the TMZ-associated transitions clustering at CpC and CpT dinucleotides.28 In a study of 23 patients, the mutation burden in recurrent tumours of patients treated with TMZ was up to 10-fold higher than in cancers of individuals not exposed to TMZ, and 98% of this mutation load were ‘TMZ-type' transitions.81 The study also revealed that TMZ exposure influenced not only the type of mutation but also the identity of the driver genes mutated in the tumours. In other words, this study suggests that a risk factor can participate in determining not only which types of mutations appear in the tumour, but also which genes become dysfunctional and drive the cancer process.
Orphan signatures and the call for more mutagenesis studies in experimental models
Genome-wide mutation data have unveiled ‘orphan' signatures undecipherable with the experimental and epidemiological data currently at hand, providing a major incentive for further targeted experimental work to decode enigmatic patterns and link them to causes of cancer. The oesophageal adenocarcinoma-linked mutation profile characterized by T to G substitutions at NpTpT is an example of a profile not readily linked to the major risk factors for the cohort in which the signature was observed, namely physical inactivity, obesity and gastro-intestinal reflux.82 This illustrates how a mutational signature per se reveals little about its author. Without hypotheses on the nature of the cancer risk factor from epidemiological and patient exposure data, and without experimental information on the mutagenic and chemical properties of carcinogens or endogenous mutational processes, a signature is undecipherable. A key demonstration of the convergence of multiple lines of information to establish cause was provided by the example cited above linking AA exposure to the unusual tumour A to T mutational signature. The only clues the signature could have provided entirely on its own were that: (a) the transversions were probably induced by an external agent, as this base substitution is a universally rare type of sequence change, and (b) the inducing agent probably generated bulky adducts on DNA bases, because these lead to transcription-coupled repair and thus a strand bias in the mutations that persist unrepaired. It was the confluence of experimental studies, epidemiology and patient exposure information that provided the necessary basis upon which a plausible cause of this signature was derived. Information on pro-mutagenic DNA adducts and other DNA lesions as well as the mutation spectra they generate in experimental systems have been essential factors in the assignment of signatures to risk factors.
The genome-wide impact of carcinogens and endogenous enzymes on DNA sequences can be efficiently captured in animal models, lower organisms and in cell-based in vitro assays.76, 83, 84, 85, 86, 87, 88, 89, 90 For example, exposure of normal murine embryonic fibroblasts (MEF) to known human carcinogens and sequencing of clones following immortalization is a rapid procedure that can generate mutational signatures corresponding to signatures in human tumours from patients exposed to the same agents (Figure 2).91, 92 This simple experimental procedure93, 94 is also suited to investigation of signatures linked to endogenous mutational processes. As proof of principle, we compared mutational signatures in immortalized MEF clones derived from MEFs isolated from mice harbouring an activation-induced cytidine deaminase (AID) transgene against signatures in non-transgenic mice, and demonstrated the expected excess of AID signature mutations in the clones derived from AID-expressing mice.92 AID, a hypermutator enzyme that promotes antibody diversity, causes off-target mutations in B-cell lymphomas and possibly other cancer types when inappropriately expressed.95, 96 Another source of experimentally induced genome-wide mutation patterns is potentially available from past in vivo toxicology projects. There is an untapped reservoir of archived tumour samples from animal carcinogen tests that can be mined using robust protocols for extraction and NGS of DNA derived from formalin-fixed, paraffin-embedded tumours already developed for human studies,77, 97, 98 allowing immediate access to information from this valuable source.
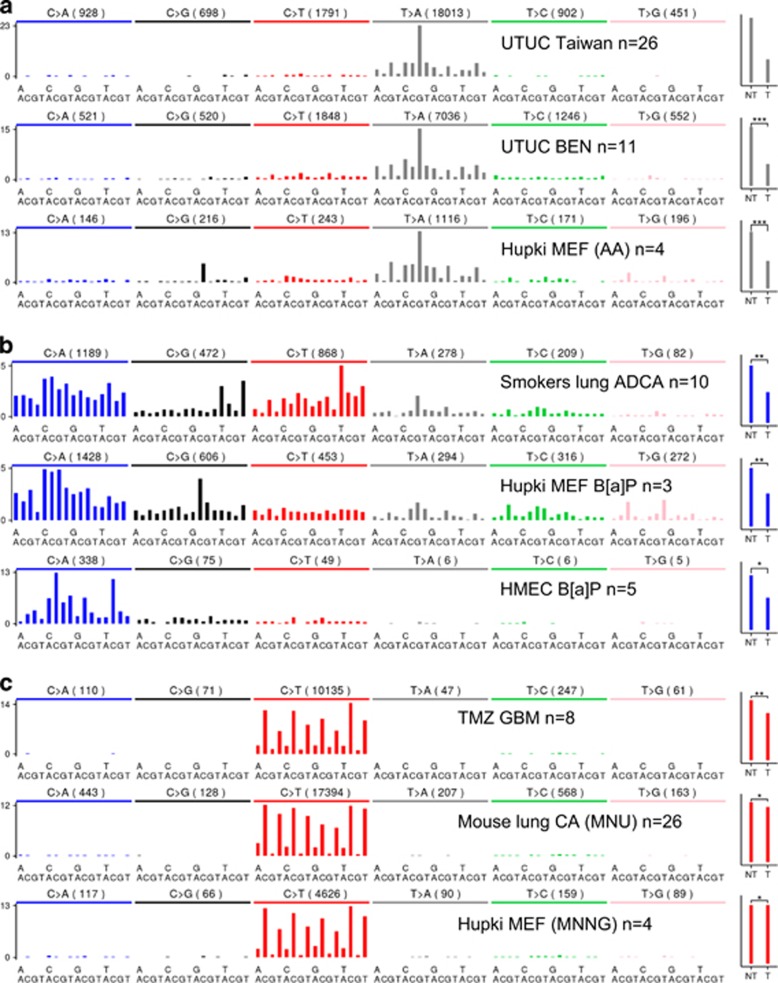
Figure 2.
A carcinogen's fingerprint in human tumour DNA can be reproduced in experimental systems. Mutation distribution spectra (showing frequency of base substitution type and context) from exome sequencing of primary human tumours, cells exposed in culture, or tumours of exposed mice. (a) Upper panels: spectra in upper urinary tract urothelial carcinomas (UTUC) of patients from Taiwan, China and from Balkan Endemic nephropathy (BEN) regions of Europe, two populations known to be exposed to AA.75, 76, 97 The lower panel shows that exposure of Hupki MEF to AA92 induces a similar mutational profile. Pooled data from multiple samples are shown for each data set. (b) Mutational spectra observed in lung adenocarcinomas (ADCA) of heavy smokers (upper panel) have features in common with spectra in Hupki MEF92 (middle panel) and human mammary epithelial cells (HMEC, lower panel) exposed to B[a]P,83 a tobacco carcinogen. (c) Spectra attributable to alkylation agents; upper panel: temozolomide treatment-related glioblastoma (TMZ GBM);81 middle panel: lung carcinoma of mice treated with methylnitrosourea (MNU);90 lower panel: Hupki MEF cells treated with methylnitrosoguanidine (MNNG).92 The bar graphs to the right show strand bias ratios. Strand bias reflects transcription-coupled repair of chemically damaged DNA bases (NT, non-transcribed strand; T, transcribed strand). Asterisks indicate χ2 test P-values for strand bias significance (*P<10E−5; **P<10E−20; ***P<10E−320; P=0 for UTUC Taiwan, in top panel of (a)). Note the less pronounced transcriptional strand bias ratios associated with the effects of alkylating agents.
Concluding remarks
Mutational signature analysis clearly incriminates environmental factors in shaping tumour mutation spectra. Risk factor-linked diversity in mutational signatures provides a framework for establishing which and to what extent certain factors do indeed contribute to the mutation burden of a tumour. The diversity is likely to be even more evident when well-designed international comparisons of mutation profiles are conducted, for example, with studies that take advantage of unusually high rates of incidence of specific tumour types in relatively restricted geographic areas (for example, gallbladder cancer in Chile). New tools for de-convoluting inherent genetic components and external factors in migrant studies are now at hand.31, 99 Heterogeneity of mutation signatures in a single cancer type implies that a one-size-fits-all approach to early detection biomarkers and molecular therapies requires refinement.
The resounding discovery from NMF-based analysis of NGS data that specific endogenous enzymatic processes appear responsible for prominent mutational signatures in a broad variety of cancers sends out a research call to identify environmental or lifestyle factors that could act by proxy, stimulating the endogenous mutators. It is important to know whether and which avoidable factors regulate these endogenous mutational processes in the natural history of cancer. An interdisciplinary approach that harnesses epidemiology, experimental models, NGS and mathematical analysis of mutations should meet these challenges.
Acknowledgments
We acknowledge these funding sources: INCa-INSERM 2015 Plan Cancer grant to JZ; LBA is supported through the J. Robert Oppenheimer Fellowship at Los Alamos National Laboratory.
The authors declare no conflict of interest.
References
- Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science 2011; 331: 1553–1558. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell 2013; 153: 17–37. [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G et al. Patterns of somatic mutation in human cancer genomes. Nature 2007; 446: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature 2009; 458: 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LAJr, Kinzler KW. Cancer genome landscapes. Science 2013; 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012; 150: 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brash DE. UV signature mutations. Photochem Photobiol 2015; 91: 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brash DE, Rudolph JA, Simon JA, Lin A, McKenna GJ, Baden HP et al. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci USA 1991; 88: 10124–10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene 2002; 21: 7435–7451. [DOI] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2010; 2: a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2010; 2: a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell 2009; 137: 413–431. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Hergenhahn M, Yang Q, Bartsch H, Wang ZQ, Hainaut P. New approaches to understanding p53 gene tumor mutation spectra. Mutat Res 1999; 431: 199–209. [DOI] [PubMed] [Google Scholar]
- Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science 1996; 274: 430–432. [DOI] [PubMed] [Google Scholar]
- Miller JH. Carcinogens induce targeted mutations in Escherichia coli. Cell 1982; 31: 5–7. [DOI] [PubMed] [Google Scholar]
- Wogan GN. Aflatoxins as risk factors for hepatocellular carcinoma in humans. Cancer Res 1992; 52: 2114s–2118s. [PubMed] [Google Scholar]
- Grollman AP, Shibutani S, Moriya M, Miller F, Wu L, Moll U et al. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc Natl Acad Sci USA 2007; 104: 12129–12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein M, Moriya M, Grollman AP, Olivier M. Analysis of TP53 mutation spectra reveals the fingerprint of the potent environmental carcinogen, aristolochic acid. Mutat Res 2013; 753: 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu IC, Metcalf RA, Sun T, Welsh JA, Wang NJ, Harris CC. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature 1991; 350: 427–428. [DOI] [PubMed] [Google Scholar]
- Bressac B, Kew M, Wands J, Ozturk M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature 1991; 350: 429–431. [DOI] [PubMed] [Google Scholar]
- Montesano R, Hainaut P, Wild CP. Hepatocellular carcinoma: from gene to public health. J Natl Cancer Inst 1997; 89: 1844–1851. [DOI] [PubMed] [Google Scholar]
- McCann J, Choi E, Yamasaki E, Ames BN. Detection of carcinogens as mutagens in the Salmonella/microsome test: assay of 300 chemicals. Proc Natl Acad Sci USA 1975; 72: 5135–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 1989; 244: 217–221. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science 1991; 253: 49–53. [DOI] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013; 3: 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature 1999; 401: 788–791. [DOI] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV et al. Signatures of mutational processes in human cancer. Nature 2013; 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Illingworth CJ, Campbell PJ, Mustonen V. EMu: probabilistic inference of mutational processes and their localization in the cancer genome. Genome Biol 2013; 14: R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi Y, Tremmel G, Miyano S, Stephens M. A simple model-based approach to inferring and visualizing cancer mutation signatures. PLoS Genet 2015; 11: e1005657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015; 47: 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage P, Doll R. The age distribution of cancer and a multi-stage theory of carcinogenesis. Br J Cancer 1954; 8: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Jones PH, Wedge DC, Sale JE, Campbell PJ, Nik-Zainal S et al. Clock-like mutational processes in human somatic cells. Nat Genet 2015; 47: 1402–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature 1993; 362: 709–715. [DOI] [PubMed] [Google Scholar]
- Wu S, Powers S, Zhu W, Hannun YA. Substantial contribution of extrinsic risk factors to cancer development. Nature 2016; 529: 43–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 2015; 347: 78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136: E359–E386. [DOI] [PubMed] [Google Scholar]
- Stewart BW, Wild CP (eds). World cancer report 2014. International Agency for Research on Cancer: Lyon, France; Geneva, Switzerland, 2014. [Google Scholar]
- Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet 2014; 15: 585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Siu HC, Leung SY, Stratton MR. A mutational signature in gastric cancer suggests therapeutic strategies. Nat Commun 2015; 6: 8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlien A, Campbell BB, de Borja R, Alexandrov LB, Merico D, Wedge D et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet 2015; 47: 257–262. [DOI] [PubMed] [Google Scholar]
- Smith HC, Bennett RP, Kizilyer A, McDougall WM, Prohaska KM. Functions and regulation of the APOBEC family of proteins. Semin Cell Dev Biol 2012; 23: 258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 2013; 45: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Roberts SA, Klimczak LJ, Sterling JF, Saini N, Malc EP et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet 2015; 47: 1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell 2002; 10: 1247–1253. [DOI] [PubMed] [Google Scholar]
- Kazanov MD, Roberts SA, Polak P, Stamatoyannopoulos J, Klimczak LJ, Gordenin DA et al. APOBEC-induced cancer mutations are uniquely enriched in early-replicating, gene-dense, and active chromatin regions. Cell Rep 2015; 13: 1103–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 2013; 494: 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K et al. Mutational processes molding the genomes of 21 breast cancers. Cell 2012; 149: 979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohler S, Da Silva Filho MI, Johansson R, Enquist-Olsson K, Henriksson R, Hemminki K et al. Impact of functional germline variants and a deletion polymorphism in APOBEC3A and APOBEC3B on breast cancer risk and survival in a Swedish study population. J Cancer Res Clin Oncol 2015; 142: 273–276. [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S, Wedge DC, Alexandrov LB, Petljak M, Butler AP, Bolli N et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat Genet 2014; 46: 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Gordenin DA. Clustered and genome-wide transient mutagenesis in human cancers: hypermutation without permanent mutators or loss of fitness. Bioessays 2014; 36: 382–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev 2014; 24: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses in the causation of human cancers - a brief historical account. Virology 2009; 384: 260–265. [DOI] [PubMed] [Google Scholar]
- Rebhandl S, Huemer M, Greil R, Geisberger R. AID/APOBEC deaminases and cancer. Oncoscience 2015; 2: 320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CJ, Xu T, Guo K, Griffin LM, Westrich JA, Lee D et al. APOBEC3A functions as a restriction factor of human papillomavirus. J Virol 2015; 89: 688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep 2014; 7: 1833–1841. [DOI] [PubMed] [Google Scholar]
- Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat Rev Cancer 2003; 3: 276–285. [DOI] [PubMed] [Google Scholar]
- Marnett LJ, Plastaras JP. Endogenous DNA damage and mutation. Trends Genet 2001; 17: 214–221. [DOI] [PubMed] [Google Scholar]
- Fedeles BI, Freudenthal BD, Yau E, Singh V, Chang SC, Li D et al. Intrinsic mutagenic properties of 5-chlorocytosine: A mechanistic connection between chronic inflammation and cancer. Proc Natl Acad Sci USA 2015; 112: E4571–E4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan P, Wild CP. Genomics of Cancer and a New Era for Cancer Prevention. PLoS Genet 2015; 11: e1005522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014; 346: 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014; 346: 256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 2015; 22: 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012; 366: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015; 348: 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Becq J, Warren A, Cheetham RK, Northen H, O'Donovan M et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett's esophagus and esophageal adenocarcinoma. Nat Genet 2015; 47: 1038–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver JM, Ross-Innes CS, Shannon N, Lynch AG, Forshew T, Barbera M et al. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat Genet 2014; 46: 837–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhou Y, Cheng C, Cui H, Cheng L, Kong P et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet 2015; 96: 597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan VG, Ebert PJ, Ting JC, Lim E, Wong SS, Teo AS et al. Whole-genome sequencing of asian lung cancers: second-hand smoke unlikely to be responsible for higher incidence of lung cancer among Asian never-smokers. Cancer Res 2014; 74: 6071–6081. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP. How the environment shapes cancer genomes. Curr Opin Oncol 2015; 27: 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011; 333: 1157–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014; 507: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering CR, Zhang J, Neskey DM, Zhao M, Jasser SA, Wang J et al. Squamous cell carcinoma of the oral tongue in young non-smokers is genomically similar to tumors in older smokers. Clin Cancer Res 2014; 20: 3842–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grollman AP. Aristolochic acid nephropathy: harbinger of a global iatrogenic disease. Environ Mol Mutagen 2013; 54: 1–7. [DOI] [PubMed] [Google Scholar]
- Hoang ML, Chen CH, Sidorenko VS, He J, Dickman KG, Yun BH et al. Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci Transl Med 2013; 5: 197ra102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon SL, Pang ST, McPherson JR, Yu W, Huang KK, Guan P et al. Genome-wide mutational signatures of aristolochic acid and its application as a screening tool. Sci Transl Med 2013; 5: 197ra101. [DOI] [PubMed] [Google Scholar]
- Jelakovic B, Castells X, Tomic K, Ardin M, Karanovic S, Zavadil J. Renal cell carcinomas of chronic kidney disease patients harbor the mutational signature of carcinogenic aristolochic acid. Int J Cancer 2015; 136: 2967–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scelo G, Riazalhosseini Y, Greger L, Letourneau L, Gonzalez-Porta M, Wozniak MB et al. Variation in genomic landscape of clear cell renal cell carcinoma across Europe. Nat Commun 2014; 5: 5135. [DOI] [PubMed] [Google Scholar]
- Poon SL, Huang MN, Choo Y, McPherson JR, Yu W, Heng HL et al. Mutation signatures implicate aristolochic acid in bladder cancer development. Genome Med 2015; 7: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JS et al. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun 2014; 5: 5696. [DOI] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 2014; 343: 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet 2013; 45: 478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson PL, Vrba L, Stampfer MR, Futscher BW. Exome-wide mutation profile in benzo[a]pyrene-derived post-stasis and immortal human mammary epithelial cells. Mutat Res Genet Toxicol Environ Mutagen 2014; 775-776: 48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flibotte S, Edgley ML, Chaudhry I, Taylor J, Neil SE, Rogula A et al. Whole-genome profiling of mutagenesis in Caenorhabditis elegans. Genetics 2010; 185: 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov AY, Quispe-Tintaya W, Gorbacheva T, White RR, Vijg J. High-throughput sequencing in mutation detection: a new generation of genotoxicity tests? Mutat Res 2015; 776: 136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier B, Cooke SL, Weiss J, Bailly AP, Alexandrov LB, Marshall J et al. C. elegans whole-genome sequencing reveals mutational signatures related to carcinogens and DNA repair deficiency. Genome Res 2014; 24: 1624–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar D, Latil M, Boeckx B, Lambrechts D, Blanpain C. Genomic landscape of carcinogen-induced and genetically induced mouse skin squamous cell carcinoma. Nat Med 2015; 21: 946–954. [DOI] [PubMed] [Google Scholar]
- Segovia R, Tam AS, Stirling PC. Dissecting genetic and environmental mutation signatures with model organisms. Trends Genet 2015; 31: 465–474. [DOI] [PubMed] [Google Scholar]
- Tam AS, Chu JS, Rose AM. Genome-wide mutational signature of the chemotherapeutic agent mitomycin C in Caenorhabditis elegans. G3 (Bethesda) 2015; 6: 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westcott PM, Halliwill KD, To MD, Rashid M, Rust AG, Keane TM et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature 2015; 517: 489–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Kucab JE, Morganella S, Glodzik D, Alexandrov LB, Arlt VM et al. The genome as a record of environmental exposure. Mutagenesis 2015; 30: 763–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Weninger A, Ardin M, Huskova H, Castells X, Vallee MP et al. Modelling mutational landscapes of human cancers in vitro. Sci Rep 2014; 4: 4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Belharazem D, Muehlbauer KR, Nedelko T, Knyazev Y, Hollstein M. Mutagenesis of human p53 tumor suppressor gene sequences in embryonic fibroblasts of genetically-engineered mice. Genet Eng (NY) 2007; 28: 45–54. [DOI] [PubMed] [Google Scholar]
- Liu Z, Hergenhahn M, Schmeiser HH, Wogan GN, Hong A, Hollstein M. `Human tumor p53 mutations are selected for in mouse embryonic fibroblasts harboring a humanized p53 gene. Proc Natl Acad Sci USA 2004; 101: 2963–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Shivarov V, Strout MP. The role of activation-induced cytidine deaminase in lymphomagenesis. Curr Opin Hematol 2012; 19: 292–298. [DOI] [PubMed] [Google Scholar]
- Pettersen HS, Galashevskaya A, Doseth B, Sousa MM, Sarno A, Visnes T et al. AID expression in B-cell lymphomas causes accumulation of genomic uracil and a distinct AID mutational signature. DNA Repair (Amst) 2015; 25: 60–71. [DOI] [PubMed] [Google Scholar]
- Castells X, Karanovic S, Ardin M, Tomic K, Xylinas E, Durand G et al. Low-coverage exome sequencing screen in formalin-fixed paraffin-embedded tumors reveals evidence of exposure to carcinogenic aristolochic acid. Cancer Epidemiol Biomarkers Prev 2015; 24: 1873–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedegaard J, Thorsen K, Lund MK, Hein AM, Hamilton-Dutoit SJ, Vang S et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One 2014; 9: e98187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 2014; 46: 1267–1273. [DOI] [PubMed] [Google Scholar]