Abstract
The aim of this study was to elucidate the epidemiology of third generation cephalosporin resistant Samonella isolates from pork of a slaughterhouse in China and the features of transferable elements carrying blaCTX-M genes. One hundred and twenty-six (7.3%) Salmonella isolates were identified; S. Derby and S. Rissen were the most two prevalent serotypes. Among these isolates 20 (15.8%) were resistant to third generation cephalosporins and nine of them carried blaCTX-M-27. S1-PFGE and replicon typing of blaCTX-M-27-carrying plasmids showed that seven were untypeable plasmids of about 104 Kb and two were IncP plasmids of about 300 Kb. Complete sequence analysis of one PBRT-untypeable plasmid showed it was a P1-like bateriophage, named SJ46, which contained a non-phage-associated region with several mobile elements, including Tn1721, ISEcp1B and IS903D. The other six 104 Kb PBRT-untypeable blaCTX-M-27-carrying plasmids also harboured the same phage-insertion region of SJ46 suggesting that they were the same P1-like bacteriophage. PFGE profiles of the parental strains revealed both potential vertical and horizontal spread of this P1-like blaCTX-M-27-containing element. Additionally, the representative gene of the P1 family bacteriophage, repL, was detected in 19.0% (24/126) of the isolates. This study indicated a potential role of P1-family bacteriophage in capture and spread of antimicrobial resistance in pathogens.
Salmonella is one of the most important causes of foodborne infections worldwide1. Salmonella has the ability to colonize the guts of healthy pigs, which can serve as carriers of this organism. When admitted to slaughterhouses, asymptomatic pigs are a potential risk for Salmonella contamination of pork meat and for Salmonella infections in human2.
For the treatment of salmonellosis, third generation cephalosporins (3GCs) are frequently used in human and veterinary medicine3. The emergence of Salmonella spp. isolates resistant to these drugs is of increasing public health concern4,5. Resistance to cephalosporins is principally by acquisition of plasmid carrying extended-spectrum β-lactamase (ESBL) genes, the most prevalent of which are the CTX-M-type ESBLs6. The increasing identification of clinical isolates containing ESBLs have led to a growing interest in researching the genetic elements responsible for their emergence and dissemination. Plasmids are considered as the most common and important element to spread ESBLs between bacterial strains. The mobilization or transfer of antimicrobial resistance gene by bacteriophages has been documented for various bacterial species, including the transfer of erythromycin resistance genes in Streptococcus pyogenes and Clostridium difficile7,8, and the transduction of β-lactamase genes in E. coli and Salmonella9,10. The horizontal spread of antimicrobial resistance genes mediated by phages and is now thought to be much more frequent than previously believed11,12. Bacteriophages have the ability to shape the bacterial microbiome in any environment. Through specialized or generalized transduction, bacteriophages can transfer genes that are advantageous to their microbial hosts, in turn promoting their own survival and dissemination13.
Bacteriophage P1, which was isolated in 1951 by Luigi Bertani, infects and lysogenizes Escherichia coli and several other enteric bacteria as independent low-copy-number plasmid-like elements14. The acquisition of the ESBL gene blaSHV-2 by a P1-family bacteriophage have been characterized in E. coli, however, there is no P1-like phage carrying blaCTX-M-type ESBL genes reported in clinical isolates of Salmonella11. In this study, we estimated the contamination of pork meat by Salmonella and reported the discovery of the transduction of blaCTX-M-27 gene by a P1-like bacteriophage in Salmonella isolated from pork from a slaughterhouse in China.
Results
Prevalence of Salmonella spp. in pork of slaughterhouse
A total of 126 (7.3%) Salmonella strains were isolated from 1728 pork samples from a large-scale slaughterhouse in Guangdong, China, during the period of April 2013 to April 2014. Three main serotypes were identified: S. Derby (66.7%, 84/126), S. Rissen (19.8%, 25/126) and S. Indiana (11.1%, 14/126). Three (2.4%) isolates could only be identified as Salmonella belonging to serotype of group B which can agglutinate with O-antiserum factor 4, however they were not Salmonella I 4, [5], 12:i-.
Resistance to cephalosporins and ESBL gene detection
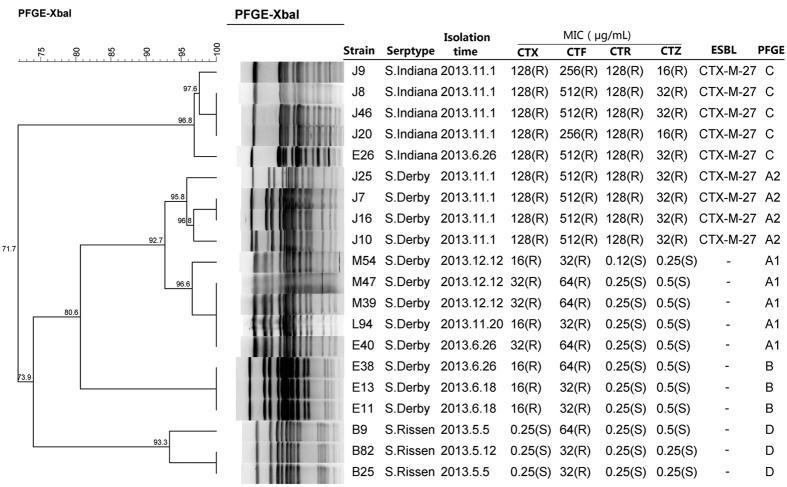
Among the 126 Salmonella isolates tested, 20 (15.8%) were resistant to third-generation cephalosporins (3GCs). The resistance rate of ceftiofur, cefotaxime, ceftriaxone and ceftazidime was 15.8%, 13.5%, 7.14% and 7.14%, respectively (Fig. 1).
Figure 1. PFGE profiles of twenty 3GC-resistant Salmonella isolated from pork in slaughterhouse in China.
MIC, minimal inhibitory concentration; CTX, cefotaxime; CTF, ceftiofur; CTR, ceftriazone; CTZ, ceftazidime; ESBL, extended-spectrum β-lactamase; PFGE, pulsed-field gel electrophoresis. “-”, not detected; “R”, resistant; “S”, susceptible.
Among the twenty 3GCs-resistant isolates, nine (strain J7, J8, J9, J10, J16, J20, J25, J46 and E26) contained ESBL-encoding gene blaCTX-M-27. No other ESBL genes were detected, which was consistent with the results of double disk synergy test. The 9 blaCTX-M-27-positive strains were highly resistant to all four tested drugs except for ceftazidime (only 1-fold higher than the resistance break point [16 ug/mL] of ceftazidime). It is noteworthy that all blaCTX-M-27 positive strains were recovered from pork samples collected on the same day, and 5 of them were S. Indiana and 4 were S. Derby. Surprisingly, the remaining 11(8 S. Derby and 3 S. Rissen) 3GCs-resistant isolates had none of the ESBL-encoding genes tested and did not produce phenotypically ESBLs. All of the 11 non-ESBL-producing 3GCs-resistant strains were susceptible to ceftazidime and ceftriazone, but resistant to ceftiofur. Eight (72.7%) of them were resistant to both ceftaxime and cetiofur. However, their resistant levels to ceftaxime and cetiofur were lower than those of strains producing CTX-M-27 (Fig. 1).
Pulsed-Field Gel Electrophoresis (PFGE)
The twenty 3GC-resistant Salmonella isolates were grouped into four XbaI-PFGE clusters designated A, B, C and D (Fig. 1). All isolates in each cluster had the same serotype. Cluster A harboured 9 S. Derby strains (92.7% genetic similarity). In addition, cluster A could be subgrouped into cluster A1 and cluster A2. Cluster A1 (95.8% genetic similarity) harboured five isolates, while cluster A2 (95.8% genetic similarity) was represented by four isolates containing blaCTX-M-27. Cluster B harboured three S. Derby strains. Cluster C harboured five S. Indiana containing blaCTX-M-27. Cluster D harboured three S. Rissen (93.3% genetic similarity).
Sizes and replicon types of plasmids carrying bla CTX-M gene
All plasmids carrying blaCTX-M-27 did not transfer by conjugation. Nine transformants bearing blaCTX-M-27 positive plasmid were obtained by electroporation. S1-PFGE and southern blot hybridization experiments confirmed that blaCTX-M were located on plasmids with two different sizes, which were about 104 Kb (n = 7, strain J7, J8, J16, J20, J25, J46 and E26) and 300 Kb (n = 2, strain J9 and J10), respectively (see Fig. S1 in the Supplemental Material).
Plasmid replicon typing revealed that both ~300 Kb plasmids were of IncP type. Surprisingly, the replicon types of the remaining seven ~104 Kb plasmids could not be determined by PCR-based replicon typing (PBRT).
Sequence analysis of a bla CTX-M-27 carrying plasmid
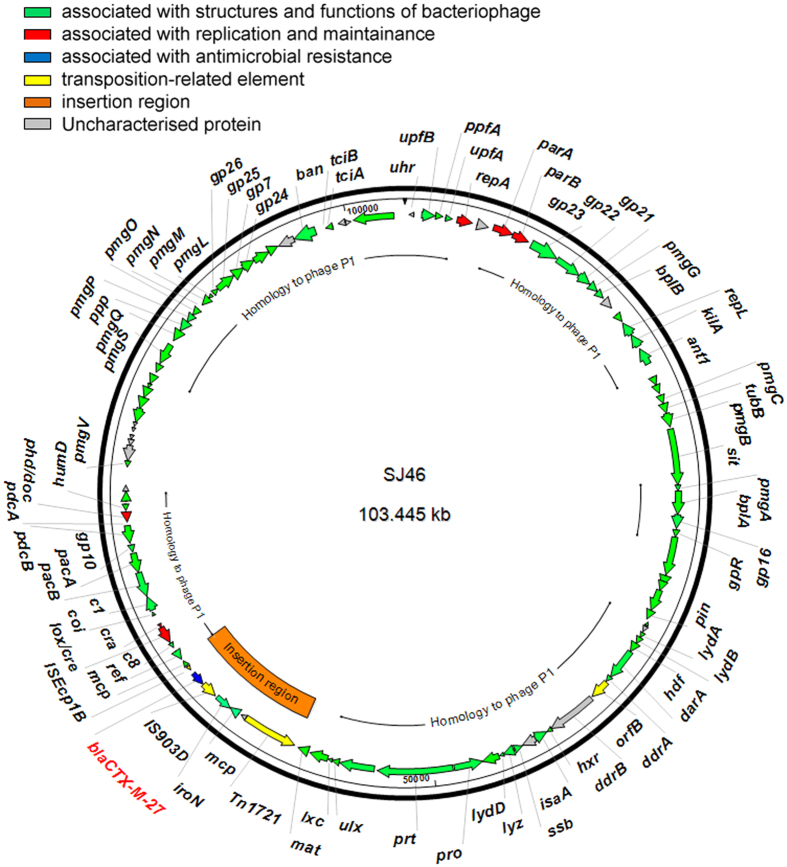
Gene sequencing and assembling of the plasmid from strain J46 revealed a 103,445 bp plasmid, named SJ46 (GenBank acceession no. KU760857). The G + C content of SJ46 is 48.59%, and was predicted to harbor 122 protein coding sequences (Table S1) and three tRNA for asparagine, threonine and methionine. Protein-coding genes account for 90% of SJ46. The results of gene annotations revealed that SJ46 turned out to be a P1-like bacteriophage, containing genes associated with DNA replication (repA), partition (parA and parB), recombination (lox/cre, cra, ref, cin), putative morphogenetic function (pmgA, pmgB, pmgC, pmgG, pmgL, pmgM, pmgN, pmgO, pmgP, pmgQ, pmgS, pmgV), cell lysis (repL, lydA, lydB, lydD, lyz, c1, coi, lxc), phage structural protein (gpR, gp7, gp16, gp21, gp22, gp23, gp24, gp25, gp26, tubB, bplA, bplB, prt, pro, mat) and DNA packaging protein (pacA and pacB) of P1 bacteriophage and genes of other function that have been reported in P1 bacteriophage genome (Fig. 2 and Table S1)15.
Figure 2. Circular map of SJ46.
The coloured arrows represent ORFs and their direction of transcription.
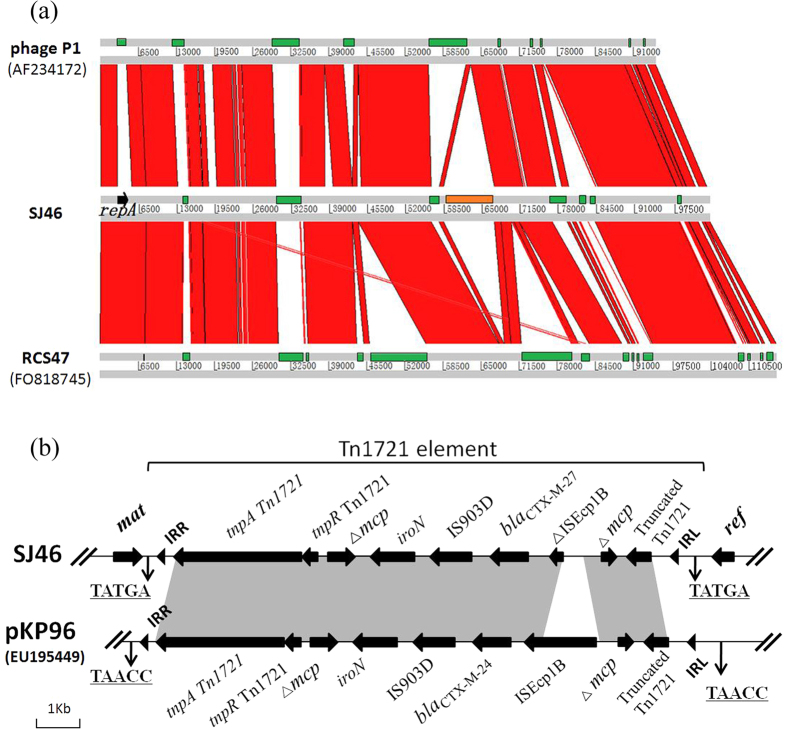
BLAST comparisons of the protein sequences against the GenBank databases showed that SJ46 displayed a high level identity to the P1 bacteriophage genome (GenBank accession no. AF234172.1) and P1-like bancterophage RCS47 carrying SHV-2 (GenBank accession no. FO818745.1). We found that 72% of SJ46 was common to the sequence of phage P1 bacteriophage, with the shared sequences being 98% identical and that 78% was common to the P1-like bacteriophage RCS47 carrying blaSHV-2, with the shared sequences being 99% identical (Fig. 3a). P1 bacteriophages lysogenize their hosts as autonomous plasmid-like elements and the circle format of the SJ46 was represented in Fig. 2.
Figure 3.
(a) Sequence synteny comparisons between SJ46, bacteriophage P1 (AF234172), and bacteriophage RCS47 (FO818745) as determined with the Artemis comparison tool. The green rectangles are specific genomic regions. Strand conservations are showed in red and the inserted region is indicated in orange. (b) Gene environment representation of blaCTX-M-27 in SJ46 and gene structure comparison of the blaCTX-M-containing region of SJ46 and pKP96. Black arrows represent ORFs and their direction. IRs of the respective mobile elements are shown as black triangles. Nucleotide letters with underlining represent a direct duplication (DR).
SJ46 has acquired an insertion of foreign DNA, which contains a 8,644-bp blaCTX-M-27-containing region (Fig. 3a and b). It is flanked by two 5 bp direct repeat sequences (TATGA), which indicated the occurrence of a transposition event. The left inverted repeat (IRL) and the right inverted repeat (IRR) are present at the junction between inserted DNA and phage DNA. The inserted fragment containing disrupted ISEcpIB, blaCTX-M-27, IS903D and a putative iron outer membrane receptor gene iroN interrupts the methyl-accepting chemotaxis gene mcp in Tn1721. This structure was inserted between a phage particle maturation gene (mat) upstream and a recombination enhancement gene (ref) downstream. The 8.6 Kb region is similar to a blaCTX-M-24 containing region on plasmid pKP96 in Klebsiella pneumonia from a sputum specimen in the First Affiliated Hospital, College of Medicine, Zhejiang University in China in 200216 (Fig. 3b).
Prevalence of P1-like bacteriophage in Salmonella isolates
We detected repL, the lytic replication gene of P1 family phage, in 24 of the 126 Salmonella isolates from pork in this study. The gene was also detected both in seven parental strains producing CTX-M-27 (strain E26, J7, J8, J16, J20, J25 and J46) and their transformants.
S1-PFGE and southern blot assay revealed that the sizes of the blaCTX-M-27 plasmids contained in these seven strains were the same, then we assumed that these mobile elements were similar to SJ46. Thus, the presence of phage sequences in the parental strain and transformants was confirmed by specific PCR using primers at the both ends of the phage-like region where one primer is in the phage-like region and the other is in the insertion resistance region. The results showed that all seven ~104 Kb blaCTX-M-27-carrying plasmids contained the phage-insertion region structure of SJ46, which suggested that they might be the same. Interestingly, blaCTX-M-27-carrying P1-like bacteriophage was evidenced in three Salmonella Indiana strains (J8, J20 and J46) and three Salmonella Derby strains (J7, J16 and J25) isolated from samples collected on the same day (November 1st, 2013). According to their PFGE profiles the three Salmonella Indiana strains were genetically similar as were two of the three Salmonella Derby suggesting, on one hand clonal spread of strains carrying blaCTX-M-27-P1-like bacteriophage and, on the other hand horizontal transfer of the blaCTX-M-27-P1-like bacteriophage in Salmonella strains of different serotypes.
Discussion
In addition to the impact of economic losses due to the salmonellosis for the swine industry, it is also a potential threat to human health. Several studies have indicated the role that the slaughter process can play in the spread and dissemination of Salmonella17. In the present study, Salmonella were detected in 7.3% (126/1728) of all pork samples. This was lower than the 68.9% reported by Y. Li et al. for swab samples from pigs after evisceration in slaughterhouses in Yangzhou, China18. This value is also lower than that of fresh slaughtered chicken meat from Shanxi (30%), Jiangsu (37.5%) and Guangdong (43.3%), China19.
S. Derby strains have been frequently isolated from pigs and pork in other countries, such as the UK20, France and Europe21. It was worrying that S. Derby was also the most common serotype isolated from infants and pre-school age children in China22. It was noteworthy that Salmonella Typhimurium or Enteritidis were always detected as the second or third most prevalent serotype in China. However, in our study, S. Rissen was the second prevalent serotype. Globally, S. Rissen is an infrequently reported serotype, but it is among the top three serotypes found in pigs and pork products in Spain and in some Southeast Asian countries23,24. S. Rissen was also recognised as a pathogen in outbreaks of human salmonellosis in Italy and USA25,26. The prevalence of S. Rissen in pork is of concern because it has been responsible for sporadic human infections in China22. This is the first report about the prevalence of S. Rissen in pork meat from China.
Nearly 16% (20/126) of isolates exhibited resistance to third-generation cephalosporins. Of these, 45% (9/20) were positive for CTX-M-27 β-lactamase and no other ESBL-encoding genes were detected. CTX-M-65, CTX-M-55 and CTX-M-14 used to be the most prevalent ESBLs detected in E. coli from food-producing animals, but CTX-M-27 was only sporadically detected in E. coli27. There is little literature reporting the prevalence of CTX-M-27 in Salmonella strains in China until recent times when a study detected blaCTX-M-27 as the most dominant ESBL genes in Salmonella Typhimirium and Indiana isolated from pigs and Chicken28. The high prevalence of blaCTX-M-27 in Salmonella from pork meat should be a cause of concern which will be a potential threat to public health.
Eleven (55%) of the 20 3GCs-resistant Salmonella isolates harboured no ESBLs genes. All of them were resistant to cetiofur and eight of them to ceftaxime. These data suggested that 3GCs resistance in these strains was mediated by other mechanisms, such as the absence of out membrane proteins (eg. OmpC and OmpF) and overexpression of efflux pump AcrAB-TolC. In Salmonella, OmpF and OmpC are the major porins, and AcrB is also the major, constitutive multidrug pump29,30. Most cephalosporins and penicillins are substrates of AcrB30. It has been reported that active efflux cannot increase the MICs of β-lactams (such as ampicillin and cephalothin) in E. coli, however, if porin permeability is decreased, efflux would produce a more visible effect31. Interestingly, one study showed that mutants lacking the wider-channel OmpF and producing only the more restrictive OmpC porin become much more resistant to good substrates of AcrB, such as benzylpenicillin or cephaloram, but not to poor substrates of AcrB, such as cefazolin and cephaloridine31. Laboratory selection using ceftazidime or ceftibuten, starting from an E. coli strain containing a TEM-1-producing plasmid, also resulted in the loss of OmpF or OmpF and OmpC32. The absence of OmpF was also seen in a ceftazidime-resistant E. coli strain33. However, changes in porins and efflux have been rarely reported for β-lactam-resistant Salmonella isolates. The present study did not show the roles of outer membrane proteins and efflux pumps in cephalosporin resistance in Salmonella isolates, however, it provides basis of further research.
Susceptible bacteria may develop resistance to antibiotics through multiple and complex mechanisms, such as mutation and horizontal gene transfer. Horizontal gene transfer can be mediated by mobile genetic elements such as insertion sequences, transposons, integrative conjugative elements, plasmids and bacteriophages, which are involved in bacterial acquisition and recombination of foreign DNA34. Bacteriophages (phages), viruses that infect bacteria, can act as vehicles for horizontal exchange of genetic information, can genetically modify their host by insertion of their DNA into the bacterial genome, and can carry genes that encode new functions or modify existing ones35. A recent metagenomic analysis reveals that bacteriophages are reservoirs of antibiotic resistance genes, including genes encoding ATP-binding cassette (ABC) and resistance-nodulation-cell division (RND) proteins, phosphotransferases, β-lactamases and plasmid-mediated quinolone resistance36.
In the present study, sequence analysis of one ~104 Kb blaCTX-M-27-carrying plasmid, named SJ46, revealed that SJ46 was a P1-like bacteriophage bearing an 8,644-bp blaCTX-M-27-containing region. It was the first time that a blaCTX-M-type gene was evidenced on a P1-like bacteriophage in 3GC-resistant Salmonella. RepA, ParA and ParB of SJ46, are the same as those of a P1-like bacteriophage RCS47 detected in E. coli in 2002 in Paris from an adult patient with a urinary infection11. RepA of SJ46 only has 42% identity to the RepA of P1 phage (P1 c1-100 Tn9, accession No. AF234172.1) and has no similarities with those of any other replicon proteins from known Inc group plasmids. According to the analysis by Billard-Pomares11, the partitioning proteins of RCS47, ParA and ParB, are identical to those of P7 phage, but distantly related to those of P1 phage, which may make it possible to avoid incompatibility with other mobile elements.
The 8.6 Kb inserted element contained a disrupted ISEcpIB, blaCTX-M-27, IS903D and a putative iron outer membrane receptor gene iron and interrupts the methyl-accepting chemotaxis gene mcp in Tn1721. A similar structure has been described in the transfer of blaCTX-M-19 and blaCTX-M-24 by plasmids in E. coli and Klebsiella pneumonia, respectively16,37, which suggested that mobilization of ESBL genes are linked to the ISEcp1B-associated transposition element. The studies of the genetic environments of blaCTX-M in the ST131 E. coli isolates from hospitals in the Kyoto and Shiga regions of Japan showed that almost all of the CTX-M-27-H30R subgroup had an IS26- ΔISEcp1- blaCTX-M-27- ΔIS903D- IS26-like structure, with variation in the length of ΔIS903D38,39. In our study, the environment of blaCTX-M-27 had a Tn1721-ISEcp1B-IS903D-ΔTn1721-like structure, and ISEcpIB was disrupted by a part of Tn1721 with complete IRL and 5 bp DRs (Fig. 3b), and suggested that blaCTX-M-27 was transferred to the phage together with ISEcp1B and IS903D from a plasmid by Tn1721 element.
Additionally, the phage-insertion region of SJ46 was also detected in the other six blaCTX-M-27-carrying plasmids. The PFGE profiles of 3GC-resistant Salmonella strains revealed both vertical (clonal) and horizontal spread of the bacteriophage P1-like element among Salmonella spp. Moreover, repL, which is the lytic replication gene of P1 family phage, was detected in 19.0% of the Salmonella isolates. This suggests that this P1-related prophage is common in Salmonella from China.
A further limitation of the study is represented by the fact that we did not obtain the sequences of the other six P1 bacteriophage-like plasmids carrying blaCTX-M-27. However, their lengths expected on the basis of the S1-PFGE results were similar to that of SJ46 and the conserved elements expected in the P1-family bacteriophage were identified.
In conclusion, high rates (15.8%) of third generation cephalosporin resistant Salmonella, of which serotypes have been also detected in human infections, were detected in meat from pig carcase from a slaughterhouse in Guangzhou, China. A P1-like bacteriophage SJ46 was found to be related to the transfer of the ESBL gene blaCTX-M-27 together with a Tn1721 element. Recently, plasmid bearing a blaCTX-M-15 gene and with chimeric characteristics consisting of pIP1206-like backbone and lysogenized phage P1-like sequences was characterized. It was postulated to have resulted from recombination between an E. coli plasmid backbone, a blaCTX-M-15 bearing region and a lysogenized phage P1-like sequence40. The high prevalence of a P1-family phage in Salmonella isolates suggest that the abilities of P1-like phages to transfer resistance genes need to be explored more thoroughly. The findings of our study also suggest that plasmid sequencing is necessary to discover the elements involved in the dissemination of antimicrobial resistance genes.
Materials and Methods
Sample collection and bacteriological analysis
A total of 1728 pork specimens were obtained between April 2013 and April 2014 from one large-scale slaughterhouse, in Guangdong, China. About 40–50 randomly selected carcasses were sampled weekly before splitting. Samples of 150 g were taken from the inside of two hind legs of each carcass using a sterile knife and placed into a sterile plastic bag. All samples were placed in a low-temperature foam box and dispatched within 3 h to the laboratory. If immediate dispatch was not possible for some reason, samples were stored in a refrigerator at 4 °C until the time of dispatch. About 25 g of meat samples cut into pieces was put into 200 mL buffered peptone water (BPW), which were then incubated at 37 °C for 12 h. One milliliter aliquot of BPW cultures were transfered to 10 mL of selenite cysteine broth and incubated at 37 °C for 24 h. The enriched content was streaked on bismuth sulphite agar and xylose lysine desoxycholate agar and incubated at 37 °C for a further 24 h. Presumptive Salmonella colonies were identified with API20E systems (BioMerieux, Beijing, China) and were serotyped using Salmonella specific O and H antigens by the slide agglutination test (S&A Company, Bangkok, Thailand).
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was conducted using the agar dilution method in accordance with the standards and guidelines described by the Clinical and Laboratory Standards Institute (CLSI). Isolates were tested for sensitivity to cefotaxime, ceftiofur, ceftazidime, ceftriaxone. Escherichia coli ATCC 25922 was used as quality control and the susceptibility (or resistance) of each isolate was determined according to the 2015 CLSI recommendations41.
Double disk synergy test
β-lactamase and ESBL-producing isolates were screened according to the double-disk synergy test method recommended by the CLSI combined with antimicrobial disks (cefotaxime 30 μg, cefotaxime-clavulanate 30 μg/10 μg, ceftazidime 30 μg, and ceftazidime-clavulanate 30 μg/10 μg) according to the manufacturer’s instructions (CLSI, 2015). Phenotypic presence of β-lactamases of the isolates was determined by detecting diameter enhancement. If the enhancement value was >5 mm, the isolate was considered to be presumptive β-lactamases producers (CLSI 2015).
Amplification of β-lactamase-encoding genes
Strains showing resistance to cefotaxime (with MIC ≥ 4 μg/ml) were screened for the presence β-lactamase genes (blaTEM, blaSHV, blaOXA, blaCTX-M blaCMY-2) by PCR using the primers and conditions described previously42. The obtained DNA amplicons were submitted to BGI Life Tech Co., Ltd. (Beijing, China) for sequencing and sequences were compared with those included in the GenBank database by using the BLAST algorithm (www.ncbi.nlm.nih.gov) and at www.lahey.org/Studies/ in order to identify specific β-lactamase genes.
Molecular typing
The genetic relatedness of the isolates was determined by pulsed-field gel electrophoresis (PFGE) using a CHEF-MAPPER System (Bio-Rad Laboratories, Hercules, CA), as previously described4. Comparison of PFGE patterns was conducted with BioNumerics software (Applied Maths, Sint-Martens-Latem,Belgium) using the Dice similarity coefficient.
Determination of replicon types and sizes of plasmids
PCR-based replicon typing (PBRT) was performed on transformants using primers as described previously43. The sizes of plasmids carrying blaCTX-M was detected by S1-PFGE and southern blot assay. PFGE with S1 nuclease (Takara Biotechnology, Dalian, China) digestion of whole genomic DNA was performed for all transformants. After southern transfer to a Hybond-N+ membrane (GE Healthcare, Little Chalfont, United Kingdom), the plasmids were probed with the blaCTX-M gene (DIG High Prime DNA Labeling and Detection Starter Kit I, Roche Applied Science, Mannheim, Germany), as described previously27.
Sequencing and analysis of a bla CTX-M-27-carrying plasmid
The plasmid from one of the CTX-M-27-carrying isolate J46, was extracted using the Qiagen Plasmid Midi Kit (Qiagen, Germany) and transferred by electroporation in to E. coli strain DH5α to yield strain TJ46. Plasmid DNA was purified from TJ46 with the Qiagen Plasmid Midi Kit (Qiagen, Germany) and was sequenced by the Roche 454 Genome Sequencer FLX system. Sequence reads were assembled by SOAP denovo software. The GeneMarKS software (http://topaz.gatech.edu/) was used to identify putative open reading frames44. The nucleotide and amino acid sequences were analyzed and compared through BLAST queries against the GenBank database.
Prevalence investigation of P1-like bacteriophage in Salmonella isolates
The presence of a P1-like bacteriophage in Salmonella isolated in this study was evaluated by detecting the present of the lytic replication gene of P1 family phage, repL, with the primers RepL-fw (5′-CCAATCAACCGTCGTTCGTG-3′) and RepL-rev (5′-TAAGCATATTTCCGCGCTGC-3′)11. The PCR method consisted of 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 45 s. This was followed by an additional 10-min extension at 72 °C. The presence of the insertion sequence on the other nontypeable plasmids carrying blaCTX-M-27 gene was detected using the following primers: IS-fw (AGAATCATCGC CGAAGGGCTGTAACTGGTTTT) and IS-rev (GCGAACATCATCCGTTGCACT CTCTTTGT).
Statiscal analysis
For statiscal analysis, we carried out Fisher’s exact tests, and P values of <0.05 were considered significant.
Additional Information
How to cite this article: Yang, L. et al. Characterization of a P1-like bacteriophage carrying CTX-M-27 in Salmonella spp. resistant to third generation cephalosporins isolated from pork in China. Sci. Rep. 7, 40710; doi: 10.1038/srep40710 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Laura JV Piddock (University of Birmingham, UK) for reading this manuscript and providing helpful feedback. This work was supported in part by the 973 Program [under Grant No. 2013CB127203], the Program for Changjiang Scholars and Innovative Research Team in University [No. IRT13063] and the National Natural Science Foundation of China [31272602].
Footnotes
Author Contributions L.Y. performed experiments, analyzed the data and wrote manuscript; W.L., G.Z.J. and W.H.Z. performed experiments; H.Z.D. and Y.H.L. analyzed the data and edited the manuscript; Z.L.Z. and H.X.J. designed the experiments, analyzed the data, edit manuscript and coordinated the whole project.
References
- Todd E. C. Epidemiology of foodborne diseases: a worldwide review. World Health Stat Q. 50, 30–50 (1997). [PubMed] [Google Scholar]
- Kerouanton A. et al. First Complete Genome Sequence of a Salmonella enterica subsp. enterica Serovar Derby Strain Associated with Pork in France. Genome Announce 3, 821–829 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collignon P. & Aarestrup F. M. Extended-spectrum β-lactamases, food, and cephalosporin use in food animals. Clin Infect Dis 44, 1391–1392 (2007). [DOI] [PubMed] [Google Scholar]
- Jiang H. X. et al. Multiple transmissible genes encoding fluoroquinolone and third-generation cephalosporin resistance co-located in non-typhoidal Salmonella isolated from food-producing animals in China. Int. J. Antimicrob. Agents 43, 242–247 (2014). [DOI] [PubMed] [Google Scholar]
- Noda T. et al. Increase in Resistance to extended-spectrum cephalosporins in Salmonella Isolated from retail chicken products in Japan. PloS one 10, e0116927 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton R., Jose M. G. A. & Galan J. C. CTX-M enzymes: origin and diffusion. Front Microbiol 3, 1–19 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh S. et al. Phage ϕC2 mediates transduction of Tn6215, encoding erythromycin resistance, between Clostridium difficile strains. MBio 4, e00840–00813 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyder S. L. & Streitfeld M. M. Transfer of erythromycin resistance from clinically isolated lysogenic strains of Streptococcus pyogenes via their endogenous phage. J Infect Dis 138, 281–286 (1978). [DOI] [PubMed] [Google Scholar]
- Smith H. W. Ampicillin resistance in Escherichia coli by phage infection. Nature 238, 205–206 (1972). [DOI] [PubMed] [Google Scholar]
- Schmieger H. & Schicklmaier P. Transduction of multiple drug resistance of Salmonella enterica serovar Typhimurium DT104. FEMS Microbiol Lett 170, 251–256 (1999). [DOI] [PubMed] [Google Scholar]
- Billard-Pomares T. et al. Characterization of a P1-like bacteriophage carrying an SHV-2 extended-spectrum β-lactamase from an Escherichia coli strain. Antimicrob Agents Chemother 58, 6550–6557 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniesa M., Colomer-Lluch M. & Jofre J. Potential impact of environmental bacteriophages in spreading antibiotic resistance genes. Future Microbiol 8, 739–751 (2013). [DOI] [PubMed] [Google Scholar]
- Modi S. R., Lee H. H., Spina C. S. & Collins J. J. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Chandler M. & Caro L. Replication of the prophage P1 during the cell cycle of Escherichia coli. Mol Gen Genet 152, 71–76 (1977). [DOI] [PubMed] [Google Scholar]
- Łobocka M. B. et al. Genome of bacteriophage P1. J Bacteriol 186, 7032–7068 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P. et al. Complete nucleotide sequence of pKP96, a 67 850 bp multiresistance plasmid encoding qnrA1, aac(6′)-Ib-cr and blaCTX-M-24 from Klebsiella pneumoniae. J Antimicrob Chemother 62, 1252–1256 (2008). [DOI] [PubMed] [Google Scholar]
- Swanenburg M. Salmonella in the pork production chain: sources of Salmonella on pork. (Utrecht Univ., 2000). [Google Scholar]
- Li Y. et al. Salmonella isolated from the slaughterhouses and correlation with pork contamination in free market. Food Control 59, 591–600 (2016). [Google Scholar]
- Zhu J. et al. Prevalence and quantification of Salmonella contamination in raw chicken carcasses at the retail in China. Food Control 44, 198–202 (2014). [Google Scholar]
- Piras F., Brown D. J., Meloni D., Mureddu A. & Mazzette R. Investigation of Salmonella enterica in Sardinian slaughter pigs: prevalence, serotype and genotype characterization. Int J Food Microbiol 151, 201–209 (2011). [DOI] [PubMed] [Google Scholar]
- Authority E. F. S. The European Union summary report on trends and sources of zoonoses, zoonotic agents and foodborne outbreaks in 2013. EFSa J 13, 3991 (2015). [Google Scholar]
- Cui S. et al. Characterization of Salmonella enterica isolates from infants and toddlers in Wuhan, China. J Antimicrob Chemother 63, 87–94 (2009). [DOI] [PubMed] [Google Scholar]
- Hendriksen R. S. et al. Antimicrobial resistance and molecular epidemiology of Salmonella Rissen from animals, food products, and patients in Thailand and Denmark. Foodborne Pathog Dis 5, 605–619 (2008). [DOI] [PubMed] [Google Scholar]
- Argüello H., Rubio P. & Carvajal A. Occurrence and epidemiology of Salmonella enterica in two slaughterhouses and cutting plants in Spain. In: the 40th International Conference on the Epidemiology and Control of Biological, Chemical and Physical Hazards in Pigs and Pork (2011).
- Boschi T. et al. Cluster of cases of Salmonella enterica serotype Rissen infection in a general hospital, Italy, 2007. Zoonoses Public Health 57, 518–522 (2010). [DOI] [PubMed] [Google Scholar]
- Higa J. Outbreak of Salmonella Rissen associated with ground white pepper: the Epi investigation. Union International Investigation Meeting Presentation, Quarterly Scientific Seminar, California. California Department of Public Health Jan 6 (2011).
- Yang L. et al. Co-prevalance of PMQR and 16S rRNA methylase genes in clinical Escherichia coli isolates with high diversity of CTX-M from diseased farmed pigeons. Vet Microbiol 178, 238–245 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang W.-H. et al. CTX-M-27 producing Salmonella enterica serotypes Typhimurium and Indiana are prevalent among food-producing animals in China. Front Microbiol 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A., Chabbert Y. A. & Semonin O. Role of porin proteins OmpF and OmpC in the permeation of beta-lactams. Antimicrob Agents Chemothe 22, 942–948 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H., Basina M., Nguyen V. & Rosenberg E. Y. Multidrug Efflux pump AcrAB of Salmonella typhimurium excretes only those β-lactam antibiotics containing lipophilic side chains. J Bacteriol 180, 4686–4692 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder K. J., Nikaido H. & Matsuhashi M. Mutants of Escherichia coli that are resistant to certain beta-lactam compounds lack the OmpF porin. Antimicrob Agents Chemothe 20, 549–552 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli M. et al. In vitro selection and characterization of mutants in TEM-1-producing Escherichia coli by ceftazidime and ceftibuten. J Chemothe 19, 123–126 (2007). [DOI] [PubMed] [Google Scholar]
- Palasubramaniam S., Subramaniam G., Muniandy S. & Parasakthi N. Extended-spectrum β-lactam resistance due to AmpC hyperproduction and CMY-2 coupled with the loss of OmpK35 in Malaysian strains of Escherichia coli and Klebsiella pneumoniae. Microb. Drug Resist. 13, 186–190 (2007). [DOI] [PubMed] [Google Scholar]
- Marti E., Variatza E. & Balcazar J. L. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol 22, 36–41 (2014). [DOI] [PubMed] [Google Scholar]
- Dobrindt U., Hochhut B., Hentschel U. & Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Revs Microbiol 2, 414–424 (2004). [DOI] [PubMed] [Google Scholar]
- Subirats J., Sànchez-Melsió A., Borrego C. M., Balcázar J. L. & Simonet P. Metagenomic analysis reveals that bacteriophages are reservoirs of antibiotic resistance genes. Int J Antimicrob Agents 48, 163–167 (2016). [DOI] [PubMed] [Google Scholar]
- Poirel L., Lartigue M.-F., Decousser J.-W. & Nordmann P. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob Agents Chemother 49, 447–450 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartigue M.-F., Poirel L. & Nordmann P. Diversity of genetic environment of blaCTX-M genes. FEMS Microbiology Letters 234, 201–207 (2004). [DOI] [PubMed] [Google Scholar]
- Matsumura Y. et al. CTX-M-27-and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. Journal of Antimicrobial Chemotherapy 70, 1639–1649 (2015). [DOI] [PubMed] [Google Scholar]
- Shin J. & Ko K. S. A plasmid bearing the blaCTX-M-15 gene and phage P1-like sequences from a sequence type 11 Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 59, 6608–6610 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute, Twenty-Fifth Informational Supplement M100-S25, CLSI, Wayne. PA (2015).
- Jiang H.-X. et al. Prevalence and characteristics of β-lactamase and plasmid-mediated quinolone resistance genes in Escherichia coli isolated from farmed fish in China. J. Antimicrob Chemother 67, 2350–2353 (2012). [DOI] [PubMed] [Google Scholar]
- Carattoli A. et al. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Meth. 63, 219–228 (2005). [DOI] [PubMed] [Google Scholar]
- Lukashin A. V. & Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26, 1107–1115 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.