Abstract
Post-translational modification by ubiquitin and ubiquitin-like proteins (UbLs) is fundamental for maintaining protein homeostasis. Efficient isolation of UbL conjugates is hampered by multiple factors, including cost and specificity of reagents, removal of UbLs by proteases, distinguishing UbL conjugates from interactors, and low quantities of modified substrates. Here we describe bioUbLs, a comprehensive set of tools for studying modifications in Drosophila and mammals, based on multicistronic expression and in vivo biotinylation using the E. coli biotin protein ligase BirA. While the bioUbLs allow rapid validation of UbL conjugation for exogenous or endogenous proteins, the single vector approach can facilitate biotinylation of most proteins of interest. Purification under denaturing conditions inactivates deconjugating enzymes and stringent washes remove UbL interactors and non-specific background. We demonstrate the utility of the method in Drosophila cells and transgenic flies, identifying an extensive set of putative SUMOylated proteins in both cases. For mammalian cells, we show conjugation and localization for many different UbLs, with the identification of novel potential substrates for UFM1. Ease of use and the flexibility to modify existing vectors will make the bioUbL system a powerful complement to existing strategies for studying this important mode of protein regulation.
Protein homeostasis contributes to the natural equilibrium within the cells, and its disruption is often the cause or consequence of multiple diseases. The balance is achieved in many instances through post-translational modifications (PTMs) by ubiquitin (Ub) and ubiquitin-like (UbL) proteins, controlling the function, localization or stability of target proteins. Ub is a 76-aa polypeptide that can modify target proteins through the process of ubiquitination, the attachment of an activated Ub moiety through a C-terminal glycine to a lysine or selected other residues in the target substrate1. The process involves the activation of Ub by an E1 enzyme, the transfer of the active moiety to an E2 conjugating enzyme and, in many instances, the cooperation of an E3 ligase that binds both the E2-bound Ub and the substrate. Deubiquitinases (DUBs) can revert the modification, conferring flexibility and regulation to the process2. Although most commonly associated to protein degradation by the proteasome, more recently ubiquitination has been related to a wide variety of cellular processes, including protein trafficking and DNA repair among others. Ub itself can be ubiquitinated in any of its seven lysines or the initiating methionine, forming chains that can adopt different conformations. This constitutes a complex code that can lead modified proteins to different outcomes3.
Ub is the most conserved protein found in all eukaryotes. Approximately 20 proteins have been identified that are related to Ub, known as UbLs4,5. Some have recognizable sequence homology with Ub, while more divergent UbLs share similar structural features. All of them share the beta-grasp fold characteristic of Ub and all participate in processes similar to ubiquitination, suggesting a common ancestry to this family of proteins4. Among the UbLs, the one that shares the highest homology with Ub is NEDD8 (NEural precursor cell-expressed, Developmentally Downregulated 8). While thousands of Ub targets have been identified, the reported number of NEDDylated proteins is lower. Among those, the cullins are RING E3 ligases that link NEDDylation to the ubiquitination of a wide spectrum of targets that participate in many cellular processes6.
The Small Ub-like MOdifier, SUMO, is one of the most studied UbLs7. In vertebrates, there are five different SUMO genes, SUMO1-5. SUMO2 and SUMO3 are almost identical and share 50% identity with SUMO1. All SUMOs use the same E1 and E2 enzymes in the process of SUMOylation, and can participate in forming polySUMO or mixed Ub-SUMO chains. SUMO4 seems to be a pseudogene or it is not processed8,9, while the recently reported SUMO5 shows tissue-specificity and participates in nuclear body formation10. In yeast and Drosophila there is a single SUMO homologue, Smt3. SUMOylation has been related to transcriptional repression and response to cellular stresses such as DNA damage.
Other UbLs are less well characterized. ISG15 (Interferon-Stimulated Gene 15) is induced by interferons secreted by virus-infected cells and participates in the anti-viral immune response11. FAT10 (HLA-F Adjacent Transcript 10, also called UBD) is expressed in immune cells and can also be induced in other cell types by interferon gamma or TNF-alpha. FAT10 can mediate Ub-independent proteasomal degradation12,13. Neither ISG15 nor FAT10 are conserved in lower eukaryotes. Both are composed of two UbL modules, whereas other UbLs have a single module. The Ub-Fold Modifier-1, UFM1, is conserved in metazoans and plants. It has a role in erythroid and megakaryocyte development, homeostasis of the endoplasmic reticulum (ER) and vesicle trafficking14. ATG8 and ATG12 are involved in the regulation of autophagy. ATG8 is a lipid modifier that is conjugated to phosphatidylethanolamine and participates in autophagosome biogenesis. There are 6 ATG8 orthologues in humans, classified as GABARAP1-2 and MAP1LC3A-B. ATG12 is conjugated to at least one other protein in the outer part of the autophagosome membrane, where this complex acts as the E3 ligase for Atg815. FAU (Finkel-Biskis-Reilly murine sarcoma virus, ubiquitously expressed) is synthesized as a fusion protein with the ribosomal protein S30. In macrophages, FAU inhibits lipopolysaccharide-induced signalling and phagocytosis, having an immunoregulatory role16. The Ub Related Modifier 1, URM1, is conserved in eukaryotes and its structure resembles that of ancient prokaryotic sulfur carriers. Sulfur donors involved in the biosynthetic pathway of thiamine (vitamin B1) and molybdopterin (MPT) in prokaryotes, ThiS and MoaD, have a certain homology with Ub and contain a beta-grasp fold. Therefore, URM1 might represent a bridge between the prokaryotic sulfur carriers and the eukaryotic UbLs17.
Given the variety of UbLs that exist, the different types of monotypic chains that they can potentially form, and that there can even exist hybrid chains containing different types of UbLs, a complex UbL code may underlie numerous biological and cellular processes. This has prompted researchers to develop different systems to isolate UbL-modified proteins using cultured cells, model organisms, and clinical tissue samples. Most approaches are modelled on those successfully used to study Ub, such as using epitope-tagged Ub (with 6xHIS or HA tags). Approaches to study endogenous Ub modifications include monoclonal antibodies directed against the di-Gly residual peptide found on Ub substrates after deubiquitination, or molecular traps formed from tandem Ub-binding domains (TUBEs)18,19.
Although more challenging due to its low abundance, SUMOylation has been extensively studied using similar approaches. Different single or double epitope tags fused to the N-terminus of SUMO have been used to enrich for conjugates. Examples include 6x-10xHIS, FLAG, Myc, 6xHIS-FLAG, 6xHIS-HA, FLAG-TEV and ProtA-TEV-CBP for use in cell culture as well as transgenic organisms20. DeSUMOylating activity is high in cell lysates, so chemical inhibitors or ideally denaturing conditions are used during capture of conjugates to preserve SUMOylation. Using exogenous tagged SUMO also allows the use of SUMO mutants that may be defective in chain formation (e.g. SUMO2-K11R) or optimized for mass spectrometry analysis (MS; e.g. SUMO2-T90R)21. To analyse endogenous SUMO conjugates, a method called PRISM has been developed for the identification of SUMOylation sites based on the chemical blockade of all free lysines, followed by treatment with a SUMO-specific protease and subsequent biotin-tagging of ‘freed’ lysines and identification by high-resolution MS22. Monoclonal antibodies with mapped epitopes in SUMO1 or SUMO2/3 allow the identification of SUMOylated proteins in endogenous conditions23. Finally, SUMO-traps have been developed to enrich and identify poly- or multi-monoSUMOylated proteins24,25. Identification of other UbL-modifications has been performed using exogenous epitope-tagged versions or using antibodies to detect endogenous substrates26,27,28,29,30,31.
Variations between these approaches can identify different targets due to diverse levels of stringency, solubility, expression levels, and antibody types and their affinities. Furthermore, antibody and molecular trap approaches are ideal but require additional expertise and expense for affinity resins. Using an in vivo biotinylation approach, it was previously demonstrated that a short N-terminal epitope (Bio or AviTag)32 on Ub is efficiently biotinylated in cultured cells and in living organisms (flies/mice) by using the E. coli BirA enzyme, and that biotinylated Ub (bioUb) is incorporated efficiently into substrates33,34. Lysis under denaturing conditions allows inactivation of UbL isopeptidases and the high affinity biotin-streptavidin interaction allows stringent washes, yielding pure UbL-modified proteins with almost no background from non-covalent interactors and non-specific contaminants. Here, using a modular multicistronic expression platform35, we have generated a set of tools to study each UbL modification under stringent conditions, suitable for MS or validation by Western blotting, in an accessible and affordable way. We applied the system to various UbL modifications in cell culture and in vivo, identifying known and potentially novel substrates. The bioUbL system can complement existing tools to study UbL modifications to understand their diversity, regulation, and roles in biological systems.
Results and Discussion
Generation of the bioUbL system in multicistronic vectors
When applying the BirA/Bio tagging system to other UbLs, we modelled the initial vectors on the bioUb-BirA fusion, which relied on the capacity of endogenous DUBs to process the tandem bioUb-BirA precursor for the separation of the bioUb and the biotinylating enzyme33,34. However, when this design was attempted for SUMO, a large proportion remained unprocessed, perhaps due to failure of endogenous SUMO proteases to access the cleavage site. We reasoned that co-expression of bioUbLs and BirA separately could improve this, using a multicistronic vector platform35. The Ac5-STABLE2 vector encodes three ORFs separated by viral 2A sequences, and were designed to allow modular exchange of the promoter and ORF elements. For the first ORF, we placed the biotinylation-target peptide (Bio) followed by sites to insert UbL cassettes. For the second ORF, we inserted BirA, encoding the enzyme that transfers biotin to the Bio epitope. To enhance expression, in some cases a human codon-optimized version was used (BirAopt)36. For the third ORF, we inserted cassettes to allow stable cell line selection (e.g. puro, puromycin resistant cassette) or selection with either drugs or fluorescence or both (GFPpuro, GFP-puro fusion protein). Also, in some cases the third ORF position was exchanged for the respective E2 conjugating enzyme for the UbL being studied, which enhanced the recovery of bioUbL conjugates. The convenience to express the respective E2 along with the bioUbL should be analysed with caution, as it will be discussed in the Conclusion.
The resulting bioUbL-2A-BirAopt (with puro, GFPpuro, or E2 conjugase; see Supplementary Data S1 for a summary of the vectors generated) is self-processed via 2A sequences (Fig. 1A, step 1), the BirA enzyme recognizes the Bio sequence (step 2), and transfers biotin to the lysine within the Bio sequence (step 3). The small C-terminal 2A remnant is removed during the activation step by endogenous UbL isopeptidases and, through action of the respective E2 conjugases and E3 ligases, the resulting bioUbL is incorporated into protein targets (X, Y, step 4). Modified proteins are purified by streptavidin-based affinity chromatography (step 5) and analysed by Western blot or MS (step 6). Importantly, lysis and pulldown are performed in denaturing conditions that inactivate UbL isopeptidases, selecting for recovery of true covalent UbL conjugates, while non-covalent UbL interactors are reduced. Since the components are genetically encoded, the system can be applied in both cell culture and transgenic organisms.
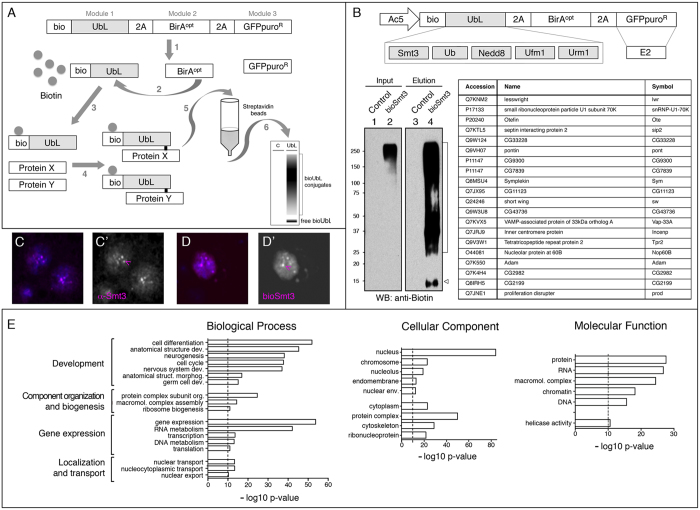
Figure 1. Isolation and identification of bioSmt3 conjugates in Drosophila cultured cells.
(A) Schematic representation of the bioUbL strategy. See text for details. (B) Up: schematic representation of the bioUbL vectors for Drosophila cells. Left: the blot shows the enrichment in bioSmt3 conjugates in the elution panel using anti-biotin antibodies (lane 4; bracket). Arrowhead indicates free bioSmt3 in lane 4. Right: top twenty bioSmt3-modified proteins based on raw intensity. (C,D) bioSmt3 (pAc510x-bioSmt3-Lwr) visualized with fluorescently labeled streptavidin (D) localizes in a similar way than the endogenous Smt3 in S2R+ cells (C). Nuclei were marked with DAPI (blue). Green and red channels are shown independently in black and white (C’, D’). (E) GO analysis for biological process, cellular component and molecular function of the selected bioSmt3-conjugated protein set (n = 1054).
bioUbL vectors for the analysis of PTMs in insect cells
As a proof of concept, to study UbLs different than Ub using the in vivo biotinylation approach, we chose to isolate and identify SUMOylated proteins in Drosophila cultured cells. Drosophila S2R+ cells were transfected with the Ac5-bioSmt3-Lwr vector or with Ac5-FC-GP vector as a control, which contains Flag-Cherry instead of bioSmt3. The vectors were appropriately expressed in cells (Supplementary Fig. S1). After pulldown we saw a notable enrichment of biotinylated proteins in the sample (Fig. 1B), as well as a prominent band corresponding to the properly processed bioSmt3 fusion.
Drosophila Smt3 localization is mainly nuclear and, as in the case of the SUMO mammalian homologs, Smt3 concentrates into discrete subnuclear domains (Fig. 1C). To confirm the localization of the biotin-conjugation system, we analysed bioSmt3 in the cells using streptavidin conjugated to a fluorescent dye (Fig. 1D). The comparison of the immunofluorescence experiments revealed that bioSmt3 localized in a way similar to the endogenous Smt3. In both cases, the proteins were mainly nuclear and localized in nuclear bodies, suggesting the proper subnuclear accumulation of the bioSUMOylated targets.
Compared to ubiquitination, modification by SUMO occurs at lower levels in cells in response to defined stimuli. For a particular target (which might be weakly expressed) only a small proportion of the cellular pool may get SUMOylated. We reasoned that supplying Lesswright (Lwr), the Drosophila homolog of SUMO E2 conjugase along with bioSmt3 (which must compete with endogenous Smt3) would allow detection of more conjugates while using less cellular lysate. To do this, we replaced the selectable marker GFPpuro by lwr. While we observed an appreciable increase in bioSmt3 conjugates (Supplementary Fig. S1), we caution that exogenous E2 expression might produce non-physiological SUMO modifications (see Conclusions). To further augment expression levels, we modified the Ac5 promoter to include 10 copies of the Upstream Activation Sequence, UAS, just before the transcriptional start site. This sequence is recognized by the yeast transcriptional activator Gal4. In the absence of Gal4, this modified Ac5 drives expression at lower levels than unmodified Ac5, but in the presence of Gal4, bioSmt3 levels were significantly higher. S2R+ cells were transfected with the Ac510x-bioSmt3-Lwr vector, or with Ac510x-FC-Lwr as a control, in presence or absence of the pAc5-Gal4 vector. After the pulldown, total protein eluates were resolved on a SDS-PAGE and analysed by MS. We identified 1341 putative SUMOylated proteins, of which 1054, corresponding to 1042 different genes, were selected based on an 4:1 intensity ratio between the experimental samples and the controls (see Methods; Supplementary Data S2; Supplementary Fig. S2).
The Gene Ontology (GO) analysis revealed the enrichment of four biological process categories: development, component organization, gene expression or localization and transport (Fig. 1E, Supplementary Data S2). The majority of the proteins were localized in the nucleus, including proteins associated to the chromosomes, nucleolus or nuclear envelope. Cytoplasmic proteins were enriched for those classified as cytoskeletal and protein complex components. With respect to molecular function, we found enrichment of proteins that regulate gene expression, considered to be the main function of cellular SUMOylation, but also binding proteins (DNA, RNA, protein), underlining a role for SUMO in forming and regulating higher-order complexes.
Most of the identified proteins are functionally related, as revealed by the STRING analysis of interconnected networks (Supplementary Fig. S1). Cytoscape MCODE analysis showed 26 interaction clusters that confirm the implication in gene expression regulation of the identified SUMO targets (transcription, RNA processing and transport, splicing, translation), as well as vesicle-mediated transport, proteasome-mediated Ub-dependent protein catabolic process, or cell cycle (Supplementary Data S2).
549 proteins (52%) of the proteins found in S2 cells were found previously SUMOylated in other Drosophila experiments done using different systems and developmental stages: Drosophila embryos, 37%37 and immune challenged S2 cells, 46%38 (Supplementary Fig. S1). This is a substantial overlap considering the different experimental settings among studies (whole embryos versus cultured cells, unstimulated versus stimulated cells). Interestingly, this percentage of overlap is lower when we compare our list with the identified targets of bioUb in either whole embryos or adult eyes39, supporting the specificity of the bioSmt3 system.
Among the 20 bioSmt3-conjugates that showed highest intensities (all absent from control samples; Supplementary Data S2), some are involved in modulation of gene expression, involving transcription regulation (Pontin, Pont; CG7839; CG2982), translation initiation (Adam), polyadenylation (Symplekin, Sym), RNA binding/processing (CG11123; Nucleolar protein at 60B, Nop60B) and splicing (small ribonucleoprotein particle U1 subunit 70 K, snRNP-U1-70K). Others have been linked to nuclear functions, such as heterochromatic centromere and chromosomal segregation (Proliferation disrupter, Prod), mitosis (Inner centromere protein, Incenp) or components of the nuclear envelope (Otefin, Ote). In the cytoplasm, represented functions include transport and trafficking (Short wing, Sw; VAMP-associated protein of 33 kDa orthologue A, Vap-33A) and protein folding (Tetratricopeptide repeat protein 2, Tpr2). Among those 20, 18 were previously identified as targets of SUMOylation (Supplementary Data S2)37,38. In addition, the human homologues of 9 of them were also reported to be SUMOylated (UBE21, snRNP-U1-70K, Pont, CG7839, Sym, CG11123, Vap-33A, Nop60B and Adam), suggesting a conserved role for SUMOylation in the function of these proteins.
In addition to Smt3, the Drosophila genome encodes other UbL proteins. These include single homologs of NEDD8 (Nedd8), UFM1 (CG34191), and URM1 (CG33276). These genes were used to create bioUbL versions and expressed in S2R+ cells (Supplementary Data S1) and expression was confirmed in S2R+ cells. Together with bioUb-BirA (adapted from UAS-bioUb-BirA)33,34, these vectors will facilitate analysis of any Ub- or UbL-modification in Drosophila cells.
Validation of bioSmt3-conjugates in Drosophila cells
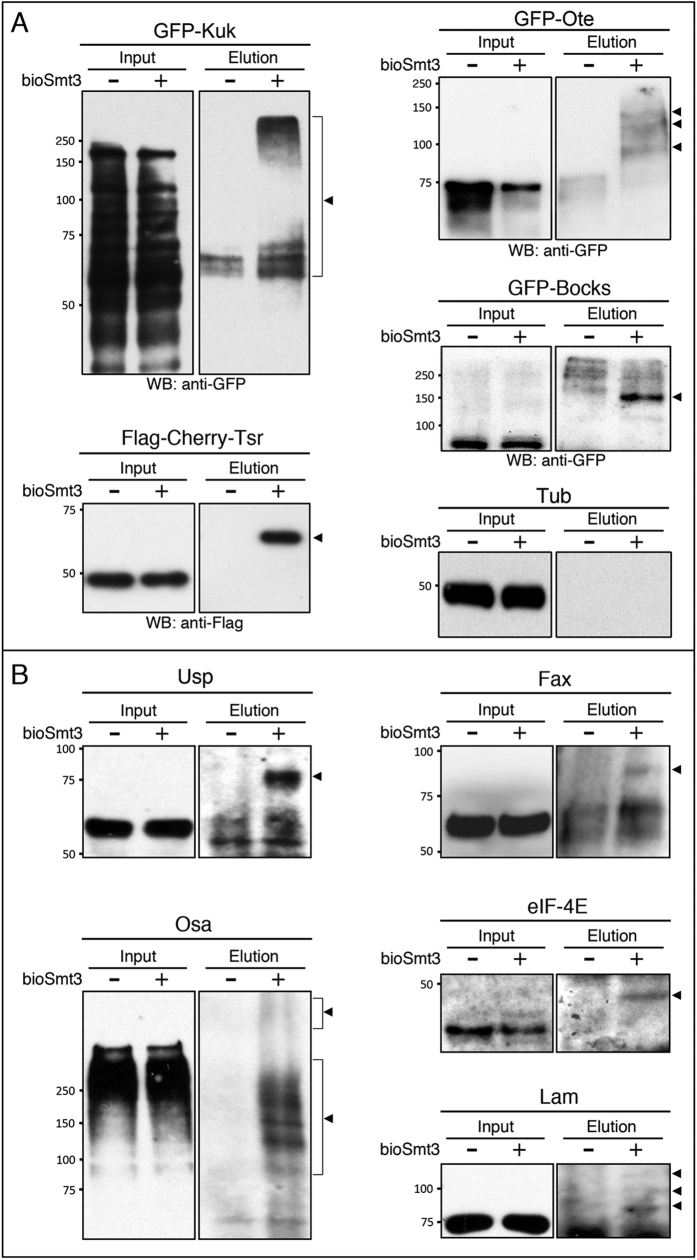
In order to validate our SUMOylation results, we chose Ote, Kugelkern (Kuk) and Bocksbeutel (Bocks), all components of the nuclear envelope, which is a structure enriched for SUMOylated proteins40,41,42 and is a described GO category represented by 34 MS hits in our list (Supplementary Data S2). Twinstar (Tsr) is the Drosophila orthologue of the actin destabilizing factor cofilin. Previous work has demonstrated SUMOylation of human cofilin43,44. Interestingly, we note that one of the identified SUMOylation sites of human cofilin (K13) overlaps with its reported nuclear export sequence (VIKVFNDMKV)45, which might have important consequences on localization for a small protein like Tsr (17 kDa). Tagged versions of these proteins (GFP- or FLAG-) were expressed in S2R+ cells, with and without bioSmt3. After the NeutrAvidin pulldowns, Western blots revealed the SUMOylated forms in the elution panels only in presence of bioSmt3 (arrowheads in Fig. 2A). Endogenous tubulin was used as a control for the input.
Figure 2. Validation of bioSmt3-modified proteins.
(A) Anti-Flag or anti-GFP Western blot of pulldowns performed in S2R+ cells transfected respectively with GFP-kuk, GFP-Ote, GFP-bocks or Flag-Cherry-tsr, together with pAc5-GAL4 and pAc510x-bioSmt3-Lwr (bioSmt3) (+) or Ac510x-FC-Lwr (−) as control. In the elution panels, arrowheads indicate the modified forms of each protein. Tubulin was used as a loading control. Molecular weight markers are shown to the left. (B) Western blot of pulldowns performed in S2R+ cells. Specific antibodies against endogenous proteins were used: Ultraspiracle (Usp), the Trithorax group protein Osa, Eukaryotic initiation factor 4E (eIF-4E), Failed axon connections (Fax) and Lamin (Lam). In elutions, arrowheads indicate the modified forms of the respective proteins. Molecular weight markers are shown to the left.
Despite the small proportion of SUMOylated fraction in the general pool of a given protein, we were able to validate bioSmt3 conjugation to some endogenous proteins by using available antibodies (Fig. 2B). Three of the validated proteins have been previously identified as SUMOylated proteins: Ultraspiracle (Usp), a nuclear receptor involved in steroid signalling46, the transcription factor Osa37,47, the intermediate filament Lamin (Lam, also a key nuclear envelope component)38 and the eukaryotic initiation factor 4E (eIF-4E)48. One of the validated proteins was not previously described to be SUMOylated in Drosophila, the Glutathione S-transferase involved in axonogenesis Failed axon connections (Fax), and thus is the first Fax homolog to be identified as a target of SUMOylation.
In vivo application of bioSmt3 in transgenic Drosophila
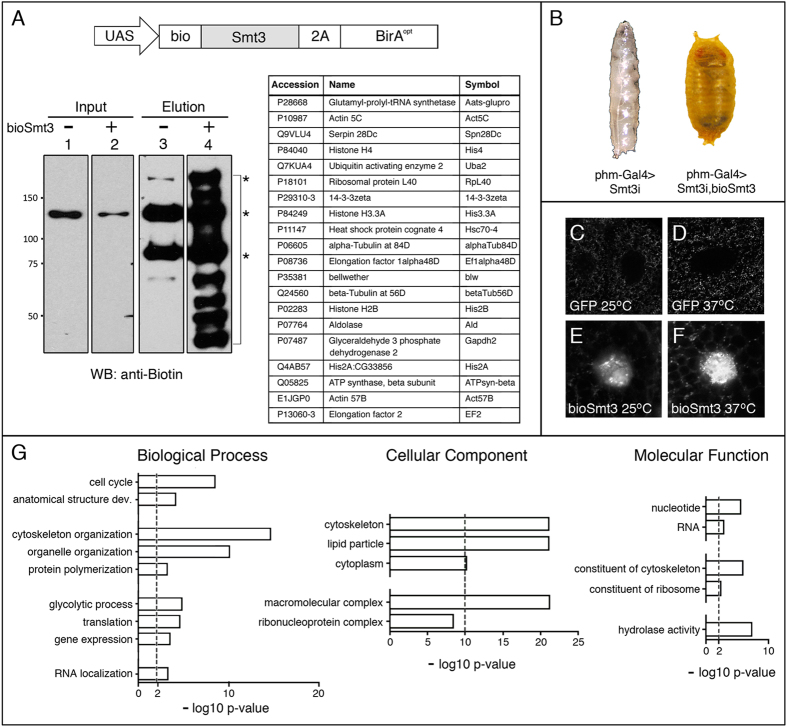
We applied the bioUbL technology in vivo by generating transgenic flies expressing the transgene bioSmt3 (Fig. 3A). To test the functionality of the bioSmt3 transgene in vivo, we analysed its capacity to rescue the silencing of endogenous smt3 in steroidogenic tissues at larval stages. Tissue-specific knockdown of smt3 in the prothorathic gland (PG) blocks development at the end of larval stages due to a reduction in the steroid levels49,50. This is achieved using a PG-specific GAL4 driver, together with UAS-Smt3 RNAi (hereinafter called smt3i). Interestingly, by introducing an RNAi-resistant bioSmt3 transgene in the smt3i background, larvae underwent normal metamorphosis and generated fertile adults (Fig. 3B). This indicates that bioSmt3 can complement at least this specific role for SUMOylation during development, which may also be the case in other tissues. Importantly, by increasing the bioSmt3/endogenous-Smt3 ratio, the chances of recovering rare bioSmt3 conjugates were increased.
Figure 3. In vivo isolation and identification of bioSmt3 conjugates.
(A) Up, schematic representation of the bioSmt3 vector used for Drosophila transgenesis. Down, the blot shows the enrichment of bioSmt3 conjugates in the elution panel using anti-biotin antibodies (lane 4, bracket; strain used: hs-Gal4; UAS-bioSmt3). Asterisks indicate the three known endogenously biotinylated proteins in Drosophila. (B) BioSmt3 can substitute for endogenous Smt3. Left, larvae silenced for smt3 in the PG, arrested in development at the end of L3 larval stage. Right, the developmental arrest is rescued by the expression of bioSmt3 in the same genetic background. (C–F). Confocal pictures of cells of the salivary glands of larvae expressing bioSmt3 (E,F) or GFP (C,D). bioSmt3 is detected by fluorescently-labeled streptavidin. Heat treatment (37 °C) increased the bioSmt3-positive bodies. (G) GO analysis for biological process, cellular component and molecular function of the selected 140 bioSmt3-conjugated proteins.
We analysed the subcellular distribution of bioSmt3 conjugates in vivo by using fluorescent streptavidin in salivary glands from larvae expressing bioSmt3 (hs-Gal4 > UAS-smt3i, p-bioSmt3) or from control larvae (hs-Gal4 > UAS-GFP). Larval food was enriched with biotin. After using heat shock (hs) at 37 °C to both activate Gal4 and stimulate SUMOylation, we observed the accumulation of bioSmt3 in nuclear bodies, most likely conjugated to proteins (Fig. 3C–F). The biotinylated proteins showed a pattern similar to that observed for endogenous Smt3 using antibodies.
For the MS experiments, we collected larvae of hs-Gal4 > UAS-Smt3i, p-bioSmt3 or hs-Gal4 > UAS-Smt3i, pUAS-BirAopt genotypes. The E2 conjugase Lwr was not exogenously expressed in these experiments. To enrich the proportion of proliferating tissues, which exhibit a higher abundance of SUMOylated proteins51, we dissected and processed the anterior part of the larval body where the brain and most of the imaginal discs are located. After pulldown, we confirmed the enrichment of bioSmt3 conjugates (Fig. 3A). Protein eluates were resolved on SDS-PAGE and analysed by MS. We identified 251 proteins as putative SUMOylation targets in vivo (Supplementary Data S3, Supplementary Fig. S3), of which 140 were selected on the basis of a 4:1 intensity increase over controls.
The GO analysis (Fig. 3G) revealed common terms with the analysis in cultured cells, such as gene expression, translation or RNA, with a higher proportion of proteins localized in the cytoplasm, with the categories of lipid particle or cytoskeleton organization being strongly represented. The STRING analysis revealed a network of proteins, where 10 clusters could be identified (Supplementary Fig. S4, Supplementary Data S3). Among those, the most prominent is associated to the GO term translation, followed by others such as microtubule cytoskeleton, proteasome-mediated Ub-dependent protein catabolic process, chromatin assembly or disassembly or protein folding, among others. Some of the clusters reflect terms related to the methodology used in this experiment, such as the heat response used to induce expression and SUMOylation, or substrate adhesion, perhaps overrepresented due to the presence of the salivary glands in the lysed tissues. Approximately 50% of the proteins identified in vivo were also present in the list of bioSUMOylated proteins obtained from cultured cells and 34 or 37% were present in the lists of SUMOylated protein identified in Drosophila embryos or immune challenged S2 cells, respectively (Supplementary Fig. S4)37,38.
Among the 20 best-scoring bioSmt3 conjugates (Supplementary Data S3), we found parallels to our S2R+ dataset, for example proteins involved in different aspects of gene regulation, as chromatin and transcription (Histones His3.3A and His2B, CG7839), mRNA polyadenylation (Protein 1 of cleavage and polyadenylation factor 1, Pcf11) and translation (Eukaryotic initiation factor 4a, eIF-4a, and eIF4AIII), as well as cytoskeleton and protein trafficking (TER94, CG7852, Tubulins betaTub60D and betaTub85D, or Wings up A, WupA). 70% of these proteins (14/20) were identified as SUMOylated either previously or in this work in S2R+ cells. Also, the human homologues of many (17/20) have been independently identified as SUMO targets. CG42671, CG30069 and WupA were not previously identified as Smt3 targets. The human homolog of WupA the muscle-enriched TNNI2 (troponin I2, fast skeletal type) has never been identified as a possibly SUMOylated protein, which may reflect that most SUMO proteomics studies are done in cells rather than using whole tissues.
The notable difference in the number of putative SUMO conjugates when comparing the MS list obtained from S2R+ cells (1054) with the list obtained from larvae (140), is likely due to experimental differences. For example, cells yield better and more uniform lysates than tissues, biotin availability in vivo may be limiting and a lower quantity of in vivo material was processed compared to cells. Also, in flies we used RNAi to increase the bioSmt3/wild-type Smt3 ratio, while in S2R+ cells, we co-expressed the E2 conjugase lwr along with bioSmt3 to enhance the process of SUMO conjugation. These aspects might affect the yield of SUMOylated proteins in both cases and likely requires optimization for specific applications. Nonetheless, our results show that the bioSmt3 system can be efficiently applied to identify potential SUMOylated proteins in cultured cells and in vivo (Figs 1 and 3; Supplementary Data S2, S3).
bioUbL vectors for the analysis of PTMs in mammalian cells
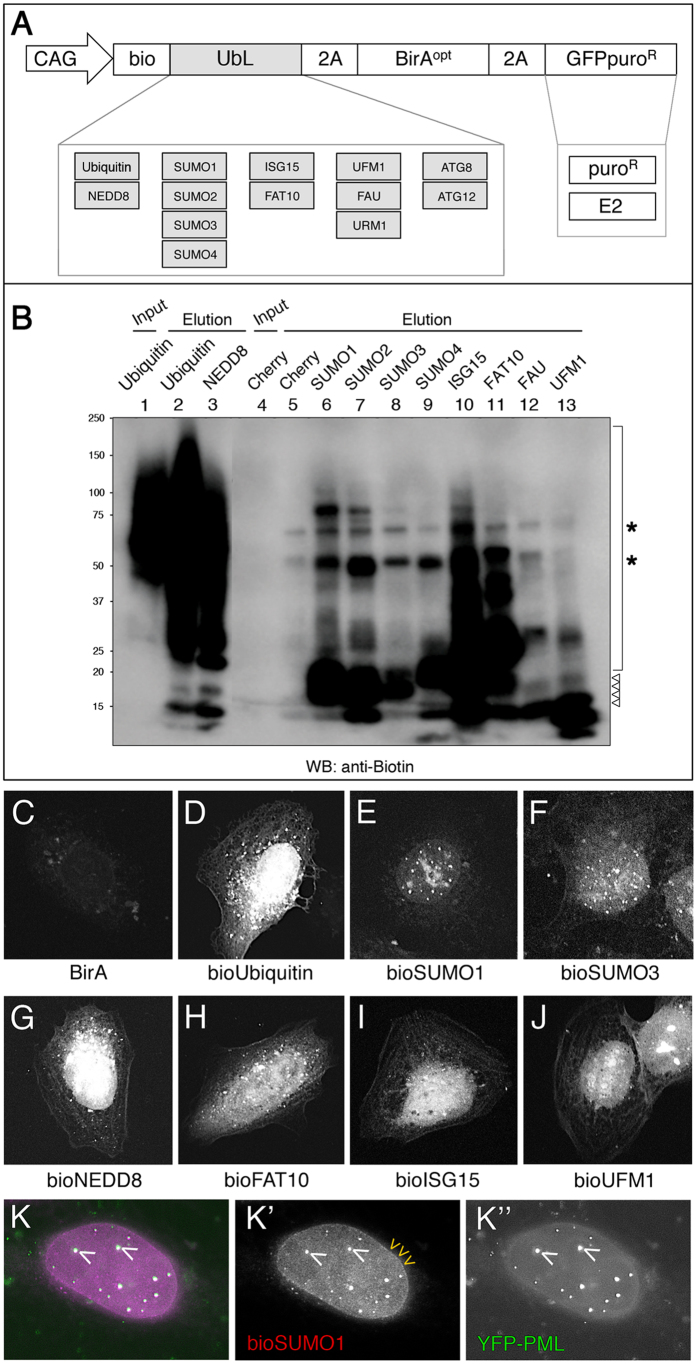
Encouraged by the Drosophila results, we turned to mammalian cells to explore whether the system could work equally well and to broaden its applications. The modularity of the pAc5-STABLE2-neo vector facilitated the switch from the insect-specific promoter Ac5 to the versatile mammalian promoter CAG (Fig. 4A). We substituted the Drosophila smt3 gene with the following mammalian UbLs: Ub, NEDD8, SUMO1-4, FAT10, ISG15, UFM1, FAU, and URM1 (Supplementary Data S1). All of these UbLs are thought to conjugate to protein targets through C-terminal glycine(s). Vectors expressing the autophagy-related ATG12 and ATG8 family (GABARAPs and LC3s) were also constructed, although these UbLs are thought to modify lipids more than proteins. Some variants of UbLs were also cloned (Supplementary Data S1). In most cases, the vectors carry a GFPpuro cassette to allow drug- or fluorescence-based selection, although other cassettes (resistance to puromycin, blasticidin, hygromycin) are possible. In some cases, the selection cassette is exchanged for the corresponding E2 enzyme to increase the efficiency of conjugation or address enzyme specificity (Fig. 4A). Using the modular architecture, other combinations could be envisioned, such as E1 activating enzymes, E3 ligases, substrates, and pathway modulators. Caution should be exercised since exogenous expression of E2 or indeed any other pathway enzyme may produce non-physiological modifications. Additional 2A-ORFs could also be inserted to expand the number of components for specific needs.
Figure 4. Isolation and localization of bioUbL-conjugates in mammalian cells.
(A) Schematic representation of the bioUbL plasmid collection. (B) BioUbL-conjugates revealed by anti-biotin Western blot. Pulldowns were performed in parallel using HEK 293FT cells expressing the different bioUbLs. Conjugates are indicated with a bracket. Asterisks indicate endogenously biotinylated proteins. Arrowheads indicate free bioUbLs. Molecular weight markers are shown to the left. (C–J) Cellular distribution of different bioUbLs. U2OS cells transfected with plasmids expressing the indicated bioUbLs (D–J) or BirA alone as a control (C). Conjugates are visualized using fluorescently-labeled streptavidin. (K) White nuclear bodies (white arrowheads indicate two examples) reflect the colocalization of bioSUMO1 (purple) with YFP-PML (green). Yellow arrowheads indicate the localization of bioSUMO1 in the nuclear membrane. (K’) and (K”) show independently the green and purple channels in black and white, respectively.
We tested the incorporation of the different bioUbLs to target proteins in HEK 293FT cells. The same number of cells were transfected in parallel with the different bioUbL constructs and NeutrAvidin pulldowns were analysed by anti-biotin Western blot (Fig. 4B). Although the cells were not submitted to additional stress (heat-shock, proteasome inhibition, etc), most of the tested bioUbLs were incorporated into target proteins. As expected, the relative amount of conjugated proteins varied depending on the modifier. For instance, bioUb, bioNEDD8 and bioISG15 showed the greatest level of incorporation, with some distinct conjugates being seen for bioSUMOs and bioFAT10. The bioUFM1 and bioFAU incorporation was negligible. This could be due to small amount of cells/lysate used, limiting expression levels of enzymes (E1/E2/E3) needed to incorporate the respective bioUbLs, or necessity of a particular stimulus to activate conjugation.
Since biotinylation occurs in vivo, we analysed the cellular distribution of the different bioUbLs by using fluorescent streptavidin (Fig. 4C–J). The different bioUbLs showed distinct localization patterns, suggesting that distribution in part may reflect localization of endogenous conjugates. Most of the UbLs are predominantly nuclear, although not exclusively, as seen with bioUb, bioNEDD8 or bioFAT10 (Fig. 4D,G,H). The bioSUMOs tested showed accumulation in nuclear bodies PML-positive, with bioSUMO1 showing further enrichment in the nuclear envelope (Fig. 4K); these structures are known to be enriched in SUMOylated proteins. The unique and characteristic patterns observed for each bioUbL suggests incorporation into a specific set of target proteins, further supporting the specificity of the bioUbL system.
Assessment of bioSUMOs in human cells
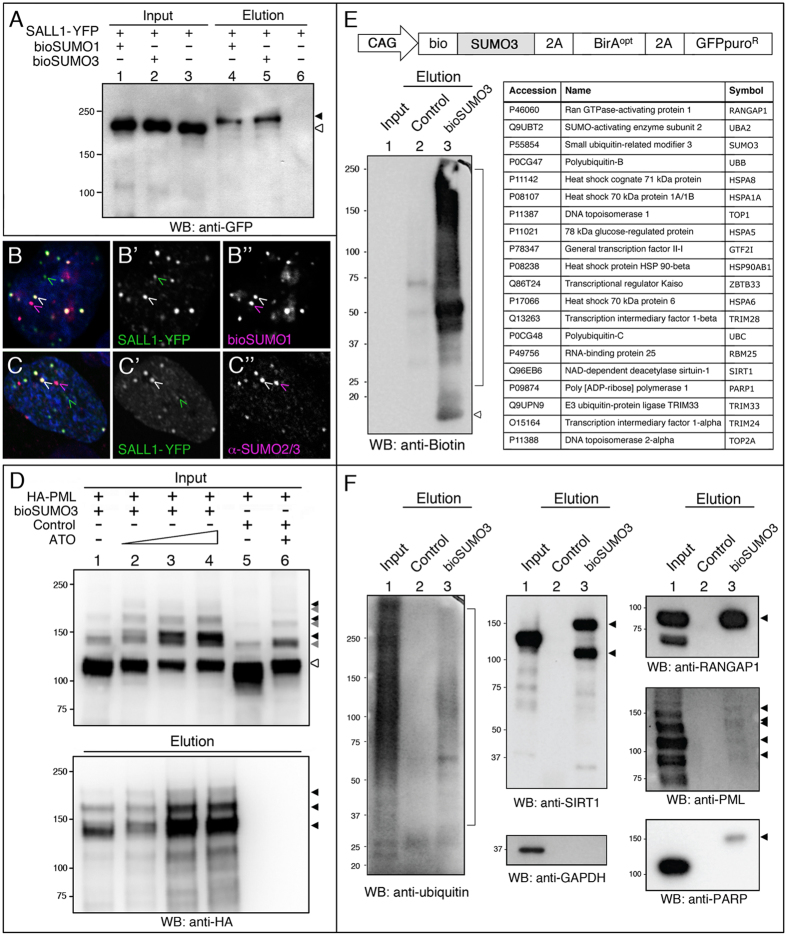
To further evaluate the suitability of the system in mammalian cells, we chose to test SUMOylation of a specific target protein. The human transcription factor SALL1 was previously described to be SUMOylated in vitro52. By co-transfecting human SALL1 fused to YFP with bioSUMO1 or bioSUMO3 into HEK 293FT, we were able to confirm the SUMOylation of SALL1 in cells (Fig. 5A). We also observed the partial colocalization of the protein with bioSUMO1 (Fig. 5B) in a similar way than with the endogenous SUMO2/3 as revealed with the anti-SUMO antibodies (Fig. 5C).
Figure 5. Isolation and identification of bioSUMO3-conjugates in mammalian cells.
(A) Western blot of pulldowns from HEK 293FT cells showing that the transcription factor SALL1 fused to YFP was SUMOylated in presence (+) of bioSUMO1 (lane 4) or bioSUMO3 (lane 5; bioSUMO1-BirA or bioSUMO3-BirA, respectively). Black arrowhead indicates the modified SALL1-YFP in the elution panel (lanes 4, 5), which is shifted in comparison to the non-modified SALL1-YFP in the input panel (white arrowhead, lanes 1–3). Molecular weight markers are shown to the left. (B,C) Partial colocalization between SALL1-YFP (green) and bioSUMO1 (purple) (bioSUMO1-BirA-UBC9) in U2OS cells (B) or with endogenous SUMO2/3 (C, purple). White arrowheads indicate colocalization. Nuclei were stained with DAPI (blue). (B’,C”) Green and purple channels are shown independently in black and white. (D) SUMOylation of PML by bioSUMO3 (bioSUMO3-BirA-GP) increases after ATO treatment. bioSUMO3-modified PML (black arrowheads) can be detected by anti-HA Western blot in the input (upper panel, lanes 1–4) and the elution (lower panel, lanes 1–4; NeutrAvidin pulldown), while the level of the non-modified form of PML is reduced (input panel, lanes 1–6; white arrowhead). Note that modification of PML by endogenous SUMO is also visible in the input panel after ATO treatment (lanes 5 and 6, grey arrowheads). Control indicates cells transfected with BirA-GP. Molecular weight markers are shown to the left. (E) Up: schematic representation of the bioSUMO3 vector for mammalian cells. Left: the Western blot shows the enrichment in bioSUMO3 conjugates in the elution panel using anti-biotin antibodies (lane 3, bracket). Arrowhead indicates free bioSUMO3. Right: bioSUMO3-modified proteins identified by nLC MS/MS on Orbitrap. (F) Validation of bioSUMO3-modified proteins. Specific antibodies against endogenous proteins were used: Ub, SIRT1, RANGAP1, PML and PARP. GAPDH is shown as a control in the input panel. In the elution panels, arrowheads indicate the modified forms of the respective proteins and the bracket indicates the ubiquitinated proteins (lanes 3). Molecular weight markers are shown to the left.
PML SUMOylation is enhanced by the treatment with arsenic trioxide (ATO), triggering its polyubiquitination and subsequent degradation by the proteasome. Co-transfection of PML and bioSUMO3 was used to test the ATO response. As expected, after treatment we detected a decrease in the levels of unmodified PML in HEK 293FT cells (Fig. 5D, input panels, white arrowhead). In pulldowns, the levels of biotinylated PML increased, as expected from an increased incorporation of bioSUMO3 provoked by ATO (Fig. 5D, elution panels, black arrowheads; Control lanes in input panels show endogenous SUMOylation).
Besides testing for bioSUMOylation of particular proteins, we performed a small-scale MS experiment using HEK 293FT cells expressing bioSUMO3 (without E2 conjugase) or BirA alone. NeutrAvidin pulldowns were subjected to gel electrophoresis and stained with SYPRO-Ruby. Eleven prominent bands were excised and submitted for in-gel digestion and further identified by nLC MS/MS in Orbitrap. Twenty main proteins were identified in those bands, with all of them being previously described as SUMO targets, confirming the validity of the system (Fig. 5E). By Western blot analysis, we were able to observe bioSUMO3-modified forms for several of these targets, including Sirtuin 1 (SIRT1), Ran GTPase-activating protein 1 (RANGAP1), PML and Poly [ADP-ribose] polymerase 1 (PARP) (Fig. 5F). These experiments demonstrated that the bioSUMO system was sufficiently sensitive to detect the SUMOylated fraction of these proteins without stressing the cells. Interestingly, we identified Ub in the pilot MS experiment (Fig. 5E) and furthermore, we detected Ub in the pulldown lane of the Western blot (Fig. 5F), which suggests that either multiple PTMs (independent SUMO3 and Ub) or mixed-chain formation (SUMO3-Ub chains) can occur on substrates.
As seen with the Drosophila bioSmt3 vector, inclusion of the SUMO E2 enzyme UBC9 in the bioSUMO vectors led to a general increase the levels of modified targets. In the case of the bioNEDD8 vector, the inclusion of the corresponding E2 UBC12 both increased the level of modified proteins and changed the overall pattern (Supplementary Fig. S5). Specifically, the presence of UBC12 favours the lower molecular weight conjugates (two asterisks) in contrast to the higher molecular weight ones (one asterisk). Since NEDD8 overexpression has been reported to lead to misincorporation into non-habitual substrates by Ub pathway enzymes53, co-introduction of the NEDD8 E2 may suppress this effect. In addition, bioNEDDylation is drastically reduced in presence of MLN4924, an inhibitor of NEDD8 activating enzyme NAE1 (Supplementary Fig. S5), confirming that bioNEDD8 is processed by the same enzymes as the endogenous NEDD8. CUL3, a known NEDDylated protein, can be captured using the bioNEDD8 vector and detected using a specific antibody (Supplementary Fig. S5).
Isolation and identification of bioUFM1 conjugates in human cells
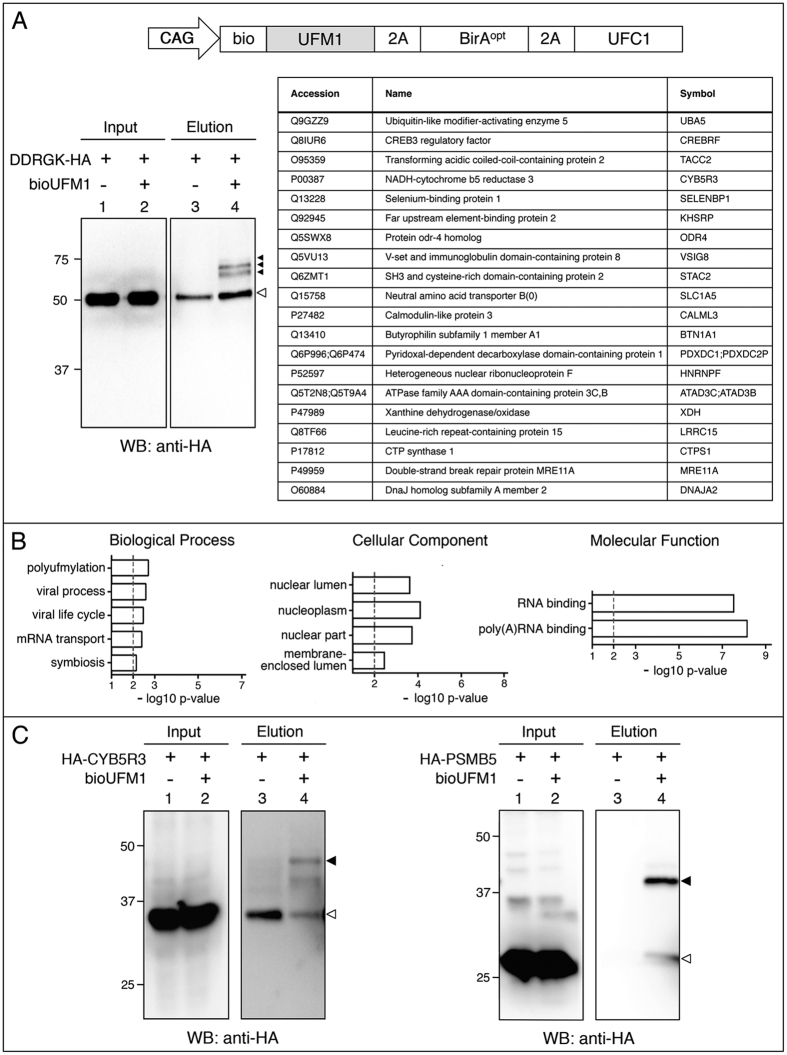
To further test the bioUbL approach in mammalian cells, we chose one of the less-studied UbLs, UFM1, to enrich for potential conjugates. To increase the efficiency of bioUFM1-conjugation, we co-expressed UFC1, the specific E2 enzyme involved in UFMylation (Fig. 6A). Few UFM1-conjugated proteins have been described, but perhaps the best characterized was DDRGK domain-containing protein 1 (DDRGK1, also known as UFBP1)54,55. A role for DDRGK1 as a co-factor of UFMylation has been postulated30, and knockout mice also display defects in embryonic erythroid development and adult hematopoiesis56. Using co-expression in HEK 293FT cells, we validated bioUFMylation of DDRGK1 (Fig. 6A).
Figure 6. Isolation and identification of bioUFM1-conjugates in mammalian cells.
(A) Up: schematic representation of the bioUFM1 vector for mammalian cells. Below, left: Validation of HA-DDRGK. Right, top twenty bioUFM1-modified proteins. (B) GO analysis for biological process, cellular component and molecular function of the selected bioUFM1-conjugated protein set (n = 82). (C) Validation of bioUFM1-modified proteins CYB5R3 and PSMB5 fused to HA tag. In the elution panels (lanes 3 and 4), black arrowheads indicate the modified forms of the respective proteins. Residual non-specific interactions of non-modified forms are indicated by white arrowheads. Molecular weight markers are shown to the left.
To identify potential UFMylated proteins, we performed a larger-scale experiment using the bioUFM1/UFC1 construct in HEK 293FT cells. Since ER stress can upregulate the expression of UFM1 and UFMylation enzymes55, cells were treated with thapsagargin, a chemical inducer of ER stress (via inhibition of the ER Ca2+ ATPase pump). Neutravidin pulldowns were resolved by SDS-PAGE and analysed by MS. After normalization to remove background hits and curation, we identified 82 proteins that are potential UFMylated targets (see Methods; Supplementary Data S4, Supplementary Fig. S6). Among these hits, we did not detect peptides from DDRGK1, but this could be due to low expression levels of this protein in HEK 293FT.
Although UFMylation has been largely studied in the context of ER function, GO analysis showed that there was a significant enrichment in categories linked to RNA, such as mRNA transport (biological process) RNA, polyA-RNA binding (molecular function) or nucleoplasm (cellular component) (Fig. 6B; Supplementary Data S4). This is consistent with the observed nuclear localization of bioUFM1 in U2OS cells (Fig. 4J). The STRING analysis revealed an interconnected network of proteins, where 2 clusters were identified related with RNA splicing and ribonucleotide biosynthetic process (Supplementary Fig. S7, Supplementary Data S4). Of note, some proteins in our hit list are detected together in stable protein complexes (e.g. SLC1A5:CYB5R3, RTN4:ILF3, ILF3:SRRM2, PFAS:PPP6R3, SF3B1:ZNF326; DDX21:PARP1)57, or as yeast two-hybrid interactors (e.g. RTN4:SNX1)58, or as functional interactors (PARP1:MRE11)59,60. Since purification of bioUFM1 conjugates is under denaturing conditions and non-covalent interactions via UFM1 should be non-existent, we speculate that UFMylation may occur on multiple members of protein complexes, as it has been shown for SUMOylation and DNA repair complexes61. Interestingly, ODR4 (also called C1ORF27) is a homolog of C. elegans odr4, an ER-associated transmembrane protein found in complex with odr8, the C. elegans orthologue of UFSP2 (UFM1-specific protease 2)62.
Out of the 82 selected proteins, 47 have been reported as targets of SUMOylation and 45 as targets of ubiquitination (Supplementary Fig. S7, Supplementary Data S4). 32 are targets both of SUMOylation and ubiquitination. It is possible that these different UbL modifications might compete for the same target lysines or that they might take place at the same time. Peptides of Ub were also detected in the MS analysis, suggesting independent UFMylation and ubiquitination of the same substrate or, alternatively, the presence of UFM1-Ub mixed chains. Increases in scale and sensitivity are necessary to reveal the sites of UFM1 conjugation on substrates.
As expected, the co-expressed E2 UFC1 was identified. In pilot experiments that expressed only bioUFM1, endogenous UFC1 was identified (data not shown). Among the next 20 best-scoring bioUFM1 conjugates, we identified UBA5, the E1 enzyme that activates UFM1. Peptides from UFL1, the reported UFM1 E3 ligase, were not identified, but again could be due to expression levels and scale of the experimental setup. We found proteins involved in splicing and RNA trafficking (KHSRP- KH-type splicing regulatory protein, HNRNPF- heterogeneous nuclear ribonucleoprotein F), meiotic recombination and double-strand break (DSB) repair (MRE11A- MRE11 meiotic recombination 11 homolog A) and protein transport (SELENBP1- selenium binding protein 1).
We chose two MS hits to verify our results. Due to links of UFM1 to erythroid development and hematopoiesis, we chose cytochrome B5 reductase 3 (CYB5R3), since mutations can give rise to methemoglobinemia, characterized by the accumulation of blue-tinted methemoglobin in the blood, which gives the patients an unusual blue-tinted skin colour. Furthermore, CYB5R3 is ER-associated, specifically on the cytoplasmic side of the outer ER membrane63. A second interesting hit is PSMB5, one of the core beta subunits of the 20S proteasome. Mutations or overexpression of PSMB5 can lead to resistance to the proteasome inhibitor bortezomib, a chemotherapeutic agent for multiple myeloma and mantle cell lymphoma. Using bioUFM1 and exogenous UFC1 in HEK 293FT cells, we were able to detect UFMylated forms of both CYB5R3 and PSMB5, even without using ER stress conditions (Fig. 6C). Further functional assessment of the role of UFMylation for these targets will require additional effort.
Taken together, the bioUFM1 approach is an efficient method for the isolation and MS identification of potential UFMylated proteins in mammalian cells, as well as for the validation of specific targets. Due to the role of UFMylation in erythroid development uncovered through mouse genetic studies, exploration of these targets or discovery of new ones could be pursued in erythroleukemic or erythroid progenitor cell lines.
Conclusions
Here we report the development of an efficient system to facilitate the study of potential UbL-modified proteins in both mammalian and Drosophila cells, as well as transgenic flies, that is suited for MS-based proteomics, validation and analysis of specific UbL-conjugated targets. This system represents multiple advantages with respect to other available UbL purification systems. The modular vectors represent a flexible platform to facilitate assembly of different elements into a single plasmid, even allowing in vivo biotinylation of non-UbL proteins of interest. Depending on the configuration, the vectors can be used for transient transfections, with FACS or drug selection allowing for enrichment, or for stable cell line generation. For bioUbLs, the possibility to co-express corresponding E2 conjugases or E3 ligases may enhance sensitivity and perhaps enrich for specific substrates of particular E2s or E3s. In the studies reported here, we have co-expressed E2 conjugases in the case of bioSmt3 in cells (but not in flies), and in the case of bioUFM1 (but not bioSUMO3). Expression of the E2 conjugases and/or E3 ligases, together with the bioUbL construct, might give rise to non-physiological modifications in cells, but this approach might facilitate the identification of novel targets below the sensitivity of other methods, or address substrate preferences for particular E3s or E2/E3 combinations. The co-expression strategy has been widely used in the literature for various UbLs, as it can be seen in some recent examples30,64,65,66,67,68,69,70. With caution in mind, we could not find specific reports showing that exogenous UBC9 or UFC1 drives unspecific modification of substrates. Related to this point, no gross developmental or physiological abnormalities were observed in mice that overexpress Ubc9 systemically, despite the overall increase in SUMOylation levels71. By overexpressing the bioUbL, we are likely saturating the existing endogenous E2 and only a certain amount of the bioUbL gets loaded (i.e. Lwr-bioSmt3). By supplying additional E2, cells will be able to make more Lwr-bioSmt3, which would lead to more SUMOylation and higher yield of lower abundance targets (so, more overall IDs in the MS analysis). UBC9 is involved in the selection of SUMO targets through its direct interaction with the SUMO consensus sites present in those targets; therefore, exogenous UBC9 expression might circumvent the need of E3 ligases or other stimulatory mechanisms (reviewed in ref. 7), but might not result in random SUMOylation of exposed lysines. However, the exogenous expression of E2/E3 enzymes might not reflect the physiological condition of a particular cell type/developmental stage/stress situation. The choice and convenience to supply exogenous E2 or not can be decided by researchers for their specific experimental settings, and secondary validations by complementary techniques is encouraged before embarking on detailed studies. All bioUbL vectors are provided without co-expression of E2s, while some of them co-express E2s, so that users can choose and compare when necessary.
To compare and contrast different approaches to UbL modifications, we highlight some advantages of the bioUbL system. The strong binding between biotin and streptavidin allows for purification under denaturing conditions, which maximizes solubility of the proteome and inactivates UbL “removal” enzymes (i.e. deubiquitinases, deSUMOylases, etc.). Likewise, strong binding allows high stringency washes that reduce non-specific background. Compared to the use of polyHis-tagged UbLs, only three endogenously biotinylated carboxylases are recovered as contaminants and are easily identified by MS. While antibody-based immunopurification of endogenous UbL-conjugates is ideal23, specific antibodies and commercial resins do not exist for all UbLs. Regarding the identification of UbL modification sites, use of antibodies that recognize branched-peptide remnants after trypsin digestion of UbL-modified proteins have been used to identify thousands of modification sites (Ub/NEDD8/ISG1572; modified SUMO73), but remains problematic due to expense, set-up, and specificity. Also, this approach is not yet available for all UbL modifications. Using SUMO as an example, identification of SUMO conjugation sites has been facilitated by introducing mutations close to the C-terminal di-Gly, allowing trypsin or LysC to generate short MS-compatible branched peptides21,74. The bioUbLs can be mutated as necessary to apply this strategy to SUMO and if needed, to other UbLs to facilitate conjugation site identification. In conjunction with antibodies to recognize an endogenous protein of interest, the bioUbLs can be used to quickly validate potential modifications by Western blotting. With co-expression of tagged proteins and appropriate scale-up, bioUbLs may also allow study of modification sites by MS, after single-step or two-step purifications. Subcellular localization of bioUbLs can be visualized using fluorescent streptavidin, although this application is limited in that one cannot distinguish between unconjugated and conjugated pools, nor localize in situ UbL modification of specific proteins. The application of bioUbLs in transgenic animals (as demonstrated for bioUB in Drosophila and mice)33,34,39 may allow tissue- or temporal-specific approaches to analyse UbL modifications. Use of the bioUbL system can provide either a global overview or a specific picture of the UbL modifications in most cell types and is compatible with drug treatments, environmental stress, and manipulations of other genes (overexpression, RNAi, CRISPR/Cas9 knockout or activation, etc.). We envision that it will complement existing techniques based on its high versatility, reliability and accessibility to most laboratories.
Taken together, these features make the bioUbL system a good choice for researchers interested in UbL modifications, either for validation when using proteins of interest or for explorative proteomics analysis.
Methods
Generation of vectors
All vectors (Supplementary Data S1) were based on Ac5-STABLE2-neo that contains three separate ORFs (Module1, Module 2, Module3; Fig. 1A) separated by 2A sequences35 (https://www.addgene.org/32426/). A codon-optimized version of BirA (BirAopt)36 was cloned into Module 2. In some cases, a C-terminal V5 epitope tag was added to facilitate detection (BirAoptV5). The Bio peptide (AviTag: MLNDIFEAQKIEWHE)32, was fused to a degenerated Drosophila smt3 sequence resistant to RNAi (EMBL database accession number FN539078)75 to generate bioSmt3, and cloned into Module 1. A cassette encoding GFP-puromycin fusion (GFPpuro) or puromycin alone (puro) was cloned into Module 3. Additional bioUbL vectors were generated by substituting the smt3 sequence. To generate the mammalian vectors, the Drosophila Actin5C (Ac5) promoter was substituted by the composite promoter CAG76. bioUbL vectors and cloning strategies are summarized in Supplementary Data S1.
For the generation of CMV-SALL1-YFP, the SALL1 ORF was amplified by high-fidelity PCR and cloned into EcoRI-SalI sites of pEYFP-N1. For GFP-kuk, GFP-Ote and GFP-bocks, the respective genes were amplified with stop codons from S2R+ cDNA by high-fidelity PCR and cloned into Ac5-STABLE1-neo. In the case of Flag-Cherry-tsr, the GFP module in Ac5-STABLE2-neo was substituted by the Tsr ORF amplified from S2R+ cDNA. For HA-CYB5R3, HA-PSMB5, and DDRGK1-2xHA, ORFs were amplified by high-fidelity PCR from 293FT or HeLa cDNA, and cloned into CB6-HA or CMV-2xHA. All vectors were checked by sequencing.
Isolation of biotinylated substrates in mammalian and Drosophila cells
HEK 293FT cells (human embryonic kidney; Invitrogen) were maintained at 37 °C in DMEM (Dulbecco’s modified Eagle’s medium; Gibco) supplemented with 10% FBS, 1% penicillin/streptomycin (GIBCO). Transient transfections were performed using the calcium phosphate method in complete medium supplemented with biotin (50 μM). Cells were harvested 48–72 hours after transfection. For the isolation of bioSUMO3-conjugates coupled with MS, 10 × 10 cm dishes were transfected with bioSUMO3-GP or FC-GP as control. For the isolation of UFM1-conjugates coupled with LC-MS, 10 × 10 cm dishes were transfected with bioUFM1-UFC1 or BirA-puro.
Drosophila S2R+ cells were obtained from DGRC (https://dgrc.cgb.indiana.edu) and cultured at 25 °C in Drosophila Schneider’s medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Gibco). Transfections were performed using calcium phosphate in 5 × 10 cm dishes with 2 μg of pAC5-Gal4 (Addgene #24344), and 8 μg of Ac510x-bioSmt3-Lwr or Ac510x-FC-Lwr in complete medium supplemented with 50 μM biotin.
Transfected cells were collected after 48–72 hours (HEK 293FT) or 72–96 hours (S2R+), washed with phosphate buffered saline (PBS) and resuspended in lysis buffer [0.5 ml/10 cm dish; 8 M urea, 1% SDS, 50 mM N-ethylmaleimide, 1× protease inhibitor cocktail (Roche) in PBS]. Sonication was performed as needed to reduce sample viscosity. To reduce urea concentration, the samples were diluted by adding binding buffer (3 M urea, 1 M NaCl, 0.25% SDS; 0.5 volume for HEK 293FT, or 3 volumes for S2R+). Incubation was done using 100 μl suspension of high-capacity NeutrAvidin-agarose beads (Thermo Scientific) overnight at room temperature (RT). Washes were done according to Franco et al.33: 2× WB1, 3× WB2, 1× WB3, 3× WB4, 1× WB1, 1× WB5 and 3× WB6 [WB1: 8 M urea, 0.25% SDS in PBS; WB2: 6 M guanidine hydrochloride in PBS; WB3: 6.4 M urea, 1 M NaCl, 0.2% SDS in PBS (pre-warmed to 37 °C); WB4: 4 M urea, 1 M NaCl, 10% isopropanol, 10% ethanol, 0.2% SDS in PBS; WB5: 8 M urea, 1% SDS in PBS; WB6: 2% SDS in PBS]. Samples were eluted in 100 μl of 4× Laemmli sample buffer with 100 mM DTT by two cycles of heating (5 minutes; 99 °C), with vortexing in between. For MS analysis, the bead slurry was transferred to a Vivaclear Mini 0.8 μm PES filter (Sartorius) and spun to recover bead-free eluate.
Isolation of biotinylated substrates in vivo
Flies were raised at 25 °C on standard Drosophila medium supplemented with biotin (50 μM). Strains: w;pUAS-BirA33; w;UAS-Smt3i50; y1w*;P{Gal4-Hsp70.PB}31-1/CyO;TM6B,Tb+ (called here hs-Gal4), Bloomington Drosophila Stock Center, BDSC#1822; w;phm-Gal4, UAS-mCD8::GFP/TM6B, Tb (called here phm-Gal4)77; w;UAS-nls-eGFP/CyO (BDSC #4775); Information about other strains can be found in FlyBase (http://flybase.bio.indiana.edu). The plasmid pJFRC81-bioSmt3 (backbone: Addgene #36432) was used for generating transgenic flies (BestGene).
Crosses between w;UAS-Smt3i,p-bioSmt3 and phm-Gal4 were performed at 25 °C. Crosses with hs-Gal4 were done at 25 °C and w;UAS-Smt3i/+;p-bioSmt3/hs-Gal4 larvae were selected at L2 instar. As controls, we used w; UAS-Smt3i/+;pUAS-BirA/hs-Gal4 larvae. Approximately 200 larvae were collected per condition and three heat shock events were performed at 37 °C during 30 minutes each. Dissections were performed in cold PBS and the lysis was done by using 1 ml of lysis buffer [8 M urea, 1% SDS, 50 mM N-ethylmaleimide in PBS and protease inhibitor mixture (Roche)]. Larval tissues were homogenized, sonicated, and after clearing by centrifugation at 4 °C, the supernatants were used for NeutrAvidin pulldowns performed as described above for Drosophila cells using 100 μl of suspension beads, using the same binding buffer dilution and washing/elution protocol.
Mass Spectrometry
Pulldown elutions were separated by SDS-PAGE and stained with Brilliant Blue G-Colloidal Concentrate (Sigma) according to manufacturer’s instructions. Gel bands were excised from the whole gel lane, destained and proteins were in-gel digested with trypsin (sequencing grade, Promega) overnight as previously described78. The resulting peptide mixtures were extracted, desalted, and concentrated prior to online nanoLC-MS/MS analysis using and EASY nLC1000 system (Proxeon) connected to a Q-Exactive orbitrap (Thermo Scientific) through a nanoelectrospray ion source79. All raw LC-MS files were processed with MaxQuant software (version 1.4.0.3, www.maxquant.org) and search against species-specific Uniprot protein sequence databases and common contaminants using the Andromeda peptide search engine with a false discovery rate of 0.01 at both peptide and protein level. In the case of bioSUMO3, a nano Acquity (Waters) coupled on-line to an Orbitrap-XL (ThermoFisher) was used. Orbitrap raw MS files were processed with Proteome Discoverer (version 1.2) and searched with Mascot (Matrix Science).
The lists of proteins identified by MS were analysed as follows. First, contaminants and proteins identified by only one peptide were eliminated. Then, only those proteins with at least four-fold higher iBAQ values (label-free intensity-based absolute protein quantification) in the experiment samples versus the controls were considered as positive hits. Calculation was done taking into account a baseline for the control, which corresponds to the minimum iBAQ values registered in each control of every experiment.
Significantly enriched Gene Ontology annotation terms were determined using VLAD (http://proto.informatics.jax.org/prototypes/vlad-1.0.3/), InnateDB80 and G:profiler81. Graphical representation in Supplementary Figs S2, S3 and S6 was done using Prism GraphPad and eulerApe v382. Network association analysis was done using STRING database83 and analysed using Cytoscape MCODE clusters and Cluster maker Visualizations84,85.
Western Blotting
Input, flowthrough (FT) and elution samples were collected at different steps of the pulldown protocol. Elution samples were prepared as described above. Input and FT samples were adjusted to 4× Laemmli buffer. After boiling, proteins were separated by SDS-PAGE (BioRad) and blotted using wet transfer to PVDF membranes (Millipore). Membranes were blocked in 1× PBS with 0.1% Tween-20 (PBS-T) and 5% non-fat dry milk for 1 hour. When anti-biotin antibody was used, 1× Casein Blocking Buffer in PBS (Sigma #B6429) was used. Primary antibodies were used in blocking buffer (1 hour at RT, or overnight at 4 °C) as follows: mouse monoclonal anti-Usp (1:200)86; mouse monoclonal anti-Osa (1:50, DSHB); mouse monoclonal anti-Lamin Dm0 (1:500, DSHB #ADL84.12); rabbit polyclonal anti-Fax (1:1000)87; rabbit polyclonal anti-eIF4E (1:1000)88; rabbit polyclonal anti-SIRT1 (1:1000, Cell Signaling Technology #D739); mouse monoclonal anti-RanGAP1C-5 (1:1000, SantaCruz #sc-28322); rabbit polyclonal anti-PARP (1:1000, Cell Signaling Technology #9542); rabbit polyclonal anti-PML (1:1000, Bethyl Laboratories); rabbit polyclonal anti-Ub (1:1000, Sigma #U5379); rabbit polyclonal anti-Cullin 3 (1:1000, Cell Signaling Technology #2759); mouse monoclonal anti-Flag M2 (1:1000, Sigma #F3165); mouse monoclonal anti-GFP (1:1000, Roche #11814460001); mouse monoclonal anti-HA (1:1000, Sigma #H3663); mouse monoclonal anti-actin AC-74 (1:5000, Sigma #A2228); HRP conjugated anti-biotin (1:1000, Cell Signaling Technology #7075); HRP conjugated anti-tubulin (1:5000, Proteintech #66031); HRP conjugated anti-GAPDH (1:5000, Proteintech #60004).
After three washes with PBS-T, blots were incubated for one hour in blocking buffer with secondary antibodies: HRP-conjugated anti-mouse (1:5000, Jackson ImmunoResearch); HRP-conjugated anti-rabbit (1:5000, Jackson ImmunoResearch). Membranes were washed again three times in PBS-T and then developed using chemiluminescence with Clarity Western ECL substrate (Bio-Rad) or SuperSignal West Femto substrate (Thermo).
Immunostainings
Drosophila S2R+ cells were transfected with calcium phosphate in 6 well-plates using 2 μg of pAc5-bioSmt3-GP or pAC5-Gal4 (Addgene #24344). After 3 days, cells were placed on coverslips treated with concanavalin A (Sigma) and, once attached (4 hours), fixation was performed with 4% paraformaldehyde followed by permeabilization with 0.1% Triton X-100 in 1x PBS.
Drosophila larvae were dissected in 1x PBS and fixed for 20 minutes in 4% paraformaldehyde, washed three times in PBT (PBS, 0.3% Triton X-100) for 20 minutes, and blocked in PBT + 1% BSA for one hour.
U2OS cells were plated directly in 24-well plates with acid-washed coverslips and transfected using jetPEI (Polyplus) or Effectene (Qiagen) according to the manufacturer’s instructions. For the localization of different bioUbLs, 1 μg of bioUbL-GP or 1 μg of BirA-puro as control were used; for the localization of SALL1, 750 ng of CMV-SALL1-YFP and 250 ng of bioSUMO1-2A-BirA-2A-UBC9 were used.
Antibodies and dyes: DAPI to label nuclei (1:15000, Sigma); Alexa Fluor 594 conjugated-streptavidin (1:200, Jackson ImmunoResearch) to visualize biotinylated proteins; mouse monoclonal anti-GFP (1:500, Roche); rabbit polyclonal anti-Smt3 (1:150)89; rabbit polyclonal anti-SUMO2/3 (1:100, Eurogentec #AV-SM23-0100). Secondary antibodies: anti-rabbit Alexa-568, anti-mouse Alexa-488 and anti-rabbit Alexa-488 (1:200, Molecular Probes). Samples were mounted in Prolong Gold antifade reagent (Molecular Probes). Confocal images were taken with a Leica DM IRE2 confocal microscope and images were processed using Adobe Photoshop CS5 software.
Additional Information
How to cite this article: Pirone, L. et al. A comprehensive platform for the analysis of ubiquitin-like protein modifications using in vivo biotinylation. Sci. Rep. 7, 40756; doi: 10.1038/srep40756 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank A. Courey, A. Carracedo, M. L. Martínez-Chantar, E. Liebl and P. Lasko and the Bloomington Drosophila Stock Center for reagents. We are grateful to the Proteomics Platform at CIC bioGUNE, member of ProteoRed-ISCIII and CIBERehd. RB thanks the Spanish MINECO (BFU2011-25986, BFU2014-52282-P) and the Consolider Program (BFU2014-57703-REDC), the Departments of Education and Industry of the Basque Government (PI2012/42), and the Bizkaia County. We also thank the UPStream consortium (ITN program PITN-GA-2011-290257, EU). This article is based upon work from COST Action (PROTEOSTASIS BM1307), supported by COST (European Cooperation in Science and Technology). UM thanks the Spanish MINECO (SAF2013-44782P). Work at Novo Nordisk Foundation Center for Protein Research (CPR) is funded in part by a generous donation from the Novo Nordisk Foundation (Grant number NNF14CC0001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions L.P. and J.S. performed the majority of experiments, unless specified otherwise. W.X. performed validation of endogenous bioSmt3 substrates. ARdS performed validation of bioSUMO3 substrates. F.E. performed MS and analysed data for bioSUMO3. J.O.S. and J.V.O. performed MS and analysed MS data for bioSmt3 and bioUFM1. M.G. performed cell culture and pulldown experiments. C.P. performed Drosophila crosses and immunostainings. J.R. and U.M. assisted in GO analysis and data interpretation. M.S.R. provided reagents and assisted in data interpretation. R.B. and J.S. conceived and directed the project and with L.P., wrote the manuscript. All authors reviewed and approved the final manuscript.
References
- Kleiger G. & Mayor T. Perilous journey: a tour of the ubiquitin-proteasome system. Trends in cell biology 24, 352–359, doi: 10.1016/j.tcb.2013.12.003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague M. J. et al. Deubiquitylases from genes to organism. Physiological reviews 93, 1289–1315, doi: 10.1152/physrev.00002.2013 (2013). [DOI] [PubMed] [Google Scholar]
- Komander D. & Rape M. The ubiquitin code. Annual review of biochemistry 81, 203–229, doi: 10.1146/annurev-biochem-060310-170328 (2012). [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature 458, 422–429, doi: 10.1038/nature07958 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen A. G. & Ploegh H. L. Ubiquitin-like proteins. Annual review of biochemistry 81, 323–357, doi: 10.1146/annurev-biochem-093010-153308 (2012). [DOI] [PubMed] [Google Scholar]
- Watson I. R., Irwin M. S. & Ohh M. NEDD8 pathways in cancer, Sine Quibus Non. Cancer cell 19, 168–176, doi: 10.1016/j.ccr.2011.01.002 (2011). [DOI] [PubMed] [Google Scholar]
- Flotho A. & Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annual review of biochemistry 82, 357–385, doi: 10.1146/annurev-biochem-061909-093311 (2013). [DOI] [PubMed] [Google Scholar]
- Guo D. et al. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nature genetics 36, 837–841, doi: 10.1038/ng1391 (2004). [DOI] [PubMed] [Google Scholar]
- Wei W. et al. A stress-dependent SUMO4 sumoylation of its substrate proteins. Biochemical and biophysical research communications 375, 454–459, doi: 10.1016/j.bbrc.2008.08.028 (2008). [DOI] [PubMed] [Google Scholar]
- Liang Y. C. et al. SUMO5, a Novel Poly-SUMO Isoform, Regulates PML Nuclear Bodies. Scientific reports 6, 26509, doi: 10.1038/srep26509 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y. J., Yoo H. M. & Chung C. H. ISG15 and immune diseases. Biochimica et biophysica acta 1802, 485–496, doi: 10.1016/j.bbadis.2010.02.006 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelzer C. & Groettrup M. FAT10: Activated by UBA6 and Functioning in Protein Degradation. Sub-cellular biochemistry 54, 238–246, doi: 10.1007/978-1-4419-6676-6_19 (2010). [DOI] [PubMed] [Google Scholar]
- Hipp M. S., Kalveram B., Raasi S., Groettrup M. & Schmidtke G. FAT10, a ubiquitin-independent signal for proteasomal degradation. Molecular and cellular biology 25, 3483–3491, doi: 10.1128/MCB.25.9.3483-3491.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y. & Xu X. UFMylation: A Unique & Fashionable Modification for Life. Genomics Proteomics Bioinformatics, doi: 10.1016/j.gpb.2016.04.001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J. & Klionsky D. J. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO reports 9, 859–864, doi: 10.1038/embor.2008.163 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh T., Nakamura M., Nabika T., Yoshimura Y. & Tanigawa Y. Ubiquitin-like polypeptide inhibits the proliferative response of T cells in vivo. Immunobiology 200, 140–149, doi: 10.1016/S0171-2985(99)80039-4 (1999). [DOI] [PubMed] [Google Scholar]
- Wang F., Liu M., Qiu R. & Ji C. The dual role of ubiquitin-like protein Urm1 as a protein modifier and sulfur carrier. Protein Cell 2, 612–619, doi: 10.1007/s13238-011-1074-6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerpe R. et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO reports 10, 1250–1258, doi: 10.1038/embor.2009.192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Paige J. S. & Jaffrey S. R. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nature biotechnology 28, 868–873, doi: 10.1038/nbt.1654 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eifler K. & Vertegaal A. C. Mapping the SUMOylated landscape. The FEBS journal 282, 3669–3680, doi: 10.1111/febs.13378 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammsalu T. et al. Proteome-wide identification of SUMO2 modification sites. Science signaling 7, rs2, doi: 10.1126/scisignal.2005146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks I. A., D’Souza R. C., Chang J. G., Mann M. & Vertegaal A. C. System-wide identification of wild-type SUMO-2 conjugation sites. Nature communications 6, 7289, doi: 10.1038/ncomms8289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J. et al. Detecting endogenous SUMO targets in mammalian cells and tissues. Nature structural & molecular biology 20, 525–531, doi: 10.1038/nsmb.2526 (2013). [DOI] [PubMed] [Google Scholar]
- Bruderer R. et al. Purification and identification of endogenous polySUMO conjugates. EMBO reports 12, 142–148, doi: 10.1038/embor.2010.206 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva-Ferrada E. et al. Analysis of SUMOylated proteins using SUMO-traps. Scientific reports 3, 1690, doi: 10.1038/srep01690 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xirodimas D. P. et al. Ribosomal proteins are targets for the NEDD8 pathway. EMBO reports 9, 280–286, doi: 10.1038/embor.2008.10 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. et al. A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. Journal of proteome research 7, 1274–1287, doi: 10.1021/pr700749v (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman J. A. & Shiekhattar R. Analysis of Nedd8-associated polypeptides: a model for deciphering the pathway for ubiquitin-like modifications. Biochemistry 45, 3014–3019, doi: 10.1021/bi052435a (2006). [DOI] [PubMed] [Google Scholar]
- Zhao C., Denison C., Huibregtse J. M., Gygi S. & Krug R. M. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proceedings of the National Academy of Sciences of the United States of America 102, 10200–10205, doi: 10.1073/pnas.0504754102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. M. et al. Modification of ASC1 by UFM1 is crucial for ERalpha transactivation and breast cancer development. Molecular cell 56, 261–274, doi: 10.1016/j.molcel.2014.08.007 (2014). [DOI] [PubMed] [Google Scholar]
- Van der Veen A. G. et al. Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proceedings of the National Academy of Sciences of the United States of America 108, 1763–1770, doi: 10.1073/pnas.1014402108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett D., Kovaleva E. & Schatz P. J. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein science: a publication of the Protein Society 8, 921–929, doi: 10.1110/ps.8.4.921 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco M., Seyfried N. T., Brand A. H., Peng J. & Mayor U. A novel strategy to isolate ubiquitin conjugates reveals wide role for ubiquitination during neural development. Molecular & cellular proteomics: MCP 10, M110 002188, doi: 10.1074/mcp.M110.002188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lectez B. et al. Ubiquitin profiling in liver using a transgenic mouse with biotinylated ubiquitin. Journal of proteome research 13, 3016–3026, doi: 10.1021/pr5001913 (2014). [DOI] [PubMed] [Google Scholar]
- Gonzalez M. et al. Generation of stable Drosophila cell lines using multicistronic vectors. Scientific reports 1, 75, doi: 10.1038/srep00075 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechold U., Gilbert C. & Ogryzko V. Codon optimization of the BirA enzyme gene leads to higher expression and an improved efficiency of biotinylation of target proteins in mammalian cells. Journal of biotechnology 116, 245–249, doi: 10.1016/j.jbiotec.2004.12.003 (2005). [DOI] [PubMed] [Google Scholar]
- Nie M., Xie Y., Loo J. A. & Courey A. J. Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PloS one 4, e5905, doi: 10.1371/journal.pone.0005905 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handu M. et al. SUMO-Enriched Proteome for Drosophila Innate Immune Response. G3 (Bethesda) 5, 2137–2154, doi: 10.1534/g3.115.020958 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez J. et al. Proteomic Analysis of the Ubiquitin Landscape in the Drosophila Embryonic Nervous System and the Adult Photoreceptor Cells. PloS one 10, e0139083, doi: 10.1371/journal.pone.0139083 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R., Gerace L. & Melchior F. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. The Journal of cell biology 140, 259–270 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T. et al. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proceedings of the National Academy of Sciences of the United States of America 101, 8551–8556 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H. & Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. The Journal of biological chemistry 275, 6252–6258 (2000). [DOI] [PubMed] [Google Scholar]
- Golebiowski F. et al. System-wide changes to SUMO modifications in response to heat shock. Science signaling 2, ra24, doi: 10.1126/scisignal.2000282 (2009). [DOI] [PubMed] [Google Scholar]
- Tammsalu T. et al. Proteome-wide identification of SUMO modification sites by mass spectrometry. Nature protocols 10, 1374–1388, doi: 10.1038/nprot.2015.095 (2015). [DOI] [PubMed] [Google Scholar]
- Munsie L. N., Desmond C. R. & Truant R. Cofilin nuclear-cytoplasmic shuttling affects cofilin-actin rod formation during stress. Journal of cell science 125, 3977–3988, doi: 10.1242/jcs.097667 (2012). [DOI] [PubMed] [Google Scholar]
- Wang J., Wang S. & Li S. Sumoylation modulates 20-hydroxyecdysone signaling by maintaining USP protein levels in Drosophila. Insect Biochem Mol Biol 54, 80–88, doi: 10.1016/j.ibmb.2014.09.003 (2014). [DOI] [PubMed] [Google Scholar]
- Monribot-Villanueva J. et al. TnaA, an SP-RING protein, interacts with Osa, a subunit of the chromatin remodeling complex BRAHMA and with the SUMOylation pathway in Drosophila melanogaster. PloS one 8, e62251, doi: 10.1371/journal.pone.0062251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Vatsyayan J., Gao C., Bakkenist C. J. & Hu J. HDAC2 promotes eIF4E sumoylation and activates mRNA translation gene specifically. The Journal of biological chemistry 285, 18139–18143, doi: 10.1074/jbc.C110.131599 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamillo A. et al. Scavenger receptors mediate the role of SUMO and Ftz-f1 in Drosophila steroidogenesis. PLoS genetics 9, e1003473, doi: 10.1371/journal.pgen.1003473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamillo A. et al. Smt3 is required for Drosophila melanogaster metamorphosis. Development 135, 1659–1668, doi: 10.1242/dev.020685 (2008). [DOI] [PubMed] [Google Scholar]
- Kanakousaki K. & Gibson M. C. A differential requirement for SUMOylation in proliferating and non-proliferating cells during Drosophila development. Development 139, 2751–2762, doi: 10.1242/dev.082974 (2012). [DOI] [PubMed] [Google Scholar]
- Netzer C., Bohlander S. K., Rieger L., Muller S. & Kohlhase J. Interaction of the developmental regulator SALL1 with UBE2I and SUMO-1. Biochemical and biophysical research communications 296, 870–876 (2002). [DOI] [PubMed] [Google Scholar]
- Hjerpe R., Thomas Y. & Kurz T. NEDD8 overexpression results in neddylation of ubiquitin substrates by the ubiquitin pathway. Journal of molecular biology 421, 27–29, doi: 10.1016/j.jmb.2012.05.013 (2012). [DOI] [PubMed] [Google Scholar]
- Komatsu M. et al. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J 23, 1977–1986, doi: 10.1038/sj.emboj.7600205 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire K. et al. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PloS one 6, e18517, doi: 10.1371/journal.pone.0018517 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y. et al. UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development. PLoS genetics 11, e1005643, doi: 10.1371/journal.pgen.1005643 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havugimana P. C. et al. A census of human soluble protein complexes. Cell 150, 1068–1081, doi: 10.1016/j.cell.2012.08.011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland T. et al. A proteome-scale map of the human interactome network. Cell 159, 1212–1226, doi: 10.1016/j.cell.2014.10.050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant H. E. et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J 28, 2601–2615, doi: 10.1038/emboj.2009.206 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M. et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123, 1213–1226, doi: 10.1016/j.cell.2005.09.038 (2005). [DOI] [PubMed] [Google Scholar]
- Jentsch S. & Psakhye I. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu Rev Genet 47, 167–186, doi: 10.1146/annurev-genet-111212-133453 (2013). [DOI] [PubMed] [Google Scholar]
- Chen C., Itakura E., Weber K. P., Hegde R. S. & de Bono M. An ER complex of ODR-4 and ODR-8/Ufm1 specific protease 2 promotes GPCR maturation by a Ufm1-independent mechanism. PLoS genetics 10, e1004082, doi: 10.1371/journal.pgen.1004082 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahian F., Sepehrizadeh Z., Moghimi B. & Mirzaei S. A. Human cytochrome b5 reductase: structure, function, and potential applications. Crit Rev Biotechnol 34, 134–143, doi: 10.3109/07388551.2012.732031 (2014). [DOI] [PubMed] [Google Scholar]
- Antunica-Noguerol M. et al. The activity of the glucocorticoid receptor is regulated by SUMO conjugation to FKBP51. Cell death and differentiation 23, 1579–1591, doi: 10.1038/cdd.2016.44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig T. J., Anderson D., Evans A. J., Girach F. & Henley J. M. SUMOylation of Syntaxin1A regulates presynaptic endocytosis. Scientific reports 5, 17669, doi: 10.1038/srep17669 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T. et al. SUMOylated NKAP is essential for chromosome alignment by anchoring CENP-E to kinetochores. Nature communications 7, 12969, doi: 10.1038/ncomms12969 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. H., Liu S. Y. & Lee E. H. SUMO modification of Akt regulates global SUMOylation and substrate SUMOylation specificity through Akt phosphorylation of Ubc9 and SUMO1. Oncogene 35, 595–607, doi: 10.1038/onc.2015.115 (2016). [DOI] [PubMed] [Google Scholar]
- Fan J. B. et al. Identification and characterization of a novel ISG15-ubiquitin mixed chain and its role in regulating protein homeostasis. Scientific reports 5, 12704, doi: 10.1038/srep12704 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Hsiang T. Y., Kuo R. L. & Krug R. M. ISG15 conjugation system targets the viral NS1 protein in influenza A virus-infected cells. Proceedings of the National Academy of Sciences of the United States of America 107, 2253–2258, doi: 10.1073/pnas.0909144107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. F., Wee S., Gunaratne J., Lane D. P. & Bulavin D. V. Isg15 controls p53 stability and functions. Cell Cycle 13, 2200–2210, doi: 10.4161/cc.29209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. J. et al. Elevated global SUMOylation in Ubc9 transgenic mice protects their brains against focal cerebral ischemic damage. PloS one 6, e25852, doi: 10.1371/journal.pone.0025852 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Molecular cell 44, 325–340, doi: 10.1016/j.molcel.2011.08.025 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoliatte F. et al. Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nature communications 5, 5409, doi: 10.1038/ncomms6409 (2014). [DOI] [PubMed] [Google Scholar]
- Galisson F. et al. A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Molecular & cellular proteomics: MCP 10, M110 004796, doi: 10.1074/mcp.M110.004796 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez J. et al. Sumoylation modulates the activity of Spalt-like proteins during wing development in Drosophila. The Journal of biological chemistry 285, 25841–25849, doi: 10.1074/jbc.M110.124024 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki J. et al. Expression vector system based on the chicken beta-actin promoter directs efficient production of interleukin-5. Gene 79, 269–277 (1989). [DOI] [PubMed] [Google Scholar]
- Mirth C., Truman J. W. & Riddiford L. M. The role of the prothoracic gland in determining critical weight for metamorphosis in Drosophila melanogaster. Current biology: CB 15, 1796–1807 (2005). [DOI] [PubMed] [Google Scholar]
- Lundby A. & Olsen J. V. GeLCMS for in-depth protein characterization and advanced analysis of proteomes. Methods Mol Biol 753, 143–155, doi: 10.1007/978-1-61779-148-2_10 (2011). [DOI] [PubMed] [Google Scholar]
- Kelstrup C. D., Young C., Lavallee R., Nielsen M. L. & Olsen J. V. Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. Journal of proteome research 11, 3487–3497, doi: 10.1021/pr3000249 (2012). [DOI] [PubMed] [Google Scholar]
- Breuer K. et al. InnateDB: systems biology of innate immunity and beyond–recent updates and continuing curation. Nucleic acids research 41, D1228–1233, doi: 10.1093/nar/gks1147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J., Arak T. & Vilo J. g:Profiler–a web server for functional interpretation of gene lists (2011 update). Nucleic acids research 39, W307–315, doi: 10.1093/nar/gkr378 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micallef L. & Rodgers P. eulerAPE: drawing area-proportional 3-Venn diagrams using ellipses. PloS one 9, e101717, doi: 10.1371/journal.pone.0101717 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic acids research 43, D447–452, doi: 10.1093/nar/gku1003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader G. D. & Hogue C. W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4, 2 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. H. et al. clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics 12, 436, doi: 10.1186/1471-2105-12-436 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson A. M. et al. DNA binding and heteromerization of the Drosophila transcription factor chorion factor 1/ultraspiracle. Proceedings of the National Academy of Sciences of the United States of America 89, 11503–11507 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates J. et al. Enabled and Capping protein play important roles in shaping cell behavior during Drosophila oogenesis. Dev Biol 333, 90–107, doi: 10.1016/j.ydbio.2009.06.030 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance P. E., Miron M., Raught B., Sonenberg N. & Lasko P. Phosphorylation of eukaryotic translation initiation factor 4E is critical for growth. Molecular and cellular biology 22, 1656–1663 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M., Bhaskar V., Fernandez J. & Courey A. J. Drosophila Ulp1, a nuclear pore-associated SUMO protease, prevents accumulation of cytoplasmic SUMO conjugates. The Journal of biological chemistry 279, 43805–43814 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.