Abstract
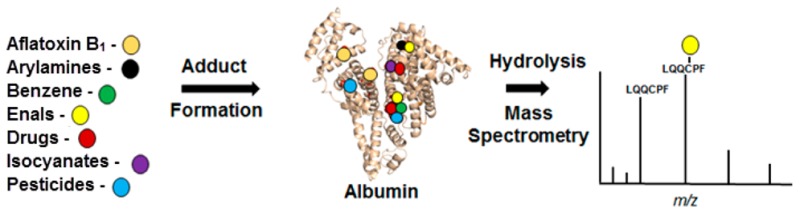
Serum albumin (Alb) is the most abundant protein in blood plasma. Alb reacts with many carcinogens and/or their electrophilic metabolites. Studies conducted over 20 years ago showed that Alb forms adducts with the human carcinogens aflatoxin B1 and benzene, which were successfully used as biomarkers in molecular epidemiology studies designed to address the role of these chemicals in cancer risk. Alb forms adducts with many therapeutic drugs or their reactive metabolites such as β-lactam antibiotics, acetylsalicylic acid, acetaminophen, nonsteroidal anti-inflammatory drugs, chemotherapeutic agents, and antiretroviral therapy drugs. The identification and characterization of the adduct structures formed with Alb have served to understand the generation of reactive metabolites and to predict idiosyncratic drug reactions and toxicities. The reaction of candidate drugs with Alb is now exploited as part of the battery of screening tools to assess the potential toxicities of drugs. The use of gas chromatography-mass spectrometry, liquid chromatography, or liquid chromatography-mass spectrometry (LC-MS) enabled the identification and quantification of multiple types of Alb xenobiotic adducts in animals and humans during the past three decades. In this perspective, we highlight the history of Alb as a target protein for adduction to environmental and dietary genotoxicants, pesticides, and herbicides, common classes of medicinal drugs, and endogenous electrophiles, and the emerging analytical mass spectrometry technologies to identify Alb-toxicant adducts in humans.
1. Albumin Synthesis, Structure, and Function
Albumin (Alb) is the most abundant protein in human serum.1 Alb is synthesized in hepatocytes, first as a prepro-albumin, which contains 609 amino acids. The N-terminal signal peptide (positions 1–18) is removed in the lumen of the endoplasmic reticulum to produce pro-albumin, which is subsequently cleaved (positions 19–24) in the Golgi vesicles to produce the mature Alb, a single polypeptide chain of 585 residues (positions 25–609) with a molecular weight of 66 438 Da.1,2 In healthy adults, Alb synthesis occurs almost exclusively in hepatocytes (∼0.2 g/kg body weight per day) and accounts for 10% of total liver protein synthesis.3 The mean concentration of Alb in the plasma of adults is approximately 43 mg/mL (∼0.6 mM), accounting for 50%–60% of total plasma proteins in healthy adults.1 The catabolism of Alb takes place primarily in muscle, skin, and the liver, resulting in a serum half-life of approximately 19–25 days.1
Alb (UniProtKB - P02768) comprises 62 Glu, 62 Ala, 61 Leu, 59 Lys, 41 Val, 36 Asp, 35 Cys, 31 Phe, 28 Thr, 24 Ser, 24 Arg, 24 Pro, 20 Gln, 18 Tyr, 17 Asn, 16 His, 12 Gly, 6 Met, 8 Ile, and a single Trp residue.2 The acidic amino acid residues exceed the basic ones, resulting in a high net negative net charge at physiological pH (62 Glu + 36 Asp versus 59 Lys + 24 Arg) which facilitates the solubility of Alb. Thirty-four of the Cys residues form 17 intramolecular disulfide bridges; the only free Cys residue is situated at position 34. The reduced form of Alb (Alb-SH) is also known as human mercaptalbumin. The disulfide bridges significantly contribute to the stability of Alb and its long biological lifetime.1 This pattern of disulfide bridges occurs for Alb in all vertebrates.1,2 The Cys34 residue of Alb resides in a microenvironment close to three ionizable residues, Asp38, His39, and Tyr84, which affect the ionization state of Cys34 resulting in an unusually low pKa value of ∼6.5 compared to pKa values of about 8.0–8.5 for Cys residues in many other proteins or peptides.4,5 As a result, the Cys34 of Alb is present predominantly as the thiolate anion at physiological pH. The low pKa for Alb-Cys34 explains its high reactivity with many oxidants and electrophiles.6 Approximately 70%–80% of total plasma Alb in adults contains the reduced sulfhydryl group of Cys34 with the remainder present as reversible mixed disulfides with low-molecular-weight thiols, such as cysteine, homocysteine, cysteinylglycine, or glutathione.7−11 The Cys34 of Alb is a potent scavenger of free radicals, and the sulfenic acid of Alb-Cys34-OH is thought to be a central intermediate in the formation of mixed disulfide Alb species in vivo.12 The S-thiolated oxidized form of Alb (Alb-SSR) is called nonmercaptalbumin. The Cys34 residue of Alb covalently binds a wide number of endogenous ligands, metal ions, and nitric oxide.13−15 Because of its high abundance, metal-binding capacity, and redox properties, the Cys34 of Alb is the predominant antioxidant in plasma.1 The Met87, Met123, Met298, Met329, Met446, and Met548 residues of Alb also contribute to antioxidant activity.13
The secondary structure of Alb is dominated by α-helices (68%), without any β-sheets. Alb contains three homologous domains designated as I (amino acid residues, 1–195), II (196–383), and III (384–585) (Figure 1). Each domain is further divided into subdomains A and B, which are composed of six and four α-helices, respectively. These subdomains possess common structural motifs, which are comparable in the amino acid sequence and in the secondary and tertiary structure.13 The tertiary structure of Alb is arranged in a globular heart-shaped conformation (Figure 1) and largely maintained even in the presence of a wide variety of ligands.16,17
Figure 1.
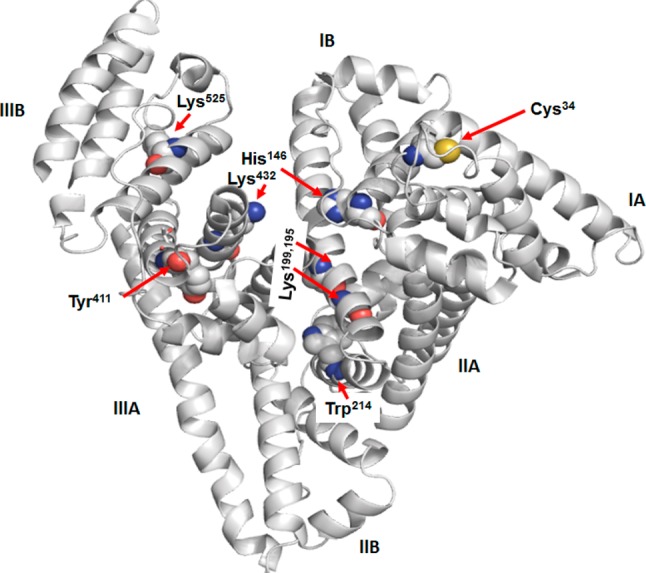
Three-dimensional structure of Alb (1A06.pdb) with the subdomains (I–III) and depiction of several reactive nucleophilic amino acid residues, which are displayed as spheres. The structure was obtained with PyMOL software.
Alb is critical for maintaining colloidal osmotic pressure and reversibly binds hormones, nonesterified long chain fatty acids, cholesterol, billirubin, hemin, and many xenobiotic compounds, including therapeutic drugs.1,18 Many commonly used drugs contain acidic or basic functional groups, such as warfarin, diazepam, and ibuprofen. These drugs usually bind noncovalently at one of two primary binding sites designated as I and II, which are located in subdomains IIA and IIIA, respectively.16,19 The noncovalent binding of endogenous biochemicals and many drugs is driven by their hydrophobic, hydrogen bonding, π–π, and ionic interactions with the different microenvironments of Alb. Site I is often referred to as the warfarin site, and typically large, heterocyclic, and negatively charged drugs bind to this site, whereas small, aromatic, carboxylic acid drugs such as diflunisal and ibuprofen, and basic drugs, including diazepam that exist mainly in the un-ionized form at neutral pH bind with high affinity to site II.18 The ability of drugs to reversibly bind to Alb improves their solubility, and as result, Alb plays a major role in their transport throughout the human body. Some of the drug binding properties of Alb can also influence the sites of covalent adduction of toxicants.20 A number of excellent reviews have been published on Alb from structure to function and drug binding properties of Alb.1−3,13,18,21
2. Introduction to Human Biomonitoring and History of Protein Adducts of Toxicants
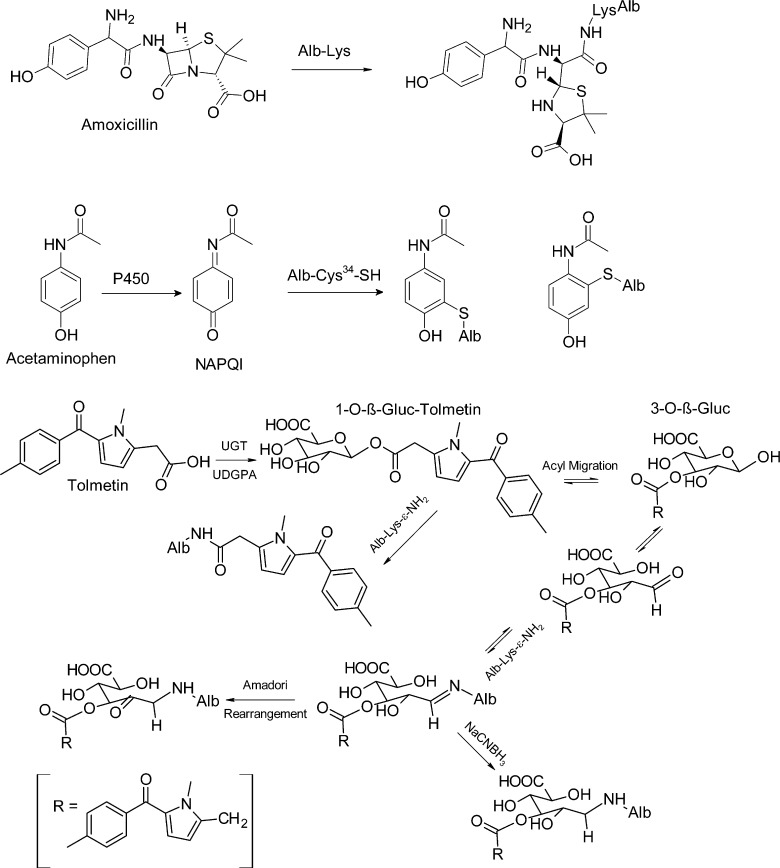
Many xenobiotics or their metabolites can react with proteins and DNA to form covalent adducts. Some protein chemical adduction products can lead to allergenic and other toxic effects,22,23 while DNA adduct formation can lead to mutations and the initiation of cancer.24 The role of DNA adducts in chemical carcinogenesis was established in the 1960s and 1970s.25−28 The covalent binding of drugs to proteins as a mechanism of toxicity emerged in the early 1970s with a series of studies conducted with acetaminophen, using covalent binding of the 14C-radiolabeled molecule to liver proteins.29−31 These studies showed that reactive electrophilic intermediates, largely products of cytochrome P450 metabolism,32,33 damage proteins and can lead to toxicity.
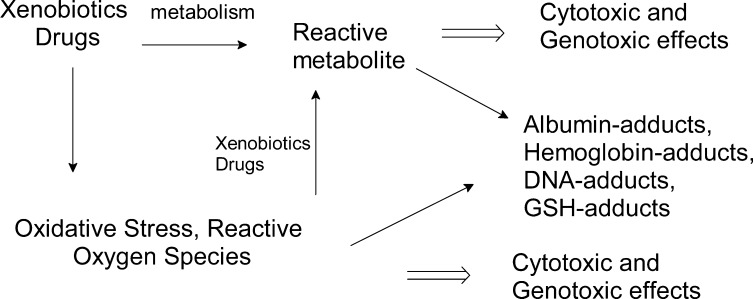
During the 1970s–1980s, investigations rapidly advanced on the characterization of reactive metabolites of carcinogens and other xenobiotics with DNA, blood proteins, and low molecular weight compounds such as glutathione in biological systems.34−39 The development of analytical methods to measure protein toxicant/carcinogen adducts in humans advanced by the late 1970s and 1980s, when laboratories employed adducts to hemoglobin (Hb) and Alb to assess carcinogen exposure.37,40−43 Since these hallmark studies, the mechanisms of enzymatic bioactivation of chemicals and the analytical methods used to measure protein toxicant adduct formation have evolved and are routinely applied to the preclinical safety assessment of drugs. Alb is used as a target protein to prescreen potentially toxic drug-candidates.23,44,45 The enzymatic formation of reactive metabolites is a major topic of research in pharmacology and toxicology.46,47 The formation of reactive metabolites from different functional groups of xenobiotics that react with proteins and other deleterious effects are shown in Figure 2.
Figure 2.
Alb adducts as biomarkers of exposure to reactive metabolites of drugs and toxicants.
The paradigm of biomonitoring of toxicants and their ensuing biological effects in risk assessment of chemicals has been the roadmap of mechanistic studies for two generations of researchers. The chemical and biological end points include (1) external exposure: air, skin, food, and water. (2) Internal exposure: e.g., parent compound or metabolite thereof in urine or blood. (3) Biologically effective dose: e.g., protein- and DNA-adducts. (4) Early biological effects: e.g., micronuclei, p53 mutations. (5) Late biological effects: e.g., altered cell structure and/or function, and (6) tumor or other diseases.48 In chemical carcinogenesis, the detection of DNA-adducts is of primary importance in the identification of chemical exposures that may contribute to the etiology of cancer.49
The measurement of DNA adducts in target organs of potential carcinogens is the most direct method to assess the genotoxic potential of a chemical. However, such studies are usually not feasible in humans because of the lack of accessible tissues. Therefore, researchers have sought readily available biospecimens and alternative biomarkers that may serve as surrogate measures of DNA damage.50,51 In 1974, Lars Ehrenberg demonstrated that there was a correlation between DNA adducts formed in target organs of mice given ethylene oxide and Hb adducts in blood.50 Subsequently, the N-terminal valine of Hb was shown to react with ethylene oxide and then to a number of other alkylating agents.52 The reactivity of Hb with aromatic amines and production of methemoglobin by genotoxic arylhydroxylamines has also been well studied and reviewed.53,54 Pereira et al. showed a linear relationship between Hb-adducts and DNA-adducts for 2-acetylaminofluorene in mice and rats.55 Hb adduct formation for many aromatic amines occurs as a sulfinamide linkage formed by reaction of the arylnitroso intermediates with the β-Cys93 chain of Hb.56−60 These adducts undergo hydrolysis with base or acid in vitro, and the liberated amines can be assayed by GC methods.61 The determination of arylamine-Hb adducts is a well established biomonitoring method and performed in many laboratories worldwide.42,52,62,64 In 1961, the toxic mold Aspergillus flavus containing aflatoxin B1 (AFB) was discovered to be responsible for the outbreak of deaths of turkeys consuming contaminated meal in Britain.63,65 The structure of AFB was elucidated by George Büchi,66 and its metabolism and DNA-adduct and protein adduct formation were largely carried out at the Massachusetts Institute of Technology in Cambridge (MA, USA) by Gerald Wogan and his colleagues.63,67,68
Biomonitoring of protein-adducts of carcinogens is an alternative and sometimes superior approach to the measurement of DNA adducts for assessing exposure.52,57,69 Thus far, adducts of toxicants formed with Hb have been more widely used in human biomonitoring studies than Alb adducts;6,52 however, there are advantages in employing Alb as a target protein for certain classes of toxicants. The hepatocyte is the major site where Alb is synthesized2 and also a major cell-type where many electrophilic metabolites of toxicants are formed.70 Consequently, the formation of Alb adducts does not require the reactive electrophiles to be transported across the cell membrane or to reach the blood compartment (where Hb is located). Moreover, some short-lived toxicants may undergo solvolysis before they can react with Hb, which is synthesized in reticulocytes and located in erythrocytes, and thus reducing chances of adduct formation. Indeed, the levels of Alb adducts formed with the liver carcinogen AFB and carcinogenic heterocyclic aromatic amines (HAA) are far greater than the levels of adducts formed with Hb, a fact making Alb adducts superior biomarkers, at least for these carcinogens.71−73 Many studies have characterized Alb adducts formed in vitro with environmental, dietary, and tobacco genotoxicants, lipid peroxide products, drug medications, and pesticides, among others.74,75 However, reports on other Alb adduct studies in humans are relatively few.6
Protein adducts of toxicants represent a marker of the biologically effective dose. However, regulatory agencies often conduct risk assessment on the toxic effects of chemicals based on toxicity (or carcinogenicity) data from experimental laboratory animal studies and extrapolate the dose at the level of minimal risk to the external dose of the chemicals from the air, water, or food supply, or through dermal adsorption.76−78 The use of Alb adducts as reliable biomarkers of exposure assessment requires that the adducts are stable and follow the predictable kinetics of removal, and ideally, the half-life of Alb adduct should be the same as unmodified Alb. The use of Alb adducts (or Hb adducts) as biomarkers in cancer risk assessment requires knowledge on the relationship between Alb and DNA adduct formation, which occurs in a distant target organ.
3. Generation of Reactive Metabolites and Identification of Protein Adducts in Vitro
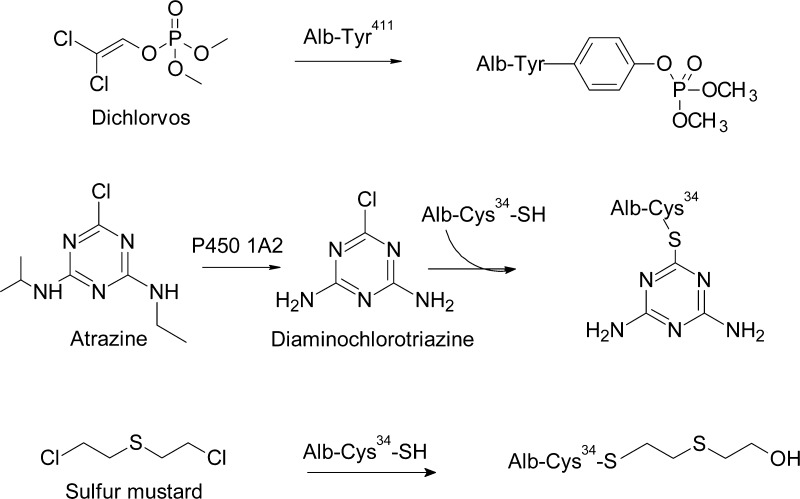
Hb with a life span of ∼120 days and Alb with a half-life of 19–25 days are the major proteins in blood.1,52 Both proteins react with a wide range of genotoxicants, xenobiotics, and endogenous electrophiles, which have been characterized by mass spectrometry.52,69,74,75 The structures of reactive short-lived intermediates of xenobiotic chemicals have been deduced from the structural characterization of their stable adducts formed with Hb and Alb in vitro.57,69,74,75,79 The formation of covalent protein adducts is thought to be driven not only by the abundance, accessibility, and relative reactivities of the different amino acid residues but also by the tertiary structure of the proteins and noncovalent binding of the metabolite with different receptor regions of proteins, in which the initial noncovalent positioning can direct the sites of toxicant adduct formation.80,81 Several nucleophilic amino acid residues of Hb (N-terminal Val) or Alb (Cys34, His146, and Tyr411) are particularly reactive toward certain classes of chemicals, and some adducts have served as biomarkers of toxicants in molecular epidemiology studies.6,52,57
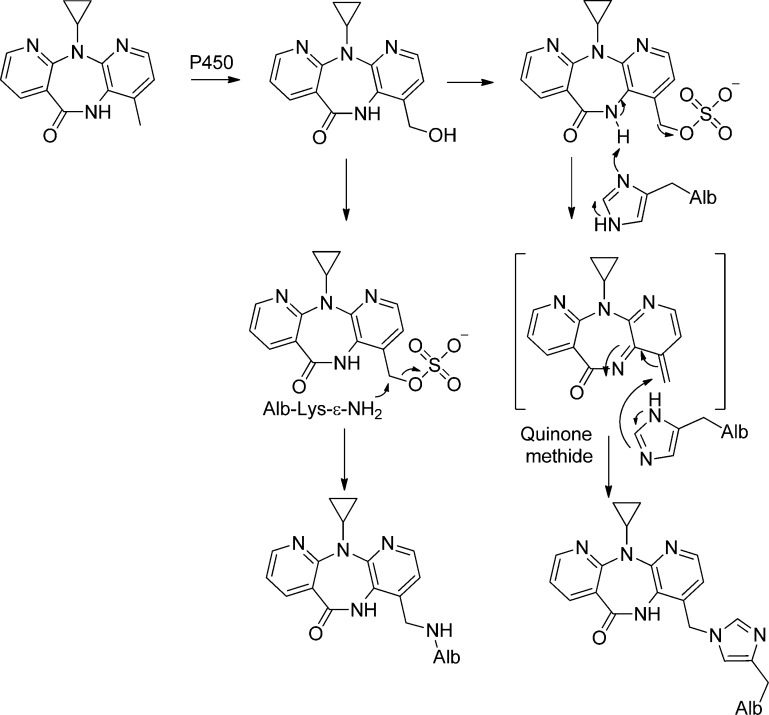
As a common first approach, Alb adducts can be characterized ex vivo by reacting plasma or purified Alb with the biologically reactive intermediate, for example, at a ratio of chemical to protein of 1:1, or 10:1 (see Table S1). Some chemicals, for examples, isothiocyanates, isocyanates, β-lactam antibiotics, and enals do not require metabolic transformation and directly react to form covalent adducts with Alb, whereas many drugs or procarcinogens require metabolism to form reactive electrophiles, which bind to Alb. Microsomes fortified with NADH produce a variety of reactive intermediates by cytochrome P450s such as short-lived alkylating agents, aldehydes, epoxides, quinones, arylhydroxlamines, arylnitroso compounds, and thiophene S-oxides, among others, which can be trapped by Alb.23,32,33,82 Microsomal mediated UDP-glucuronosyltransferases catalyze the bioactivation of NSAIDS by the formation of acyl glucuronide (Gluc) drug conjugates, which react with Lys residues of Alb.83−85 A number of reactive intermediates also can be prepared chemically.86,87
Commercial albumin preparations often contain very high levels of mixed disulfides at Cys34, and adduct formation can be underestimated at this site unless Alb is pretreated with β-mercaptoethanol,88 which selectively reduces Cys34 mixed disulfides without disruption of the internal Alb Cys disulfide bonds.21,88 The reactivity of commercial Alb with some electrophiles may differ from that of Alb in plasma because the fatty acids and/or other endogenous ligands removed during the processing of commercial Alb can alter the conformation of the protein and affect drug interactions and reactivities.16−18 The reaction of Alb with a high molar excess of toxicants can lead to the formation of adducts which do not normally occur at the lower concentrations of the chemicals present in vivo. Therefore, once the major sites of Alb adduction are elucidated in vitro, kinetic experiments should be performed with limiting amounts of toxicant to identify the most reactive amino acid residues; these are the sites which may be expected to form adducts in vivo.89−91 The exact position of the adduct formation with Alb has not been elucidated in all biomonitoring studies. For example, the molecular epidemiology studies in populations exposed to AFB were successful without knowing the specific binding site(s) of AFB with Lys residues of Alb. The elucidation of the binding sites of xenobiotics to proteins is useful for the elucidation of the potential immunogenic properties of the adduct92 or to develop strategies to measure multiple adducts after isolating a peptide of Alb with a nucleophilic hot-spot such as Cys34.6,93,94
4. Albumin Purification and Hydrolysis Conditions for Adduct Studies
The isolation of Alb is based on its solubility, it is negatively charged at pH > 5, its affinity for binding hydrophobic substances, and its unusual stability has been reviewed.95 Alb is isolated from plasma using fractional protein precipitation with ethanol or ammonium sulfate. The solubility of Alb, like that of most proteins, is minimal at its isoelectric point near pH 5. The solubility increases many fold just one pH unit away from its isoelectric point. The Cohn Method 6 is the major procedure for the commercial fractionation of plasma proteins using ethanol and pH adjustments. The term Fraction V for nearly pure (>96%) Alb is used since Alb precipitates in the fifth step of the procedure.96
4.1. Fractional Precipitation and Affinity Blue Purification Methods
The principal methods used for the purification of Alb in biomonitoring studies are summarized in the following paragraph. Bechtold performed a multistep precipitation method with increasing amounts of ethanol to isolate Alb from plasma.97 The Alb isolated was >95% pure, as measured by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). Other methods used an increasing amount of ammonium sulfate instead of ethanol as the precipitating agent. Rappaport et al.98 added a saturated solution of ammonium sulfate to the plasma until the final concentration of ammonium sulfate at 2.5 M (63% of saturation). The immunoglobulins, which formed a white precipitate, were removed by centrifugation, and the Alb was isolated from small molecular weight proteins by size exclusion chromatography with Sephadex G-25. The recovery of Alb was estimated to be 70%, and the purity of the Alb was estimated to be 92% pure by SDS–PAGE. In later studies by Rappaport and co-workers,99,100 Alb was obtained from plasma by adding a solution of saturated ammonium sulfate dropwise until a final concentration of 50% was achieved. This mixture was then centrifuged to remove the immunoglobulins. The supernatant was dialyzed (12 000–14 000 molecular weight cut off (MWC) membranes), against deionized water and then lyophilized to a constant weight. Another ammonium sulfate precipitation scheme was used by Wild et al.,101 where saturated ammonium sulfate (0.75 mL) was slowly added to plasma or serum (0.5 mL) on ice. The precipitated immunoglobulins were removed by centrifugation. The supernatant was removed to a clean tube and acetic acid added to adjust to pH 5, and the precipitated Alb was collected by centrifugation. A yield of approximately 10 mg (ca. 50%) of Alb was routinely obtained with a purity >95% as judged by SDS–PAGE. Recently, a rapid method was developed to obtain about 0.1 mg of Alb with a 72–75% purity.94 Plasma (5 μL) was added to 60 μL of 50% methanol and incubated at room temperature for 15 min with constant agitation. The samples were centrifuged to remove precipitates and immediately diluted with four volumes of digestion buffer, and digested with trypsin.94
Affinity chromatography with Cibacron blue F3G A, a sulfonated polyaromatic dye, which selectively binds human Alb over other serum proteins, has been used to isolate Alb from plasma.102−104 Plasma samples are diluted with low salt buffer and applied to the affinity dye bound to various supports obtained from commercial vendors. The salt concentration is increased to disrupt the binding and elute the Alb fraction. Sometimes, intermediate salt concentrations have been used to increase the purity of Alb.102,105 In some instances, the fraction containing Alb is dialyzed against water in dialysis tubing (MWC 12–14 kDa) or further purified by ultracentrifugal filtration to remove low molecular weight proteins (10 or 30 kDa MWC).99,106
Young et al.107 compared different methods to purify Alb: precipitation with ethanol according to the procedure of Bechtold97 or by different commercially available albumin purification kits. The authors achieved the highest purity (88%) with Hi-Trap Blue columns (Cibacron Blue is the affinity dye).107 In other studies using affinity chromatography based on the dye Cibacron blue, higher purities were achieved when the samples were dialyzed with membranes of 12–30 kDa MWC.99,106 Therefore, smaller proteins, which decrease the purity of Alb (see SDS–PAGE107), were eliminated. Careful application of these affinity blue columns generally yields Alb with a purity >95%.20 A 1 mL HiTrap blue column can retain more than 20 mg of Alb.107 The methods used to elute proteins from the HiTrap blue columns may not be quantitative.108 Therefore, with multiple use, the affinity dye may gradually lose its capacity to retain Alb, resulting in lower recoveries of Alb. In summary, the highest throughput to purify Alb is achieved by the fractional precipitation methods using ammonium sulfate without final precipitation of Alb;98,99,109 however, the highest levels of purity are achieved with Cibacron blue dye in conjunction with a low MWC filter.
4.2. Enzymatic Digestion of Albumin
4.2.1. Pronase
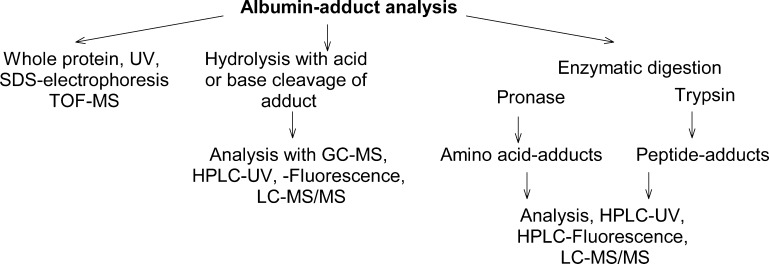
Prior to the advent of electrospray ionization mass spectrometry (ESI-MS) and its coupling to HPLC,110,111 many Alb adducts were assayed after acid or base hydrolysis or by proteolysis with pronase, and the adducted amino acids or the carcinogen hydrolysis products were detected by GC-MS, HPLC with UV, fluorescence, or radioactive detection (Figure 3).20,71,88,109,112−114 LC-MS methods69,115 have supplanted many of these earlier analytical methods. Pronase (synonym: actinase E and pronase E), a mixture of a broad spectrum of proteases from Streptomyces griseus, has often been used to digest Alb. Mono-, di-, tri-, or tetra-peptides containing adducts were recovered, depending upon the structure of the adduct and conditions of digestion.20,88,116−118 Different pronase preparations are commercially available.
Figure 3.
Approaches to measure Alb adducts.
Delatour et al.119 compared different approaches to digest bovine Alb using 6 M HCl at 110 °C, pronase digest for 24 or 48 h (Alb/pronase = 30:1), or a cocktail of enzymes (pepsin, pronase, aminopeptidase, and prolidase). The HCl-based hydrolysis yielded a digestion efficiency of 95%, while only 25 and 75% efficiencies were achieved with pronase and the cocktail of enzymes, respectively, when the digestion efficiency was measured by amino acid analysis. These data on pronase digestion are not comparable to many other studies, which used much larger amounts of Pronase in relation to Alb. Sabbioni et al.120 performed experiments with different ratios of Alb/pronase 10:1, 3:1, and 1:1 at pH 7.4. The highest recovery of the main adduct of AFB with Alb was obtained after the enzymatic hydrolysis using an Alb/pronase ratio of 3:1. For the analysis of isocyanate adducts formed with Alb, enzymatic hydrolyses were performed at pH 8.9 or at pH 7.4 with a Alb/pronase ratio of 3:1. The recovery of adducts obtained at pH 8.9 were about 2-fold higher than that at pH 7.4.121 Young et al.107 tested if the method of Alb purification had an influence on the amount of adducts per mg of Alb determined by LC-MS/MS. Plasma was incubated with sulfur mustard. Alb was isolated using the different methods described in Young et al.107 Large differences were observed in the levels of the sulfur mustard adduct formed with Cys34 of Alb: S-[2-[(hydroxyethyl)thio]ethyl]Cys-Pro-Phe (HETE-CPF). Therefore, the presence of peptide fragments from other plasma proteins (PP) than Alb appeared to have caused an ion suppression matrix and interfered with the quantification of adducted peptides originating from Alb.
4.2.2. Albumin Denaturation
Alb contains 17 intrachain disulfide bonds, which makes the protein resistant to denaturation.2,17 For digestion with trypsin or other serine proteases, Alb is commonly denatured by heat treatment at 50–60 °C, and internal disulfide bonds are reduced with dithiothreitol or tris(2-carboxyethyl)phosphine in the presence of denaturants, followed by alkylation of the newly formed sulfhydryl groups with iodoacetamide, to expose as many sites as possible prior to proteolytic digestion.122,123 The proteolytic digestion of Alb produces polypeptides of various lengths according to the specificity of the enzymes employed. Missed cleavages of peptides can occur during the digestion due to the solvent buffer conditions, nonspecific chemical or enzymatic cleavage of proteins, post-translational modifications of proteins, or by the introduction of bulky toxicants, which can affect the efficiency of enzyme digestion.122,123 These conditions of protein denaturation also may destroy thermal labile adducts. Depending upon the location of the adduct and its linkage within Alb, the denaturation of Alb may not be required for quantitative recovery of the adduct by proteolysis. For example, the sulfonamide adduct formed by reaction of Cys34 of Alb with the N-oxidized metabolite of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), a carcinogen formed in cooked meat,124 was quantitatively recovered from Alb by digestion with a mixture of trypsin and chymotrypsin without prior reduction of the disulfide bonds.125 Very recently, the digestion of Alb with trypsin using pressure cycling was shown to efficiently recover the T3 peptide A21LVLIAFAQYLLVLIAFAQYLQQCPFEDHVK40 without prior denaturation and reduction of internal disulfide bonds.94
4.2.3. Trypsin and Other Proteases
The proteolysis of proteins, including Alb adducts in MS-based proteomics, is most commonly done with trypsin as an alternative to digestion with protease mixtures. The selective digestion of Alb with single proteases can permit the identification of the specific sites of toxicant adduction by performing collision induced dissociation (CID) of the peptides to form the b-ion, y-ion, and a-ion series.126,127 The identification of common sites or “hot-spots” (see Supporting Information, Table S1) of adduct formation can be used to establish a database of nucleophilic sites of Alb for specific classes of toxicants and serve as a basis to develop strategies for adductomics. Trypsin is a serine protease and cleaves peptides on the C-terminal side of Lys and Arg residues except for when a proline residue is on the carboxyl side of the cleavage site in which case the cleavage will not occur. Trypsin digestion yields an optimal average peptide length of ∼14 amino acids, with a well-defined positive charge at the N-terminus, and the peptides often exist as doubly [M+2H]2+ or triply charged species [M+3H]3+ which favors the formation of y-ions by ESI/MS.128,129 Other serine proteases including Lys-C, which retains activity in strong denaturants, (cleaves Lys following Pro to decrease missed cleavages); chymotrypsin (cleaves C-terminal of Phe, Tyr, Leu, Trp, and Met); Glu-C (cleaves C-terminal of Glu and Asp); and pepsin, an aspartate protease (cleaves C-terminal of Tyr, Phe, and Trp) have also been employed. The selection of proteases depends upon the site of toxicant adduction, the specificity of the protease, and the conditions including the buffers and denaturation reagents required for digestion.129 Different proteases yield different peptide sequences, some of which may prove to be more sensitive toward ESI than tryptic peptides.130 The responses of signals of peptides under ESI can vary by up to 50-fold.125,131,132 In some instances, a combination of trypsin plus chymotrypsin may be used for proteolysis since some modified Lys residues are not recognized by trypsin, resulting in a missed-cleavage and an increase in the molecular weight and the length of the adducted peptides so that the peptides are not suitable for LC–MS analysis.81
5. Analytical MS-Based Instruments and Scanning Techniques to Characterize and Quantitate Albumin Adducts
Different types of MS instrumentation have been employed to study protein adducts. The most commonly used MS instruments include triple quadrupole, ion trap, hybrid triple quadrupole/linear ion trap instrument MS (QTRAP MS), QTOF MS, and Orbitrap. A variety of these instruments are available commercially from vendors such as Thermo Fisher Scientific, Agilent, Waters, Bruker, Sciex, and Shimadzu. The capabilities of MS instrumentation, including strengths and weaknesses, have been discussed in other reviews133−136 and only briefly highlighted here. Fourier transform ion cyclotron instruments, which can reach resolution in excess of 106, is commonly used in top-down proteomics of high molecular weight proteins, but reports of its use in protein adduct studies are limited because of the great expense of the instruments and is not discussed here.
5.1. Triple Quadrupole Mass Spectrometry (TQ MS)
The titled instruments are commonly used for targeted tandem mass spectrometry (MS/MS) both for small molecules and proteomics.137,138 Triple quadrupole mass spectrometers (TQ MS) contain two mass filtering quadrupoles (Q1 and Q3) and a third quadrupole (q2), which serves as a collision cell or ion guide (q2) and is positioned between the two mass filters. The collision cell is filled with an inert gas (typically, nitrogen or argon). The ions in Q1 are accelerated into the collision cell where they collide with the neutral gas molecules to produce the fragment ions, or in the case of peptides, the b-ion, y-ion, and a-ion type fragment ion series using nomenclature described by Roepstorff and Fohlman.126,127 This technique is termed CID and is the most commonly used mechanism to fragment peptides or proteins in the gas phase. In targeted MS/MS experiments, termed selected reaction monitoring (SRM) mode, Q1 and Q3 are set at fixed specific masses, allowing only a specific a fragment ion from a precursor ion in Q1 to be detected in Q3. This method is extremely sensitive with a short duty cycle, permitting the simultaneous quantitative measurements of numerous analytes or peptides. However, complex protein digest mixtures can contain many peptides with the same or similar precursor mass-to-charge ratio (i.e., ± 1.5 m/z) and result in false positives. Criteria on data reliability and validation of the analytical measurements of peptides by TQ MS have been recommended.139
5.2. Quadrupole Ion Trap (IT) Mass Spectrometry
The titled instruments use a quadrupole electric field to capture charged ions within a large m/z range (e.g., 100–4000). Ion motions are subjected to the quadrupolar field. The IT applies additional electric potential along the axial axis to stop ions exiting the device, achieving a “trapping” effect. Following the trapping period, the accumulated ions can be selectively ejected out of the trap and detected based on their m/z value, allowing for mass analysis. In tandem MS, IT permits ion storage, isolation, fragmentation, and sequential ejection to occur in a time-dependent approach (“tandem in-time”), in contrast to the TQ MS described above where three quadrupoles are connected sequentially to perform a single MS2 scan in a “tandem-in-space” manner. Since the fragmentation in the IT is performed in a time-dependent manner, the process of isolation and excitation can be repeated continuously, resulting in multistage MSn scanning, which provide extensive mass spectral characterization and sequencing of peptides.140,141 The QTRAP platform from Sciex, is an hybrid tandem MS instrument which can function as either a dedicated triple quadrupole MS quantitative measurement or as a highly sensitive linear ion trap mass spectrometer. This MS instrument has been used extensively in small molecule and macromolecule applications,137,142 and has been used to identify a number of Alb adducts formed with drugs.143,144
5.3. Time of Flight Mass Spectrometry
(TOF MS) is a method by which the mass-to-charge (m/z) of a molecule is determined through a time measurement. Ions are accelerated by an electric field of known strength, resulting in ions of the same charge state having the same kinetic energy. The ions enter a 1–2 m long drift tube and travel toward the ion detector. The velocity of the ion and time-of-flight required to reach the detector depends on the m/z and velocity is inversely proportional to the m/z. Thus, ions of higher m/z travel more slowly than the ions of lower mass, and ions are separated in space as a function of their velocities. TOF MS provide full spectra and accurate masses with resolution of greater than 10,000 (resolving power R = mass of the second peak/delta mass of the peaks necessary for separation at mass M). TOF MS are often coupled Matrix-assisted laser desorption/ionization(MALDI) or electrospray ion sources and used for characterization of intact or chemically modified peptides.133 A hybrid instrument consisting of a quadrupole mass filter, a collision cell, and a TOF mass analyzer (QTOF) is commonly used in proteomics, where the precursor ions are selected in the quadrupole and sent to the collision cell for fragmentation. The product ions generated are detected by the TOF MS. Because of their high resolution accurate mass measurements combined with tandem MS capabilities for peptide sequencing, QTOF MS instruments are often used in proteomics.145
5.4. Orbitrap Instruments
This HRAMS from Thermo Scientific has been on the market for over a decade.146 The newer instruments have mass resolution exceeding 100,000 and mass accuracies within 1 ppm. The Orbitrap is widely used in the field of proteomics for both bottom-up and top-down proteomic applications. In its current configuration, packets of ions are injected into the Orbitrap via the quadrupole of the C-trap. In the Orbitrap, ions oscillate around a central spindle-like electrode in the axial dimension. The ion oscillations are detected using image current and are transformed into mass spectra by Fourier transformation.146,147
5.5. Top-down and Bottom-up MS
Both MS approaches have been used to characterize post-translationally or chemically modified proteins (Figure 4).148,149 In top-down proteomics, the intact protein and its covalent modifications are analyzed by high mass accuracy, mass resolving power instruments, such as by Fourier transform ion cyclotron instruments or Orbitrap instruments.150 Top-down proteomics has been commonly used for the study of protein isoforms and their post-translational modifications.150−152 However, the screening of Alb toxicant adducts, by top-down tandem MS approaches, has not been largely used, in part because of the large size of Alb (66.5kDa), which makes MS/MS analyses and full sequence coverage of the adducted peptides challenging. Moreover, the level of modification of Alb by most toxicants is generally low and occurs at 0.1% or less in vivo (mol of toxicant per mol of Alb) and severely restricts the usefulness of top-down proteomics. However, top-down proteomic approaches have been commonly used to determine the reactivity of Alb with certain drugs and toxicants in vitro. The Alb toxicant adduct mixture is directly infused into the MS, and the spectrum of the multiply mass to charge envelope of Alb is deconvoluted to obtain the molecular mass of the modified protein.153 In humans, top-down approaches have been used to characterize Alb adduction products to several endogenous biochemicals, including mixed disulfide adducts formed at the Cys34 residue with cysteine, homocysteine, and glutathione.7−11 The Cys34 is also a major target of Alb for adduction with nitric oxide,9,11 and Cys34 oxidation to form the sulfenic, sulfinic, and sulfonic acids haa been characterized.12,154 In addition, glycated species8 or truncated Alb products have been identified, where the first two amino acids (Asp-Ala) are lost from the N-terminus, and leucine is lost from the C-terminus of Alb.10,11 These chemical modifications of Alb have been detected by direct infusion of the purified Alb solution employing ESI with TQ MS or QTOF MS instruments.7−11 However, proteolytic digestion of the Alb followed by bottom-up proteomics was required to locate the specific sites of adduction. Bottom-up proteomics is by far the most commonly used method to identify and elucidate the sites of covalent modification of the majority of Alb toxicant adducts.
Figure 4.
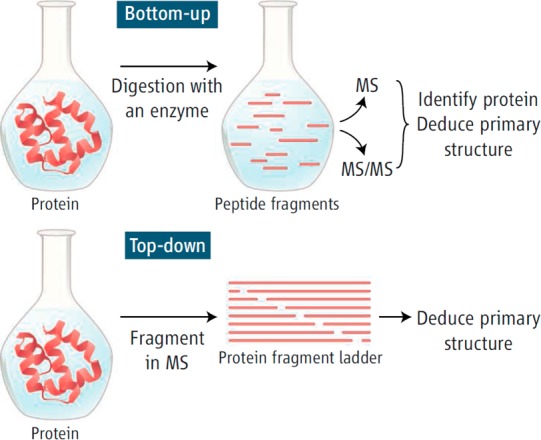
Top-down and bottom-up MS. Reprinted with permission from ref (149). Copyright 2006 AAAS.
5.6. LC-MS Scanning Methods. Targeted and Untargeted Scanning Methods
Targeted SRM, data-dependent acquisition (DDA), or information-dependent acquisition (IDA), and data-independent (DIA) acquisition methods are the principal MS scanning methods used to detect peptides and offer complementary capabilities for adduct analyses.155 SRM methods are routinely conducted with TQ MS instruments. However, because of the complexity of protein digests, even a digest of a single protein such as Alb can contain many isobaric interferences and restrict the usage of the TQ MS for the characterization of peptide-toxicant adducts.132,137,138,156 Specific guidelines have been proposed for the validation of peptide measurements by TQ MS.139
With recent QTOF MS instruments, up to 25 MS/MS spectra can be obtained per duty cycle, and chemically modified peptides can be searched by combination of precursor ions and their characteristic b- and y-product ions. However, for slower scanning ion trap based instruments, DDA or IDA is often used for screening. Generally, the top 5 or 10 ions in abundance eluting from the column into the MS undergo CID and are used to characterize the peptides. These ions can be placed on an exclusion list for a finite period of time, and then the ensuing top 5 or 10 ions are scanned. DDA is biased toward abundant peptides; peptides of low abundance, such as those adducted with toxicants, can be missed in complex biological samples.
In contrast to DDA or IDA scanning methods, DIA is performed by scanning all ions over a selected m/z range, generally a consecutive series of windows of m/z of 5 or 10, which enhances sensitivity and minimizes the space-charging effects that can occur in the ion trap.140,141 The product ion spectra are then acquired on the consecutive series of m/z windows. This generates full product ion spectra of all precursor peptide ions. DIA may be optimally suited for scanning multiple peptide adducts for which there are common peptide sequences and sites of adduct modification, and the peptide adducts that have the same charge state as the unmodified peptides.157 Ideally, the adducts are stable upon CID, and the peptide adducts and unmodified peptides undergo CID to generate, common b-ion and y-ion series from the N- and C-terminus, respectively, up to the site of modification. The full scan precursor ions are then matched to the fragment ions to calculate the mass of the modification, and the modified fragment ions are overlaid with the unmodified fragment ions to verify the mass of the precursor ion calculated. Porter and Bereman157 developed this DIA approach with a high mass-measurement accuracy Orbitrap (<5 ppm) to screen for adducts formed in vitro with Alb at the Cys34. Thirty-six features were observed, which represented oxidation products and mixed disulfide adducts of Cys34. Putative Hb adducts formed at the Cys93 beta chain were also characterized by DIA scanning.157
5.7. Characterization of Peptide Adduct Sequences
The toxicant modified peptides can be screened by predicting the adducts formed from in silico proteolytic digests of Alb and by setting chemical modifications of toxicants as variable modifications on Lys, His, Cys, or other nucleophilic amino acids. Online software such as ProteinProspector (http://prospector.ucsf.edu/prospector/mshome.htm) can facilitate these data mining processes by calculating for the b-ion, y-ion, and a-ion series.
MALDI MS is used to identify differences between untreated and toxicant-treated Alb samples, following proteolysis to rapidly identify chemically modified peptides. However, MALDI MS does not identify the specific site of modification of the peptide, and tandem MS methods are required to fragment peptides, followed by sequencing of the b-ion and y-ion series to locate the precise site of modification.158 The adducted peptides can be mined by manual de novo sequencing or employing search algorithms such as Sequest,159 Mascot160,161 TagRecon, or Myrimatch.162,163 However, some peptides modified with toxicants may not undergo CID in a manner similar to that of the nonadducted peptide, and the fragmentation pattern may not lead to predictable b-ion and y-ion series but produce a prominent cleavage at the linkage of the toxicant moiety. Moreover, some prominent peptide adducts, such as those occurring at Y*411TK,91,130,164 possess too few unmodified b-ions and y-ions and can be missed by search algorithms.91 Therefore, careful manual inspection of the product ion spectra and the search for unique fragment ions attributed to the toxicant can help to identify adducted peptide precursor ions.81,91,165
6. Albumin Adducts Formed with Toxicants and Carcinogens
Over the past decade, a considerable amount of work has been published on Alb adducts formed with toxicants and characterized from in vitro reactions.6,69,74,75,134,166 The chemicals and sites of adduction to Alb modified in vitro are reported in the text and in Table S1 (Supporting Information). The Alb adducts found in vivo are presented in Tables 1 and 2 and in the text. Some of the prevalent exposures to common toxicants and approaches to measure these Alb adducts and their occurrence in humans are described in the following paragraphs.
Table 1. Albumin Adducts Found in Vivo.
| compound | analysis | workup | albumin adduct |
|---|---|---|---|
| Aflatoxin B1 (AFB) | HPLC-fa, ELISA LC-MS/MS | Pronase | The major adduct = AFB-Lys in rats and human (Figure 8).71,101,115,120,203,205,207 Trypsin digest of in vitro modified bovine Alb yielded adducts with Lys455 and Lys548.201 |
| Aflatoxin G1 (AFG) | HPLC-f, ELISA | Pronase | Determination of the Alb adduct with lysine in rats: AFG-Lys.209 |
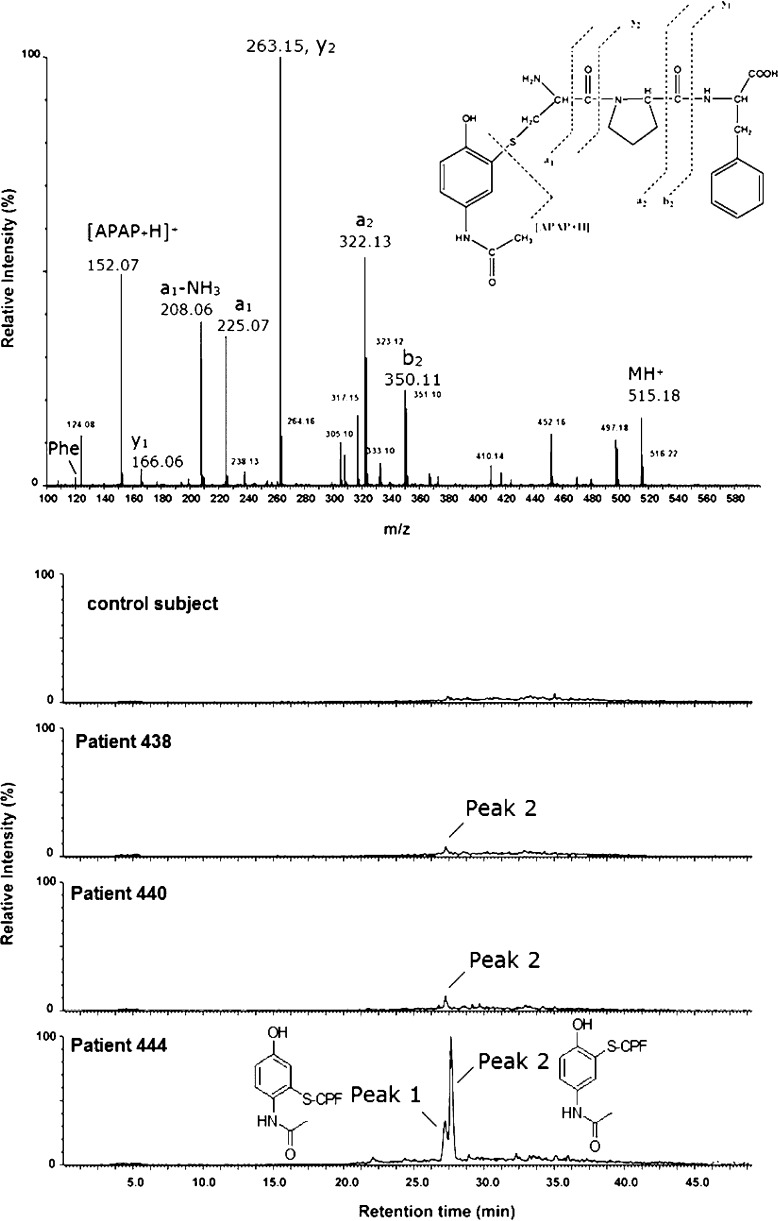
| N-Acetyl-4-aminophenol | LC-MS/MS | Pronase | N-Acetyl-p-benzoquinoneimine-Cys-Pro-Phe in humans (Figure 15,16).116 |
| 4,4′-Methylenediphenyl diisocyanate (MDI) | LC-MS/MS | Pronase | MDI-Lys and AcMDI-Lys in rats,121 and humans (Figure 8).106 |
| 2,4- and 2,6-toluene diisocyanate (TDI) | LC-MS/MS | Pronase | 3A4MP-Lys, 5A2MP-Lys, and 3A2MP-Lys in humans (Figure 8).232 |
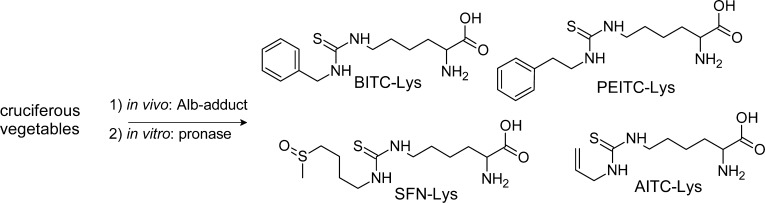
| Isothiocyanates (ITC) released from glucusinolates | LC-MS/MS | Pronase | Phenethyl-ITC-Lys, benzyl-ITC-Lys, allyl-ITC-Lys, sulforaphane-Lys (Figure 13),269,270 and 1-methoxy-3-indolylmethyl glucosinolate Nτ-(1-methoxy-3-indolylmethyl)-His adducts in mice.323 |
| Benzene (B) | GC-MS | Raney-nickel derivatization | Raney-nickel cleaves Cys-bound benzene (B): 1,2-BQ-Cys → catechol,3241,4-BQ-Cys → hydroquinone (Figure 5)324 Hydrolysis and derivatization of benzene oxide (BO)-Cys, 1,2-BQ-Cys, and 1,4-BQ-Cys yields phenyltrifluorothioacetate,99O,O′,S-tris-trifluoroacetyl-catechol,172 and O,O′,S-tris-trifluoroacetyl-hydroquinone, respectively. |
| Pentachlorophenol | GC-MS | Raney-nickel | Tetrachloro-1,4-benzoquinone-Cys → Raney-nickel, adducts in rats.325 |
| Styrene | GC-MS | Raney-nickel | Adducts of styrene-7,8-oxide with Cys, Raney-nickel cleavage yields 1-phenylethanol and 2-phenylethanol in rats and workers.98,185 |
| Naphthalene | GC-MS | Hydrolysis, derivatization | Reaction with methanesulfonic acid and trifluoroacetic acidanhydride yields derivatives of 1-sulfanyl- dihydronaphthalene-2-ol (NPOS1), 2-sulfanyl-1,2-dihydronaphthalene-1-ol (NPOS2), 4-sulfanyl-1,2-naphthalene-1,2-diol (1,2-NPQ-4S), and 2-sulfanyl-1,4-naphthalene-1,4-diol (1,4-NPQ-2S) (Figure S1).179 NPOS1 is the major adduct in rats,179 mice,100 1,2-NPQ, and 1,4NPQ adducts in humans.181 |
| Tetrachloroethene (PER)326 | GC-MS | Hydrolysis, derivatization | Rats exposed to 40 ppm PER for 6 h and 0.35–0.48 pmol N-(dichloroacetyl)-l-(dichloroacetyl)-l-lysine/mg plasma proteins (PP). |
| Sulfur mustard | LC-MS/MS | Pronase, Trypsin | Pronase treatment of Alb, S-[2-[(hydroxyethyl)thio]ethyl];Cys-Pro-Phe was found in human samples (Figure 12).315 The tryptic fragment ALVLIAFAQYLQQCPFEDHVK of in vitro modified Alb contains the cysteine adduct.327,328 Detection of this adduct did not succeed in human samples.258 |
| Chlorpyrifos and/or diazinon329 | LC-MS/MS | Pronase | Tyrosine diethylphosphothioate and tyrosine diethylphosphoro-adduct in a patient that had ingested chlorpyrifos.330 |
| Dichlorvos130 | LC-MS/MS | Pepsin | In two suicidal patients using dichlorvos, tyrosine-dimethoxyphosphate (Tyr411) was identified in Alb peptides VRY411TKKVPQVSTPTL and LVRY411TKKVPQVSTPTL. |
| Sarin, soman, tabun, and cyclosarin247 | LC-MS/MS | Pronase | Tyrosine-adduct in guinea pigs. |
| Alcohol | ELISA | Immune response to acetaldehyde-human serum Alb adduct among healthy subjects related to alcohol intake.331 | |
| Oxidative stress: 3-nitro-tyrosine332 | LC-MS/MS | Acid hydrolysis | 3-Nitro-tyrosine was found in Alb from rats. |
| Oxidative stress: malondialdehyde49 | LC-MS/MS | In patients with idiopathic pulmonary arterial hypertension, pulmonary hypertension,and sickle cell anemia LC-MS/MS adduct with Lys-159. | |
| Oxidative stress: malondialdehyde299 | UV/vis | Derivatization whole protein | Ischemia/reperfusion damage of Alb in patients; protein carbonylation was measured after dervatization with 2,4-dinitrophenylhydrazine (DNPH).308 |
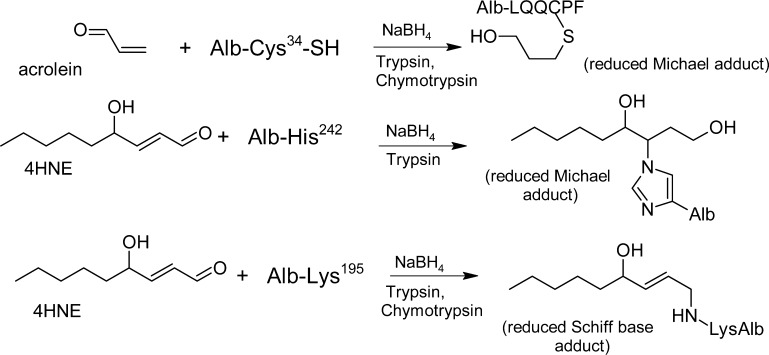
| Oxidative stress: acrolein299 | LC-MS/MS | Trypsin + chymotrypsin | Ischemia/reperfusion damage of Alb in patients; the adduct-level of LQQC(acrolein)PF in Alb increased from 0.6 ± 0.4% to 2.3 ± 0.7%; after 10 min of reperfusion. |
| Oxidative stress: ischemia modified Alb (IMA)333,334 | Cobalt-binding test | IMA relates to the decreased binding capacity of Alb for cobalt. In 283 healthy subjects, IMA ranged from 52.8 to 116.6 U/mL. Cut-off value for normal vs high IMA = 85 U/mL. For nonpathologic conditions, IMA is ca. 1–2% of the total Alb concentration and 6–8% in patients with ischemia. | |
| Oxidative stress: carbonylation; chronic arthritis | UV/vis | Derivatization whole protein | Carbonylated plasma proteins of children with different forms of juvenile chronic arthritis determined after derivatization with DNPH; carbonylation level was significantly higher than that in the healthy group (1.36 ± 0.68 vs 0.81 ± 0.16 nmol carbonyl/mg of protein).335 |
| Oxidative stress: carbonylation; chronic renal failure | UV/vis | Derivatization whole protein | Carbonylation levels in patients with chronic renal failure (13.7 ± 4.5 μmol/L) measured after derivatization with DNPH was higher than that in normal volunteers (0.76 ± 0.51 μmol/L), and higher than that in patients on chronic maintenance hemodialysis (16.95 ± 2.62 μmol/L).336 |
| Oxidative stress: cysteinylation | LC-MS/MS | Trypsin + chymotrypsin | Cysteinylation measured as LQQC(Cys)PF is significantly increased in end stage renal disease patients.7 |
HPLC-fluorescence (HPLC-f).
Table 2. Adduct Formation with Albumin (Alb) and/or Plasma Proteins (PP) in Rodents Exposed to Arylamines and Nitroarenes.
| PB-indexa total 14C or 3H-label | PB-indexa hydrolyzable adduct | Dose [mmol/kg] | % of dose boundb | Adduct ratio Hb/PPe | Adduct ratio Hb/Albe | |
|---|---|---|---|---|---|---|
| Benzidine | 1632 (PP)i | ND (PP)186 | 0.0011 | 0.360% | 0.5186 | |
| 3,3′-Dichlorobenzidine (DCBz) | 529 (PP)i | 138 (PP)186 | 0.05 | 0.166% | 0.2186 | |
| 1-Nitropyrene | 181 (PP)337 | adductg,337 | 0.00004–0.004 | 0.04%337 | 0.25338 | |
| Nitrobenzene | 136 (PP)61 | 79 (PP)186 | 0.2 | 0.0300% | 7.6186 | |
| 4-Chloroaniline (4CA) | 82 (PP)339 | 19.7 (PP)186 | 0.014 | 0.0181% | 29.3i,186 | |
| Acetanilide | 70 (PP)61 | 11.2 (PP)186 | 0.15 | 0.0154% | 2.5186 | |
| 2-Aminofluorene | 72 (PP)340 | 0.5 | 0.0159% | 2.38f,340 | ||
| 2-Nitrofluorene | 5.0 (PP)340 | 0.5 | 0.0011% | 0.32f,340 | ||
| 1-Aminopyrene | 0.44 (PP)340 | 0.5 | 0.0001% | 0.73f,340 | ||
| 2-Aminonaphthalene | 0.2 (PP)340 | 0.5 | 0.00004% | 50f,340 | ||
| 2-Nitronaphthalene | NDh(PP)340 | only Hb-adduct340 | ||||
| Aniline | ND (PP)340 | 0.5 | only Hb-adductf,h,340 42 in humanf,341 | |||
| 2-Methylaniline (2MA) | 297 (Alb)193 | ND (Alb)193 | 0.466 | 0.0291% | 0.94193 | |
| 4,4′-Methylenebis(2-chloroaniline) (MOCA)192 | 279 (Alb)192 | ND (Alb)192 | 0.0037 | 0.027% | 1.0192 | |
| 2-Amino-3-methylimidazo [4,5-f]quinoline (IQ)88,191 | 223 (Alb) | 22.3 (Alb) | 0.15 | 0.022%d | 0.33–0.288 | |
| 4-Aminobiphenyl (4ABP)c220 | 204 (Alb)c,j | 0.012 | 0.02% | 25189 |
Protein binding (PB)-index: (pmol compound/mg protein)/(mmol compound/kg body weight).
Calculated with the assumptions: per kg rat there are 0.98 g Alb and or 2.205g plasma proteins (PP).342
After pronase digestion: isolation of the adduct with Trp214 (Ala-Trp-Ala-Val).20
After pronase digestion: N2-(Pro-Tyr-cysteinesulfinyl-)IQ; 0.014–0.043% of the dose bound to Alb = experimental value.
Total binding (= hydrolyzable plus nonhydrolyzable) to Hb devided by total binding to Alb and/or PP.
Ratio of hydrolyzable adducts.
1-Acetylamino-X,Y-diacetoxy-pyrenes.337
ND = not detected
Neumann186 used a PB-index expressed as (mmol compound/mol protein)/(mmol/kg body weight). A molecular weight of 68000 was taken for the calculations.339 Therefore, (106/68000)·[(mmol compound/mol PP)/(mmol compound/kg body weight)] yields the values expressed as [(pmol compound/mg PP)/(mmol compound/kg body weight)]. Differences from the experimental results are due to rounding errors.
This value was calculated from the percentage of the dose bound to Alb and from the dose.
6.1. Benzene
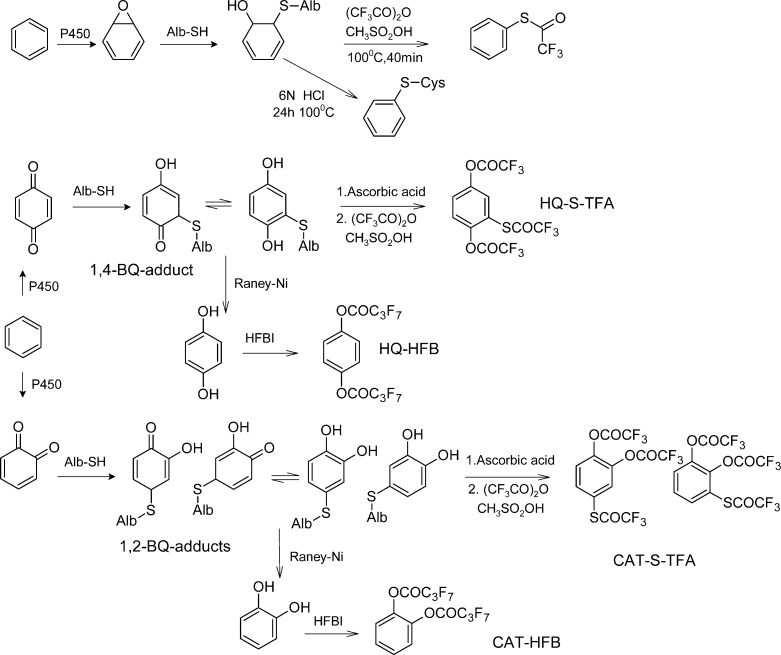
Benzene is an important industrial and environmental chemical that causes leukemia in humans and various cancers in experimental laboratory animals.167,168 The mechanism of its carcinogenicity is thought to involve the DNA damage induced by one or more metabolites, including benzene oxide (BO), 1,2-benzoquinone (1,2-BQ), and 1,4-benzoquinone (1,4-BQ), which are produced by P450 2E1.169,170 These metabolites can react with blood proteins to form adducts (Figure 5, Table 1).171,172 In the following, the studies of Alb-adducts found in vivo are summarized. Bechtold et al.97 found Alb-adducts of benzene formed with Cys34 in rats and humans. The adduct, S-phenylcysteine (SPC), was determined by isotope dilution GC-MS. Alb adducts were found in F344/N rats exposed by gavage to 0–10,000 μmol/kg benzene. The adduct levels increased with the dose. At a dose of 1000 μmol/kg, the adduct levels increased sublinearly. SPC was found also in humans occupationally exposed to 0–23 ppm benzene.
Figure 5.
Alb adducts of benzene, heptafluorobutyrylimidazole (HBFI).99,109,179
Waidyanatha et al.109 developed a method to detect mono-S-substituted cysteinyl adducts of 1,2- and 1,4-BQ in Alb. Alb was treated with trifluoroacetic anhydride and methanesulfonic acid. The resulting isomers of O,O′,S-tris-trifluoroacetyl-hydroquinone and -catechol were determined by GC-MS. F344 rats received a single oral dose of 50–400 mg [13C6]benzene/kg, in order to avoid the additional adduct-level from the ubiquitous [12C6]benzene. In Alb, a dose-related increase in both [13C6]1,2- and [13C6]1,4-BQ adducts was observed. The adduct level of [13C6]1,4-BQ-Alb was much larger than the adduct level of [13C6]1,2-BQ-Alb. The background [12C6] adducts of 1,2- and 1,4-BQ in F344 rats were 2.7 and 11.4 nmol/g. In comparison, the background levels of 1,2- and 1,4-BQ adducts in commercial human Alb (n = 10) were 1.6 and 8.9 nmol/g Alb.
The stability of BO-Alb and 1,4-BQ-Alb was investigated in rats given a single oral dose of isotope labeled benzene.173 BO and 1,4-BQ adducts with Alb both decayed with rates consistent with those of Alb turnover in the rat. The half-life for 1,4-BQ-Alb (2.5 days) was shorter than that for BO-Alb (3.1 days), suggesting some instability of 1,4-BQ-Alb. Results of a limited time course study of 11 human subjects174 indicated moderate chemical instability of 1,4-BQ-Alb (half-life = 13.5 days compared with 19–25 days for normal Alb turnover), whereas no evidence of instability of BO-Alb was observed.
Alb adducts BO-Alb and 1,4-BQ-Alb were investigated among 134 workers exposed to benzene and 51 unexposed controls in Tianjin, China.174 Concentrations of both adducts increased with benzene exposure. Adduct levels were less than proportional to benzene exposure, suggesting saturable P450 2E1 metabolism of benzene. This was confirmed in follow-up studies involving exposed workers.172,175 The transition from linear to saturable metabolism began at approximately 1 ppm. Adduct levels were generally lower in older workers. The ratio of 1,4-BQ-Alb:BO-Alb decreased with age and increased with alcohol consumption. This indicates that factors affecting P450 2E1 metabolism exerted a greater role on the production of 1,4-BQ than BO. The nonlinearity of the benzene adduct formation was confirmed in later studies.176
In summary, the benzene adducts with Alb have been found in exposed workers and in controls. Therefore, benzene exposure is ubiquitous.177
6.2. Naphthalene
Naphthalene is an important industrial chemical, which has recently been shown to cause tumors of the respiratory tract in rodents.178 The reactive metabolites of naphthalene are similar to the metabolites of benzene. Naphthalene-1,2-oxide (NPO), 1,2-naphthoquinone (1,2-NPQ), and 1,4-naphthoquinone (1,4-NPQ) are the major reactive metabolites of naphthalene (Supporting Information, Figure S1 and Table 1). These metabolites were reported to form cysteinyl adducts with Alb of F344 rats.179 1-Sulfanyl- dihydronaphthalene-2-ol (NPOS1) and 2-sulfanyl-1,2-dihydronaphthalene-1-ol (NPOS2) are the Alb adducts formed with NPO. Sulfanyl-1,2-naphthalene-1,2-diol (1,2-NPQ-4S) and 2-sulfanyl-1,4-naphthalene-1,4-diol (1,4-NPQ-2S) are the Cys adducts resulting from the reaction of 1,2-NPQ and 1,4-NPQ with Alb, respectively (Figure S1, Supporting Information). Alb is hydrolyzed and derivatized with trifluoroacetic anhydride and methanesulfonic acid to obtain S-naphthalen-1-yl trifluoroethanethioate (NPO1-S-TFA), and S-naphthalen-2-yl trifluoroethanethioate (NPO2-S-TFA), 2-[(trifluoroacetyl)sulfanyl]naphthalene-1,4-diyl bis(trifluoroacetate) (1,4-NPQ-S-TFA), and 4-[(trifluoroacetyl)sulfanyl]naphthalene-1,2-diyl bis(trifluoroacetate) (1,2-NPQ-S-TFA) from NPOS1, NPOS2, 1,4-NPQ-2S, and 1,2-NPQ-2S, respectively. Cysteinyl adducts of Alb with NPO and 1,2- and 1,4-NPQ were produced in a dose-dependent manner. Of the two structural isomers resulting from NPO, levels of NPO1 adducts were greater than those of NPO2 adducts in Alb. 1,2-NPQ-Alb was present in larger amounts than 1,4-NPQ-Alb. The shape of the dose–response curves was sublinear at doses above 200 mg of naphthalene per kg body weight. Low background levels of 1,2-NPQ-Alb and 1,4-NPQ-Alb were found in control animals. However, NPO-Alb adducts were not detected in control animals.
The stability of cysteinyl adducts of NPO, 1,2-NPQ, and 1,4-NPQ were investigated in Alb of male F344 rats following a single administration of two different doses (400 or 800 mg naphthalene per kg body weight).180 The half-lives of NPO-Alb and 1,2-NPQ-Alb were approximately 2 days and 1 day, respectively. The normal half-life of Alb in the rat is 2.5–3 days. Therefore, especially the 1,2-NPQ-Alb adduct is unstable.
1,2-NPQ- and 1,4-NPQ-Alb were detected in human subjects (n = 22).181 The median levels of 1,2-NPQ-Alb were 268 and 203 (pmol/g) in male (n = 11) and female (n = 11) subjects, respectively. The median levels of 1,4-NPQ-Alb were 45.0 and 38.9 pmol/g in male and female subjects, respectively.
6.3. Styrene
Styrene is used in the production of resins and plastics.182 Styrene-7,8-oxide (SO) is the primary metabolite of styrene that forms adducts with DNA and proteins.183 SO is both mutagenic and carcinogenic in animals.182 Alb adducts of SO were measured in 48 workers exposed to both styrene and SO in a boat manufacturing plant.184 Personal exposures to both substances were measured repeatedly over the course of 1 year. Cysteine containing adducts and carboxylic acid adducts of SO with Alb were determined. Alb proteins were subjected to base hydrolysis to release styrene glycol (SG), representing carboxylic acid-bound SO. Alb was treated with Raney-nickel to release 1-phenylethanol (1-PE) and 2-phenylethanol (2-PE), representing cysteine-bound SO. The mean levels of 1-PE-Alb, 2-PE-Alb, and SG-Alb were 0.29, 1.68, and 1.8 (nmol/g Alb), respectively. Similar levels of 1-PE-Alb (0.6 nmol/g Alb) and 2-PE-Alb (2.84 nmol/g Alb) were found in workers exposed to styrene in the reinforced-plastics industry and in unexposed subjects.185
6.4. Aromatic Amines
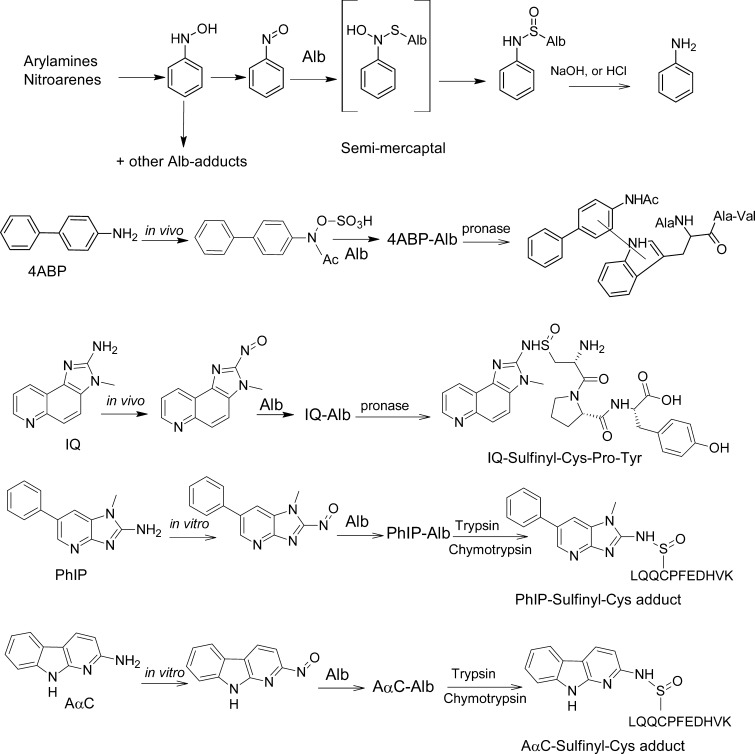
Alb adducts of aromatic amines have been determined only in a few studies. In general, only Hb-adducts were determined in human biomonitoring studies because many aromatic amines bind to Hb at higher levels than to PP186 (Table 2). An exception to this rule appears, for example, with the HAA PhIP, which preferentially binds to Alb in humans.73 The oxidation of the exocyclic amine groups of arylamines and heterocyclic arylamines is primarily catalyzed by P450s, to form the N-hydroxylated metabolites, which are reactive intermediates that bind to DNA and proteins.64 Alb adduct formation of primary arylamines was investigated in rodents (Figure 6 and Table 2). However, direct comparison of arylamine adduct formation with Hb and PP has been determined only in a few studies. Neumann’s group186 investigated the formation Hb and PP adducts after giving female Wistar rats several radiolabeled arylamines (Table 2). In general, Hb-binding is higher; however, for the bicyclic arylamines 3,3′-dichlorobenzidine and benzidine, the binding was higher with PP than with Hb. In the case of 4CA, binding to Hb was over 30-fold larger than that to PPs. In all other cases, the differences of adduct formation were below a factor of 3-fold.
Figure 6.
Formation of acid labile Alb adducts of aromatic amines and HAA,88,91,165,186,192,195 and the nonhydrolyzable adduct of 4ABP.20
Most of the arylamine Hb adducts are labile and undergo hydrolysis in vitro, by mild acid or base, to form the arylamines. The amount of amines recovered by this hydrolysis treatment ranged between 32 to 93% of the bound Hb-adduct.186 For arylamine adducts with PP, the amounts hydrolyzed varied from 0 to 58% of the bound adduct.186 According to our current knowledge of arylamine adduct formation, the hydrolyzable fraction is derived from the reaction products of the arylnitroso-derivatives yielding aryl sulfinamide adducts with cysteine.53,187 The nonhydrolyzable fraction probably results from the reaction products of activated N-hydroxyarylamines or their nitrenium/carbenium ion species with other amino acids. Protein adducts formed with nitrosoarenes are generally higher with Hb than with PP. However, the absolute values for nonhydrolyzable adducts are higher with PP than with Hb. Therefore, it appears that nitroso-derivatives are a minor component in the plasma and are likely generated from the arylhydroxlamines within the erythrocytes.54,188 This can be explained by the fact that N-hydroxyarylamines can be readily transformed to nitrosoarenes by a co-oxidation reaction with oxyHb in the erythrocytes.54,188 In the experiments with radiolabeled 2-amino-3-methylimdazo[4,5-f]quinoline (IQ), 4,4′-methylenebis(2-chloroaniline) (MOCA) and 4ABP, Alb was isolated (Table 2). In the case of 4ABP, the binding to Hb was 25-fold higher than that to Alb.56,189 Therefore, human biomonitoring studies were performed using Hb-adducts.56,189,190 IQ binding was higher to Alb than to Hb.88,191 For MOCA (dose = 1 mg/kg), the total (nonhydrolyzable and hydrolyzable adduct) binding ratio between MOCA-Hb and MOCA-Alb was 1 (Table 2). For 2MA (dose = 50 mg/kg), the total (nonhydrolyzable and hydrolyzable adduct) binding ratio between 2MA-Hb and 2MA-Alb was 0.94 (Table 2).193 The percentage of adduct hydrolyzable with base was 54%, 0%, 63%, and 0% for MOCA-Hb, MOCA-Alb, 2MA-Hb, and 2MA-Alb, respectively.56,189,190,192 In the experiments with 2MA, Alb was isolated by fractional acid precipitation; under such conditions, sulfinamide adducts are partially cleaved.56 In the same study, animals were treated with different amounts of 2MA. 2MA-Alb binding was not linear; but 2MA-Hb binding increased in a linear dose-dependent manner. The biological half-lives of 2MA bound to Alb or Hb were observed to be 2.6 and 12.3 days, respectively, after rats were given a single dose of [14C]2MA.194
The structure of the major nonhydrolyzable Alb adduct formed in rats exposed to 4ABP was elucidated by 1H NMR and MS.20 Serum Alb was isolated from male Sprague–Dawley rats dosed by gavage at 27 h after administration of [3H]4ABP. Pronase digestion of the purified Alb yielded a mixture of radiolabeled materials, which were resolved into five major components by reverse phase liquid chromatography. From detailed UV, 1H NMR, and mass spectral analyses, four of these components were determined to be 4ABP, 4-N-hydroxy-4-acetyl-ABP, and two other metabolites, all of which are presumed to be noncovalently associated with Alb. The fifth component, however, resulted from covalent bond formation and was identified as a tetrapeptide containing 3-tryptophanyl-4-acetyl-ABP, the amino acid sequence of which was H2N-Ala-Trp-Ala-Val (Figure 6).20 Subsequently, the sulfate esters of several carcinogenic arylhydroxamic acids, including N-sulfonyloxy-N-acetyl-4-aminobiphenyl, N-hydroxy-N-acetyl-2-aminofluorene, and N-hydroxy-N,N′-diacetylbenzidine were reported to bind to the sole Trp214 residue of human Alb in vitro.79 The cooked meat carcinogen PhIP and the non-nucleoside reverse transcriptase inhibitor Nevaripine also form adducts at the Trp214 of human Alb in vitro.195−197 Since rat and human Alb contain only a single tryptophan residue at Trp214 situated in a hydrophobic drug-binding site, its high selectivity for carcinogen binding suggests a unique role for Alb in the detoxification and/or transport of ultimate carcinogenic/toxic metabolites.80
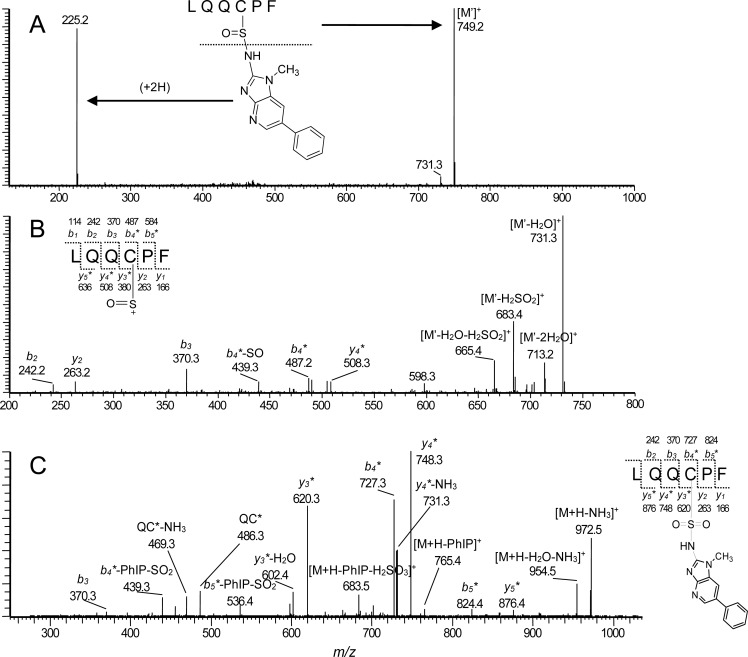
The structure of the acid-labile sulfinamide Alb adduct of IQ, a carcinogenic HAA,124 formed in rat was characterized by 1H NMR and MS.88 Alb of [3H]-IQ male-treated Sprague–Dawley rats was isolated and digested with pronase. Multiple adducts were detected by HPLC with liquid scintillation counting. About 10% of the adducted material was characterized as the tripeptide C*PY containing N2-cysteinylsulfinyl-IQ adduct (Figure 7). More recently, PhIP and 2-amino-9H-pyrido[2,3-b]indole (AαC), another HAA carcinogen present in tobacco smoke and well-done cooked meats,124 were shown to react with the Cys34 of human Alb to form sulfinamide and sulfonamide adducts in vitro (Figure 6).91,165,195 These adducts were sufficiently stable to characterize the following trypsin/chymotrypsin digestion as LQQC*PF or the missed-cleavage peptide LQQC*PFEDHVK by LC-MS/MS (Figure 7). An acid-labile Alb adduct of PhIP was reported in a cohort in Italy.198 This adduct may be the sulfinamide linked adduct of PhIP characterized in vitro.91,195
Figure 7.
(A) ESI product ion spectra of LQQC*PF (C-[S=O]-PhIP) sulfinamide ([M + H]+ at m/z 973.3), (B) second generation product ion spectrum of the ion at m/z 749.2 [M + H – PhIP]+, and (C) LQQC*PF (C-[SO2]-PhIP) sulfonamide ([M + H]+ at m/z 989.5. Adapted from ref (91). Copyright 2012 American Chemical Society. Adapted with permission from ref (125). Copyright 2014 Elsevier.
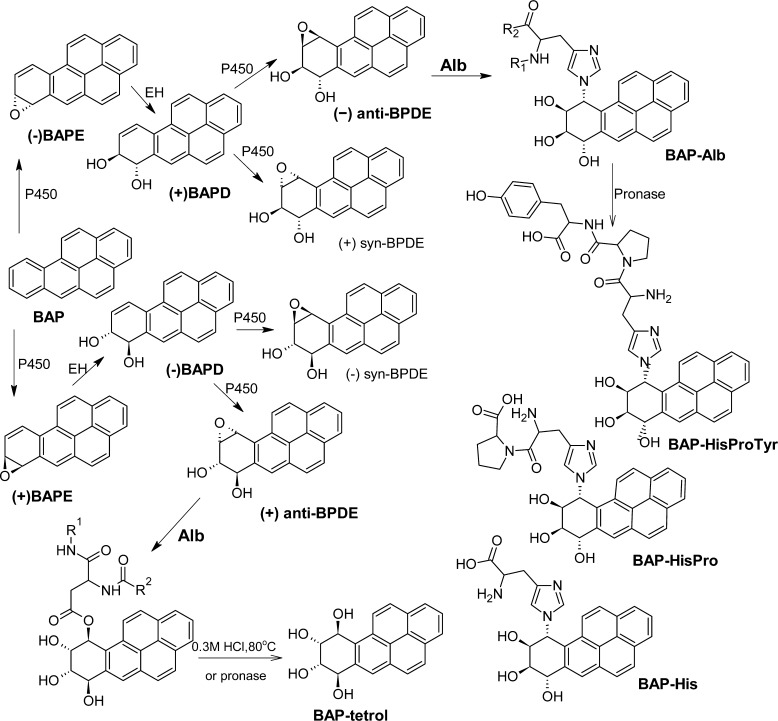
6.5. Aflatoxin B1 (AFB)
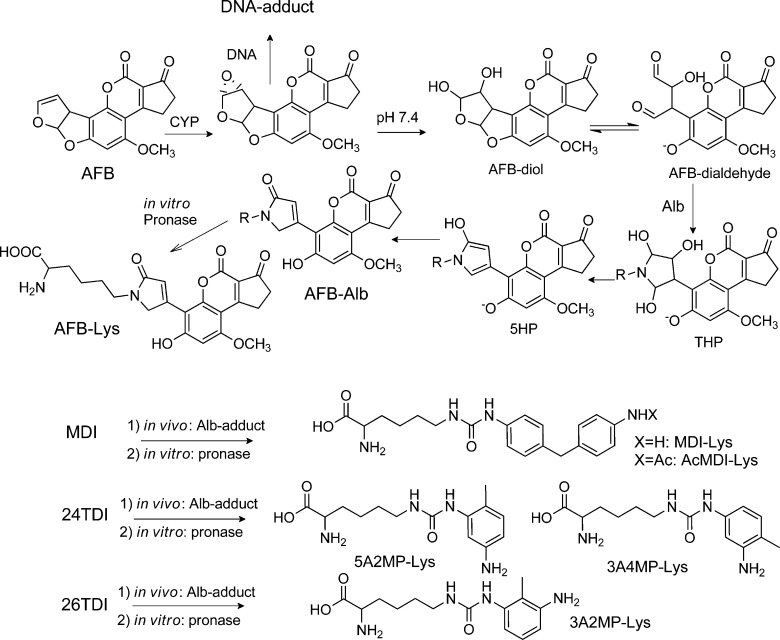
AFB is a fungal toxicant and a potent animal carcinogen found as a contaminant in various staple food crops in many underdeveloped countries.199 Positive associations have been reported between dietary AFB exposure and the incidence of hepatocellular carcinoma in Asia and Africa and were greatly strengthened by the application of validated biomarkers, which included DNA and Alb adducts, and a characteristic mutation spectrum in the p53 tumor suppressor gene that is linked to a DNA adduct of AFB.24,63,72,114 AFB undergoes metabolism by P450 enzymes to the 8,9-epoxide, which has a central role in DNA and protein adduct formation. The AFB exo-8,9-epoxide hydrolyzes rapidly to the dihydrodiol (t1/2 1 s at 23 °C).200 The AFB dihydrodiol, rearranges the AFB dialdehyde (Figure 8), which reacts with Alb to generate a Lys adduct.71,120 The sites of adduction were reported to occur at Lys455 and Lys548 (Lys431 and Lys524 of the mature protein) of bovine albumin.201 The sites of adduction to human Alb have not been reported, although human Alb contains Lys residues at these homologous sites as bovine Alb.
Figure 8.
Alb adducts of AFB71,120 and isocyanates121 found in vivo. The phenolic OH of AFB-Lys and AFB-dialdehyde is deprotonated at pH 7.4 (bathochromic shift of the UV-spectra pH 4.0 to pH 7.4).71,101
6.5.1. Correlation of AFB-Adducts with DNA and Albumin
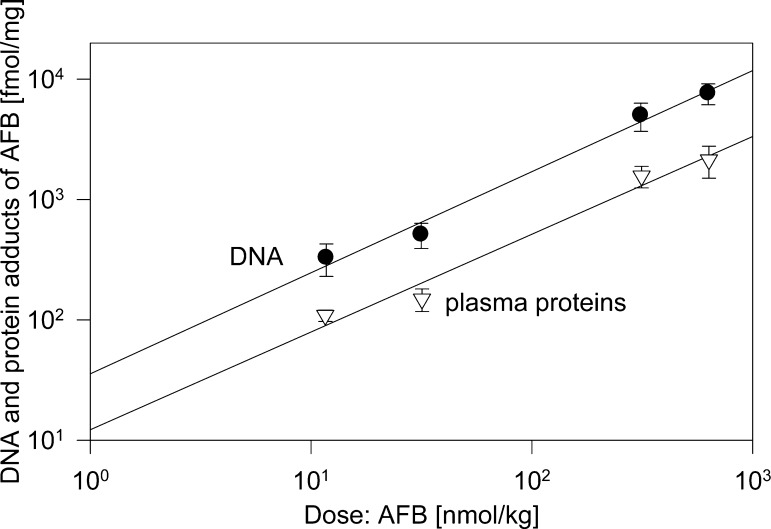
Wild et al.202 studied the binding of AFB to PP and liver DNA in male Wistar rats (Figure 9 and Table 1). A constant ratio was found between levels of AFB bound to PP and that bound to liver DNA 24 h after a single dose.101 In total, 0.98% to 2.15% of the administered dose was bound to the PP at this time point. In the chronic study, binding of AFB to PP was 3-fold higher than that after a single dose, and DNA-adducts increased by 2.5-fold. The DNA and PP adducts levels reached a plateau between days 7 and 14 of treatment. Fractionation of the PPs showed that all detectable bound AFB was associated with a single protein corresponding to Alb. Thus, a constant ratio was observed between the concentration of Alb-bound AFB and that bound to liver DNA, the target organ for carcinogenesis by AFB.
Figure 9.
DNA- and plasma protein-adducts of AFB in rats.202 In a separate experiment, it was shown that among the plasma proteins only Alb formed adducts with AFB.202
6.5.2. Stability of the AFB-Albumin Adduct and Elucidation of Adduct Structure
The radioactivity associated with Alb following administration of 14C-AFB to rats was cleared with a half-life of 2.5 days, which is not significantly different from the half-life of unmodified Alb in the rat.71 The product isolated from a pronase digest of in vivo modified Alb was identical to the synthetic product. The synthetic product was obtained by the acylase catalyzed deacetylation of the reaction product of Nα-acetyl-l-lysine with AFB-dibromo and characterized by UV, fluorescence, 1H NMR, 13C NMR,120 and fast atom bombardment mass spectrometry.71 AFB-dibromo (in vitro) or AFB-epoxide (in vivo) react first with water to yield the same intermediate, the AFB-diol. The dialdehyde reacts with Nε of lysine to form a Schiff base or a 2,4,5-trihydroxy-pyrrolidine (THP) (Figure 8). THP loses two water molecules to yield the 5-hydroxy-pyrrole (5HP), which is known to tautomerize to 5-oxo-3-pyrroline and 5-oxo-2-pyrroline in a ratio of 9 to 1.120 The 1H NMR and 13C NMR spectra supported the proposed structure with the coumarin unit attached to the 3 position of 5-oxo-3-pyrroline (AFB-Lys).120 This structure was confirmed 12 years later by Guengerich et al.201
AFB adducts with other proteins? Organ perfusion experiments were performed with livers from male Wistar rats.203 The perfusate contained an AFB dose equivalent to 8 mg/kg body wt and was recirculated for 2–2.5 h. Alb, synthesized and excreted by the perfused liver during this time, was isolated, digested with pronase, and analyzed for AFB-Lys. The experiment was repeated by adding histones, globulin, or Alb to the perfusate. The presence of these proteins in the perfusate did not substantially alter the binding of AFB, to Alb (721 ± 197 pmol/mg Alb), indicating that that metabolism of AFB in the perfused liver is unaltered by the protein additions. AFB-Lys. These findings indicate that in the perfused liver secreted protein-reactive AFB is produced and suggests that AFB metabolites react in the cell and in the recirculating system with Alb.
Okoye et al.204 reported in an experiment with rats treated simultaneously with [3H]AFB and l-[14C]leucine that the majority (>90%) of rat serum AFB-Alb adduct was formed by a modification of the protein at the time of its synthesis in the hepatocyte. This finding further indicates that Alb adducts are a dosimeter for degree of modification of DNA in hepatocytes.
In human samples, AFB-Lys was not found in Alb depleted plasma, demonstrating that binding occurred almost exclusively to Alb.203 Therefore, whole serum was digested with pronase, and the adducts were purified using an AFB- monoclonal antibody immunoaffinity chromatography and quantified by HPLC with fluorescence detection. It should be noted that the recovery of AFB-Lys adduct is lower from digestion in whole plasma than in the presence of purified Alb.205 This was shown by spiking rat serum in to both a solution of human serum Alb and in to human serum. The recovery of AFB-Lys was 78% ± 10% in the presence of serum.
6.5.3. Quantification of AFB-Albumin Adducts by Immunological, Fluorescence, and LC-MS Methods in Human Studies
Adducts of AFB with Alb have been measured by ELISA,206 HPLC-fluorescence,71,120,203 or by isotope dilution mass spectrometry (IDMS).115,207,208 The results obtained from quantitation with ELISA and with HPLC-fluorescence were compared in several studies.101,114,209 Wild et al.101 observed a 10-fold higher level of AFB-Alb adducts when measured by ELISA than by HPLC-fluorescence in human sera from Kenya,101 while the difference in sera from rats treated with AFB was 4- to 5-fold greater than the level measured by HPLC-fluorescence. Scholl compared the performance of the ELISA-method, HPLC-fluorescence method, and the IDMS method using samples from an acute aflatoxicosis outbreak in Kenya.115 The Deming regression slopes for the HPLC-fluorescence and ELISA concentrations as a function of the IDMS concentrations were 0.71 (r2 = 0.95) and 3.3 (r2 = 0.96). When the samples were classified as cases or controls, based on clinical diagnosis, all methods were predictive of the outcome (p < 0.01). IDMS was the most sensitive technique, and HPLC-fluorescence was the least sensitive method.
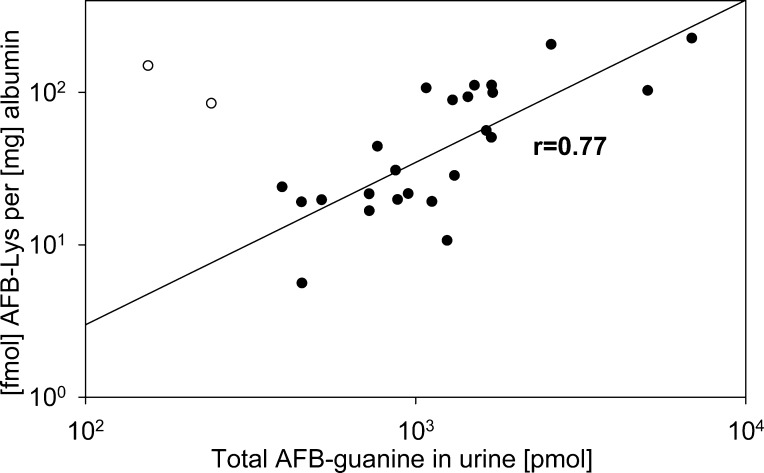
China and Africa have the highest intake of AFB and the highest incidences of liver cancer.210 Molecular epidemiological studies in populations exposed to AFB have been summarized recently.63,210−212 Studies were undertaken to explore the relationship between dietary intake of AFBs, the serum Alb adducts, the hepatitis B virus-carrier status, and the excretion of the major AFB-DNA adduct and other metabolites into the urine of chronically exposed people. The levels of Alb adduct AFB-Lys correlated well with the amounts of AFB-DNA adduct excreted in urine (Figure 10).203,205 Interestingly, the levels of AFB-Lys was about 4 times lower in females than in males with a similar AFB-intake in the Chinese cohort.205 AFB-Lys adducts were assessed in populations of the United States during the Health and Nutrition Examination Survey 1999–2000. About 1% of the U.S. population had detectable levels (≥0.02 μg/L) of AFB-Lys.213 Therefore, it appears that AFB exposure is mainly a health concern for developing countries.
Figure 10.
Correlation between the albumin adduct AFB-Lys and urinary AFB-guanine in Chinese males.203,205 Two outliers (white circles) were not included in the regression analysis.
6.5.4. Comparison of Albumin Adducts with Individual Susceptibilities and Cancer Prevention Measures
Dithiolethiones, including oltipraz and the unsubstituted molecule 1,2-dithiole-3-thione, are potent inhibitors of AFB-induced hepatic tumorigenesis in rats.214−216 In rats fed 1,2-dithiole-3-thione, the overall reduction in the levels of hepatic DNA adducts, urinary AFB-guanine, and AFB-Alb adducts over the 2-week exposure period were 76%, 62%, and 66%, respectively.214 Egner et al.215 confirmed that long-term intervention with oltipraz produced a reduction in levels of the AFB-Alb biomarker at all times throughout AFB exposure. This parallel reduction of urinary AFB-guanine and serum AFB-Alb adducts levels relative to target organ DNA adduct burden implies that these biomarkers are noninvasive dosimeters, which can be used to evaluate the efficacy of chemoprotective interventions.63,211,212
6.6. Isocyanates
Isocyanates are considered one of the main causes of occupational asthma.217,218 Isocyanate-induced asthma results usually from repeated exposure during which sensitization occurs. Alb adducts have been linked to the mechanism of occupational asthma caused by isocyanates.219 Isocyanate–protein adducts trigger both immune responses and are probably the antigenic basis for isocyanate asthma.219 In the past, Alb or plasma protein samples of isocyanate exposed workers were analyzed after treating the samples for several hours with base or acid.102,105,220 Such treatments cleave many adducts nonspecifically.221 However, with these harsh methods of hydrolysis it was not possible to determine if the biomarkers resulted from exposure to isocyanates or to the corresponding aromatic amines. In order to perform biomonitoring studies, putative reaction products of MDI with amino acids were synthesized in vitro.106,222 A method was developed to measure Alb adducts with isocyanates.106,121,222,223 Lysine adducts of MDI (= MDI-Lys and AcMDI-Lys) were found in Alb of exposed subjects (Figure 8). Alb was digested with pronase and analyzed with LC-MS/MS in the presence of the isotope labeled adduct standards. The major adduct MDI-Lys was found in factory workers and construction site workers.106,224−226 This new biomonitoring procedure will allow assessment of suspected exposure sources and may contribute to the identification of individuals who are particularly vulnerable for developing bronchial asthma and other respiratory diseases after exposure to isocyanates. Recently, Alb was modified in vitro with isocyanates,92,227−230 The in vitro modifications with MDI, TDI, and HDI yielded adduction sites with several Lys (Supporting Information, Table S1). The experiments were performed with a high ratio (1:1, 5:1, 10:1, and 40:1) of isocyanates (MDI, HDI, TDI) to Alb.228−230 The high concentrations used for the experiment greatly exceeded the levels of human exposure. In industrial workers, the median adduct level of MDI-Alb was 30 fmol/mg Alb (= 0.002 mol MDI-Lys/mol Alb).106 This adduct level is 300-fold less than the levels (0.59 mol/mol Alb) obtained from the in vitro experiment with the lowest dose of MDI229 (MDI/Alb = 1:1). Therefore, such in vitro experiments should be performed with a ratio of isocyanate to protein equal to 1:10 and 1:100 in order to obtain values which are close to the in vivo situation and for reliable development of analytical methodology.
In order to determine adducts of toluene diisocyanates (TDI) with blood proteins, several reaction products of 2,4-toluene diisocyanate (24TDI) and 2,6- toluene diisocyanate (26TDI) with amino acids were synthesized in vitro.231 Thereafter, adducts with Alb were determined in workers accidentally exposed to 24TDI and 26TDI (Figure 8).232 After digestion with pronase, the covalent adduct with lysine was identified.232 This work presents a new procedure for the determination of isocyanate-specific Alb adducts. Isotope dilution mass spectrometry was used to measure the adducts in Alb present in workers exposed to TDI. 24TDI and 26TDI formed adducts with lysine (Figure 8): 3A4MP-Lys, 5A2MP-Lys, and 3A2MP-Lys.102,220 In future studies, this method of hydrolysis with pronase can be applied to measure TDI-exposures in workers.
In patients with kidney disease, carbamylation reaction products with lysine of Alb were found.233 Carbamylation is a nonenzymatic post-translational protein modification mediated by cyanate, a dissociation product of urea.
6.7. Polycyclic Aromatic Hydrocarbons (PAHs)
Polycyclic aromatic hydrocarbons (PAHs) are carcinogens that are ubiquitous in the environment. Many methods were developed to assess the exposure to PAHs through DNA and protein adducts.113,234 Benzo[a]pyrene (BAP) is one prototypical PAH with major carcinogenic potential. BAP forms DNA-adducts, Hb-adducts, and Alb-adducts. Enzyme-linked immunosorbent assay (ELISA) and physical–chemical methods have been used to quantify the internal exposure to PAHs. ELISA is nonspecific due to cross-reactivity of the antibody with other PAHs. In the majority of studies, it was not possible to distinguish between exposed and nonexposed individuals using the ELISA method.113,234 The BAP Alb adduct levels obtained with ELISA were several orders of magnitude higher than those determined by physicochemical methods such as GC-MS, LC-MS, or HPLC-fluorescence or −UV detection. Alb adducts of (+)-anti-BAP-diol epoxide (BPDE) were analyzed in animal and human samples. Human serum Alb was reacted with the (+)-enantiomer (+anti-BPDE) (Figure 11) and (−)-enantiomers (-anti-BPDE) of r-7,t-8-dihydroxy-t-9,t-10-epoxy-7,8,9,10-tetrahydro-BAP (anti-BPDE).235,236 The +anti-BPDE forms carboxylic ester adducts with Alb at either Asp187 or Glu188, while the −anti-BPDE forms Nτ-histidine adducts at His146, His-BPDE, BPDE-His-Pro, and BPDE-His-Pro-Tyr. Alb was digested with pronase and analyzed with HPLC and line induced fluorescence (HPLC-LIF). In human samples, Özbal et al.237 found BPDE-His-Pro adducts using HPLC-LIF. Alb was digested with pronase and purified with immunoaffinity columns. The adduct levels ranged from <0.04 to 0.77 fmol/mg Alb in nonsmoking and smoking individuals in the general population. However, the study could not distinguish between the different types of exposure to BAP. The BPDE-Alb adducts were detected in 28% of the potentially exposed subjects.238,239 In a limited number of studies, adducts from BPDE bound as esters to Alb (assumed to be (+)-anti-BPDE adducts) have been measured by MS methods as tetrols, after their release from Alb by mild hydrolysis. Cohorts without occupational exposure showed mean adduct levels in the range 0.01–0.1 fmol/mg Alb. When BAP personal monitoring data were used as the criterion of exposure,239,240 no correlation was found with the levels of BPDE-Alb adducts. Undetected sources of PAH, such as the diet, might markedly distort the exposure profile generated with individual air sampling data and affect the frequency and levels of protein biomarkers.
Figure 11.
Alb-adducts of BAP; EH = epoxide hyrolase.235,236,241,343 BAP-tetrols were obtained after pronase235,236 digestion or acid hydrolysis.240
A LC-MS/MS method employing TQ MS was developed to measure adducts of BPDE formed at His146 with in vitro modified human Alb.241 This method was also applied to measure adducts at the homologous sequence in BAP-exposed mice (100 mg/kg of body weight for 1, 3, 7, and 28 days). BPDE-His adducts (BPDE-His-Pro and BPDE-His-Pro-Tyr) were recovered from pronase digestion of mouse Alb. The level was 400 times lower (calculated per gram macromolecule) than the level of BPDE-dG adduct in liver-DNA.241 The specific and stable BPDE adducts to His in Alb are potential biomarkers of in vivo dose of BPDE, though this requires a considerable improvement in the sensitivity of the LC-MS/MS method.
6.8. Pesticides and Nerve Agents
Organophosphates (OPs) are a class of chemicals that are widely used as pesticides in agriculture. Many OP pesticides possess structural features in common to the nerve agents sarin and soman, which phosphylate a serine residue in the active site Acetylcholinesterase (AChE), resulting in inactivation of the enzyme as the mode of toxicity.118,164,242 OPs also phosphylate Lys, and Tyr residues of proteins, resulting neurotoxicity, but independent of AChE activity.243 The underlying mechanisms for OP neurotoxicity and the target proteins, are still unclear, but the mode of action most likely involves chemical modification of proteins. Many OPs react with Alb.130,243−245 The Tyr411 is the primary residue of human Alb that reacts with OPs and nerve agents. Adduct formation has been characterized for dichlorvos, chlorpyrifos oxon, diisopropylfluorophosphate, and fluorophosphinate biotin. When Alb or plasma was treated in vitro with these OPs, additional adductions were identified at Tyr138, Tyr148, Tyr150, Tyr161, Ser232, Ser287, Tyr401, and Tyr452 of Alb.130,246−250 (Figure 12, Table 1, and Supporting Information, Table S1). Human Alb also possesses pseudoesterase activity through some of its nucleophilic amino acid residues. Esterase activity with p-nitrophenyl acetate has been attributed to the reactivity at Tyr411, which is the first amino acid residue of Alb to undergo acetylation.251 The selective reactivity of Tyr411 for these toxic electrophiles has been explained in part by the microenvironment of Alb surrounding this amino acid. Tyr411 is situated in a binding pocket with Arg410 in the center of the drug-binding site II in subdomain IIIA of Alb. The phenolic OH group of tyrosine has a pKa of ∼10, whereas the pKa of the phenolic OH group of Tyr411 is lowered to 7.9 due to the electrostatic interaction with guanidinium ω-NH of Arg410, resulting in increased reactivity of Tyr411 with OPs and nerve agents.252 Some OPs also form adducts at several Lys residues of Alb.243 The Try411 of Alb was shown to be covalently modified in the serum of humans poisoned by dichlorvos. The dimethoxyphosphate adducted Tyr411 was identified in the serum of two patients who had attempted suicide with dichlorvos. The serum (10 μL) was digested with pepsin, and the Alb peptides VRY*411TKKVPQVSTPTL and LVRY411TKKVPQVSTPTL were purified by offline HPLC, and then adducts were detected by LC-MS/MS.130
Figure 12.
6.9. Atrazine
2-Chloro-4-(ethylamino)-6-(isopropylamino)-s-triazine) (Atrazine) is one of the most commonly used herbicides in the United States and used mainly for corn production.253 In 2004, the application of atrazine was banned in the European Union. In the United States, atrazine and its metabolites are well dispersed in the environment and frequently detected in groundwater.254 Atrazine is still one of the worlds’ most heavily used herbicides, and due to its persistence, the safety of its usage remains a matter of controversy.255 Atrazine undergoes metabolism primarily by P4501A2 to form diaminochlorotriazene,256 which forms adducts with the Cys34 residue of rat and human Alb in vitro.257 (Figure 12) The Alb adduct was characterized by MS/MS, following digestion with thermolysin, and the sequence was characterized as the single missed cleavage peptide LQQC*PFEDH (the theoretical thermolysin digest is LQQCP). To our knowledge, this adduct has not been reported in humans.
6.10. Sulfur Mustard
Bis(2-chloroethyl) sulfide, commonly known as mustard gas, is a cytotoxic and vesicant chemical warfare agent and reacts with human Alb. Following trypsin digestion, the tryptic peptide containing Cys34 from amino acid residues from 21 to 41 was identified as a prominent site of adduction.258,259 MS/MS sequencing showed that the adduct formed at Cys34. A sensitive TQ MS method was devised by digesting the Alb with pronase and measurement of the tripeptide containing adduct S-[2-[(Hydroxyethyl)thio]ethyl]-Cys-Pro-Phe.258 The incomplete digestion of Alb by pronase to yield *Cys-Pro-Phe adducts of several HAA also has been reported.88,260 The S-[2-[(Hydroxyethyl)thio]ethyl]-Cys-Pro-Phe adduct was successfully measured with only 3 mg of Alb purified from blood samples of Iranian victims of the Iran–Iraq war (1986), by TQ MS in the SRM mode. The limit of detection was reportedly 10-fold lower than that of the Edman degradation method to detect N-terminal Val adducts of Hb.258
7. Adduct Formation of Chemopreventive Agents
7.1. Isothiocyanates
Isothiocyanates (ITCs) found in cruciferous vegetables possess cancer preventive activity in animals, and increased dietary intake of ITCs has been shown to be associated with a reduced cancer risk in humans.261 ITCs exert their cancer chemopreventive action by multiple mechanisms.261−264 In cells, protein adducts account for most of the total cellular ITC uptake at 4 h after treatment.265 The major urinary excretion products of ITCs in humans are N-acetyl cysteine conjugates, which are rapidly eliminated in urine within 24 h.263 The measurement of these urinary metabolites may provide the exposure history of the last 24 h, However, the mercapturic acids of ITC are not stable, and stable biomarkers of ITC are required for large epidemiological and pharmacological studies. The effect of a drug is related to the drug concentration at the steady state level in the blood. However, for ITCs, the biologically active intermediate released from the cruciferous vegetables at intake is not stable. ITCs are released from glucosinolates (GLs), by the enzyme myrosinase, which occur in the same plants, separated cellularly from the GLs. Myrosinase mixes with the GL and effects the hydrolysis when tissues of plants are damaged or chewed.266,267 It has been shown that ITCs do not accumulate in the blood after chronic doses of cruciferous vegetables.268 Stable, biological markers, which accumulate over time, are needed for clinical studies. Therefore, the measurement of stable blood protein adducts of ITC can provide an estimate of the dose necessary to reach the therapeutic effect. A method was developed to determine stable reaction products of ITCs with Alb in humans.269,270 The ITCs formed adducts with lysine of Alb269 (Figure 13): BITC-Lys, PEITC-Lys, SFN-Lys, and AITC-Lys. The adduct levels were quantified by IDMS with a TQ MS, using the corresponding ITC-[13C615N2]Lys as internal standards (Figure 14). The applicability of the method was tested with biological samples obtained from different human studies.269,270 In humans consuming garden cress, watercress, or broccoli, the BITC-Lys, PEITC-Lys, and SFN-Lys were released from Alb after enzymatic digestion of the proteins and quantified by LC-MS/MS. This new analytical method will enable quantification of ITC adducts in blood proteins from large prospective studies investigating the role of dietary chemoprotective agents in cancer risk.
Figure 13.
Alb adducts of isothiocyanates269 found in vivo.
Figure 14.
LC-MS/MS analyses of Alb digests obtained from (A) mouse control, (B) mouse exposed to phenylethylisothiocyanate-(PEITC)-AcCys, (C) human exposed to watercress, and (D) the PEITC-Lys standard compound. The right panels show the chromatogram of the IS, PEITC-[13C615N2]Lys, which was added at the beginning of digestion procedure. PEITC-Lys and PEITC-[13C615N2]Lys were detected with the MRM transitions m/z 308.1/145.0 and 316.1/153.0, respectively.269
8. Adduct Formation with Therapeutic Drugs
Many therapeutic drugs of diverse structures and mechanisms of action undergo metabolism by Phase I or Phase II drug-metabolizing enzymes. Some of the pathways of metabolism can give rise to electrophilic, short-lived intermediates, which can covalently bind to various proteins (Figure 15–17).82,144,271,272
Figure 15.
Figure 17.
The covalent binding sites of therapeutic drugs to Alb are summarized in Supporting Information, Table S1. In contrast to the studies described in the former sections 6.1–6.8 and 7.1, most studies described in this section are qualitative. The detected adducts were not available as synthetic standards in order to perform a quantitation of the adduct levels.
8.1. β-Lactam Antibiotics
Allergenicity is a common response toward a wide-spectrum of antibiotics.273 The β-lactam antibiotics, such as amoxicillin (AX), are among the most widely prescribed drugs and frequently elicit allergic reactions.274,275 The covalent adduction of AX, other antibiotics, and drugs to proteins is considered a key process for the allergic response.274,275 In the case of AX, covalent modification occurs through nucleophilic attack of the β-lactam ring and adduction by the ε-amino group of Lys in proteins (Figure 15). This process gives rise to a haptenized penicilloyl–protein adduct, which can elicit an immune response.274−276 Alb was the major plasma protein found to react with AX in vitro.81 A comprehensive study showed that Lys190, Lys199, Lys351, Lys432, Lys541, and Lys545 (Table S1, Supporting Information) were the major sites of reaction of AX with Alb ex vivo in plasma.81,277 Interestingly, the sites of Lys adduction can vary widely between subjects treated with the structurally related β-lactam antibiotic benzylpenicillin and was found to form multiple adducts in vivo with Alb in human subjects at Lys190, Lys195, Lys432, and Lys541.278
8.2. Nonsteroidal Anti-inflammatory drugs (NSAIDS)
UDP-glucuronosyltransferases (UGTs) are important conjugation enzymes and catalyze the glucuronidation of numerous drugs, endobiotics, and procarcinogens.279 The UGT-mediated glucuronide (Gluc) conjugation of chemicals is generally considered as a mechanism of detoxication, and the Gluc conjugates are generally less biologically or chemically reactive than their parent compounds.85 However, many carboxylic acid containing drugs including NSAIDS are metabolized by UGTs to form β-1-O-acyl glucuronides (AGs), which are able to form adducts with nucleophiles in proteins and may contribute to toxicity of NSAIDS.276 Evidence for the covalent binding of AGs to Alb by MALDI TOF MS and by CID with a four-sector tandem mass spectrometer was provided by Ding et al.83,280 Tolmetin glucuronide (TG) was shown to bind to Alb via transacylation reactions to form adducts with Lys residues via displacement of the glucuronic acid moiety. Alternatively, a second mechanism of TG adduct formation involves migration of the acyl group to the 2-, 3-, or 4-hydroxyl groups of the sugar moiety; tautomerization of the pyranose ring to its aldose form; and condensation of the aldehyde group of the ring-opened tautomer with a lysine ε-amino group on Alb to form an imine (Figure 15). Lys195, Lys199, and Lys541 were the most abundant sites for TG adduct formation. The glycated imine adduct predominated over the transacylated adduct, when limiting amounts of TG were reacted with Alb.
Alb has been shown to be the major target for many AGs in plasma because of its abundance and the presence of multiple accessible nucleophilic amino acid side chains. The relative contributions of transacylated and glycated adducts vary as a function of the structures of the AGs. In humans, both transacylated and glycated adducts of patients taking diclofenac were reported to form at Lys195 and Lys199 residues, when assayed by multiple reaction monitoring with a QTRAP MS.281 Some NSAIDS containing the carboxylic acid moiety undergo bioactivation to form electrophilic acyl-coenzyme A thioester derivatives (acyl-CoAs); some of these conjugates exhibit greater reactivity with Alb than AGs to produce the same transacylated adducts.276,282,283
8.3. Acetaminophen
Acetaminophen is primarily metabolized by conjugating enzymes to form stable Gluc and sulfate conjugates.284 At therapeutic doses, acetaminophen is viewed as a safe drug. However, a small proportion of the drug is bioactivated by P450s to produce the highly reactive N-acetyl-p-benzoquinoneimine (NAPQI) intermediate,285 which is detoxicated by conjugation with glutathione (Figure 15). At elevated doses, NAPQI alkylates proteins via a Michael addition reaction with nucleophilic cysteine thiol groups and induces toxicity.30,31,286 Hoffman and co-workers initially reported that NAPQI adducted to bovine Alb via a thioether linkage formed between the Cys34 and the C-3 position of NAPQI.287 More recently, Damsten and colleagues developed a bioanalytical scheme to characterize the adducts formed between Alb and NAPQI in humans.116 The Cys34 residue was also identified as a major site of adduction with human Alb, and two regioisomer C*PF tripeptide NAPQI adducts were identified by TQ MS, following proteolysis of the Alb with pronase E. This method was then successfully applied to detect the adducts in subjects who had taken high levels of acetaminophen (Figure 16).116 The identity of the adduct was confirmed by its product ion spectrum (Figure 16). Cooke and colleagues showed that the single amino acid C*-NAPQI adduct could be recovered from Alb following digestion with Protease type XIV from Streptomyces griseus. An alternative approach employed pepsin to digest NAPQI-Alb adducts in the mouse and also showed that the Cys34 was a major site of adduct formation.117
Figure 16.
LC-MS/MS analysis of NAPQI-CPF adducts in human serum samples (MH+ at m/z 515.2) obtained after the pronase digest. Ion chromatograms of summed ions: m/z 152.1, 208.1, 225.1, 322.2, 323.2, and 351.2 in the control subject, patient 438, patient 440, and patient 444. The analyses of the 23-, 4-, and 25-h time point samples of patients 438, 440, and 444, respectively, are shown. The upper panel shows the product ion spectrum of the NAPQI-CPF adduct obtained from the reaction of synthetic NAPQI with synthetic tripeptide CPF. The product ion spectra of the NAPQI-CPF adducts corresponding to regioisomers of NAPQI-CPF (peak 1 and peak 2) in patient 444 are shown. Adapted with permission from ref (116). Copyright 2007 American Society for Pharmacology and Experimental Therapeutics.
8.4. Nevirapine (NVP)
NVP is a non-nucleoside reverse transcriptase inhibitor used against human immunodeficiency virus type-1 (HIV-1). Despite its clinical efficacy, NVP is associated with a variety of toxic responses that include hepatotoxicity and skin rash, which are thought to be attributed to the reactive metabolites that bind to proteins.288 One important bioactivation pathway involves the P450 oxidation of the 4-methyl substituent to 4-hydroxymethyl-NVP (12-OH-NVP). This metabolite has been proposed to either undergo further oxidation to produce the quinone methide or undergo an esterification mediated by sulfotransferases to produce 12-sulfoxyl-NVP; both are reactive metabolites and can adduct to macromolecules (Figure 17).22,288 The benzylic sulfate derivative, catalyzed by sulfotransferase 1A1*1 in the skin, is thought to be responsible for protein adduct formation and the development of skin rash in rodents and humans.22 In contrast, the majority of protein binding in rodent liver model appears to be attributed to the quinone methide, which is the direct P450 oxidation product of NVP.288 Single amino acid adducts were synthesized with 12-mesyloxy-NVP.289 Meng et al. examined the reactivity of 12-sulfoxy-NVP with Alb and found that at a 50-fold mol excess, the drug bound selectively to His146, His242, His338, and Cys34, but no Lys adducts were detected (Table S1 of the Supporting Information).143 When equimolar concentration of 12-sulfoxy-NVP was reacted with Alb, His146 was the primary site of adduct formation. Antunes and co-workers used a 100-fold mol excess of 12-mesyloxy-NVP and found adducts at Trp214, Cys34, and several Lys residues.143,197 The differences sites of Alb adduct formation reported by these two groups may be attributed to the differences in the structures of the reactive intermediates, reaction conditions, or methods of MS analysis. The tryptic peptide R145H*PYFYAPELLFFAK159 adducted with NVP at His146 was detected in four of the nine patients undergoing therapy with a dose of either 200 mg twice daily or 400 mg once daily for at least 2 weeks, by multiple reaction monitoring with the QTRAP MS.143 The reaction of 12-sulfoxyl-NVP with proteins may occur through direct nucleophilic substitution. An alternative mechanism for Alb adduct formation was proposed to occur via a concerted elimination–addition mechanism, where the His146 residue or some other nearby basic residues in Alb acted as a general base, abstracting the proton from amide and promoting the elimination of the sulfoxyl group to form a quinone methide intermediate (Figure 17).143
9. Endogenous Electrophiles
Protein modification by reactive carbonyl species, particularly those produced by peroxidation of polyunsaturated fatty acids, are thought to play an important role in the etiology and/or progression of cardiovascular, diabetic, and neurodegenerative diseases.3,290,291 Acrolein, among the 2-alkenals, is by far the strongest electrophile.290,292 Acrolein is a byproduct of lipid peroxidation292 and also found at high levels in tobacco smoke with levels ranging from 51–223 μg per cigarette.293 Early studies reported oxidative damage and carbonylation of proteins by the Western blotting technique, where oxidized proteins were derivatized with dinitrophenylhydrazine, separated by SDS–gel electrophoresis, and screened with antibodies against the dinitrophenyl groups.294 Investigations by Uchida and co-workers using mass spectrometry showed that the reaction of acrolein with protein Lys of bovine Alb results in β-substituted propanals (R-NH–CH2–CH2–CHO), to form the Nε-(3-formyl-3,4-dehydropiperidino)lysine adduct, and the stable adduct Nε-(3-methylpyridinium)lysine is prominently formed in oxidized low-density lipid proteins.196,295
PP show differences in susceptibility to oxidative modification: Alb was reported to be the main protein in plasma carbonylated in chronic kidney disease and hemodialysis patients, when assessed by Western blotting.296 Increased levels of carbonylated plasma proteins have been observed in smokers, with Alb being the major carbonylated protein in bronchoalveolar lavage fluid of elder smokers.13,297 Certain α,β-unsaturated aldehydes present in the mainstream cigarette smoke extract induced Alb carbonylation.297 Many α,β-unsaturated aldehydes, including acrolein and crotonaldehyde, selectively reacted with the sulfhydryl group of Cys, the imidazole group of His, and the ε-amino group of Lys (Table S1 of the Supporting Information).90,292,295,298 These electrophiles depleted the Alb Cys34 free thiol and markedly decreased the levels of free Lys residues by formation of covalent carbonyl adducts. Acrolein and crotonaldehyde were reported to form Michael adducts at Cys34, His39, Lys351, and Lys525, and acrolein formed a Schiff base with Lys541 and Lys545297 (Figure 18). The binding sites of endogenous compounds with Alb and the sites of oxidative stress in Alb have been summarized in Table S1 (Supporting Information).
Figure 18.
Identification of Michael adducts and Schiff base adducts of acrolein and 4HNE formed with Alb. Alb was treated with NaBH4 to stabilize adducts prior to proteolysis.5,89,299,344
Aldini et al. established a method employing TQ MS and the precursor ion scan mode to monitor Michael adducts formed at the Cys34 of Alb.5 Following treatment with NaBH4, the modified Alb was digested with a mixture of trypsin and chymotrypsin to produce LQQC*PF-adducts, which were monitored by precursor ion scan mode using unmodified y2 (m/z 263.1), b2 (m/z 242.1), and b3 (m/z 370.2) ions to identify the m/z of the adducted peptides [M + H]+. Further characterization of the reactivity of Alb toward 4-hydroxy-trans-2-nonenal (HNE), by ion trap mass spectrometry, also showed that Cys34 and Lys199 were the most reactive adduction sites of Alb in plasma reactions with limiting concentrations of HNE and resulted in the formation of a Michael and Schiff base adducts, respectively (Figure 17).90 Alb showed high scavenging activity for HNE. The rate constant of HNE trapping by albumin was 50.61 (1.89 M–1 s–1) and that of Cys34 (29.37 M–1 s–1) was an order of magnitude higher than that of GSH (3.81 M–1 s–1). In contrast to these findings, Szapacs and co-workers reported that the HNE formed a Michael adduct at His246 as the primary site for adduct formation with Alb.89 The low predicted pKa value His246 was proposed to account for its high reactivity toward HNE. The different preferred sites of HNE binding to Alb reported by these two laboratories may be attributed to different reactions conditions employed for study: Aldini conducted reactions of HNE with plasma, followed by Alb purification and proteolysis with a mixture of trypsin and chymotrypsin, whereas Szapacs reacted HNE with purified Alb and digested the modified protein with trypsin. The data-dependent scanning modes opposed to the targeted MS/MS conditions used in these two studies also may have detected different adducted peptides. By employing the precursor ion scanning mode, Witort and colleagues showed that acrolein was the main reactive carbonyl species that adducted to Cys34 of Alb in vivo, and the adduct level increased by almost 4-fold during ischemia-reperfusion injury in patients undergoing hepatectomy.299
Using an HRAMS Orbitrap, Goto reported that oxidation (+16 Da) occurred at the Tryp214 of the tryptic peptide sequence A213WAVAR218 and glycation (+162 Da) at the Lys525 residue of the tryptic peptide K525QTALVELVK534 of pooled Alb from healthy subjects.300
10. Challenges and Emerging Technologies in Measuring Albumin–Toxicant Adducts in Humans
10.1. Multiple Exposures and Adductomics
Urban pollution, tobacco smoke, diet, some medicines, pesticides, and endogenously produced electrophiles are thought to contribute to the etiology of cancer, cardiovascular, neurotoxicity, and other diseases.6,227,24,290 Thus, biomarkers of a large number of toxicants are required for risk assessment of global exposure.6,301 With the advances made in hyphenated mass spectrometric instrumentation over the past decade, particularly HRAMS with tandem and multistage scanning, and the advanced proteomic technologies, it is now possible to identify and locate the sites of xenobiotic adduction to Alb. Thus, there is considerable incentive to examine the reactivity of Alb with other environmental toxicants to assess human exposure to multiple classes of hazardous chemicals. Biomonitoring of protein adducts is part of the exposome initiative.302,303 However, techniques employing protein adducts to simultaneously measure exposure to multiple toxicants have not advanced rapidly mainly because of the shortage of analytical methods to assay protein adducts derived from low levels of chemical exposures in complex mixtures.154,304 Is it feasible to determine hundreds of Alb-adducts at the same time and create an exposome database? Unmodified, or post-translational modifications,305 glycation products,306,307 and oxidative products of proteins308−311 can interfere with the quantification of Alb adducts formed with environmental and occupational toxicants and xenobiotics. The levels of Alb-adducts of xenobiotics are usually 10- to 1000-fold lower than the levels of endogenous adducts, and many adducts formed with dietary and environmental carcinogens can occur at even lower levels. The digests of Alb-carcinogen adducts obtained by pronase or trypsin contain vast amounts of excess unmodified amino acids or peptides, which can result in ion suppression effects or contain isobaric chemicals that interfere with the LC-MS analysis.312 Therefore, selective enrichment procedures are required to isolate Alb-adducts for MS measurement. HRAMS instruments are expected to play an important role in Alb adductomics.
A number of the Alb adduct structures described in this perspective have been deduced by MS; however, the structures of some adducts are ambiguous. For rigorous analysis, the chemical identities of new adducts should be established by spectroscopic characterization that includes 1H NMR and 13C NMR, combined with mass spectral data, and other spectral criteria to fully support identification. The reliable identification and quantitation of adducts in humans require fully characterized standards and stable isotopically labeled internal standards to fulfill the requirements of validated measurements of toxicants in humans.
A number of Alb-adducts formed with drugs or endogenous electrophiles have been identified in humans.94,116,130,134,143,281,299 Many of these adducts were characterized as tryptic peptides, and the sites of adduction were determined by peptide sequencing using LC-MS/MS. The dosages of drugs exceeded 100 mg or greater, and levels of endogenous electrophiles in plasma can exceed micromolar concentrations.313 These high levels of chemical exposures greatly facilitated the analyses. In contrast, the exposures to environmental, tobacco, and dietary carcinogens often occur at amounts of μg or less per day, resulting in considerably lower levels of chemical modification of Alb. For example, the cooked meat carcinogen, PhIP, has been reported to form adduct(s) with Alb in humans at a level of ∼500 pg PhIP/g Alb or approximately ∼1.5 PhIP molecules bound per 10(7) molecules of albumin, based on accelerator mass spectrometry measurement.73 The measurement of Alb-carcinogen adducts at these low levels of modification remains a challenging endeavor even for the most sensitive of HRAMS instruments. In the case of the LQQC*PFEDHVK sulfinamide adduct of PhIP, the signal of response of the parent amine released by acid hydrolysis of adducted peptide was more than 10-fold superior to that of the intact peptide adduct, when assayed by LC-MS/MS.125,195 The relative sensitivities of peptide adduct responses are expected to be dependent on the basicity of the toxicant and the peptide sequences. Systematic studies have not examined the signals of response of peptide adducts versus the hydrolyzed carcinogen/toxicant products under ESI conditions, and such studies merit investigation for devising optimal approaches to measure Alb adducts in humans.
Thus far, the successes in measuring Alb-carcinogen adducts in humans have largely been with those adducts that are cleaved from Alb by acid or base treatment (i.e., Cys-BQ or BAP tetraols), or by the extensive digestion of Alb with a mixture of proteases to produce mono-amino acid adducts (AFB-Lys adducts). The physicochemical properties of these covalently adducted amino acids or carcinogen hydrolysis products are sufficiently distinct from nonmodified amino acids or peptides that selective enrichment procedures could be developed to isolate and assay these Alb adduction products. The use of trypsin or other specific proteases to digest Alb produces defined peptides where sites of the toxicant adduction can be precisely located by MS/MS sequencing. These types of analyses provide information about the sites of adduction to Alb, which are usually lost when digestions are done by pronase or acid/base hydrolysis, unless adducts are formed and retained to the solitary Cys34 or Tryp214 residues of Alb.
Proteases, such as trypsin, can produce long peptides such as the T3 tryptic peptide A21LVLIAFAQYLQQCPFEDHVK40, whereas pronase digestion yields mono-, di-, or tripetide Cys containing adducts. The logD values (pH dependent octanol – water partition coefficient) of modified and unmodified peptides are often very similar if the adducted chemical is small such as sulfur mustard: the maximum calculated (MarvinSketch, www.chemaxon.com) logD for S-[2-[(hydroxyethyl)thio]ethyl]Cys-Pro-Phe (HETE-CPF) is −1.97 at pH 5.5 versus the unmodified CPF at logD = −1.94. However, adducts formed with benzene and 4ABP (sulfonamide-adduct with sulfur of Cys) yield logD values of −0.17 and −0.12, respectively, at pH 5.5. For adducts of single amino acids, the influence of the adduct on the logD is even larger than the effect on a tri- or oligopeptide containing adducts. For Cys adducts formed with longer peptides such as the T3 peptide, the influence of the adduct on the logD values is considerably smaller. For example, for a sulfonamide adduct of 4-ABP with Cys of the T3-fragment the highest logD value is at −10.3 (pH 4.0 = pH with the maximum level of the logD), compared to the unmodified T3-fragment with a logD of −10.4 at pH 4.0. Thus, more facile enrichment and chromatographic separations of small molecular weight peptide adducts may be achieved than for the corresponding tryptic adducts, where the influence of the adduct on the logD is greatly diminished. For the analysis of MDI-Alb adducts in workers, the single amino acid adduct (MDI-Lys) released after pronase digestion can be detected at lower levels106 than the MDI-peptide fragment released after trypsin digestion.314 In the case of the Alb adduct of sulfur mustard, the tryptic T3 adducted peptide could not be found in human samples,258 whereas the C*PF sulfur mustard adduct, obtained by pronase digestion, was identified in human samples.315 Prolonged gradients may be required for complete resolution of adducts obtained from tryptic digests of Alb.93,154 Multiple strategies will be required for adduct enrichment and will be influenced by the structures of the toxicants, the chemical stabilities of the linkages, and the site(s) of adduction to Alb.
10.2. Subadductomics Approaches to Screen for Albumin Adducts
Subadductomic approaches to measure certain classes of carcinogens have already been achieved. For example, a dozen aromatic amines have been simultaneously monitored by their common site of adduction at the β-Cys93 residue of Hb in the form of the arylsulfinamide linkage,79,190 and a number of alkylating agent adducts formed at the N-terminal Val of Hb also can be measured.52,316 However, novel screening approaches are needed to biomonitor other classes of toxicant adducts formed with Alb. One approach under development by Rappaport and colleagues is the selective enrichment of peptide adducts formed at Cys34 of Alb by means of a thiol affinity resin.153 This technique removes the large excess of nonmodified mercaptalbumin from the tryptic digest, resulting in an enriched fraction of tryptic peptide adducts at the Cys34 of the T3 peptide A21LVLIAFAQYLQQC*PFEDHVK40. This enrichment procedure, followed by DDA with an Orbitrap, led to the detection of 43 features at T3, including Cys34 sulfinic and sulfonic acids, several mixed disulfides, and small molecular weight alkylating agents.93,94,154 The screening technique detected a presumed adduct of acrylonitrile, but it failed to detect T3 adducts of other prominent carcinogens formed in tobacco smoke such as acrylamide, benzene and butadiene, among others.94 Another enrichment approach has been to design a polyclonal anti-T3 antibody of Alb that could be employed to capture Cys34 adducts in tryptic digests of Alb.317 These enrichment methods and bioanalytical approaches are promising, but they require further improvements in LC-MS analyses and more comprehensive scanning methods, such as DIA, particularly for those adducts formed at low abundance. One possible lead may be to further digest the enriched T3 peptide adducts with Pronase to obtain smaller molecular weight peptide adducts, which may be more sensitive toward ESI and also provide characteristic fragment ions associated with the toxicant.
The Tyr138, His146, Lys199, Lys351, Tyr411, and Lys525 represent other nucleophilic hotspots of Alb and react with different classes of electrophiles. Antibodies raised against Alb peptides containing these nucleophilic sites could be used for enrichment of adducted peptides by immunoaffinity chromatography. Depending upon the specificity and affinity of the antibodies, the antibodies may be used to either enrich adducted peptides (if they are recognized) or deplete the digest of vast excess of nonmodified peptides to facilitate the analyses of adducts.
Biotinylated probes with different functional groups have been employed to enrich several different classes of adducts including enals.318 This approach may be exploited to examine specific classes of Alb adducts as another subadductome approach. Offline HPLC prepurification of Alb digests and the collection of specific regions of the chromatograms can be devised to enrich certain classes of peptide adducts prior to LC-MS. However, other techniques and chemistries must be devised to selectively isolate peptide adducts from multiple classes of toxicants.
10.3. Human Albumin Transgenic Mouse Model
Many studies have employed rodent models to investigate the reactivity of toxicants with Alb. However, the differences between the sequences of human and rodent serum Alb can result in the formation of different adducts or adducts with nonhomologous sequences, and thus, the rodent biomarker is not directly transferable to human studies. For example, the differences in sequence surrounding the Cys34 of human and mouse Alb lead to different sized peptides recovered by proteases. Recently, a human Alb-transgenic mouse model was created.319 Immunoblot and mass spectrometry analyses showed that the transgenic human Alb protein was a full-length, mature protein that coexisted with endogenous mouse Alb. Further improvement in this model in which only the human Alb gene is produced is under development. This “humanized” mouse model can be used to examine human Alb adduct formation in mice exposed to multiple common environmental, tobacco, dietary, medicinal, and other toxicants. Such studies could never be conducted in humans because of the potential toxicities of these complex mixture of chemicals. The humanized Alb mouse model represents a significant advance in screening tools for Alb adductomics biomarker research.
10.4. Identification of Unknown Adducts in Humans
The unambiguous identification of toxicants adducted to peptides is another challenge in protein adductomics. MS/MS sequencing of Alb peptides allows for identification of the peptide and location of the site of adduct formation. However, the CID of peptide adducts often does not result in appreciable cleavage of the toxicants and generation of their fragment ions. Therefore, the analyst only has the addition of the mass to the peptide as information to decipher the identity of the toxicant. The employment of multistage scanning (MSn) with ion trap instruments may be used for the spectral characterization of some types of adducts which contain basic functional ionizable groups.165,320 The use of high-energy collision dissociation can fragment the covalent links formed between some toxicants and Alb and provide additional mass spectral data to characterize the structure of the covalent linkages.165,195
Analytical challenges remain in the acquisition of high quality product ion spectra of peptide adducts present at low levels in a complex proteolytic digest. Programs such as Skyline,321 DIA Umpire,322 and others under development will be required to deconvolute spectral data to identify precursor and product ions of peptide adducts.155 However, the identification of unknown peptide–toxicant adducts at low abundance will be demanding, particularly because there are no databases available for this purpose.6,154 Chemical databases and molecular weights of masses added to peptides can be searched by online tools to facilitate the identification of putative adducted masses to Alb, including The National Institute of Standards (http://webbook.nist.gov/chemistry/); Unimod protein modifications for mass spectrometry (www.unimod.org); The Association for Biomedical Resource Facilities (https://abrf.org/delta-mass); The Human Metabolome Database (www.hmdb.ca/metabolites); PubChem (http://pubchem.ncbi.nlm.nih.gov); the US Environmental Protect Agency CompTox Dashboard (https://comptox.epa.gov/dashboard); the NIH LiverTox database (http://livertox.nih.gov/); m/z cloud Advance mass spectral database (https://www.mzcloud.org/); and Institute of Chemical Sciences and Engineering (http://www.chemcalc.org/). Ultimately, the proposed toxicants or their metabolites must be reacted with the peptides, and the resulting adducts sequenced by MS/MS and MSn, and coeluted with the Alb adducts formed in vivo for unequivocal identification.
11. Conclusions
Many carcinogen adducts formed with human Alb samples have been measured following chemical hydrolysis or pronase digestion of Alb. Such hydrolysis procedures often yield the released parent compounds, oxidized metabolites, or small molecular weight peptide adducts, which can be facilely isolated from the bulk of Alb and analyzed. Proteases such as trypsin or a combination of trypsin/chymotrypsin have been used to digest Alb adducts formed with many xenobiotics. The MS analyses have revealed that the sites of adduct formation with Alb are driven not only by the reactivity of nucleophilic amino acid sites and their accessibility but also by the noncovalent interactions of the toxicant within the different microenvironments of Alb, which guide the sites of toxicant adduction. As more sites of adduction of toxicants to Alb are characterized, a clearer understanding of the relationship between the toxicant structures and their preferred site of adduct formation with Alb may become apparent and facilitate the development of multiadductomic approaches to nucleophilic hot-spot sites within Alb. Such studies are underway with adducts formed at Cys34 of Alb.6,93,94 Exposome-based studies on Alb adducts will undoubtedly require the development of multiple types of bioanalytical schemes for digestion of Alb and enrichment of adducts, and their ensuing measurements by different MS instruments.
Glossary
Abbreviations
- AChE
acetylcholinesterase
- acyl-CoAs
acyl-coenzyme A thioester derivatives
- AFB
aflatoxin B1
- AFG
aflatoxin G1;
- AGs
β-1-O-acyl glucuronides
- Alb
albumin
- 4ABP
4-aminobiphenyl
- AαC
2-amino-9H-pyrido[2,3-b]indole
- atrazine
2-chloro-4-(ethylamino)-6-(isopropylamino)-s-triazine)
- APAP
acetaminophen
- AX
amoxicillin
- B
benzene
- BO
benzene oxide
- 1,2-BQ
1,2-benzoquinone
- 1,4-BQ
1,4-benzoquinone
- BAP
benzo[a]pyrene
- BPDE
BAP-diol epoxide
- anti-BPDE
r-7,t-8-dihydroxy-t-9,t-10-epoxy-7,8,9,10-tetrahydro-BAP
- 4CA
4-chloroaniline
- CID
collision induced dissociation
- DcBz
3,3′-dichlorobenzidine
- DDA
data dependent acquisition
- DIA
data-independent acquisition
- ESI-MS
electrospray ionization mass spectrometry
- ELISA
enzyme-linked immunosorbent assay
- Gluc
glucuronide
- GSH
glutathione
- HAA
heterocyclic aromatic amines
- Hb
hemoglobin
- HNE
4-hydroxy-trans-2-nonenal
- HETE-CPF
S-[2-[(hydroxyethyl)thio]ethyl]Cys-Pro-Phe
- HRAMS
high resolution accurate mass spectrometry
- IDA
information dependent acquisition
- IDMS
isotope dilution mass spectrometry
- IT
ion trap
- LC-MS
liquid chromatography-mass spectrometry
- MALDI
matrix-assisted laser desorption/ionization
- m/z
mass-to-charge
- MSn
multistage scanning
- MDI
4,4′-methylenediphenyl diisocyanate
- MOCA
4,4′-methylenebis(2-chloroaniline)
- MWC
molecular weight cut off
- 2MA
2-methylaniline
- NPO
naphthalene-1,2-oxide
- NPOS1
1-sulfanyl-dihydronaphthalene-2-ol
- NPOS2
2-sulfanyl-dihydronaphthalene-1-ol
- NPO1-S-TFA
S-naphthalen-1-yl trifluoroethanethioate
- NPO2-S-TFA
S-naphthalen-2-yl trifluoroethanethioate
- 1,2-NPQ-S-TFA
4-[(trifluoroacetyl)sulfanyl]naphthalene-1,2-diyl bis(trifluoroacetate
- 1,4-NPQ-S-TFA
2-[(trifluoroacetyl)sulfanyl]naphthalene-1,4-diyl bis(trifluoroacetate)
- 1,2-NPQ
1,2-naphthoquinone
- 1,4-NPQ
1,4-naphthoquinone
- NSAIDS
nonsteroidal anti-inflammatory drugs
- Nvp
Nevirapine
- OPs
organophosphates
- P450
cytochrome P450
- PAHs
polycyclic aromatic hydrocarbons
- PBI
protein binding index
- 1-PE
1-phenylethanol
- 2-PE
2-phenylethanol
- PP
plasma proteins
- NVP
nevirapine
- PAHs
polycyclic aromatic hydrocarbons
- PEITC
phenylethyl isothiocyanate
- PhIP
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- QTOF MS
quadrupole time-of-flight mass spectrometry
- QTRAP MS
hybrid triple quadrupole/linear ion trap mass spectrometer
- IQ
2-amino-3-methylimidazo[4,5-f]quinoline
- IT
quadrupole ion trap (IT)
- SRM
selected reaction monitoring
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SPC
S-phenylcysteine
- SG
styrene glycol
- SO
styrene-7,8-oxide
- TOF MS
time-of-flight mass spectrometry
- TDI
toluene diisocyanate
- 24TDI
2,4-toluene diisocyanate
- 26TDI
2,6-toluene diisocyanate
- TG
tolmetin glucuronide
- THP
2,4,5-trihydroxy-pyrrolidine
- TQ MS
triple quadrupole mass spectrometry
- UGTs
UDP-glucuronosyltransferases
Biographies
Professor Dr. Gabriele Sabbioni obtained his Ph.D. in organic chemistry at the University of Bern. After his Ph.D., he worked at the University of Toronto, Massachusetts Institute of Technology, University of Würzburg, University of Munich, and Tulane University. Presently, he is working at the Institute of Environmental and Occupational Toxicology in Airolo and at the Alpine Institute of Chemistry and Toxicology in Olivone (both in Switzerland), and he is teaching at the University of Munich. He performed QSAR studies of arylamines and nitroarenes. He synthesized and analyzed several DNA and protein adducts of xenobiotics and used them in biomonitoring studies.
Dr. Rob Turesky, Ph.D., is a Professor in the Department of Medicinal Chemistry, and Director of the Masonic Cancer Center’s Analytical Biochemistry shared resource, a mass spectrometry facility devoted to the cancer and chemoprevention programs at the University of Minnesota. His research is focused on the biochemical toxicology of hazardous chemicals in the environment, diet, and tobacco. The laboratory characterizes pathways of metabolism of genotoxicants and establishes biomarkers, including metabolites, protein-, and DNA adducts. Mass spectrometric methods are applied to measure these biomarkers in molecular epidemiology studies that seek to understand the role of chemical exposures in the origin of cancer.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.chemrestox.6b00366.
In vitro modification of human serum albumin (Alb) with chemicals and determination of the reacting amino acid by MS in tryptic digests and Alb adducts of naphthalene (PDF)
The research conducted in the Turesky laboratory was supported by Grants R01CA122320, RO1CA134700, and R01CA134700-03S1 from the National Cancer Institute and the Family Smoking Prevention and Tobacco Control Act, and mass spectrometry was carried out in the Analytical Biochemistry Shared Resource of the Masonic Cancer Center, University of Minnesota, funded in part by Cancer Center Support Grant CA-077598.
The authors declare no competing financial interest.
Supplementary Material
References
- Peters T., Jr. (1996) All about Albumin. Biochemistry, Genetics, and Medical applications, Academic Press, San Diego, CA. [Google Scholar]
- Peters T. Jr. (1985) Serum albumin. Adv. Protein Chem. 37, 161–245. 10.1016/S0065-3233(08)60065-0. [DOI] [PubMed] [Google Scholar]
- Colombo G.; Clerici M.; Giustarini D.; Rossi R.; Milzani A.; Dalle-Donne I. (2012) Redox albuminomics: oxidized albumin in human diseases. Antioxid. Redox Signaling 17, 1515–1527. 10.1089/ars.2012.4702. [DOI] [PubMed] [Google Scholar]
- Stewart A. J.; Blindauer C. A.; Berezenko S.; Sleep D.; Tooth D.; Sadler P. J. (2005) Role of Tyr84 in controlling the reactivity of Cys34 of human albumin. FEBS J. 272, 353–362. 10.1111/j.1742-4658.2004.04474.x. [DOI] [PubMed] [Google Scholar]
- Aldini G.; Regazzoni L.; Orioli M.; Rimoldi I.; Facino R. M.; Carini M. (2008) A tandem MS precursor-ion scan approach to identify variable covalent modification of albumin Cys34: a new tool for studying vascular carbonylation. J. Mass Spectrom. 43, 1470–1481. 10.1002/jms.1419. [DOI] [PubMed] [Google Scholar]
- Rappaport S. M.; Li H.; Grigoryan H.; Funk W. E.; Williams E. R. (2012) Adductomics: Characterizing exposures to reactive electrophiles. Toxicol. Lett. 213, 83–90. 10.1016/j.toxlet.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regazzoni L.; Del Vecchio L.; Altomare A.; Yeum K. J.; Cusi D.; Locatelli F.; Carini M.; Aldini G. (2013) Human serum albumin cysteinylation is increased in end stage renal disease patients and reduced by hemodialysis: mass spectrometry studies. Free Radical Res. 47, 172–180. 10.3109/10715762.2012.756139. [DOI] [PubMed] [Google Scholar]
- Kleinova M.; Belgacem O.; Pock K.; Rizzi A.; Buchacher A.; Allmaier G. (2005) Characterization of cysteinylation of pharmaceutical-grade human serum albumin by electrospray ionization mass spectrometry and low-energy collision-induced dissociation tandem mass spectrometry. Rapid Commun. Mass Spectrom. 19, 2965–2973. 10.1002/rcm.2154. [DOI] [PubMed] [Google Scholar]
- Beck J. L.; Ambahera S.; Yong S. R.; Sheil M. M.; de J. J.; Ralph S. F. (2004) Direct observation of covalent adducts with Cys34 of human serum albumin using mass spectrometry. Anal. Biochem. 325, 326–336. 10.1016/j.ab.2003.10.041. [DOI] [PubMed] [Google Scholar]
- Bar-Or D.; Bar-Or R.; Rael L. T.; Gardner D. K.; Slone D. S.; Craun M. L. (2005) Heterogeneity and oxidation status of commercial human albumin preparations in clinical use. Crit. Care Med. 33, 1638–1641. 10.1097/01.CCM.0000169876.14858.91. [DOI] [PubMed] [Google Scholar]
- Kawakami A.; Kubota K.; Yamada N.; Tagami U.; Takehana K.; Sonaka I.; Suzuki E.; Hirayama K. (2006) Identification and characterization of oxidized human serum albumin. A slight structural change impairs its ligand-binding and antioxidant functions. FEBS J. 273, 3346–3357. 10.1111/j.1742-4658.2006.05341.x. [DOI] [PubMed] [Google Scholar]
- Carballal S.; Radi R.; Kirk M. C.; Barnes S.; Freeman B. A.; Alvarez B. (2003) Sulfenic acid formation in human serum albumin by hydrogen peroxide and peroxynitrite. Biochemistry 42, 9906–9914. 10.1021/bi027434m. [DOI] [PubMed] [Google Scholar]
- Fanali G.; di Masi A.; Trezza V.; Marino M.; Fasano M.; Ascenzi P. (2012) Human serum albumin: from bench to bedside. Mol. Aspects Med. 33, 209–290. 10.1016/j.mam.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Bal W.; Sokolowska M.; Kurowska E.; Faller P. (2013) Binding of transition metal ions to albumin: sites, affinities and rates. Biochim. Biophys. Acta, Gen. Subj. 1830, 5444–5455. 10.1016/j.bbagen.2013.06.018. [DOI] [PubMed] [Google Scholar]
- Stamler J. S.; Jaraki O.; Osborne J.; Simon D. I.; Keaney J.; Vita J.; Singel D.; Valeri C. R.; Loscalzo J. (1992) Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc. Natl. Acad. Sci. U. S. A. 89, 7674–7677. 10.1073/pnas.89.16.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X. M.; Carter D. C. (1992) Atomic structure and chemistry of human serum albumin. Nature 358, 209–215. 10.1038/358209a0. [DOI] [PubMed] [Google Scholar]
- Sugio S.; Kashima A.; Mochizuki S.; Noda M.; Kobayashi K. (1999) Crystal structure of human serum albumin at 2.5 A resolution. Protein Eng., Des. Sel. 12, 439–446. 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- Ghuman J.; Zunszain P. A.; Petitpas I.; Bhattacharya A. A.; Otagiri M.; Curry S. (2005) Structural basis of the drug-binding specificity of human serum albumin. J. Mol. Biol. 353, 38–52. 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- Sudlow G.; Birkett D. J.; Wade D. N. (1975) The characterization of two specific drug binding sites on human serum albumin. Mol. Pharmacol. 11, 824–832. [PubMed] [Google Scholar]
- Skipper P. L.; Obiedzinski M. W.; Tannenbaum S. R.; Miller D. W.; Mitchum R. K.; Kadlubar F. F. (1985) Identification of the major serum albumin adduct formed by 4-aminobiphenyl in vivo in rats. Cancer Res. 45, 5122–5127. [PubMed] [Google Scholar]
- Peters T. Jr. (1970) Serum albumin. Adv. Clin. Chem. 13, 37–111. 10.1016/S0065-2423(08)60385-6. [DOI] [PubMed] [Google Scholar]
- Sharma A. M.; Novalen M.; Tanino T.; Uetrecht J. P. (2013) 12-OH-nevirapine sulfate, formed in the skin, is responsible for nevirapine-induced skin rash. Chem. Res. Toxicol. 26, 817–827. 10.1021/tx400098z. [DOI] [PubMed] [Google Scholar]
- Baillie T. A. (2009) Approaches to the assessment of stable and chemically reactive drug metabolites in early clinical trials. Chem. Res. Toxicol. 22, 263–266. 10.1021/tx800439k. [DOI] [PubMed] [Google Scholar]
- Loeb L. A.; Harris C. C. (2008) Advances in chemical carcinogenesis: a historical review and prospective. Cancer Res. 68, 6863–6872. 10.1158/0008-5472.CAN-08-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. A. (1970) Carcinogenesis by chemicals: an overview--G. H. A. Clowes memorial lecture. Cancer Res. 30, 559–576. [PubMed] [Google Scholar]
- Miller E. C. (1978) Some current perspectives on chemical carcinogenesis in humans and experimental animals: Presidential address. Cancer Res. 38, 1479–1496. [PubMed] [Google Scholar]
- Boyland E. (1986) The metabolism of foreign compounds and the induction of cancer. Xenobiotica 16, 899–913. 10.3109/00498258609038973. [DOI] [PubMed] [Google Scholar]
- Dipple A.; Lawley P. D.; Brookes P. (1968) Theory of tumour initiation by chemical carcinogens: dependence of activity on structure of ultimate carcinogen. Eur. J. Cancer 4, 493–506. 10.1016/0014-2964(68)90005-4. [DOI] [PubMed] [Google Scholar]
- Potter W. Z.; Davis D. C.; Mitchell J. R.; Jollow D. J.; Gillette J. R.; Brodie B. B. (1973) Acetaminophen-induced hepatic necrosis. 3. Cytochrome P-450-mediated covalent binding in vitro. J. Pharmacol. Exp. Ther. 187, 203–210. [PubMed] [Google Scholar]
- Jollow D. J.; Mitchell J. R.; Potter W. Z.; Davis D. C.; Gillette J. R.; Brodie B. B. (1973) Acetaminophen-induced hepatic necrosis. II. Role of covalent binding in vivo. J. Pharmacol. Exp. Ther. 187, 195–202. [PubMed] [Google Scholar]
- Mitchell J. R.; Jollow D. J.; Potter W. Z.; Davis D. C.; Gillette J. R.; Brodie B. B. (1973) Acetaminophen-induced hepatic necrosis. I. Role of drug metabolism. J. Pharmacol. Exp. Ther. 187, 185–194. [PubMed] [Google Scholar]
- Guengerich F. P.; Shimada T. (1991) Oxidation of toxic and carcinogenic chemicals by human cytochrome P-450 enzymes. Chem. Res. Toxicol. 4, 391–407. 10.1021/tx00022a001. [DOI] [PubMed] [Google Scholar]
- Guengerich F. P. (2001) Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem. Res. Toxicol. 14, 611–650. 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- van Welie R. T.; van Dijck R. G.; Vermeulen N. P.; van Sittert N. J. (1992) Mercapturic acids, protein adducts, and DNA adducts as biomarkers of electrophilic chemicals. Crit. Rev. Toxicol. 22, 271–306. 10.3109/10408449209146310. [DOI] [PubMed] [Google Scholar]
- Kadlubar F. F.; Miller J. A.; Miller E. C. (1976) Microsomal N-oxidation of the hepatocarcinogen N-methyl-4-aminoazobenzene and the reactivity of N-hydroxy-N-methyl-4-aminoazobenzene. Cancer Res. 36, 1196–1206. [PubMed] [Google Scholar]
- Henderson R. F.; Bechtold W. E.; Bond J. A.; Sun J. D. (1989) The use of biological markers in toxicology. Crit. Rev. Toxicol. 20, 65–82. 10.3109/10408448909017904. [DOI] [PubMed] [Google Scholar]
- Metzleb M.; Neumann H. G. (1971) [Importance of chemico-biological interactions for the toxic and carcinogenic effect of aromatic amines. IV. Metabolic patterns of trans-4-demethylaminostilbene, cis-4-dimethylaminostilbene and 4-dimethylaminobibenzyl in liver, kidney and excretion products of the rat]. Z. Krebsforsch. Klin. Onkol. 76, 16–39. 10.1007/BF00304284. [DOI] [PubMed] [Google Scholar]
- Beland F. A.; Beranek D. T.; Dooley K. L.; Heflich R. H.; Kadlubar F. F. (1983) Arylamine-DNA adducts in vitro and in vivo: their role in bacterial mutagenesis and urinary bladder carcinogenesis. Environ. Health Perspect. 49, 125–134. 10.1289/ehp.8349125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beland F. A.; Kadlubar F. F. (1985) Formation and persistence of arylamine DNA adducts in vivo. Environ. Health Perspect. 62, 19–30. 10.1289/ehp.856219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer P. B.; Neumann H.-G.; Henschler D. (1987) Estimation of exposure of man to substances reacting covalently with macromolecules. Arch. Toxicol. 60, 251–260. 10.1007/BF01234663. [DOI] [PubMed] [Google Scholar]
- Calleman C. J.; Ehrenberg L.; Jansson B.; Osterman-Golkar S.; Segerback D.; Svensson K.; Wachtmeister C. A. (1978) Monitoring and risk assessment by means of alkyl groups in hemoglobin in persons occupationally exposed to ethylene oxide. J. Environ. Pathol. Toxicol. 2, 427–442. [PubMed] [Google Scholar]
- Skipper P. L.; Tannenbaum S. R. (1990) Protein adducts in the molecular dosimetry of chemical carcinogens. Carcinogenesis 11, 507–518. 10.1093/carcin/11.4.507. [DOI] [PubMed] [Google Scholar]
- Neumann H. G. (1984) Analysis of hemoglobin as a dose monitor for alkylating and arylating agents. Arch. Toxicol. 56, 1–6. 10.1007/BF00316343. [DOI] [PubMed] [Google Scholar]
- Gillette J. R. (1974) A perspective on the role of chemically reactive metabolites of foreign compounds in toxicity. II. Alterations in the kinetics of covalent binding. Biochem. Pharmacol. 23, 2927–2938. 10.1016/0006-2952(74)90267-6. [DOI] [PubMed] [Google Scholar]
- Baillie T. A. (2006) Future of toxicology-metabolic activation and drug design: challenges and opportunities in chemical toxicology. Chem. Res. Toxicol. 19, 889–893. 10.1021/tx060062o. [DOI] [PubMed] [Google Scholar]
- Park B. K.; Boobis A.; Clarke S.; Goldring C. E.; Jones D.; Kenna J. G.; Lambert C.; Laverty H. G.; Naisbitt D. J.; Nelson S.; Nicoll-Griffith D. A.; Obach R. S.; Routledge P.; Smith D. A.; Tweedie D. J.; Vermeulen N.; Williams D. P.; Wilson I. D.; Baillie T. A. (2011) Managing the challenge of chemically reactive metabolites in drug development. Nat. Rev. Drug Discovery 10, 292–306. 10.1038/nrd3408. [DOI] [PubMed] [Google Scholar]
- Baillie T. A. (2016) Chemically Reactive Versus Stable Drug Metabolites: Role in Adverse Drug Reactions. In RSC Drug Discovery Series 202–226. 10.1039/9781782622376-00202. [DOI] [Google Scholar]
- Commission of the European Communities, International Programme on Chemical Safety, World Health Organization Regional Ofice for Europe, Institute of Occupational Health (Finland) (1988) Indicators for Assessing Exposure and Biological Effects of Genotoxic Chemicals: Consensus and Technical Reports, Proceedings of an International Workshop on Indicators in Human Biological Materials for Assessing Exposure to and/or Biological Effects of Genotoxic Chemicals, Luxembourg, 6−9 July 1987, EUR 11642 EN, Office for Official Publications of the European Communities, Luxembourg, 1988; pp 1−191. [Google Scholar]
- Balbo S.; Turesky R. J.; Villalta P. W. (2014) DNA Adductomics. Chem. Res. Toxicol. 27, 356–366. 10.1021/tx4004352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethrenberg L.; Hiesche K. D.; Osterman-Golkar S.; Wenneberg I. (1974) Evaluation of genetic risks of alkylating agents: tissue doses in the mouse from air contaminated with ethylene oxide. Mutat. Res., Fundam. Mol. Mech. Mutagen. 24, 83–103. 10.1016/0027-5107(74)90123-7. [DOI] [PubMed] [Google Scholar]
- Wogan G. N.; Gorelick N. J. (1985) Chemical and biochemical dosimetry of exposure to genotoxic chemicals. Environ. Health Perspect. 62, 5–18. 10.1289/ehp.85625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornqvist M.; Fred C.; Haglund J.; Helleberg H.; Paulsson B.; Rydberg P. (2002) Protein adducts: quantitative and qualitative aspects of their formation, analysis and applications. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 778, 279–308. 10.1016/S1570-0232(02)00172-1. [DOI] [PubMed] [Google Scholar]
- Eyer P., and Gallemann D. (1996) Reactions of Nitrosoarenes with SH Groups, In The Chemistry of Amino, Nitroso, Nitro and Related Groups, pp 999–1039, John Wiley & Sons, Ltd., New York. [Google Scholar]
- Kiese M. (1974) Methemoglobinemia: A Comprehensive Treatise, CRC Press, Inc., Cleveland, OH. [Google Scholar]
- Pereira M. A.; Lin L. H.; Chang L. W. (1981) Dose-dependency of 2-acetylaminofluorene binding to liver DNA and hemoglobin in mice and rats. Toxicol. Appl. Pharmacol. 60, 472–478. 10.1016/0041-008X(81)90333-1. [DOI] [PubMed] [Google Scholar]
- Green L. C.; Skipper P. L.; Turesky R. J.; Bryant M. S.; Tannenbaum S. R. (1984) In vivo dosimetry of 4-aminobiphenyl in rats via a cysteine adduct in hemoglobin. Cancer Res. 44, 4254–4259. [PubMed] [Google Scholar]
- Skipper P. L.; Peng X.; SooHoo C. K.; Tannenbaum S. R. (1994) Protein adducts as biomarkers of human carcinogen exposure. Drug Metab. Rev. 26, 111–124. 10.3109/03602539409029787. [DOI] [PubMed] [Google Scholar]
- Skipper P. L.; Tannenbaum S. R.; Ross R. K.; Yu M. C. (2003) Nonsmoking-related arylamine exposure and bladder cancer risk. Cancer Epidemiol. Biomarkers Prev. 12, 503–507. [PubMed] [Google Scholar]
- Neumann H. G.; Albrecht O.; van D. C.; Zwirner-Baier I. (1995) Macromolecular adducts caused by environmental chemicals. Clin. Chem. Lab. Med. 41, 1835–1840. [PubMed] [Google Scholar]
- Ringe D.; Turesky R. J.; Skipper P. L.; Tannenbaum S. R. (1988) Structure of the single stable hemoglobin adduct formed by 4-aminobiphenyl in vivo. Chem. Res. Toxicol. 1, 22–24. 10.1021/tx00001a003. [DOI] [PubMed] [Google Scholar]
- Albrecht W.; Neumann H.-G. (1985) Biomonitoring of aniline and nitrobenzene. Arch. Toxicol. 57, 1–5. 10.1007/BF00286566. [DOI] [PubMed] [Google Scholar]
- Sabbioni G.; Jones C. R. (2002) Biomonitoring of arylamines and nitroarenes. Biomarkers 7, 347–421. 10.1080/13547500210147253. [DOI] [PubMed] [Google Scholar]
- Turesky R. J.; Le Marchand L. (2011) Metabolism and biomarkers of heterocyclic aromatic amines in molecular epidemiology studies: lessons learned from aromatic amines. Chem. Res. Toxicol. 24, 1169–1214. 10.1021/tx200135s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler T. W.; Roebuck B. D.; Wogan G. N.; Groopman J. D. (2011) Aflatoxin: a 50-year odyssey of mechanistic and translational toxicology. Toxicol. Sci. 120 (Suppl 1), S28–48. 10.1093/toxsci/kfq283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbitt B. F.; O’Kelly J.; Sargeant K.; Sheridan A. (1962) Aspergillus flavus and turkey X disease. Toxic metabolites of Aspergillus flavus. Nature 195, 1062–1063. 10.1038/1951062a0. [DOI] [PubMed] [Google Scholar]
- Buechi G.; Foulkes D. M.; Kurono M.; Mitchell G. F.; Schneider R. S. (1967) Total synthesis of racemic aflatoxin B1. J. Am. Chem. Soc. 89, 6745–6753. 10.1021/ja01001a062. [DOI] [PubMed] [Google Scholar]
- Sporn M. B.; Dingman W.; Phelps H. L.; Wogan G. N. (1966) Aflatoxin B1: Binding to DNA in vitro and alteration of RNA metabolism in vivo. Science 151, 1539–1541. 10.1126/science.151.3717.1539. [DOI] [PubMed] [Google Scholar]
- Busby W. F. Jr.; Wogan G. N. (1984) Aflatoxins. ACS Monogr. 182, 945–1136. [Google Scholar]
- Liebler D. C. (2002) Proteomic approaches to characterize protein modifications: new tools to study the effects of environmental exposures. Environ. Health Perspect. 110 (Suppl 1), 3–9. 10.1289/ehp.02110s113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillouzo A.; Morel F.; Langouët S.; Maheo K.; Rissel M. (1997) Use of hepatocyte cultures for the study of hepatotoxic compounds. J. Hepatol. 26 (Suppl 2), 73–80. 10.1016/S0168-8278(97)80499-0. [DOI] [PubMed] [Google Scholar]
- Sabbioni G.; Skipper P. L.; Buchi G.; Tannenbaum S. R. (1987) Isolation and characterization of the major serum albumin adduct formed by aflatoxin B1 in vivo in rats. Carcinogenesis 8, 819–824. 10.1093/carcin/8.6.819. [DOI] [PubMed] [Google Scholar]
- Gan L. S.; Skipper P. L.; Peng X. C.; Groopman J. D.; Chen J. S.; Wogan G. N.; Tannenbaum S. R. (1988) Serum albumin adducts in the molecular epidemiology of aflatoxin carcinogenesis: correlation with aflatoxin B1 intake and urinary excretion of aflatoxin M1. Carcinogenesis 9, 1323–1325. 10.1093/carcin/9.7.1323. [DOI] [PubMed] [Google Scholar]
- Dingley K. H.; Curtis K. D.; Nowell S.; Felton J. S.; Lang N. P.; Turteltaub K. W. (1999) DNA and protein adduct formation in the colon and blood of humans after exposure to a dietary-relevant dose of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Cancer Epidemiol. Biomarkers Prev. 8, 507–512. [PubMed] [Google Scholar]
- Rubino F. M.; Pitton M.; Di Fabio D.; Colombi A. (2009) Toward an ″omic″ physiopathology of reactive chemicals: thirty years of mass spectrometric study of the protein adducts with endogenous and xenobiotic compounds. Mass Spectrom. Rev. 28, 725–784. 10.1002/mas.20207. [DOI] [PubMed] [Google Scholar]
- Yang X.; Bartlett M. G. (2016) Identification of protein adduction using mass spectrometry: Protein adducts as biomarkers and predictors of toxicity mechanisms. Rapid Commun. Mass Spectrom. 30, 652–664. 10.1002/rcm.7462. [DOI] [PubMed] [Google Scholar]
- Clewell H. J.; Tan Y. M.; Campbell J. L.; Andersen M. E. (2008) Quantitative interpretation of human biomonitoring data. Toxicol. Appl. Pharmacol. 231, 122–133. 10.1016/j.taap.2008.04.021. [DOI] [PubMed] [Google Scholar]
- Hays S. M.; Aylward L. L.; LaKind J. S.; Bartels M. J.; Barton H. A.; Boogaard P. J.; Brunk C.; DiZio S.; Dourson M.; Goldstein D. A.; Lipscomb J.; Kilpatrick M. E.; Krewski D.; Krishnan K.; Nordberg M.; Okino M.; Tan Y. M.; Viau C.; Yager J. W.; Biomonitoring Equivalents Expert W. (2008) Guidelines for the derivation of Biomonitoring Equivalents: report from the Biomonitoring Equivalents Expert Workshop. Regul. Toxicol. Pharmacol. 51, S4–15. 10.1016/j.yrtph.2008.05.004. [DOI] [PubMed] [Google Scholar]
- LaKind J. S.; Aylward L. L.; Brunk C.; DiZio S.; Dourson M.; Goldstein D. A.; Kilpatrick M. E.; Krewski D.; Bartels M. J.; Barton H. A.; Boogaard P. J.; Lipscomb J.; Krishnan K.; Nordberg M.; Okino M.; Tan Y. M.; Viau C.; Yager J. W.; Hays S. M.; Biomonitoring Equivalents Expert W. (2008) Guidelines for the communication of Biomonitoring Equivalents: report from the Biomonitoring Equivalents Expert Workshop. Regul. Toxicol. Pharmacol. 51, S16–26. 10.1016/j.yrtph.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Tannenbaum S. R.; Skipper P. L.; Wishnok J. S.; Stillwell W. G.; Day B. W.; Taghizadeh K. (1993) Characterization of various classes of protein adducts. Environ. Health Perspect. 99, 51–55. 10.1289/ehp.939951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skipper P. L. (1996) Influence of tertiary structure on nucleophilic substitution reactions of proteins. Chem. Res. Toxicol. 9, 918–923. 10.1021/tx960028h. [DOI] [PubMed] [Google Scholar]
- Ariza A.; Garzon D.; Abanades D. R.; de los Ríos V.; Vistoli G.; Torres M. J.; Carini M.; Aldini G.; Perez-Sala D. (2012) Protein haptenation by amoxicillin: high resolution mass spectrometry analysis and identification of target proteins in serum. J. Proteomics 77, 504–520. 10.1016/j.jprot.2012.09.030. [DOI] [PubMed] [Google Scholar]
- Stepan A. F.; Walker D. P.; Bauman J.; Price D. A.; Baillie T. A.; Kalgutkar A. S.; Aleo M. D. (2011) Structural alert/reactive metabolite concept as applied in medicinal chemistry to mitigate the risk of idiosyncratic drug toxicity: a perspective based on the critical examination of trends in the top 200 drugs marketed in the United States. Chem. Res. Toxicol. 24, 1345–1410. 10.1021/tx200168d. [DOI] [PubMed] [Google Scholar]
- Ding A.; Ojingwa J. C.; McDonagh A. F.; Burlingame A. L.; Benet L. Z. (1993) Evidence for covalent binding of acyl glucuronides to serum albumin via an imine mechanism as revealed by tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 90, 3797–3801. 10.1073/pnas.90.9.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benet L. Z.; Spahn-Langguth H.; Iwakawa S.; Volland C.; Mizuma T.; Mayer S.; Mutschler E.; Lin E. T. (1993) Predictability of the covalent binding of acidic drugs in man. Life Sci. 53, PL141–146. 10.1016/0024-3205(93)90279-C. [DOI] [PubMed] [Google Scholar]
- Ritter J. K. (2000) Roles of glucuronidation and UDP-glucuronosyltransferases in xenobiotic bioactivation reactions. Chem.-Biol. Interact. 129, 171–193. 10.1016/S0009-2797(00)00198-8. [DOI] [PubMed] [Google Scholar]
- Baertschi S. W.; Raney K. D.; Stone M. P.; Harris T. M. (1988) Preparation of the 8,9-epoxide of the mycotoxin aflatoxin B1: the ultimate carcinogenic species. J. Am. Chem. Soc. 110, 7929–7931. 10.1021/ja00231a083. [DOI] [Google Scholar]
- Westra J. G. (1981) A rapid and simple synthesis of reactive metabolites of carcinogenic aromatic amines in high yield. Carcinogenesis 2, 355–357. 10.1021/tx00027a002. [DOI] [PubMed] [Google Scholar]
- Turesky R. J.; Skipper P. L.; Tannenbaum S. R. (1987) Binding of 2-amino-3-methylimidazo[4,5-f]quinoline to hemoglobin and albumin in vivo in the rat. Identification of an adduct suitable for dosimetry. Carcinogenesis 8, 1537–1542. 10.1093/carcin/8.10.1537. [DOI] [PubMed] [Google Scholar]
- Szapacs M. E.; Riggins J. N.; Zimmerman L. J.; Liebler D. C. (2006) Covalent adduction of human serum albumin by 4-hydroxy-2-nonenal: kinetic analysis of competing alkylation reactions. Biochemistry 45, 10521–10528. 10.1021/bi060535q. [DOI] [PubMed] [Google Scholar]
- Aldini G.; Vistoli G.; Regazzoni L.; Gamberoni L.; Facino R. M.; Yamaguchi S.; Uchida K.; Carini M. (2008) Albumin is the main nucleophilic target of human plasma: a protective role against pro-atherogenic electrophilic reactive carbonyl species?. Chem. Res. Toxicol. 21, 824–835. 10.1021/tx700349r. [DOI] [PubMed] [Google Scholar]
- Peng L.; Dasari S.; Tabb D. L.; Turesky R. J. (2012) Mapping serum albumin adducts of the food-borne carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by data-dependent tandem mass spectrometry. Chem. Res. Toxicol. 25, 2179–2193. 10.1021/tx300253j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo P.; Wisnewski A. V.; Lummus Z.; Cartier A.; Malo J. L.; Boulet L. P.; Bernstein D. I. (2007) Diisocyanate conjugate and immunoassay characteristics influence detection of specific antibodies in HDI-exposed workers. Clin. Exp. Allergy 37, 1095–1102. 10.1111/j.1365-2222.2007.02745.x. [DOI] [PubMed] [Google Scholar]
- Li H.; Grigoryan H.; Funk W. E.; Lu S. S.; Rose S.; Williams E. R.; Rappaport S. M. (2011) Profiling Cys34 adducts of human serum albumin by fixed-step selected reaction monitoring. Mol. Cell. Proteomics 10, M110.004606. 10.1074/mcp.M110.004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan H.; Edmands W. M.; Lu S. S.; Yano Y.; Regazzoni L.; Iavarone A. T.; Williams E. R.; Rappaport S. M. (2016) An adductomics pipeline for untargeted analysis of modifications to Cys34 of human serum albumin. Anal. Chem. 88, 10504–10512. 10.1021/acs.analchem.6b02553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters T., Jr. (1995) 7-Practical Aspects: Albumin in the Laboratory, In All About Albumin, pp 285–318, Academic Press, San Diego, CA. [Google Scholar]
- Cohn E. J.; Hughes W. L. Jr.; Weare J. H. (1947) Preparation and properties of serum and plasma proteins. XIII. Crystallization of serum albumins from ethanol-water mixtures. J. Am. Chem. Soc. 69, 1753–1761. 10.1021/ja01199a051. [DOI] [PubMed] [Google Scholar]
- Bechtold W. E.; Willis J. K.; Sun J. D.; Griffith W. C.; Reddy T. V. (1992) Biological markers of exposure to benzene: S-phenylcysteine in albumin. Carcinogenesis 13, 1217–1220. 10.1093/carcin/13.7.1217. [DOI] [PubMed] [Google Scholar]
- Rappaport S. M.; Ting D.; Jin Z.; Yeowell-O’Connell K.; Waidyanatha S.; McDonald T. (1993) Application of Raney nickel to measure adducts of styrene oxide with hemoglobin and albumin. Chem. Res. Toxicol. 6, 238–244. 10.1021/tx00032a014. [DOI] [PubMed] [Google Scholar]
- Lindstrom A. B.; Yeowell-O’Connell K.; Waidyanatha S.; McDonald T. A.; Golding B. T.; Rappaport S. M. (1998) Formation of hemoglobin and albumin adducts of benzene oxide in mouse, rat, and human blood. Chem. Res. Toxicol. 11, 302–310. 10.1021/tx9701788. [DOI] [PubMed] [Google Scholar]
- Waidyanatha S.; Rappaport S. M. (2008) Hemoglobin and albumin adducts of naphthalene-1,2-oxide, 1,2-naphthoquinone and 1,4-naphthoquinone in Swiss Webster mice. Chem.-Biol. Interact. 172, 105–114. 10.1016/j.cbi.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Wild C. P.; Jiang Y. Z.; Sabbioni G.; Chapot B.; Montesano R. (1990) Evaluation of methods for quantitation of aflatoxin-albumin adducts and their application to human exposure assessment. Cancer Res. 50, 245–251. [PubMed] [Google Scholar]
- Sepai O.; Henschler D.; Czech S.; Eckert P.; Sabbioni G. (1995) Exposure to toluenediamines from polyurethane-covered breast implants. Toxicol. Lett. 77, 371–378. 10.1016/0378-4274(95)03320-3. [DOI] [PubMed] [Google Scholar]
- Travis J.; Bowen J.; Tewksbury D.; Johnson D.; Pannell R. (1976) Isolation of albumin from whole human plasma and fractionation of albumin-depleted plasma. Biochem. J. 157, 301–306. 10.1042/bj1570301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherbarrow R. J.; Dean P. D. (1980) Studies on the mechanism of binding of serum albumins to immobilized cibacron blue F3G A. Biochem. J. 189, 27–34. 10.1042/bj1890027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepai O.; Henschler D.; Sabbioni G. (1995) Albumin adducts, hemoglobin adducts and urinary metabolites in workers exposed to 4,4′-methylenediphenyl diisocyanate. Carcinogenesis 16, 2583–2587. 10.1093/carcin/16.10.2583. [DOI] [PubMed] [Google Scholar]
- Sabbioni G.; Dongari N.; Kumar A. (2010) Determination of a new biomarker in subjects exposed to 4,4′-methylenediphenyl diisocyanate. Biomarkers 15, 508–515. 10.3109/1354750X.2010.490880. [DOI] [PubMed] [Google Scholar]
- Young C. L.; Woolfitt A. R.; McWilliams L. G.; Moura H.; Boyer A. E.; Barr J. R. (2005) Survey of albumin purification methods for the analysis of albumin-organic toxicant adducts by liquid chromatography-electrospray ionization-tandem mass spectrometry. Proteomics 5, 4973–4979. 10.1002/pmic.200402081. [DOI] [PubMed] [Google Scholar]
- Di Girolamo F.; Righetti P. G. (2011) Plasma proteomics for biomarker discovery: a study in blue. Electrophoresis 32, 3638–3644. 10.1002/elps.201100307. [DOI] [PubMed] [Google Scholar]
- Waidyanatha S.; Yeowell-O’Connell K.; Rappaport S. M. (1998) A new assay for albumin and hemoglobin adducts of 1,2- and 1,4-benzoquinones. Chem.-Biol. Interact. 115, 117–139. 10.1016/S0009-2797(98)00067-2. [DOI] [PubMed] [Google Scholar]
- Fenn J. B.; Mann M.; Meng C. K.; Wong S. F.; Whitehouse C. M. (1989) Electrospray ionization for mass spectrometry of large biomolecules. Science 246, 64–71. 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- Meng C. K.; Fenn J. B. (1990) Analyzing organic molecules with electrospray mass spectrometry. Am. Biotechnol. Lab. 8, 54–60. [PubMed] [Google Scholar]
- Hoffmann K. J.; Streeter A. J.; Axworthy D. B.; Baillie T. A. (1985) Structural characterization of the major covalent adduct formed in vitro between acetaminophen and bovine serum albumin. Chem.-Biol. Interact. 53, 155–172. 10.1016/S0009-2797(85)80093-4. [DOI] [PubMed] [Google Scholar]
- Boysen G.; Hecht S. S. (2003) Analysis of DNA and protein adducts of benzo[a]pyrene in human tissues using structure-specific methods. Mutat. Res., Rev. Mutat. Res. 543, 17–30. 10.1016/S1383-5742(02)00068-6. [DOI] [PubMed] [Google Scholar]
- Wild C. P.; Hudson G. J.; Sabbioni G.; Chapot B.; Hall A. J.; Wogan G. N.; Whittle H.; Montesano R.; Groopman J. D. (1992) Dietary intake of aflatoxins and the level of albumin-bound aflatoxin in peripheral blood in The Gambia, West Africa. Cancer Epidemiol. Biomarkers Prev. 1, 229–234. [PubMed] [Google Scholar]
- McCoy L. F.; Scholl P. F.; Sutcliffe A. E.; Kieszak S. M.; Powers C. D.; Rogers H. S.; Gong Y. Y.; Groopman J. D.; Wild C. P.; Schleicher R. L. (2008) Human aflatoxin albumin adducts quantitatively compared by ELISA, HPLC with fluorescence detection, and HPLC with isotope dilution mass spectrometry. Cancer Epidemiol., Biomarkers Prev. 17, 1653–1657. 10.1158/1055-9965.EPI-07-2780. [DOI] [PubMed] [Google Scholar]
- Damsten M. C.; Commandeur J. N.; Fidder A.; Hulst A. G.; Touw D.; Noort D.; Vermeulen N. P. (2007) Liquid chromatography/tandem mass spectrometry detection of covalent binding of acetaminophen to human serum albumin. Drug Metab. Dispos. 35, 1408–1417. 10.1124/dmd.106.014233. [DOI] [PubMed] [Google Scholar]
- LeBlanc A.; Shiao T. C.; Roy R.; Sleno L. (2014) Absolute quantitation of NAPQI-modified rat serum albumin by LC-MS/MS: monitoring acetaminophen covalent binding in vivo. Chem. Res. Toxicol. 27, 1632–1639. 10.1021/tx500284g. [DOI] [PubMed] [Google Scholar]
- Noort D.; Benschop H. P.; Black R. M. (2002) Biomonitoring of exposure to chemical warfare agents: a review. Toxicol. Appl. Pharmacol. 184, 116–126. 10.1006/taap.2002.9449. [DOI] [PubMed] [Google Scholar]
- Delatour T.; Fenaille F.; Parisod V.; Richoz J.; Vuichoud J.; Mottier P.; Buetler T. (2007) A comparative study of proteolysis methods for the measurement of 3-nitrotyrosine residues: Enzymatic digestion versus hydrochloric acid-mediated hydrolysis. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 851, 268–276. 10.1016/j.jchromb.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Sabbioni G. (1990) Chemical and physical properties of the major serum albumin adduct of aflatoxin B1 and their implications for the quantification in biological samples. Chem.-Biol. Interact. 75, 1–15. 10.1016/0009-2797(90)90018-I. [DOI] [PubMed] [Google Scholar]
- Kumar A.; Dongari N.; Sabbioni G. (2009) New isocyanate-specific albumin adducts of 4,4′-methylenediphenyl diisocyanate (MDI) in rats. Chem. Res. Toxicol. 22, 1975–1983. 10.1021/tx900270z. [DOI] [PubMed] [Google Scholar]
- Kinter M., and Sherman N. E. (2000) Protein Sequencing and Identification Using Tandem Mass Spectrometry, Wiley-Interscience, New York. [Google Scholar]
- Ham A. J., Caprioli R. M., and Gross M. L. (2005) The Encyclopedia of Mass Spectrometry, Vol. 2 Biological Applications Part A: Peptides and Proteins, pp 10–17, Elsevier Ltd., San Diego, CA. [Google Scholar]
- Sugimura T.; Wakabayashi K.; Nakagama H.; Nagao M. (2004) Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci. 95, 290–299. 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L.; Turesky R. J. (2014) Optimizing proteolytic digestion conditions for the analysis of serum albumin adducts of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, a potential human carcinogen formed in cooked meat. J. Proteomics 103, 267–278. 10.1016/j.jprot.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepstorff P.; Fohlman J. (1984) Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 11, 601. 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- Papayannopoulos I. A. (1995) The interpretation of collision-induced dissociation tandem mass spectra of peptides. Mass Spectrom. Rev. 14, 49–73. 10.1002/mas.1280140104. [DOI] [Google Scholar]
- Burkhart J. M.; Schumbrutzki C.; Wortelkamp S.; Sickmann A.; Zahedi R. P. (2012) Systematic and quantitative comparison of digest efficiency and specificity reveals the impact of trypsin quality on MS-based proteomics. J. Proteomics 75, 1454–1462. 10.1016/j.jprot.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Giansanti P.; Tsiatsiani L.; Low T. Y.; Heck A. J. (2016) Six alternative proteases for mass spectrometry-based proteomics beyond trypsin. Nat. Protoc. 11, 993–1006. 10.1038/nprot.2016.057. [DOI] [PubMed] [Google Scholar]
- Li B.; Ricordel I.; Schopfer L. M.; Baud F.; Megarbane B.; Nachon F.; Masson P.; Lockridge O. (2010) Detection of adduct on tyrosine 411 of albumin in humans poisoned by dichlorvos. Toxicol. Sci. 116, 23–31. 10.1093/toxsci/kfq117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulevich S. E.; Frey B. L.; Kreitinger G.; Smith L. M. (2010) Alkylating tryptic peptides to enhance electrospray ionization mass spectrometry analysis. Anal. Chem. 82, 10135–10142. 10.1021/ac1019792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P.; Aebersold R. (2012) Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods 9, 555–566. 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- Watson T. J., and Sparkman O. D. (2007) Introduction to Mass Spectrometry. Instrumentation, Applications and Atrategies for Data Interpretation, Vol. 4th ed., John Wiley & Sons, Ltd., Chichester, England. [Google Scholar]
- Colzani M.; Aldini G.; Carini M. (2013) Mass spectrometric approaches for the identification and quantification of reactive carbonyl species protein adducts. J. Proteomics 92, 28–50. 10.1016/j.jprot.2013.03.030. [DOI] [PubMed] [Google Scholar]
- Tretyakova N.; Villalta P. W.; Kotapati S. (2013) Mass spectrometry of structurally modified DNA. Chem. Rev. 113, 2395–2436. 10.1021/cr300391r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demartini D. R. (2013) A Short Overview of the Components in Mass Spectrometry Instrumentation for Proteomics Analyses, Tandem Mass Spectrometry - Molecular Characterization (Coelho A. V., Ed.) InTech, Rijeka, Croatia.
- Hopfgartner G.; Varesio E.; Tschappat V.; Grivet C.; Bourgogne E.; Leuthold L. A. (2004) Triple quadrupole linear ion trap mass spectrometer for the analysis of small molecules and macromolecules. J. Mass Spectrom. 39, 845–855. 10.1002/jms.659. [DOI] [PubMed] [Google Scholar]
- Gillette M. A.; Carr S. A. (2013) Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat. Methods 10, 28–34. 10.1038/nmeth.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr S. A.; Abbatiello S. E.; Ackermann B. L.; Borchers C.; Domon B.; Deutsch E. W.; Grant R. P.; Hoofnagle A. N.; Huttenhain R.; Koomen J. M.; Liebler D. C.; Liu T.; MacLean B.; Mani D. R.; Mansfield E.; Neubert H.; Paulovich A. G.; Reiter L.; Vitek O.; Aebersold R.; Anderson L.; Bethem R.; Blonder J.; Boja E.; Botelho J.; Boyne M.; Bradshaw R. A.; Burlingame A. L.; Chan D.; Keshishian H.; Kuhn E.; Kinsinger C.; Lee J. S.; Lee S. W.; Moritz R.; Oses-Prieto J.; Rifai N.; Ritchie J.; Rodriguez H.; Srinivas P. R.; Townsend R. R.; Van Eyk J.; Whiteley G.; Wiita A.; Weintraub S. (2014) Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol. Cell. Proteomics 13, 907–917. 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. V.; Schwartz J. C.; Griep-Raming J.; Nielsen M. L.; Damoc E.; Denisov E.; Lange O.; Remes P.; Taylor D.; Splendore M.; Wouters E. R.; Senko M.; Makarov A.; Mann M.; Horning S. (2009) A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769. 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J. C.; Senko M. W.; Syka J. E. P. (2002) A two-dimensional quadrupole ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 13, 659–669. 10.1016/S1044-0305(02)00384-7. [DOI] [PubMed] [Google Scholar]
- Sandra K.; Devreese B.; Van Beeumen J.; Stals I.; Claeyssens M. (2004) The Q-Trap mass spectrometer, a novel tool in the study of protein glycosylation. J. Am. Soc. Mass Spectrom. 15, 413–423. 10.1016/j.jasms.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Meng X.; Howarth A.; Earnshaw C. J.; Jenkins R. E.; French N. S.; Back D. J.; Naisbitt D. J.; Park B. K. (2013) Detection of drug bioactivation in vivo: mechanism of nevirapine-albumin conjugate formation in patients. Chem. Res. Toxicol. 26, 575–583. 10.1021/tx4000107. [DOI] [PubMed] [Google Scholar]
- Tailor A.; Waddington J. C.; Meng X.; Park B. K. (2016) Mass spectrometric and functional aspects of drug–protein conjugation. Chem. Res. Toxicol. 10.1021/acs.chemrestox.6b00147. [DOI] [PubMed] [Google Scholar]
- Beck S.; Michalski A.; Raether O.; Lubeck M.; Kaspar S.; Goedecke N.; Baessmann C.; Hornburg D.; Meier F.; Paron I.; Kulak N. A.; Cox J.; Mann M. (2015) The impact II, a very high-resolution quadrupole time-of-flight instrument (QTOF) for deep shotgun proteomics. Mol. Cell. Proteomics 14, 2014–2029. 10.1074/mcp.M114.047407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliuk S.; Makarov A. (2015) Evolution of Orbitrap Mass Spectrometry Instrumentation. Annu. Rev. Anal. Chem. 8, 61–80. 10.1146/annurev-anchem-071114-040325. [DOI] [PubMed] [Google Scholar]
- Savaryn J. P.; Toby T. K.; Kelleher N. L. (2016) A researcher’s guide to mass spectrometry-based proteomics. Proteomics 16, 2435. 10.1002/pmic.201600113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siuti N.; Kelleher N. L. (2007) Decoding protein modifications using top-down mass spectrometry. Nat. Methods 4, 817–821. 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chait B. T. (2006) Chemistry. Mass spectrometry: bottom-up or top-down?. Science 314, 65–66. 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- Ahlf D. R.; Compton P. D.; Tran J. C.; Early B. P.; Thomas P. M.; Kelleher N. L. (2012) Evaluation of the compact high-field orbitrap for top-down proteomics of human cells. J. Proteome Res. 11, 4308–4314. 10.1021/pr3004216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel T. E.; Aryal U. K.; Hengel S. M.; Baker E. S.; Kelly R. T.; Robinson E. W.; Smith R. D. (2012) Mass spectrometry-based proteomics: existing capabilities and future directions. Chem. Soc. Rev. 41, 3912–3928. 10.1039/c2cs15331a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Ge Y. (2011) Comprehensive analysis of protein modifications by top-down mass spectrometry. Circ.: Cardiovasc. Genet. 4, 711. 10.1161/CIRCGENETICS.110.957829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk W. E.; Li H.; Iavarone A. T.; Williams E. R.; Riby J.; Rappaport S. M. (2010) Enrichment of cysteinyl adducts of human serum albumin. Anal. Biochem. 400, 61–68. 10.1016/j.ab.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan H.; Li H.; Iavarone A. T.; Williams E. R.; Rappaport S. M. (2012) Cys34 adducts of reactive oxygen species in human serum albumin. Chem. Res. Toxicol. 25, 1633–1642. 10.1021/tx300096a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law K. P.; Lim Y. P. (2013) Recent advances in mass spectrometry: data independent analysis and hyper reaction monitoring. Expert Rev. Proteomics 10, 551–566. 10.1586/14789450.2013.858022. [DOI] [PubMed] [Google Scholar]
- Lange V.; Picotti P.; Domon B.; Aebersold R. (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 4, 222. 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter C. J.; Bereman M. S. (2015) Data-independent-acquisition mass spectrometry for identification of targeted-peptide site-specific modifications. Anal. Bioanal. Chem. 407, 6627–6635. 10.1007/s00216-015-8819-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Person M. D.; Monks T. J.; Lau S. S. (2003) An integrated approach to identifying chemically induced posttranslational modifications using comparative MALDI-MS and targeted HPLC-ESI-MS/MS. Chem. Res. Toxicol. 16, 598–608. 10.1021/tx020109f. [DOI] [PubMed] [Google Scholar]
- Yates J. R. 3rd; Eng J. K.; McCormack A. L.; Schieltz D. (1995) Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67, 1426–1436. 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- Creasy D. M.; Cottrell J. S. (2002) Error tolerant searching of uninterpreted tandem mass spectrometry data. Proteomics 2, 1426–1434. . [DOI] [PubMed] [Google Scholar]
- Creasy D. M.; Cottrell J. S. (2004) Unimod: Protein modifications for mass spectrometry. Proteomics 4, 1534–1536. 10.1002/pmic.200300744. [DOI] [PubMed] [Google Scholar]
- Dasari S.; Chambers M. C.; Codreanu S. G.; Liebler D. C.; Collins B. C.; Pennington S. R.; Gallagher W. M.; Tabb D. L. (2011) Sequence tagging reveals unexpected modifications in toxicoproteomics. Chem. Res. Toxicol. 24, 204–216. 10.1021/tx100275t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb D. L.; Fernando C. G.; Chambers M. C. (2007) MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 6, 654–661. 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.; Nachon F.; Froment M. T.; Verdier L.; Debouzy J. C.; Brasme B.; Gillon E.; Schopfer L. M.; Lockridge O.; Masson P. (2008) Binding and hydrolysis of soman by human serum albumin. Chem. Res. Toxicol. 21, 421–431. 10.1021/tx700339m. [DOI] [PubMed] [Google Scholar]
- Pathak K. V.; Bellamri M.; Wang Y.; Langouet S.; Turesky R. J. (2015) 2-Amino-9H-pyrido[2,3-b]indole (AalphaC) adducts and thiol oxidation of serum albumin as potential biomarkers of tobacco smoke. J. Biol. Chem. 290, 16304–16318. 10.1074/jbc.M115.646539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayre L. M.; Lin D.; Yuan Q.; Zhu X.; Tang X. (2006) Protein adducts generated from products of lipid oxidation: focus on HNE and one. Drug Metab. Rev. 38, 651–675. 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- Snyder R.; Witz G.; Goldstein B. D. (1993) The toxicology of benzene. Environ. Health Perspect. 100, 293–306. 10.1289/ehp.93100293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (2009) Chemical Agents and Related Occupations, IARC Monogr. Eval. Carcinog. Risks Hum, Vol. 100F39–163. [Google Scholar]
- Guengerich F. P.; Kim D. H.; Iwasaki M. (1991) Role of human cytochrome P-450 IIE1 in the oxidation of many low molecular weight cancer suspects. Chem. Res. Toxicol. 4, 168–179. 10.1021/tx00020a008. [DOI] [PubMed] [Google Scholar]
- Seaton M. J.; Schlosser P. M.; Bond J. A.; Medinsky M. A. (1994) Benzene metabolism by human liver microsomes in relation to cytochrome P450 2E1 activity. Carcinogenesis 15, 1799–1806. 10.1093/carcin/15.9.1799. [DOI] [PubMed] [Google Scholar]
- Monks T. J.; Butterworth M.; Lau S. S. (2010) The fate of benzene-oxide. Chem.-Biol. Interact. 184, 201–206. 10.1016/j.cbi.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport S. M.; Waidyanatha S.; Yeowell-O’Connell K.; Rothman N.; Smith M. T.; Zhang L.; Qu Q.; Shore R.; Li G.; Yin S. (2005) Protein adducts as biomarkers of human benzene metabolism. Chem.-Biol. Interact. 153–154, 103–109. 10.1016/j.cbi.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Troester M. A.; Lindstrom A. B.; Kupper L. L.; Waidyanatha S.; Rappaport S. M. (2000) Stability of hemoglobin and albumin adducts of benzene oxide and 1,4-benzoquinone after administration of benzene to F344 rats. Toxicol. Sci. 54, 88–94. 10.1093/toxsci/54.1.88. [DOI] [PubMed] [Google Scholar]
- Rappaport S. M.; Waidyanatha S.; Qu Q.; Shore R.; Jin X.; Cohen B.; Chen L. C.; Melikian A. A.; Li G.; Yin S.; Yan H.; Xu B.; Mu R.; Li Y.; Zhang X.; Li K. (2002) Albumin adducts of benzene oxide and 1,4-benzoquinone as measures of human benzene metabolism. Cancer Res. 62, 1330–1337. [PubMed] [Google Scholar]
- Rappaport S. M.; Yeowell-O’Connell K.; Smith M. T.; Dosemeci M.; Hayes R. B.; Zhang L.; Li G.; Yin S.; Rothman N. (2002) Non-linear production of benzene oxide-albumin adducts with human exposure to benzene. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 778, 367–374. 10.1016/S0378-4347(01)00457-1. [DOI] [PubMed] [Google Scholar]
- Lin Y. S.; Vermeulen R.; Tsai C. H.; Waidyanatha S.; Lan Q.; Rothman N.; Smith M. T.; Zhang L.; Shen M.; Li G.; Yin S.; Kim S.; Rappaport S. M. (2007) Albumin adducts of electrophilic benzene metabolites in benzene-exposed and control workers. Environ. Health Perspect. 115, 28–34. 10.1289/ehp.8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. S.; McKelvey W.; Waidyanatha S.; Rappaport S. M. (2006) Variability of albumin adducts of 1,4-benzoquinone, a toxic metabolite of benzene, in human volunteers. Biomarkers 11, 14–27. 10.1080/13547500500382975. [DOI] [PubMed] [Google Scholar]
- Preuss R.; Angerer J.; Drexler H. (2003) Naphthalene--an environmental and occupational toxicant. Int. Arch. Occup. Environ. Health 76, 556–576. 10.1007/s00420-003-0458-1. [DOI] [PubMed] [Google Scholar]
- Waidyanatha S.; Troester M. A.; Lindstrom A. B.; Rappaport S. M. (2002) Measurement of hemoglobin and albumin adducts of naphthalene-1,2-oxide, 1,2-naphthoquinone and 1,4-naphthoquinone after administration of naphthalene to F344 rats. Chem.-Biol. Interact. 141, 189–210. 10.1016/S0009-2797(02)00048-0. [DOI] [PubMed] [Google Scholar]
- Troester M. A.; Lindstrom A. B.; Waidyanatha S.; Kupper L. L.; Rappaport S. M. (2002) Stability of hemoglobin and albumin adducts of naphthalene oxide, 1,2-naphthoquinone, and 1,4-naphthoquinone. Toxicol. Sci. 68, 314–321. 10.1093/toxsci/68.2.314. [DOI] [PubMed] [Google Scholar]
- Lin P. H.; Chen D. R.; Wang T. W.; Lin C. H.; Chuang M. C. (2009) Investigation of the cumulative tissue doses of naphthoquinones in human serum using protein adducts as biomarker of exposure. Chem.-Biol. Interact. 181, 107–114. 10.1016/j.cbi.2009.05.016. [DOI] [PubMed] [Google Scholar]
- International Agency of Cancer Research (1994) Styrene. IARC Monogr. Eval. Carcinog. Risks Hum, Vol. 60, pp 233–320. [PMC free article] [PubMed] [Google Scholar]
- Phillips D. H.; Farmer P. B. (1994) Evidence for DNA and protein binding by styrene and styrene oxide. Crit. Rev. Toxicol. 24 (Suppl), S35–46. 10.3109/10408449409020139. [DOI] [PubMed] [Google Scholar]
- Yeowell-O’Connell K.; Jin Z.; Rappaport S. M. (1996) Determination of albumin and hemoglobin adducts in workers exposed to styrene and styrene oxide. Cancer Epidemiol. Biomarkers Prev. 5, 205–215. [PubMed] [Google Scholar]
- Fustinoni S.; Colosio C.; Colombi A.; Lastrucci L.; Yeowell-O’Connell K.; Rappaport S. M. (1998) Albumin and hemoglobin adducts as biomarkers of exposure to styrene in fiberglass-reinforced-plastics workers. Int. Arch. Occup. Environ. Health 71, 35–41. 10.1007/s004200050247. [DOI] [PubMed] [Google Scholar]
- Neumann H. G.; Birner G.; Kowallik P.; Schutze D.; Zwirner-Baier I. (1993) Hemoglobin adducts of N-substituted aryl compounds in exposure control and risk assessment. Environ. Health Perspect. 99, 65–69. 10.1289/ehp.939965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazanis S.; McClelland R. A. (1992) Electrophilic intermediate in the reaction of glutathione and nitroso arenes. J. Am. Chem. Soc. 114, 3052–3059. 10.1021/ja00034a043. [DOI] [Google Scholar]
- Kiese M. (1966) The biochemical production of ferrihemoglobin-forming derivatives from aromatic amines, and mechanisms of ferrihemoglobin formation. Pharmacol. Rev. 18, 1091–1161. [PubMed] [Google Scholar]
- Skipper P. L.; Green L. C.; Bryant M. S.; Tannenbaum S. R.; Kadlubar F. F. (1984) Monitoring exposure to 4-aminobiphenyl via blood protein adducts. IARC Sci. Publ. 143–150. [PubMed] [Google Scholar]
- Bryant M. S.; Vineis P.; Skipper P. L.; Tannenbaum S. R. (1988) Hemoglobin adducts of aromatic amines: associations with smoking status and type of tobacco. Proc. Natl. Acad. Sci. U. S. A. 85, 9788–9791. 10.1073/pnas.85.24.9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turesky R. J. (1986) 2-Amino-3-methylimidazo [4,5-f] quinoline: Metabolism in the rat and identification of a specific protein adduct, Ph.D. Thesis, Massachusetts Institute of Technology, Cambridge, MA, U.S.A. [Google Scholar]
- Sabbioni G.; Neumann H. G. (1990) Quantification of haemoglobin binding of 4,4′-methylenebis(2-chloroaniline) (MOCA) in rats. Arch. Toxicol. 64, 451–458. 10.1007/BF01977626. [DOI] [PubMed] [Google Scholar]
- DeBord D. G.; Swearengin T. F.; Cheever K. L.; Booth-Jones A. D.; Wissinger L. A. (1992) Binding characteristics of ortho-toluidine to rat hemoglobin and albumin. Arch. Toxicol. 66, 231–236. 10.1007/BF02307167. [DOI] [PubMed] [Google Scholar]
- Cheever K. L.; DeBord D. G.; Swearengin T. F.; Booth-Jones A. D. (1992) ortho-toluidine blood protein adducts: HPLC analysis with fluorescence detection after a single dose in the adult male rat. Toxicol. Sci. 18, 522–531. 10.1093/toxsci/18.4.522. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Peng L.; Bellamri M.; Langouet S.; Turesky R. J. (2015) Mass spectrometric characterization of human serum albumin adducts formed with N-oxidized metabolites of 2-amino-1-methylphenylimidazo[4,5-b]pyridine in human plasma and hepatocytes. Chem. Res. Toxicol. 28, 1045–1059. 10.1021/acs.chemrestox.5b00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K.; Kanematsu M.; Sakai K.; Matsuda T.; Hattori N.; Mizuno Y.; Suzuki D.; Miyata T.; Noguchi N.; Niki E.; Osawa T. (1998) Protein-bound acrolein: potential markers for oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 95, 4882–4887. 10.1073/pnas.95.9.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes A. M.; Godinho A. L.; Martins I. L.; Oliveira M. C.; Gomes R. A.; Coelho A. V.; Beland F. A.; Marques M. M. (2010) Protein adducts as prospective biomarkers of nevirapine toxicity. Chem. Res. Toxicol. 23, 1714–1725. 10.1021/tx100186t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magagnotti C.; Orsi F.; Bagnati R.; Celli N.; Rotilio D.; Fanelli R.; Airoldi L. (2000) Effect of diet on serum albumin and hemoglobin adducts of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in humans. Int. J. Cancer 88, 1–6. . [DOI] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (1993) Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins, IARC Monogr. Eval. Carcinog. Risks Hum, Vol. 56, pp 165–242.8411620 [Google Scholar]
- Guengerich F. P.; Johnson W. W. (1999) Kinetics of hydrolysis and reaction of aflatoxin B1 exo-8,9-epoxide and relevance to toxicity and detoxication. Drug Metab. Rev. 31, 141–158. 10.1081/DMR-100101911. [DOI] [PubMed] [Google Scholar]
- Guengerich F. P.; Arneson K. O.; Williams K. M.; Deng Z.; Harris T. M. (2002) Reaction of aflatoxin B(1) oxidation products with lysine. Chem. Res. Toxicol. 15, 780–792. 10.1021/tx010156s. [DOI] [PubMed] [Google Scholar]
- Wild C. P.; Garner R. C.; Montesano R.; Tursi F. (1986) Aflatoxin B1 binding to plasma albumin and liver DNA upon chronic administration to rats. Carcinogenesis 7, 853–858. 10.1093/carcin/7.6.853. [DOI] [PubMed] [Google Scholar]
- Sabbioni G.; Ambs S.; Wogan G. N.; Groopman J. D. (1990) The aflatoxin-lysine adduct quantified by high-performance liquid chromatography from human serum albumin samples. Carcinogenesis 11, 2063–2066. 10.1093/carcin/11.11.2063. [DOI] [PubMed] [Google Scholar]
- Okoye Z. S.; Riley J.; Judah D. J.; Neal G. E. (1990) The in vivo site of formation of a carcinogen-serum albumin adduct. Biochem. Pharmacol. 40, 2560–2562. 10.1016/0006-2952(90)90100-Y. [DOI] [PubMed] [Google Scholar]
- Sabbioni G., and Sepai O. (1998) Determination of Human Exposure to Aflatoxins, in Mycotoxins in Agriculture and Food Safety (Sinha K. K., and Bhatnagar D., Eds.) pp 183–226, Marcel Dekker, Inc., New York. [Google Scholar]
- Chapot B., and Wild C. P. (1991) ELISA for quantification of aflatoxin-albumin and their application to human exposure assessment, in Techniques in Diagnostic Pathology (Warthol M., van Velzer D., and Bullock G. R., Eds.) pp 135–155, Academic Press, London. [Google Scholar]
- McCoy L. F.; Scholl P. F.; Schleicher R. L.; Groopman J. D.; Powers C. D.; Pfeiffer C. M. (2005) Analysis of aflatoxin B1-lysine adduct in serum using isotope-dilution liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 19, 2203–2210. 10.1002/rcm.2045. [DOI] [PubMed] [Google Scholar]
- Scholl P. F.; Turner P. C.; Sutcliffe A. E.; Sylla A.; Diallo M. S.; Friesen M. D.; Groopman J. D.; Wild C. P. (2006) Quantitative comparison of aflatoxin B1 serum albumin adducts in humans by isotope dilution mass spectrometry and ELISA. Cancer Epidemiol., Biomarkers Prev. 15, 823–826. 10.1158/1055-9965.EPI-05-0890. [DOI] [PubMed] [Google Scholar]
- Sabbioni G.; Wild C. P. (1991) Identification of an aflatoxin G1-serum albumin adduct and its relevance to the measurement of human exposure to aflatoxins. Carcinogenesis 12, 97–103. 10.1093/carcin/12.1.97. [DOI] [PubMed] [Google Scholar]
- Wild C. P., Miller J. D., and Groopman J. D. (2015) Mycotoxin Control in Low- and Middle-Income Countries, in IARC Working Group Report 9, International Agency for Research on Cancer, Lyon, France. [PubMed] [Google Scholar]
- Groopman J. D.; Kensler T. W.; Wild C. P. (2008) Protective interventions to prevent aflatoxin-induced carcinogenesis in developing countries. Annu. Rev. Public Health 29, 187–203. 10.1146/annurev.publhealth.29.020907.090859. [DOI] [PubMed] [Google Scholar]
- Wogan G. N.; Kensler T. W.; Groopman J. D. (2012) Present and future directions of translational research on aflatoxin and hepatocellular carcinoma. A review. Food Addit. Contam., Part A 29, 249–257. 10.1080/19440049.2011.563370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleicher R. L.; McCoy L. F.; Powers C. D.; Sternberg M. R.; Pfeiffer C. M. (2013) Serum concentrations of an aflatoxin-albumin adduct in the National Health and Nutrition Examination Survey (NHANES) 1999–2000. Clin. Chim. Acta 423, 46–50. 10.1016/j.cca.2013.04.011. [DOI] [PubMed] [Google Scholar]
- Groopman J. D.; DeMatos P.; Egner P. A.; Love-Hunt A.; Kensler T. W. (1992) Molecular dosimetry of urinary aflatoxin-N7-guanine and serum aflatoxin-albumin adducts predicts chemoprotection by 1,2-dithiole-3-thione in rats. Carcinogenesis 13, 101–106. 10.1093/carcin/13.1.101. [DOI] [PubMed] [Google Scholar]
- Egner P. A.; Gange S. J.; Dolan P. M.; Groopman J. D.; Munoz A.; Kensler T. W. (1995) Levels of aflatoxin-albumin biomarkers in rat plasma are modulated by both long-term and transient interventions with oltipraz. Carcinogenesis 16, 1769–1773. 10.1093/carcin/16.8.1769. [DOI] [PubMed] [Google Scholar]
- Kensler T. W.; Groopman J. D.; Roebuck B. D. (1998) Use of aflatoxin adducts as intermediate endpoints to assess the efficacy of chemopreventive interventions in animals and man. Mutat. Res., Fundam. Mol. Mech. Mutagen. 402, 165–172. 10.1016/S0027-5107(97)00294-7. [DOI] [PubMed] [Google Scholar]
- Karol M. H.; Dean J. H. (1986) Respiratory effects of inhaled isocyanates. Crit. Rev. Toxicol. 16, 349–379. 10.3109/10408448609037467. [DOI] [PubMed] [Google Scholar]
- Baur X. (1986) Asthma caused by isocyanates. Allergologie 9, 487–496. [Google Scholar]
- Raulf-Heimsoth M.; Baur X. (1998) Pathomechanisms and pathophysiology of isocyanate-induced diseases--summary of present knowledge. Am. J. Ind. Med. 34, 137–143. . [DOI] [PubMed] [Google Scholar]
- Lind P.; Skarping G.; Dalene M. (1996) Biomarkers of toluene diisocyanate and thermal degradation products of Polyurethane, with special reference to the sample preparation. Anal. Chim. Acta 333, 277–283. 10.1016/0003-2670(96)00291-7. [DOI] [Google Scholar]
- Sabbioni G.; Lamb J. H.; Farmer P. B.; Sepai O. (1997) Reactions of 4-methylphenyl isocyanate with amino acids. Biomarkers 2, 223–232. 10.1080/135475097231599. [DOI] [PubMed] [Google Scholar]
- Sabbioni G.; Dongari N.; Schneider S.; Kumar A. (2012) Synthetic approaches to obtain amino acid adducts of 4,4′-methylenediphenyl diisocyanate. Chem. Res. Toxicol. 25, 2704–2714. 10.1021/tx300347e. [DOI] [PubMed] [Google Scholar]
- Sabbioni G.; Hartley R.; Henschler D.; Hollrigl-Rosta A.; Koeber R.; Schneider S. (2000) Isocyanate-specific hemoglobin adduct in rats exposed to 4, 4′-methylenediphenyl diisocyanate. Chem. Res. Toxicol. 13, 82–89. 10.1021/tx990096e. [DOI] [PubMed] [Google Scholar]
- Sabbioni G.; Dongari N.; Kumar A.; Baur X. (2016) Determination of albumin adducts of 4,4′-methylenediphenyl diisocyanate after specific inhalative challenge tests in workers. Toxicol. Lett. 260, 46–51. 10.1016/j.toxlet.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Sabbioni G.; Dongari N.; Sepai O.; Kumar A. (2016) Determination of albumin adducts of 4,4′-methylenediphenyl diisocyanate in workers of a 4,4′-methylenedianiline factory. Biomarkers 21, 731–738. 10.3109/1354750X.2016.1172117. [DOI] [PubMed] [Google Scholar]
- Sabbioni G.; Vanimireddy L. R.; Lummus Z. L.; Bernstein D. I. (2016) Comparison of biological effects with albumin adducts of 4,4′-methylenediphenyl diisocyanate in workers. Arch. Toxicol. 10.1007/s00204-016-1846-0. [DOI] [PubMed] [Google Scholar]
- Wisnewski A. V.; Liu J.; Redlich C. A. (2010) Antigenic changes in human albumin caused by reactivity with the occupational allergen diphenylmethane diisocyanate. Anal. Biochem. 400, 251–258. 10.1016/j.ab.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettick J. M.; Siegel P. D. (2012) Comparative analysis of aromatic diisocyanate conjugation to human albumin utilizing multiplexed tandem mass spectrometry. Int. J. Mass Spectrom. 309, 168–175. 10.1016/j.ijms.2011.09.015. [DOI] [Google Scholar]
- Mhike M.; Chipinda I.; Hettick J. M.; Simoyi R. H.; Lemons A.; Green B. J.; Siegel P. D. (2013) Characterization of methylene diphenyl diisocyanate-haptenated human serum albumin and hemoglobin. Anal. Biochem. 440, 197–204. 10.1016/j.ab.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhike M.; Hettick J. M.; Chipinda I.; Law B. F.; Bledsoe T. A.; Lemons A. R.; Nayak A. P.; Green B. J.; Beezhold D. H.; Simoyi R. H.; Siegel P. D. (2016) Characterization and comparative analysis of 2,4-toluene diisocyanate and 1,6-hexamethylene diisocyanate haptenated human serum albumin and hemoglobin. J. Immunol. Methods 431, 38–44. 10.1016/j.jim.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbioni G.; Hartley R.; Schneider S. (2001) Synthesis of adducts with amino acids as potential dosimeters for the biomonitoring of humans exposed to toluenediisocyanate. Chem. Res. Toxicol. 14, 1573–1583. 10.1021/tx010053+. [DOI] [PubMed] [Google Scholar]
- Sabbioni G.; Gu Q.; Vanimireddy L. R. (2012) Determination of isocyanate specific albumin-adducts in workers exposed to toluene diisocyanates. Biomarkers 17, 150–159. 10.3109/1354750X.2011.645166. [DOI] [PubMed] [Google Scholar]
- Kalim S.; Karumanchi S. A.; Thadhani R. I.; Berg A. H. (2014) Protein carbamylation in kidney disease: pathogenesis and clinical implications. Am. J. Kidney Dis. 64, 793–803. 10.1053/j.ajkd.2014.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafferlein H. U.; Marczynski B.; Mensing T.; Bruning T. (2010) Albumin and hemoglobin adducts of benzo[a]pyrene in humans--analytical methods, exposure assessment, and recommendations for future directions. Crit. Rev. Toxicol. 40, 126–150. 10.3109/10408440903283633. [DOI] [PubMed] [Google Scholar]
- Day B. W.; Doxtader M. M.; Rich R. H.; Skipper P. L.; Singh K.; Dasari R. R.; Tannenbaum S. R. (1992) Human serum albumin-benzo[a]pyrene anti-diol epoxide adduct structure elucidation by fluorescence line narrowing spectroscopy. Chem. Res. Toxicol. 5, 71–76. 10.1021/tx00025a012. [DOI] [PubMed] [Google Scholar]
- Day B. W.; Skipper P. L.; Zaia J.; Singh K.; Tannenbaum S. R. (1994) Enantiospecificity of covalent adduct formation by benzo[a]pyrene anti-diol epoxide with human serum albumin. Chem. Res. Toxicol. 7, 829–835. 10.1021/tx00042a017. [DOI] [PubMed] [Google Scholar]
- Ozbal C. C.; Skipper P. L.; Yu M. C.; London S. J.; Dasari R. R.; Tannenbaum S. R. (2000) Quantification of (7S,8R)-dihydroxy-(9R,10S)-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene adducts in human serum albumin by laser-induced fluorescence: implications for the in vivo metabolism of benzo[a]pyrene. Cancer Epidemiol. Biomarkers Prev. 9, 733–739. [PubMed] [Google Scholar]
- Pastorelli R.; Guanci M.; Cerri A.; Minoia C.; Carrer P.; Negri E.; Fanelli R.; Airoldi L. (2000) Benzo(a)pyrene diolepoxide-haemoglobin and albumin adducts at low levels of benzo(a)pyrene exposure. Biomarkers 5, 245–251. 10.1080/135475000413791. [DOI] [PubMed] [Google Scholar]
- Pastorelli R.; Cerri A.; Rozio M.; Dell’Omo M.; Muzi G.; Abbritti G.; Fanelli R.; Airoldi L. (2001) Benzo(a)pyrene diolepoxide adducts to albumin in workers exposed to polycyclic aromatic hydrocarbons: association with specific CYP1A1, GSTM1, GSTP1 and EHPX genotypes. Biomarkers 6, 357–374. 10.1080/13547500110044267. [DOI] [PubMed] [Google Scholar]
- Pastorelli R.; Guanci M.; Cerri A.; Negri E.; La Vecchia C.; Fumagalli F.; Mezzetti M.; Cappelli R.; Panigalli T.; Fanelli R.; Airoldi L. (1998) Impact of inherited polymorphisms in glutathione S-transferase M1, microsomal epoxide hydrolase, cytochrome P450 enzymes on DNA, and blood protein adducts of benzo(a)pyrene-diolepoxide. Cancer Epidemiol. Biomarkers Prev. 7, 703–709. [PubMed] [Google Scholar]
- Westberg E.; Hedebrant U.; Haglund J.; Alsberg T.; Eriksson J.; Seidel A.; Tornqvist M. (2014) Conditions for sample preparation and quantitative HPLC/MS-MS analysis of bulky adducts to serum albumin with diolepoxides of polycyclic aromatic hydrocarbons as models. Anal. Bioanal. Chem. 406, 1519–1530. 10.1007/s00216-013-7540-7. [DOI] [PubMed] [Google Scholar]
- Crow B. S.; Pantazides B. G.; Quinones-Gonzalez J.; Garton J. W.; Carter M. D.; Perez J. W.; Watson C. M.; Tomcik D. J.; Crenshaw M. D.; Brewer B. N.; Riches J. R.; Stubbs S. J.; Read R. W.; Evans R. A.; Thomas J. D.; Blake T. A.; Johnson R. C. (2014) Simultaneous measurement of tabun, sarin, soman, cyclosarin, VR, VX, and VM adducts to tyrosine in blood products by isotope dilution UHPLC-MS/MS. Anal. Chem. 86, 10397–10405. 10.1021/ac502886c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockridge O.; Schopfer L. M. (2010) Review of tyrosine and lysine as new motifs for organophosphate binding to proteins that have no active site serine. Chem.-Biol. Interact. 187, 344–348. 10.1016/j.cbi.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan H.; Li B.; Xue W.; Grigoryan M.; Schopfer L. M.; Lockridge O. (2009) Mass spectral characterization of organophosphate-labeled lysine in peptides. Anal. Biochem. 394, 92–100. 10.1016/j.ab.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S. J.; Carr J.; Carlson J. E.; Tong L.; Xue W.; Li Y.; Schopfer L. M.; Li B.; Nachon F.; Asojo O.; Thompson C. M.; Hinrichs S. H.; Masson P.; Lockridge O. (2008) Five tyrosines and two serines in human albumin are labeled by the organophosphorus agent FP-biotin. Chem. Res. Toxicol. 21, 1787–1794. 10.1021/tx800144z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.; Schopfer L. M.; Hinrichs S. H.; Masson P.; Lockridge O. (2007) Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry assay for organophosphorus toxicants bound to human albumin at Tyr411. Anal. Biochem. 361, 263–272. 10.1016/j.ab.2006.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams N. H.; Harrison J. M.; Read R. W.; Black R. M. (2007) Phosphylated tyrosine in albumin as a biomarker of exposure to organophosphorus nerve agents. Arch. Toxicol. 81, 627–639. 10.1007/s00204-007-0191-8. [DOI] [PubMed] [Google Scholar]
- Li B.; Schopfer L. M.; Grigoryan H.; Thompson C. M.; Hinrichs S. H.; Masson P.; Lockridge O. (2009) Tyrosines of human and mouse transferrin covalently labeled by organophosphorus agents: a new motif for binding to proteins that have no active site serine. Toxicol. Sci. 107, 144–155. 10.1093/toxsci/kfn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noort D.; Hulst A. G.; van Zuylen A.; van Rijssel E.; van der Schans M. J. (2009) Covalent binding of organophosphorothioates to albumin: a new perspective for OP-pesticide biomonitoring?. Arch. Toxicol. 83, 1031–1036. 10.1007/s00204-009-0456-5. [DOI] [PubMed] [Google Scholar]
- John H.; Breyer F.; Thumfart J. O.; Hochstetter H.; Thiermann H. (2010) Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) for detection and identification of albumin phosphylation by organophosphorus pesticides and G- and V-type nerve agents. Anal. Bioanal. Chem. 398, 2677–2691. 10.1007/s00216-010-4076-y. [DOI] [PubMed] [Google Scholar]
- Lockridge O.; Xue W.; Gaydess A.; Grigoryan H.; Ding S. J.; Schopfer L. M.; Hinrichs S. H.; Masson P. (2008) Pseudo-esterase activity of human albumin: slow turnover on tyrosine 411 and stable acetylation of 82 residues including 59 lysines. J. Biol. Chem. 283, 22582–22590. 10.1074/jbc.M802555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed N.; Dobler D.; Dean M.; Thornalley P. J. (2005) Peptide mapping identifies hotspot site of modification in human serum albumin by methylglyoxal involved in ligand binding and esterase activity. J. Biol. Chem. 280, 5724–5732. 10.1074/jbc.M410973200. [DOI] [PubMed] [Google Scholar]
- Grube A., Donaldson D., Kiely T., and Wu L. (2011) Pesticides Industry Sales and Usage 2006 and 2007 Market Estimates, Biological and Economic Analysis Division, Office of Pesticide Programs, Office of Chemical Safety and Pollution Prevention, U.S. Environmental Protection Agency Washington, DC . [Google Scholar]
- Gammon D. W.; Aldous C. N.; Carr W. C. Jr.; Sanborn J. R.; Pfeifer K. F. (2005) A risk assessment of atrazine use in California: human health and ecological aspects. Pest Manage. Sci. 61, 331–355. 10.1002/ps.1000. [DOI] [PubMed] [Google Scholar]
- Jablonowski N. D.; Schaffer A.; Burauel P. (2011) Still present after all these years: persistence plus potential toxicity raise questions about the use of atrazine. Environ. Sci. Pollut. Res. 18, 328–331. 10.1007/s11356-010-0431-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode M.; Stobe P.; Thiede B.; Schuphan I.; Schmidt B. (2004) Biotransformation of atrazine in transgenic tobacco cell culture expressing human P450. Pest Manage. Sci. 60, 49–58. 10.1002/ps.770. [DOI] [PubMed] [Google Scholar]
- Dooley G. P.; Hanneman W. H.; Carbone D. L.; Legare M. E.; Andersen M. E.; Tessari J. D. (2007) Development of an immunochemical detection method for atrazine-induced albumin adducts. Chem. Res. Toxicol. 20, 1061–1066. 10.1021/tx700083v. [DOI] [PubMed] [Google Scholar]
- Noort D.; Hulst A. G.; de Jong L. P.; Benschop H. P. (1999) Alkylation of human serum albumin by sulfur mustard in vitro and in vivo: mass spectrometric analysis of a cysteine adduct as a sensitive biomarker of exposure. Chem. Res. Toxicol. 12, 715–721. 10.1021/tx9900369. [DOI] [PubMed] [Google Scholar]
- Noort D.; Fidder A.; Degenhardt-Langelaan C. E.; Hulst A. G. (2008) Retrospective detection of sulfur mustard exposure by mass spectrometric analysis of adducts to albumin and hemoglobin: an in vivo study. J. Anal. Toxicol. 32, 25–30. 10.1093/jat/32.1.25. [DOI] [PubMed] [Google Scholar]
- Reistad R.; Frandsen H.; Grivas S.; Alexander J. (1994) In vitro formation and degradation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) protein adducts. Carcinogenesis 15, 2547–2552. 10.1093/carcin/15.11.2547. [DOI] [PubMed] [Google Scholar]
- Hecht S. S. (2000) Inhibition of carcinogenesis by isothiocyanates. Drug Metab. Rev. 32, 395–411. 10.1081/DMR-100102342. [DOI] [PubMed] [Google Scholar]
- Kensler T. W.; Egner P. A.; Agyeman A. S.; Visvanathan K.; Groopman J. D.; Chen J. G.; Chen T. Y.; Fahey J. W.; Talalay P. (2012) Keap1-nrf2 signaling: a target for cancer prevention by sulforaphane. Top. Curr. Chem. 329, 163–177. 10.1007/128_2012_339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway C. C.; Yang Y. M.; Chung F. L. (2002) Isothiocyanates as cancer chemopreventive agents: their biological activities and metabolism in rodents and humans. Curr. Drug Metab. 3, 233–255. 10.2174/1389200023337496. [DOI] [PubMed] [Google Scholar]
- Morris M. E., and Telang U. (2006) Isothiocyanates and Cancer Prevention, in Nutrition and Cancer Prevention (Awad A. T., and Bradford P. G., Eds.) pp 435–453, CRC/Taylor & Francis, Boca Raton, FL. [Google Scholar]
- Mi L.; Wang X.; Govind S.; Hood B. L.; Veenstra T. D.; Conrads T. P.; Saha D. T.; Goldman R.; Chung F. L. (2007) The role of protein binding in induction of apoptosis by phenethyl isothiocyanate and sulforaphane in human non-small lung cancer cells. Cancer Res. 67, 6409–6416. 10.1158/0008-5472.CAN-07-0340. [DOI] [PubMed] [Google Scholar]
- Bones A. M.; Rossiter J. T. (2006) The enzymic and chemically induced decomposition of glucosinolates. Phytochemistry 67, 1053–1067. 10.1016/j.phytochem.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Holst B.; Williamson G. (2004) A critical review of the bioavailability of glucosinolates and related compounds. Nat. Prod. Rep. 21, 425–447. 10.1039/b204039p. [DOI] [PubMed] [Google Scholar]
- Hanlon N.; Coldham N.; Gielbert A.; Sauer M. J.; Ioannides C. (2009) Repeated intake of broccoli does not lead to higher plasma levels of sulforaphane in human volunteers. Cancer Lett. 284, 15–20. 10.1016/j.canlet.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Kumar A.; Sabbioni G. (2010) New biomarkers for monitoring the levels of isothiocyanates in humans. Chem. Res. Toxicol. 23, 756–765. 10.1021/tx900393t. [DOI] [PubMed] [Google Scholar]
- Kumar A.; Vineis P.; Sacerdote C.; Fiorini L.; Sabbioni G. (2010) Determination of new biomarkers to monitor the dietary consumption of isothiocyanates. Biomarkers 15, 739–745. 10.3109/1354750X.2010.517567. [DOI] [PubMed] [Google Scholar]
- Zhou S.; Chan E.; Duan W.; Huang M.; Chen Y. Z. (2005) Drug bioactivation, covalent binding to target proteins and toxicity relevance. Drug Metab. Rev. 37, 41–213. 10.1081/DMR-200028812. [DOI] [PubMed] [Google Scholar]
- Kalgutkar A. S.; Gardner I.; Obach R. S.; Shaffer C. L.; Callegari E.; Henne K. R.; Mutlib A. E.; Dalvie D. K.; Lee J. S.; Nakai Y.; O’Donnell J. P.; Boer J.; Harriman S. P. (2005) A comprehensive listing of bioactivation pathways of organic functional groups. Curr. Drug Metab. 6, 161–225. 10.2174/1389200054021799. [DOI] [PubMed] [Google Scholar]
- Uetrecht J.; Naisbitt D. J. (2013) Idiosyncratic adverse drug reactions: current concepts. Pharmacol. Rev. 65, 779–808. 10.1124/pr.113.007450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariza A.; Montanez M. I.; Perez-Sala D. (2011) Proteomics in immunological reactions to drugs. Curr. Opin. Allergy Clin. Immunol. 11, 305–312. 10.1097/ACI.0b013e3283489ae5. [DOI] [PubMed] [Google Scholar]
- Faulkner L.; Meng X.; Park B. K.; Naisbitt D. J. (2014) The importance of hapten-protein complex formation in the development of drug allergy. Curr. Opin. Allergy Clin. Immunol. 14, 293–300. 10.1097/ACI.0000000000000078. [DOI] [PubMed] [Google Scholar]
- Skonberg C.; Olsen J.; Madsen K. G.; Hansen S. H.; Grillo M. P. (2008) Metabolic activation of carboxylic acids. Expert Opin. Drug Metab. Toxicol. 4, 425–438. 10.1517/17425255.4.4.425. [DOI] [PubMed] [Google Scholar]
- Garzon D.; Ariza A.; Regazzoni L.; Clerici R.; Altomare A.; Sirtori F. R.; Carini M.; Torres M. J.; Perez-Sala D.; Aldini G. (2014) Mass spectrometric strategies for the identification and characterization of human serum albumin covalently adducted by amoxicillin: ex vivo studies. Chem. Res. Toxicol. 27, 1566–1574. 10.1021/tx500210e. [DOI] [PubMed] [Google Scholar]
- Meng X.; Jenkins R. E.; Berry N. G.; Maggs J. L.; Farrell J.; Lane C. S.; Stachulski A. V.; French N. S.; Naisbitt D. J.; Pirmohamed M.; Park B. K. (2011) Direct evidence for the formation of diastereoisomeric benzylpenicilloyl haptens from benzylpenicillin and benzylpenicillenic acid in patients. J. Pharmacol. Exp. Ther. 338, 841–849. 10.1124/jpet.111.183871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette C. (2003) Pharmacogenomics of human UDP-glucuronosyltransferase enzymes. Pharmacogenomics J. 3, 136–158. 10.1038/sj.tpj.6500171. [DOI] [PubMed] [Google Scholar]
- Ding A.; Zia-Amirhosseini P.; McDonagh A. F.; Burlingame A. L.; Benet L. Z. (1995) Reactivity of tolmetin glucuronide with human serum albumin. Identification of binding sites and mechanisms of reaction by tandem mass spectrometry. Drug Metab. Dispos. 23, 369–376. [PubMed] [Google Scholar]
- Hammond T. G.; Meng X.; Jenkins R. E.; Maggs J. L.; Castelazo A. S.; Regan S. L.; Bennett S. N.; Earnshaw C. J.; Aithal G. P.; Pande I.; Kenna J. G.; Stachulski A. V.; Park B. K.; Williams D. P. (2014) Mass spectrometric characterization of circulating covalent protein adducts derived from a drug acyl glucuronide metabolite: multiple albumin adductions in diclofenac patients. J. Pharmacol. Exp. Ther. 350, 387–402. 10.1124/jpet.114.215079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell M.; Breitholtz K.; Isin E. M.; Jurva U.; Weidolf L. (2015) Significantly different covalent binding of oxidative Metabolites, Acyl Glucuronides, and S-Acyl CoA Conjugates Formed from xenobiotic carboxylic acids in human liver microsomes. Chem. Res. Toxicol. 28, 886–896. 10.1021/tx500514z. [DOI] [PubMed] [Google Scholar]
- Olsen J.; Li C.; Skonberg C.; Bjornsdottir I.; Sidenius U.; Benet L. Z.; Hansen S. H. (2007) Studies on the metabolism of tolmetin to the chemically reactive acyl-coenzyme A thioester intermediate in rats. Drug Metab. Dispos. 35, 758–764. 10.1124/dmd.106.013334. [DOI] [PubMed] [Google Scholar]
- David Josephy P. (2005) The molecular toxicology of acetaminophen. Drug Metab. Rev. 37, 581–594. 10.1080/03602530500205200. [DOI] [PubMed] [Google Scholar]
- Dahlin D. C.; Miwa G. T.; Lu A. Y.; Nelson S. D. (1984) N-acetyl-p-benzoquinone imine: a cytochrome P-450-mediated oxidation product of acetaminophen. Proc. Natl. Acad. Sci. U. S. A. 81, 1327–1331. 10.1073/pnas.81.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeschke H.; Gores G. J.; Cederbaum A. I.; Hinson J. A.; Pessayre D.; Lemasters J. J. (2002) Mechanisms of hepatotoxicity. Toxicol. Sci. 65, 166–176. 10.1093/toxsci/65.2.166. [DOI] [PubMed] [Google Scholar]
- Hoffmann K. J.; Streeter A. J.; Axworthy D. B.; Baillie T. A. (1985) Identification of the major covalent adduct formed in vitro and in vivo between acetaminophen and mouse liver proteins. Mol. Pharmacol. 27, 566–573. [PubMed] [Google Scholar]
- Sharma A. M.; Li Y.; Novalen M.; Hayes M. A.; Uetrecht J. (2012) Bioactivation of nevirapine to a reactive quinone methide: implications for liver injury. Chem. Res. Toxicol. 25, 1708–1719. 10.1021/tx300172s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes A. M. M.; Godinho A. L. A.; Martins I. L.; Justino G. C.; Beland F. A.; Marques M. M. (2010) Amino acid adduct formation by the nevirapine metabolite, 12-hydroxynevirapine—A possible factor in nevirapine toxicity. Chem. Res. Toxicol. 23, 888–899. 10.1021/tx900443z. [DOI] [PubMed] [Google Scholar]
- Aldini G.; Orioli M.; Carini M. (2011) Protein modification by acrolein: relevance to pathological conditions and inhibition by aldehyde sequestering agents. Mol. Nutr. Food Res. 55, 1301–1319. 10.1002/mnfr.201100182. [DOI] [PubMed] [Google Scholar]
- Stadtman E. R. (2006) Protein oxidation and aging. Free Radical Res. 40, 1250–1258. 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- Esterbauer H.; Schaur R. J.; Zollner H. (1991) Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radical Biol. Med. 11, 81–128. 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (2002) Tobacco smoke and involuntary smoking. IARC Monogr. Eval. Carcinog. Risks Hum., vol 83, pp 73–75.. [PMC free article] [PubMed] [Google Scholar]
- Shacter E.; Williams J. A.; Lim M.; Levine R. L. (1994) Differential susceptibility of plasma proteins to oxidative modification: examination by western blot immunoassay. Free Radical Biol. Med. 17, 429–437. 10.1016/0891-5849(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Maeshima T.; Honda K.; Chikazawa M.; Shibata T.; Kawai Y.; Akagawa M.; Uchida K. (2012) Quantitative analysis of acrolein-specific adducts generated during lipid peroxidation-modification of proteins in vitro: identification of N(tau)-(3-propanal)histidine as the major adduct. Chem. Res. Toxicol. 25, 1384–1392. 10.1021/tx3000818. [DOI] [PubMed] [Google Scholar]
- Himmelfarb J.; McMonagle E. (2001) Albumin is the major plasma protein target of oxidant stress in uremia. Kidney Int. 60, 358–363. 10.1046/j.1523-1755.2001.00807.x. [DOI] [PubMed] [Google Scholar]
- Colombo G.; Aldini G.; Orioli M.; Giustarini D.; Gornati R.; Rossi R.; Colombo R.; Carini M.; Milzani A.; Dalle-Donne I. (2010) Water-Soluble alpha,beta-unsaturated aldehydes of cigarette smoke induce carbonylation of human serum albumin. Antioxid. Redox Signaling 12, 349–364. 10.1089/ars.2009.2806. [DOI] [PubMed] [Google Scholar]
- Gan J. C.; Oandasan A.; Ansari G. A. S. (1991) In vitro covalent modification of serum albumin by acrolein. Chemosphere 23, 939–947. 10.1016/0045-6535(91)90098-X. [DOI] [Google Scholar]
- Witort E.; Capaccioli S.; Becatti M.; Fiorillo C.; Batignani G.; Pavoni V.; Piccini M.; Orioli M.; Carini M.; Aldini G.; Lulli M. (2016) Albumin Cys34 adducted by acrolein as a marker of oxidative stress in ischemia-reperfusion injury during hepatectomy. Free Radical Res. 50, 831–839. 10.1080/10715762.2016.1179736. [DOI] [PubMed] [Google Scholar]
- Goto T.; Murata K.; Lee S. H.; Oe T. (2013) Complete amino acid sequencing and immunoaffinity clean-up can facilitate screening of various chemical modifications on human serum albumin. Anal. Bioanal. Chem. 405, 7383–7395. 10.1007/s00216-013-7146-0. [DOI] [PubMed] [Google Scholar]
- Wild C. P. (2012) The exposome: from concept to utility. Int. J. Epidemiol. 41, 24–32. 10.1093/ije/dyr236. [DOI] [PubMed] [Google Scholar]
- Wild C. P. (2005) Complementing the genome with an ″exposome″: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol., Biomarkers Prev. 14, 1847–1850. 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- Wild C. P.; Scalbert A.; Herceg Z. (2013) Measuring the exposome: a powerful basis for evaluating environmental exposures and cancer risk. Environ. Mol. Mutagen. 54, 480–499. 10.1002/em.21777. [DOI] [PubMed] [Google Scholar]
- Carlsson H.; von Stedingk H.; Nilsson U.; Tornqvist M. (2014) LC-MS/MS screening strategy for unknown adducts to N-terminal valine in hemoglobin applied to smokers and nonsmokers. Chem. Res. Toxicol. 27, 2062–2070. 10.1021/tx5002749. [DOI] [PubMed] [Google Scholar]
- Doll S.; Burlingame A. L. (2015) Mass spectrometry-based detection and assignment of protein posttranslational modifications. ACS Chem. Biol. 10, 63–71. 10.1021/cb500904b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnaby O. S.; Cerny R. L.; Clarke W.; Hage D. S. (2011) Comparison of modification sites formed on human serum albumin at various stages of glycation. Clin. Chim. Acta 412, 277–285. 10.1016/j.cca.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vistoli G.; De Maddis D.; Cipak A.; Zarkovic N.; Carini M.; Aldini G. (2013) Advanced glycoxidation and lipoxidation end products (AGEs and ALEs): an overview of their mechanisms of formation. Free Radical Res. 47 (Suppl 1), 3–27. 10.3109/10715762.2013.815348. [DOI] [PubMed] [Google Scholar]
- Colombo G.; Clerici M.; Garavaglia M. E.; Giustarini D.; Rossi R.; Milzani A.; Dalle-Donne I. (2016) A step-by-step protocol for assaying protein carbonylation in biological samples. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 1019, 178–190. 10.1016/j.jchromb.2015.11.052. [DOI] [PubMed] [Google Scholar]
- Fedorova M.; Bollineni R. C.; Hoffmann R. (2014) Protein carbonylation as a major hallmark of oxidative damage: Update of analytical strategies. Mass Spectrom. Rev. 33, 79–97. 10.1002/mas.21381. [DOI] [PubMed] [Google Scholar]
- Frolov A.; Hoffmann R. (2010) Identification and relative quantification of specific glycation sites in human serum albumin. Anal. Bioanal. Chem. 397, 2349–2356. 10.1007/s00216-010-3810-9. [DOI] [PubMed] [Google Scholar]
- Aldini G.; Domingues M. R.; Spickett C. M.; Domingues P.; Altomare A.; Sanchez-Gomez F. J.; Oeste C. L.; Perez-Sala D. (2015) Protein lipoxidation: Detection strategies and challenges. Redox Biol. 5, 253–266. 10.1016/j.redox.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessome L. L.; Volmer D. A. (2006) Ion Suppression: A Major Concern in Mass Spectrometry. LCGC North Am. 24, 498–510. [Google Scholar]
- Sharmin S.; Sakata K.; Kashiwagi K.; Ueda S.; Iwasaki S.; Shirahata A.; Igarashi K. (2001) Polyamine cytotoxicity in the presence of bovine serum amine oxidase. Biochem. Biophys. Res. Commun. 282, 228–235. 10.1006/bbrc.2001.4569. [DOI] [PubMed] [Google Scholar]
- Luna L. G.; Green B. J.; Zhang F.; Arnold S. M.; Siegel P. D.; Bartels M. J. (2014) Quantitation of 4,4′-methylene diphenyl diisocyanate human serum albumin adducts. Toxicol. Rep. 1, 743–751. 10.1016/j.toxrep.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andacht T. M.; Pantazides B. G.; Crow B. S.; Fidder A.; Noort D.; Thomas J. D.; Blake T. A.; Johnson R. C. (2014) An enhanced throughput method for quantification of sulfur mustard adducts to human serum albumin via isotope dilution tandem mass spectrometry. J. Anal. Toxicol. 38, 8–15. 10.1093/jat/bkt088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel S.; Evans-Johnson J. A.; Georgieva N. I.; Boysen G. (2013) Exposure profiling of reactive compounds in complex mixtures. Toxicology 313, 145–150. 10.1016/j.tox.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M. K.; Grigoryan H.; Iavarone A. T.; Rappaport S. M. (2014) Antibody enrichment and mass spectrometry of albumin-Cys34 adducts. Chem. Res. Toxicol. 27, 400–407. 10.1021/tx400337k. [DOI] [PubMed] [Google Scholar]
- Liebler D. C. (2008) Protein damage by reactive electrophiles: targets and consequences. Chem. Res. Toxicol. 21, 117–128. 10.1021/tx700235t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J.; Wang Y.; Turesky R. J.; Kluetzman K.; Zhang Q. Y.; Ding X. (2016) Novel transgenic mouse model for studying human serum albumin as a biomarker of carcinogenic exposure. Chem. Res. Toxicol. 29, 797–809. 10.1021/acs.chemrestox.5b00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L.; Turesky R. J. (2011) Mass spectrometric characterization of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine N-oxidized metabolites bound at Cys34 of human serum albumin. Chem. Res. Toxicol. 24, 2004–2017. 10.1021/tx2003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B.; Tomazela D. M.; Shulman N.; Chambers M.; Finney G. L.; Frewen B.; Kern R.; Tabb D. L.; Liebler D. C.; MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968. 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou C. C.; Tsai C. F.; Teo G.; Chen Y. J.; Nesvizhskii A. I. (2016) Untargeted, spectral library-free analysis of data independent acquisition proteomics data generated using Orbitrap mass spectrometers. Proteomics 16, 2257. 10.1002/pmic.201500526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barknowitz G.; Engst W.; Schmidt S.; Bernau M.; Monien B. H.; Kramer M.; Florian S.; Glatt H. (2014) Identification and quantification of protein adducts formed by metabolites of 1-methoxy-3-indolylmethyl glucosinolate in vitro and in mouse models. Chem. Res. Toxicol. 27, 188–199. 10.1021/tx400277w. [DOI] [PubMed] [Google Scholar]
- McDonald T. A.; Waidyanatha S.; Rappaport S. M. (1993) Measurement of adducts of benzoquinone with hemoglobin and albumin. Carcinogenesis 14, 1927–1932. 10.1093/carcin/14.9.1927. [DOI] [PubMed] [Google Scholar]
- Waidyanatha S.; McDonald T. A.; Lin P. H.; Rappaport S. M. (1994) Measurement of hemoglobin and albumin adducts of tetrachlorobenzoquinone. Chem. Res. Toxicol. 7, 463–468. 10.1021/tx00039a027. [DOI] [PubMed] [Google Scholar]
- Pahler A.; Volkel W.; Dekant W. (1999) Quantitation of N epsilon-(dichloroacetyl)-L-lysine in proteins after perchloroethene exposure by gas chromatography-mass spectrometry using chemical ionization and negative ion detection following immunoaffinity chromatography. J. Chromatogr. A 847, 25–34. 10.1016/S0021-9673(99)00276-9. [DOI] [PubMed] [Google Scholar]
- Noort D.; Fidder A.; Hulst A. G.; de Jong L. P.; Benschop H. P. (2000) Diagnosis and dosimetry of exposure to sulfur mustard: development of a standard operating procedure for mass spectrometric analysis of haemoglobin adducts: exploratory research on albumin and keratin adducts. J. Anal. Toxicol. 20 (Suppl 1), S187–192. [DOI] [PubMed] [Google Scholar]
- Liu C.; Liang L.; Xiang Y.; Yu H.; Zhou S.; Xi H.; Liu S.; Liu J. (2015) An improved method for retrospective quantification of sulfur mustard exposure by detection of its albumin adduct using ultra-high pressure liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 407, 7037–7046. 10.1007/s00216-015-8842-8. [DOI] [PubMed] [Google Scholar]
- van der Schans M. J.; Hulst A. G.; van der Riet-van Oeveren D.; Noort D.; Benschop H. P.; Dishovsky C. (2013) New tools in diagnosis and biomonitoring of intoxications with organophosphorothioates: case studies with chlorpyrifos and diazinon. Chem.-Biol. Interact. 203, 96–102. 10.1016/j.cbi.2012.10.014. [DOI] [PubMed] [Google Scholar]
- Li B.; Eyer P.; Eddleston M.; Jiang W.; Schopfer L. M.; Lockridge O. (2013) Protein tyrosine adduct in humans self-poisoned by chlorpyrifos. Toxicol. Appl. Pharmacol. 269, 215–225. 10.1016/j.taap.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanazzi V.; Schiliro T.; Carraro E.; Gilli G. (2013) Immune response to acetaldehyde-human serum albumin adduct among healthy subjects related to alcohol intake. Environ. Toxicol. Pharmacol. 36, 378–383. 10.1016/j.etap.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Delatour T.; Richoz J.; Vouros P.; Turesky R. J. (2002) Simultaneous determination of 3-nitrotyrosine and tyrosine in plasma proteins of rats and assessment of artifactual tyrosine nitration. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 779, 189–199. 10.1016/S1570-0232(02)00370-7. [DOI] [PubMed] [Google Scholar]
- Sbarouni E.; Georgiadou P.; Voudris V. (2011) Ischemia modified albumin changes - review and clinical implications. Clin. Chem. Lab. Med. 49, 177–184. 10.1515/CCLM.2011.037. [DOI] [PubMed] [Google Scholar]
- Sitar M. E.; Aydin S.; Cakatay U. (2013) Human serum albumin and its relation with oxidative stress. Clin. Lab. 59, 945–952. 10.7754/Clin.Lab.2012.121115. [DOI] [PubMed] [Google Scholar]
- Renke J.; Popadiuk S.; Korzon M.; Bugajczyk B.; Woźniak M. (2000) Protein carbonyl groups’ content as a useful clinical marker of antioxidant barrier impairment in plasma of children with juvenile chronic arthritis. Free Radical Biol. Med. 29, 101–104. 10.1016/S0891-5849(00)00288-4. [DOI] [PubMed] [Google Scholar]
- Himmelfarb J.; McMonagle E.; McMenamin E. (2000) Plasma protein thiol oxidation and carbonyl formation in chronic renal failure. Kidney Int. 58, 2571–2578. 10.1046/j.1523-1755.2000.00443.x. [DOI] [PubMed] [Google Scholar]
- el-Bayoumy K.; Johnson B.; Partian S.; Upadhyaya P.; Hecht S. S. (1994) In vivo binding of 1-nitropyrene to albumin in the rat. Carcinogenesis 15, 119–123. 10.1093/carcin/15.1.119. [DOI] [PubMed] [Google Scholar]
- van Bekkum Y. M.; van den Broek P. H.; Scheepers P. T.; Noordhoek J.; Bos R. P. (1999) Biological fate of [14C]-1-nitropyrene in rats following intragastric administration. Chem.-Biol. Interact. 117, 15–33. 10.1016/S0009-2797(98)00095-7. [DOI] [PubMed] [Google Scholar]
- Birner G. (1989) Zur Hämoglobinbindung aromatischer Amine und Ihre Bedeutung für Expositionskontrolle und Risikobeurteilung im Rahmen des Biologischen Monitoring, Ph.D. Thesis, Julius-Maximilians-Universität Würzburg, Würzburg, Germany. [Google Scholar]
- Suzuki J.; Meguro S.; Morita O.; Hirayama S.; Suzuki S. (1989) Comparison of in vivo binding of aromatic nitro and amino compounds to rat hemoglobin. Biochem. Pharmacol. 38, 3511–3519. 10.1016/0006-2952(89)90122-6. [DOI] [PubMed] [Google Scholar]
- Thier R.; Lewalter J.; Selinski S.; Bolt H. M. (2001) Biological monitoring in workers in a nitrobenzene reduction plant: haemoglobin versus serum albumin adducts. Int. Arch. Occup. Environ. Health 74, 483–488. 10.1007/s004200100250. [DOI] [PubMed] [Google Scholar]
- Waynforth H. B. (1980) Experimental and Surgical Technique in the Rat. Academic Press, London. [Google Scholar]
- Westberg E. A.; Singh R.; Hedebrant U.; Koukouves G.; Souliotis V. L.; Farmer P. B.; Segerback D.; Kyrtopoulos S.; Tornqvist M. A. (2015) Adduct levels from benzo[a]pyrenediol epoxide: Relative formation to histidine in serum albumin and to deoxyguanosine in DNA in vitro and in vivo in mice measured by LC/MS-MS methods. Toxicol. Lett. 232, 28–36. 10.1016/j.toxlet.2014.09.019. [DOI] [PubMed] [Google Scholar]
- Aldini G.; Gamberoni L.; Orioli M.; Beretta G.; Regazzoni L.; Maffei F. R.; Carini M. (2006) Mass spectrometric characterization of covalent modification of human serum albumin by 4-hydroxy-trans-2-nonenal. J. Mass Spectrom. 41, 1149–1161. 10.1002/jms.1067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.