Abstract
The proper maintenance of potassium homeostasis is crucial for cell viability. Among the major determinants of potassium uptake in the model organism Saccharomyces cerevisiae are the Trk1 high affinity potassium transporter and the functionally redundant Hal4 (Sat4) and Hal5 protein kinases. These kinases are required for the plasma membrane accumulation of not only Trk1 but also several nutrient permeases. Here, we show that overexpression of the target of rapamycin complex 1 (TORC1) effector NPR1 improves hal4 hal5 growth defects by stabilizing nutrient permeases at the plasma membrane. We subsequently found that internal potassium levels and TORC1 activity are linked. Specifically, growth under limiting potassium alters the activities of Npr1 and another TORC1 effector kinase, Sch9; hal4 hal5 and trk1 trk2 mutants display hypersensitivity to rapamycin, and reciprocally, TORC1 inhibition reduces potassium accumulation. Our results demonstrate that in addition to carbon and nitrogen, TORC1 also responds to and regulates potassium fluxes.
Keywords: endocytosis, intracellular trafficking, potassium transport, target of rapamycin (TOR), yeast
Introduction
The target of rapamycin (TOR)4 pathway is known to respond to several nutrients, such as carbon and nitrogen, but whether potassium levels influence this pathway is unknown. Potassium is a key nutrient controlling several important biophysical parameters that have profound effects on cellular physiology, and the proper regulation of cellular ion homeostasis is a requirement for all living organisms. Among its many physiological functions, potassium is fundamental for the regulation of cellular volume, intracellular pH, maintenance of membrane potential, and protein synthesis (1–3). Trk1 is the high affinity transporter responsible for potassium uptake in the model organism Saccharomyces cerevisiae. Two functionally redundant protein kinases, Hal4 (Sat4) and Hal5, are positive regulators of the Trk1 transporter (4). More specifically, they regulate plasma membrane stability of the Trk1 transporter and other nutrient permeases, such as Can1, Mup1, Fur4, or Hxt1 (5, 6) by an unknown mechanism. We have observed that upon removal of potassium supplementation, these ion and nutrient transporters aberrantly accumulate in the vacuole. This instability of ion and nutrient transporters leads to alterations in the uptake and metabolism of carbon and nitrogen in hal4 hal5 mutants.
This function of the Hal4 and Hal5 kinases in the regulation of plasma membrane protein stability is similar to the roles of other members of the NPR/HAL5 family, including Ptk2 and Npr1. Ptk2 has been shown to regulate the activity of the Pma1 H+-ATPase, presumably by mediating the phosphorylation of serine 899 (7, 8). Npr1 has been shown to be an effector of the TOR pathway (see below) and to regulate the trafficking of several amino acid permeases including Gap1, Bap2, and Tat2 (9–12). Npr1-dependent phosphorylation is proposed to affect Rsp5-dependent ubiquitylation of these permeases, consequently influencing the amount of these proteins present in the plasma membrane.
RSP5 is an essential gene that encodes a HECT-type E3 ubiquitin ligase of the Nedd4 family that is responsible for the ubiquitylation of many ion and nutrient transporter proteins (13). Rsp5 requires, in many cases, specific adaptors for recognition and subsequent ubiquitylation of the target protein (14, 15). So far, 19 Rsp5 adaptor proteins have been described, among which there are 14 ART (arrestin-related trafficking) (16, 17). This E3 ubiquitin ligase/adaptor system is necessary for the regulation of ion and nutrient transporters and, therefore, plays a key role in nutrient homeostasis.
As mentioned, one of the central signal transduction pathways controlling nutrient homeostasis in eukaryotic organisms is the TOR pathway. Two separate signaling branches are controlled by two multiprotein complexes, termed TOR complex 1 (TORC1) and TOR complex 2 (TORC2) (18). TORC1 is rapamycin-sensitive, and in yeast it contains Kog1, Lst8, Tco89, and either Tor1 or Tor2 (19, 20). The physiological processes controlled by this signaling branch include synthesis and degradation of both proteins and mRNA, ribosome biogenesis, autophagy, and nutrient transport (21). Under favorable growth conditions, the TORC1 pathway is active, and cells maintain active ribosome biogenesis, protein translation, and nutrient uptake. However, when TORC1 activity is inhibited by the addition of rapamycin, under conditions of carbon or nitrogen starvation or in response to several stress conditions, protein translation dramatically decreases, stress responsive transcription factors are activated, and autophagy is induced (18, 21–23). TORC1 has been shown to be constitutively localized to the cytoplasmic face of the membrane of the vacuole, the nutrient reservoir in yeast, via its association with the EGO complex (24, 25). TORC1 regulates diverse aspects of cell physiology primarily via two major effector branches, the AGC family kinase Sch9 and Tap42-associated type 2A protein phosphatases (18). Sch9 is a direct substrate of TORC1 (25), and monitoring Sch9 phosphorylation serves as a convenient proxy to assess TORC1 activity. The mechanisms by which TORC1 signals to Tap42-PP2A complexes are less well understood (18).
TORC1 modulates nutrient homeostasis in part by regulating the complement of nutrient permeases at the plasma membrane via the Tap42-PP2A effector Npr1. Npr1 is a protein kinase activated upon TORC1 inhibition presumably by Tap42-PP2A-mediated dephosphorylation. Activated Npr1 phosphorylates a subset of Rsp5 adaptor proteins, including Ldb19 (hereafter referred to by its alias, Art1), Aly2 (Art3), Aly1 (Art6), Bul1, and Bul2 (26–28). The phosphorylation status of these adaptor proteins influences the Rsp5-mediated ubiquitylation of specific transporter proteins and thus participates in the regulation of the intracellular trafficking of nutrient transporters both from the plasma membrane to the vacuole via the multivesicular body (MVB) pathway and from the Golgi to the plasma membrane (26–28).
In this study we present data demonstrating that the TORC1 effector Npr1 is involved in the Hal4- and Hal5-mediated regulation of nutrient transporters. Specifically, Npr1 activity is low in hal4 hal5 cells, and NPR1 overexpression partially rescues hal4 hal5 phenotypes, which are themselves likely caused by accumulation of the Npr1-regulated ART protein, Art1. Moreover, we present evidence for a reciprocal regulation between potassium and the TORC1 signaling pathway by showing potassium-responsive changes in the phosphorylation status of both Npr1 and Sch9, rapamycin sensitivity of mutants with reduced internal potassium, and a decrease in potassium accumulation in rapamycin-treated cells.
Results
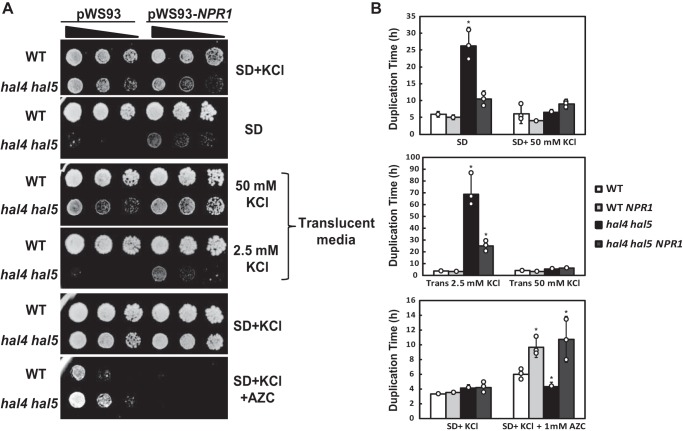
The TORC1 effector Npr1 is a protein kinase that has been implicated in the regulation of Rsp5-mediated permease degradation (9, 10, 26–28). More specifically, this kinase has been shown to phosphorylate members of the ART family, and in the case of Art1 this phosphorylation reduces its activity as an Rsp5 adaptor, leading to an increase in the plasma membrane accumulation of amino acid permeases, such as Can1 and Mup1 (27). We previously reported that in strains lacking the protein kinases Hal4 and Hal5, several plasma membrane transport proteins are aberrantly accumulated in the vacuole (5, 6). We tested whether the overexpression of NPR1 and the corresponding phospho-inhibition of ART proteins would improve the growth phenotypes observed for the hal4 hal5 mutant. As reported, the hal4 hal5 mutant presents a marked growth defect in SD media that is ameliorated by supplementing with KCl (4, 5). SD medium contains ∼7 mm KCl, but because of the high amount of ammonia, which is a competitive inhibitor of Trk1, this medium is relatively poor in potassium. Translucent medium is a yeast nitrogen base (YNB)-based medium that has been reformulated to contain negligible amounts of potassium (15 μm), which enables more precise studies of potassium-dependent phenotypes (29).
As shown in Fig. 1, A and B, NPR1 acts as a multicopy suppressor of the growth defect of the hal4 hal5 mutant in media with limiting potassium concentrations (both SD and translucent). The overexpression of NPR1 did not lead to wild type growth levels of the hal4 hal5 mutant, but it clearly improved the growth of this mutant in limiting potassium conditions.
FIGURE 1.
NPR1 overexpression rescued the growth defects of the hal4 hal5 mutant. A, serial dilutions of the indicated strains (BY4741 background) containing the empty plasmid (pWS93) or the NPR1 multicopy plasmid (pWS93-NPR1) were spotted onto SD medium supplemented or not with 0.1 m KCl onto potassium-free Translucent medium supplemented with 50 mm (replete) or 2.5 mm (limiting) KCl or onto potassium-supplemented SD media without (SD+KCl) or with 1 mm AZC (SD+KCl+AZC). Similar results were obtained in the W303-1A genetic background. B, the duplication times were determined for the same strains used in A in the indicated media as described under “Experimental Procedures.” The bars represent the average value for three independent determinations, the circles represent each individual data point, and the error bars show the S.D. Similar results were observed in two different experiments. Asterisks (*) indicate statistical significance (t test) with a p value < 0.05.
As mentioned above, the hal4 hal5 mutant has also been shown to have reduced levels of several amino acid permeases, which can be conveniently monitored using toxic analogs of amino acids, such as azetidine-2-carboxylic acid (AZC), a toxic analogue of proline (30). We observe that the hal4 hal5 mutant is more tolerant than the wild type control to AZC. The AZC tolerance observed in these mutants suggests that, as with other amino acid transporters, proline-specific permeases are present at reduced levels at the cell surface as compared with the wild type strain. The fact that NPR1 overexpression reduces this tolerance suggests that expression of this kinase restores the levels of amino acid permeases, as would be expected from previously published data showing that Npr1 regulates the plasma membrane accumulation of several amino acid permeases. Interestingly, NPR1 overexpression decreased the growth of both the wild type and the hal4 hal5 mutant in the presence of AZC, presumably by leading to a marked increase in the accumulation of transporters of this toxic proline analogue. For example, Npr1 is known to regulate not only Art1 but also other ART family members, Aly1 and Aly2, which are involved in the delivery of Gap1 to the plasma membrane (26, 31). Because this general amino acid permease is known to transport AZC, this likely explains the increased sensitivity of both strains.
We directly tested the hypothesis that Npr1 overexpression can ameliorate the amino acid transporter instability observed in hal4 hal5 mutants by monitoring the accumulation of the high affinity methionine permease, Mup1. We chose this permease based on our previous observations that Mup1 stability is acutely responsive to loss of HAL4 and HAL5 and that this permease is regulated by Art1 (6, 27). As shown in Fig. 2, A–C, NPR1 overexpression significantly increased the plasma membrane accumulation of Mup1 in hal4 hal5 mutants. As observed in Fig. 2A, upon removal of potassium, the Mup1-GFP signal was detected almost exclusively in the vacuole. However, upon overexpression of Npr1 in the hal4 hal5 mutant, we observed an increase in Mup1-GFP at the plasma membrane both in the presence and absence of potassium supplementation. These observations are quantified and confirmed by immunodetection in Fig. 2, B and C, respectively. We observe that the number of cells with Mup1-GFP in the plasma membrane increased 2-fold in potassium grown cells and upon potassium depletion in hal4 hal5 cells overexpressing NPR1. This observation is confirmed by the immunodetection of Mup1 in potassium-depleted hal4 hal5 cells expressing NPR1 (compare the third and fourth lanes, Fig. 2C). Moreover, we confirmed this effect of Npr1 overexpression on Mup1 stability in both the BY4741 and the W303-1A genetic backgrounds.
FIGURE 2.
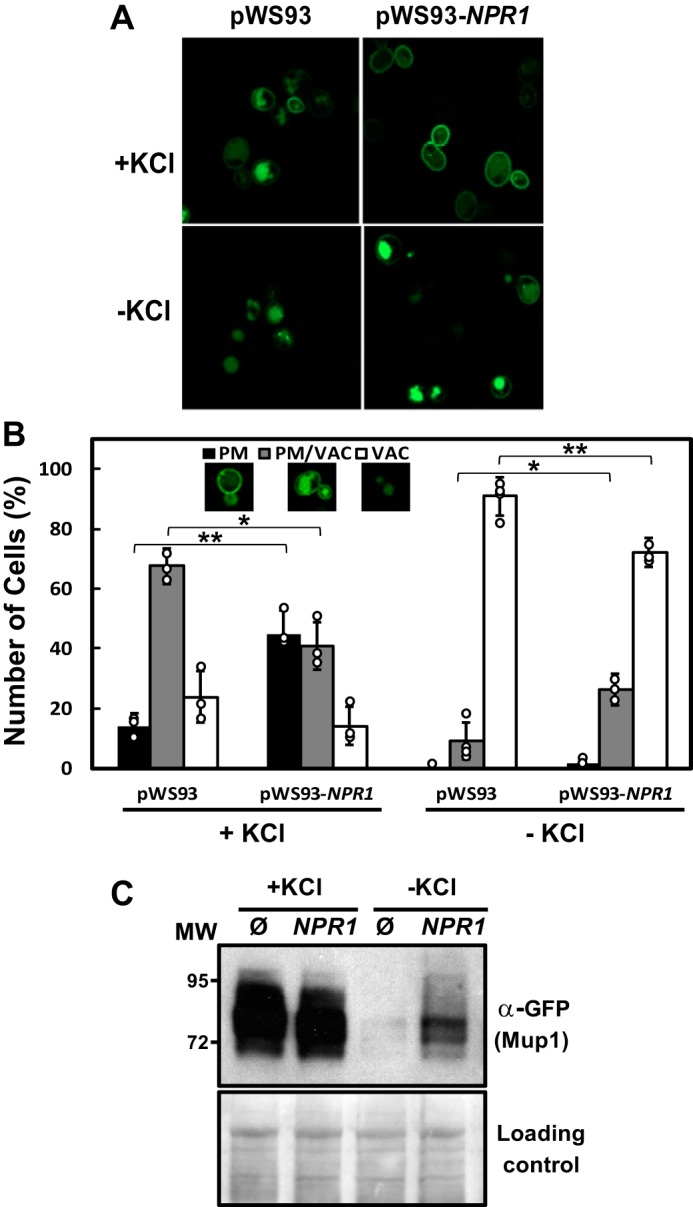
NPR1 overexpression increased plasma membrane stability of the Mup1 permease in the hal4 hal5 mutant. The BY4741 hal4 hal5 mutant was transformed with the centromeric plasmid containing a Mup1-GFP fusion and an empty plasmid or an NPR1-containing multicopy plasmid. The indicated strains were grown to mid-log phase in SD medium supplemented with 0.1 m KCl and then transferred to low potassium medium for 2 h (−KCl) or medium supplemented with 0.1 m KCl (+KCl). A, confocal microscopy images of Mup1-GFP localization in the hal4 hal5 mutant under the indicated experimental conditions. B, quantification of the number of cells that show Mup1 exclusively in the plasma membrane (PM), in the plasma membrane, and in the vacuole (PM/VAC) and strictly in the vacuole under the indicated experimental conditions. More than 50 cells were analyzed for each condition in four independent experiments. The bars represent the average value for the independent experiments, the circles represent each individual data point, and the error bars show the S.D. C, immunodetection of Mup1 present in the insoluble fraction isolated from the indicated strains. Asterisks (*) indicate statistical significance (t test) with a p value < 0.05. Double asterisks (**) indicate statistical significance with p value < 0.01.
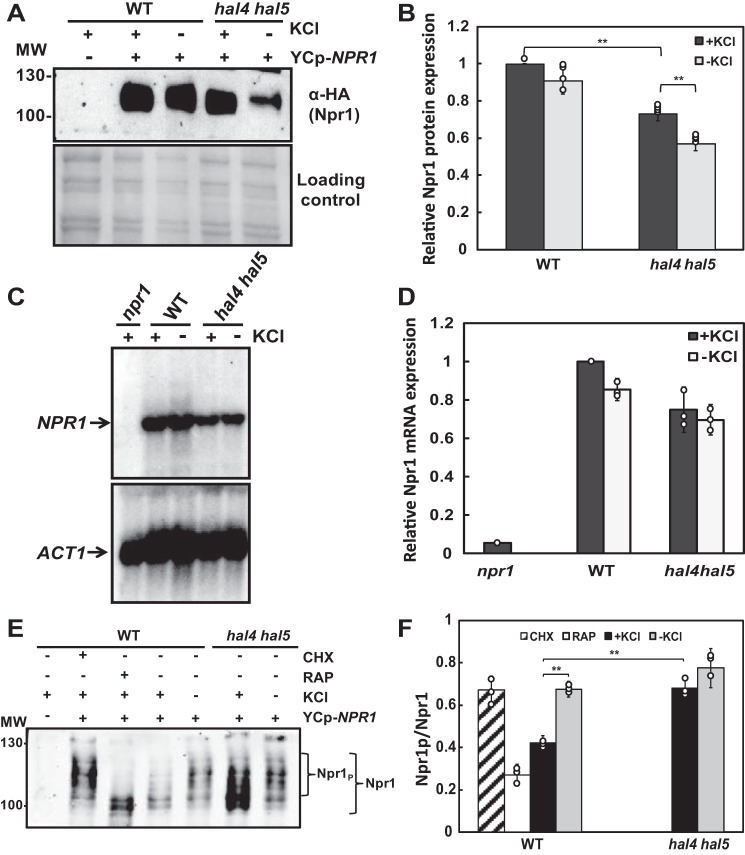
These results suggest that one defect of the hal4 hal5 mutant may be a decrease in Npr1 activity. To test this possibility, we examined the accumulation of Npr1 at both the protein and mRNA levels. As shown in Fig. 3, A–D, the amount of Npr1 protein, but not mRNA, was significantly decreased in hal4 hal5 mutants. As shown in Fig. 3A, the amount of Npr1 protein was reduced in hal4 hal5 mutants, especially upon removal of potassium. We quantified this decrease in several clones and observed a 40% reduction in Npr1 protein accumulation in the hal4 hal5 mutant (Fig. 3B). No significant differences were observed in the accumulation of NPR1 mRNA, suggesting that the regulation is not transcriptional (Fig. 3, C and D).
FIGURE 3.
Npr1 protein expression levels were reduced, whereas Npr1 phosphorylation levels were increased in hal4 hal5 mutant strains. A, the indicated BY4741 strains were transformed with a plasmid expressing the Npr1-HA fusion protein, grown to mid-log phase in SD medium supplemented with 0.1 m KCl, and incubated for 30 min in medium supplemented (+) or not (−) with 0.1 m KCl. The amount of Npr1 was analyzed using the anti-HA antibody (top panel), and the amount of protein present in each sample is shown in the Direct blue-stained filter (bottom panel). B, Npr1-HA quantification of the experiment described in panel A. Npr1 protein levels of WT cells grown in potassium supplemented media (+KCl) were used as the reference value. The bars represent the average value for the independent experiments, the circles represent each individual data point, and the error bars show the S.D. C, total RNA was extracted from the indicated strains, and conditions are described in panel A. Northern analysis was performed using a specific probe corresponding to nucleotides 1 to 658 of NPR1 (top panel). The membrane was re-probed with ACT1 as a loading control (bottom panel). D, the NPR1 mRNA signal was normalized using ACT1, and the results are expressed as relative induction of NPR1 using the value corresponding to the WT+KCl sample as the reference value. The bars represent the average value for the independent experiments, the circles represent each individual data point, and the error bars show the S.D. E, the indicated strains from panel A were grown in Translucent medium supplemented with 50 mm KCl. Cells were treated with rapamycin (RAP) (200 ng/ml), cycloheximide (CHX) (25 μg/ml), or incubated for 30 min without potassium and further analyzed for Npr1 electrophoretic mobility. F, Npr1 phosphoshift quantification was performed by densitometry. The position of the Npr1 phosphorylation bands was defined using WT cells treated with cycloheximide. Npr1P = phosphorylated Npr1. The bars represent the average value for the independent experiments, the circles represent each individual data point, and the error bars show the S.D. Double asterisks (**) indicate statistical significance with a p value < 0.01. Similar results were observed in the W303-1A background.
Several reports have thoroughly demonstrated that the level of Npr1 phosphorylation is controlled by the TORC1 protein kinase complex, increasing its phosphorylation level under conditions of TORC1 activation and becoming dephosphorylated upon TORC1 inhibition (10, 27, 32). Moreover, these studies have shown that TORC1-dependent phosphorylation of Npr1 leads to decreased Nrp1 activity. Therefore, we next examined the relative levels of Npr1 phosphorylation in wild type (WT) and hal4 hal5 strains grown in potassium-replete media and after incubation of the cells in low potassium media. As controls, we treated the cells with both cycloheximide (TORC1 activator) and rapamycin (TORC1 inhibitor). We observed that in the hal4 hal5 mutant there is an increase in the proportion of Npr1 that is phosphorylated, as the migration pattern is very similar to that observed upon cycloheximide treatment both in the presence and absence of potassium (compare the second lane with the sixth and seventh lanes, Fig. 3E). We quantified the relative phosphorylation state of Npr1 in each of the conditions tested in multiple clones. As observed in Fig. 3F, the ratio of phosphorylated Npr1 is very similar in wild type strains treated with the TORC1 activator and in the strains lacking the Hal4 and Hal5 kinases. As stated, this phosphorylation has been correlated with a decrease in Npr1 activity and thus is consistent with the observations described above suggesting that Npr1 activity is reduced in hal4 hal5 mutants (10, 32). Interestingly, this increased phosphorylation is also observed in low potassium conditions in the wild type strain (fifth lane, Fig. 3E). This observation will be discussed in more detail below.
Previous reports demonstrate that Npr1 phosphorylates and inactivates Art1 (27). This phosphorylation can be observed as a band shift, with the upper band of the Art1 doublet corresponding to the post-translationally modified form (bottom panel, Fig. 4B and Ref. 33). Therefore, we hypothesized that the decrease in Npr1 activity observed in hal4 hal5 mutants would lead to an increase in the accumulation of active Art1, thus providing a plausible explanation for the aberrant degradation of Art1-dependent cargos, like Can1 and Mup1, observed in this mutant. A schematic diagram depicting this model is shown in Fig. 4A. In support of this hypothesis, we observed a marked increase in the accumulation of the non-phosphorylated, active form of the Art1 protein and a decrease in the proportion of phosphorylated Art1 in hal4 hal5 mutants, which is reverted upon NPR1 overexpression (Fig. 4, B and C). This result shows a correlation with decreased Npr1 activity and increased accumulation of active Art1. Our data provide a mechanistic explanation for the previously reported hal4 hal5 phenotypes, which implicate the TORC1 effector kinase Npr1 and the Rsp5/adaptor network in the aberrant degradation of amino acid permeases.
FIGURE 4.
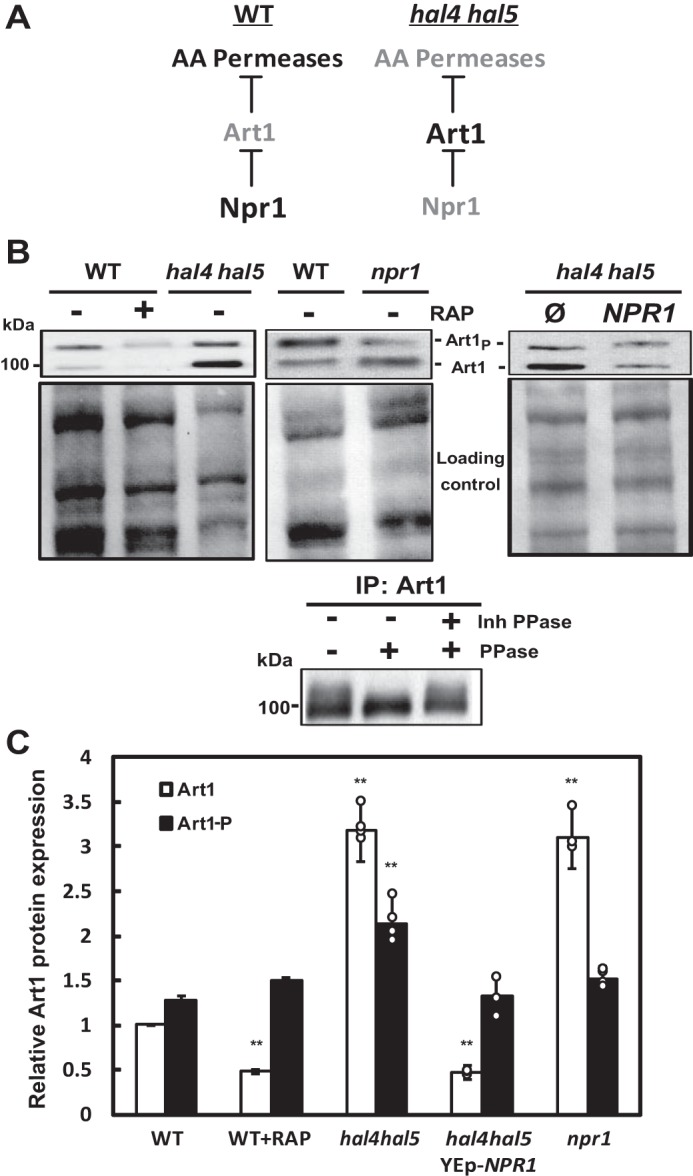
Art1 protein levels were increased in hal4 hal5 mutant strains. A, schematic representation of the proposed model relating Npr1 and Art1 activity to permease degradation in the hal4 hal5 mutant. B, the indicated BY4741 strains were transformed with a plasmid expressing the Art1-HA fusion protein and grown to mid-log phase in SD medium supplemented with 0.1 m KCl. The WT strain was incubated for 30 min with 200 ng/ml of rapamycin (WT+RAP). The amount and electrophoretic mobility of Art1 was analyzed using the anti-HA antibody (top panel), and the amount of protein present in each sample is shown in the Direct blue-stained filter below each Western blot. The bottom panels show the change in the electrophoretic mobility of immunoprecipitated Art1 in the presence and absence of phosphatase treatment. The wild type strain expressing Art1-HA was grown to mid-log phase and treated for 30 min with 200 ng/ml rapamycin. Art1 was immunoprecipitated and split into three aliquots that were treated as indicated in the figure. C, quantification of Art1 and phosphorylated Art1 (Art1-P) from conditions described in panel A. Non-phosphorylated Art1 protein levels of WT cells grown in potassium-supplemented media (+KCl) are used as the reference value. The bars represent the average value for the independent experiments, the circles represent each individual data point, and the error bars show the S.D. Double asterisks (**) indicate statistical significance (t test) with p value < 0.01.
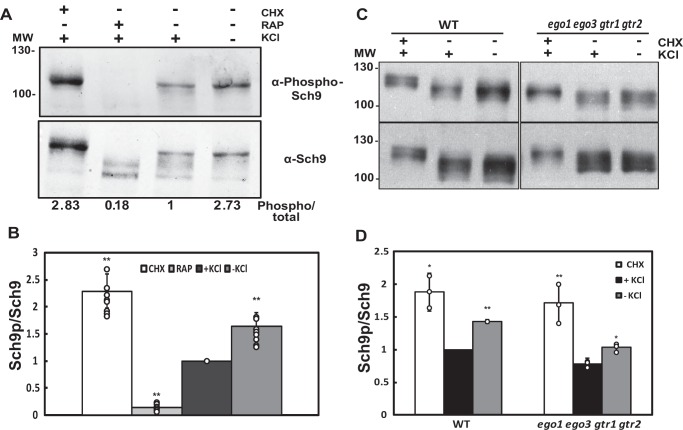
As mentioned above, a very interesting observation that can be extracted from the data presented in Fig. 3, E and F, is that Npr1 phosphorylation increases in response to limiting potassium conditions, not only in the hal4 hal5 mutant but also in the wild type strain (compare the fourth and fifth lanes in Fig. 3E). This result suggests that it is the low potassium that activates the TORC1 signaling pathway. To further corroborate this observation, we tested the phosphorylation of another TORC1 effector, Sch9, which is a direct substrate of TORC1, and its phosphorylation is used as a read-out to measure TORC1 activity. As in the case of Npr1, we used cycloheximide to activate TORC1 and rapamycin to inhibit the complex. In this way we can observe the hyper- and hypophosphorylated state of Sch9. Using a phospho-specific antibody, we observed that the levels of phosphorylation of Sch9 also increase in response to limiting potassium concentrations (Fig. 5, A and B). These results are in good agreement with the data presented regarding the phosphorylation of Npr1 tested under the same conditions (Fig. 3. E and F). Taken together, these results indicate that low potassium concentrations enhance the activation of TORC1.
FIGURE 5.
Levels of Sch9 phosphorylation increased in response to limiting potassium conditions. A, BY4741 cells were grown in Translucent medium supplemented with 50 mm KCl. Cells were treated with rapamycin (RAP) (200 ng/ml) or cycloheximide (CHX) (25 μg/ml) or incubated for 30 min without potassium and further analyzed for Sch9 phosphorylation by immunodetection employing the phospho-specific Sch9S758 and pan-Sch9 antibodies. B, quantification of Sch9 phosphorylation levels from experiments performed as in panel A were normalized using Sch9 total protein, and the results are expressed as relative phosphorylation of Sch9 considering WT cells grown in potassium-supplemented Translucent medium (+KCl) as the reference value. Results correspond to the mean values ± S.D. of independent clones. Sch9p = phosphorylated Sch9. Similar results were also observed in the W303-1A background. C, TB50a control (WT) and ego1 ego3 gtr1 gtr2 strains were analyzed as described in A. A representative experiment is shown. D, quantification of Sch9 phosphorylation levels was performed as in B. The bars represent the average value for the independent experiments, the circles represent each individual data point, and the error bars show the S.D. Asterisks (*) indicate statistical significance (t test) with a p value < 0.05. Double asterisks (**) indicates statistical significance with a p value < 0.01.
In the case of other nutrients, like nitrogen, starvation leads to TORC1 inactivation (18). Here, we observe that in conditions of suboptimal potassium concentrations (hal4 hal5 mutant) or during the initial stages of decreased external potassium, TORC1 substrates are hyperphosphorylated. At least two hypotheses can be proposed to explain this response: 1) TORC1 activation is required to mobilize potassium from the vacuole to maintain adequate levels of this cation, or 2) TORC1 activation favors the uptake of potassium from the extracellular media to ameliorate the potassium deficit.
To begin to study these possibilities, we next examined whether this signaling pathway requires the EGO complex. Previous results in yeast and higher eukaryotes have shown that the EGO complex (Regulator complex in higher eukaryotes) functions upstream of TORC1 to mediate amino acid signaling and that in mammals this complex is required for localization of the TORC1 complex to the vacuole (24, 34–36). Therefore, we tested whether the EGO complex, which regulates some activities of TORC1, is required for the Sch9 phosphorylation observed upon potassium starvation. As observed in Fig. 5, C and D, the increased Sch9 phosphorylation observed in low potassium medium does not require the EGO complex, as the levels of Sch9 phosphorylation are very similar in the mutant lacking the four components of the EGO complex (ego1 ego2 gtr1 gtr2) and the isogenic wild type control. This experiment does not formally rule out a role for the vacuole in this response, as additional experiments would be required to fully address this question, but it does show that this mechanism of TORC1 activation does not require the EGO complex.
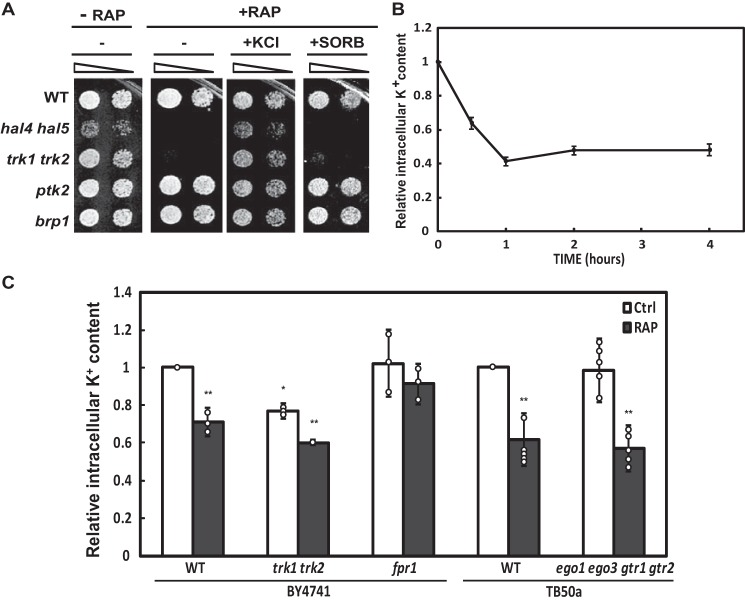
To experimentally test the second hypothesis postulating that the TORC1 complex is involved in maintaining internal potassium concentrations, we tested the rapamycin sensitivity of strains defective in high affinity potassium transport. We reasoned that if TORC1 activity is necessary to maintain internal potassium concentrations, mutants with lower internal potassium may be more sensitive to TORC1 inhibition. We observed that both the hal4 hal5 and trk1 trk2 mutants are sensitive to rapamycin (Fig. 6A).
FIGURE 6.
Rapamycin treatment inhibited the growth of high affinity potassium uptake mutants and lowered internal potassium accumulation. A, the indicated BY4741 strains were grown to saturation in YPD medium, serially diluted, and spotted onto plates of YPD and YPD supplemented with 5 ng/ml rapamycin with or without potassium supplementation (0.5 m) (+KCl) or the addition of the osmotically equivalent amount of sorbitol (0.8 m) (+SORB). Growth was recorded after 3 to 4 days of incubation at 28 °C. This experiment was performed in three different biological replicates. B, cells from wild type yeast (BY4741) were grown to mid-log phase in YPD, 200 ng/ml rapamycin (RAP) were added, and samples were taken at the indicated times for the determination of the intracellular potassium concentration. The average values of triplicate determinations are represented in the graph. The error bars represent the S.D. Essentially identical results were obtained in three independent experiments. All data points present a p value > 0.01, as compared with the control sample. Similar results were observed in the W303-1A background. C, cells from wild type yeast (BY4741), trk1 trk2 and fpr1 mutants and the ego1 ego3 gtr1 gtr2 strain and its wild type control (TB50a) were grown to mid-log phase in YPD, and 200 ng/ml of rapamycin (RAP) or the vehicle (control) was added during 60 min. Samples were taken for the determination of the intracellular potassium concentration. The value obtained for WT cells treated with the vehicle (control) was set as the reference value. The bars represent the average values, the circles represent each individual data point, and the error bars show the S.D. Asterisks (*) indicate statistical significance (t test) with a p value < 0.05. Double asterisks (**) indicate a p value < 0.01.
To determine whether this sensitivity involves internal acidification or decreased amino acid uptake, known to occur in these low internal potassium mutants, we tested the rapamycin sensitivity of two other mutant strains. The brp1 mutant lacks most of the PMA1 promoter, leading to decreased expression of the H+-ATPase, whereas the activity of Pma1 is decreased in ptk2 mutants. In both cases, the cytosol is more acidic and amino acid uptake is reduced due to the decrease in the proton gradient (7, 36). Neither of these mutants was sensitive to rapamycin (Fig. 6A).
This analysis allows us to discard a role for decreased internal pH and decreased amino acid uptake in this phenotype, suggesting that it is the low internal potassium concentrations of the hal4 hal5 and trk1 trk2 mutants that led to rapamycin sensitivity. Further supporting this interpretation, we observe that the addition of external potassium (but not the osmotic equivalent of sorbitol) clearly rescues the growth of the hal4 hal5 and trk1 trk2 mutants in the presence of rapamycin (Fig. 6A).
Taken together, these results suggest that the hal4 hal5 and trk1 trk2 mutants are sensitive to rapamycin due to inhibition of residual potassium uptake known to take place in these mutant strains (37). To directly test whether rapamycin treatment affects potassium accumulation, we measured this parameter in the wild type strain treated with rapamycin (Fig. 6B). We observe a marked decrease in potassium accumulation within 30 min, which reached its lowest level after 1 h. These decreased internal potassium levels were maintained over the duration of the assay (4 h).
To determine whether this phenotype was caused by TORC1 inhibition and not a nonspecific effect of rapamycin toxicity, we next tested the strain lacking the FPR1 gene, which encodes the protein necessary for the formation of the rapamycin-Tor1/2 complex (38, 39). We included the trk1 trk2 mutant in this analysis to test whether the presence of the high affinity potassium transporters influenced this phenotype. We observed that rapamycin treatment decreased the internal potassium accumulation in both wild type and trk1 trk2 strains but had no effect in the fpr1 mutant (Fig. 6C). The amount of potassium in untreated trk1 trk2 strains is reduced compared with the wild type control, as would be expected, and the addition of rapamycin led to a further decrease in the amount of internal potassium in this mutant, in agreement with the growth profile presented above for the trk1 trk2 mutant. These results show that inhibition of TORC1 reduces potassium accumulation in a Trk1/Trk2-independent fashion.
Finally, as we did for Sch9 activation, we tested whether the EGO complex plays a role in the decreased potassium accumulation upon rapamycin treatment. In Fig. 6C we showed that rapamycin treatment led to a decrease in potassium accumulation in mutants lacking the gene encoding the EGO complex (ego1 ego2 gtr1 gtr2). This result suggests that this complex is not strictly required for this phenotype, as observed for Sch9 phosphorylation. It also confirms the rapamycin-mediated decrease in internal potassium in a second genetic background (TB50a). We also tested the W303-1A background and observed similar results (data not shown).
Discussion
The results presented here reveal a connection between potassium homeostasis and TORC1 activity. On one hand, we found that two TORC1 effectors, Npr1 and Sch9, are hyperphosphorylated when internal potassium concentrations are constitutively low or in response to a shift to low potassium medium. On the other hand, we also found that inhibition of TORC1 activity reduces potassium accumulation independently of the Trk1 and Trk2 high affinity transporters. Thus, we conclude that TORC1 performs an unanticipated and potentially conserved role in potassium homeostasis.
The regulation of potassium homeostasis is crucial for many biophysical parameters, such as membrane potential and internal pH, and previous studies have shown that mutations leading to alterations in the activity of the two major transport systems governing membrane potential in the model yeast S. cerevisiae have clear effects on many aspects of nutrient uptake. More specifically, mutations that decrease Pma1 or Trk1 activity led to decreased amino acid uptake, whereas mutations that led to overactivation of either system increase uptake by creating a more favorable proton gradient for the symport of amino acids (4, 7, 40, 41). Some of the mutations characterized directly affect the transport protein by direct deletion of either the promoter region, the coding sequence, or by deletion of enzymes that post-translationally regulate the transporters, such as Ptk2 or Ppz1. In other cases, such as Hal4 and Hal5, the mechanism of action is more complex. This mutant is known to have reduced internal potassium via regulation of Trk1 and Trk2, but how these kinases regulate potassium homeostasis is still being defined. Our previous studies have demonstrated that in strains lacking the Hal4 and Hal5 kinases, Trk1 and several other nutrient transporters aberrantly accumulate in the vacuole in the absence of potassium supplementation (5, 6).
To begin to define the function of the Hal4 and Hal5 kinases in the maintenance of transporter plasma membrane stability, we first determined whether this regulation involved Rsp5-mediated endocytosis. In our previous data using the hal4 hal5 npi1 triple mutant, latrunculin A-mediated inhibition of endocytosis, and mutated versions of Rsp5 support a role for the Hal4 and Hal5 kinase in regulating the ubiquitin-mediated endocytosis of nutrient transporters (5).5
Here, we report that Npr1 acts as a multicopy suppressor of the hal4 hal5 mutant. Moreover, the accumulation of the Npr1 kinase is reduced in these mutants, and the phosphorylation levels of the remaining Npr1 protein are also increased. Phosphorylation of Npr1 has been shown to correlate with decreased activity of the kinase; therefore, the levels of active Npr1 are markedly reduced in the hal4 hal5 mutant (10, 32). Moreover, we observed that a decrease in Npr1 activity leads to an over-accumulation of the active form of the Rsp5 adaptor, Art1. Taken together, our data indicate that the aberrant degradation of nutrient transporters, such as Can1 or Mup1, observed in the hal4 hal5 mutant is explained, at least in part, by deficient Npr1 activity. Deficient Npr1 activity would lead to an increase in Rsp5-mediated ubiquitylation of amino acid permeases by increasing the interaction of Rsp5 with its adaptor protein, Art1. This interpretation is directly supported by the data presented here showing that Mup1 plasma membrane accumulation increases in hal4 hal5 mutants overexpressing Npr1 and provides a mechanistic explanation for the observed phenotypes.
One very interesting observation stemming from the studies described above is that in low potassium medium, the phosphorylation levels of Npr1 increase not only in the hal4 hal5 mutants but also in the wild type strain. Importantly, we show that the phosphorylation levels of an additional TORC1 effector, Sch9, are also increased in response to low potassium. However, we did not obtain any evidence that Sch9 is required for potassium uptake or accumulation, as the growth pattern of strains lacking SCH9 is not altered by external potassium concentrations and the rapamycin-dependent decrease in potassium accumulation was similar in the wild type and sch9 mutant.6
Previous experiments examining the subcellular localization of the TORC1-responsive Gln3 transcription factor, which is regulated by Npr1, also support this finding (42). These authors report that in mutants lacking HAL4 or HAL5, which have lower internal potassium, Gln3 nuclear localization is reduced, whereas in mutants lacking PPZ1, which have higher intracellular potassium, Gln3 accumulates in the nucleus (4, 40). These data add support to a model in which low intracellular potassium would favor TORC1 activity, impeding Gln3 nuclear localization, whereas internal potassium accumulation would lead to a decrease in TORC1 activity, inducing Gln3 translocation to the nucleus. This same model explains the hal4 hal5 mutant phenotypes that are rescued by NPR1 overexpression discussed above. More specifically, the lower internal potassium concentration in hal4 hal5 mutants would lead to TORC1 activation and, consequently, Npr1 phosphorylation. This inhibition of Npr1 activity leads to an accumulation of active Art1 and subsequent Rsp5-mediated ubiquitylation and vacuolar accumulation of a subset of amino acid permeases. Thus, this model provides a unifying explanation for several previously reported phenomena.
In the case of other nutrients, such as nitrogen, starvation induces TORC1 inactivation. Our data suggest that in conditions of limiting potassium, TORC1 activity is favored. Recent evidence suggests that different stress or starvation conditions have varying effects on TORC1 signaling (43) so that the view that starvation leads to TORC1 activation may be too simplistic. Also, cycloheximide treatment is also known to activate TORC1, presumably by increasing intracellular amino acids. Thus, one possibility is that low internal potassium favors the exit of amino acids stored in the vacuole to increase availability in the cytosol. It may be that TORC1 activation is required to ameliorate the more acute problem of low internal potassium or another parameter that is caused by this deficit. Further experiments are needed to clarify this issue. However, this line of investigation has lead to the more important observation that TORC1 inhibition has a pronounced effect on internal potassium concentrations. The physiological effect of this inhibition is reflected in the marked sensitivity displayed by the trk1 trk2 and hal4 hal5 mutants to rapamycin.
The results discussed above suggest that rapamycin treatment decreases potassium accumulation independently of Trk1 and Trk2, which we prove experimentally by showing that rapamycin treatment decreases internal potassium concentrations in both the wild type strain and in the trk1 trk2 mutant. Importantly, this effect is specific for TORC1 inhibition and not an unrelated effect of rapamycin, as the fpr1 mutant, which lacks the gene encoding the peptidyl-prolyl cis-trans isomerase necessary for rapamycin to bind to and inhibit TORC1, is unaffected (38, 44). Further studies are required to determine the molecular mechanism involved, but several possibilities exist. For example, TORC1 could inhibit the outward rectifying Tok1 channel. Upon TORC1 inhibition, Tok1 would become activated, and internal potassium would be released. However, as the tok1 mutant presents no growth phenotype in the presence of rapamycin and rapamycin treatment of tok1 strains leads to the same decrease in internal potassium (data not shown), this possibility appears unlikely.
Alternatively, TORC1 could activate Trk1-Trk2-independent potassium uptake. There is no consensus in the literature as to how potassium uptake is mediated in the absence of Trk1 and Trk2. Some studies suggest that a channel activity denominated NSC1 is responsible, but this channel has yet to be characterized at the molecular level (45, 46). Other candidates for these uptake systems include nonspecific uptake by the Qdr2 drug/H+ antiporter, glucose permeases, and the Kch1 and Kch2 (Prm6) potassium transporter proteins (47–50).
Yet another possibility is TORC1-mediated regulation of the plasma membrane H+-ATPase Pma1 or the vacuolar H+-ATPase (V-ATPase) complex. In this scenario, TORC1 inhibition would lead to a decrease in H+-ATPase activity of one or both proteins, which would decrease the cytosolic pH and the membrane potential. This change in the membrane potential would reduce potassium uptake through the nonspecific uptake systems described above. It has been reported that cytosolic pH regulates TORC1 activity via the V-ATPase (51). Our data indicate that a reciprocal regulation may exist between TORC1 and the V-ATPase if the rapamycin-induced decrease in potassium accumulation is indeed mediated by a decrease in V-ATPase activity. Further studies will determine whether this is the case and, if so, whether this is the regulatory mechanism responsible for the TORC1-dependent effect on potassium homeostasis presented here.
Our work uncovers a functional relationship between potassium homeostasis and TORC1 activity. As potassium is the most abundant alkali metal found in many organisms and it controls several aspects of cellular physiology, its connection with the TORC1 pathway may be important not only in the model yeast, S. cerevisiae, but also in many other organisms including pathogenic fungi and higher eukaryotes.
Experimental Procedures
Yeast Strains, Culture Conditions, and Reagents
All single mutant strains from the BY4741 background used in this work were obtained from the EUROSCARF collection. Gene disruptions were confirmed by sequencing PCR products of the regions flanking the kanMX insertion. The BY4741 trk1 trk2 mutant strain was kindly provided by Dr. Hana Sychrová (52). The BY4741 hal4 hal5 strain has been described previously (4, 6). The EGO complex mutant is derived from the TB50a background (HS195–2.2d gtr1::hph ego1::KanMX gtr2::hph ego3::KanMX). YPD contained 2% glucose, 2% peptone, and 1% yeast extract. Minimal medium (SD) contained 2% glucose, 0.7% yeast nitrogen base (Difco) without amino acids, 50 mm succinic acid adjusted to pH 5.5 with Tris, and the nutritional components required by the strains with appropriate auxotrophic markers. Experiments in limiting potassium conditions were performed by growing the cells in Translucent medium supplemented with 50 mm KCl, and then cells were transferred to Translucent K+-free medium with or without KCl for the indicated times. Growth assays were performed on solid media by spotting serial dilutions of saturated cultures onto plates with the indicated composition. Images were taken after 3–5 days of growth. The growth in liquid media was recorded using a Bioscreen C analyzer (ThermoFisher) using a 600-nm filter every 15 min during a 48-h period. The calculation of doubling times was carried out as described (53). Rapamycin was purchased from LC Laboratories (Woburn, MA), cycloheximide was from Applichem Inc., and AZC was from Sigma.
Plasmids Used in This Work
The YEp-MUP1-GFP plasmid was previously described (54). The YCp-NPR1 plasmid used in this work was generated from pRS415-promNPR1-NPR1-3xHA (27) by replacing LEU2 marker with URA3 marker. The pWS93-NPR1 plasmid (also referred in this work as YEp-NPR1) was constructed by inserting the SmaI/SalI NPR1 genomic fragment into the same sites in the pWS93 vector (55). The pAG416-ART1-3xHA plasmid was constructed by homologous recombination in yeast using the pAG416GPD vector (Gateway). The ART1 coding sequence was amplified by PCR from genomic DNA using the following primers: Art1-recomb-5′, ACCAGAACTTAGTTTCGACGGATTCTAGAACTAGTGGATCCCATGGCATTTTCACGTCTTACATC, and Art1-recomb-3′, CATTCCACCTAAGCTTGATATCGAATTCCTGCAGCCCATCACCTGGGTTATTCTATTGGAATCTAG. The plasmid was linearized using NotI, dephosphorylated, and co-transformed into the art1 mutant with the ART1 PCR fragment. Positive clones were selected first by growth in media without uracil and then by Western blotting analysis. Plasmids were recovered from candidate clones, transformed into Escherichia coli, purified, and confirmed by sequencing. The pRS411 plasmid (CEN/ARS, MET15) was used for complementing methionine auxotrophy in BY4741 hal4 hal5 strain in the experiment described in Fig. 2, A–C (56).
Protein Extraction and Cell Fractionation
The indicated yeast strains were grown in potassium-supplemented minimal medium to an optical density at 660 nm of 0.5, and cells were incubated for the indicated times in medium with or without potassium supplementation, harvested by centrifugation, and frozen at −70 °C. Cells were resuspended in homogenization buffer (50 mm Tris (pH 7.6), 0.1 m KCl, 5 mm EDTA, 5 mm dithioerythritol, 20% (w/v) sucrose, and protease inhibitor mixture (Roche Diagnostics) and lysed by vortexing with glass beads. The lysate was collected after centrifugation for 5 min at 500 × g at 4 °C. The crude extract was separated into soluble and particulate fractions by centrifugation for 30 min at 16,000 × g at 4 °C. The particulate fraction was resuspended directly in Laemmli sample buffer.
To monitor the changes in the phosphorylation state of Npr1, Art1, and Sch9, cells were grown in potassium supplemented SD or Translucent medium, treated as indicated in the corresponding figure legend, mixed with TCA (final concentration 6%), and put on ice for at least 5 min before cells were collected by centrifugation, washed twice with cold acetone, and dried in a SpeedVac. Cell lysis was done in 100 μl of urea buffer (25 mm Tris (pH 6.8), 5 mm EDTA, 6 m urea, 1% SDS) with 200 μl of glass beads in a bead beater (5 times for 45 s) with subsequent heating for 10 min at 65 °C.
Immunoprecipitation and Phosphatase Treatment
The wild type strain expressing Art1-HA was grown in YPD media to mid-log phase and treated with 200 ng/ml rapamycin for 30 min. Cells were collected and lysed as described above, and Art1-HA was immunoprecipitated using anti-HA antibodies (Covance) and Protein A/G magnetic beads (ThermoFisher). Three aliquots were prepared. The first was left untreated, and the other two aliquots were treated for 30 min at 37 °C with 40 units of alkaline phosphatase (Roche Diagnostics) in the presence or absence of PhosSTOP phosphatase inhibitor (Roche Diagnostics).
Western and Immunoblotting
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and immunoblotted with either a monoclonal anti-HA antibody (1:5000) (Covance) for Npr1 and Art1 detection or anti-GFP (1:10,000) (Roche Diagnostics) for Mup1 detection. Immunoreactive bands were visualized using the ECLPlus chemiluminescence system and horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences). When necessary, membranes were stripped by incubation for 30 min at 50 °C in a buffer containing 100 mm 2-mercaptoethanol, 2% sodium dodecyl sulfate, and 62.5 mm Tris-HCl (pH 6.7) followed by extensive washes and re-probed as described above. Transfer quality and total protein levels were monitored by Direct blue 71 (Sigma) staining of the membrane as described (57).
The Li-Cor infrared fluorescent system was used for quantitative Western blotting. Sch9 and Sch9S758 antibodies were incubated in PBS 0.01% Tween 20 (PBST) supplemented with 5% BSA. Washing steps were performed using PBST. After 1 h of blocking in PBST 5% BSA, membranes were probed overnight with primary antibodies. Membranes were washed three times with PBST, and anti-Sch9S758 and anti-Sch9 were detected with donkey anti-rabbit and goat anti-rabbit secondary antibodies coupled to the infrared dyes IRDye680RD, respectively (Li-Cor, NE). After three washes, fluorescence was detected using the Odysseys IR imaging system (Li-Cor). Signal quantification was done using Image Studio Lite v 4 software from LI-COR.
Confocal Microscopy
Fluorescence images were obtained for live cells using the Zeiss 780 confocal microscope with excitation at 488 nm and detection at 510–550 nm (objective: plan-apochromat 40×/1.3 oil DIC M27, ZEN 2012 software).
Northern Blot Analysis
Total RNA was isolated from 50 ml of yeast cells that were grown to mid-log phase in SD supplemented with 0.1 m KCl. Cells were washed twice with SD medium and resuspended to the same minimal medium without potassium supplement (−KCl) or containing 0.1 m KCl (+KCl). Approximately 20 μg of RNA per lane was separated in formaldehyde gels and transferred onto nylon membranes (Hybond-N; Amersham Biosciences). Radioactively labeled probes were hybridized in PSE buffer (300 mm sodium phosphate (pH 7.2), 7% sodium dodecyl sulfate, 1 mm EDTA). Probes used were as follows: a PCR fragment representing nucleotides 1–658 of the NPR1 gene and nucleotides 518–1182 of ACT1, amplified from chromosomal yeast DNA. Signal quantification was done using Fujifilm BAS-1500 phosphorimaging.
Measurement of Intracellular Potassium Concentrations
Measurements of intracellular potassium were performed essentially as described (4). Briefly, cells were grown in YPD to an optical density at 660 nm of 0.6–0.7 and treated with 200 ng/ml rapamycin or the equivalent vehicle (control) during 60 min at 28 °C. Aliquots of 10 ml were taken at the indicated times, centrifuged for 5 min at 500 × g and 4 °C, and washed twice with 10 ml of ice-cold washing solution (20 mm MgCl2 and iso-osmotic sorbitol). The cell pellets were resuspended with 1 ml of cold washing solution, centrifuged again, and resuspended in 0.5 ml of 20 mm MgCl2. Ions were extracted by heating the cells for 15 min at 95 °C. After centrifugation, aliquots of the supernatant were analyzed with an atomic absorption spectrometer (Varian) in flame emission mode. The amount of potassium (nmol/mg fresh weight) was calculated for each sample, and the data were normalized using the untreated WT strain as the reference value.
Author Contributions
C. P., R. L., and L. Y. designed the experiments. C. P., A. F.-B., and L. Y. carried out the experiments. C. P., R. L. and L. Y. wrote the manuscript.
This work was supported by Spanish Ministry of Science and Innovation (Madrid, Spain) Grant BFU2011-30197-C03-03 (to L. Y.). The authors declare that they have no conflicts of interest with the contents of this article.
C. Primo and L. Yenush, unpublished results.
A. Ferri and L. Yenush, unpublished results.
- TOR
- target of rapamycin
- ART
- arrestin-related trafficking
- TORC1
- TOR complex 1
- AZC
- l-azetidine-2-carboxylic acid
- V-ATPase
- vacuolar H+-ATPase
- SD
- synthetic defined.
References
- 1. Merchan S., Bernal D., Serrano R., and Yenush L. (2004) Response of the Saccharomyces cerevisiae Mpk1 mitogen-activated protein kinase pathway to increases in internal turgor pressure caused by loss of Ppz protein phosphatases. Eukaryot. Cell 3, 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Page M. J., and Di Cera E. (2006) Role of Na+ and K+ in enzyme function. Physiol. Rev. 86, 1049–1092 [DOI] [PubMed] [Google Scholar]
- 3. Lubin M., and Ennis H. L. (1964) On the role of intracellular potassium in protein synthesis. Biochim. Biophys. Acta 80, 614–631 [DOI] [PubMed] [Google Scholar]
- 4. Mulet J. M., Leube M. P., Kron S. J., Rios G., Fink G. R., and Serrano R. (1999) A novel mechanism of ion homeostasis and salt tolerance in yeast: the Hal4 and Hal5 protein kinases modulate the Trk1-Trk2 potassium transporter. Mol. Cell. Biol. 19, 3328–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pérez-Valle J., Jenkins H., Merchan S., Montiel V., Ramos J., Sharma S., Serrano R., and Yenush L. (2007) Key role for intracellular K+ and protein kinases Sat4/Hal4 and Ha15 in the plasma membrane stabilization of yeast nutrient transporters. Mol. Cell. Biol. 27, 5725–5736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pérez-Valle J., Rothe J., Primo C., Martínez Pastor M., Ariño J., Pascual-Ahuir A., Mulet J. M., Serrano R., and Yenush L. (2010) Hal4 and Hal5 protein kinases are required for general control of carbon and nitrogen uptake and metabolism. Eukaryot. Cell 9, 1881–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goossens A., de La Fuente N., Forment J., Serrano R., and Portillo F. (2000) Regulation of yeast H+-ATPase by protein kinases belonging to a family dedicated to activation of plasma membrane transporters. Mol. Cell. Biol. 20, 7654–7661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eraso P., Mazón M. J., and Portillo F. (2006) Yeast protein kinase Ptk2 localizes at the plasma membrane and phosphorylates in vitro the C-terminal peptide of the H+-ATPase. Biochim. Biophys. Acta 1758, 164–170 [DOI] [PubMed] [Google Scholar]
- 9. De Craene J. O., Soetens O., and Andre B. (2001) The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J. Biol. Chem. 276, 43939–43948 [DOI] [PubMed] [Google Scholar]
- 10. Schmidt A., Beck T., Koller A., Kunz J., and Hall M. N. (1998) The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 17, 6924–6931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grenson M., and Dubois E. (1982) Pleiotropic deficiency in nitrogen-uptake systems and derepression of nitrogen-catabolic enzymes in npr-1 mutants of Saccharomyces cerevisiae. Eur. J. Biochem. 121, 643–647 [DOI] [PubMed] [Google Scholar]
- 12. Omura F., and Kodama Y. (2004) The N-terminal domain of yeast Bap2 permease is phosphorylated dependently on the Npr1 kinase in response to starvation. FEMS Microbiol. Lett. 230, 227–234 [DOI] [PubMed] [Google Scholar]
- 13. Rotin D., Staub O., and Haguenauer-Tsapis R. (2000) Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 176, 1–17 [DOI] [PubMed] [Google Scholar]
- 14. Léon S., and Haguenauer-Tsapis R. (2009) Ubiquitin ligase adaptors: regulators of ubiquitylation and endocytosis of plasma membrane proteins. Exp. Cell Res. 315, 1574–1583 [DOI] [PubMed] [Google Scholar]
- 15. Novoselova T. V., Zahira K., Rose R. S., and Sullivan J. A. (2012) Bul proteins, a nonredundant, antagonistic family of ubiquitin ligase regulatory proteins. Eukaryot. Cell 11, 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lauwers E., Erpapazoglou Z., Haguenauer-Tsapis R., and André B. (2010) The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 20, 196–204 [DOI] [PubMed] [Google Scholar]
- 17. Aubry L., and Klein G. (2013) True arrestins and arrestin-fold proteins: a structure-based appraisal. Prog. Mol. Biol. Transl. Sci. 118, 21–56 [DOI] [PubMed] [Google Scholar]
- 18. Loewith R., and Hall M. N. (2011) Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189, 1177–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., and Hall M. N. (2002) Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457–468 [DOI] [PubMed] [Google Scholar]
- 20. Reinke A., Anderson S., McCaffery J. M., Yates J. 3rd, Aronova S., Chu S., Fairclough S., Iverson C., Wedaman K. P., and Powers T. (2004) TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 279, 14752–14762 [DOI] [PubMed] [Google Scholar]
- 21. De Virgilio C., and Loewith R. (2006) Cell growth control: little eukaryotes make big contributions. Oncogene 25, 6392–6415 [DOI] [PubMed] [Google Scholar]
- 22. Crespo J. L., and Hall M. N. (2002) Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 66, 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wullschleger S., Loewith R., and Hall M. N. (2006) TOR signaling in growth and metabolism. Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 24. Binda M., Péli-Gulli M. P., Bonfils G., Panchaud N., Urban J., Sturgill T. W., Loewith R., and De Virgilio C. (2009) The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 35, 563–573 [DOI] [PubMed] [Google Scholar]
- 25. Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., Broach J. R., De Virgilio C., Hall M. N., and Loewith R. (2007) Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26, 663–674 [DOI] [PubMed] [Google Scholar]
- 26. O'Donnell A. F., Apffel A., Gardner R. G., and Cyert M. S. (2010) α-Arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol. Biol. Cell 21, 3552–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. MacGurn J. A., Hsu P. C., Smolka M. B., and Emr S. D. (2011) TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell 147, 1104–1117 [DOI] [PubMed] [Google Scholar]
- 28. Merhi A., and André B. (2012) Internal amino acids promote Gap1 permease ubiquitylation via TORC1/Npr1/14-3-3-dependent control of the Bul arrestin-like adaptors. Mol. Cell. Biol. 32, 4510–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Navarrete C., Petrezsélyová S., Barreto L., Martínez J. L., Zahrádka J., Ariño J., Sychrová H., and Ramos J. (2010) Lack of main K plus uptake systems in Saccharomyces cerevisiae cells affects yeast performance in both potassium-sufficient and potassium-limiting conditions. FEMS Yeast Res. 10, 508–517 [DOI] [PubMed] [Google Scholar]
- 30. Hoshikawa C., Shichiri M., Nakamori S., and Takagi H. (2003) A nonconserved Ala401 in the yeast Rsp5 ubiquitin ligase is involved in degradation of Gap1 permease and stress-induced abnormal proteins. Proc. Natl. Acad. Sci. U.S.A. 100, 11505–11510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crapeau M., Merhi A., and André B. (2014) Stress conditions promote yeast Gap1 permease ubiquitylation and down-regulation via the arrestin-like Bul and Aly proteins. J. Biol. Chem. 289, 22103–22116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gander S., Bonenfant D., Altermatt P., Martin D. E., Hauri S., Moes S., Hall M. N., and Jenoe P. (2008) Identification of the rapamycin-sensitive phosphorylation sites within the Ser/Thr-rich domain of the yeast Npr1 protein kinase. Rapid Commun. Mass Spectrom. 22, 3743–3753 [DOI] [PubMed] [Google Scholar]
- 33. MacGurn J. A., Hsu P.-C., and Emr S. D. (2012) Ubiquitin and membrane protein turnover: from cradle to grave. Annu. Rev. Biochem. 81, 231–259 [DOI] [PubMed] [Google Scholar]
- 34. Kim E., Goraksha-Hicks P., Li L., Neufeld T. P., and Guan K. L. (2008) Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., and Sabatini D. M. (2008) The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., and Sabatini D. M. (2011) mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science 334, 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Madrid R., Gómez M. J., Ramos J., and Rodríguez-Navarro A. (1998) Ectopic potassium uptake in trk1 trk2 mutants of Saccharomyces cerevisiae correlates with a highly hyperpolarized membrane potential. J. Biol. Chem. 273, 14838–14844 [DOI] [PubMed] [Google Scholar]
- 38. Lorenz M. C., and Heitman J. (1995) TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J. Biol. Chem. 270, 27531–27537 [DOI] [PubMed] [Google Scholar]
- 39. Heitman J., Movva N. R., and Hall M. N. (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253, 905–909 [DOI] [PubMed] [Google Scholar]
- 40. Yenush L., Mulet J. M., Ariño J., and Serrano R. (2002) The Ppz protein phosphatases are key regulators of K+ and pH homeostasis: implications for salt tolerance, cell wall integrity, and cell cycle progression. EMBO J. 21, 920–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barreto L., Canadell D., Petrezsélyová S., Navarrete C., Maresová L., Peréz-Valle J., Herrera R., Olier I., Giraldo J., Sychrová H., Yenush L., Ramos J., and Ariño J. (2011) A genomewide screen for tolerance to cationic drugs reveals genes important for potassium homeostasis in Saccharomyces cerevisiae. Eukaryot. Cell 10, 1241–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hirasaki M., Horiguchi M., Numamoto M., Sugiyama M., Kaneko Y., Nogi Y., and Harashima S. (2011) Saccharomyces cerevisiae protein phosphatase Ppz1 and protein kinases Sat4 and Hal5 are involved in the control of subcellular localization of Gln3 by likely regulating its phosphorylation state. J. Biosci. Bioeng. 111, 249–254 [DOI] [PubMed] [Google Scholar]
- 43. Hughes Hallett J. E., Luo X., and Capaldi A. P. (2014) State transitions in the TORC1 signaling pathway and information processing in Saccharomyces cerevisiae. Genetics 198, 773–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Heitman J., Movva N. R., Hiestand P. C., and Hall M. N. (1991) FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 88, 1948–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bihler H., Slayman C. L., and Bertl A. (1998) NSC1: a novel high-current inward rectifier for cations in the plasma membrane of Saccharomyces cerevisiae. FEBS Lett. 432, 59–64 [DOI] [PubMed] [Google Scholar]
- 46. Bihler H., Slayman C. L., and Bertl A. (2002) Low-affinity potassium uptake by Saccharomyces cerevisiae is mediated by NSC1, a calcium-blocked non-specific cation channel. Biochim. Biophys. Acta 1558, 109–118 [DOI] [PubMed] [Google Scholar]
- 47. Ko C. H., Liang H., and Gaber R. F. (1993) Roles of multiple glucose transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 638–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wright M. B., Ramos J., Gomez M. J., Moulder K., Scherrer M., Munson G., and Gaber R. F. (1997) Potassium transport by amino acid permeases in Saccharomyces cerevisiae. J. Biol. Chem. 272, 13647–13652 [DOI] [PubMed] [Google Scholar]
- 49. Vargas R. C., García-Salcedo R., Tenreiro S., Teixeira M. C., Fernandes A. R., Ramos J., and Sá-Correia I. (2007) Saccharomyces cerevisiae multidrug resistance transporter Qdr2 is implicated in potassium uptake, providing a physiological advantage to quinidine-stressed cells. Eukaryot. Cell 6, 134–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stefan C. P., Zhang N., Sokabe T., Rivetta A., Slayman C. L., Montell C., and Cunningham K. W. (2013) Activation of an essential calcium signaling pathway in Saccharomyces cerevisiae by Kch1 and Kch2, putative low-affinity potassium transporters. Eukaryot. Cell 12, 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dechant R., Saad S., Ibáñez A. J., and Peter M. (2014) Cytosolic pH regulates cell growth through distinct GTPases, Arf1 and Gtr1, to promote Ras/PKA and TORC1 activity. Mol. Cell 55, 409–421 [DOI] [PubMed] [Google Scholar]
- 52. Petrezselyova S., Zahradka J., and Sychrová H. (2010) Saccharomyces cerevisiae BY4741 and W303-1A laboratory strains differ in salt tolerance. Fungal Biol. 114, 144–150 [DOI] [PubMed] [Google Scholar]
- 53. Warringer J., and Blomberg A. (2003) Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast 20, 53–67 [DOI] [PubMed] [Google Scholar]
- 54. Teis D., Saksena S., and Emr S. D. (2008) Ordered assembly of theESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell 15, 578–589 [DOI] [PubMed] [Google Scholar]
- 55. Song W., and Carlson M. (1998) Srb/mediator proteins interact functionally and physically with transcriptional repressor Sfl1. EMBO J. 17, 5757–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., and Boeke J. D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 57. Zeng L., Guo J., Xu H. B., Huang R., Shao W., Yang L., Wang M., Chen J., and Xie P. (2013) Direct Blue 71 staining as a destaining-free alternative loading control method for Western blotting. Electrophoresis 34, 2234–2239 [DOI] [PubMed] [Google Scholar]