Abstract
Glutathione degradation plays an important role in glutathione and redox homeostasis, and thus it is imperative to understand the enzymes and the mechanisms involved in glutathione degradation in detail. We describe here ChaC2, a member of the ChaC family of γ-glutamylcyclotransferases, as an enzyme that degrades glutathione in the cytosol of mammalian cells. ChaC2 is distinct from the previously described ChaC1, to which ChaC2 shows ∼50% sequence identity. Human and mouse ChaC2 proteins purified in vitro show 10–20-fold lower catalytic efficiency than ChaC1, although they showed comparable Km values (Km of 3.7 ± 0.4 mm and kcat of 15.9 ± 1.0 min−1 toward glutathione for human ChaC2; Km of 2.2 ± 0.4 mm and kcat of 225.2 ± 15 min−1 toward glutathione for human ChaC1). The ChaC1 and ChaC2 proteins also shared the same specificity for reduced glutathione, with no activity against either γ-glutamyl amino acids or oxidized glutathione. The ChaC2 proteins were found to be expressed constitutively in cells, unlike the tightly regulated ChaC1. Moreover, lower eukaryotes have a single member of the ChaC family that appears to be orthologous to ChaC2. In addition, we determined the crystal structure of yeast ChaC2 homologue, GCG1, at 1.34 Å resolution, which represents the first structure of the ChaC family of proteins. The catalytic site is defined by a fortuitous benzoic acid molecule bound to the crystal structure. The mechanism for binding and catalytic activity of this new enzyme of glutathione degradation, which is involved in continuous but basal turnover of cytosolic glutathione, is proposed.
Keywords: crystal structure, Michaelis-Menten, protein expression, protein structure, X-ray crystallography, γ-glutamylcyclotransferase fold, ChaC proteins, ChaC2, GCG1, glutathione degradation
Introduction
Glutathione is an essential metabolite in almost all eukaryotic cells; the only exceptions are the mitochondrial protozoans (1). In addition to the essential role in mitochondrial iron-sulfur biogenesis (2), glutathione plays many other roles that include its role as the principal redox buffer (2, 3) in sulfur storage and transport, xenobiotic, metal, and ROS8 detoxification (4, 5) and its ability to regulate protein function through glutathionylation (6). Glutathione is essential in eukaryotes, and knocking out glutathione biosynthesis is embryonically lethal in mammals (7). Furthermore, glutathione depletion is a hallmark of apoptosis (8) and low glutathione levels have been strongly correlated with several disease states (9). However, high levels of glutathione are also deleterious to the cell (10–13). Thus, it is important for the cell to ensure that the levels of glutathione are tightly regulated. Glutathione degradation and turnover is an important factor in glutathione homeostasis (14). However, for several decades, only a single enzyme, γ-glutamyltranspeptidase, was shown to be responsible for glutathione degradation, but its effects were on non-cytosolic pools of glutathione (15). Recent studies have led to the discovery of two new enzymes for cytosolic glutathione degradation. The first is the Dug enzyme consisting of the (Dug2p-Dug3p)2 enzyme complex that can specifically break down glutathione to Cys-Gly and glutamate (16, 17). This pathway is exclusively present in yeast and fungi. The second pathway involves the ChaC1 enzyme, a member of the ChaC family of γ-glutamylcyclotransferases that can specifically degrade glutathione to 5-oxoproline and Cys-Gly (10) and is induced under conditions of ER stress in the cell (10, 18). Members of the ChaC family have been found in organisms ranging from the unicellular bacteria and yeast to the higher multicellular eukaryotes such as Drosophila melanogaster, mouse, and humans. In Escherichia coli and yeast, a single homologue of ChaC has been found to exist. However, in the higher multicellular eukaryotes, two ChaC homologues are present, ChaC1 and ChaC2. Although the mouse and human ChaC1 function as glutathione-degrading enzymes (10, 19), the function of the mammalian ChaC2 proteins has not been explored.
In this manuscript we have investigated the function of ChaC2 and determined that it functions in glutathione degradation similar to ChaC1 proteins. However, the ChaC2 proteins are characterized by a far lower catalytic efficiency than ChaC1 and, unlike ChaC1, are constitutively expressed. Furthermore, the single ChaC homologue present in lower eukaryotes and prokaryotes appears to be functionally equivalent to the ChaC2 rather than the ChaC1. In addition, we have solved the crystal structure of the ChaC2 homologue in yeast. This is the first crystal structure to be determined for any member of the ChaC family of proteins and thus provides insights into the mechanism of action of ChaC proteins.
Results
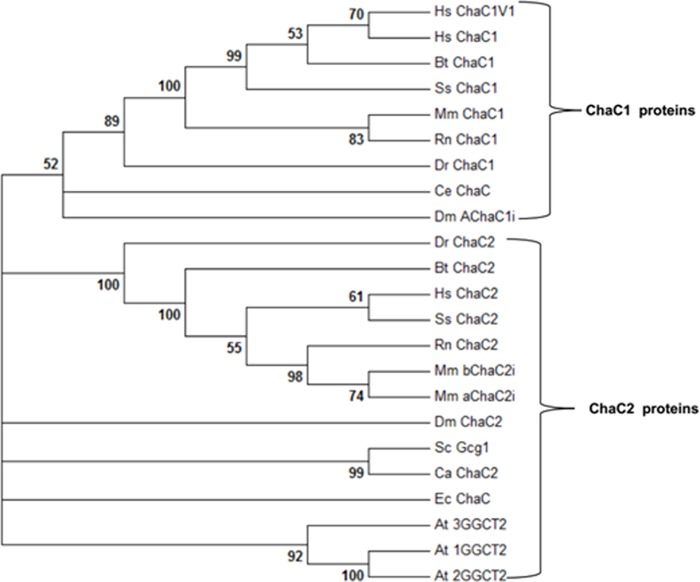
Sequence Analysis and Phylogeny Suggests ChaC1 and ChaC2 Represent Two Distinct Members of the ChaC Family
The amino acid sequence of mouse ChaC1 and mouse ChaC2 displays ∼50% identity between them. Similarly, human ChaC1 shows 50% identity to the human ChaC2. In contrast, when mouse ChaC1 was compared with human ChaC1, there was an 88% sequence identity, and mouse ChaC2 compared with human ChaC2 reveals an ∼94% identity between the proteins (data not shown). This suggested that the ChaC1 and ChaC2 proteins might represent two distinct members of the ChaC family. To investigate this further we built a phylogenetic tree with members of the ChaC family proteins from different organisms. The phylogenetic tree strongly suggests that the ChaC family consists of two different branches represented by the ChaC1 and ChaC2 proteins (Fig. 1). Interestingly, we observed that the higher eukaryotes had both ChaC1 and ChaC2 members, whereas some of the lower eukaryotes such as Caenorhabditis elegans, yeast such as Saccharomyces cerevisiae, fungi, and unicellular protozoans such as Tetrahymena thermophila and bacteria such as E. coli had a single ChaC.
FIGURE 1.
NJ-based phylogenetic tree of ChaC1 and ChaC2 proteins. The NJ-based tree was created using MEGA6 software with 1000 bootstrapping. Two distinct members of ChaC family are marked, and bootstrap values are indicated respectively. Dm, D. melanogaster; Hs, Homo sapiens; Mm, Mus musculus; Dr, Danio rerio; Rn, Rattus norvegicus; Ce, C. elegans; At, A. thaliana; Dm, Drosophila melanogaster; Sc, S. cerevisiae; Ca, Candida albicans; Ss, Sus scrofa; Ec, E. coli; Bt, Bos taurus. Hs ChaC1, AAH19625.1; Hs ChaC1V1,NP_077016.2; Hs ChaC2, NP_001008708.1; Sc Gcg1, NP_011090.3, Mm aChaC2i, NP_080803.1; Mm bChaC2i, NP_001277596.1; Dm ChaC2, NP_651176.1; Ca ChaC2, NP_503578.1; Bt ChaC1, NP_001092352.1; Bt ChaC2,NP_001068996.1; Ss ChaC1, XP_005659810.1; Mm ChaC1, NP_081205.1; Rn ChaC1, NP_001020187.1; Dr ChaC1, NP_001103596.2; BtChaC2, NP_001068996.1; Ss ChaC2, XP_003125197.1; Ec ChaC, NP_415736.2; Rn ChaC1, NP_001166908.1; Rn ChaC2, NP_001020187.1; At 3GGCT2, NP_564490.1; At 2GGCT2, NP_567871.1; At1GGCT2, NP_197994.1; Ce ChaC, NP_503578.1; Dr ChaC2, NP_001025128.1.
The Human ChaC2 Functions Specifically in Glutathione Degradation
To make a preliminary evaluation into the function of ChaC2, we cloned the human ChaC2 and evaluated its function by simple growth-based functional assays that evaluate glutathione degradation in yeast. ChaC2 expressed downstream of the yeast constitutive TEF promoter in the centromeric p416TEF vector was transformed into the yeast S. cerevisiae ABC 1723, and the transformants were evaluated for their growth on either the sulfur-containing dipeptides γ-glutamylmethionine and γ-glutamylcysteine or the tripeptide, γ-glutamyl-cysteinyl-glycine (glutathione), as sulfur source. This strain is an organic sulfur auxotroph due to the met15Δ (leads to a defect in inorganic sulfur assimilation pathway) and cannot utilize glutathione due to the presence of the deletions ecm38Δ (γ-glutamyltranspeptidase gene) and dug3Δ (component of the DUG enzyme). The strain thus allows a check of the glutathione degrading capability of ChaC2 proteins. We observed that the human ChaC2 expressed in yeast could allow growth only on glutathione but could not grow on either of the two γ-glutamyl dipeptides, γ-Glu-Met or γ-Glu-Cys. Human ChaC1 was also evaluated on these different substrates and was found to act with a similar specificity on glutathione as had been observed with the mouse ChaC1 (Fig. 2A). To examine the relative efficiencies with which they acted on glutathione within the cell, the human ChaC2 and human ChaC1 bearing constructs were expressed in a yeast strain with a defect in glutathione biosynthesis to see how they retarded growth on low glutathione supplementation. Both the proteins led to retarded growth of these strains, although human ChaC1 was more efficient than ChaC2 in depleting glutathione levels in vivo as seen from the growth patterns (Fig. 2B). To further evaluate the effects of human ChaC2 and ChaC1 in vivo, we estimated the reactive oxygen species (ROS) levels in cells transformed with either human ChaC2 or ChaC1. The overexpression of human ChaC1 led to an 80% increase in ROS levels, whereas with human ChaC2 it was only a 28% increase in ROS levels (Fig. 2C). These results indicate that along with the increased depletion of glutathione that occurred with ChaC1 overexpression, a corresponding increase in ROS was also seen, whereas in the less efficient ChaC2, a lower depletion of glutathione and a consequent lower increase in ROS was observed. To confirm that the increased ROS was a consequence of lower glutathione levels, we also directly estimated glutathione levels in cells that had ongoing glutathione biosynthesis. ChaC1-overexpressing cells showed a significantly lower glutathione level as compared with controls, whereas ChaC2-expressing cells showed only a small decrease in glutathione levels (Fig. 2D).
FIGURE 2.
A, in vivo growth assay in S. cerevisiae strain to check glutathione degradation function of human ChaC1 and ChaC2; S. cerevisiae strain ABC1723 was transformed with p416TEF harboring human ChaC1 and ChaC2. The transformants were serially diluted and spotted on SD medium plates containing glutathione or methionine or γ-Glu-Met as a sulfur source. B, in vivo growth assay in S. cerevisiae strain ABC1195 (MATα his3Δ1 lys2Δ0 met15Δ0 ura3Δ0 gsh1 Δ::LEU2) to check glutathione-degrading efficiencies. Transformants were grown in SD media containing GSH, serially diluted, and spotted on SD plate containing 20 μm GSH. C, ROS levels in yeast cells (ABC1195) expressing human ChaC1 and ChaC2; ROS levels were measured using DCF-DA probe. D, glutathione levels of yeast BY 4741 transformed with human ChaC1 and ChaC2. Lysates from the yeast transformants were used for glutathione estimation as explained under “Experimental Procedures.” The two datasets for each sample from two independent experiments were obtained and plotted using graph pad prism software. *, indicates p value < 0.05.
The Purified Human and Mouse ChaC2 Proteins Degrade Glutathione at Lower Efficiency Than Their ChaC1 Homologues in Vitro
To evaluate the glutathione degradation capability of the ChaC2 proteins more rigorously, the human and mouse ChaC2 proteins were expressed in E. coli and purified as described under “Experimental Procedures.” The purified proteins on SDS- PAGE displayed a size of 22 kDa and 21 kDa, respectively, for the human and mouse proteins (data not shown), close to their predicted molecular size. In gel filtration studies the ChaC2 showed a single monomeric peak (data not shown). Enzyme assays to evaluate the efficiency of human ChaC2 on different γ-glutamyl compounds (compared with ChaC1) revealed that in in vitro human ChaC2 also displayed catalytic activity against glutathione but did not show any activity against either oxidized glutathione or γ-Glu-Cys (data not shown). The kinetics of the purified proteins were determined using the Dug1p-coupled assay. Human ChaC2 displayed Michaelis-Menten kinetics and showed a Km of 3.7 ± 0.4 mm and kcat of 15.9 ± 1.0 min−1 toward glutathione (Fig. 3A), whereas mouse ChaC2 similarly displayed Michaelis-Menten kinetics with a Km of 3 ± 0.40 mm and kcat of 7.6 ± 0.5 min−1 toward glutathione (data not shown). Thus, the kinetic parameters of human and mouse ChaC2 were comparable, and their Km values appeared to be physiologically relevant considering glutathione is found at millimolar levels in the cell. In comparison, the human ChaC1 showed a Km of 2.2 ± 0.4 mm and kcat of 225.2 ± 15 min−1 toward glutathione (Fig. 3B). As reported earlier, the mouse ChaC1 had a Km of 3.13 ± 0.40 mm (10) and kcat of 391 ± 3.1 min−1. Thus, although the Km of both human ChaC2 and human ChaC1 were comparable and within the physiological range of glutathione concentrations in the cell, the kcat of ChaC2 protein showed 10–20-fold lower catalytic efficiency than the human ChaC1 protein.
FIGURE 3.
Michaelis-Menten plot of human ChaC2 (A) and human ChaC1 (B) for glutathione. Human ChaC2 and human ChaC1 proteins were used for kinetic studies. Different concentrations of glutathione ranging from 0.6 mm to 15 mm were used. A Dug1p-coupled assay was used to estimate cysteine released as described under “Experimental Procedures.” Datasets of three independent experiments were analyzed using nonlinear regression (by Graphpad prism5).
Human ChaC2 Is a Cytosolic, Constitutively Expressed Protein
To experimentally determine the localization of human ChaC2, anti-human ChaC2 antibodies were used to immunostain natively expressed ChaC2 in HEK cells. The results indicated that ChaC2 is localized in the cytosol (Fig. 4).
FIGURE 4.
Cytosolic expression of human ChaC2. Expression of human ChaC2 in cytosol was confirmed by fluorescent microscopy. HEK cells immunostained with anti-ChaC2 antibody displayed staining throughout cytoplasm. DAPI counterstaining was done to show the location of nucleus.
To examine if the human ChaC2 expression was induced during ER stress similar to what had been observed with the mouse and human ChaC1 proteins, we subjected HEK cells to the ER stress-inducing agent tunicamycin. ChaC1 and ChaC2 mRNA levels were measured using real time quantitative PCR, and protein levels were measured using Western blot analysis (Fig. 5, A and B). We observed that ER stress induces the expression of human ChaC1 significantly, as reported earlier, but no significant changes were observed in the case of human ChaC2.
FIGURE 5.
Human ChaC1, but not human ChaC2, is induced specifically by ER stress inducers. Cells (HEK293) were cultured in untreated media (control) and in media containing tunicamycin. RNA (A) and protein levels (B) were measured at different time intervals using real time PCR and Western blot analysis respectively. Regulation of human ChaC1 and ChaC2 under sulfur starvation conditions (C and D); cells (HEK293) were cultured in media that lacks sulfur sources. RNA (C) and protein (D) levels were measured at different time intervals using real time PCR and Western blot analysis. The data set obtained from three independent experiments were plotted using Graphpad prism5 software (*, indicates a p value <0.05).
Glutathione degradation also serves as a source of sulfur for the cells. Hence, we investigated the effect of sulfur starvation on human ChaC1 and ChaC2 expression by real-time PCR analysis. As illustrated in Fig. 5C, we found that sulfur starvation up to 18 h significantly increased ChaC1 transcription by ∼10-fold, but on further extension beyond 18 h the levels started decreasing, which may be due to a loss of cell viability. In comparison, in the case of human ChaC2 there was no significant change in the levels. We also evaluated the expression levels at the protein level by Western blot analysis where human ChaC1 levels increased significantly at 18–24 h of exposure, but only a small increase in the levels of human ChaC2 was observed (Fig. 5D).
In human breast and ovarian cancer cells, human ChaC1 has been shown to be up-regulated. We, therefore, examined whether human ChaC2 was found at higher levels in cancer cells. We carried out a comparison of human ChaC1 and ChaC2 RNA levels in HEK (non-tumorigenic) cells and various tumorigenic cell lines of different tissue origin. Higher levels of ChaC1 transcripts were present in all the tumorigenic cell lines as compared with HEK cells, even without the need for induction. In comparison to this, levels of human ChaC2 transcripts in tumorigenic cell lines were comparable to, or slightly less than the levels seen in HEK cells (Fig. 6).
FIGURE 6.

Comparison of human ChaC1 and ChaC2 expression in different cell lines. Expression levels were compared between HEK293, a non-tumorigenic cell line and tumorigenic cell lines A549 (lung adenocarcinoma), Hep3b (hepatocellular carcinoma), MCF7 (breast adenocarcinoma), and IMR32 (neuroblastoma) cell lines by measuring RNA levels using real time quantitative PCR. The experiments were carried out in duplicate (*, indicates a p value <0.05).
E. coli ChaC and Yeast GCG1 Also Function as ChaC2-like Proteins
During the sequence and phylogenetic analysis it was evident that many of the unicellular eukaryotes and prokaryotes had a single homologue of the ChaC family. Moreover, sequence comparisons with the S. cerevisiae GCG1 protein (ORF YER163c; Yeast ChaC homologue) indicated that it had greater similarity to the ChaC2 proteins. This was also reflected in the kinetic analysis. We previously investigated the kinetics of the yeast GCG1 along with the mouse ChaC1 (10). The yeast GCG1 has been shown to be cytosolic (20). The Km of GCG1 was found to be 1.5 ± 0.20 mm (10), whereas the kcat had a value of 50.8 min−1. The kcat was 9-fold lower than the kcat of mouse ChaC1. The significance of this difference in kcat was not recognized at that point because the ChaC1 and ChaC2 had not been defined as two distinct members of ChaC family earlier. In the case of few other organisms, such as T. thermophila and E. coli, which also had a single homologue, it was not immediately clear from sequence similarities if the protein was a member of the ChaC1 or ChaC2 families. We decided to evaluate one of these proteins, the E. coli ChaC for its kinetic parameters. E. coli ChaC was earlier shown to specifically degrade glutathione (10), but its kinetic parameters were never evaluated. The purified E. coli ChaC protein was used to determine the kinetic parameters. The protein displayed Michaelis-Menten kinetics, and the Km and kcat toward glutathione was found to be 3.1 ± 0.91 mm and 13 ± 0.27 min−1, respectively (data not shown). The Km being within the physiological range of glutathione suggested that the activity was physiologically relevant, whereas the kcat indicates a lower catalytic efficiency than mouse and human ChaC1 but comparable efficiency to the mouse and human ChaC2. This suggested that the E. coli ChaC was functionally orthologous to the ChaC2 protein family.
Crystallographic Studies and Quality of the Yeast ChaC2 Homolog (GCG1) Model
To gain mechanistic insights we initiated the crystal structure determination of ChaC2 protein. The attempt to crystallize the human ChaC2 was unsuccessful. However, the crystallization of yeast GCG1, a homologue of human ChaC2, was successful, and we were able to collect X-ray diffraction data up to 2.2 Å from an in-house X-ray source. Despite many attempts we could not solve the structure by molecular replacement using the close homologs that were available in the Protein Data Bank (PDB) as templates. Therefore, we decided to determine the structure by anomalous dispersion method by incorporating selenomethionine residues in the GCG1 protein. Because the native GCG1 protein contained only three methionine residues, including the initial N-terminal methionine, we mutated an additional four residues to methionine (S42M, L77M, A141M, and V176M) to increase the chance of solving the structure by single or multiple anomalous dispersion (SAD or MAD) methods. The native GCG1 protein was overexpressed, purified, and crystallized in P1 space group with unit cell parameters a = 41.78 Å, b = 62.02 Å, c = 61.70 Å, and α = 113.71°, β = 89.89°, and γ = 101.80°. The calculation of Matthew's coefficient (21) and solvent content suggested two molecules in the asymmetric unit, with the values corresponding to 2.7 Å3 Da−1 and 55%, respectively. Similarly, the semet-GCG1-mutant overexpressed in the presence of selenomethionine was purified and crystallized in P42212 space group with unit cell parameters of a = b = 110.55 Å and c = 42.59 Å. Assuming one molecule per asymmetric unit, the calculated Matthew's coefficient was 2.5 Å3 Da−1 with 51% solvent content.
Initially, the crystal structure of semet-GCG1-mutant was solved by the SAD method with one molecule in the asymmetric unit, and the model was further refined using PHENIX suite of programs with final Rwork and Rfree (22) of 0.17 and 0.21, respectively. The model of semet-GCG1-mutant consists of 224 residues (of 232), 142 water molecules, and 1 succinic acid molecule. The crystal structure of native GCG1 was solved by the molecular replacement method using the structure of semet-GCG1-mutant as template. The crystal structure of GCG1 showed two molecules in the asymmetric unit. The GCG1 model was further refined using the PHENIX suite of programs with Rwork and Rfree converged to 0.16 and 0.19, respectively. The final GCG1 model consisted of 445 residues (of 464), 441 solvent molecules, 2 benzoic acid molecules, and 2 phosphate ions. The initial N-terminal 5 residues and the residues from 210 to 214 were not included in the model due to lack of electron density. The calculated buried area (434 Å2) at the interface using the PISA (23) server indicated that the two molecules in the asymmetric unit did not form any stable oligomer, which was consistent with our gel-filtration studies, and thus, GCG1 exists as monomer in solution. The final refinement statistics for native and semet-GCG1-mutant structures are shown in Table 1.
TABLE 1.
Data collection and refinement statistics
Single crystal was used for collecting each dataset. Se, selenium.
| Semet-GCG1-mutant | GCG1-benzoic acid complex | |
|---|---|---|
| Data collection | Se SAD | |
| Resolution range (Å) | 50.00-1.75 (1.78-1.75)a | 50.00-1.34 (1.39-1.34) |
| Space group | P42212 | P1 |
| Unit cell parameters | ||
| a, b, c (Å) | 110.55, 110.55, 42.59 | 41.78, 62.02, 61.70 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 113.71, 89.89, 101.80 |
| Total no. of reflections | 907,878 | 1,049,615 |
| Unique reflections | 25,966 | 103,046 |
| Mosaicity range (°) | 0.5–0.7 | 0.7–1.1 |
| Rmergeb | 0.10 (0.93) | 0.08 (0.84) |
| Overall I/σ(I) | 41.9 (2.1) | 23.6 (2.0) |
| Completeness (%) | 96.3 (76.5) | 84.5 (32.3) |
| Redundancy | 34.9 (15.2) | 10.2 (6.0) |
| Rpimc | 0.02 (0.23) | 0.03 (0.33) |
| CC1/2 | 0.98 (0.84) | 0.96 (0.83) |
| Refinement | ||
| Resolution (Å) | 37.40–1.75 | 30.14–1.34 |
| No. reflections | 25,799 | 103,046 |
| Rwork/Rfree | 0.17/0.21 | 0.16/0.19 |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.015 | 0.012 |
| Bond angles (°) | 1.21 | 1.18 |
| Ramachandran plot | ||
| Most favored region (%) | 99.56 | 97.95 |
| Additionally allowed region (%) | 0.44 | 2.05 |
| Number of residues | 224 (of 232) | 445 (of 464) |
| Ligand name (number) | Succinic acid (1) | Benzoic acid (2) |
| Ion name (number) | Phosphate (2) | |
| Number of atoms | ||
| Protein | 1,835 | 3,526 |
| Ligand/ion | 8/0 | 18/10 |
| Solvent molecules | 142 | 441 |
| B-factors (Å2) | ||
| Wilson factor | 25.03 | 20.58 |
| Protein | 35.96 | 33.11 |
| Ligand/phosphate ion | 29.74 | 23.95/22.12 |
| Solvent molecules | 49.23 | 48.96 |
| PDB ID | 5HWI | 5HWK |
a Values in parentheses are for the highest resolution shell.
b Rmerge = ΣhklΣi Ii(hkl)|− 〈I(hkl)〉|/ΣhklΣiIi(hkl), where I(hkl) is the intensity of reflection hkl.
c Rpim = Σhkl[n/(n - 1)]1/2Σi Ii(hkl) − 〈I(hkl)〉 /ΣhklΣi Ii(hkl), where n is the multiplicity, other variables as defined for Rmerge.
Overall Three-dimensional Structure of GCG1
The crystal structure of GCG1 revealed that it belonged to the γ-glutamylcyclotransferase-like fold despite its low sequence similarity with other members of this family. The overall topology of GCG1 is shown in Fig. 7 and mainly consists of seven antiparallel β-strands and six α-helices. Earlier, the γ-glutamylcyclotransferase fold was defined with the features consisting of a β-barrel topology with two strands “crossing over,” a highly conserved helix, a binding cavity formed at one side of the β-barrel, a loop after strand β1 containing a conserved (V/A)YG(S/T) motif, a conserved tyrosine in strand β4, and an aromatic residue in strand β5 (24). Essentially, all these features were present in a GCG1 model with the β-barrel topology formed by strands β1-β2-β5-β6-β7 with crossing over strands β6 and β7, a highly conserved helix α-2, the loop after strand β1 containing a 12-GYGS-15 motif, a conserved tyrosine (Tyr-119) in β6 strand, and an aromatic residue (Tyr-161) in strand β7, thus establishing the γ-glutamylcyclotransferase fold for GCG1.
FIGURE 7.

Crystal structure of GCG1. Stereo diagram showing the overall fold of GCG1 in schematic form. The secondary structures are shown in cyan, pink, and white for helices, strands, and loops, respectively. The benzoic acid (green) and phosphate ion (orange) are shown in stick model. The figure was generated using PyMOL (42).
Catalytic Site of GCG1
The crystal structure of GCG1, during the refinement, consistently showed an electron density in the difference Fourier map >3.0 σ level. The electron density was modeled with a benzoic acid according to the shape of the electron density and refined further. The refinement parameters for the benzoic acid were within the limits and showed well defined electron density for it (Fig. 8A). To confirm the presence of benzoic acid, the GCG1was subjected to mass spectrometric analysis as described under “Experimental Procedures.” The mass spectrometric data (data not shown) showed a peak at 122.0823 Da that corresponded to benzoic acid (molecular mass 122.12 Da). Although we did not add any benzoic acid during the purification or crystallization process, it was possible that it might originate from culture media or from trace elements present in the chemicals used for protein purification. A similar observation was also reported in the other crystal structures (25, 26). The benzoic acid interacts with GCG1 mainly through hydrogen-bond and hydrophobic interactions (Fig. 8B). The O2 of benzoic acid interacts with backbone nitrogen atoms of Tyr-13. Moreover, O2 also interacts with the carbonyl oxygen of Leu-11 and backbone nitrogen of Tyr-161 through a water molecule (W2). The O1 of benzoic acid interacts with backbone nitrogen atoms of Gly-14 and Leu-16. Similarly, the C3 of benzoic acid interacts with CE1 of Tyr-119 through hydrophobic interaction.
FIGURE 8.
Catalytic site of GCG1. A, stereo diagram showing the omit map overlaid on the catalytic site of GCG1. The omit map was generated by deleting the benzoic acid (BEZ) from the model and refined with simulated annealing. The resultant Fourier (pale cyan) and difference Fourier (salmon) maps contoured at 1.5 and 3.0 σ level, respectively, are displayed around the catalytic site. The benzoic acid is shown in green. The electron density for water molecules are not shown for clarity purpose. B, stereo diagram showing the hydrogen bond interactions between benzoic acid (green) and residues from GCG1 (white). The side chain for some residues is not shown for clarity purpose. C, stereo diagram showing the omit map overlaid on the catalytic site of semet-GCG1-mutant. The omit map was generated by deleting the succinic acid (SIN) from the model and refined with simulated annealing. The resultant Fourier (pale cyan) and difference Fourier (salmon) maps were displayed at 1.0 and 3.0 σ levels, respectively. The succinic acid is shown in yellow. D, stereo diagram showing the hydrogen bond interactions between succinic acid (yellow) and residues from semet-GCG1-mutant (white). E, superposition of benzoic acid bound with GCG1 (green), succinic acid bound with GCG1 (yellow), and 5-oxo-proline bound with GGACT (light pink). The inset shows the close view of superimposed ligands of benzoic acid (green), succinic acid (yellow), and 5-oxo-proline (light pink).
Interestingly, the crystal structure of semet-GCG1-mutant also showed electron density at the same position as benzoic acid in native GCG1 structure. However, we could model this electron density only with succinic acid (Fig. 8C), as benzoic acid was not compatible with the shape of the electron density, and 1 m succinic acid was used for the crystallization of semet-GCG1-mutant protein. The succinic acid in the model occupies a similar position as that of benzoic acid, and its interactions were also conserved (Fig. 8D). The O1 of succinic acid interacted with backbone nitrogen of Gly-14, Ser-15, and Leu-16, whereas O2 interacted with the backbone nitrogen of Tyr-13. In addition, O2 also interacted with carbonyl oxygen of Leu-11 and backbone nitrogen of Tyr-161 through a water molecule. The O3 of succinic acid interacted with backbone nitrogen and hydroxyl group of Ser-15. It also interacted with hydroxyl group of Tyr-196. Finally the O4 of succinic acid interacted with OE1 and OE2 of Glu-115 along with hydroxyl group of Ser-15, thus stabilizing its interaction with GCG1.
The superposition of GCG1 with γ-glutamylaminecyclotransferase (GGACT) (24) revealed that the benzoic acid and succinic acid occupied the same position as that of 5-oxoproline (a product of GCG1 and GGACT) in GGACT (Fig. 8E). Therefore, the crystal structure of GCG1 represents the benzoic acid-bound form, and the site where benzoic acid is bound is presumably the catalytic site of GCG1.
Comparison of GCG1 with Other Members of the γ-Glutamylcyclotransferase Family
Although the similarity search using GCG1 sequence did not reveal any close homolog structure available in the PDB database, the similarity search using the structure of GCG1 in PDBeFOLD server (27) revealed similar structures that belong to the γ-glutamylcyclotransferase family. The GCG1 structure showed close similarity with γ-glutamylcyclotransferase (PDB ID 3JUC) (28) and γ-glutamylaminecyclotransferase (PDB ID 3CRY) (24) with a root mean square deviation of ∼2.5 Å for 135 Cα atoms (Fig. 9A). However, the superposition of these structures revealed that they were similar only up to the β-barrel region, where the γ-glutamyl moiety binds to the protein. Apart from this region each protein had a different number of α-helices and β-strands that decorate the β-barrel topology.
FIGURE 9.
Comparison of members of γ-glutamylcyclotransferase. A, stereo diagram showing the superposition GCG1 (green), γ-glutamylcyclotransferase (cyan), and γ-lutamylaminecyclotransferase (light pink). The benzoic acid (green) bound at the catalytic site is shown in stick model. B, multiple sequence alignment of GCG1 and its homologues. Identical residues were shown in a red background, whereas similar residues were shown in a yellow background. The secondary structure of GCG1 was marked on the top of the alignment. The residues were aligned using T-coffee software and merged with secondary structure using ESPript (43). C, stereo diagram in schematic form showing the superposition of GCG1 (green) and semet-GCG1-mutant (yellow). The subtle conformational changes of the loop regions are marked with arrows. The benzoic acid (green) and succinic acid (yellow) bound at the catalytic site of GCG1 and semet-GCG1-mutant are shown in stick model.
Discussion
The studies described here demonstrate a separate pathway in mammalian cells for constitutive, cytosolic degradation of glutathione that is mediated by the ChaC2 protein. The ChaC2 proteins were shown to specifically degrade glutathione to yield 5-oxoproline and Cys-Gly. Earlier, the ChaC proteins were grouped as one family. However, it is clear from both sequence and function that ChaC1 and ChaC2 proteins represent distinct branches in the ChaC family. The discovery that the ChaC2 protein acts on cytosolic glutathione in a manner distinct from ChaC1 makes it the third enzyme capable of degrading glutathione in mammalian cells, the others being the ChaC1 and the γ-glutamyltranspeptidase enzymes. With the yeast S. cerevisiae and the plant Arabidopsis thaliana also having multiple pathways for glutathione degradation (29), it is clear that glutathione degradation, which has until now been relatively poorly investigated, has a far more important role to play in glutathione homeostasis. Studies in yeast have in fact led to conclude that glutathione degradation is the key element in glutathione homeostasis (14).
In contrast to what has been previously considered, studies on yeast have shown that glutathione degradation occurs continuously and that glutathione has a relatively short half-life. Under stress conditions, glutathione was found to be turned over rapidly with a half-life of 60 min, but surprisingly, even under un-stressed conditions glutathione was turned over with a half-life of ∼90 min (14). The human and mouse ChaC2 proteins described are likely to participate in a similar continuous and constant, housekeeping function of glutathione degradation in mammalian cells.
Lower eukaryotes and especially the unicellular eukaryotes have only a single ChaC member. Sequence and functional evaluation indicated that the single ChaC member in these lower eukaryotes is orthologous to the low efficiency ChaC2 protein. Thus, it is likely that the ChaC2 enzyme involved in constitutive, low level turnover is the ancestral enzyme, whereas the ChaC1 proteins present exclusively in higher eukaryotes that might have evolved with higher catalytic efficiency for carrying out acute glutathione turnover required under stress conditions.
Despite their importance in maintaining glutathione homeostasis, no structural information hitherto is available for any member of the ChaC family proteins. Therefore, to gain mechanistic insights into the functioning of ChaC family, we have determined the crystal structure of GCG1 from yeast, a ChaC2 homologue at 1.34 Å resolution. The crystal structure of GCG1 reveals a γ-glutamylcyclotransferase-like fold despite its low sequence similarity.
Earlier studies with γ-glutamylcyclotransferase (24) have proposed a Glu residue to act as a general acid/base where the α-amino group of l-γ-glutamyl moiety of the substrate is deprotonated by the Glu followed by nucleophilic attack of this amine onto the side chain amide carbon atom leading to formation of an oxyanion intermediate. This oxyanion eventually collapses to form 5-oxoproline. The protonated Glu donates a hydrogen ion to the amine of the α-linked amino portion of the substrate. We speculate that the GCG1 will also follow the same catalytic mechanism to convert the glutathione into 5-oxoproline and Cys-Gly peptide, as the Glu residue (corresponding to Glu-115 in GCG1) is conserved throughout the ChaC family (Fig. 9B). Importantly, the mutation of E115A in GCG1 completely abolished its catalytic activity (10). Notably, the recently identified Botch (30), which is identical to mouse ChaC1, has been shown to possess a γ-glutamylcyclotransferase activity that acts on the Notch protein. However, the glutamate on a protein does not have a free α-amino group that is required for deprotonation, and thus the mechanism of Notch inactivation is yet unknown and needs to be established.
To understand the differential catalytic efficiency shown by ChaC2 as compared with ChaC1, we compared the sequence of ChaC1 with the GCG1 sequence and structure. The GCG1 structure, although it shows structural similarity with other γ-glutamylcyclotransferase family proteins, this similarity is extended only up to the β-barrel topology and a α-helix, a conserved region across all members of this family (Fig. 9A). The other secondary structures and loops beyond this β-barrel topology are not conserved and may contribute to the specificity and possible differences in catalytic efficiency of the two proteins. In addition, apart from small sequence variations between ChaC1 and ChaC2, an initial N-terminal region of ∼25 amino acids is missing in the ChaC2 and other ChaC proteins compared with ChaC1 (Fig. 9B). This region may partly contribute to higher catalytic efficiency shown by ChaC1. However, the contribution of other residues that vary between ChaC1 and ChaC2 are not ruled out, and further experiments are required to determine their role in displaying different catalytic efficiencies.
Interestingly, the crystal structure of native GCG1 and semet-GCG1-mutant clearly shows an additional electron density at the catalytic site that could be modeled as benzoic acid and succinic acid, respectively. Although, the interactions of these two ligands with the protein are largely conserved (Fig. 8, B and E), the superposition of these two structures revealed a conformational change observed in a loop (Loop1) formed by the residues 110YLNVREQNGY119 (Fig. 9C). Based on this observed conformational change, we propose that the Loop1 of GCG1 may play a role in acquiring its specificity. However, additional experiments and GCG1 structures along with substrate are required to validate our prediction.
In summary, we have demonstrated that the ChaC2 protein of mammals and its ortholog in lower eukaryotes and prokaryotes is responsible for the continuous, but basal, turnover of cytosolic glutathione. and thus it represents an additional pathway in glutathione degradation.
Experimental Procedures
Chemicals and Reagents
All the chemicals and regents used were either analytical grade or molecular biology grade. A gel extraction kit, plasmid preparation kits, and Ni-NTA resins were obtained from Qiagen. Oligonucleotides were synthesized from Sigma and IDT. All the media components were obtained from Difco. Glutathione, cysteine, ninhydrin, methionine, sorbitol, and GSSG were obtained from Sigma. Amplex red kit for glutamate estimation was obtained from Roche Applied Science. Restriction enzymes, Vent DNA polymerase, and other DNA modifying enzymes were obtained from New England BioLabs. PAGE protein markers were obtained from MBI Fermentas.
Strains, Media, and Growth Conditions
E. coli DH5α was used as a cloning host, and BL21, BL21 DE3 plysS and BL21 Rosetta were used as an expression host. The S. cerevisiae ABC1723 (MATα his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ecm38Δ::KanMX4 dug3), ABC 1195 (MATα his3Δ1 lys2Δ0 met15Δ0 ura3Δ0 gsh1 Δ::leu2), and BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) were used for genetic complementation, ROS, and glutathione assays, respectively. The yeast strains were maintained at YPD (yeast extract, peptone, and dextrose). For genetic complementation assays synthetic defined (SD) minimal media containing YNB (yeast nitrogen base), ammonium sulfate, and dextrose supplemented with histidine, leucine, lysine, and methionine (80 mg/liter) were used. Glutathione was used at a concentration of 300 μm. Yeast transformations were carried out using lithium acetate method (31).
Cell Culture
Cell lines were obtained from NCCS-Pune and maintained in DMEM and RPMI 1640 supplemented with 10% fetal bovine serum, penicillin (100units/ml), and streptomycin (100 μg/ml; Invitrogen) at 37 °C in a humidified atmosphere with 5% CO2.
Bioinformatics Analysis
Human ChaC2 and ChaC1 sequences were obtained from the NCBI database and analyzed for homologues in different organisms using BLASTp. Multiple sequence analysis was performed using T-coffee software (32). The multiple sequence data file was then used to obtain a neighbor joining-based phylogenetic tree with MEGA6 software (33).
Cloning and Expression of Human ChaC1, Human/Mouse ChaC2, E. coli ChaC, Yeast GCG1, and Semet-GCG1-mutant Proteins
The human ChaC1/ChaC2 and mouse ChaC2 genes were PCR-amplified from cDNA clones that were obtained from SAF laboratories and subcloned into the yeast expression vector p416TEF using primers as mentioned in Table 2. The genes were cloned at the BamH1 and XhoI sites. Additionally, the mouse ChaC2, human ChaC2, and yeast GCG1 were PCR-amplified using a C-terminal His8-tag reverse primer and cloned in the pET23d vector. Similarly, the E. coli ChaC (amplified from E. coli genomic DNA), yeast GCG1 mutant (S42M, L77M, A141M, and V176M), and human ChaC1 were cloned in pET23a vector. For the expression of proteins the constructs carrying the mouse ChaC2, human ChaC2, human ChaC1, yeast GCG1, GCG1-mutant, and E. coli ChaC were transformed in E. coli BL21 (DE3), E. coli BL21 Rosetta, E. coli BL21 (DE3) pLysS, E. coli BL21 (DE3), E. coli BL21 Rosetta, and E. coli BL21 (DE3) pLysS, respectively.
TABLE 2.
Primers used in this study
| Name | Sequences (5′-3′) |
|---|---|
| HChaC1bamH1F | TTCTACGGATCCATGAAGCAGGAGTCTGCAGCCCCGAAC |
| HChaC1 Xho1R | GTTCTTCTCGAGTCACACCAGCGCCAGAGCCTGC |
| HChaC2 bamH1F | CATGTTAGGATCC ATGTGGGTTTTTGGTTACGGGTC |
| HChaC2 Xho1R | ACTTGATTCTCGAG TTATATGCAATTGAGGTTCTGT |
| MChaC2bamH1F | CGTGCAGGATCCATGTGGGTGTTTGGTTATGGGTCC |
| MChaC2XhoIR | CGTGCAGGATCCATGTGGGTGTTTGGTTATGGGTCC |
| HChaC1NdeIF | AGCTATCATATGAAGCAGGAGTCTGCAGCC |
| HChaC1XhoI8XHisR | CTATGGCTCGAGTTAGTGGTGGTGGTGGTGGTGGTGGTGCACCAGCGCCAGAGCCTGC |
| HChaC2NcoIF | GCTTCGTCCATGGCCATGTGGGTTTTTGGTTACGG |
| HChaC2XhoI8XHisR | CTATGGCTCGAGTTAGTGGTGGTGGTGGTGGTGGTGGTGTATGCAATTGAGGTTCTGTTTC |
| MChaC2NcoIF | CGTGCTAGTCCATGGCCATGTGGGTGTTTGGTTATGGGTCC |
| MChaC2XhoI8XHisR | CTATGCCTCGAGTTAGTGGTGGTGGTGGTGGTGGTGGTGTTTCCTTCTAAGCGTTCCTTTAC |
| E. coli ChaCNdeIF | AGTTCGCATATGATAACGCGTGATTTCTTGATG |
| E. coli ChaCXhoI8XHisR | ATTGATCTCGAGTCAGTGGTGGTGATGGTGATGATGATGGGCGAATCCCGGACGTAACACACC |
| GCG1NdeIF | ACTTGTCATATGACTAATGACAACAGTGGTATCTGG |
| GCG1Xho1R | ACTGTTCTCGAGTTAGTGGTGGTGGTGGTGGTGGTGGTGCCT |
| GCG18XHisXhoIR | AATTATCTCGAGTTAGTGGTGGTGGTGGTGGTGGTGGTGCCTGTATTTATTTACAGTCTCCAATAG |
| GCG1,124AMF | CGGTTCTGGCAAATGTCCACTGATCATAGGGGTACACC |
| GCG1,124AM | GGTGTACCCCTATGATCAGTGGACATTTGCCAGAACC |
| GCG1,230LMR | TTGAAGAATGTGAATTATGATAGTGAAAGTGCACC |
| GCG1,230LMF | GGTGCACTTTCACTATCATAATTCACATTCTTCAA |
| GCG1,271VMF | CCAGGATCCCGACGACTTAATGACCATAGGGGTAGTG |
| GCG1,271VMR | CACTACCCCTATGGTCATTAAGTCGTCGGGATCCTGG |
| GCG1,421AMF | CGAGGCAGAATTGGGTGAAATGTTAGAACAGTTGCCTCG |
| GCG1,421AMR | CGAGGCAACTGTTCTAACATTTCACCCAATTCTGCCTCG |
| GCG1,526VMF | CGTCGGACCAGAGACCATGGATGAAACTGCGAAAGTAATTG |
| GCG1,526VMR | CAATTACTTTCGCAGTTTCATCCATGGTCTCTGGTCCGACG |
For the expression of proteins in E. coli, the primary cultures were grown overnight in LB media containing ampicillin (100 μg/ml). Chloramphenicol (35 μg/ml) was also added when E. coli strains carried dual selection. The secondary cultures bearing the mouse ChaC2, human ChaC1/ChaC2, and yeast GCG1 (10) clones were inoculated at 0.05 A600 nm and allowed to grow until the A600 nm reached 0.5–0.6 (1 A600 nm = 2 × 107 cells). The cultures were induced with 0.5 mm IPTG and kept at 18 °C for 16 h. For E. coli ChaC, the protocol was slightly modified because the majority of the E. coli ChaC protein entered into inclusion bodies. Therefore, the secondary culture was allowed to grow until 0.3 A600 nm, and 3% ethanol was added to the culture along with 0.5 mm IPTG. The culture was then allowed to grow at 18 °C with 180 rpm for 15 h. For the expression and labeling of GCG1-mutant with selenomethionine (semet-GCG1-mutant), a single colony carrying pET23a-GCG1-mutant was inoculated in 50 ml of minimal media (Molecular Dimensions, Suffolk, UK) containing l-methionine supplemented with 100 μg/ml ampicillin and 35 μg/ml chloramphenicol and incubated at 37 °C for 16 h. The overnight grown culture was harvested, and the pellet was washed three times with distilled water, and finally resuspended in 1 ml of water. The resuspended pellet was inoculated in 500 ml of prewarmed minimal media containing l-selenomethionine. The cells were grown until the A600 nm reached a value of 0.5 followed by the addition 0.1 m sorbitol and 0.1 mm IPTG, and the cells were grown further for 16 h at 16 °C. All the cells were harvested by spinning the culture at 10,000 rpm for 10 min at 4 °C.
Purification of Human ChaC1, Human/Mouse ChaC2, E. coli ChaC, Yeast GCG1, and Semet-GCG1-mutant Proteins
The harvested cell pellets were lysed by sonication in a buffer containing 50 mm Tris, pH 8, 300 mm NaCl, and 1 mm PMSF. The cell lysates were centrifuged at 15,000 × g for 1 h at 4 °C, and the supernatant obtained was loaded over Ni-NTA columns that were pre-equilibrated with 50 mm Tris, pH 8.0, 300 mm NaCl buffer (buffer A). The Ni-NTA columns were washed with 5 column volumes of buffer A containing 20 mm imidazole. The bound proteins were eluted in buffer A containing 200–250 mm imidazole and dialyzed overnight against 50 mm Tris, pH 8.0, 300 mm NaCl using a 10-kDa cutoff membrane except yeast GCG1 and semet-GCG1-mutant, which were dialyzed with 100 mm Tris and 200 mm NaCl. The GCG1 and semet-GCG1-mutant proteins were further purified by gel filtration chromatography using a pre-packed Sephacryl S-200 column (16/600 mm, GE Healthcare) that was pre-equilibrated with 100 mm Tris, pH 8.0, 200 mm NaCl buffer. In addition, the gel filtration chromatography using a pre-packed Superdex 200 column (10/300 mm) (GE Healthcare) was also carried out to estimate the molecular weight and oligomeric nature of Ni-NTA-purified human ChaC1, human ChaC2, mouse ChaC2, and E. coli ChaC proteins. The peaks obtained were analyzed against a standard curve that was established using protein standards (GE Healthcare). The purity of proteins were checked by SDS-PAGE, and the concentration of proteins were estimated by the Bradford method (34) at each step of purification.
In Vivo Functional Complementation Assays in S. cerevisiae
The transformants were grown overnight in SD medium containing methionine or glutathione. Primary culture was used to inoculate secondary culture and allowed to grow until 0.6 A600 nm. The cells were harvested, washed, and diluted in sterile water at 0.2 A600 nm. Serial dilutions were then made, and 10 μl of cell suspension was spotted on plates containing SD medium with or without glutathione.
Assay of γ-Glutamylcyclotransferase Activity on Glutathione, γ-Glu-Cys, Using Dug1p-coupled Assay
Glutathione degradation activity of ChaC2 proteins were performed using a Dug1-coupled assay as described earlier (10). 5 μg of human ChaC2, 2.5 μg of mouse ChaC2, and 10 μg of E. coli ChaC were incubated with 0.2–15 mm GSH in 50 μl of reaction mix containing 50 mm Tris, pH 8, and 5 mm DTT for 30 min at 37 °C. The reaction was terminated thereafter by heating at 95 °C for 5 min. 5 μg of recombinant Dug1p and 20 μm MnCl2 was added to each reaction and incubated further for 1 h at 37 °C. Cysteine release was estimated using a ninhydrin-based method (35). The kinetic parameters were obtained by plotting data using graph pad prism 5 software.
Assay of γ-Glutamylcyclotransferase Activity on GSSG and Glutathione Using 5-Oxoprolinase-coupled Assay
The activity against GSSG was checked by measuring the 5-oxoproline released during the reaction using 2.5 μg of human ChaC2 and human ChaC1 proteins. The 5-oxoproline was estimated using 5-oxoprolinase coupled assay as described earlier (10).
Determination of Intracellular ROS Generation
Intracellular ROS generation was assessed using 5-(and-6)-chloromethyl-2,7-dichlorodihydrofluorescein diacetate, (CM-H2DCFDA) (Molecular Probes). WT yeast cells ABC1195 transformed with human ChaC1 and human ChaC2 cloned in p416TEF expression vector were grown in minimal media containing 10 μm glutathione. 0.8 A600 nm cells were pelleted, washed, and loaded with CM-H2DCFDA (10 μm) for 30 min. Cells were washed with PBS, and fluorescence (excitation 493 nm, emission 527 nm) was then measured. To confirm that DCF was able to detect increased oxidative stress, cells in control medium were treated with 100 μm H2O2, and it was taken as the positive control.
Confocal microscopy was also done to show dose-dependent changes in DCF levels in cells exposed to H2O2. Cells were plated onto poly-l-lysine-coated coverslips and loaded with CM-H2DCFDA (10 μm) 30 min. After fixing with 4% paraformaldehyde, cells were washed with PBS and mounted using Vectashield mounting medium and stored at 4 °C in dark until imaging. Data were normalized to values obtained from normoxic controls.
Determination of Intracellular Glutathione Levels
The S. cerevisiae BY4741 strains transformed with different plasmids were grown to an A600 nm of 0.6–0.8, harvested, and resuspended to an A600 nm of 10. Cells were lysed in 800 μl of KPE buffer (0.1 m potassium phosphate buffer and 5 mm EDTA, pH 7.5) containing 0.6% sulfosalicylic acid and 0.1% Triton X-100 using the glass bead lysis method. The supernatant was used for the estimation of glutathione by DTNB method according to the protocol by Rahman et al. (36).
RNA Isolation and Quantitative Real-time PCR
Cultured cells were exposed to sulfur stress by growing in sulfur and cysteine-free DMEM for different time periods. Total RNA was extracted using an RNeasy kit (Qiagen) and quantified using a spectrophotometer at 260 nm. 500 ng of total RNA was reverse-transcribed to cDNA using a Superscript II kit (Takara) with oligo(dT) primer. In the real-time PCR step, PCR reactions were performed in triplicate with 1 μl of cDNA/20-μl reaction and primers specific for human ChaC1 and human ChaC2 using SYBR Premix ExTaq in an Eppendorf RealPlex Mastercycler. Thermal cycling was initiated at 94 °C for 30 s followed by 40 cycles of PCR (94 °C for 5 s and 68 °C for 30 s). β-Actin was used as an endogenous control. The Ct method was applied to normalize expression relative to the actin gene.
Western Blot Analysis
For Western blot analysis cells were lysed in buffer containing 150 mm NaCl, 1.0% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mm Tris, pH 8.0. Protein concentration was estimated by the Bradford reagent. Proteins (50 μg) were electrophoresed on a 15% SDS-polyacrylamide gel and transferred to nitrocellulose membrane (Millipore). The membrane was blocked with 5% BSA and probed with monoclonal antibody to ChaC1 (catalog #AV42623, Sigma) and ChaC2 (catalog #SAB2104121, Sigma) at 1:750 dilutions and incubated at 4 °C overnight followed by incubation with secondary antibody for 2 h at 37 °C. Immunoreactive bands were visualized using the enhanced chemiluminescence detection system (Amersham Biosciences) following the manufacturer's protocol.
Immunocytochemistry for in Vivo Localization
Cultured cells were plated onto poly-l-lysine-coated coverslips and fixed with 4% paraformaldehyde after exposing cells for 48 h to tunicamycin. Cells were blocked with 5% BSA and then incubated with monoclonal antibody to ChaC2 (1:100; Sigma) at 4 °C overnight and then incubated in goat anti-rabbit Alexa Fluor 488 (1:20000; Invitrogen) for 30 min. For control, primary antibody was not added in the above treatment. After washing in PBS, slides were mounted using Vectashield mounting medium and stored at 4 °C in the dark until imaging. Images were captured using confocal microscope (Zeiss LSM 780).
Mass Spectrometry Analysis of the Purified Protein
The purified GCG1 protein used for crystallographic studies was subjected to mass spectrometric analysis to confirm the presence of bound benzoic acid with the purified protein. The intact protein mass was analyzed using WATERS SYNAPT G2S QTOF mass spectrophotometer in electrospray ionization positive mode. The mass range and scan rate were set to record m/z from 50 to 2000 daltons in resolution mode. The sample was directly infused with a flow rate of 100 μl/min, capillary voltage of 3 KV, cone voltage of 25 V at 120 °C source temperature, and dissolvation temperature of 350 °C. Raw data obtained was analyzed using mass lynx software.
Crystallization, Data Collection, and Structure Determination of GCG1
The yeast ChaC2 homolog, GCG1 (20 mg/ml), and semet-GCG1-mutant (12 mg/ml) in 100 mm Tris, pH 8.0, 200 mm NaCl were used to set up various commercial crystallization screens by sitting drop vapor diffusion method with 96-well plates (Molecular Dimensions). Each protein drop with 1 μl of protein and 1 μl of precipitant was equilibrated against 60 μl of reservoir solution followed by incubation at 20 °C. The good quality crystals for native GCG1 appeared in 0.2 m ammonium phosphate, 20% PEG 3350, pH 4.7, and 15% glycerol after seeding. For semet-GCG1-mutant, quality crystals were obtained in 1 m succinic acid, pH 7.0, 0.1 m HEPES, pH 7.0, and 1% w/v PEG-MME 2000 after seeding.
The X-ray diffraction data for native GCG1 and semet-GCG1-mutant were collected using synchrotron radiation available at BM14 beamline (European Synchrotron Radiation Facility (ESRF), Grenoble, France) equipped with MAR Mosaic CCD detector. The semet-GCG1-mutant crystal was soaked in 20% (v/v) glycerol in mother liquor for 10 s and immediately flash-cooled in liquid nitrogen before data collection. Total 991 images were collected for semet-GCG1-mutant crystals at wavelength of 0.9787 Å. For native GCG1, the glycerol (15%) used in crystallization experiment was sufficient for cryoprotection, and therefore, the crystal was directly flash-cooled in liquid nitrogen. A total of 2000 images were collected for native GCG1 at wavelength of 0.9763 Å. Both the datasets were indexed, integrated, and scaled using HKL2000 (44) suite of programs.
The crystal structure of semet-GCG1-mutant was solved by SAD method using AUTOSOL as implemented in PHENIX (37). Initially, 4 of 7 selenium atoms (at residue numbers 42, 141, 176, and 207) were located. The other 3 selenium atoms anticipated at residue numbers 1, 77, and 210 were not located by the program. Moreover, refining these residues as selenomethionine at these positions did not fit well, and therefore, these residues were refined as methionine only. The incomplete incorporation of selenomethionine in the protein may be due to the usage of non methionine-auxotrophic strain for the expression of semet-GCG1-mutant protein. Nevertheless, the initial phases obtained from four selenium atoms were improved by density modification, and the modified map showed excellent electron density. Using the modified map, the automatic model building was able to build around 200 residues of 232 residues. Further refinement and model building were carried out in PHENIX and COOT (37, 38), respectively. The three-dimensional structure thus obtained for the semet-GCG1-mutant was used as a template for the structure determination of native GCG1 by molecular replacement method using PHASER (39) as implemented in CCP4 (40) suite. The PHASER with default parameters gave a single solution with two molecules in the asymmetric unit. Further refinement was done using rigid body and restrained refinement using REFMAC5 (41) followed by refinement in PHENIX. The manual model building was done using COOT. The GCG1 model was subjected to simulating annealing refinement using PHENIX for further improvement and to minimize any model bias. All the atoms were refined with anisotropic temperature factors. The model building and refinement were carried out iteratively until the Rwork and Rfree were converged.
Author Contributions
A. Kaur purified and characterized the human enzymes and made the mutants. A. C. performed the in vivo experiments. R. G. and R. S. performed the experiments related to purification, crystallization, data collection, and structure analysis of GCG1. S. K. supervised the analysis and structural characterization of GCG1. A. K. B. analyzed and supervised the experiments related to enzyme characterization and in vivo experiments. A. Kumar purified and characterized the mouse ChaC2 protein. All authors contributed in writing and preparation of manuscript.
Acknowledgments
We thank the EMBL staff Dr. Hassan Belrhali and Dr. Babu A. Manjasetty for providing support on the beamline and EMBL-DBT for providing access to the BM14 beamline at the ESRF. We also thank the Department of Biotechnology, Government of India for providing financial assistance to R. G. for collecting data at BM14.
The authors declare that they have no conflicts of interest with the content of this article.
The atomic coordinates and structure factors (codes 5HWI and 5HWK) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ROS
- reactive oxygen species
- ER
- endoplasmic reticulum
- SAD
- single anomalous dispersion
- GGACT
- γ-glutamylaminecyclotransferase
- Ni-NTA
- nickel-nitrilotriacetic acid
- SD
- synthetic defined
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- semet
- selenomethionine
- DCF
- dichlorodihydrofluorescein
- CM-H2DCFDA
- 5-(and-6)-chloromethyl-2,7-dichlorodihydrofluorescein diacetate.
References
- 1. Fahey R. C., Newton G. L., Arrick B., Overdank-Bogart T., and Aley S. B. (1984) Entamoeba histolytica: a eukaryote without glutathione metabolism. Science 224, 70–72 [DOI] [PubMed] [Google Scholar]
- 2. Meister A., and Anderson M. E. (1983) Glutathione. Annu. Rev. Biochem. 52, 711–760 [DOI] [PubMed] [Google Scholar]
- 3. Penninckx M. J., and Elskens M. T. (1993) Metabolism and functions of glutathione in micro-organisms. Adv. Microb. Physiol. 34, 239–301 [DOI] [PubMed] [Google Scholar]
- 4. Arthur J. R. (2000) The glutathione peroxidases. Cell. Mol. Life Sci. 57, 1825–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hayes J. D., Flanagan J. U., and Jowsey I. R. (2005) Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 45, 51–88 [DOI] [PubMed] [Google Scholar]
- 6. Dalle-Donne I., Rossi R., Colombo G., Giustarini D., and Milzani A. (2009) Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem. Sci. 34, 85–96 [DOI] [PubMed] [Google Scholar]
- 7. Winkler A., Njålsson R., Carlsson K., Elgadi A., Rozell B., Abraham L., Ercal N., Shi Z. Z., Lieberman M. W., Larsson A., and Norgren S. (2011) Glutathione is essential for early embryogenesis: analysis of a glutathione synthetase knockout mouse. Biochem. Biophys. Res. Commun. 412, 121–126 [DOI] [PubMed] [Google Scholar]
- 8. Franco R., and Cidlowski J. A. (2009) Apoptosis and glutathione: beyond an antioxidant. Cell Death Differ. 16, 1303–1314 [DOI] [PubMed] [Google Scholar]
- 9. Townsend D. M., Tew K. D., and Tapiero H. (2003) The importance of glutathione in human disease. Biomed. Pharmacother. 57, 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumar A., Tikoo S., Maity S., Sengupta S., Sengupta S., Kaur A., and Bachhawat A. K. (2012) Mammalian proapoptotic factor ChaC1 and its homologues function as γ-glutamyl cyclotransferases acting specifically on glutathione. EMBO Rep. 13, 1095–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar C., Igbaria A., D'Autreaux B., Planson A. G., Junot C., Godat E., Bachhawat A. K., Delaunay-Moisan A., and Toledano M. B. (2011) Glutathione revisited: a vital function in iron metabolism and ancillary role in thiol-redox control. EMBO J. 30, 2044–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Srikanth C. V., Vats P., Bourbouloux A., Delrot S., and Bachhawat A. K. (2005) Multiple cis-regulatory elements and the yeast sulphur regulatory network are required for the regulation of the yeast glutathione transporter, Hgt1p. Curr. Genet. 47, 345–358 [DOI] [PubMed] [Google Scholar]
- 13. Rajasekaran N. S., Connell P., Christians E. S., Yan L.-J., Taylor R. P., Orosz A., Zhang X. Q., Stevenson T. J., Peshock R. M., Leopold J. A., Barry W. H., Loscalzo J., Odelberg S. J., and Benjamin I. J. (2007) Human αB-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell 130, 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baudouin-Cornu P., Lagniel G., Kumar C., Huang M. E., and Labarre J. (2012) Glutathione degradation is a key determinant of glutathione homeostasis. J. Biol. Chem. 287, 4552–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tate S. S., and Meister A. (1981) γ-Glutamyl transpeptidase: catalytic, structural, and functional aspects. Mol. Cell Biochem. 39, 357–368 [DOI] [PubMed] [Google Scholar]
- 16. Kaur H., Kumar C., Junot C., Toledano M. B., and Bachhawat A. K. (2009) Dug1p Is a Cys-Gly peptidase of the γ-glutamyl cycle of Saccharomyces cerevisiae and represents a novel family of Cys-Gly peptidases. J. Biol. Chem. 284, 14493–14502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ganguli D., Kumar C., and Bachhawat A. K. (2007) The alternative pathway of glutathione degradation is mediated by a novel protein complex involving three new genes in Saccharomyces cerevisiae. Genetics 175, 1137–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mungrue I. N., Pagnon J., Kohannim O., Gargalovic P. S., and Lusis A. J. (2009) CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J. Immunol. 182, 466–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crawford R. R., Prescott E. T., Sylvester C. F., Higdon A. N., Shan J., Kilberg M. S., and Mungrue I. N. (2015) Human CHAC1 protein degrades glutathione, and mRNA induction is regulated by the transcription factors ATF4 and ATF3 and a bipartite ATF/CRE regulatory element. J. Biol. Chem. 290, 15878–15891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fujiwara S., Kawazoe T., Ohnishi K., Kitagawa T., Popa C., Valls M., Genin S., Nakamura K., Kuramitsu Y., Tanaka N., and Tabuchi M. (2016) RipAY, a plant pathogen effector protein, exhibits robust γ-glutamyl cyclotransferase activity when stimulated by eukaryotic thioredoxins. J. Biol. Chem. 291, 6813–6830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kantardjieff K. A., and Rupp B. (2003) Matthews coefficient probabilities: improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 12, 1865–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brünger A. T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 23. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 24. Oakley A. J., Coggan M., and Board P. G. (2010) Identification and characterization of γ-glutamylamine cyclotransferase, an enzyme responsible for γ-glutamyl-ϵ-lysine catabolism. J. Biol. Chem. 285, 9642–9648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steegborn C., Danot O., Huber R., and Clausen T. (2001) Crystal structure of transcription factor MalT domain III: a novel helix repeat fold implicated in regulated oligomerization. Structure 9, 1051–1060 [DOI] [PubMed] [Google Scholar]
- 26. Hwang J., Nguyen L. T., Jeon Y. H., Lee C. Y., and Kim M. H. (2015) Crystal structure of fully oxidized human thioredoxin. Biochem. Biophys. Res. Commun. 467, 218–222 [DOI] [PubMed] [Google Scholar]
- 27. Krissinel E., and Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 28. Oakley A. J., Yamada T., Liu D., Coggan M., Clark A. G., and Board P. G. (2008) The identification and structural characterization of C7orf24 as γ-glutamyl cyclotransferase an essential enzyme in the γ-glutamyl cycle. J. Biol. Chem. 283, 22031–22042 [DOI] [PubMed] [Google Scholar]
- 29. Kumar S., Kaur A., Chattopadhyay B., and Bachhawat A. K. (2015) Defining the cytosolic pathway of glutathione degradation in Arabidopsis thaliana: role of the ChaC/GCG family of γ-glutamyl cyclotransferases as glutathione-degrading enzymes and AtLAP1 as the Cys-Gly peptidase. Biochem. J. 468, 73–85 [DOI] [PubMed] [Google Scholar]
- 30. Chi Z., Byrne S. T., Dolinko A., Harraz M. M., Kim M. S., Umanah G., Zhong J., Chen R., Zhang J., Xu J., Chen L., Pandey A., Dawson T. M., and Dawson V. L. (2014) Botch is a γ-glutamyl cyclotransferase that deglycinates and antagonizes Notch. Cell Rep. 7, 681–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ito H., Fukuda Y., Murata K., and Kimura A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Notredame C., Higgins D. G., and Heringa J. (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 33. Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 35. Gaitonde M. (1967) A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem. J. 104, 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rahman I., Kode A., and Biswas S. K. (2006) Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 1, 3159–3165 [DOI] [PubMed] [Google Scholar]
- 37. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 39. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version. 1.2r3pre Ed., Schrödinger, L., LLC, New York [Google Scholar]
- 43. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]