Abstract
Targeted inhibitors of the human epidermal growth factor receptor 2 (HER2), such as trastuzumab and lapatinib, are among the first examples of molecularly targeted cancer therapy and have proven largely effective for the treatment of HER2-positive breast cancers. However, approximately half of those patients either do not respond to these therapies or develop secondary resistance. Although a few signaling pathways have been implicated, a comprehensive understanding of mechanisms underlying HER2 inhibitor drug resistance is still lacking. To address this critical question, we undertook a concerted approach using patient expression data sets, HER2-positive cell lines, and tumor samples biopsied both before and after trastuzumab treatment. Together, these methods revealed that high expression and activation of a specific subset of receptor tyrosine kinases (RTKs) was strongly associated with poor clinical prognosis and the development of resistance. Mechanistically, these RTKs are capable of maintaining downstream signal transduction to promote tumor growth via the suppression of cellular senescence. Consequently, these findings provide the rationale for the design of therapeutic strategies for overcoming drug resistance in breast cancer via combinational inhibition of the limited number of targets from this specific subset of RTKs.
Keywords: breast cancer, cellular senescence, drug resistance, insulin-like growth factor (IGF), receptor tyrosine kinase, human epidermal growth factor receptor 2 (HER2), targeted therapy
Introduction
Breast cancer is the most common invasive cancer in women, accounting for more than 40,000 deaths in the United States per year (1). The HER2-positive subtype comprises ∼20% of all breast cancers and is defined as displaying overexpression of the human epidermal growth factor receptor 2 (HER2) protein or amplification of the ERBB2 gene, as assayed by immunohistochemistry or fluorescence in situ hybridization, respectively. HER2 is a transmembrane protein that heterodimerizes with and activates other members of the ErbB family of receptor tyrosine kinases, resulting in increased cell growth and proliferation (2). In addition to breast cancer, HER2 overexpression or ERBB2 gene amplification occurs in several other human malignancies, including ovarian, stomach, and uterine cancers (3), where it is also associated with recurrence and poor prognosis (4). Targeted HER2 inhibitors, including the monoclonal antibodies trastuzumab (trade name Herceptin) and pertuzumab, the EGFR/HER2 inhibitor lapatinib, and the antibody-drug conjugate trastuzumab emtansine (T-DM1) have been the standard of care for HER2-positive breast cancer since the Food and Drug Administration approval of trastuzumab in 1998 (5).
Although targeted HER2 inhibition has proven largely effective for the treatment of HER2-positive breast cancers, approximately 40–60% of all patients either do not respond to treatment or respond initially but eventually acquire secondary resistance (6, 7). Several anticancer drug resistance mechanisms have been identified, including HER2 proteolysis, Mucin-4 overexpression, and loss of the PTEN phosphatase (8). However, these mechanisms are either not easily targetable or are unlikely to account for resistance in the majority of patients. In addition, several receptor tyrosine kinases (RTKs)3 have been implicated to play a role in bypassing HER2 inhibition (9), and similar bypass resistance mechanisms have been observed in other cancer types, such as EGFR-positive lung cancer and BRAF mutant melanoma (10). However, it is currently unclear which RTKs are the most important mediators of resistance to HER2 inhibition, because there has not been a comprehensive study of all RTKs to identify those specifically associated with drug resistance in HER2-positive breast cancer patients.
To address this issue, we have undertaken a systematic bioinformatic analysis of the expression of 49 human RTKs as it relates to HER2 breast cancer patient survival using a database composed of 22 publicly available data sets. Of the RTKs in which high expression was associated with poor patient survival, only a distinct subset of RTKs was able to functionally confer drug resistance when overexpressed in HER2-positive breast cancer cells. Moreover, using a panel of HER2-positive breast tumors, we observed that the expression of these RTKs was specifically elevated only in patient tumors that did not respond to trastuzumab therapy. Importantly, those RTKs were not universally up-regulated by every tumor, because different combinations of these RTKs were found to have the ability to drive resistance in individual patients and cell lines. However, for each tumor and cell line analyzed, at least one of these receptors was associated with insensitivity to HER2 inhibition, suggesting that those RTKs might serve as viable targets for overcoming drug resistance. Mechanistically, these RTKs appear to confer resistance by maintaining signaling flux through both the PI3K/Akt and RAS/RAF/MAPK pathways and inhibiting the onset of cellular senescence. Thus, cellular senescence probably represents an important biological process underlying the clinical efficacy of HER2 inhibition.
Results
Bioinformatic Identification and Experimental Validation of Specific RTKs Mediating Trastuzumab Resistance
One proposed mechanism for the development of resistance to targeted cancer therapy involves utilization of alternate signaling molecules by cancer cells to bypass pharmacological inhibition and maintain signaling output (11–13). To identify RTKs that might mediate drug resistance in HER2-positive breast cancer, we conducted an unbiased bioinformatic screen of all human RTKs by analyzing gene expression data of HER2-positive patients from an integrative database consisting of 22 publicly available data sets. RTKs most highly implicated in drug resistance from this analysis included MST1R (also called RON), TYRO3, IGF1R, EPHB3, PDGFRb, EPHB2, and ROR2 (Fig. 1A). Of these, MST1R and IGF1R have been previously reported to play roles in resistance to HER2 inhibition (14–17). Surprisingly, high expression of MET (hepatocyte growth factor receptor), which has been implicated in resistance to EGFR inhibitors in colorectal cancer (18), was associated with better clinical outcome in HER2-positive breast cancer (Fig. 1A). It should be noted that these gene expression data were acquired from patient tumors before therapy, and thus this analysis might not identify all of the RTKs capable of mediating resistance. Nonetheless, these results implicate a subset of RTKs as molecules that function to worsen patient outcome in HER2-positive breast cancer and thus serve as an entry point for more detailed studies on the role of these RTKs in anti-HER2 drug resistance.
FIGURE 1.
Bioinformatic identification and experimental validation of RTKs mediating trastuzumab resistance. A, forest plot of the concordance index estimates of survival risk for each RTK in HER2 patients. Data are ordered and presented as the mean ± S.E. Higher concordance index numbers predict worse outcome. The most significant p values are also shown; *, p < 0.05; **, p < 0.01; ***, p < 0.001. B–D, SKBR3, BT474, or AU565 breast cancer cells engineered to stably express the indicated RTKs were cultured for 2 days in increasing amounts of lapatinib, and then cell viability was measured and plotted as a function of drug concentration. Error bars, S.E. Results are summarized in E. NR, no rescue; P, partial rescue; R, rescue. F–H, Kaplan-Meier curves of IGF1R, TYRO3, and PDGFRb in each of the HER2 molecular subtype. Gene expression is split on the median into low and high classifications, and data are presented as the probability of relapse-free survival or distant metastasis-free survival versus time in months.
To assess whether these candidate RTKs could confer resistance to HER2 inhibition in an experimental setting, we stably expressed each of these candidate RTKs individually in three HER2-positive cell lines known to be sensitive to HER2 inhibition (supplemental Fig. S1A) (19). Following 3-day treatment with increasing amounts of the EGFR/HER2 inhibitor lapatinib, we quantified cell viability using a luminescence-based assay. As expected, the viability of cells expressing the control vector was strongly impaired by HER2 inhibition, with an IC50 < 100 nm for all three cell lines assayed (Fig. 1, B–D). Four of the seven candidate RTKs had no measurable effect on cell viability when expressed in any of the three cell lines, indicating that these receptors are unable to confer resistance in this experimental setting. However, ectopic expression of IGF1R, TYRO3, or PDGFRb was found to robustly improve breast cancer cell viability in the presence of lapatinib, although total HER2 levels were not significantly altered in these cells (supplemental Fig. S1B). Similar results were observed using the anti-HER2 monoclonal antibody trastuzumab (supplemental Fig. S1C), suggesting that breast cancer cells use analogous molecular mechanisms to establish resistance to either lapatinib or trastuzumab, although alternative resistance mechanisms, such as immune system engagement, might differ for these two drugs (9). Interestingly, the response of each of the three HER2-positive cell lines analyzed was differentially affected by forced expression of these RTKs. Whereas IGF1R was most effective at rescuing BT474 cell viability, TYRO3 provided the strongest rescue of the SKBR3 and AU565 cell lines. PDGFRb expression resulted in partial rescue of two of the three drug-sensitive cell lines assayed (data summarized in Fig. 1E). Taken together, these results provide evidence that only a specific subset of RTKs, including IGF1R, TYRO3, and PDGFRb, can confer resistance to HER2 inhibition in breast cancer, albeit to different extents for different tumor cell lines, although we might have missed those RTKs that could exert their effect only in an in vivo setting, particularly if they function primarily in immune cells.
To provide another line of support for the correlation between clinical outcome of HER2-positive patients and the expression pattern of a distinct subset of RTKs, we conducted further analysis of a breast cancer metaset composed of 333 unique HER2-positive patient samples. Expression of each RTK was split on the median into low and high classifications, Kaplan-Meier curves were plotted, and p values were calculated using the log-rank method (Kaplan-Meier curves for the expression of all RTKs are shown in supplemental Fig. S2). This analysis confirmed that the expression of a similar subset of RTKs, such as IGF1R and TYRO3, is strongly correlated with clinical outcome for HER2-positive breast cancer (Fig. 1, F and G). PDGFRb also appeared to be associated with patient survival, although this result was less statistically significant (p = 0.06), perhaps because different tumors utilize this RTK to various extents to generate resistance, as was observed for different breast cancer cell lines cultured in vitro (Fig. 1H). Based on these bioinformatic and experimental results, we concluded that this subset of RTKs, including IGF1R, TYRO3, and PDGFRb, warrants further study as receptors that influence HER2-positive breast cancer patient survival, possibly through drug resistance mechanisms.
The Subset of RTKs Mediating Resistance Is Up-regulated in Resistant Tumors and Cell Lines as a Result of HER2 Inhibition
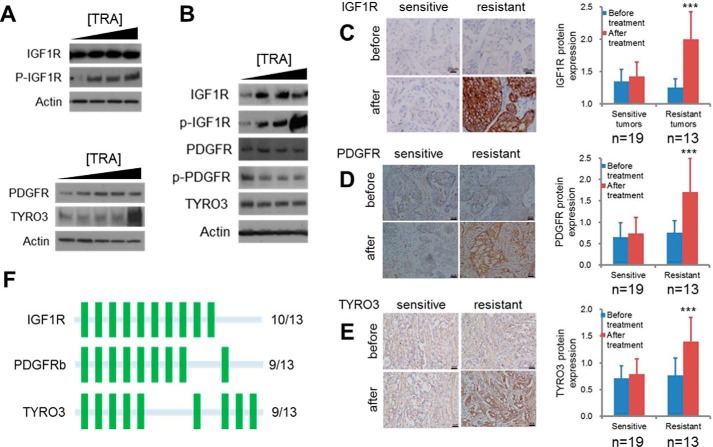
Because ectopic expression of this subset of RTKs was able to promote resistance to HER2 inhibition in breast cancer cells grown in culture (Fig. 1, B–E), we wondered whether these cells might up-regulate these specific RTKs during the acquisition of drug resistance. To explore this, we compared sensitive BT474 cells with a resistant derivative that was generated by culturing BT474 cells in the presence of lapatinib to select for resistant clones (rBT474) (20). Subsequent studies revealed that rBT474 cells do not respond to trastuzumab, and basal expression of IGF1R, PDGFRb, and TYRO3 was higher in the resistant cells compared with the parental line, whereas HER2 expression levels were relatively unchanged (supplemental Fig. S3, A and B). To investigate this further, we cultured rBT474 cells in increasing concentrations of trastuzumab for 72 h and measured both the total and phosphorylated (active) forms of these RTKs. Interestingly, low levels of trastuzumab led to elevated levels of IGF1R, phospho-IGF1R, and PDGFRb, whereas TYRO3 expression was increased only at the highest concentration (Fig. 2A). We also observed increased expression of IGF1R, phospho-IGF1R, and PDGFRb, but not TYRO3, in an independent, intrinsically resistant cell line, MDA-MB-361, as a result of sustained HER2 inhibition (Fig. 2B), implicating the induction of these RTKs as a common response utilized by drug-resistant breast cancers to combat HER2 inhibition.
FIGURE 2.
RTKs mediating resistance are up-regulated in drug resistant tumors and cell lines as a result of HER2 inhibition. A and B, IGF1R, PDGFRb, and TYRO3 expression levels increase as a result of trastuzumab (TRA) treatment in drug-resistant breast cancer cells. rBT474 (A) or MDA-MB-361 (B) cells were cultured for 72 h in the presence of 1, 3, or 10 μm trastuzumab, after which cells were harvested and protein lysates were immunoblotted using the indicated antibodies. C–E, representative images and quantification of IGF1R, PDGFRb, and TYRO3 immunohistochemistry in resistant and sensitive tumors before and after neoadjuvant trastuzumab therapy. Samples were obtained from HER2-positive breast cancers by mammotome biopsy before therapy and from surgery after treatment. n = 19 sensitive tumors; 13 resistant tumors. Error bars, S.D.; ***, p < 0.01. See supplemental Table S3 for exact p values. F, frequency of RTK overexpression after HER2 inhibition based on immunohistochemistry analysis of unresponsive patient tumors. A green bar represents up-regulation of the indicated RTK in an individual tumor.
The increased RTK expression that was observed to result from HER2 inhibition might be caused by acute up-regulation (feedback activation) or instead reflect the selection of a subpopulation of cells with higher expression (21, 22). To explore this, we measured both total and activated IGF1R in MDA-MB-361 cells at multiple time points following HER2 inhibition. This analysis revealed different kinetics for the observed increases in total and phosphorylated IGF1R, with IGF1R phosphorylation appearing 1–3 h after HER2 inhibition and total IGF1R increasing at ∼72 h (supplemental Fig. S3C), suggesting that both direct IGF1R activation and long term feedback up-regulation or selection for cells expressing high levels of IGF1R could play roles in the development of resistance.
We next determined whether these observations concerning cancer cells grown in culture were directly correlated with clinical outcomes for breast cancer patients. To this end, we obtained tumor samples from 32 HER2-positive breast cancer patients before and after neoadjuvant trastuzumab therapy (supplemental Table S1). Samples obtained before treatment were acquired by mammotome breast biopsy, which is a standard procedure used for diagnostic purposes. Of the 32 tumors analyzed, 19 were classified as responsive, whereas the remaining 13 were categorized as trastuzumab-resistant based on the existence of a stable or progressive disease (supplemental Fig. S4). This response rate (59.4%) is similar to previously reported response rates for trastuzumab therapy (6, 7). Consistent with the results from the cell culture studies, immunohistochemical staining using antibodies specific for this subset of RTKs, IGF1R, PDGFRb, and TYRO3, revealed that both sensitive and resistant tumors expressed relatively low levels of these RTKs before treatment (Fig. 2, C–E). Whereas targeted therapy had no significant effect on receptor expression in responsive tumors, recalcitrant tumors displayed significantly elevated IGF1R, PDGFRb, and TYRO3 protein levels after therapy, suggesting that these RTKs might promote resistance in the clinical setting. Importantly, this response was not uniform across all patient samples, with individual tumors inducing various combinations of these RTKs, indicative of heterogeneity in patient response to treatment (Fig. 2F and supplemental Table S2). Nonetheless, each of the 13 resistant tumors analyzed was found to significantly up-regulate at least one of these three RTKs, which supports their potential to function as therapeutic targets for the treatment of trastuzumab-resistant breast cancer.
Activity of the Distinct Subset of RTKs Is Necessary to Sustain Major Intracellular Signal Transduction Pathways in Response to HER2 Inhibition
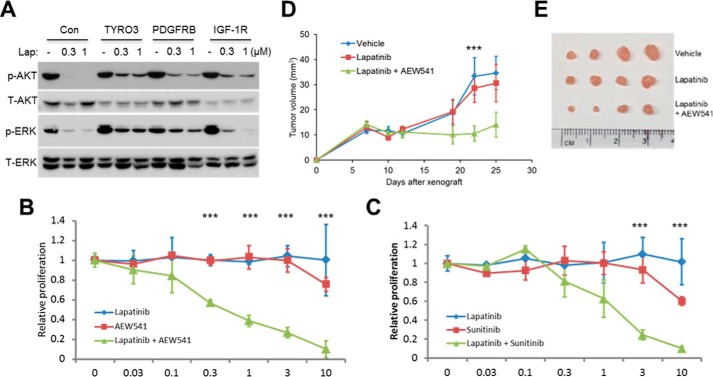
Various RTKs are known to regulate mammalian cell growth and proliferation by activating a shared repertoire of intracellular signaling cascades, most prominently the PI3K/Akt and Ras/Raf/ERK pathways. Based on our findings that this subset of RTKs could promote resistance to HER2 inhibition, we hypothesized that this specific group of RTKs might be utilized by breast cancer cells to maintain these signaling pathways at their original strength and spectrum when HER2 activity is blocked. In contrast, those RTKs incapable of conferring resistance, such as EPHB3, EPHB2, and ROR2 (Fig. 1, B–E), might not be able to adequately compensate for the loss in signaling input to the downstream pathways in the context of HER2 inhibition. To test this, we utilized trastuzumab-sensitive HER2-positive cell lines engineered to stably express these RTKs. As shown in Fig. 3A, 24-h lapatinib treatment of the vector control SKBR3 cells resulted in a strong reduction in the phosphorylated (active) forms of Akt and ERK, as measured by immunoblotting. Cells expressing TYRO3 or PDGFRb, however, were able to maintain robust levels of these phosphorylated proteins in the presence of HER2 inhibition, a result indicating that these specific RTKs are capable of sustaining signaling flux through these pathways to promote resistance. In contrast, we found that EPHB2 and EPHB3, RTKs that were previously shown to be incapable of conferring lapatinib resistance in this cell line, were unable to restore the loss of Akt and ERK signaling resulting from HER2 inhibition (supplemental Fig. S5A). Signaling activity of an additional sensitive cell line, AU565, was rescued by forced expression of TYRO3 and IGF1R but not PDGFRb (supplemental Fig. S5B), a result that correlates with the ability of these RTKs to rescue the viability of this cell line in the presence of HER2 inhibition (Fig. 1D). Therefore, there is a close relationship between the ability of a specific RTK to confer drug resistance and its capacity to maintain flux through important downstream signaling pathways. Moreover, these results suggest that robust activation of major intracellular signaling cascades is necessary for a specific RTK to promote the development of resistance to anti-HER2-based therapy.
FIGURE 3.
IGF1R, TYRO3, and PDGFRb activity is necessary to sustain signal transduction and tumor cell proliferation in response to HER2 inhibition. A, SKBR3 cells expressing TYRO3, IGF1R, or PDGFRb were treated with lapatinib (Lap) for 24 h, and then proteins were resolved by SDS-PAGE and immunoblotted using antibodies specific for the indicated signaling proteins. B, synergistic inhibition of trastuzumab-resistant cell growth by HER2 and IGF1R chemical inhibitors. rBT474 cells were cultured for 72 h in the presence of increasing concentrations of lapatinib, AEW541, or lapatinib plus AEW541 and then metabolically labeled with 2 μCi of [3H]thymidine for 16 h, after which DNA was precipitated and thymidine incorporation was measured by scintillation counting. C, rBT474 cells were cultured as in B but treated with the PDGFRb inhibitor sunitinib instead of AEW541. Error bars, S.E.; ***, p < 0.05. D and E, combined HER2 and IGF1R inhibition delays mammary tumor formation in mice. Immunodeficient BALB/c nude mice were orthotopically injected with rBT474 breast cancer cells, and, after allowing 1 week for tumor formation, were treated daily with vehicle, lapatinib, or lapatinib plus AEW541. Tumor sizes were measured twice weekly using vernier calipers until day 25, at which point mice were sacrificed, and dissected tumors were photographed (E). Mean tumor volumes ± S.E. are shown in D; ***, p < 0.05 (n = 4 mice/group).
Efficacy of Combinational RTK Inhibition in Overcoming Resistance
If this subset of RTKs is important in the development of anticancer drug resistance, then treatment with pharmacological inhibitors targeting these RTKs is predicted to confer susceptibility to HER2 inhibition. To explore this, we cultured rBT474 cells in the presence of lapatinib, the IGF1R-specific kinase inhibitor NVP-AEW54118 (AEW541), or a combination of both small molecules. AEW541 has been previously shown to possess >10-fold increased inhibitory activity against IGF1R compared with other tyrosine kinases, including HER2 and PDGFR (23). As expected, HER2 inhibition in SKBR3 breast cancer cells resulted in elevated levels of phosphorylated IGF1R, which was abrogated by administration of AEW541 (supplemental Fig. S5C). More importantly, whereas treatment with either inhibitor alone had no significant effect on cell growth, the combination of lapatinib and AEW541 produced a dose-dependent decrease in cell proliferation (IC50 ∼300 nm; Fig. 3B). Similar results were obtained using a chemical inhibitor targeting PDGFRb (sunitinib; Fig. 3C). It should be noted that, unlike AEW541, sunitinib is not specific for its target, but can also inhibit other RTKs, such as VEGF receptors (24). However, in support of the notion that this compound is impeding breast cancer cell proliferation by specifically blocking PDGFRb, similar assays revealed that pharmacological inhibition of c-Met or c-Kit, RTKs not predicted to be associated with resistance (Fig. 1A), had no measurable effect on cell viability either alone or in combination with lapatinib. Together, these data indicate that combinational inhibition of specific RTKs could be an effective strategy for overcoming resistance to HER2 inhibition.
To determine whether this combinational therapy could affect mammary tumor formation in vivo, we employed a mouse model of drug-resistant breast cancer by injecting rBT474 breast cancer cells into the mammary fat pad of immunodeficient BALB/c nude mice. Because AEW541 is currently the most selective chemical inhibitor that is widely available for our three targets, we initially chose to focus on IGF1R inhibition for these in vivo studies. After allowing 1 week for tumor formation, we began to administer either lapatinib or lapatinib combined with AEW541 by daily oral gavage. Consistent with our in vitro results, lapatinib treatment alone had no measurable effect on tumor progression compared with vehicle. In contrast, combined HER2/IGF1R inhibition resulted in significantly reduced tumor burdens in these animals (Fig. 3, D and E). These results demonstrate the potential therapeutic efficacy of pharmacologically targeting members of this distinct subset of RTKs to circumvent resistance.
As mentioned above, pharmacological inhibitors specifically targeting PDGFRb and TYRO3 are not widely available. Recently, several clinical trials have examined the effects of PDGFR inhibition by imatinib on the progression of various types of breast cancer (25, 26). However, in addition to both PDGFRa and PDGFRb, imatinib is known to inhibit other tyrosine kinases, including Abl and c-Kit, which might account for the toxicity observed in some of these studies (25, 26). Moreover, all of these clinical trials with imatinib have been conducted on advanced or metastatic breast cancer and were not focused on the HER2 subtype. Therefore, future studies examining combinational PDGFRb inhibition for drug-resistant HER2-positive breast cancer are warranted. Efforts to develop selective TYRO3 pharmacological inhibitors suitable for in vivo drug studies are also currently under way, and an important future objective will be to examine the efficacy of combinational IGF1R, PDGFRb, and TYRO3 inhibition on HER2-positive tumor growth, especially in the context of drug resistance.
Increased Expression of Inhibitors of Cellular Senescence IGFBP3 and IGFBP5 Is Associated with Responsiveness to HER2 Inhibition
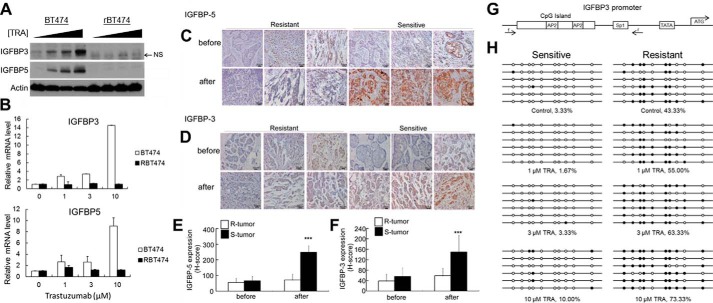
Based on the finding that synthetic compounds blocking IGF1R signaling impede drug-resistant tumor formation, we tested whether endogenous biological IGF1R inhibitors might play some role in conferring sensitivity to HER2 inhibition. Insulin-like growth factor-binding proteins (IGFBPs) are secreted proteins that prevent the initiation of IGF1R signaling by sequestering extracellular IGF1 such that it is unable to bind and activate IGF1R (27). To determine whether IGFBPs are involved in responsiveness to HER2 inhibition, we used immunoblotting and quantitative real-time PCR (qPCR) to detect changes in IGFBP expression upon trastuzumab treatment. This approach revealed that the expression of two IGFBP family members, IGFBP3 and IGFBP5, was induced in a dose-dependent manner in BT474 cells at both the protein and mRNA levels but remained undetectable in rBT474 cells after 72 h of treatment (Fig. 4, A and B). Immunohistochemistry of patient tumor samples further revealed that the expression of both IGFBP3 and IGFBP5 was elevated only in sensitive tumors following treatment (Fig. 4, C–F). These results suggest that one mechanism mediating responsiveness to HER2 inhibition is negative regulation of IGF1R signaling via induction of IGFBP3 and IGFBP5.
FIGURE 4.
Increased expression of IGFBP3 and IGFBP5 is associated with responsiveness to HER2 inhibition. A and B, expression of IGFBP3 and IGFBP5 in sensitive and resistant breast cancer cells after HER2 inhibition. BT474 or resistant BT474 (rBT474) breast cancer cells were cultured in the presence of 1, 3, or 10 μm trastuzumab for 72 h. IGFBP3 and IGFBP5 protein and mRNA levels were detected by immunoblotting (A) and qPCR (B), respectively. NS, nonspecific band. C–F, representative images (C and D) and quantification (E and F) of IGFBP immunohistochemistry in resistant (R) and sensitive (S) tumors before and after neoadjuvant trastuzumab therapy. The expression of both IGFBPs selectively increased only in responsive tumors. Samples were obtained from HER2-positive breast cancers by mammotome biopsy before therapy and from surgery after treatment. Error bars, S.D.; ***, p < 0.01. p values are provided in supplemental Table S3. G, structure of the IGFBP3 promoter. Locations of the primers used for bisulfite sequencing, TATA box, start codon, and transcription factor binding sites are indicated. H, methylation analysis of the IGFBP3 promoter in BT474 and rBT474 cells. BT474 (sensitive) or rBT474 (resistant) cells were cultured for 72 h in the presence of increasing concentrations of trastuzumab, after which genomic DNA was purified for bisulfite sequencing.
Because IGFBP3 and IGFBP5 are regulated by HER2 inhibition at the mRNA level (Fig. 4B), we hypothesized that molecular mechanisms underlying this regulation might include that at the transcriptional level, and such information could be useful in distinguishing sensitive from resistant cells. Previous reports have highlighted a role for promoter methylation in the transcriptional control of both IGFBP3 and IGFBP5. The IGFBP3 promoter contains a large CpG island close to the transcription start site (Fig. 4G), and hypermethylation of this region is associated with low IGFBP3 expression and poor clinical outcome in lung, ovarian, and colorectal cancers (28–30). The gene encoding IGFBP5 has also been shown to contain a weak CpG island within its first exon (31). These studies suggest that DNA methylation might be an important mechanism for controlling the levels of these two proteins within breast cancer cells. To explore this possibility, we examined the potential link of this epigenetic modification and breast cancer sensitivity to trastuzumab using bisulfite sequencing. As illustrated in Fig. 4H, the sensitive BT474 cell line contained low levels of IGFBP3 promoter methylation both before and after treatment, which is consistent with the observed induction of IGFBP3 mRNA. In contrast, IGFBP3 methylation was markedly higher in rBT474 cells resistant to HER2 inhibition and was further elevated by trastuzumab treatment, which might function to lock IGFBP3 expression in a transcriptionally repressed state. Although we do not know the precise molecular mechanism responsible for regulating IGFBP3 promoter methylation, a previous study has indicated that its functional significance is correlated with the mutational status of p53, which is a known activator of IGFBP3 transcription (30). Although examining this interaction with p53 in breast cancer cells will require further investigation, our results suggest that methylation of the IGFBP3 promoter could serve as a possible biomarker for predicting resistance in HER2-positive breast cancers.
Responsiveness to Anti-HER2 Therapy Is Associated with Breast Cancer Cellular Senescence
Although HER2 inhibition is therapeutically effective for ∼50% of all breast cancers that overexpress HER2 (32), the most critical cellular and molecular processes responsible for trastuzumab's clinical efficacy remain controversial and are not completely understood (33). IGFBP3 and IGFBP5 are known to promote cellular senescence (34–37), which is defined as a state of irreversible cell cycle arrest that plays a tumor-suppressive role in various types of cancer (38–40). Molecularly, senescent cells often express a common set of markers, including the cell cycle inhibitors p16 and p21 and the lysosomal enzyme β-galactosidase (senescence-associated β-galactosidase, SA-βgal) (41). Because we have previously observed that sustained inhibition of another ErbB family member, EGFR, is sufficient to induce senescence in normal lung and mammary epithelial cells (42), we were prompted to investigate whether HER2 inhibition could also promote senescence in breast cancer.
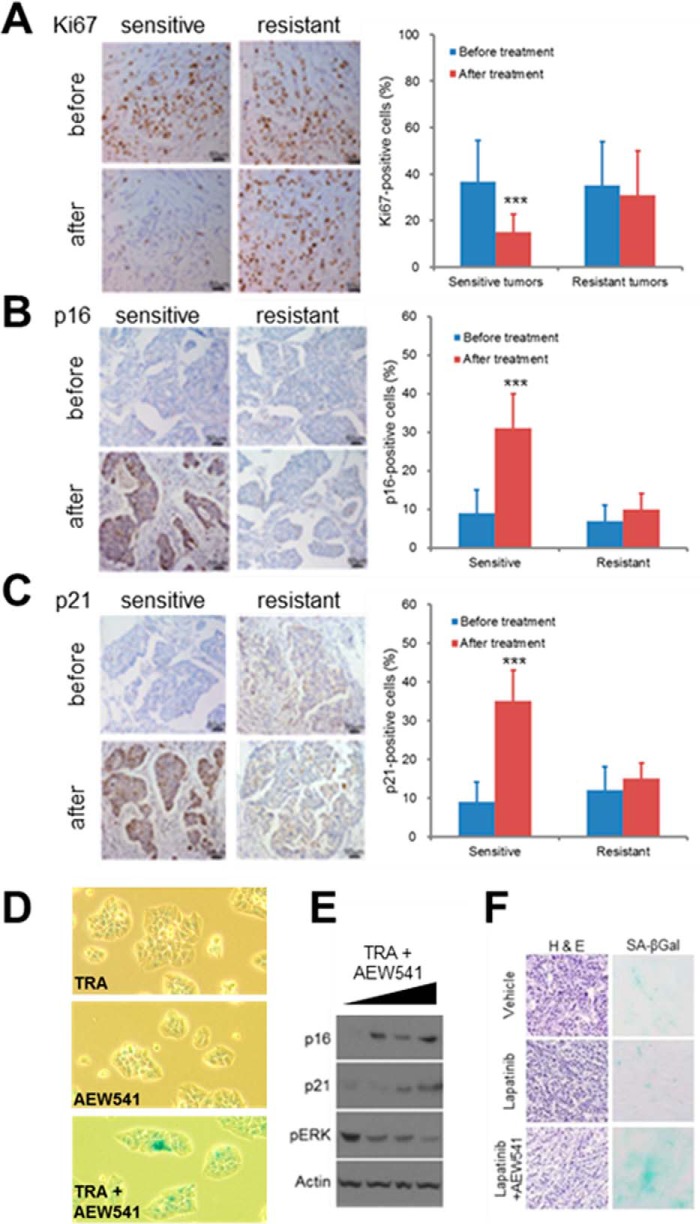
To determine whether cellular senescence might underlie the ability of HER2 inhibitors to impede breast tumor progression, we cultured drug-sensitive BT474 and SKBR3 breast cancer cells for 3 days in the presence of increasing concentrations of trastuzumab. This procedure led to the appearance of morphologically altered, non-dividing cells that expressed p16 and p21 (supplemental Fig. S6, A and B). These results suggest that breast cancer cells sensitive to HER2 inhibition might be dependent upon HER2 activity to escape senescence and maintain proliferation. To study this in a more clinically relevant context, we used the same group of 32 patient tumors to assess cellular senescence as it relates to patient outcome. As expected, tumors that responded to targeted therapy contained sharply reduced expression of the proliferation marker Ki67 (Fig. 5A). Moreover, sensitive tumors also displayed elevated levels of p16 and p21 expression (Fig. 5, B and C), indicating that HER2 inhibition impaired human breast cancer progression by inducing tumor cell senescence. Importantly, recalcitrant tumors did not express increased levels of these senescence markers upon completion of the treatment regimen (Fig. 5, B and C). These results provide support for the notion that cellular senescence is an important biological mechanism underlying susceptibility to HER2 inhibition, although induction of apoptosis could also be involved.
FIGURE 5.
Effective anti-HER2 therapy results in tumor cell senescence. A–C, representative images and quantification of Ki67 (A), p16 (B), and p21 (C) immunohistochemistry in resistant and sensitive tumors before and after neoadjuvant trastuzumab therapy. Samples were obtained from HER2-positive breast cancers by mammotome biopsy before therapy and from surgery after treatment. Error bars, S.D.; ***, p < 0.01. Exact p values can be found in supplemental Table S3. D–F, combined pharmacological inhibition of HER2 and IGF1R leads to cellular senescence in drug-resistant breast cancer cells. MDA-MB-361 cells were treated for 72 h with trastuzumab (TRA) and AEW541 as indicated, and cellular senescence was assayed by SA-βgal staining (D) and immunoblotting cell lysates with antibodies specific for the senescence-associated proteins p16 and p21 (E). F, rBT474 breast cancer cells were grown as tumor xenografts in BALB/c nude mice (n = 4/group). Following 1 week of tumor establishment, mice were treated with either vehicle, lapatinib (50 mg/kg/day), or lapatinib combined with AEW541 (both at 50 mg/kg/day) for 18 days. Frozen tumor sections were stained with hematoxylin and eosin to examine tumor histology or for SA-βgal activity to detect cellular senescence.
In addition to IGFBPs, senescent cells are known to produce and secrete a diverse assortment of proteins, including cytokines, chemokines, growth factors, adhesion molecules, and extracellular proteases (43, 44). This response has been termed the senescence-associated secretory phenotype (SASP) and can have pleiotropic effects on neighboring cells within the microenvironment (44). To further investigate the relationship between drug sensitivity and cellular senescence, we measured the expression levels of various factors that have been implicated in SASP in either sensitive or resistant BT474 cells treated with trastuzumab. Strikingly, many of these transcripts were significantly elevated in the sensitive BT474 cells that appeared to senesce in response to HER2 inhibition but displayed no change or even reduced expression in resistant cells (supplemental Fig. S6C). These results provide further support for the role of cellular senescence in responsiveness to HER2 inhibition and implicate the involvement of cell-extrinsic factors in determining drug sensitivity of HER2-positive breast cancer.
As shown earlier, targeted inhibition of IGF1R or PDGFRb confers sensitivity to breast cancer cells that are resistant to HER2 inhibition alone (Fig. 3, C and D). To assess whether this combination therapy leads to cellular senescence, we first treated resistant breast cancer cells with trastuzumab and AEW541 and measured senescence using the molecular markers p16, p21, and SA-βgal. Indeed, MDA-MB-361 cells treated with both drugs expressed increased levels of both cell cycle inhibitors and also stained positive for SA-βgal activity (Fig. 5, D and E). Moreover, mammary tumor sections from mice treated with both lapatinib and AEW541 also contained elevated SA-βgal activity (Fig. 5F), supporting the involvement of senescence in mediating tumor responsiveness in vivo. To examine the secretory profile of cells induced to senesce via combined HER2/IGF1R inhibition, we performed SASP profiling of drug-resistant cells treated with either the combination of both inhibitors or trastuzumab alone. Interestingly, several of the same secreted proteins were induced by this combinational treatment as in sensitive cells rendered to senesce by HER2 inhibition alone (supplemental Fig. S6D). In contrast, most SASP transcript levels were unaltered in resistant cells treated only with trastuzumab, providing further evidence that IGF1R inhibition confers susceptibility to HER2 inhibition by promoting cellular senescence via both cell-intrinsic and -extrinsic mechanisms. Finally, to determine whether PDGFRb signaling is also involved in suppressing cellular senescence in the context of HER2-positive breast cancer, we treated resistant cells with trastuzumab alone or trastuzumab in combination with the PDGFRb inhibitor sunitinib or imatinib. Similarly to IGF1R inhibition, PDGFRb antagonism led to increased expression of multiple cell cycle inhibitors, including p15, p21, and p27, as well as elevated SA-βgal activity (supplemental Fig. S6, E–G). We conclude that, among this subset of RTKs, both IGF1R and PDGFRb confer resistance to HER2 inhibition by suppressing the cellular senescence process.
Discussion
Here, employing a systematic approach with both expression analysis of human breast cancers and experimental manipulation of RTK expression and activity, we have demonstrated that only a distinct subset of RTKs is involved in the generation of resistance to targeted therapy against HER2 in breast cancer, although most, if not all, RTKs possess the ability to activate very similar downstream signaling pathways. Among this specific subset of RTKs, IGF1R has been implicated in bypassing HER2 inhibition (14–16). However, these previous studies did not directly investigate the effects of trastuzumab therapy on IGF1R in patient tumors and were instead mainly limited to cell culture models of breast cancer. Both TYRO3 and PDGFRb were previously unknown to be involved in anti-HER2 drug resistance. More broadly, our results highlight the ability of resistant tumors to utilize different combinations of RTKs belonging to this subset to achieve resistance, indicating complexity and heterogeneity in response to therapy, a notion also supported by a previous study (11) demonstrating that different growth factors display differential abilities to mediate resistance in different cancer cell lines. This notwithstanding, each of the resistant cell lines and tumors analyzed in our study was found to up-regulate at least one of these distinct RTKs, suggesting that we have identified molecular mechanisms that account for resistance in a high percentage of HER2-positive patients. Therefore, our study reveals both widespread and tumor-specific responses involved in the establishment of anticancer drug resistance.
Our findings that breast cancer cells utilize alternate RTKs to bypass targeted inhibition and establish chemoresistance have clear parallels with previous work implicating RTK switching in the epithelial-to-mesenchymal transition (EMT). This phenomenon has been studied most thoroughly in the context of non-small cell lung cancer, wherein independent studies have demonstrated that lung cancer cells can undergo an EMT program to up-regulate either AXL or IGF1R to bypass targeted EGFR inhibition (45, 46). Another recent study showed that epithelial lung tumors utilize ERBB3 to drive signal transduction through the PI3K signaling axis, whereas mesenchymal tumor cells down-regulate ERBB3 and instead maintain signaling flux via up-regulation of PIK3CA, resulting in reduced growth factor dependence (47). These results raise the possibility that the increased utilization of IGF1R, TYRO3, and PDGFRb by drug-resistant breast cancer cells observed in our study is associated with an unidentified EMT program. If this is the case, then, by up-regulating distinct RTK subsets to drive resistance, anti-HER2 targeted therapy might also act to drive EMT or select for a mesenchymal subpopulation within the tumor.
In sum, the results reported here reveal several important considerations that should guide the development of new targeted therapies for drug-resistant tumors. First, given the heterogeneity in molecular mechanisms employed by tumors to establish resistance, a single targeted therapy will be unlikely to produce a lasting response against resistant tumors, and combinational therapies will instead be required. Second, because the RTK subset was up-regulated in patient tumors and cell lines only after anti-HER2 therapy, these molecules will be unlikely to serve as prognostic markers, making it difficult to predict the specific molecular mechanisms that an individual tumor might employ to generate resistance. Although mixtures containing inhibitors targeting all three RTKs might be utilized to overcome this problem, it is possible that additional RTKs or other molecular pathways not revealed in our study could also play important roles in mediating resistance. As an alternative approach, immunotherapy might be used in the future to surmount the problem of resistance to targeted cancer therapies, because this treatment utilizes a fundamentally different mechanism compared with conventional targeted therapies (48). Indeed, clinical trials using antibodies targeting the PD-1/PD-L1 pathway in triple-negative breast cancer are currently under way. In conclusion, although the subset of RTKs identified in this study could guide the development of targeted therapies for drug-resistant breast cancer, our study also reveals that tumor heterogeneity produces significant complexities that should be considered in future efforts to overcome anticancer drug resistance.
Experimental Procedures
Cell Lines and Reagents
All breast cancer cell lines used in this study were obtained from the Duke Cell Culture Facility, with the exception of rBT474, which was established by the laboratory of Neil Spector. BT474, rBT474, SKBR3, AU565, MDA-MB-361, and T47D cells were cultured in RPMI 1640 supplemented with 10% FBS and 1% penicillin/streptomycin.
Lapatinib (catalog no. CT-LP002) was purchased from ChemieTek. Trastuzumab was obtained from the Duke Cancer Center Pharmacy and Neil Spector with the assistance of Gerry Blobe and Wenle Xia. NVP-AEW541 was provided by Novartis. Sunitinib (catalog no. S1042), imatinib (catalog no. S2475), and JNJ-38877605 (catalog no. S1114) were purchased from Selleck Chemicals. c-Met/RON dual kinase inhibitor (catalog no. 448104) was purchased from Millipore. ISCK03 (catalog no. 355981) was purchased from Santa Cruz Biotechnology, Inc.
Ectopic RTK Expression
RTK lentiviral expression plasmids (pLX304 vector) were a gift from Dr. Kris Wood. HER2-positive breast cancer cells were infected overnight and then selected in 1 μg/ml puromycin for 2 days. Cells in 96-well plates (2000 cells/well) were then treated with increasing concentrations of the indicated RTK inhibitor for an additional 2 days, after which cell viability was measured using the Cell Titer-Glo luminescent cell viability assay (Promega G7571). Viability curves were generated using GraphPad Prism. Results are reported from at least triplicate samples as the mean ± S.D. Protein expression was verified by immunoblotting using an anti-V5 antibody (Abcam 9116).
Western Blotting
After the indicated treatment, proteins were extracted using a 30-min incubation of cells in cold 50 mm Tris-HCl (pH 7.4), 50 mm NaCl, 0.5% Nonidet P-40, 1 mm DTT, 100 μm phenylmethylsulfonyl fluoride, 1× protease inhibitor mixture (Sigma catalog no. P2714), and 1× phosphatase inhibitor mixture (Thermo Scientific catalog no. 1861277). Protein concentrations were determined by a Bio-Rad protein assay, and 30 μg of total protein was loaded in each lane of an SDS-polyacrylamide gel. Primary antibodies were diluted in 1% (w/v) nonfat dry milk in Tris-buffered saline with 0.05% Tween 20 (TBST) and incubated with PVDF membranes at 4 °C with gentle shaking overnight.
Primary antibodies used for immunoblotting are as follows. Phospho-Akt (Ser-473, 4060), Akt (4691), ERK (4695), phospho-IGF1R (Tyr-980, 4568), IGF1R (9750), phospho-PDGFRb (Tyr-740, 3168), TYRO3 (5585), p21 (2947), and p27 (2552) were from Cell Signaling. Phospho-ERK (Tyr-204, 7383), PDGFRb (339), IGFBP5 (6006), p53 (126), and p15 (612) were from Santa Cruz Biotechnology. Anti-V5 was from Abcam (9116). Anti-EPHB2 was from R&D Systems (AF467). Anti-EPHB3 was from Abnova (H00002049-M01). Anti-IGFBP3 was from BD Biosciences (611504). Anti-β-actin was from Sigma (A2228).
Survival Analysis
A breast cancer metaset was derived from 25 publicly available data sets comprising 4885 unique patient samples. The raw data were downloaded from GEO, normalized with fRMA, and batch-corrected using the COMBAT algorithm within R (49). Each tumor was then classified into PAM50 (50) molecular subtypes using Genefu (51). Gene expression was split on the median into low and high classifications, and Kaplan-Meier curves were plotted in R. Reported p values were calculated using the log-rank method. Forest plots and concordance index estimates were generated using the survcomp (52) package. Data sets used were GSE10780, GSE11121, GSE12093, GSE12276, GSE1456, GSE16391, GSE16446, GSE17705, GSE17907, GSE19615, GSE20194, GSE2034, GSE20685, GSE20711, GSE2109, GSE21653, GSE22093, GSE24185, GSE25066, GSE3494, GSE5460, GSE6532, GSE6532, GSE7390, and GSE9195.
HER2+ Breast Cancer Patient Study
Paired paraffin-embedded samples from 32 breast cancer tissues were collected before and after neoadjuvant trastuzumab therapy. All patients underwent preoperative neoadjuvant therapy with 3–4 cycles of the triweekly TCH regimen (Taxotere, 75 mg/m2; carboplatin, AUC (area under the curve) 6 mg/ml/min; and Herceptin, 8 mg/kg loading dose followed by 6 mg/kg every 3 weeks). The clinical response was assessed based on the RECIST (Response Evaluation Criteria in Solid Tumors). Antibodies used for immunohistochemistry are as follows: IGF1R (Cell Signaling, catalog no. 3027, 1:500), PDGFRb (Santa Cruz Biotechnology, sc-339, 1:500), TYRO3 (Novus, NBP2-23725, 1:100), IGFBP3 (Abcam, ab76001, 1:50), IGFBP5 (Santa Cruz Biotechnology, sc-6006, 1:500), Ki67 (Abcam, ab66155, 1:100), p16 (Abcam, ab189302, 1:500), and p21 (Abcam, ab54562, 3 μg/ml). All of those antibodies have been validated for immunohistochemistry staining. t test and the χ2 test were used to compare continuous and categorical variables of two groups. All pathological biomarkers were pathologically reviewed and quantified independently by two breast pathologists. These two pathologists were blinded to the clinical outcome.
Thymidine Incorporation Assay
After growth on a 24-well plate for 1 day, cancer cells were treated as indicated and then metabolically labeled with [3H]thymidine (2 μCi/well) overnight. Following three washes with PBS, cellular macromolecules were precipitated by a 15-min incubation of cells on ice with 5% trichloroacetic acid and then suspended in 0.5 m NaOH, 0.5% SDS for liquid scintillation counting using a Beckman LS 6000SC counter.
Mouse Xenografts and in Vivo Drug Studies
rBT474 tumor cell xenografts were established through the 200-μl orthotopic injection of cancer cell suspension (3 × 107 cells/ml) into the fourth gland of the mammary fat pad of immunodeficient BALB/c nude mice (4 mice/group). Tumor volume was monitored twice per week, and after 1 week, mice were randomly assigned to treatment with vehicle (25 mm l-(+)-tartaric acid, 0.5% Tween 80 by daily gavage), lapatinib (50 mg/kg/day in vehicle by oral gavage), or laptatinib combined with AEW541 (50 mg/kg/day of each in vehicle by oral gavage). Lapatinib and AEW541 were dissolved in DMSO, mixed with vehicle, and stored at −80 °C in single aliquots. Tumor volumes were calculated using the formula S × S × L × 0.52, where S and L are the short and long caliper measurements, respectively. Animals were sacrificed and tumors were dissected at 25 days after the fat pad injection. Tumors were embedded in O.C.T. compound, sectioned at 10 μm using a Leica CM3050 S cryostat, and stained for SA-βgal activity or with hematoxylin and eosin.
Quantitative RT-PCR Analysis
Total RNA was extracted using TRIzol reagent (Ambion) and converted into cDNA with the iScript cDNA synthesis kit (Bio-Rad). PCR was performed on a MasterCycler RealPlex4 real-time PCR system (Eppendorf) using specific primer pairs for senescence-associated mRNAs.
Methylation Analysis
The methylation status of the CpG dinucleotides within the CpG island of the IGFBP3 promoter was analyzed. BT474 and rBT474 cells were treated for 72 h with the indicated concentrations of trastuzumab, and then genomic DNA was purified using a genomic DNA extraction kit (Qiagen). A bisulfite sequencing assay was performed on 1.0 μg of bisulfite-treated genomic DNA from each sample. Bisulfite conversion was performed using the MethylDetector bisulfite modification kit (Active Motif) according to the manufacturer's instructions. The fragments of interest were amplified using Platinum TaqDNA Polymerase High Fidelity (Invitrogen) with the following primer pair: forward, 5′-TGGGTATATTTTGGTTTTTGTAGA-3′; reverse, 5′-AAAACCAAAATAACCCAAAACAC-3′. PCR products were gel-purified and cloned into the pCR4-TOPO TA vector (Invitrogen). Individual bacterial colonies were picked and sequenced using the M13 reverse primer (5′-GTTTTCCCAGTCACGAC-3′) to measure DNA methylation.
SA-βgal Assays
SA-βgal activity was detected using the methods of Debacq-Chainiaux et al. (53). Cells were first washed twice with PBS and then fixed using 2% formaldehyde and 0.2% glutaraldehyde in PBS for 3 min. Following three washes in PBS, cells were stained using 40 mm citric acid/sodium phosphate buffer (pH 6.0), 5 mm potassium ferrocyanide, 5 mm potassium ferracyanide, 150 mm NaCl, 2 mm MgCl2, and 1 mg/ml X-Gal, typically overnight at 37 °C. Microscopic analyses were performed using an Olympus CK40 microscope with a DP20 camera.
Statistical Analysis
Data are presented as the mean ± S.E. or mean ± S.D. To determine p values, two-tailed Student's t tests were performed (unless otherwise indicated). p < 0.05 was considered statistically significant. For survival curves, Kaplan-Meier analysis was used, with statistical comparison among curves performed using the log-rank test.
Study Approval
All experiments involving mice were approved by the Duke Institutional Animal Care and Use Committee and followed all state and federal rules and regulations. All specimens were obtained with written informed consent of the patients and approved by the institutional ethical committee for clinical research.
Author Contributions
P. B. A. and R. C. conducted most of the experiments, analyzed the results, and wrote most of the paper. J. S. J. and D. P. M. performed the initial bioinformatic analysis of RTK expression in HER2-positive breast cancer. C. G. and E. S. performed the immunohistochemical staining of HER2-positive tumor tissues. L. Y., P. Y., and X. X. provided assistance with mouse xenograft experiments. Y. D. and G. J. M. provided assistance with immunoblotting and qPCR. X.-F. W. conceived the idea for the project and wrote the paper with P. B. A.
Supplementary Material
Acknowledgments
We thank Neil Spector, Wenle Xia, Gerry Blobe, and the Duke Cancer Center Pharmacy for providing trastuzumab used in this study. We thank Kris Wood for providing lentiviral RTK expression plasmids and helpful discussion.
This work was supported by National Institutes of Health Grants CA154586 and CA186800 (to X.-F. W.), CA174643 (to D. P. M.), and CA059365 (to P. B. A.) and Department of Defense Grant BC151189 (to X.-F. W.). This work was also supported by 973 Projects 2010CB912800 and 2011CB504203 from the Ministry of Science and Technology of China; Natural Science Foundation of China Grants 81672594, 81230060, 81261140373, 81272893, and 81472466; National S&T Major Special Project on New Drug Innovation of China Grant 2011ZX09102-010-02); and Guangdong Province Grants 2014A03036003, S2012030006287, 2014A030310378, and 2016A050502018 (to E. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables S1–S3 and Figs. S1–S6.
- RTK
- receptor tyrosine kinase
- EGFR
- EGF receptor
- PDGFR
- PDGF receptor
- IGFBP
- insulin-like growth factor-binding protein
- qPCR
- quantitative real-time PCR
- SA-βgal
- senescence-associated β-galactosidase
- EMT
- epithelial-to-mesenchymal transition.
References
- 1. Jemal A., Center M. M., DeSantis C., and Ward E. M. (2010) Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol. Biomarkers Prev. 19, 1893–1907 [DOI] [PubMed] [Google Scholar]
- 2. Citri A., and Yarden Y. (2006) EGF-ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell Biol. 7, 505–516 [DOI] [PubMed] [Google Scholar]
- 3. Moasser M. M. (2007) The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene 26, 6469–6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Slamon D. J., Clark G. M., Wong S. G., Levin W. J., Ullrich A., and McGuire W. L. (1987) Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235, 177–182 [DOI] [PubMed] [Google Scholar]
- 5. Hynes N. E., and Lane H. A. (2005) ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer 5, 341–354 [DOI] [PubMed] [Google Scholar]
- 6. Slamon D. J., Leyland-Jones B., Shak S., Fuchs H., Paton V., Bajamonde A., Fleming T., Eiermann W., Wolter J., Pegram M., Baselga J., and Norton L. (2001) Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 344, 783–792 [DOI] [PubMed] [Google Scholar]
- 7. Romond E. H., Perez E. A., Bryant J., Suman V. J., Geyer C. E. Jr., Davidson N. E., Tan-Chiu E., Martino S., Paik S., Kaufman P. A., Swain S. M., Pisansky T. M., Fehrenbacher L., Kutteh L. A., Vogel V. G., et al. (2005) Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N. Engl. J. Med. 353, 1673–1684 [DOI] [PubMed] [Google Scholar]
- 8. Nahta R., and Esteva F. J. (2006) HER2 therapy: molecular mechanisms of trastuzumab resistance. Breast Cancer Res. 8, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vu T., and Claret F. X. (2012) Trastuzumab: updated mechanisms of action and resistance in breast cancer. Front. Oncol. 2, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Groenendijk F. H., and Bernards R. (2014) Drug resistance to targeted therapies: deja vu all over again. Mol. Oncol. 8, 1067–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wilson T. R., Fridlyand J., Yan Y., Penuel E., Burton L., Chan E., Peng J., Lin E., Wang Y., Sosman J., Ribas A., Li J., Moffat J., Sutherlin D. P., Koeppen H., et al. (2012) Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 487, 505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Niederst M. J., and Engelman J. A. (2013) Bypass mechanisms of resistance to receptor tyrosine kinase inhibition in lung cancer. Sci. Signal. 6, re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alexander P. B., and Wang X. F. (2015) Resistance to receptor tyrosine kinase inhibition in cancer: molecular mechanisms and therapeutic strategies. Front. Med. 9, 134–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harris L. N., You F., Schnitt S. J., Witkiewicz A., Lu X., Sgroi D., Ryan P. D., Come S. E., Burstein H. J., Lesnikoski B. A., Kamma M., Friedman P. N., Gelman R., Iglehart J. D., and Winer E. P. (2007) Predictors of resistance to preoperative trastuzumab and vinorelbine for HER2-positive early breast cancer. Clin. Cancer Res. 13, 1198–1207 [DOI] [PubMed] [Google Scholar]
- 15. Lu Y., Zi X., Zhao Y., Mascarenhas D., and Pollak M. (2001) Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (Herceptin). J. Natl. Cancer Inst. 93, 1852–1857 [DOI] [PubMed] [Google Scholar]
- 16. Lu Y., Zi X., and Pollak M. (2004) Molecular mechanisms underlying IGF-I-induced attenuation of the growth-inhibitory activity of trastuzumab (Herceptin) on SKBR3 breast cancer cells. Int. J. Cancer 108, 334–341 [DOI] [PubMed] [Google Scholar]
- 17. Wang Q., Quan H., Zhao J., Xie C., Wang L., and Lou L. (2013) RON confers lapatinib resistance in HER2-positive breast cancer cells. Cancer Lett. 340, 43–50 [DOI] [PubMed] [Google Scholar]
- 18. Bardelli A., Corso S., Bertotti A., Hobor S., Valtorta E., Siravegna G., Sartore-Bianchi A., Scala E., Cassingena A., Zecchin D., Apicella M., Migliardi G., Galimi F., Lauricella C., Zanon C., et al. (2013) Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 3, 658–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Neve R. M., Chin K., Fridlyand J., Yeh J., Baehner F. L., Fevr T., Clark L., Bayani N., Coppe J. P., Tong F., Speed T., Spellman P. T., DeVries S., Lapuk A., Wang N. J., et al. (2006) A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10, 515–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xia W., Bacus S., Hegde P., Husain I., Strum J., Liu L., Paulazzo G., Lyass L., Trusk P., Hill J., Harris J., and Spector N. L. (2006) A model of acquired autoresistance to a potent ErbB2 tyrosine kinase inhibitor and a therapeutic strategy to prevent its onset in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 103, 7795–7800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chandarlapaty S., Sawai A., Scaltriti M., Rodrik-Outmezguine V., Grbovic-Huezo O., Serra V., Majumder P. K., Baselga J., and Rosen N. (2011) AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 19, 58–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chakrabarty A., Sánchez V., Kuba M. G., Rinehart C., and Arteaga C. L. (2012) Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proc. Natl. Acad. Sci. U.S.A. 109, 2718–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. García-Echeverrá C., Pearson M. A., Marti A., Meyer T., Mestan J., Zimmermann J., Gao J., Brueggen J., Capraro H. G., Cozens R., Evans D. B., Fabbro D., Furet P., Porta D. G., Liebetanz J., et al. (2004) In vivo antitumor activity of NVP-AEW541: a novel, potent, and selective inhibitor of the IGF-IR kinase. Cancer Cell 5, 231–239 [DOI] [PubMed] [Google Scholar]
- 24. Roskoski R., Jr. (2007) Sunitinib: a VEGF and PDGF receptor protein kinase and angiogenesis inhibitor. Biochem. Biophys. Res. Commun. 356, 323–328 [DOI] [PubMed] [Google Scholar]
- 25. Yardley D. A., Burris H. A. 3rd, Markus T., Spigel D. R., Greco F. A., Mainwaring M., Waterhouse D. M., Webb C. D., and Hainsworth J. D. (2009) Phase II trial of docetaxal plus imatinib mesylate in the treatment of patients with metastatic breast cancer. Clin. Breast Cancer 9, 237–242 [DOI] [PubMed] [Google Scholar]
- 26. Modi S., Seidman A. D., Dickler M., Moasser M., D'Andrea G., Moynahan M. E., Menell J., Panageas K. S., Tan L. K., Norton L., and Hudis C. A. (2005) A phase II trial of imatinib mesylate monotherapy in patients with metastatic breast cancer. Breast Cancer Res. Treat. 90, 157–163 [DOI] [PubMed] [Google Scholar]
- 27. Clemmons D. R. (1997) Insulin-like growth factor binding proteins and their role in controlling IGF actions. Cytokine Growth Factor Rev. 8, 45–62 [DOI] [PubMed] [Google Scholar]
- 28. Chang Y. S., Wang L., Liu D., Mao L., Hong W. K., Khuri F. R., and Lee H. Y. (2002) Correlation between insulin-like growth factor-binding protein-3 promoter methylation and prognosis of patients with stage I non-small cell lung cancer. Clin. Cancer Res. 8, 3669–3675 [PubMed] [Google Scholar]
- 29. Wiley A., Katsaros D., Fracchioli S., and Yu H. (2006) Methylation of the insulin-like growth factor binding protein-3 gene and prognosis of epithelial ovarian cancer. Int. J. Gynecol. Cancer 16, 210–218 [DOI] [PubMed] [Google Scholar]
- 30. Kawasaki T., Nosho K., Ohnishi M., Suemoto Y., Kirkner G. J., Fuchs C. S., and Ogino S. (2007) IGFBP3 promoter methylation in colorectal cancer: relationship with microsatellite instability, CpG island methylator phenotype, and p53. Neoplasia 9, 1091–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rivenbark A. G., Jones W. D., Risher J. D., and Coleman W. B. (2006) DNA methylation-dependent epigenetic regulation of gene expression in MCF-7 breast cancer cells. Epigenetics 1, 32–44 [DOI] [PubMed] [Google Scholar]
- 32. Hudis C. A. (2007) Trastuzumab: mechanism of action and use in clinical practice. N. Engl. J. Med. 357, 39–51 [DOI] [PubMed] [Google Scholar]
- 33. Spector N. L., and Blackwell K. L. (2009) Understanding the mechanisms behind trastuzumab therapy for human epidermal growth factor receptor 2-positive breast cancer. J. Clin. Oncol. 27, 5838–5847 [DOI] [PubMed] [Google Scholar]
- 34. Kim K. S., Kim M. S., Seu Y. B., Chung H. Y., Kim J. H., and Kim J. R. (2007) Regulation of replicative senescence by insulin-like growth factor-binding protein 3 in human umbilical vein endothelial cells. Aging Cell 6, 535–545 [DOI] [PubMed] [Google Scholar]
- 35. Kim K. S., Seu Y. B., Baek S. H., Kim M. J., Kim K. J., Kim J. H., and Kim J. R. (2007) Induction of cellular senescence by insulin-like growth factor binding protein-5 through a p53-dependent mechanism. Mol. Biol. Cell 18, 4543–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Elzi D. J., Lai Y., Song M., Hakala K., Weintraub S. T., and Shiio Y. (2012) Plasminogen activator inhibitor 1-insulin-like growth factor binding protein 3 cascade regulates stress-induced senescence. Proc. Natl. Acad. Sci. U.S.A. 109, 12052–12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kojima H., Kunimoto H., Inoue T., and Nakajima K. (2012) The STAT3-IGFBP5 axis is critical for IL-6/gp130-induced premature senescence in human fibroblasts. Cell Cycle 11, 730–739 [DOI] [PubMed] [Google Scholar]
- 38. Kuilman T., Michaloglou C., Mooi W. J., and Peeper D. S. (2010) The essence of senescence. Genes Dev. 24, 2463–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lowe S. W., Cepero E., and Evan G. (2004) Intrinsic tumour suppression. Nature 432, 307–315 [DOI] [PubMed] [Google Scholar]
- 40. Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A. J., Barradas M., Benguría A., Zaballos A., Flores J. M., Barbacid M., Beach D., and Serrano M. (2005) Tumour biology: senescence in premalignant tumours. Nature 436, 642. [DOI] [PubMed] [Google Scholar]
- 41. Rodier F., and Campisi J. (2011) Four faces of cellular senescence. J. Cell Biol. 192, 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alexander P. B., Yuan L., Yang P., Sun T., Chen R., Xiang H., Chen J., Wu H., Radiloff D. R., and Wang X. F. (2015) EGF promotes mammalian cell growth by suppressing cellular senescence. Cell Res. 25, 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Coppé J. P., Patil C. K., Rodier F., Sun Y., Muñoz D. P., Goldstein J., Nelson P. S., Desprez P. Y., and Campisi J. (2008) Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6, 2853–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Coppé J. P., Desprez P. Y., Krtolica A., and Campisi J. (2010) The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 5, 99–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Byers L. A., Diao L., Wang J., Saintigny P., Girard L., Peyton M., Shen L., Fan Y., Giri U., Tumula P. K., Nilsson M. B., Gudikote J., Tran H., Cardnell R. J., Bearss D. J., et al. (2013) An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin. Cancer Res. 19, 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhou J., Wang J., Zeng Y., Zhang X., Hu Q., Zheng J., Chen B., Xie B., and Zhang W. M. (2015) Implication of epithelial-mesenchymal transition in IGF1R-induced resistance to EGFR-TKIs in advanced non-small cell lung cancer. Oncotarget 6, 44332–44345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Salt M. B., Bandyopadhyay S., and McCormick F. (2014) Epithelial-to-mesenchymal transition rewires the molecular path to PI3K-dependent proliferation. Cancer Discov. 4, 186–199 [DOI] [PubMed] [Google Scholar]
- 48. Sharma P., and Allison J. P. (2015) Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell 161, 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kastner K., Margraf J. T., Clark T., and Streb C. (2014) A molecular placeholder strategy to access a family of transition-metal-functionalized vanadium oxide clusters. Chemistry 20, 12269–12273 [DOI] [PubMed] [Google Scholar]
- 50. Parker J. S., Mullins M., Cheang M. C., Leung S., Voduc D., Vickery T., Davies S., Fauron C., He X., Hu Z., Quackenbush J. F., Stijleman I. J., Palazzo J., Marron J. S., Nobel A. B., et al. (2009) Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27, 1160–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thompson C. A. (2015) Biosimilar filgrastim approved, but with “placeholder” generic name. Am. J. Health Syst. Pharm. 72, 592–594 [DOI] [PubMed] [Google Scholar]
- 52. Schröder M. S., Culhane A. C., Quackenbush J., and Haibe-Kains B. (2011) survcomp: an R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics 27, 3206–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Debacq-Chainiaux F., Erusalimsky J. D., Campisi J., and Toussaint O. (2009) Protocols to detect senescence-associated β-galactosidase (SA-βgal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 4, 1798–1806 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.