Abstract
Here, we report a genome-wide survey of immunoglobulin light chain (IGL) genes of torafugu (Takifugu rubripes) revealing multi-clusters spanning three separate chromosomes (v5 assembly) and 45 scaffolds (v4 assembly). Conventional sequence similarity searches and motif scanning approaches based on recombination signal sequence (RSS) motifs were used. We found that three IGL isotypes (L1, L2, and L3) exist in torafugu and that several loci for each isotype are present. The transcriptional orientations of the variable IGL (VL) segments were found to be either the same (in the L2 isotype) or opposite (in the L1 and L3 isotypes) to the IGL joining (JL) and constant (CL) segments, suggesting they can undergo rearrangement by deletion or inversion when expressed. Alignments of expressed sequence tags (ESTs) to corresponding germline gene segments revealed expression of the three IGL isotypes in torafugu. Taken together, our findings provide a genomic framework for torafugu IGL genes and show that the IG diversity of this species could be attributed to at least three distinct chromosomal regions.
The adaptive immune system (AIS) functions via a diverse repertoire of antigen receptors: immunoglobulins (IGs) and T cell receptors (TCRs). The key effectors, IGs, which are expressed only by jawed vertebrates, are primarily involved in antibody responses1. A typical IG molecule consists of two identical heavy (IGH) and two identical light (IGL) chains. Each IGL chain has two domains: a constant (C) and variable (V) domain. The IGL C domain is encoded by the constant (CL) gene. As for the V domain, individual variable (VL) and joining (JL) gene segments rearrange somatically at the DNA level to generate V-J regions, which, after transcription and translation, encode the functional V domains of IGL2. Primary diversification of IGs occurs early in B cell development during V(D)J recombination. The IG repertoire is further diversified by the action of activation-induced cytidine deaminase (AID), which catalyzes IG somatic hypermutation and class switch recombination (absent in fishes) in mammals and other tetrapods3,4. V(D)J recombination is initiated by RAG recombinase, which recognizes recombination signal sequences (RSSs) flanking each V, D, and J gene segment and cleaves DNA during V(D)J recombination5. The RSSs are composed of conserved heptamer and nonamer sequences, separated by either 12 ± 1 or 23 ± 1 base pair (bp) spacer sequences6. The V domain consists of three complementarity-determining regions (CDRs) of highly variable sequence and four framework regions (FRs) of relatively constant sequence.
Jawed vertebrate species, with the exception of chickens, ducks, and snakes, express more than one IGL isotype7,8,9. It has long been known that mammals have two distinct IGL isotypes called kappa (κ) and lambda (λ), but additional IGL isotypes have been described in other vertebrate groups. The current classification system groups all vertebrate IGLs into four main ancestral branches: kappa (mammalian κ, elasmobranch type III/NS4, teleost L1/L3/F/G, Xenopus ρ), lambda (mammalian λ, elasmobranch type II/NS3), sigma (Xenopus σ, teleost L2, elasmobranch type IV), and sigma-2 (elasmobranch type I/NS5, variant sigma-type in coelacanth)10,11.
Traditionally, different vertebrate IGL sequences are classified by: (1) sequence identity, (2) IGL gene organization, and (3) spacing of RSS heptamer and nonamer motifs that flank VL and JL12. Additionally, Criscitiello and Flajnik13 have proposed CDR lengths of corresponding VL, specifically CDR1 and CDR2, to be a valid criterion for the classification of IGL. Another IGL classification criterion using a set of 21 conserved molecular sequence markers to distinguish κ, λ, and σ IGL isotypes was later proposed by Das et al.14.
Teleost IGL genes, as those of cartilaginous fish, have been shown to be in a multi-clustered configuration15,16,17,18,19,20,21, defined as independently rearranging mini-loci consisting of few gene segments (multiple V segments, one J) and one C domain exon22. The IGL loci in teleosts form tightly linked clusters and there are significant differences in the number of loci for each isotype among species. The presence of multiple clusters on one or more chromosomes, similar to those found in zebrafish (Danio rerio), three-spined stickleback (Gasterosteus aculeatus), and medaka (Oryzias latipes), suggests a major role for cluster duplication in the generation of IG diversity in teleosts18.
Torafugu (Takifugu rubripes) has a recognizable adaptive immune system and one of the smallest genomes (~400 Mb) among vertebrates23, which makes it a good model for research in comparative immunology. Two partial annotations of torafugu IGLs have been reported18,24, revealing IGL assemblages with respect to gene segment number, cluster orientations, and organization on three scaffolds and two clones that contain L1 and L2 loci, respectively. In this work, we have scanned the torafugu genome assemblies to provide an extended annotation of torafugu IGLs as well as their genomic organization. Our research showed the identification of a third teleost IGL isotype (L3) in torafugu and an expansion of the IGL genes that were identified in previous studies.
Results
Identification of IGL genes on torafugu genomic chromosomes and scaffolds
A total of 82 IGL gene segments in torafugu were found to be localized on three different chromosomes, i.e., 2, 3, and 5, and were confined to 45 different genomic scaffolds (see annotation details in Supplementary Dataset File). Of the scaffolds, four (scaffold 10, 158, 54, and 139) were assigned to separate chromosomes, whereas most of the IGL genes could not be anchored to chromosomes. Altogether, 48 VL (Table 1 and Supplementary Dataset File), 13 JL (Table 2 and Supplementary Dataset File) and 21 CL (Fig. 1 and Supplementary Dataset File) gene segments25 (except for those that might be present in gaps and cannot be identified at present) were identified.
Table 1. Genomic features of the torafugu VL genes.
| IGLV family | VL gene | Fct | Promoter | Gene structure | RSS | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Octamer | (nt) | TATA | (nt) | L-PART1 (nt) | Intron | V-exon | 7mer | Spacer (nt) | 9mer | |||
| IGLV1 | V1a^ | Pa | — | — | — | — | — | — | 114 | CACAGTG | 12 | ACAAAAACC |
| V1b^ | Pb | ATTTGCAT | 27 | TTTAAA | 64 | 40 | 137 | 249 | — | — | — | |
| V1c^ | ORFl | ATTTGCAT | 27 | TTTAAA | 64 | 40 | 137 | 285 | CACAGTG | 12 | ACAAAAACC | |
| V1d^ | Pa | — | — | — | — | — | — | 267 | CACAGTG | 12 | ACAAAAACC | |
| V1e^ | ORFl | ATTTGCAT | 27 | TTTAAA | 64 | 40 | 137 | 291 | CACAGTG | 12 | ACAAAAACC | |
| V1f | ORFl | — | — | TTTAAA | 64 | 40 | 137 | 300 | CACAGTG | 12 | ACAAAAACC | |
| V1g^ | Pa | — | — | — | — | — | — | 300 | CACAGTG | 12 | ACAAAAACC | |
| V1h^ | ORFl | ATTTGCAT | 27 | TTTAAA | 64 | 40 | 137 | 300 | CACAGTG | 12 | ACAAAAACC | |
| V1i | Pa | — | — | — | — | — | — | 300 | CACAGTG | 12 | ACAAAAACC | |
| V1j | ORFl | ATTTGCAT | 27 | TTTAAA | 64 | 40 | 137 | 285 | CACAGTG | 12 | ACAAAAACC | |
| V1k^ | ORFl | — | — | — | — | 40 | 137 | 291 | CACAGTG | 12 | ACAAAAACC | |
| V1l^ | ORFl | ATTTGCAT | 27 | TTTAAA | — | 40 | 133 | 291 | CACAGTG | 12 | ACAAAAACC | |
| V1m^ | ORFl | ATTTGCAT | 27 | TTTAAA | 64 | 40 | 140 | 285 | CACAGTG | 12 | ACAAAAACC | |
| V1n | ORFl | ATTTGCAT | 27 | TTTAAA | 64 | 40 | 137 | 300 | CACAGTG | 12 | ACAAAAACC | |
| V1o | ORFl | ATTTGCAT | 27 | TTTAAA | 62 | 40 | 137 | 285 | CACAGTG | 12 | ACAAAAACC | |
| V1p^ | ORFl | — | — | TTTAAA | 64 | 40 | 137 | 288 | CACAGTG | 12 | ACAAAAACC | |
| V1q^ | ORFl | ATTTGCAT | 27 | TTTAAA | 62 | 40 | 137 | 300 | CACAGTG | 12 | ACAAAAACC | |
| V1r^ | Pb | ATTTGCAT | 27 | TTTAAA | 64 | 40 | 133 | 168 | — | — | — | |
| V1s^ | ORFl | — | — | — | — | 40 | 137 | 291 | CACAGTG | 12 | ACAAAAACC | |
| V1t^ | ORFl | ATTTGCAT | 27 | TTTAAA | 64 | 40 | 137 | 291 | CACAGTG | 12 | ACAAAAACC | |
| IGLV2 | V2a^ | Pf | ATGTAAAT | 107 | TATTAA | 97 | 40 | 92 | 326 | CACAGTG | 12 | ACAAAAACC |
| V2b^ | Pb | ATGTAAAT | 107 | TATTAA | 97 | 40 | 92 | 206 | — | — | — | |
| V2c^ | Ph | ATGTAAAT | 107 | TATTAA | 97 | 40 | 92 | 323 | CACAGTG | 12 | ACAAAAACC | |
| V2d^ | F | ATGCAAAT | 101 | TATTAA | 97 | 40 | 92 | 302 | CACAGTG | 12 | ACAAAAACC | |
| V2e^ | Pk | ATGTAAAT | 107 | TATTAA | 97 | 40 | 92 | 326 | CACAGTG | 12 | ACAAAAACC | |
| V2f^ | Pa | ATGTAAAT | — | — | — | — | — | 329 | CACAGTG | 12 | ACAAAAACC | |
| V2g^ | F | TTGAAAAT | 88 | TATTAA | 97 | 40 | 92 | 326 | CACAGTG | 12 | ACAAAAACC | |
| V2h^ | Pb | ATGTAAAT | 107 | TATTAA | 97 | 40 | 92 | 215 | — | — | — | |
| V2i^ | F | ATGCAAAT | 101 | TATTAA | 97 | 40 | 92 | 329 | CACAGTG | 12 | ACAAAAACC | |
| V2j^ | Pd | ATGTAAAT | 107 | TATTAA | 97 | 40 | 195 | 225 | CACAGTG | 12 | ACAAAAACC | |
| V2k^ | F | ATGTAAAT | 107 | TATTAA | 97 | 40 | 217 | 182 | CACAGTG | 12 | ACAAAAACC | |
| V2l | Pb | ATGTAAAT | 107 | TATTAA | 97 | 40 | 92 | 230 | — | — | — | |
| V2m | Pc | ATGTAAAT | 107 | TATTAA | 97 | 40 | 92 | 329 | CACAGTG | 12 | ACAAAAACC | |
| V2n | Pe | ATGTAAAT | 107 | TATTAA | 97 | 40 | 92 | 326 | CACAGTG | 12 | — | |
| V2o | Pg | — | — | TTAAAT | 97 | 40 | 92 | 326 | CACAGTG | 12 | ACAAAAACC | |
| V2p | Pi | — | — | — | — | 40 | 92 | 326 | CACAGTG | 12 | ACAAAAACC | |
| V2q | Pb | ATGTAAAT | 107 | TATTAA | 97 | 40 | 92 | 302 | — | — | — | |
| V2r | Pj | — | — | TATTAA | 97 | 40 | 92 | 329 | CACAGTG | 12 | ACAAAAACC | |
| V2s | Pa | — | — | — | — | — | 92 | 326 | CACAGTG | 12 | ACAAAACCT | |
| V2t | F | ATGTAAAT | 107 | TATTAA | 97 | 40 | 89 | 323 | CACAGTG | 12 | ACAAAAACC | |
| V2u | Pa | — | — | — | — | — | — | 329 | CACAGTG | 12 | ACAAAAACC | |
| V2v | F | — | — | — | — | 40 | 92 | 329 | CACAGTG | 12 | ACAAAAACC | |
| IGLV3 | V3a^ | F | ATTTCCAT | 38 | TTTATA | 65 | 52 | 84 | 303 | CACAGTG | 12 | ACAAACCCT |
| V3b^ | F | ATTTCCAT | 38 | TTTATA | 65 | 52 | 85 | 314 | CACAGTG | 12 | ACAAAAACT | |
| V3c^ | F | ATTTCCAT | 38 | TTTATA | 65 | 52 | 85 | 317 | CACAGTG | 12 | ACAAAAACC | |
| V3d^ | F | ATTTCCAT | 38 | TTTATA | 65 | 52 | 84 | 315 | CACAGTG | 12 | ACAAAAACC | |
| V3e | Pa | — | — | — | — | — | — | 306 | CACAGTG | 12 | ACAAAAACC | |
| V3f | Pa | — | — | — | — | — | — | 315 | CACAGTG | 12 | ACAAAAACC | |
Fct functionality, F functional, P pseudogene, ORF open reading frame, R reverse strand,
^ VL gene segments depicted in schematic diagram of the genomic loci,
aL–PART1 is missing;
b3′ truncation;
c1 nt deletion and frameshift at position 659 R; 2 nt deletion and frameshift from 637 R;
d1 nt deletion and frameshift at position 5176 R; 2 nt deletion and frameshift from 5154 R; e1 nt deletion and frameshift at position 4359; 2 nt deletion and frameshift from 4381;
f1 nt deletion and frameshift at position 4666; 2 nt deletion and frameshift from 4678;
g1 nt deletion and frameshift at position 3685 R; 2 nt deletion and frameshift from 3673 R;
h1 nt insertion and frameshift at position 540; 1 nt deletion and frameshift at position 586; 1 nt deletion and frameshift at position 608;
i6 nt deletion and frameshift from 1896; 1 nt deletion and frameshift at position 1936; 2 nt deletion and frameshift from 1955;
j1 nt insertion and frameshift at position 439 R; 4 nt deletion and frameshift from 456 R;
k2 nt deletions in CDR1-IMGT and CDR2-IMGT regions and frameshift mutations at 1418 and 1487; 4 nt deletion and frameshift from 1429; 1 nt deletion and frameshift at position 1462;
l1st-CYS replaced by Ala.
Table 2. Torafugu JL nucleotide and AA sequences with associated RSS.
| JL gene | Fct | J-Nonamer | Spacer | J-Heptamer | J region nt and AA sequences |
|---|---|---|---|---|---|
| J1a | F | GGTTTTTGT | ACGACCACTTGATGAGTTTGTAT | CACTGTG |  |
| J1b | F | GGTTTTTGT | ACGACCACTTGATGAGTTTGTAT | CACTGTG | 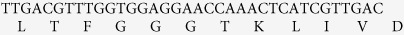 |
| J1c | F | GGTTTTTGT | ACGACCACTTGATGAGTTTGTAT | CACTGTG | 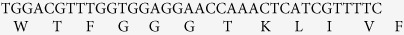 |
| J2a | F | GGTTTTTGT | ACAGCTGTGTGTACAAACTGAAT | CACTGTG |  |
| J2b | F | GGTTTTTGT | ACAGCTGTGTGTACAAACTGAAT | CACTGTG |  |
| J2c | F | GGTTTTTGT | ACAGCTGTGTGTACAAACTGAAT | CACTGTG | 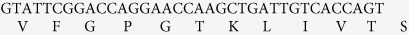 |
| J2d | F | GGTTTTTGT | ACAGCTGTGTGTACAATCTGAAT | CACTGTG |  |
| J2e | F | — | — | CACTGTG |  |
| J2f | F | GGTTTTTGT | ACAGCTGTGTGTACAAACTGAAT | CACTGTG |  |
| J2g | F | GGTTTTTGT | ACAGCTGTGTGTACAAACTGAAT | CACTGTG |  |
| J2h | F | GGTTTTTGT | ACAGCTGTGTGTACAAACTGAAT | CACTGTG | 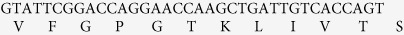 |
| J3a | F | GGTTTTTGT | ACGACCACTTGATGAGTTTGTAT | CACTGTG | 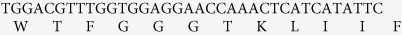 |
| J3b | F | GGTTTTTGT | ACGACCACTTGATGAGTTTGTAT | CACTGTG | 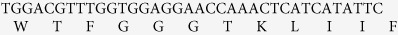 |
Fct functionality, F functional.
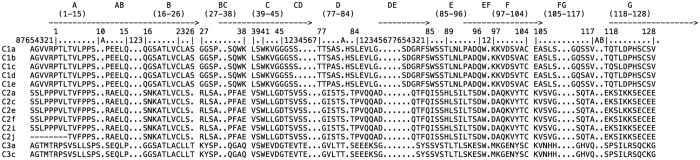
Figure 1. IMGT protein display of in-frame torafugu CL representative amino acid sequences.
The protein display is shown using IMGT header (IMGT Repertoire, http://www.imgt.org).
Identification of a third teleost IGL isotype in torafugu
Homology in the C domain is the most reliable criterion for classifying a teleost IGL isotype18. As mentioned, two IGL isotypes have been reported in torafugu: L1 and L2. Here, we used the published IGL sequences from various teleosts to search the torafugu database (http://www.fugu-sg.org/). As a result, three scaffolds (scaffold 2422, 2488, and 3698) were found to carry CL sequences that had homology (47–53% amino acid identities) with the L3 C domains of zebrafish, carp (Cyprinus carpio), and channel catfish (Ictalurus punctatus). This degree of homology in the C domain exceeds the limit used to distinguish mammalian κ and λ C domains (35–37%), thus further strengthens the identification of a torafugu L3. BLAST26 searches with the VL segments on the three scaffolds revealed similarities with L1/L3 V from other teleosts. After amino acid identity, RSS orientation is the second most common characteristic used for distinguishing IGL isotypes13. The torafugu L3 RSSs have the V12-23J motif, similar to that in mammalian κ27,28.
Type 3 IGL organization
Of the three scaffolds (2422, 2488, and 3698) that carry one L3 C sequence each, scaffold 2422 contains one each of a functional L1 V (V1c), L1 V without leader sequence (V1d), and JL (J3a); scaffold 3698 contains one JL (J3b); and scaffold 2488 contains three VL sequences that belong to L1 V (V1e) and L3 V (V3b and V3c) within the same cluster (Fig. 2). This heterogeneity suggests an organization of multiple clusters. If a region harboring one CL is considered as one cluster, at least three clusters should exist at the L3 loci. The L3 C sequences share 48–75% identity with each other at the amino acid level, which suggests their divergence from each other, while they are nonetheless distinguishable from the L1/L2 C sequences (10–31% identity in all inter-type pair-wise comparisons). The functional VL segments fall into two groups and correspond to L1 V (V1c, V1d, and V1e) and L3 V (V3b and V3c), respectively. Within a group, they are 88–92% identical at amino acid level over the VL coding sequences; between the two groups, they share 34–42% identities. All five VL segments are arranged in the opposite transcriptional orientation to their CL and JL on each individual scaffold, similar to that described for other teleost L3 genes10.
Figure 2. Overall organization of representative type 3 IGL genes.
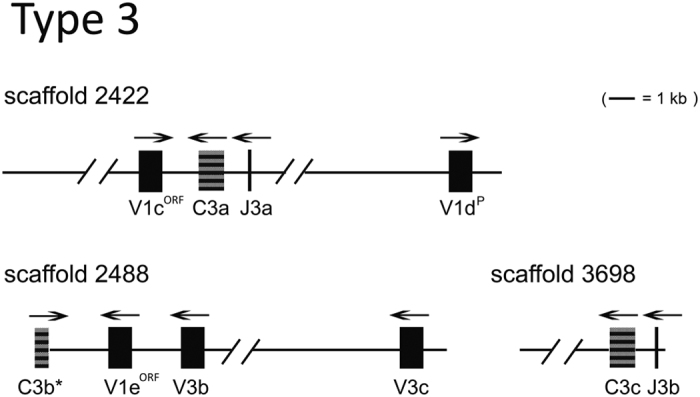
Scaffold 2422 of 14,667 bp, 2488 of 13,611 bp, and 3698 of 3784 bp, are shown to scale, with exon size exaggerated. The transcriptional polarity is indicated by overhead arrow. Each gene is labeled, and an asterisk denotes incomplete coding sequences. VP/ORF denotes pseudogene (P) or ORF sequence.
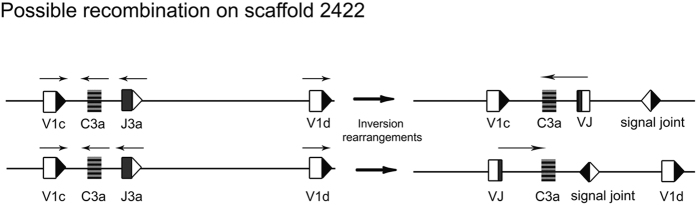
The V1d sequence was defined as a pseudogene due to the absence of a leader sequence in the current assembly. However, it may rearrange functionally to JL with its identifiable VL exon and the downstream RSS sequence. Therefore, the VL on both sides of the JL/CL will likely undergo rearrangement with C3a and J3a through inversion as in other teleosts. For example, V1d will possibly invert to join J3a, while V1c will recombine through inversion of J3a and C3a (Fig. 3).
Figure 3. Inversion rearrangements on scaffold 2422.
The transcription polarity of the rearranged VJ, at the right, is indicated by arrowheads on the top of VJ-C. The JL-RSS is indicated as a white triangle, the VL-RSS is indicated as a black triangle.
Type 2 IGL organization
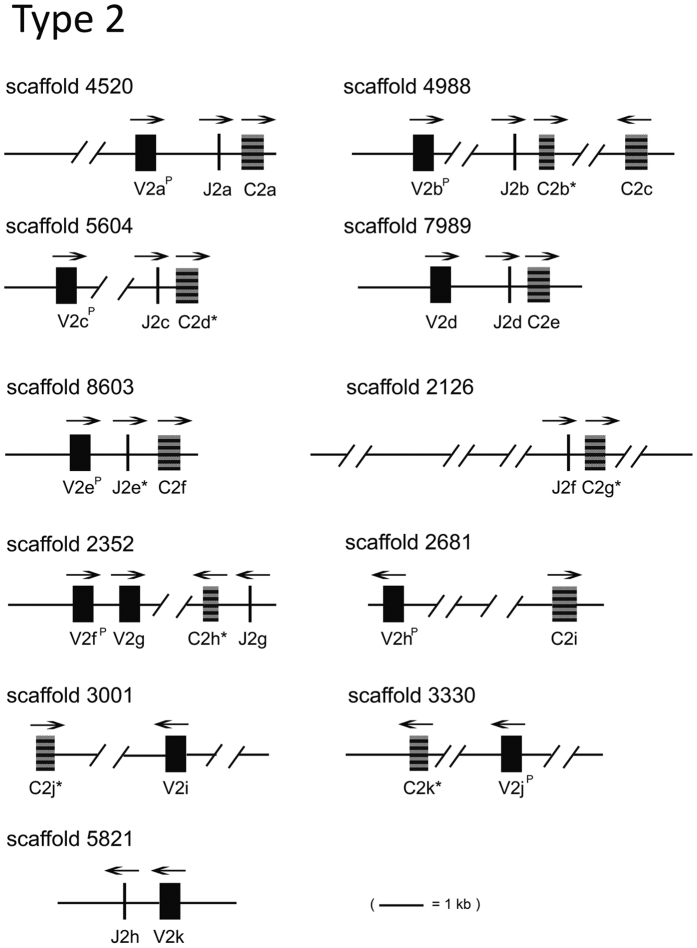
A search with L2 C sequences from various teleosts showed good matches with 10 scaffolds (scaffold 4520, 4988, 5604, 7989, 8603, 2126, 2352, 2681, 3001, and 3330) in the v4 assembly. Other scaffolds were found to contain either L2 V or J sequences (Fig. 4). The torafugu L2 loci contain 22 VL, 8 JL, and 11 CL gene segments. All 22 V-matching sequences (some were found only as fragments owing to gaps in the sequences) were summarized in Table 1. The genomic organization of L2 genes was depicted in Fig. 4. C2a, C2c, and C2i are identical with the published L2 torafugu C sequence18. Other L2 C sequences (those with complete coding sequences) are 92–99% identical with C2a in the derived amino acid sequences and only share 15–35% identity with L1/L3 C sequences, suggesting that they duplicated among themselves and diverged long ago from other types. The L2 V gene segments are either in the same or in the opposite transcriptional orientation as their corresponding JL and CL, which is topologically similar to the three-spined stickleback L2 genes on chromosome 1115. It is worthy to note that although all the scaffolds carrying VL in the opposite orientation as CL and JL are missing sequence information between VL and JL-CL (e.g., sequences in scaffold 4988, 2352, 2681, and 3001). For example, the orientation of V2f and V2g on scaffold 2352 appears to be opposite to that of C2h and J2g. However, two possibilities should be considered: (1) the gaps between these gene segments may contain novel CL and JL segments with the same orientation as V2f and V2g and (2) scaffold joining might reveal additional VL segments that are downstream of and in the same orientation as C2h and J2g. The L2 locus is most likely occupied by eleven clusters, and on average one VL segment resides in each cluster. Conventional recombination at the L2 locus would occur. For example, rearrangement between V2d and J2d on scaffold 7989 will occur by deletion of the intervening DNA to form a VLJL.
Figure 4. Overall organization of representative type 2 IGL genes.
The transcriptional polarity is indicated by overhead arrow. An asterisk denotes an incomplete coding sequence. VP/ORF denotes a pseudogene or ORF sequence.
On scaffold 54 and 139, assigned respectively to chromosome 3 and 529, only one L2 V was detected and no corresponding CL or JL could be identified, based on both v4 and v5 assemblies. The other L2 sequences identified on v4 scaffolds could not be assigned to v5 chromosome (s) due to the presence of gaps.
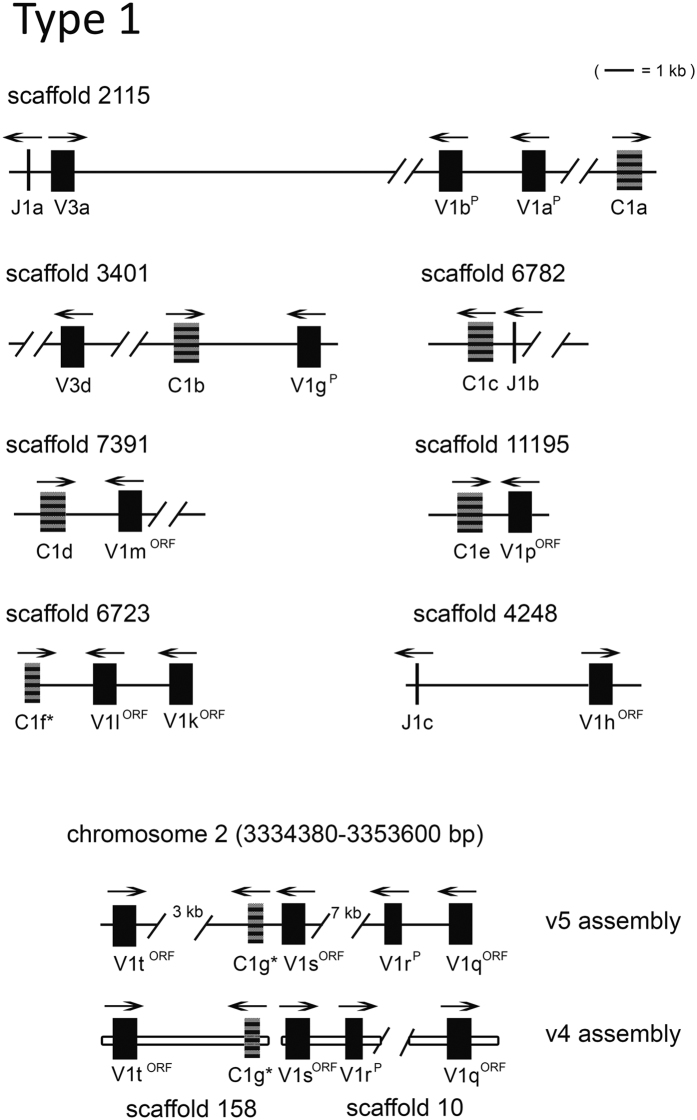
Type 1 IGL organization
L1 and L3 V sequences appear to be intermixed (discussed below). We described L1 IGL genes on at least seven genomic scaffolds (scaffolds with L1 C), thus they might operate as seven loci. As expected, L1 C sequences possess high amino acid identity (≥96%) with each other and the divergence from other types was evident (15–35% identity compared to L2/L3 C). As depicted in Fig. 5, the transcriptional polarity pattern in the L1 loci presents as VL in both orientations to JL and CL. In fact, in all but one instance (chromosome 2), the overall impression is that the L1 locus is organized as VL opposite to nearby JL and CL. On chromosome 2, four VL segments were identified, with three placed in the same transcriptional orientation to the CL (C1g) and another one in the opposite direction. On the other hand, sequences on scaffold 158 were perfectly assigned to chromosome 2, including V1t and C1g, while scaffold 10 was anchored to chromosome 2 in reverse, that is, it has the same VL segments (V1q, V1r, and V1s) in opposite directions (Fig. 5).
Figure 5. Overall organization of representative type 1 IGL genes.
The transcriptional polarity is indicated by overhead arrow. An asterisk denotes an incomplete coding sequence. VP/ORF denotes a pseudogene or ORF sequence. Scaffold 158 and 10 were assigned to chromosome 2.
IGL cluster estimation
Southern blots of torafugu genomic DNA from sperm probed with different types of CL reveal that the IGL genomic organization in this species is of the cluster type (Fig. 6). More than two bands in most digests suggest multiple IGL loci. Judging by the number of hybridizing bands, seven and three IGL loci are common in L1 and L3. For the L2 isotype, the number of clusters is lower than predicted. It is noticeable that the two bands digested by PstI are much stronger than other bands in L2 blots, which is attributable, at least in part, to the fact that there is no or limited polymorphism with PstI and many bands are hybridized at the same spot.
Figure 6. Southern blot of genomic DNA from torafugu sperm probed with torafugu IGLC.
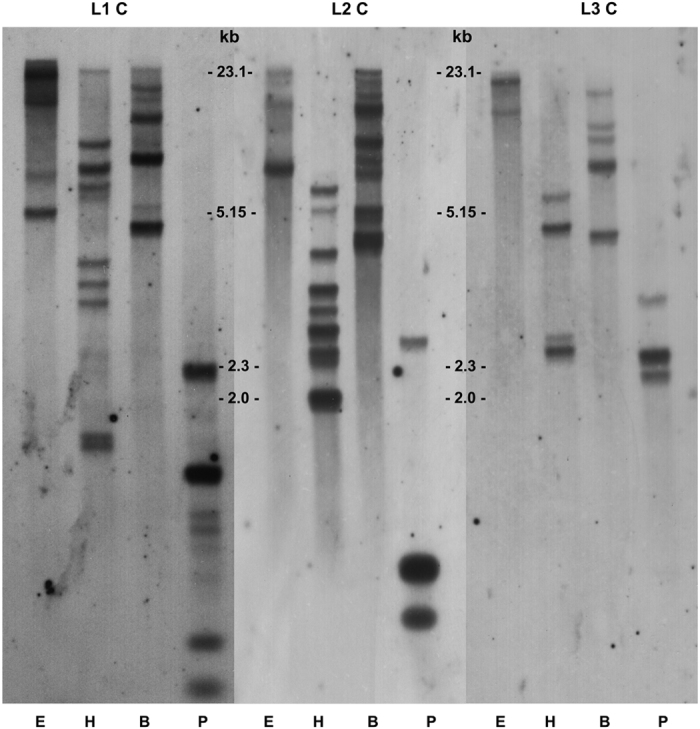
Restriction endonucleases are indicated at the bottom: EcoRI (E), HindIII (H), BamHI (B), and PstI (P). Figures are cropped and the original blots images are available in Additional File.
Phylogenetic analyses
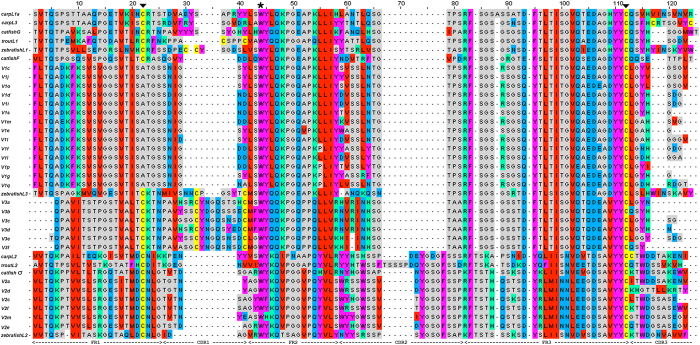
The VL domains of different teleost species and IGL isotypes were aligned (Fig. 7). Similar to the report by Criscitiello and Flajnik13, the comparison analysis revealed the conservation of a long CDR2 in L2 V (relative to other isotypes) and a long CDR1 in L3 V. The torafugu L1 V sequences were found to possess both short CDR1 and short CDR2, and were missing the key amino acid 1st-CYS in the FR1 region; this may be a torafugu-specific finding.
Figure 7. Overview window from Jalview of alignment of teleosts VL representative amino acid sequences as determined by MAFFT.
Hyphens denote gaps. FR and CDR regions are labeled according to Kabat delineation42. The conserved Tryptophan (Trp, W) in FR2 region is indicated by an asterisk. Cysteines (Cys, C) that are expected to form intra-chain disulfide bridges are indicated by solid black triangles, with the exception of torafugu IGLV1 group sequences (wherein Cys is replaced by Ala).
A phylogenetic tree was constructed based on the alignment of VL amino acid sequences from various vertebrates (Fig. 8). The torafugu L2/σ V sequences (V2a, V2c, V2d, V2e, and V2f) clustered strongly together and were distinct from the κ group (including teleost L1 and L3), which seemed to be mingled (VL sequences from the same scaffold are not necessarily in one group). Interestingly, although all the torafugu IGLV1 and IGLV3 sequences belong to the mammalian κ isotype, they clustered to separate groups. This suggests that they are probably associated with different sub-isotypes or a teleost-specific IGL isotype, as is the case in stickleback15.
Figure 8. Phylogenetic analysis of representative VL from various vertebrates.
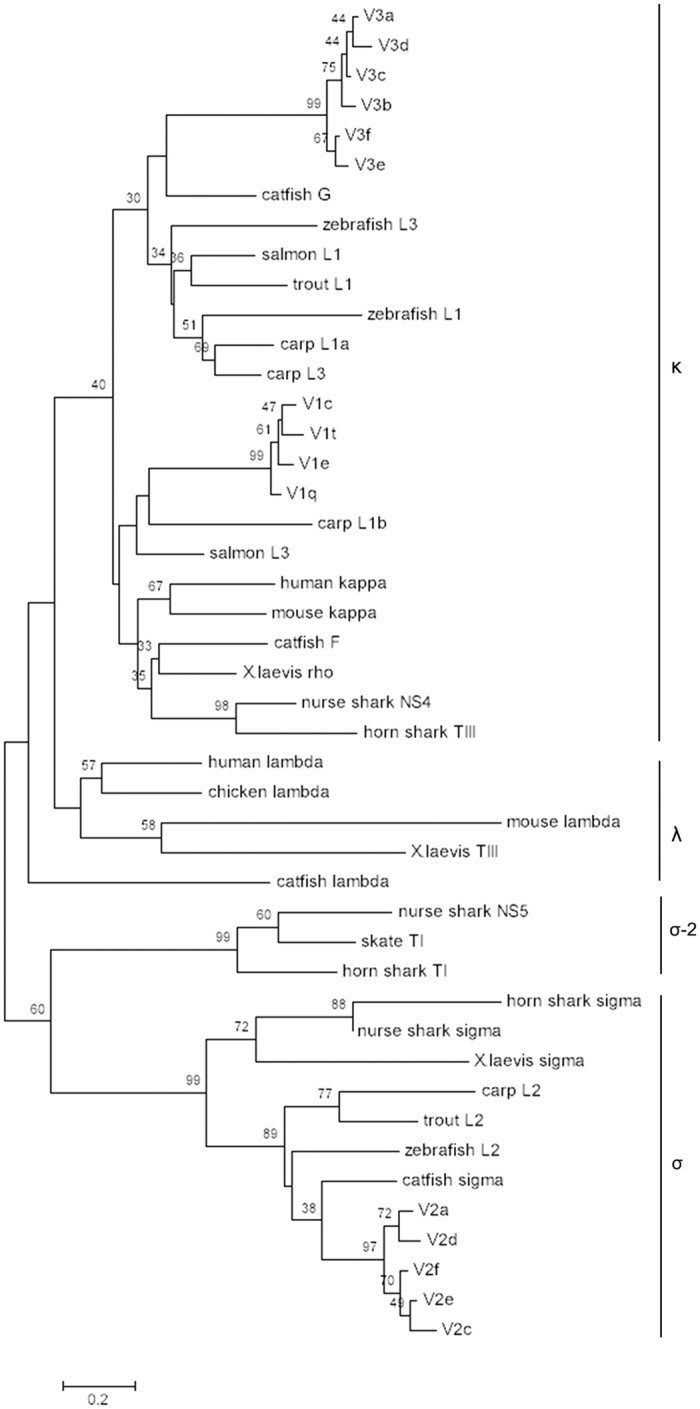
The NJ tree was constructed using MEGA 7 with 1000 bootstrap replications. GenBank accession numbers are: zebrafish L1 (AF246185); carp L1a (AB073328); carp L3 (AB073335); zebrafish L3 (AF246193); salmon (Salmo salar) L1 (AF273012); trout (Oncorhynchus mykiss) L1 (X65260); catfish G (L25533); carp L1b (AB073332); human kappa (S46371); mouse kappa (MUSIGKACN); salmon L3 (AF406956); catfish F (U25705); X. laevis (Xenopus laevis) rho (XELIGLVAA); horn shark (Heterodontus francisci) TIII (L25561); nurse shark (Ginglymostoma cirratum) NS4 (A49633); X. laevis TIII (L76575); mouse lambda (AY648665); chicken lambda (M24403); human lambda (AAA59013); catfish lambda (EU925383); nurse shark NS5 (AAV34678); skate (Leucoraja erinacea) TI (L25568); horn shark TI (X15315); horn shark sigma (EF114760); nurse shark sigma (EF114765); X. laevis sigma (S78544); carp L2 (AB091113); trout L2 (AAB41310); zebrafish L2 (AF246162); catfish sigma (EU872021).
Torafugu CL segments were compared using phylogenetic trees to evaluate the CL relationships among vertebrates (Fig. 9). None of the torafugu CL segments cluster with mammalian κ or λ IGL sequences. However, torafugu CL segments group strongly in branches with sequences belonging to the same teleost isotype (L1, L2, and L3), suggesting that teleosts share a common derivation and that three or more IGL isotypes may have been present in a teleost ancestor. A close relationship between torafugu (belonging to the Tetradontiformes order, Acanthopterygii superorder), and other species from the Perciformes order (Acanthopterygii), such as seabass (Dicentrarchus labrax), rockcod (Trematomus bernacchii), and wolffish (Anarhichas minor), is also evident from the tree. In addition, phylogenetic analysis consistently revealed the tendency of CL clustering according to taxonomic group rather than the isotype13,30. Taken together, the results of the phylogenetic analysis of the torafugu VL and CL sequences revealed different selective pressures on the two domains, wherein CL tends to cluster according to taxonomic group, while VL tends to group by isotype.
Figure 9. Phylogenetic analysis of CL from various vertebrates.
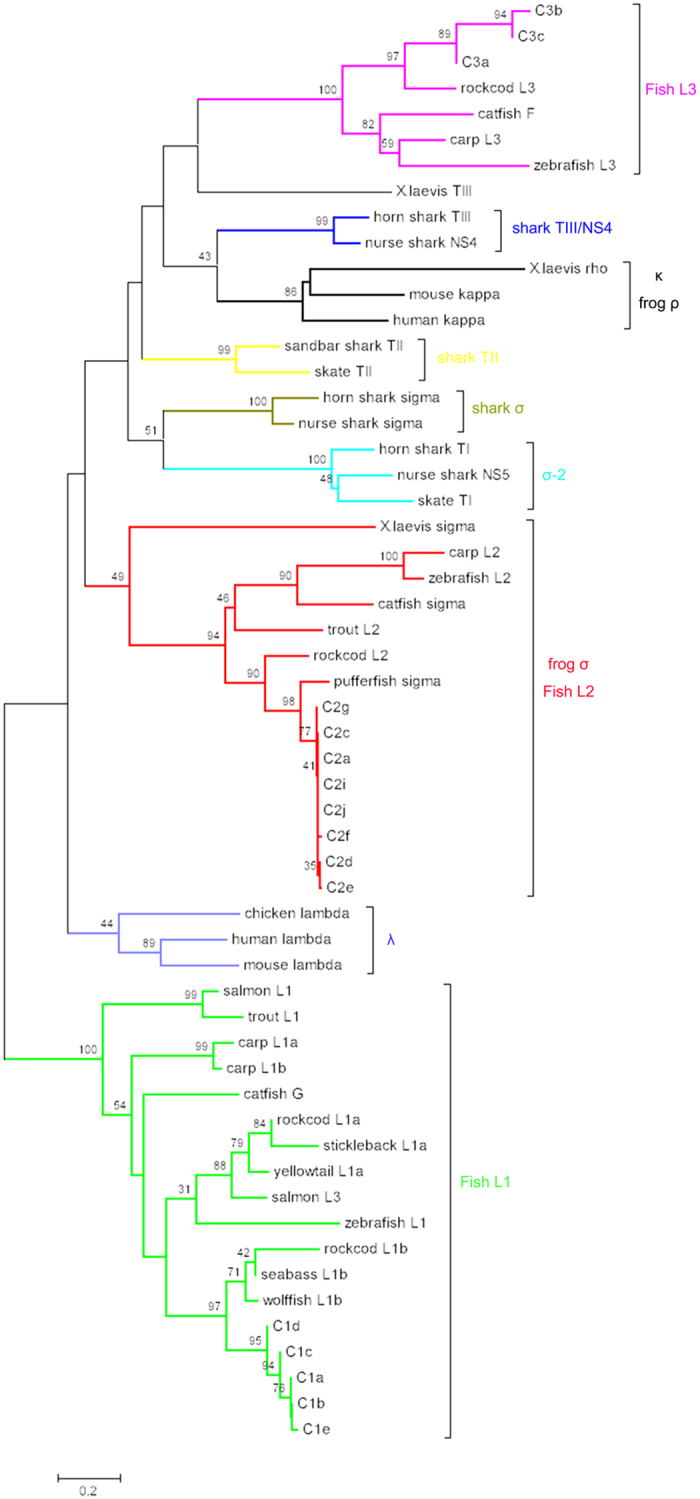
GenBank accession numbers are as follows: wolffish L1b (AF137398); rockcod L1b (DQ842622); seabass L1b (AJ400216); zebrafish L1 (AF246185); salmon L3 (AF406956); yellowtail L1a (AB062619); rockcod L1a (EF114784); stickleback L1a (AY278356); catfish G (L25533); carp L1a (AB035728); carp L1b (AB035729); salmon L1 (AF273012); trout L1 (X65260); chicken lambda (M24403); human lambda (AAH07782); mouse lambda (J00592); X. laevis sigma (S78544); carp L2 (AB103558); zebrafish L2 (AF246162); catfish sigma (EU872021); trout L2 (AAB41310); rockcod L2 (EF114785); pufferfish (Tetraodon nigroviridis) sigma (AJ575637); horn shark TI (X15315); nurse shark NS5 (AAV34681); skate TI (L25568); horn shark sigma (EF114760); nurse shark sigma (EF114765); sandbar shark (Carcharhinus plumbeus) TII (M81314); skate TII (L25566); mouse kappa (AB048524); X. laevis rho (XELIGLVAA); human kappa (M11937); carp L3 (AB035730); zebrafish L3 (AF246193); catfish F (U25705); rockcod L3 (DQ842626).
Isotype distribution was assessed for the JL segments and JL1, JL2, and JL3 sequences were distinguished (Supplementary Fig. S1). Of all JL segments identified, those belonging to L1 and L3 were most similar to each other.
Analysis of VL gene 5′ flanking regulatory sequences
We examined 5′ flanking sequences for identified VL segments to reveal possible regulatory features. The 5′ flanking region contains two conserved motifs, namely the octamer motif, which is critical to correct transcription of IGL genes, and the TATA box for the general transcription process31. As summarized in Table 1, all 5′ flanking sequences of functional VL segments exhibit considerable family-specific conservation i.e., (1) all the functional or open reading frame (ORF) segments of the IGLV1 family contain sequences completely identical to the octamer consensus (ATTTGCAT) and the TATA consensus (TTTAAA); (2) IGLV2 sequences show slightly less conserved octamer sequences and most functional members have single point variation (ATG-T/C-AAAT) in the octamer sequence; the TATA consensus (TATTAA) is well conserved across functional IGLV2 genes; (3) members of the IGLV3 family have consensus octamer (ATTTCCAT) and TATA (TTTATA) sequences.
Functionality of torafugu IGL loci
A total of fifteen torafugu EST sequences associated with IGL expression were identified from the NCBI EST database. Alignment of torafugu ESTs to concordant genomic VL segments revealed that all functional IGLV3 genes were expressed, while only one IGLV2 sequence (V2k) was expressed. Additionally, expression of all the IGLV1 sequences was observed despite the fact that they were missing the 1st-CYS in the FR1 region. Expression of all the complete CL segments was also observed with one exception: the C1d on scaffold 7391. Upon detailed examination, 9 ESTs and 6 ESTs were found to be concordant with the L2 locus and L1/L3 loci, respectively. Interestingly, ESTs associated with L2 and L3 C sequences were found to lack a VL segment, except for EST AL835785, which carried a complete VLJL-CL (L2 C). In comparison, expression of L1 C sequences was often found to be with either IGLV1 or IGLV3 sequences (Supplementary Table S1). The identity of all the retrieved ESTs to genomic VL and CL segments is 95–100%, suggesting the feasibility of using this method to assign ESTs to concordant genomic sequences.
Discussion
In the present study, we have characterized the torafugu IGL genomic organization based on available genome data sets. It has been reported that torafugu has two IGL isotypes, L1 and L2. Here, a teleost L3 isotype was newly identified, demonstrating that torafugu possesses at least three IGL isotypes. All the IGL genes have been found to be partitioned over multiple scaffolds (v4 assembly). Currently, we can only speculate that torafugu IGL genes should be assigned to three different chromosomes due to incomplete sequence information from the v5 assembly. Our observations must be taken as a step forward in the elucidation of torafugu IGL genomic organization and future studies on more complete genome assembly may help to address the current issues with gaps and false assemblies in the whole genome sequence.
During vertebrate phylogeny, IGL genes have undergone major evolutionary transitions involving genomic arrangements. One extreme example is the presence of a single IGL isotype (λ) in bird species, such as chicken and zebra finch7,32. Unlike mammalian κ and λ loci, which are often arranged in a translocon fashion, teleost IGL genes are organized in distinct clusters of (VL-JL-CL)n. Herein, we show that torafugu IGL genes are arranged in a compact multi-cluster configuration, supported by both the genomic organization and the Southern blot result. This observation is similar to that found in other teleosts, suggesting a conservation of the cluster IGL organization among teleost species.
In regard to the comparative analysis of the sequences of torafugu CL with those of other vertebrates, the relative distances are in agreement with the phylogenetic relationships. The torafugu CL share the same cluster with teleost L1, L2, and L3, respectively. Moreover, a sister-group relationship (Fig. 9) in the superorder Acanthopterygii between torafugu L1 C sequences and those of the L1b subgroup (wolffish L1b, seabass L1b, and rockcod L1b) is supported by the observed high bootstraps values. At this time, we did not find an L1a C homolog in torafugu, but if such sequences are found in the future, this would further support the hypothesis that L1a and L1b subtypes exist in the Acanthopterygii L1 isotype33. In addition, the identification of an L3 in torafugu (Acanthopterygii), together with the presence of L3 in rockcod (Acanthopterygii) and Ostariophysi (catfish, zebrafish, and carp), suggests that the divergence between L1 and L3 took place at or before the emergence of Euteleosts18.
Finally, screening of the EST database indicates that the majority of IGLV1 and IGLV3 genes are expressed. However, most of the ESTs associated with the expression of L2 C do not have a corresponding VL segment. This phenomenon has been previously described in zebrafish34 and medaka IGκ19, and it may be related to the low efficiency in eliminating aberrant IGL transcripts35.
The observation that torafugu VL and CL from different isotypes (i.e., L1 and L3) may join together to achieve potential expression at the rearrangement level is somewhat reminiscent of the previous finding in zebrafish wherein inversional VJ-rearrangements leapfrog CL occur between clusters21.It is plausible that (1) torafugu L1 and L3 clusters are close to one another in the genome, which may allow recombination between different isotypes, (2) torafugu with multiple CL on a scaffold are poised to reconstruct the IGL locus by inversional rearrangement, which can bring VL from one cluster into another, similar to that of zebrafish21. With efforts to sequence additional genomes, it will be intriguing to investigate whether the inversional inter-cluster rearrangement is teleost-specific or a commonplace in other species.
Methods
Retrieval of IGL genes from the torafugu genome
Genome builds of torafugu (assembly v4, October 2004 and assembly v5, January 2010) available from the Fugu Genome Project29 (http://www.fugu-sg.org/) were searched to locate the IGL genes. Published IGL amino acid sequences from torafugu18,24 and other teleosts15,17,20 were used as queries in TBLASTN alignments (cutoff E-value of 10−15) to retrieve relevant scaffolds and chromosomes. Genomic sequences that contain matches for both VL and CL were downloaded for further analysis. The identified genomic sequences were subsequently used as queries in BLASTN searches against the EST database at NCBI to retrieve expression data. Expression of VL genes was determined by BLAST hits using a 95% threshold identity and a 10−15 E-value threshold, while ESTs were assigned to concordant CL when a ≥99% identity was met.
Annotation of torafugu IGL
Artemis36 was used to annotate the IGL loci, including the transcriptional polarity and relative positions of VL and CL in the genomic sequences. C exons were discerned by comparing resultant genomic sequences with published IGL mRNAs. VL genes were determined based on the presence of canonical RSS (allowing 2 nucleotide mismatches), with ORFs that match for IG signature sequences using IgBLAST (www.ncbi.nlm.nih.gov/projects/igblast) and IMGT/V-QUEST37 (the Teleostei unit), and finally by pattern searches for 23RSS or 12RSS flanking ends of gene segments. To identify the JL genes, which are too short to be detected by BLAST searches, we performed pattern searches to find JL-specific RSSs among the initial genomic sequences that contain VL and CL. The pattern is a consensus RSS heptamer and a nonamer with a 22-24 bp spacer (CACAGTG-N22-24-ACAAAAACC) region. Splice sites between leader and V exons were discerned by FSPLICE (http://linux1.softberry.com/berry.phtml). Exon boundaries of VL, JL, and CL were refined by alignment with known VJ-C cDNA sequences and torafugu EST sequences (from Fugu Genome Project)38.
Nomenclature
Identified IGL genes were annotated according to the IMGT® nomenclature39. For the VL genes, all retrieved sequences without a truncation, frameshift mutation, or premature stop codon in the leader exon and the V exon, which had conserved residues (1st-CYS, conserved-TRP, and 2nd-CYS) in FR1, FR2, and FR3 regions, respectively, and possessed a proper RSS, were deemed as functional genes. For the CL and JL gene segments, retrieved sequences without frameshift mutations and internal stop codons were regarded as potentially functional genes. In addition, examination of RSS was implemented to determine putative functionality of JL.
Comparative phylogenetic studies
Phylogenetic studies were carried out using the MEGA7 program40. Multiple sequence alignments were performed using MAFFT41. The neighbor-joining (NJ) method was used to construct phylogenetic trees (pair-wise deletion, Jones-Taylor-Thornton matrix) and to enter range-activated sites by gamma parameter 2.5. Evaluation of the veracity of these trees was done by executing a bootstrap procedure of 1000 replicates.
Southern blotting
Genomic DNA from torafugu sperm (5 μg; extracted using DNeasy® Blood & Tissue Kit, Qiagen, Valencia, CA) was digested with EcoRI, HindIII, BamHI, and PstI. The digested DNA was electrophoresed on 0.8% agarose gels for 16 h and transferred onto Hybond-N+ membranes (GE Healthcare, Piscataway, NJ). Hybridizations and subsequent detection were performed according to the manufacturer’s instructions (Amersham AlkPhos Direct™, GE Healthcare). Torafugu C probes consisted of the entire CL domain of L1, L2, and L3. The probes were amplified using Platinum® Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA). The conditions for the thermal cycler were: 94 °C for 2 min, followed by 30 cycles of 94 °C for 30 s, 55 °C for 30 s, 68 °C for 1 min, and a final extension at 68 °C for 5 min (see primer details in Supplementary Table S2).
Additional Information
Accession codes: The annotation data (Supplementary Dataset File) of v4 scaffolds have been deposited in GenBank under accession numbers KU350660 to KU350678, KU359177 to KU359180, and KU365386 to KU365407.
How to cite this article: Fu, X. et al. Immunoglobulin light chain (IGL) genes in torafugu: Genomic organization and identification of a third teleost IGL isotype. Sci. Rep. 7, 40416; doi: 10.1038/srep40416 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by JSPS KAKENHI, Grant Numbers 24248034 and 25640099. The annotation data is available in NCBI under accession numbers KU350660-KU350678, KU359177-359180, and KU365386-KU365407.
Footnotes
Author Contributions S.A. conceived of and planned the study; X.F. analyzed data and wrote the manuscript; F.J.Z. performed the Southern blot experiment; S.W. supervised the study.
References
- Flajnik M. F. & Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat. Rev. Genet. 11, 1–14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc M. P. & Lefranc G. The immunoglobulin factsbook. Academic Press (2001). [Google Scholar]
- Teng G. & Papavasiliou F. N. Immunoglobulin somatic hypermutation. Annu. Rev. Genet. 41, 107–120 (2007). [DOI] [PubMed] [Google Scholar]
- Schatz D. G. & Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 11, 251–63 (2011). [DOI] [PubMed] [Google Scholar]
- Swanson P. C. The bounty of RAGs: Recombination signal complexes and reaction outcomes. Immunol. Rev. 200, 90–114 (2004). [DOI] [PubMed] [Google Scholar]
- Akira S., Okazaki K. & Sakano H. Two pairs of recombination signals are sufficient to cause immunoglobulin V-(D)-J joining. Science. 238, 1134–1138 (1987). [DOI] [PubMed] [Google Scholar]
- Das S., Mohamedy U., Hirano M., Nei M. & Nikolaidis N. Analysis of the immunoglobulin light chain genes in zebra finch: evolutionary implications. Mol. Biol. Evol. 27, 113–120 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambón-Deza F., Sánchez-Espinel C., Mirete-Bachiller S. & Magadán-Mompó S. Snakes antibodies. Dev. Comp. Immunol. 38, 1–9 (2012). [DOI] [PubMed] [Google Scholar]
- Lundqvist M. L., Middleton D. L., Radford C., Warr G. W. & Magor K. E. Immunoglobulins of the non-galliform birds: antibody expression and repertoire in the duck. Dev. Comp. Immunol. 30, 93–100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edholm E. S., Wilson M. & Bengten E. Immunoglobulin light (IgL) chains in ectothermic vertebrates. Dev. Comp. Immunol. 35, 906–915 (2011). [DOI] [PubMed] [Google Scholar]
- Saha N. R. et al. Genome complexity in the coelacanth is reflected in its adaptive immune system. J. Exp. Zool. Part B Mol. Dev. Evol. 322, 438–463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilström L. The mysterious immunoglobulin light chain. Dev. Comp. Immunol. 26, 207–215 (2002). [DOI] [PubMed] [Google Scholar]
- Criscitiello M. F. & Flajnik M. F. Four primordial immunoglobulin light chain isotypes, including λ and κ, identified in the most primitive living jawed vertebrates. Eur. J. Immunol. 37, 2683–2694 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Nikolaidis N., Klein J. & Nei M. Evolutionary redefinition of immunoglobulin light chain isotypes in tetrapods using molecular markers. Proc Natl Acad Sci USA 105, 16647–16652 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y. et al. The immunoglobulin gene loci in the teleost Gasterosteus aculeatus. Fish Shellfish Immunol. 28, 40–48 (2010). [DOI] [PubMed] [Google Scholar]
- Daggfeldt A., Bengten E. & Pilstrom L. A cluster type organization of the loci of the immunoglobulin light-chain in Atlantic cod (Gadus morhua L.) and rainbow trout (Oncorhynchus mykiss Walbaum) indicated by nucleotide sequences of cDNAs and hybridization analysis. Immunogenetics 38, 199–209 (1993). [DOI] [PubMed] [Google Scholar]
- Edholm E.-S. et al. Identification of Igσ and Igλ in channel catfish, Ictalurus punctatus, and Igλ in Atlantic cod, Gadus morhua. Immunogenetics 61, 353–370 (2009). [DOI] [PubMed] [Google Scholar]
- Hsu E. & Criscitiello M. F. Diverse immunoglobulin light chain organizations in fish retain potential to revise B cell receptor specificities. J. Immunol. 177, 2452–62 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magadán-Mompó S., Zimmerman A. M., Sánchez-Espinel C. & Gambón-Deza F. Immunoglobulin light chains in medaka (Oryzias latipes). Immunogenetics 65, 387–396 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman A. M., Romanowski K. E. & Maddox B. J. Targeted annotation of immunoglobulin light chain (IgL) genes in zebrafish from BAC clones reveals kappa-like recombining/deleting elements within IgL constant regions. Fish Shellfish Immunol. 31, 697–703 (2011). [DOI] [PubMed] [Google Scholar]
- Zimmerman A. M., Yeo G., Howe K., Maddox B. J. & Steiner L. A. Immunoglobulin light chain (IgL) genes in zebrafish: Genomic configurations and inversional rearrangements between (VL-JL-CL) gene clusters. Dev. Comp. Immunol. 32, 421–434 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman G. W., Anderson M. K. & Rast J. P. Evolution of antigen binding receptors. Annu. Rev. Immunol. 17, 109–147 (1999). [DOI] [PubMed] [Google Scholar]
- Aparicio S. et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 297, 1301–1310 (2002). [DOI] [PubMed] [Google Scholar]
- Saha N. R., Suetake H. & Suzuki Y. Characterization and expression of the immunoglobulin light chain in the fugu: Evidence of a solitaire type. Immunogenetics 56, 47–55 (2004). [DOI] [PubMed] [Google Scholar]
- Lefranc M. P. et al. IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Dev. Comp. Immunol. 29, 185–203 (2005). [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–10 (1990). [DOI] [PubMed] [Google Scholar]
- Sakano H., Hüppi K., Heinrich G. & Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature 280, 288–294 (1979). [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature 302, 575–581 (1983). [DOI] [PubMed] [Google Scholar]
- Kai W. et al. Integration of the genetic map and genome assembly of fugu facilitates insights into distinct features of genome evolution in teleosts and mammals. Genome Biol. Evol. 3, 424–442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast J. P. et al. Immunoglobulin light chain class multiplicity and alternative organizational forms in early vertebrate phylogeny. Immunogenetics 40, 83–99 (1994). [DOI] [PubMed] [Google Scholar]
- Falkner F. G. & Zachau H. G. Correct transcription of an immunoglobulin kappa gene requires an upstream fragment containing conserved sequence elements. Nature 310, 71–74 (1984). [DOI] [PubMed] [Google Scholar]
- Parvari R. et al. Analyses of chicken immunoglobulin light chain cDNA clones indicate a few germline V lambda genes and allotypes of the C lambda locus. EMBO J. 6, 97–102 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscia M. R., Giacomelli S., De Santi C., Varriale S. & Oreste U. Immunoglobulin light chain isotypes in the teleost Trematomus bernacchii. Mol. Immunol. 45, 3096–3106 (2008). [DOI] [PubMed] [Google Scholar]
- Haire R. N., Rast J. P., Litman R. T. & Litman G. W. Characterization of three isotypes of immunoglobulin light chains and T-cell antigen receptor α in zebrafish. Immunogenetics 51, 915–923 (2000). [DOI] [PubMed] [Google Scholar]
- Chemin G. et al. Multiple RNA surveillance mechanisms cooperate to reduce the amount of nonfunctional Ig κ transcripts. J. Immunol. 184, 5009–17 (2010). [DOI] [PubMed] [Google Scholar]
- Carver T. et al. Artemis and ACT: Viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24, 2672–2676 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet X., Lefranc M. P. & Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36, W503–W508 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. S. et al. Fugu ESTs: New resources for transcription analysis and genome annotation. Genome Res. 13, 2747–2753 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc M. P. WHO-IUIS Nomenclature Subcommittee for immunoglobulins and T cell receptors report. Dev. Comp. Immunol. 32, 461–463 (2008). [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G. & Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K. & Standley D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat E. A., Wu T. T., Perry H. M., Gottesman K. S. & Koeler C. Sequences of proteins of immunological interest. Analytical Biochemistry 138, 265 (1984). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.