Abstract
Whole-genome analysis of cancer specimens is commonplace, and investigators frequently share or re-use specimens in later studies. Duplicate expression profiles in public databases will impact re-analysis if left undetected, a so-called “doppelgänger” effect. We propose a method that should be routine practice to accurately match duplicate cancer transcriptomes when nucleotide-level sequence data are unavailable, even for samples profiled by different microarray technologies or by both microarray and RNA sequencing. We demonstrate the effectiveness of the method in databases containing dozens of datasets and thousands of ovarian, breast, bladder, and colorectal cancer microarray profiles and of matching microarray and RNA sequencing expression profiles from The Cancer Genome Atlas (TCGA). We identified probable duplicates among more than 50% of studies, originating in different continents, using different technologies, published years apart, and even within the TCGA itself. Finally, we provide the doppelgangR Bioconductor package for screening transcriptome databases for duplicates. Given the potential for unrecognized duplication to falsely inflate prediction accuracy and confidence in differential expression, doppelgänger-checking should be a part of standard procedure for combining multiple genomic datasets.
Sufficient germ-line sequence markers provide a “fingerprint” that can be matched uniquely in a database of genotypes (1). Publicly available human genomic data is therefore normally summarized at a level that cannot be identified uniquely to protect patient privacy. Cancer transcriptomes undergo alterations that are highly distinctive but much more difficult to identify uniquely in summarized form. Re-use of tissue specimens is widespread in clinical genomic studies, creating a “doppelgänger effect” in publicly available datasets: hidden duplicates that, if left undetected, can inflate statistical significance or apparent accuracy of genomic models when combining data from different studies (Figure 1A). The proposed method relies on exhaustive comparisons of dataset pairs and sample pairs to empirically estimate the distribution of pairwise transcriptome correlations between biological replicates within a dataset or between two datasets where potentially different profiling technologies were used. The key aspects to identifying duplicates in a pair of datasets are 1) using transcript identifiers available in both datasets, 2) batch correction (2), 3) calculating Pearson’s Correlation Coefficient (PCC) between every sample in one dataset against every sample in the other dataset, and 4) duplicate-oriented outlier detection. The background distribution of pairwise PCC values varies depending on the tissue assayed and the technologies used, and must be estimated for every dataset pair. Doppelgängers can be identified as outliers at the high end of the distribution of batch-corrected correlations. The detailed methodology of package development and validation can be found in the Supplementary Material (available online).
Figure 1.
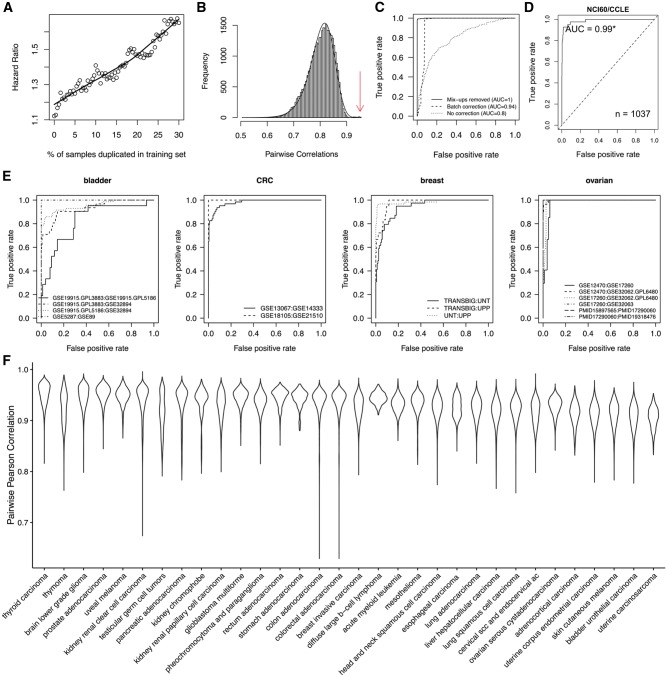
Demonstration and benchmarking of the doppelgangR method for identifying expression profiles of the same biological specimen. A) The “doppelgänger” effect: hidden duplicates can inflate the apparent accuracy of predictive and prognostic models. Models of overall survival for high-grade, serous ovarian cancer were trained and then validated in two studies containing duplicates identified by doppelgangR (see Supplementary Methods, available online). Validation set hazard ratio (HR) was calculated with duplicates incrementally removed so that between 0% and 30% cross-study duplication of samples remained. Thirty percent duplication inflates the apparent hazard ratio from 1.1 to 1.7. B) doppelgangR identifies duplicate expression profiles as outliers with unusually high pairwise correlation compared with other pairs of unrelated expression profiles. This histogram is the diagnostic plot produced by doppelgangR software, showing the best fit to the distribution of pairwise correlations, with vertical darklines showing outliers that are probable duplicates in the UNT (3) and Miller et al. breast cancer datasets (4). C) Batch correction allows RNA-seq profiles to be matched accurately to Affymetrix microarray profiles in the The Cancer Genome Atlas (TCGA) ovarian cancer dataset. True positives are tumors whose RNA-seq and microarray profiles are more highly correlated to each other than to any other profile. Batch correction increases area under the receiver operating characteristic plot (AUC) from 0.79 to AUC = 0.94, and removing 50 microarray profiles incorrectly labeled by TCGA further increases AUC to > 0.995. D and E) Benchmarking. We estimated the accuracy of the doppelgangR approach by applying it to pairs of datasets with confirmed duplicates (see Supplementary Methods, available online). D) Shows AUC for identifying the 43 cell lines present in two different panels (CCLE [n = 1037] and NCI60 [n = 59]). E) The performance on primary tumor data in four cancer types. The AUC averaged across the four cancer types is 0.97. F) Suitability to TCGA cancer types. The doppelgangR approach only works for cancer types in which expression profiles of individual tumors are sufficiently distinct. Violin plots depict distributions of Pearson Correlation Coefficients (PCCs) for all pairs of expression profiles within each TCGA dataset, in order (left to right) of increasing distinctiveness. In cancer types with high pairwise PCCs, such as thyroid carcinoma, patients have very similar expression profiles and are hard to distinguish based on expression data only. In contrast, in cancer types with low PCCs, such as bladder cancer, extensive genomic alterations generate unique expression fingerprints that make doppelgänger identification possible.
We studied databases of ovarian, breast, bladder, and colorectal cancers and of cell lines and assessed their accuracy against a “gold standard” of duplicated samples generated through further manual inspection of expression data, clinical annotations, and sample identifiers (Supplementary Table 1, available online). Confirmed doppelgängers were identified in more than half of all studies (Table 1). For example, among the 1467 breast cancer gene expression profiles, doppelgangR identifies 59 samples present in both the Sotiriou et al. (3) and Miller et al. (4) studies (Figure 1B; additional samples are duplicated by the TRANSBIG dataset, see Table 1). Although these studies were published by Belgian and Singaporean groups, respectively, careful reading of the papers reveals that their datasets shared a cohort of samples originating from Uppsala County, Sweden. Such international collaborations are beneficial to the cancer research community, but pose challenges to investigators developing independent validations and meta-analyses. In the ovarian cancer database, which we have inspected in great detail (5), we identified 17% of records as nonunique, including duplicates in different datasets originating from the same institution (6,7), between the TCGA dataset and datasets of institutions that contributed samples to the TCGA project (8,9) and within the TCGA dataset itself. In approximately 75% of duplicate pairs, samples matched by expression data had identical or compatible clinical and tumor data, but in the other 25% of cases the clinical data were discordant (10). Previous work on identifying duplicate microarray profiles has been limited to matching identical raw data files (11), and this would not identify any of these duplicates.
Table 1.
Overview of confirmed doppelgängers in all studies*
| Dataset identifier by type of cancer | Total No. samples | No. of doppelgängers | Institutional source of doppelgängers |
|---|---|---|---|
| Bladder | |||
| GSE1827, GSE13507, GSE31189, GSE31684, GSE37317, PMID: 17099711 | 570 | 0 | Various, no doppelgängers identified |
| GSE19915, GSE32894 | 490 | 84 | University Hospital of Lund, Sweden |
| GSE89, GSE5287 | 70 | 2 | Aarhus University Hospital, Denmark |
| Breast | |||
| MAINZ, NKI, VDX | 881 | 0 | Various, no doppelgängers identified |
| TRANSBIG, UNT, UPP | 586 | 78 | Uppsala County, Sweden |
| Colorectal | |||
| GSE2109, GSE3964, GSE4045, GSE11237, GSE12225, GSE12945, GSE13294, GSE26682, GSE27544, GSE28702, GSE45270, TCGA (READ) | 1275 | 0 | Various, no doppelgängers identified |
| GSE13067, GSE14333 | 364 | 41 | Royal Melbourne Hospital, Australia |
| GSE4526, GSE14095 | 225 | 37 | Teikyo University School of Medicine, Japan |
| GSE14333, GSE17538 | 754 | 569 | H. Lee Moffitt Cancer Center, USA |
| GSE18105, GSE21510, GSE21815 | 400 | 95 | Tokyo Medical and Dental University Hospital, Japan |
| GSE26906, GSE39582 | 656 | 90 | Various, France |
| GSE33113, TCGA (COAD) | 226 | 2 | Academic Medical Center, Netherlands |
| Ovarian | |||
| GSE14764, GSE19829, GSE26712, GSE30161, GSE44104, GSE49997, GSE6008, GSE6822, GSE8842, GSE9891, GSE12418, GSE13876 | 1415 | 0 | Various, no doppelgängers identified |
| E-MTAB-386, GSE18520 | 192 | 1 | Brigham and Women’s Hospital, USA |
| GSE12470, GSE17260, GSE32062, GSE32063 | 463 | 139 | Niigata University, Japan |
| GSE20565, GSE26193 | 247 | 93 | Resource Biological Center of the Institut Curie, France |
| TCGA, GSE2109, GSE51088, PMIDs: 15897565, 17290060, 19318476 | 1176 | 2 | International Genomics Consortium (IGC) |
| 10 | Cedars-Sinai Medical Center, USA | ||
| 88 | Duke University Medical Center, USA |
Gene expression data were obtained from several R/Bioconductor packages (Supplementary Methods, available online), and the listed ids are the study ids given in these packages.
In addition to identifying samples reprofiled by microarray, our approach accurately aligns microarray and log-transformed RNA-seq profiles for the same patients with area under the receiver operating characteristic curve (AUC) greater than 0.9 for 10 of 12 TCGA cancer types where both microarray and RNA-seq are available (Supplementary Table 2, available online). AUC is less than 0.9 for two cancer types among the 10 “quietest” genomes (Figure 1F), kidney renal clear cell and papillary cell carcinoma. Batch correction across datasets is critical: For example, in ovarian cancer, batch correction increases AUC from 0.80 to 0.94. Further inspection of the remaining errors reveals that almost all were because of an experimental RNA mix-up in the original TCGA Affymetrix microarray dataset, resulting in erroneously duplicated profiles attributed to different patients. Correction of the mixed-up samples increases AUC for matching RNA-seq to microarray profiles to greater than 0.995 (Figure 1C). We reported this sample mix-up to the TCGA Data Coordinating Center, which in turn removed these 50 profiles on August 25, 2015 (12).
Our approach of duplicate identification reliably works when individual tumors have distinctive expression profiles, as is the case for cell line panels (Figure 1D) and for primary tumors from breast, ovarian, bladder, and colorectal cancers (Figure 1E). We expected it to be more prone to false positives for less differentiated expression profiles such as low-grade and early-stage tumors, and, generally observed, this where sufficient numbers of annotated samples were available: Samples falsely identified as duplicates were enriched for low-grade (CRC: 95% confidence interval [CI] = 1.2 to 2.2; ovarian: 95% CI = 1.0 to 1.5) and early-stage (bladder: 95% CI = 1.3 to 4.2; CRC: 95% CI = 1.6 to 2.6). The exception was early-stage ovarian cancer samples, for which doppelgangR was extremely effective. These samples have distinctive profiles, and their rate of sharing was high, possibly because of the rarity of early-stage ovarian cancer and the high importance of specimens.
We investigated the potential for applying this method to each of the 32 TCGA cancer types based on individual distinctiveness of transcriptomic aberrations. Using log-transformed level III RNA-seq data, which are summarized at the level of gene symbols, we calculated correlations between all sample pairs (Figure 1F). Our ranking of the cancer types for transcriptome distinctiveness, based on the 99.9th percentile of correlation between expression profiles, is very similar to Lawrence et al. (13), in which tumors were ranked by mutation rate. We do not expect our method to be effective for cancer types with “quiet” genomes and high correlation between nonduplicate expression profiles, such as kidney renal papillary cell carcinoma and the cancer types to its left in Figure 1F. We also note the existence of highly distinctive expression subtypes, such as in glioblastoma multiforme IDH1 mutant vs wild-type cases (14), which produce bimodal pairwise correlations that may complicate duplicate identification if these subtypes are not separated. Several other cancer types from TCGA exhibit distinctive subtypes that should be considered (Supplementary Figure 1, available online). Known subtypes in the datasets we examined in detail did not impact performance, however, such as prevalence of estrogen receptor–positive breast cancer tumors.
We note the potential utility of the doppelgänger approach also for identifying duplicates within a single study with as few as five samples (see Supplementary Results and Supplementary Figures 2-3, available online). In the databases we studied, within-study duplication was less common than between studies, but we found likely duplicates in six ovarian cancer studies and one CRC study. These profiles were such extreme outliers in terms of similarity of gene expression profiles that they are unlikely to have originated from different tumors, assuming the samples passed basic QC metrics (eg, coverage, tumor purity, RNA integrity scores). The approach reliably grouped together healthy tissues, which have much more homogeneous expression profiles than cancer tissues. Although our benchmarking is limited to microarray and RNA-seq data, we see no reason why this approach should not work for other quantitative mRNA assays such as nanoString and multiplexed quantitative real-time polymerase chain reaction, or even proteomic or other molecular profiles, provided that biological replicates are sufficiently distinct relative to technical replicates.
As genomic databases grow and collect tumor specimens from international collaborators, the chance of inter- and intrastudy duplication increases. Analysis of duplicate samples is a substantial concern that could alter the identification of subsets of patients with clinical differences or the development of specific gene signatures. While this approach can help identify duplicate profiles even when germ-line sequences are not available, we note some limitations. Automatic setting of the threshold defining “outliers” is a difficult problem to solve generally and should be reviewed using diagnostic histograms generated by the doppelgangR package. A thorough review should take into consideration potential collaborations and/or multiple institution clinical management of individual patients, as well as suspiciously similar clinical patient data and identifiers.
Funding
This work was supported by grants from the National Cancer Institute at the National Institutes of Health (1RC4CA156551-01 and 5R01CA142832 to MB, 1R03CA191447-01A1 and U24CA180996 to LW). Additional funding was generously provided by the Julie Fund (MB).
Notes
The study funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Supplementary Material
References
- 1.Butler JM. Genetics and genomics of core short tandem repeat loci used in human identity testing. J Forensic Sci. 2006;51(2):253–265. [DOI] [PubMed] [Google Scholar]
- 2.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. [DOI] [PubMed] [Google Scholar]
- 3.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272. [DOI] [PubMed] [Google Scholar]
- 4.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102(38):13550–13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganzfried BF, Riester M, Haibe-Kains B, et al. curatedOvarianData: clinically annotated data for the ovarian cancer transcriptome. DATABASE. 2013;2013:bat013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshihara K, Tajima A, Yahata T, et al. Gene expression profile for predicting survival in advanced-stage serous ovarian cancer across two independent datasets. PLoS One. 2010;5(3):e9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshihara K, Tsunoda T, Shigemizu D, et al. High-risk ovarian cancer based on 126-gene expression signature is uniquely characterized by downregulation of antigen presentation pathway. Clin Cancer Res. 2012;18(5):1374–1385. [DOI] [PubMed] [Google Scholar]
- 8.Bonome T, Levine DA, Shih J, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68(13):5478–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waldron L, Haibe-Kains B, Culhane AC, et al. Comparative meta-analysis of prognostic gene signatures for late-stage ovarian cancer. J Natl Cancer Inst. 2014;106(5). DOI: 10.1093/jnci/dju049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheng Q, Shyr Y, Chen X. DupChecker: a bioconductor package for checking high-throughput genomic data redundancy in meta-analysis. BMC Bioinformatics. 2014;15:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.TCGA Data Coordinating Center. https://web.archive.org/web/20151116001051/https://tcga-data.nci.nih.gov/tcgafiles/ftp_auth/distro_ftpusers/anonymous/tumor/ov/cgcc/broad.mit.edu/ht_hg-u133a/transcriptome/broad.mit.edu_OV.HT_HG-U133A.Level_1.40.1007.0/README_BATCH_40.txt. Accessed May 20, 2015.
- 13.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.