Abstract
Aim:
In cancer immunotherapy, biomarkers are important for identification of responsive patients. This study was aimed to find biomarkers that predict clinical outcome of WT1 peptide vaccination.
Materials & methods:
Candidate genes that were expressed differentially between long- and short-term survivors were identified by cDNA microarray analysis of peripheral blood mononuclear cells that were extracted from 30 glioblastoma patients (discovery set) prior to vaccination and validated by quantitative RT-PCR using discovery set and different 23 patients (validation set).
Results:
SDC-4 mRNA expression levels distinguished between the long- and short-term survivors: 1-year survival rates were 64.0 and 18.5% in SDC4-low and -high patients, respectively.
Conclusion:
SDC-4 is a novel predictive biomarker for the efficacy of WT1 peptide vaccine.
Keywords: : cancer immunotherapy, DC-HIL, GBM, glioblastoma multiforme, immune checkpoint inhibitor, malignant glioma, peptide vaccine, SDC-4, Syndecan-4, WT1
Lay abstract
Recently, cancer immunotherapies are becoming a standard therapeutic option. To improve their efficacy, identification of biomarkers is important to select responsive patients. In this study, we identified SDC-4 as a biomarker to predict clinical outcome using peripheral blood mononuclear cells obtained from patients with glioblastoma, a malignant brain tumor, who were treated with WT1 peptide vaccine. With 30 samples of peripheral blood mononuclear cells prior to vaccination, 32 candidate genes were filtrated by microarray, and finally only SDC-4 was validated by RT-PCR using another 23 samples. Accordingly, 1-year survival rates were 64.0 and 18.5% in SDC4-low and -high patients, respectively.
WT1, a transcription factor, regulates many kinds of important genes that play important roles in embryogenesis, cell proliferation and differentiation [1–3]. The WT1 gene is overexpressed in leukemia and a variety of solid tumors, in which it exerts an oncogenic function [4,5]. WT1, a pan-tumor-associated antigen (TAA), was identified as the best one among 75 TAAs, based on criteria including therapeutic function, immunogenicity, oncogenicity, specificity, expression levels and percent of positive cells, expression levels in stem cell, number of patients with antigen-positive cancers, number of epitopes and cellular localization [6]. Our group and others have performed WT1-targeted cancer immunotherapy, including WT1 peptide vaccination, and WT1 peptide-pulsed [7] or WT1 mRNA-electroporated [7–9] dendritic cell (DC) therapy, and obtained a series of successful results with positive immunological and clinical responses in patients with glioblastoma multiforme (GBM) [10–14], acute myeloid leukemia [15–19], chronic myeloid leukemia [20,21], myelodysplastic syndromes [22–24], multiple myeloma [25], malignant melanoma [26], infantile rhabdomyosarcoma [27], and lung [28], breast [15], pancreatic [29,30], ovarian [31,32], uterine [23,33] and salivary gland cancers [34,35].
GBM is a malignant brain tumor with very poor prognosis. The standard therapy for the newly diagnosed GBM is surgery, followed by irradiation and chemotherapy. However, a 5-year survival rate is less than 10%, and once recurrence occurs, therapeutic options are limited [36]. In 2008, we reported promising results from a Phase II clinical study of WT1 peptide vaccination in patients with recurrent or conventional therapy-resistant GBM. In patients who received the vaccination, progression-free survival (PFS) at 6 months was 33.3%, and the median overall survival (OS) was 36.7 weeks, suggesting the therapeutic potential of WT1 peptide vaccine for GBM patients [10].
In order to improve the clinical usefulness of WT1 peptide vaccine, it is crucial not only to biologically enhance the vaccine's efficacy but also to select patients who are likely to respond to the vaccine. Identification of responders would be facilitated by the availability of reliable biomarkers that predicted the clinical outcome of patients treated with the WT1 peptide vaccine.
To date, several studies by our group and others have identified markers that are correlated with the clinical efficacy of WT1-based immunotherapy. Malignant glioma patients with higher WT1 expression levels (score 3–4) lived longer after immunotherapy than those with lower expression levels (score 1–2) [13]. An increase in the frequency of WT1-specific cytotoxic T lymphocytes (CTLs) before and after WT1 peptide vaccination was correlated with clinical response [15,17,30]. Upon repeated vaccination with WT1 peptide, an increase in the frequencies of effector memory subsets, which are important for maintenance of WT1-specific CTLs, was also identified as a predictor of responders [30]. Lymphocyte numbers in peripheral blood and antigen-specific delayed-type hypersensitivity (DTH) also predict clinical outcome in various immunotherapies [37]. However, no reliable biomarkers have yet been established that can predict the clinical outcome of immunotherapies, such as TAA-targeting cancer vaccines prior to therapy. High-throughput technologies, such as cDNA microarray analysis, have been investigated as approaches to discover biomarkers for prediction of clinical outcomes of therapeutic interventions.
In this study, we used cDNA microarrays to comprehensively analyze gene-expression profiles of peripheral blood mononuclear cells (PBMCs) with the goal of identifying biomarkers that predicted the outcome of WT1 peptide vaccination in patients with recurrent or conventional therapy-resistant GBM. The results revealed that SDC-4 mRNA expression levels prior to WT1 vaccination were a promising predictive biomarker for clinical outcome in these patients.
Materials & methods
WT1 peptide vaccine
The WT1 peptide vaccine consists of WT1-CTL epitope peptide and Montanide ISA51 adjuvant. The peptide used in this study is a modified 9-mer WT1 peptide (amino acids [aa] 235–243 CYTWNQMNL; mWT1–235), in which M was replaced by Y at the second amino acid position, an anchor position for HLA-A*24:02, of the natural WT1–235 peptide (235–243 CMTWNQMNL; nWT1–235). The binding affinity of mWT1–235 for HLA-A*24:02 is higher than that of nWT1–235, and mWT1–235 induced a much stronger CTL response against WT1-expressing tumor cells. GMP-grade WT1 peptide was purchased from Multiple Peptide Systems (CA, USA) and Peptide Institute (Osaka, Japan) as the lyophilized peptide.
Patients
Sixty patients were enrolled in a Phase II study of WT1 peptide vaccination of GBM patients, the result of which we reported previously [10,13]. Patients with recurrent or progressive GBM were eligible to be enrolled in the Phase II study if their disease was resistant to conventional chemotherapy or radiotherapy. Other inclusion criteria were as described in our previous study. Briefly, they were: first, age between 16 and 80 years; second, expression of WT1 in glioma cells as determined by immunohistochemical analysis; third, HLA-A*2402–positivity; fourth, estimated survival of more than 3 months; fifth, ECOG Performance Status Grade 0–2; sixth, no severe organ function impairment and seventh, written informed consent of the patient. All enrolled patients were histologically proven to have GBM (Grade 4) based on the WHO criteria.
PBMC samples obtained prior to WT1 vaccination were available from 53 of the 60 patients enrolled in the Phase II study; all 53 were used in this study. Thirty of the patients were randomly assigned to the discovery set, and the remaining 23 were assigned to the validation set.
The median OS of the patients in the discovery set was 347 days from the start of the vaccination and 460 days from the time of recurrence or disease progression to death or censored time points. The median PFS, which was defined as the days from the start of the vaccination to the disease progression, was 62 days. In the microarray analysis, to select differentially expressed genes (DEGs) that influence not only short-term outcome (RECIST, PFS and OS from the time of vaccination started) but also long-term outcome, the 30 patients in a discovery set were divided into two groups with OS from the time of recurrence or disease progression. The 15 patients with OS of ≥460 days were defined as long-term survivors (OS median: 1133 days, range: 480–2678 days), whereas the remaining 15 patients with OS of <460 days were defined as short-term survivors (OS median: 216 days, range: 138–458 days) (Table 1).
Table 1. . Patients’ characteristics in a discovery set.
| Characteristic | Short OS (n = 15) | Long OS (n = 15) |
Hazard ratio 95% CI |
p-value† | |
|---|---|---|---|---|---|
| Lower | Upper | ||||
| Age (years): | |||||
| – Mean (range) |
58 (25–67) |
46 (20–75) |
0.07 |
2.76 |
0.36 |
| Gender: | |||||
| – Male (%) |
9 (60) |
11 (73) |
0.35 |
3.88 |
0.93 |
| KPS: | |||||
| – Mean (range) |
75 (45–95) |
80 (55–100) |
0.005 |
0.50 |
0.009* |
| Surgical treatment (%) |
13 (87) |
15 (100) |
0.15 |
11.7 |
0.90 |
| Chemotherapy, n (%) |
14 (93) |
11 (73) |
1.26 |
26.5 |
0.021* |
| RT before vaccination (Gy): | |||||
| – Average (SD) | 67 (22.3) | 61 (8.7) | 0.03 | 13.3 | 0.96 |
†Cox proportional hazard regression with the OS from vaccination started as the time variable was used.
*Statistical significance (p < 0.05).
KPS: Karnovski performance status; OS: Overall survival; RT: Radio therapy (total absorbed dose); SD: Standard deviation.
In the verification process using discovery set, OS was the days from the first vaccination to death or censored time points which should more strictly reflect the vaccination effect. Thus, in the verification step, patients who survived ≥347 days (the median OS of the discovery set according to the immediately preceding definition) were defined as long-term survivors, whereas those who survived <340 days were defined as short-term survivors.
In the validation set, median OS from the first vaccination was 257 days. The 12 patients who survived ≥256 days were defined as long-term survivors, and the 11 patients who survived <256 days were defined as short-term survivors.
All short-term survivors in the discovery and validation sets died before a censored time point. Therefore, patient categorization of long- and short-term survivors was fixed (Supplementary Table 1). The Phase II study of WT1 peptide vaccination and cDNA microarray analysis of the blood samples was approved by the ethical review board of Osaka University Hospital.
WT1 peptide vaccination schedule
After informed consent was obtained, weekly intradermal injection of 3.0 mg of HLA-A*24:02–restricted mWT1–235 peptide emulsified with Montanide ISA51 adjuvant was initiated. Vaccinations were scheduled for 12 consecutive weeks, after which responses were evaluated by MRI. Responses were classified as complete response, partial response, stable disease and progressive disease using MRI according to the RECIST (Response Evaluation Criteria In Solid Tumors) criteria. When clinical response was obvious, WT1 vaccination was continued at 2-week intervals for the next few months, and then at 1–3-month intervals until obvious tumor progression or deterioration of the patient's condition was observed.
Blood samples
Peripheral blood was obtained from patients immediately before the first vaccination, followed by separation of PBMCs by density gradient centrifugation using Lymphocyte Separation Solution (Nacalai Tesque, Kyoto, Japan). Separated PBMCs were stored in liquid nitrogen prior to use.
RNA isolation from PBMCs & cDNA microarray analysis
Frozen PBMCs were thawed, and total RNA was isolated using the TRIzol reagent (Life Technologies, CA, USA), purified using chloroform and subjected to isopropanol and ethanol precipitation. Purified total RNA was quantitated using a NanoDrop ND-1000 (Thermo Fisher Scientific, MA, USA).
RNAs from PBMCs of the discovery set patients were sent to Toray Industries (Tokyo, Japan), which performed RNA-based cDNA microarray analysis using Human oligo chip 25k ver1.00 (Toray Industries).
Microarray analysis data were transformed using global normalization, followed by quantile normalization [38]. First, genes with intensities less than ([average intensities of blank spots] + 10 × [standard deviation of intensities of blank spots]) were excluded. To identify DEGs between long- and short-term survivors, a volcano plot of -log10(p-value) of Welch's t-test between the two groups (y-axis) versus log2(fold change) (x-axis) was made [39]. The fold change between the two groups was calculated as log2([mean signal intensities in long-term survivors]/[mean signal intensities in short-term survivors]). DEGs were selected using this volcano plot according to the following conditions: y > |x|−1, |x| >0.5 and y >1.0. To enrich for DEGs, correlations between individual gene expression intensities and either PFS or clinical responses according to RECIST criteria were examined by Pearson's and Spearman's correlation coefficient, respectively. Genes that did not exhibit significant p-values (alpha level 0.1 for Pearson's correlation coefficient and 0.2 for Spearman's correlation coefficient) were excluded from further analysis. In this series of microarray analyses, a nonstringent cut-off for the p-value was set according to MAQC guidelines [40]. Finally, genes that satisfied the following six conditions were retained for further analysis: first, average intensity ≥([average intensities of blank spots] + 10 × [standard deviation of intensities of blank spots]); second, -log10(p-value of difference between long- and short-OS groups) was greater than |log2(fold difference)|–1; third, |log2(fold difference)| was greater than 0.5; fourth, -log10 (p-value of difference between long- and short-OS groups) greater than 1.0; fifth, p-value of Pearson's correlation between PFS and signal intensities less than 0.1; and sixth, p-value of Spearman's correlation between clinical outcome and signal intensities less than 0.2.
RT-PCR
RNAs from PBMCs were reverse-transcribed into first-strand cDNA using SuperScript VILO cDNA Synthesis Kit and Master Mix (Life Technologies). Sequences of primers used for RT-PCR are provided in Supplementary Table 2. Target sequences are available from the NCBI nucleotide database [41]. Sample cDNAs were preamplified with dilute primer mixture for 15 cycles prior to loading to the BioMark 48.48 dynamic array nanofluidic chips (Fluidigm, CA, USA). RT-PCR was performed on a BioMark HD (Fluidigm). Regarding internal control genes, we quantified many control genes, including ACTB, GAPDH, RPL13, RPL18A, PPIA, TSR2 and RNA28S1. Each control gene was validated in comparison with each other, and correlation matrix was generated, followed by scoring each gene. Consequently, ACTB was determined as the most stable internal control gene among all the candidate control genes and was used as the internal control. Then, ΔCTs were calculated as ([CT value of each genes] – [CT value of ACTB]). All data concerning gene expression levels were converted to log2(1 + 2−ΔCT), and the relative expression levels were statistically analyzed.
Statistical analysis
Cox proportional hazard regression models were used to evaluate associations between patient characteristics and OS in the discovery and validation sets. In the microarray analysis, PFS was logarithmically transformed to yield a normal distribution.
In statistical analysis of the result from RT-PCR, the Jarque–Bera test was used to assess normality of variable distributions, and the F-test was used to test homogeneity of variance.
To promote statistical power, parametric analyses were conducted as much as possible. For examination of the difference between long- and short-term survivors, an appropriate transformation (e.g., logarithmic, square-root, cube-root or fourth-root transformation) was performed for individual genes to satisfy the requirement for normality. All normally distributed variables were checked for homogeneity of variance and analyzed by Student's t-test. Non-normally distributed variables were analyzed using the Mann–Whitney U-test.
The SDC-4 expression levels that discriminated responders and nonresponders (SDC-4 cut-off value) were determined using a receiver operating characteristic (ROC) curve in order to maximize the Youden's index (defined as Youden's J statistic = sensitivity + specificity -1). The ROC curve was drawn based on the cut-off value of OS, which maximized area under the ROC curve (AUC). The horizontal and vertical axes indicate 1-specificity and sensitivity for responder, respectively. Sensitivity, specificity, positive and negative predictive values, and accuracy of the prediction of OS by the SDC-4 expression levels were calculated using standard formulas. Kaplan–Meier curves and a two-sided log-rank test were used to assess differences between two groups defined by the SDC-4 cut-off value. Statistical analysis was conducted using appropriate software including JMP Pro 10.0 (SAS Institute, Inc., NC, USA) and R-commander [42].
Results
Identification of DEGs in PBMCs prior to WT1 vaccination
Patients with recurrent or conventional therapy-resistant GBM who were treated with WT1 peptide vaccine were selected as the cohort for this analysis. PBMCs were obtained prior to WT1 vaccination from 53 of 60 patients in this vaccination trial. The strategy to search useful biomarkers for prediction of clinical outcome is shown schematically in Figure 1A. Patients were assigned randomly into the discovery and validation sets (30 and 23 patients, respectively); within the validation set, patients were classified as long-term or short-term survivors (15 patients each) (Table 1). Cox proportional hazard regression models revealed that within the discovery set, there was no association between OS following vaccination and age, gender, presence or absence of prior surgical treatment, or prior total absorbed dose of radiotherapy; however, low-performance status and history of chemotherapy were indicators of worse clinical outcome (Table 1). We discussed these issues that affect clinical outcome later in the last part of the Results and Discussion section.
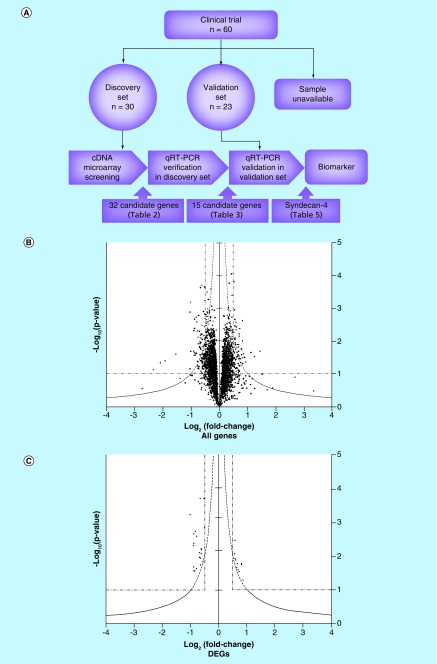
Figure 1. . Selection of candidate genes by cDNA microarray analysis.
(A) Strategy to find biomarkers is schematically shown. First, DEGs were screened by cDNA microarray and the expression levels of screened genes were verified by quantitative RT-PCR using 30 glioblastoma multiforme patients in a discovery set. Second, these verified DEGs were validated using different 23 GBM patients in a validation set. Finally, only SDC-4 was identified as a biomarker. (B) A volcano plot was generated. Each dot corresponds to one gene. X- and y-axes indicate fold change (log2[short/long]) of signal intensities of individual genes and the statistical significance (-log10[p-value]) of the difference in the signal intensities of individual genes between long- and short-term survivors, respectively. Dashed line indicates y = |x|–1, y = 1 and |x| = 0.5. (C) Thirty-two candidate DEGs were extracted as described in the text. Gene names and their statistical evaluations are shown in Table 2.
DEG: Differentially expressed gene.
To identify candidate genes that were expressed differentially between long- and short-term survivors, we performed cDNA microarray analysis on PBMCs. A volcano plot of statistical significance (-log10 p-value) versus log2 (fold change), in which each dot indicates one of a total of 25,000 genes, is shown in Figure 1B. DEGs were extracted as follows. First, we excluded genes with intensities less than ([average intensities of blank spots] + 10 × [standard deviation of intensities of blank spots]). The remaining 3037 genes were filtrated to a set of 74 genes that satisfied the following three criteria, y > |x|–1, |x| > 0.5 and y > 1.0. To further enrich for candidate genes, the correlation between the signal intensities of individual genes and either PFS or clinical response was evaluated by Pearson's correlation and Spearman's rank correlation analysis, respectively, and genes with p-values ≥ 0.1 (Pearson's) or ≥ 0.2 (Spearman's) were excluded. Ultimately, 32 genes were selected as candidates (Table 2 & Figure 1C). Of these candidate genes, 11 were highly expressed in long-term survivors (Table 2, upper); and 21 were highly expressed in short-term survivors (Table 2, lower).
Table 2. . Thirty-two candidate genes that differentially expressed between long- and short-term survivors in peripheral blood mononuclear cells prior to WT1 peptide vaccination in 30 glioblastoma multiforme patients in a discovery set.
| Gene | Fold change† | p-value‡ |
|---|---|---|
|
TLR10 |
0.85 |
0.051 |
|
KLHDC8B |
0.80 |
0.018 |
|
RALGPS2 |
0.80 |
0.017 |
|
CD79B |
0.71 |
0.029 |
|
TNFAIP8L2 |
0.69 |
0.026 |
| GCNT2 |
0.65 |
0.017 |
|
EVA1 |
0.64 |
0.022 |
|
CETN3 |
0.59 |
0.013 |
|
ROGDI |
0.58 |
0.012 |
|
CD82 |
0.57 |
0.004 |
|
IL17RA |
0.55 |
0.009 |
|
TNFSF14 |
-1.02 |
<0.001 |
|
CST1 |
-0.90 |
0.005 |
|
FBXO32 |
-0.90 |
0.021 |
|
ULBP2 |
-0.88 |
0.028 |
|
OASL |
-0.88 |
0.004 |
|
PHLDA1 |
-0.88 |
0.002 |
|
FASLG |
-0.86 |
0.028 |
|
XAGE5 |
-0.83 |
0.002 |
|
SDC4 |
-0.80 |
0.024 |
|
ZNF659 |
-0.79 |
0.003 |
|
SMAD7 |
-0.69 |
0.011 |
|
SLC7A5 |
-0.68 |
0.008 |
|
VPS37B |
-0.66 |
<0.001 |
|
HPGD |
-0.65 |
0.018 |
|
MLF1 |
-0.65 |
0.012 |
|
SKI |
-0.63 |
0.006 |
|
EXPH5 |
-0.63 |
0.009 |
|
ZC3H12A |
-0.61 |
0.007 |
|
UAP1 |
-0.57 |
0.015 |
|
CCNT1 |
-0.53 |
<0.001 |
| ITGA5 | -0.52 | 0.002 |
†Log2(long/short).
‡Two-tailed Welch's t-test.
Verification of candidate genes by quantitative RT-PCR
Expression levels of the 32 candidate genes identified in the cDNA microarray analysis of the patients in the discovery set were verified by quantitative RT-PCR.
In order to ensure a rigorous statistical analysis, individual gene expression levels obtained by quantitative RT-PCR were transformed appropriately to satisfy the requirement for normality. Distributions were checked for normality by the Jarque–Bera test, for homogeneity of variance by the F-test, and for significance by one-tailed Student's t-test (Table 3). When no transformation yielded a sufficiently normal distribution, the one-tailed Mann–Whitney U-test was used. Because one-tailed tests were used, genes with fold changes that were inverted relative to those obtained from the microarrays were automatically excluded. As a result of these analyses, 15 genes whose expression levels correlated to OS were retained, and 17 were excluded (Table 3).
Table 3. . Verification of 32 candidate genes by quantitative RT-PCR.
| Gene | Transformation | Normal distribution | Statistical method | Fold change† | p-value | Result |
|---|---|---|---|---|---|---|
|
EXPH5 |
Square root |
True |
Left-sided Student t-test |
-0.34 |
<0.001* |
Verified |
|
ZC3H12a |
Failed |
False |
Left-sided Mann–Whitney U-test |
-1.23 |
0.004* |
Verified |
|
VPS37B |
Not needed |
True |
Left-sided Student t-test |
-1.19 |
0.005* |
Verified |
|
PHLDA1 |
Square root |
True |
Left-sided Student t-test |
-1.33 |
0.007* |
Verified |
|
TNFSF14 |
4th root |
True |
Left-sided Student t-test |
-1.70 |
0.007* |
Verified |
|
UAP1 |
Square root |
True |
Left-sided Student t-test |
-1.58 |
0.013* |
Verified |
|
HPGD |
Cube root |
True |
Left-sided Student t-test |
-0.68 |
0.016* |
Verified |
|
SKI |
4th root |
True |
Left-sided Student t-test |
-0.76 |
0.016* |
Verified |
|
SDC4 |
Logarithmic |
True |
Left-sided Student t-test |
-0.35 |
0.023* |
Verified |
|
FOXO32 |
Logarithmic |
True |
Left-sided Student t-test |
-2.02 |
0.023* |
Verified |
|
SLC7A5 |
Square root |
True |
Left-sided Student t-test |
-0.97 |
0.026* |
Verified |
|
ULBP2 |
Logarithmic |
True |
Left-sided Student t-test |
-2.04 |
0.030* |
Verified |
|
CCNT1 |
Cube root |
True |
Left-sided Student t-test |
-0.60 |
0.031* |
Verified |
|
FASLG |
Cube root |
True |
Left-sided Student t-test |
-1.46 |
0.036* |
Verified |
|
OASL |
Square root |
True |
Left-sided Student t-test |
-1.04 |
0.039* |
Verified |
|
SMAD7 |
Square root |
True |
Left-sided Student t-test |
-1.53 |
0.067 |
Excluded |
|
GCNT2 |
Failed |
False |
Right-sided Mann–Whitney U-test |
3.82 |
0.189 |
Excluded |
|
EVA1 |
Not needed |
True |
Right-sided Student t-test |
0.00 |
0.220 |
Excluded |
|
ITGA5 |
Cube root |
True |
Left-sided Student t-test |
-0.76 |
0.241 |
Excluded |
|
F3 |
Failed |
False |
Left-sided Mann–Whitney U-test |
-2.11 |
0.252 |
Excluded |
|
XAGE5 |
Failed |
False |
Left-sided Mann–Whitney U-test |
-1.77 |
0.261 |
Excluded |
|
MLF1 |
Square root |
True |
Left-sided Student t-test |
-0.46 |
0.448 |
Excluded |
|
TLR10 |
Failed |
False |
Right-sided Mann–Whitney U-test |
4.73 |
0.491 |
Excluded |
|
CETN3 |
Cube root |
True |
Right-sided Student t-test |
0.05 |
0.523 |
Excluded |
|
CD82 |
Square root |
True |
Right-sided Student t-test |
-0.08 |
0.577 |
Excluded |
|
TNFAIP8L2 |
Cube root |
True |
Right-sided Student t-test |
-0.40 |
0.646 |
Excluded |
|
CD79B |
Square root |
True |
Right-sided Student t-test |
0.45 |
0.653 |
Excluded |
|
IL17RA |
Cube root |
True |
Right-sided Student t-test |
0.47 |
0.738 |
Excluded |
|
RALGPS2 |
Not needed |
True |
Right-sided Student t-test |
-0.20 |
0.744 |
Excluded |
|
KLHDC8B |
Logarithmic |
True |
Right-sided Student t-test |
0.37 |
0.821 |
Excluded |
|
ZNF659 |
Failed |
False |
Left-sided Mann–Whitney U-test |
1.42 |
0.934 |
Excluded |
| ROGD1 | Square root | True | Right-sided Student t-test | 0.07 | 0.982 | Excluded |
†Log2(mean of gene expression levels in long-term survivors/mean of gene expression levels in short-term survivors).
*Statistical significance (p < 0.05).
Verification of 15 candidate genes using patients in the validation set
Next, we prepared a validation set consisting of 23 GBM patients treated with WT1 peptide vaccine following the same protocol used in the discovery set. Characteristics of patients in the validation set are provided in Table 4. No association was detected between OS and age, gender, performance status, the presence or absence of prior surgical treatment and chemotherapy, or prior total absorbed doses on radiotherapy. Using data from the patients in the validation set, we investigated whether the expression levels of the 15 candidate genes identified in the discovery set correlated with OS using the same statistical methods as those used in the discovery set. Only expression of SDC-4 significantly (negatively) correlated with OS in the validation set (Table 5).
Table 4. . Patients’ characteristics in a validation set.
| Characteristic | Short OS (n = 11) | Long OS (n = 12) |
Hazard ratio 95% CI |
p-value† | |
|---|---|---|---|---|---|
| Lower | Upper | ||||
| Age (years): | |||||
| – Mean (range) |
48.9 (28–63) |
50.8 (30–71) |
0.14 |
5.48 |
0.90 |
| Gender: | |||||
| – Male (%) |
7 (58) |
7 (63) |
0.21 |
1.88 |
0.40 |
| KPS: | |||||
| – Mean (range) |
78 (50–100) |
84 (50–100) |
0.08 |
2.21 |
0.29 |
| Surgical treatment (%) |
10 (91) |
12 (100) |
0.06 |
19.9 |
0.86 |
| Chemotherapy, No. (%) |
9 (82) |
10 (83) |
0.11 |
2.12 |
0.28 |
| RT before vaccination (Gy): | |||||
| – Average (SD) | 57 (9.0) | 59 (2.9) | 0.005 | 2.54 | 0.13 |
†Cox proportional hazard regression with the OS from vaccination started as the time variable was used.
KPS: Karnovski performance status; OS: Overall survival; RT: Radio therapy (total absorbed dose); SD: Standard deviation.
Table 5. . Validation of 15 candidate genes using different 23 glioblastoma multiforme patients in a validation set.
| Gene | Fold change† | p-value‡ | Result |
|---|---|---|---|
|
SDC4 |
-1.85 |
0.020* |
Validated |
|
HPGD |
-0.84 |
0.076 |
Not reproducible |
|
PHLDA1 |
-0.59 |
0.154 |
Not reproducible |
|
VPS37B |
-1.81 |
0.269 |
Not reproducible |
|
OASL |
-0.21 |
0.485 |
Not reproducible |
|
TNFSF14 |
0.27 |
0.555 |
Not reproducible |
|
FASLG |
3.62 |
0.602 |
Not reproducible |
|
UAP1 |
2.75 |
0.608 |
Not reproducible |
|
SLC7A5 |
1.82 |
0.679 |
Not reproducible |
|
FOXO32 |
2.21 |
0.760 |
Not reproducible |
|
SKI |
3.80 |
0.844 |
Not reproducible |
|
EXPH5 |
2.43 |
0.869 |
Not reproducible |
|
ULBP2 |
2.65 |
0.953 |
Not reproducible |
|
ZC3H12a |
2.71 |
0.964 |
Not reproducible |
| CCNT1 | 1.09 | 0.975 | Not reproducible |
†Log2(long/short).
‡The same statistical methods as those in Table 3 were used.
*Statistical significance (p < 0.05).
Prognostic prediction of GBM patients treated with WT1 peptide vaccine using SDC-4 expression levels
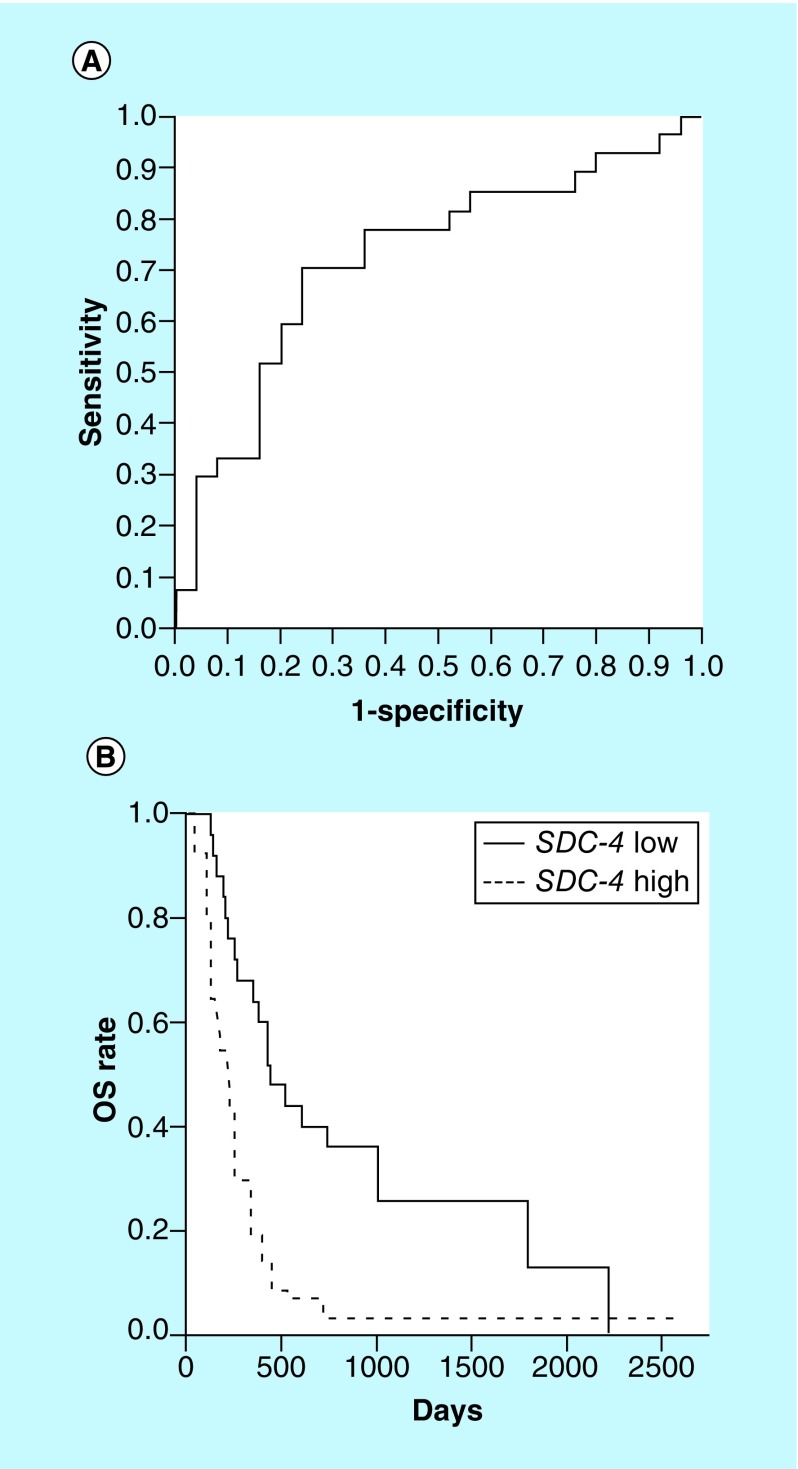
First, based on the values of OS and SDC-4 expression levels, we generated a ROC curve using all 53 patients from the discovery and validation sets (Figure 2A). Generally, area under the ROC curve (AUC) of an efficient biomarker shows high AUC, maximum of which is 1.0. Since AUC varies in accordance with cut-off values of OS, the cut-off value was selected in order to maximize the AUC; therefore, the patients were classified into two groups, ‘responders’ and ‘nonresponders’ by cut-off value of 256 days and then AUC was 0.72. The best cut-off value of SDC-4 expression levels to classify responders (OS of ≥256 days) and nonresponders (OS of <256 days) was determined to be 0.001 according to the AUC-maximized ROC curve using the Youden's index; SDC-4 expression levels ≤0.001 and >0.001 predicted responders and nonresponders, respectively. Prediction of likely responders had 70.4% sensitivity, 76.0% specificity, 76.0% positive prediction value, 70.4% negative prediction value and 73.1% accuracy; in addition, a Chi-square test demonstrated a significant difference (p < 0.001).
Figure 2. . SDC-4 is a prediction marker for overall survival.
(A) AUC-maximized receiver operating characteristic curve was generated. Optimal cut-off value, 0.001 of SDC-4 relative expression levels (log2[1 + 2−ΔCT]) discriminates between long-term (OS ≥256 days) and short-term (OS <256 days) survivors, and SDC-4 expression levels in long-term survivors are ≤0.001. Statistical capabilities are 70.4% sensitivity, 76.0% specificity, 76.0% positive predictive value, 70.4% negative predictive value and 73.1% accuracy, and a Chi-square test shows a statistical significance (p < 0.001). All the 53 patients in the discovery and validation sets are included in this analysis. (B) Kaplan–Meier curves of OS (days) of patients with ≤0.001 (SDC-4 low) and >0.001 (SDC-4 high) of SDC-4-expression levels are shown. Comparison of OS between the two groups is performed using a two-sided log-rank test, and the difference in OS is statistically significant (p < 0.001). One-year OS rates were 64.0 and 18.5% in SDC-4-low and -high patients, respectively.
OS: Overall survival.
Furthermore, we performed survival analysis using this cut-off value of SDC-4 expression level (0.001). The patients were divided into two groups, SDC-4-low (SDC-4 ≤ 0.001) and -high (SDC-4 > 0.001) groups (Figure 2B). The difference in OS between the two groups was estimated by Kaplan–Meier curves and the subsequent log-rank test. The results revealed that SDC-4-low patients survived significantly longer than SDC-4-high patients; 1-year OS rates were 64.0 and 18.5%, respectively (Figure 2B).
As shown in Table 1, low-performance status and the past history of chemotherapy were indicated to be risk factors of worse clinical outcome for the discovery set patients. Therefore, potential confounding effects of SDC-4 expression levels and baseline patient characteristics, including the two factors mentioned above, were assessed using Cox proportional hazard regression models in all the 53 patients (Supplementary Table 3). This analysis allowed us to assess the association between SDC-4 expression levels and the risk of death at any given time points while controlling for other predictors that may affect the risk of death. As a result, it was suggested that high SDC-4 expression level of SDC-4 was a significant risk factor for the worse clinical outcome (HR: 13.8; 95% CI: 1.35–84.2; p = 0.027) and a predictor of the clinical outcome independent of other risk factors.
Discussion
In this study, we demonstrated that SDC-4 expression levels in PBMCs obtained prior to WT1 vaccination were significantly and negatively correlated with OS of recurrent or conventional therapy-resistant GBM patients who were treated with WT1 peptide vaccine, and that SDC-4 expression levels were a useful biomarker for prediction of clinical outcome.
T cells are a major player in tumor immunity. T cells are activated through an interaction between T-cell receptors and antigen/MHC molecule complexes on antigen-presenting cells (APCs); that is regulated in a costimulatory or coinhibitory manner by accessory receptors. A costimulatory signal is transmitted by the interaction between CD28 receptor on T cells and CD80 or CD86 on APCs. On the other hand, coinhibitory signals are transmitted by a variety of molecules, including the interactions among CTLA-4 (cytotoxic T lymphocyte antigen-4) on T cells for CD80 and CD86 on APCs; programmed cell death-1 (PD-1) and its ligands PD-L1 and PD-L2; B- and T-lymphocyte attenuator and herpes virus entry mediator; Tim-3 (T cell immunoglobulin- and mucin domain-containing molecules 3) and Tim-3L [43,44]; and by TIGIT (T cell immunoreceptor with immunoglobulin and ITIM domain) [45]. SDC-4 is a novel type of coinhibitor distinct from those listed above.
Syndecans are type-I transmembrane heparan sulfate proteoglycans that bind to the extracellular matrix and a variety of cytokines, chemokines and growth factors, thereby modifiying their local concentration, stability and accessibility to their respective receptors; consequently, they significantly influence cell proliferation and differentiation. Mammals have four known syndecans, (SDC-1, -2, -3 and -4), of which only SDC-4 is expressed ubiquitously; expression of the other three is tissue-restricted [46]. SDC-4 is upregulated on activated T cells via activation of NF-κB [47]. In addition, it interacts with DC-associated heparan sulfate proteoglycan-dependent integrin ligand (DC-HIL) and thus mediates the coinhibitory effect of DC-HIL on T-cell activation [43]. Knockdown of SDC-4 expression enhances the T-cell response to APCs, and blockage of endogenous SDC-4 using specific antibodies or soluble SDC-4 receptor enhances T-cell reactions to syngeneic and allogeneic stimulation in vitro and exacerbated contact hypersensitivity responses in vivo [48]. Consistent with this, transplantation of SDC-4-/- T cells into sublethally γ-irradiated allogeneic mice induces hyperproliferation of infused T cells [48]. Collectively, these observations indicate that SDC-4 is the T-cell ligand through which DC-HIL mediates its coinhibitory function. On the other hand, DC-HIL also binds to SDC-4 on activated T cells and is expressed most strongly by epidermal Langerhans cells, an immature type of DC [49]. DC-HIL expression levels on CD14+ monocytes inversely correlate with allostimulatory capacity, and knockdown of DC-HIL enhances allostimulation. Deletion of DC-HIL abrogates the T-cell suppressor activity of myeloid-derived suppressor cells (MDSCs) [50]. These observations clearly suggest that the SDC-4/DC-HIL pathway is one of the most important factors in regulation of immune responses mediated by T cells, APCs and MDSCs. Most importantly, downregulation of either or both SDC-4 and DC-HIL augments T-cell-mediated-immune responses by attenuating the interaction between the two molecules and/or reducing the T-cell suppressor function of MDSCs [50].
Based on the findings described above, our present results might be interpreted as follows. Low expression of SDC-4 in PBMCs, which correlated with favorable clinical effects of WT1 peptide vaccination of GBM patients, reflected downregulated expression of SDC-4 on T cells. In addition, this downregulation of SDC-4 augmented WT1-specific immune responses to WT1 peptide vaccination by attenuating the interaction between SDC-4 and DC-HIL on monocytes (DCs), leading to improved clinical effects.
On the other hand, it was also reported that, besides T-cell activation state, various pathophysiological conditions, including bacterial endotoxin shock [51], acute pneumonia [52], Helicobacter pylori infection [53], atherosclerosis [54] and ischemic heart disease [55], are associated with SDC-4 expression levels in PBMCs. Therefore, we need to examine the influence of expression levels of SDC-4, which is expressed in various types of cells, on clinical effect of WT1 peptide vaccination from multiple pathophysiological aspects. Further studies should be needed to address this issue.
Regarding the two risk factors in Table 1 and Supplementary Table 3, low-performance status is a common risk factor for poor prognosis of GBM, and the past chemotherapy might dampen immunological competence that supports WT1 peptide-based immunotherapy. On the other hand, although old age is a common risk factor for poor prognosis of GBM as well as low-performance status, no association between OS and age was detected. Since WT1 immunotherapy is a very mild therapy without significant adverse effects (only skin erythema at the vaccine injection sites) that give the organ damages, almost all patients, regardless of ages, are tolerable to the WT1 immunotherapy and may be effective to immunotherapy. On the other hand, since chemotherapy has strong adverse effects, the patients become nontolerable to it as the patients’ ages increase, resulting in the decrease in clinical outcome. Therefore, age is not important prognostic factor in immunotherapy, but PS, which reflects the patients’ immune conditions, is a very important prognostic factor. Repeated chemotherapy gives the damages to the immune system and thus decreases the clinical effects of the following immunotherapy. Therefore, the results that previous chemotherapy but not age is a bad prognostic factor are reasonable.
To our knowledge, only one previous study [56] involved experiments similar to ours; those authors reported biomarkers that predicted the outcome of vaccination with four kinds of HLA-A-restricted peptides in patients with conventional therapy-resistant prostate cancer. In particular, they identified LRRN3, PCDH17, HIST1H4C and PGLYRP1, but not SDC-4, as biomarkers that discriminated between long- (>900 days) and short-term (≤900 days) survivors. The discrepancy in the markers identified in our study and theirs, which used a different peptide vaccine, implies that different targeted TAAs lead to different immune responses in patients. This might mean that these different immune responses are dependent upon whether T cells, DCs and MDSCs or other cell types are major players in the immune reaction, and that the major players are determined by differences in the targeted TAAs and/or types of malignancies.
Since sample size of the present study was not so large, the processes to identify the SDC-4 as a biomarker were divided into the two: discovery and validation processes. The candidate genes identified in the discovery process using 30 patients were validated in the next process using different 23 patients in a validation set, resulting in the identification of SDC-4 alone as a biomarker to predict clinical outcome of WT1 peptide vaccine. We wish to perform a prospective study composed of a larger sample size in the near future, and then we may obtain a more definite finding that supports the conclusion of the present study. In addition, a large sample size-based study may give us an opportunity to find another promising biomarker for WT1 peptide vaccination, and a combination of SDC-4 and the other biomarkers may become a more precise predictor for clinical outcome of the therapy.
Conclusion
In this study, among patients with glioblastoma who received WT1 peptide vaccine, low SDC-4 expression level in PBMCs prior to vaccination was a prognostic factor for long-term survivors. This result was compatible with previously reported immune-suppressive functions of SDC-4 expressed on T cells. These findings suggested that SDC-4 might be useful not only as a biomarker to select patients who would respond to the immunotherapy but also as a target to improve the effect of the immunotherapy.
Executive summary.
Background
WT1 overexpresses in leukemia and a variety of solid tumors and performs an oncogenic function, and WT1 protein is one of the most superior pan-tumor-associated antigens.
Glioblastoma multiforme (GBM) is a malignant brain tumor with very poor prognosis, and once recurrence occurs, therapeutic options are limited.
In 2008, we reported the promising results of a Phase II clinical study of WT1 vaccination for recurrent or conventional therapy-resistant GBM patients.
In order to improve the clinical usefulness of WT1 peptide vaccine, it is important not only to enhance clinical efficacy but also to select the responders to WT1 peptide vaccine.
Novel methods to predict responders prior to WT1 vaccination were awaited.
Materials & methods
Peripheral blood mononuclear cells from 30 GBM patients prior to WT1 vaccination in a discovery set were subjected to cDNA microarray analysis, candidate genes that differentially expressed between long- and short-term survivors were selected, followed by verification with quantitative RT-PCR.
The filtrated candidate genes were validated for the correlation between gene expression levels and clinical effects using different 23 GBM patients of a validation set.
Results
Thirty-two differentially expressed genes (DEGs) were extracted by cDNA microarray analysis using 30 GBM patients in a discovery set, and 15 DEGs were verified by quantitative RT-PCR.
The 15 DEGs were validated using 23 patients in a validation set, and only SDC-4 was identified as a biomarker for prediction of overall survival (OS).
Cut-off value of mRNA expression levels of SDC-4 that discriminated between OS of ≥256 days (responders) and OS of <56 days (nonresponders) was 0.001, and the SDC-4 expression levels of ≤0.001 and >0.001 predicted responders and nonresponders, respectively, with 70.5% sensitivity and 76.0% specificity.
One-year OS rates were 64.0 and 18.5% in SDC-4-low and -high patients, respectively.
Conclusion
SDC-4 is a biomarker to predict clinical outcome for GBM patients treated by WT1 peptide vaccine.
Supplementary Material
Acknowledgements
The authors thank the nursing teams for their care of the patients in this study, and Tomoe Umeda for coordination of clinical research.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Open access
This work is licensed under the Creative Commons Attribution 4.0 License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
References
- 1.Drummond IA, Madden SL, Rohwer-Nutter P, Bell GI, Sukhatme VP, Rauscher FJ., 3rd Repression of the insulin-like growth factor II gene by the Wilms tumor suppressor WT1. Science. 1992;257(5070):674–678. doi: 10.1126/science.1323141. [DOI] [PubMed] [Google Scholar]
- 2.Hewitt SM, Hamada S, Mcdonnell TJ, Rauscher FJ, 3rd, Saunders GF. Regulation of the proto-oncogenes BCL-2 and c-MYC by the Wilms’ tumor suppressor gene WT1. Cancer Res. 1995;55(22):5386–5389. [PubMed] [Google Scholar]
- 3.Kim J, Prawitt D, Bardeesy N, et al. The Wilms’ tumor suppressor gene (WT1) product regulates Dax-1 gene expression during gonadal differentiation. Mol. Cell. Biol. 1999;19(3):2289–2299. doi: 10.1128/mcb.19.3.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugiyama H. Wilms’ tumor gene WT1: its oncogenic function and clinical application. Int. J. Hematol. 2001;73(2):177–187. doi: 10.1007/BF02981935. [DOI] [PubMed] [Google Scholar]
- 5.Nakatsuka S, Oji Y, Horiuchi T, et al. Immunohistochemical detection of WT1 protein in a variety of cancer cells. Mod. Pathol. 2006;19(6):804–814. doi: 10.1038/modpathol.3800588. [DOI] [PubMed] [Google Scholar]
- 6.Cheever MA, Allison JP, Ferris AS, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin. Cancer Res. 2009;15(17):5323–5337. doi: 10.1158/1078-0432.CCR-09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coosemans A, Vanderstraeten A, Tuyaerts S, et al. Wilms’ tumor gene 1 (WT1)-loaded dendritic cell immunotherapy in patients with uterine tumors: a Phase I/II clinical trial. Anticancer Res. 2013;33(12):5495–5500. [PubMed] [Google Scholar]
- 8.Van Driessche A, Van de Velde AL, Nijs G, et al. Clinical-grade manufacturing of autologous mature mRNA-electroporated dendritic cells and safety testing in acute myeloid leukemia patients in a Phase I dose-escalation clinical trial. Cytotherapy. 2009;11(5):653–668. doi: 10.1080/14653240902960411. [DOI] [PubMed] [Google Scholar]
- 9.Coosemans A, Vanderstraeten A, Tuyaerts S, et al. Immunological response after WT1 mRNA-loaded dendritic cell immunotherapy in ovarian carcinoma and carcinosarcoma. Anticancer Res. 2013;33(9):3855–3859. [PubMed] [Google Scholar]
- 10.Izumoto S, Tsuboi A, Oka Y, et al. Phase II clinical trial of Wilms tumor 1 peptide vaccination for patients with recurrent glioblastoma multiforme. J. Neurosurg. 2008;108(5):963–971. doi: 10.3171/JNS/2008/108/5/0963. [DOI] [PubMed] [Google Scholar]
- 11.Hashimoto N, Tsuboi A, Chiba Y, et al. Immunotherapy targeting the Wilms’ tumor 1 gene product for patients with malignant brain tumors. Brain Nerve. 2009;61(7):805–814. [PubMed] [Google Scholar]
- 12.Chiba Y, Hashimoto N, Tsuboi A, et al. Effects of concomitant temozolomide and radiation therapies on WT1-specific T-cells in malignant glioma. Jpn. J. Clin. Oncol. 2010;40(5):395–403. doi: 10.1093/jjco/hyp196. [DOI] [PubMed] [Google Scholar]
- 13.Chiba Y, Hashimoto N, Tsuboi A, et al. Prognostic value of WT1 protein expression level and MIB-1 staining index as predictor of response to WT1 immunotherapy in glioblastoma patients. Brain Tumor Pathol. 2010;27(1):29–34. doi: 10.1007/s10014-010-0265-9. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto N, Tsuboi A, Kagawa N, et al. Wilms tumor 1 peptide vaccination combined with temozolomide against newly diagnosed glioblastoma: safety and impact on immunological response. Cancer Immunol. Immunother. 2015;64(6):707–716. doi: 10.1007/s00262-015-1674-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oka Y, Tsuboi A, Taguchi T, et al. Induction of WT1 (Wilms’ tumor gene)-specific cytotoxic T lymphocytes by WT1 peptide vaccine and the resultant cancer regression. Proc. Natl Acad. Sci. USA. 2004;101(38):13885–13890. doi: 10.1073/pnas.0405884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Tendeloo VF, Van de Velde A, Van Driessche A, et al. Induction of complete and molecular remissions in acute myeloid leukemia by Wilms’ tumor 1 antigen-targeted dendritic cell vaccination. Proc. Natl Acad. Sci. USA. 2010;107(31):13824–13829. doi: 10.1073/pnas.1008051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hashii Y, Sato-Miyashita E, Matsumura R, et al. WT1 peptide vaccination following allogeneic stem cell transplantation in pediatric leukemic patients with high risk for relapse: successful maintenance of durable remission. Leukemia. 2012;26(3):530–532. doi: 10.1038/leu.2011.226. [DOI] [PubMed] [Google Scholar]
- 18.Tsuboi A, Oka Y, Kyo T, et al. Long-term WT1 peptide vaccination for patients with acute myeloid leukemia with minimal residual disease. Leukemia. 2012;26(6):1410–1413. doi: 10.1038/leu.2011.343. [DOI] [PubMed] [Google Scholar]
- 19.Oka Y, Tsuboi A, Oji Y, Kawase I, Sugiyama H. WT1 peptide vaccine for the treatment of cancer. Curr. Opin. Immunol. 2008;20(2):211–220. doi: 10.1016/j.coi.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Narita M, Masuko M, Kurasaki T, et al. WT1 peptide vaccination in combination with imatinib therapy for a patient with CML in the chronic phase. Int. J. Med. Sci. 2010;7(2):72–81. doi: 10.7150/ijms.7.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oji Y, Oka Y, Nishida S, et al. WT1 peptide vaccine induces reduction in minimal residual disease in an Imatinib-treated CML patient. Eur. J. Haematol. 2010;85(4):358–360. doi: 10.1111/j.1600-0609.2010.01497.x. [DOI] [PubMed] [Google Scholar]
- 22.Kawakami M, Oka Y, Tsuboi A, et al. Clinical and immunologic responses to very low-dose vaccination with WT1 peptide (5 microg/body) in a patient with chronic myelomonocytic leukemia. Int. J. Hematol. 2007;85(5):426–429. doi: 10.1532/IJH97.06194. [DOI] [PubMed] [Google Scholar]
- 23.Ohno S, Kyo S, Myojo S, et al. Wilms’ tumor 1 (WT1) peptide immunotherapy for gynecological malignancy. Anticancer Res. 2009;29(11):4779–4784. [PubMed] [Google Scholar]
- 24.Oka Y, Tsuboi A, Murakami M, et al. Wilms tumor gene peptide-based immunotherapy for patients with overt leukemia from myelodysplastic syndrome (MDS) or MDS with myelofibrosis. Int. J. Hematol. 2003;78(1):56–61. doi: 10.1007/BF02983241. [DOI] [PubMed] [Google Scholar]
- 25.Tsuboi A, Oka Y, Nakajima H, et al. Wilms tumor gene WT1 peptide-based immunotherapy induced a minimal response in a patient with advanced therapy-resistant multiple myeloma. Int. J. Hematol. 2007;86(5):414–417. doi: 10.1007/BF02983998. [DOI] [PubMed] [Google Scholar]
- 26.Nishioka M, Tanemura A, Nishida S, et al. Vaccination with WT-1 (Wilms’ tumor gene-1) peptide and BCG-CWS in melanoma. Eur. J. Dermatol. 2012;22(2):258–259. doi: 10.1684/ejd.2011.1619. [DOI] [PubMed] [Google Scholar]
- 27.Ohta H, Hashii Y, Yoneda A, et al. WT1 (Wilms tumor 1) peptide immunotherapy for childhood rhabdomyosarcoma: a case report. Pediatr. Hematol. Oncol. 2009;26(1):74–83. doi: 10.1080/08880010802435500. [DOI] [PubMed] [Google Scholar]
- 28.Tsuboi A, Oka Y, Osaki T, et al. WT1 peptide-based immunotherapy for patients with lung cancer: report of two cases. Microbiol. Immunol. 2004;48(3):175–184. doi: 10.1111/j.1348-0421.2004.tb03503.x. [DOI] [PubMed] [Google Scholar]
- 29.Takahara A, Koido S, Ito M, et al. Gemcitabine enhances Wilms’ tumor gene WT1 expression and sensitizes human pancreatic cancer cells with WT1-specific T-cell-mediated antitumor immune response. Cancer Immunol. Immunother. 2011;60(9):1289–1297. doi: 10.1007/s00262-011-1033-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishida S, Koido S, Takeda Y, et al. Wilms tumor gene (WT1) peptide-based cancer vaccine combined with gemcitabine for patients with advanced pancreatic cancer. J. Immunother. 2014;37(2):105–114. doi: 10.1097/CJI.0000000000000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dohi S, Ohno S, Ohno Y, et al. WT1 peptide vaccine stabilized intractable ovarian cancer patient for one year: a case report. Anticancer Res. 2011;31(7):2441–2445. [PubMed] [Google Scholar]
- 32.Miyatake T, Ueda Y, Morimoto A, et al. WT1 peptide immunotherapy for gynecologic malignancies resistant to conventional therapies: a Phase II trial. J. Cancer Res. Clin. Oncol. 2013;139(3):457–463. doi: 10.1007/s00432-012-1348-2. [DOI] [PubMed] [Google Scholar]
- 33.Ohno S, Dohi S, Ohno Y, et al. Immunohistochemical detection of WT1 protein in endometrial cancer. Anticancer Res. 2009;29(5):1691–1695. [PubMed] [Google Scholar]
- 34.Sasabe E, Hamada F, Iiyama T, Udaka K, Sugiyama H, Yamamoto T. Wilm's tumor gene WT1 peptide immunotherapy for pulmonary metastasis from adenoid cystic carcinoma of the salivary gland. Oral Oncol. 2011;47(1):77–78. doi: 10.1016/j.oraloncology.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 35.Shirakata T, Oka Y, Nishida S, et al. WT1 peptide therapy for a patient with chemotherapy-resistant salivary gland cancer. Anticancer Res. 2012;32(3):1081–1085. [PubMed] [Google Scholar]
- 36.Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16(7):896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogi C, Aruga A. New concepts of biomarkers and clinical outcomes for therapeutic cancer vaccines in clinical trials. Immunotherapy. 2014;6(10):1025–1036. doi: 10.2217/imt.14.74. [DOI] [PubMed] [Google Scholar]
- 38.Shippy R, Fulmer-Smentek S, Jensen RV, et al. Using RNA sample titrations to assess microarray platform performance and normalization techniques. Nat. Biotechnol. 2006;24(9):1123–1131. doi: 10.1038/nbt1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat. Rev. Genet. 2006;7(1):55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 40.Shi L, Reid LH, Jones WD, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 2006;24(9):1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.NCBI nucleotide database. http://www.ncbi.nlm.nih.gov/nuccore
- 42.The R Project for Statistical Computing. www.r-project.org
- 43.Chung JS, Dougherty I, Cruz PD, Jr, Ariizumi K. Syndecan-4 mediates the coinhibitory function of DC-HIL on T cell activation. J. Immunol. 2007;179(9):5778–5784. doi: 10.4049/jimmunol.179.9.5778. [DOI] [PubMed] [Google Scholar]
- 44.Chung JS, Sato K, Dougherty Ii, Cruz PD, Jr, Ariizumi K. DC-HIL is a negative regulator of T lymphocyte activation. Blood. 2007;109(10):4320–4327. doi: 10.1182/blood-2006-11-053769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu X, Harden K, Gonzalez LC, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat. Immunol. 2009;10(1):48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- 46.Teixe T, Nieto-Blanco P, Vilella R, Engel P, Reina M, Espel E. Syndecan-2 and -4 expressed on activated primary human CD4+ lymphocytes can regulate T cell activation. Mol. Immunol. 2008;45(10):2905–2919. doi: 10.1016/j.molimm.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 47.Okuyama E, Suzuki A, Murata M, et al. Molecular mechanisms of syndecan-4 upregulation by TNF-alpha in the endothelium-like EAhy926 cells. J. Biochem. 2013;154(1):41–50. doi: 10.1093/jb/mvt024. [DOI] [PubMed] [Google Scholar]
- 48.Chung JS, Tomihari M, Tamura K, Kojima T, Cruz PD, Jr, Ariizumi K. The DC-HIL ligand syndecan-4 is a negative regulator of T-cell allo-reactivity responsible for graft-versus-host disease. Immunology. 2013;138(2):173–182. doi: 10.1111/imm.12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung JS, Yudate T, Tomihari M, Akiyoshi H, Cruz PD, Jr, Ariizumi K. Binding of DC-HIL to dermatophytic fungi induces tyrosine phosphorylation and potentiates antigen presenting cell function. J. Immunol. 2009;183(8):5190–5198. doi: 10.4049/jimmunol.0901319. [DOI] [PubMed] [Google Scholar]
- 50.Chung JS, Tamura K, Akiyoshi H, Cruz PD, Jr, Ariizumi K. The DC-HIL/syndecan-4 pathway regulates autoimmune responses through myeloid-derived suppressor cells. J. Immunol. 2014;192(6):2576–2584. doi: 10.4049/jimmunol.1301857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishiguro K, Kadomatsu K, Kojima T, et al. Syndecan-4 deficiency leads to high mortality of lipopolysaccharide-injected mice. J. Biol. Chem. 2001;276(50):47483–47488. doi: 10.1074/jbc.M106268200. [DOI] [PubMed] [Google Scholar]
- 52.Tanino Y, Chang MY, Wang X, et al. Syndecan-4 regulates early neutrophil migration and pulmonary inflammation in response to lipopolysaccharide. Am. J. Respir. Cell Mol. Biol. 2012;47(2):196–202. doi: 10.1165/rcmb.2011-0294OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith MF, Jr, Novotny J, Carl VS, Comeau LD. Helicobacter pylori and toll-like receptor agonists induce syndecan-4 expression in an NF-kappaB-dependent manner. Glycobiology. 2006;16(3):221–229. doi: 10.1093/glycob/cwj061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boyanovsky BB, Shridas P, Simons M, Van der Westhuyzen DR, Webb NR. Syndecan-4 mediates macrophage uptake of group V secretory phospholipase A2-modified LDL. J. Lipid Res. 2009;50(4):641–650. doi: 10.1194/jlr.M800450-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li J, Brown LF, Laham RJ, Volk R, Simons M. Macrophage-dependent regulation of syndecan gene expression. Circ. Res. 1997;81(5):785–796. doi: 10.1161/01.res.81.5.785. [DOI] [PubMed] [Google Scholar]
- 56.Komatsu N, Matsueda S, Tashiro K, et al. Gene expression profiles in peripheral blood as a biomarker in cancer patients receiving peptide vaccination. Cancer. 2012;118(12):3208–3221. doi: 10.1002/cncr.26636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.