Abstract
The global physiological effects of glucocorticoids are well established, and the framework of transcriptional regulation by the glucocorticoid receptor (GR) has been described. However, the genes directly under GR control that trigger these physiological effects are largely unknown. To address this issue in a single cell type, we identified glucocorticoid-responsive genes in A549 human lung adenocarcinoma cells by microarray analysis and quantitative real-time PCR. Reduction of GR expression by RNA interference diminished the effects of dexamethasone on all tested target genes, thus confirming the essential role of GR in glucocorticoid-regulated gene expression. To identify primary GR target genes, in which GR is a component of the transciptional regulatory complex, we developed a strategy that uses chromatin immunoprecipitation to scan putative regulatory regions of target genes for sites occupied by specifically bound GR. We screened 11 glucocorticoid-regulated genes, and we identified GR-binding regions for eight of them (five induced and three repressed). Thus, our approach provides a means for rapid identification of primary GR target genes and glucocorticoid-response elements, which will facilitate analyses of transcriptional regulatory mechanisms and determination of hormone-regulated gene networks.
Glucocorticoids are steroid hormones that influence cellular differentiation and developmental processes, as well as many aspects of mammalian physiology. Glucocorticoids exert their actions by means of the intracellular glucocorticoid receptor (GR), which associates with genomic glucocorticoid-response elements (GREs) upon binding to hormones (1, 2). The bound receptor functions within multicomponent transcriptional regulatory complexes, of which the composition and function are determined, at least in part, by the structure of the bound ligand, the sequence and organization of the response element, and the “menu” of available cofactors within the responsive cell. Even within a single cell type, GR can activate some genes and represses others, underscoring the importance of response elements as determinants of regulatory complex activity (1, 2).
Notably, most studies of GR-regulatory mechanisms have used synthetic reporter constructs containing multimerized idealized GREs. These systems do not accurately represent bona fide GREs at chromosomal genes, which are typically “composite elements” that recruit GRs together with other regulators and cofactors to form large, multicomponent regulatory complexes (3, 4). Within a given cell type or tissue, the physiological response to glucocorticoids is composed of a network of responsive genes, each of which is controlled by a gene-specific regulatory complex. Although hormonal ligands and genomic response elements are clearly involved in specifying the composition and function of these complexes, the details, or “rules,” that govern this important combinatorial process have not been determined.
To approach these issues, we need to identify within a single cell type a set of “primary” GR target genes. Here, we define genes at which GR occupies the GREs either by direct DNA binding or by means of protein–protein interactions with bound factors. Comparison of different regulatory complexes within that cell-specific set of genes could then reveal the determinants of specificity. Because glucocorticoids alter the expression of both primary and secondary target genes (which are modulated by the product of a primary target gene rather than by GR), an initial challenge is to distinguish between these two classes. Thus, it is essential to detect multiple primary GR targets as a first step in assessing the composition and activity of regulatory complexes assembled at different response elements.
Here, we describe a straightforward and systematic strategy to identify primary GR target genes in any cell type. In this study, we used A549 lung adenocarcinoma cells, which express functional GR and display characteristics of type II alveolar epithelial cells (5, 6). Notably, glucocorticoids play important roles in lung development and physiology (7, 8, 9) and are commonly used to treat respiratory diseases, such as asthma and chronic lung diseases, because of their potent immunosuppressive and antiinflammatory activities (10, 11). Our experimental approach was to use DNA microarrays to identify genes whose expression is modulated by glucocorticoid treatment and then to develop an efficient strategy for defining the subset of those genes that are primary targets of GR.
Materials and Methods
Microarray and Quantitative Real-Time PCR (qPCR). Methods of microarray and qPCR have been described (12). GenBank accession numbers and gene information can be found in the Source database (available at http://genome-www5.stanford.edu/cgi-bin/source/sourcesearch). All primer sequences used in this article are available by request.
RNA Interference (RNAi). The GR cDNA sequences corresponding to nucleotides 804–1,789 and the full-length enhanced GFP (EGFP) cDNA sequences were amplified by PCR and subcloned into pBluescript II KS(+) (Stratagene). The preparation of double-stranded RNA was as described in ref. 13. Small interfering RNA (siRNA) was produced as described by Yang et al. (14). The siRNA was transfected into A549 cells (in a six-well plate) by using Oligofectamin (Invitrogen) according to the manufacturer's technical manual. Cells were transfected again 24 h after the first transfection. After an additional 16–20 h, cells were treated with ethanol or dexamethasone for 4–5 h. Cells were then harvested for total RNA preparation or immunoblotting. Total RNA was prepared by using the RNeasy kit (Qiagen, Valencia, CA).
Chromatin Immunoprecipitation (ChIP). ChIP was performed as described in ref. 15, with a few modifications. First, A549 cells were cross-linked at room temperature for 10 min. Second, N499 antibody raised against GR was added to cleared chromatin extract and incubated with rotation at 4°C overnight. Last, qPCR was used to monitor ChIP results. qPCRs (35 μl) contained 1.25 units of Taq enzyme (Promega), 1× reaction buffer, 1.5 mM MgCl2, 0.5 mM dNTP (Invitrogen), 0.2× SYBR green I dye (Molecular Probes), 357 nM primer (each), and an appropriate amount of DNA. Typically, cells collected from three 150-mm plates (5 × 108 cells) could be used for 40–50 qPCRs.
Cell Lines, Plasmids, and Transfections. GR-binding regions identified from ChIP scanning were amplified by PCR. The PCR fragments were digested by appropriate restriction enzymes and subcloned into a pGL3-Basic reporter plasmid, which lacks any promoter sequences, or pGL3-Promoter, which contains SV40 TATA sequences (Promega). GR-binding regions located within the first 500 bp upstream of the transcription start site were inserted into pGL3-Basic, whereas GR-binding regions located beyond the proximal 500 bp of the transcription start site were inserted into pGL3-Promoter. QuikChange mutagenesis kit was used to make site-directed mutations according to the manufacturer's technical manual (Promega).
The culturing of A549 cells, transfection, and assay for β-galactosidase and luciferase activity were performed as described in ref. 13.
Results and Discussion
Identification of Glucocorticoid-Responsive Genes in A549 Human Lung Adenocarcinoma Cells. Total RNA of A549 cells that were treated with dexamethasone or ethanol (as a vehicle control) for 6 h was isolated for comparative expression surveys by using DNA microarrays that represent 29,778 independent human genes (12). Cycloheximide, an inhibitor of de novo protein synthesis, was added to minimize potential secondary effects and focus mainly on primary glucocorticoid target genes. Overall, 108 genes were induced, whereas 73 genes were repressed >2-fold in these microarray experiments. We then used qPCR to confirm the regulation of glucocorticoid target genes of which the full-length cDNA sequences are available. As shown in Table 1, we confirmed 50 glucocorticoid-induced and 21 glucocorticoid-repressed genes. The regulation of these target genes at 2, 6, and 24 h after induction is shown in Tables 2 and 3, which are published as supporting information on the PNAS web site.
Table 1. Glucocorticoid-regulated genes.
| Inflammation | Growth and apoptosis | Signal transduction | Metabolism | Transport | Other and unknown functions |
|---|---|---|---|---|---|
| GR-activated gene | |||||
| CCL20 (AI285199) | GADD45B (AI184305) | AKAP13 (T50096) | ANGPTL4* (T54298) | ENaCa* (AA459197) | ABHD2* (AA676225) |
| GILZ* (AA775091) | HIAP1* (AA001536) | ANKRD1 (AA488072) | B3GNT5 (AA043551) | MT-1I* (N80129) | CTEN* (AA443948) |
| FLAP* (T49652) | Kip2/p57* (AI676118) | CDC42EP3* (AA708976) | EKI2* (H77535) | SLC19A2* (AI468752) | FLJ11127* (T98201) |
| THBD* (AA256378) | MFGE8 (AA448941) | CPEB4 (N47682) | MGAM (AA894763) | SLC26A2 (AA704222) | FLJ20371 (AA682445) |
| S100P* (AA775091) | DNER (H28681) | Stomatin* (R62868) | GPR115 (AA126828) | ||
| SLUG* (H57309) | EHM2 (H94262) | GPR153 (AA777493) | |||
| hSPRY1 (AA055440) | ET-2* (AI140863) | LOC144100 (R44496) | |||
| TNFAIP3 (AA476272) | FKBP5* (W86653) | LRRC8* (AA043409) | |||
| FGD4* (AI222351) | MCJ* (AI005521) | ||||
| IHPK3* (N52903) | PPG/Serglycin* (AA278759) | ||||
| IRS2 (AA134862) | SDPR* (R09729) | ||||
| POU5F1* (AA996055) | SPINK5L3 (AA461492) | ||||
| PPP1R14C* (R43270) | TMG4 (R53734) | ||||
| RGS2* (AI675670) | |||||
| SEC14L1 (AI366724) | |||||
| TGFBR3 (N26658) | |||||
| GR-repressed gene | |||||
| COX-2* (R80217) | Cullin 1 (AA486790) | ARL8 (AA621363) | AMIGO2 (N22620) | ||
| PDE4B/2* (AA453293) | CAP3/IP9* (AA430512) | BHLHB2* (T62084) | CG1/XP28 (R80217) | ||
| CCL2* (AA425102) | FGFBP1 (AA936757) | ENC1* (H72112) | KIAA1376 (R33609) | ||
| IL-11* (AI148233) | TRIP-Br2* (AA489839) | GEM* (AA418017) | FLJ22761 (AA702464) | ||
| RDC1 (N53172) | NAV3* (AA705735) | ||||
| SNK* (AA460152) | PMP2 (R26960) | ||||
| ZIC2 (H40921) | |||||
GenBank accession numbers are given in parentheses for all genes.
Gene induction or repression is inhibited by GR RNAi. The remaining genes have not been tested
Among these 71 confirmed genes, some genes [such as ENaCα (16), FKBP5 (17), FLAP (18), and SNK (19)] have been reported to be glucocorticoid-responsive in A549 or other cell types, whereas others appeared to be newly identified targets. Overall, our results revealed several important aspects of glucocorticoid actions in A549 cells. First, we identified genes that are likely to mediate the antiproliferative, antiapoptotic, and antiinflammatory effects of glucocorticoids (Table 1). Furthermore, in addition to ENaCα, we detected another glucocorticoid-induced gene, stomatin, which may be involved in the regulation of sodium transport (20). The Caenorhabditis elegans homolog of stomatin (mec-2) acts in the same genetic pathway as two C. elegans homologs of ENaC (mec-4 and mec-10) (21), and MEC-2 protein has been shown to potentiate the transport activities of MEC-4 and MEC-10 in Xenopus oocytes (22). Finally, glucocorticoids also regulated a member of the choline/ethanolamine kinase family, EKI2 (23), which may be involved in phospholipid biosynthesis required for surfactant production. Thus, in A549 cells, it appears that elements of at least five glucocorticoid-regulated gene networks, including growth inhibition, antiapoptosis, antiinflammation, sodium transport, and surfactant production, may be operative in a single cell context. Interestingly, glucocorticoids appeared to regulate two genes in a counterintuitive manner; FLAP, which is a 5-lipoxygenase-activating protein that is involved in the production of inflammatory mediators leukotrienes (24), and CCL20, which is a chemokine (25), were glucocorticoid-induced rather than glucocorticoid-repressed. The physiological significance of these observations has not yet been examined. Notably, we also uncovered genes whose regulation by glucocorticoids had not, to our knowledge, been noted, and ≈15% of the primary GR target genes were of unknown function (Table 1).
GR Is Essential for the Regulation of Target-Gene Expression by Glucocorticoids. To assess the role of GR in the regulation of target-gene expression by glucocorticoids, we reduced the expression of GR by RNAi in A549 cells and then tested whether identified target genes remained glucocorticoid-responsive. As shown in Fig. 1A, the transfection of GR siRNA into A549 cells reduced the expression of GR protein profoundly, whereas the expression of actin was not affected. As a negative control, transfection of EGFP siRNAs into A549 cells did not influence the expression of GR (Fig. 1 A). We isolated RNA from cells transfected with either EGFP or GR siRNAs and treated with either ethanol or dexamethasone. We then compared the regulation of target-gene expression by glucocorticoids in EGFP and GR siRNA transfected cells by using qPCR. As shown in Fig. 1 B and C, GR RNAi dramatically compromised glucocorticoid responses of four (two glucocorticoid-induced and two glucocorticoid-repressed) representative glucocorticoid target genes. Overall, the glucocorticoid-regulated expression of all of the 42 confirmed target genes that we analyzed was reduced severely in GR siRNA transfected cells (Table 1), confirming the essential role of GR in glucocorticoid target-gene expression.
Fig. 1.
The effect of GR RNAi on the expression of GR target genes. (A) GR RNAi reduced GR expression in A549 cells. A549 cells were transfected with GR siRNA (1.5 μg) for 48 h. Cells were then collected and lysed. GR and actin were detected by immunoblotting with N499 and anti-actin antibody (Sigma). (B) GR RNAi reduced the induction of EKI2 and GILZ gene expression by dexamethasone. (C) GR RNAi compromised the repression of GEM and SNK gene expression by dexamethasone. Cells were transfected as described for A and treated with dexamethasone or ethanol for 4–5 h. RNA was isolated and the expression of GILZ, EKI2, SNK, and GEM was determined by qPCR. Data show a representative result from at least two independent RNAi experiments.
Using a “ChIP Scanning Assay” to Identify GREs in Glucocorticoid Target Genes. An essential step in understanding the combinatorial specificity of glucocorticoid-regulated gene transcription in vivo is the identification of multiple GREs within a single cell type that control a set of primary GR target genes. Searching for these elements by scanning for consensus or idealized binding sequences is insufficient because bona fide genomic GR-binding sites have been shown to display considerable sequence variability, and GR associates with some GREs by protein–protein interactions with non-GR DNA-bound factors rather than by direct DNA binding. Therefore, identifying primary GR target genes requires demonstration of GR occupancy and function at GREs.
To identify genomic segments occupied in vivo by GR, we developed a ChIP scanning assay that can survey ≈3 kb of DNA upstream of glucocorticoid-responsive genes rapidly and easily. In this method, we followed a standard ChIP procedure (15) of formaldehyde cross-linking of A549 cells, shearing of chromatin to 300- to 800-bp fragments, and precipitating cross-linked GR–DNA complexes with GR-specific antibody. We then performed qPCR on the precipitated DNA fragments by using for each gene six pairs of oligonucleotide primers that would amplify ≈100-bp segments. Each fragment was separated by ≈400 bp upstream of a GR-responsive gene (relative to the transcription start site) and, thus, spanned ≈2.5 kb. Given the average size of the sheared DNA fragments (≈550 bp), this arrangement of amplified sequences will scan effectively for GR occupancy in a ≈3 kb segment of upstream DNA. Because we routinely carry out qPCR in a 96-well-plate format, we can scan seven genes together with a set of standards in a single-plate assay. The assay should be readily adaptable to a 384-well-plate format, which would permit scanning of 31 genes per plate. Clearly, this procedure would miss genes with GREs outside of the 3-kb surveyed segment, but the method is highly efficient, and many regulatory elements have been shown to reside within the targeted region.
Primary GR-Induced Genes. Using this method, we scanned for GREs in eight glucocorticoid-induced and three glucocorticoid-repressed genes. As shown in Fig. 2, GR-binding regions were found in the following five glucocorticoid-induced genes: GILZ, THBD, SDPR, SLC19A2, and PPG. However, we did not detect significant GR occupancy close to the following three other glucocorticoid-activated genes: ANGPTL4, EKI2, and IHPK3. Conceivably, the GREs for these genes reside outside of the 3 kb of promoter-proximal DNA, or they may be secondary rather than primary target gene. Alternatively, the epitope detected by our GR antibody may be unavailable in the regulatory complexes assembled at these three GREs. To test whether the identified GR-binding regions are glucocorticoid-responsive, we inserted each of them into reporter constructs encoding firefly luciferase. The average amplified fragment size of qPCR is ≈100 bp; however, because sheared ChIP fragments in our experiments are 300–800 bp, the PCR fragments do not necessarily include GREs. Therefore, we subcloned segments that extend ≈300 bp upstream and downstream of each ChIP fragment into reporter plasmids. For GILZ, we subcloned a 1-kb fragment that contains 300 bp upstream from nucleotide -2,220 and 300 bp downstream from nucleotide -1,794 into a reporter (Fig. 3, pGILZ1) because two ChIP fragments that showed strong GR binding are within 500 bp (Fig. 2 A, amplified nucleotides -2,220 to -2,133 and -1,919 to -1,794, relative to the predicted transcription-initiation site).
Fig. 2.
Identifying primary GR-regulated genes and mapping their GR-binding regions. A549 cells were treated with ethanol or dexamethasone for 90 min, and ChIP-scanning experiments were performed on GILZ (A), THBD (B), SDPR (C), SLC19A2 (D), PPG (E), BHLHB2 (F), GEM (G), and SNK (H). The PCR primers for ChIP scanning that correspond to distinct regions in each promoter are indicated. The fold enrichment values for the experimental regions were determined by normalizing to the internal control hsp70 gene value. Hatched bars indicate the scanning regions, which were enriched >2-fold. Data represent the SEM of the fold enrichment (dexamethasone-treated cells divided by ethanol-treated cells) from at least three experiments.
Fig. 3.
Analysis of glucocorticoid responses of distinct GR-binding sites of glucocorticoid-induced genes. Reporter plasmids (75 ng) were cotransfected with pcDNA3-hGR (150 ng) and RSV-βGal (100 ng) into A549 cells in a 24-well plate. After 4 h, cells were washed with PBS and treated with 0.1 μM dexamethasone for an additional 16–20 h. Cells were then lysed and assayed for luciferase and β-Gal activities. Data represent the SEM of the fold induction (dexamethasone-treated cells divided by ethanol-treated cells) of luciferase activity from at least three experiments.
As shown in Fig. 3, treatment with dexamethasone induced strong luciferase expression from reporters that contain the GR-binding fragment of GILZ (pGILZ1), SLC19A 2 (pSLC19A2), or the proximal SDPR GR-binding region (pSDPR1). A reporter construct containing the distal SDPR GR-binding region did not confer dexamethasone regulation in A549 cells (Fig. 3, pSDPR2). Overall, these results confirm that GR-binding regions identified for GILZ, SDPR, and SLC19A2 contain functional GREs.
To localize precisely the GRE sequences within the fragments implicated by ChIP scanning and functional assays, we identified and mutated sequences that contain at least six of eight conserved nucleotides of the consensus GR-binding site (ACANNNTGTTNT) (26). In this article, we focus on the four sites found within the GILZ GR-binding fragment (Fig. 4A). In each case, we mutated the TGT segment because these three positions appear to be especially important for GRE activity (27). Mutating the potential sites one at a time, we found that mutation of three of the four sites resulted in reduced glucocorticoid responses (Fig. 4, pGILZ1.1, pGILZ1.3, and pGILZ1.4) and, therefore, contribute to overall GILZ GRE activity (pGILZ1.5). This analysis demonstrates that ChIP scanning efficiently identifies functional response elements.
Fig. 4.
Mutational analysis of potential GREs in GILZ GR-binding sites. (A) Within a 1-kb fragment encompassing GR-binding and GR-responsive regions (pGILZ1, Figs. 2 A and 3), four potential GREs were revealed by sequence analysis. These GREs were mutated as shown (mutated residues are underlined). (B) The effect of single or combinatorial mutations of potential GREs on glucocorticoid responses. Reporter plasmids (75 ng) were cotransfected with pcDNA3-hGR (150 ng) and RSV-βGal (100 ng) into A549 cells in a 24-well plate. After 24 h, cells were washed with PBS and treated with 0.1 μM dexamethasone for an additional 16–20 h. Cells were then lysed and assayed for luciferase and β-Gal activities. Data represent the SEM of the fold induction (dexamethasone-treated cells divided by ethanol-treated cells) of luciferase activity from at least three experiments.
Interestingly, the GR-binding regions of THBD or PPG promoters did not confer dexamethasone responsiveness when transfected into A549 cells (Fig. 3, pTHBD1, pTHBD2, and pPPG1). One interpretation of these results is that, although these regions are occupied by GR, they may require accessory regulatory regions (for PPG) or the combination of multiple GR-binding sites (for THBD) to mediate glucocorticoid responses. The former scenario would be reminiscent of the GRE that controls rat PEPCK expression, in which the isolated GR-binding sites fail to confer glucocorticoid responsiveness in a reporter assay (28), but hormonal regulation is recovered upon addition of adjacent sequences that do not themselves bind GR (29). Indeed, we found that a fragment that includes additional upstream sequences together with the GR-binding region of PPG (-1,458 to +90) displayed GRE activity (Fig. 3, pPPG2).
Overall, three of five identified primary GR-induced genes (GILZ, SDPR, and THBD) contained multiple GR-binding sites. Interestingly, neither GR-binding site of THBD conferred glucocorticoid responsiveness when tested individually, whereas one of the GR-binding fragments of SDPR was an active GRE, and both GR-binding regions of GILZ were functional. Furthermore, in addition to the GR-binding site, other regulatory regions of PPG were required for hormonal response. Thus, the mechanisms by which glucocorticoids activate these genes may be varied and distinct. Overall, most naturally occurring GREs appear to be composite elements, which typically contain binding sites for multiple DNA-binding regulators.
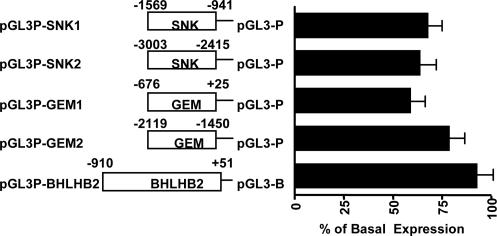
Primary GR-Repressed Genes. We also scanned the putative regulatory regions of three glucocorticoid-repressed genes (SNK, GEM, and BHLHB2) and identified by ChIP at least two GR-occupied sites associated with each gene (Fig. 2 F–H). Various GR-binding fragments from these three genes were used to create five reporter constructs to assay for functional GREs (Fig. 5). Expression from three of these reporters in transfected A549 cells was reduced significantly by treatment with dexamethasone (Fig. 5), including reporters that contained either of the GR-binding regions of SNK (Fig. 5, pGL3P-SNK1 and pGL3P-SNK2), or the proximal GR-binding region of GEM (nucleotides -676 to +25; Fig. 5, pGL3P-GEM1). In contrast, reporters containing the distal GR-binding region of GEM (nucleotides -2,119 to -1,450; Fig. 5, pGL3P-GEM2) or a GR-binding region of BHLHB2 (which contains two GR-binding regions; Fig. 5, pGL3P-BHLHB2) responded only weakly to dexamethasone. Perhaps the glucocorticoid response of BHLHB2 gene requires regulatory regions outside of the GR-binding sites, as with the PPG gene described above. In the case of GEM, the distal GR-binding site (nucleotides -2,119 to -1,450) may play an accessory role for proximal GR-binding site (nucleotides -676 to +25).
Fig. 5.
Analysis of glucocorticoid responses of distinct GR-binding sites of glucocorticoid-repressed genes. Reporter plasmids (75 ng) were cotransfected with pcDNA3-hGR (150 ng) and RSV-βGal (100 ng) into A549 cells in a 24-well plate. After 4 h, cells were washed with PBS and treated with 0.1 μM dexamethasone for an additional 16–20 h. Cells were then lysed and assayed for luciferase and β-Gal activities. Data represent the SEM of the percentage of full luciferase expression (dexamethasone-treated cells divided by ethanol-treated cells) of luciferase activity from at least three experiments.
Overall, our experiments demonstrate that ChIP scanning can efficiently identify small regions that contain active GREs functionally occupied by GR. The basis of this strategy is to screen for sequences occupied by a given regulator adjacent to genes known to be altered in their expression when that regulator is active. An alternate approach, which combines ChIP and DNA microarray analysis (called “ChIP on chip”), first screens for genomic binding sites for a particular regulator and then seeks genes linked to those sites that may be controlled by that regulator (30, 31, 32). Although ChIP on chip is powerful, it is a more complicated procedure, requires whole-genome microarrays, and tests for function at the end of the analysis. In contrast, ChIP scanning begins with function and proceeds to identification of the subset of primary target genes and analyses of regulatory sequences and complexes.
Conclusion
In this article, we identified primary GR target genes, as well as information that is critical for pursuing the physiological and pharmacological effects of glucocorticoids and deciphering fundamental aspects of the evolution and function of gene-regulatory networks. Although an analysis of the gene networks that specify glucocorticoid action in lung is beyond the scope of this study, our results identified a series of genes involved in antiproliferation, antiapoptosis, antiinflammation, and surfactant synthesis, all of which are primary glucocorticoid-controlled processes in lung epithelia. Thus, our findings provide a strong basis for determination of the gene-control circuits in lung that are governed by GR.
Both physiological studies and mechanistic investigations of network functions require that we sort the primary GR target genes from among the full set of responding genes detected on the microarrays. Hence, we developed the ChIP scanning strategy, which efficiently detects and localizes sites of GR occupancy in vivo close to responsive genes. Moreover, the approach should be general: identifying regulated genes, localizing regulator binding regions in vivo, and testing their functionality in cell-based reporter assays.
Supplementary Material
Acknowledgments
We thank Dr. Dun Yang (University of California, San Francisco) for providing an expression vector to purify recombinant GST-RNaseIII, and Drs. Rik Derynck, Brian Black, Eric Bolton, Neal Freedman and Inez Rogatsky for reviewing the manuscript. J.-C.W. was supported by an American Heart Association postdoctoral fellowship. This work was supported by grants from National Institutes of Health.
Author contributions: J.-C.W. and K.R.Y. designed research; J.-C.W., M.K.D., D.N., and D.K. performed research; C.H. contributed new reagents/analytic tools; J.-C.W., M.K.D., D.N., D.K., and K.R.Y. analyzed data; and J.-C.W. and K.R.Y. wrote the paper.
Abbreviations: siRNA, small interfering RNA; RNAi, RNA interference; GR, glucocorticoid receptor; GRE, glucocorticoid-response element; qPCR, quantitative real-time PCR; EGFP, enhanced GFP; ChIP, chromatin immunoprecipitation.
References
- 1.Yamamoto, K. R. (1985) Annu. Rev. Genet. 19, 209-252. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto, K. R. (1995) Harvey Lect. 91, 1-19. [PubMed] [Google Scholar]
- 3.Miner, J. N. & Yamamoto, K. R. (1991) Trends Biochem. Sci. 16, 423-426. [DOI] [PubMed] [Google Scholar]
- 4.Lucas, P. C. & Granner, D. K. (1992) Annu. Rev. Biochem. 61, 1131-1173. [DOI] [PubMed] [Google Scholar]
- 5.Ballard, P. L., Mason, R. J. & Douglas, W. H. (1978) Endocrinology 102, 1570-1575. [DOI] [PubMed] [Google Scholar]
- 6.Shapiro, D. L., Nardone, L. L., Rooney, S. A., Motoyama, E. K. & Munoz, J. L. (1978) Biochim. Biophys. Acta 530, 197-207. [DOI] [PubMed] [Google Scholar]
- 7.Olver, R. E., Walters, D. V. & S, M. W. (2004) Annu. Rev. Physiol. 66, 77-101. [DOI] [PubMed] [Google Scholar]
- 8.Jobe, A. H. & Ikegami, M. (2000) Annu. Rev. Physiol. 62, 825-846. [DOI] [PubMed] [Google Scholar]
- 9.Bolt, R. J., van Weissenbruch, M. M., Lafeber, H. N. & Delemarre-van de Waal, H. A. (2001) Pediatr. Pulmonol. 32, 76-91. [DOI] [PubMed] [Google Scholar]
- 10.Jobe, A. H. (2001) Semin. Neonatol. 6, 331-342. [DOI] [PubMed] [Google Scholar]
- 11.Thompson, B. T. (2003) Crit. Care Med. 31, S253-7. [DOI] [PubMed] [Google Scholar]
- 12.Rogatsky, I., Wang, J. C., Derynck, M. K., Nonaka, D. F., Khodabakhsh, D. B., Haqq, C. M., Darimont, B. D., Garabedian, M. J. & Yamamoto, K. R. (2003) Proc. Natl. Acad. Sci. USA 100, 13845-13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang, J. C., Walker, A., Blackwell, T. K. & Yamamoto, K. R. (2004) J. Biol. Chem. 279, 29270-29277. [DOI] [PubMed] [Google Scholar]
- 14.Yang, D., Buchholz, F., Huang, Z., Goga, A., Chen, C. Y., Brodsky, F. M. & Bishop, J. M. (2002) Proc. Natl. Acad. Sci. USA 99, 9942-9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nissen, R. M. & Yamamoto, K. R. (2000) Genes Dev. 14, 2314-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itani, O. A., Auerbach, S. D., Husted, R. F., Volk, K. A., Ageloff, S., Knepper, M. A., Stokes, J. B. & Thomas, C. P. (2002) Am. J. Physiol. 282, L631-L641. [DOI] [PubMed] [Google Scholar]
- 17.Vermeer, H., Hendriks-Stegeman, B. I., van der Burg, B., van Buul-Offers, S. C. & Jansen, M. (2003) J. Clin. Endocrinol. Metab. 88, 277-284. [DOI] [PubMed] [Google Scholar]
- 18.Uz, T., Dwivedi, Y., Savani, P. D., Impagnatiello, F., Pandey, G. & Manev, H. (1999) J. Neurochem. 73, 693-699. [DOI] [PubMed] [Google Scholar]
- 19.Simmons, D. L., Neel, B. G., Stevens, R., Evett, G. & Erikson, R. L. (1992) Mol. Cell. Biol. 12, 4164-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mannsfeldt, A. G., Carroll, P., Stucky, C. L. & Lewin, G. R. (1999) Mol. Cell. Neurosci. 13, 391-404. [DOI] [PubMed] [Google Scholar]
- 21.Huang, M., Gu, G., Ferguson, E. L. & Chalfie, M. (1995) Nature 378, 292-295. [DOI] [PubMed] [Google Scholar]
- 22.Goodman, M. B., Ernstrom, G. G., Chelur, D. S., O'Hagan, R., Yao, C. A. & Chalfie, M. (2002) Nature 415, 1039-1042. [DOI] [PubMed] [Google Scholar]
- 23.Lykidis, A., Wang, J., Karim, M. A. & Jackowski, S. (2001) J. Biol. Chem. 276, 2174-2179. [DOI] [PubMed] [Google Scholar]
- 24.Peters-Golden, M. & Brock, T. G. (2003) Prostaglandins Leukotrienes Essent. Fatty Acids 69, 99-109. [DOI] [PubMed] [Google Scholar]
- 25.Schutyser, E., Struyf, S. & Van Damme, J. (2003) Cytokine Growth Factor Rev. 14, 409-426. [DOI] [PubMed] [Google Scholar]
- 26.Chen, L., Finnerty, C., Gustafson, W. C., Bush, C. R., Chi, P., Guo, H., Luxon, B., Fields, A. P. & Thompson, E. A. (2003) Recent Prog. Horm. Res. 58, 155-174. [DOI] [PubMed] [Google Scholar]
- 27.Nordeen, S. K., Suh, B. J., Kuhnel, B. & Hutchison, C. D. (1990) Mol. Endocrinol. 4, 1866-1873. [DOI] [PubMed] [Google Scholar]
- 28.Scott, D. K., Stromstedt, P. E., Wang, J. C. & Granner, D. K. (1998) Mol. Endocrinol. 12, 482-491. [DOI] [PubMed] [Google Scholar]
- 29.Imai, E., Stromstedt, P. E., Quinn, P. G., Carlstedt-Duke, J., Gustafsson, J. A. & Granner, D. K. (1990) Mol. Cell. Biol. 10, 4712-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren, B., Cam, H., Takahashi, Y., Volkert, T., Terragni, J., Young, R. A. & Dynlacht, B. D. (2002) Genes Dev. 16, 245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinmann, A. S., Yan, P. S., Oberley, M. J., Huang, T. H. & Farnham, P. J. (2002) Genes Dev. 16, 235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buck, M. J. & Lieb, J. D. (2004) Genomics 83, 349-360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.