Abstract
Elucidating the mechanism of over and under expression of proteins is critical in developing a better understanding of cancer. Multiple techniques are used to examine differential expression of proteins in cells and assess changes in protein expression in response to therapies such as radiation. Reduced expression can be caused by protein inactivation, mRNA instability, or reduced transcription. The following protocol was used to determine the mechanism for the reduced expression of an antiapoptotic factor, survivin, in normal tissues in response to radiation and the defect in cancer cells that prevents this reduction. We also examined ways to overcome survivin over expression in cancer cells in order to sensitize them to radiation. We will focus on the use of antisense oligonucleotides, cell cycle analysis, and luciferase reporter genes.
Keywords: Survivin, Antisense oligonucleotides, Luciferase
Introduction
Regulators of cell death are commonly mutated in numerous cancers. Survivin, a member of the inhibitors of apoptosis family (IAP), is known to decrease apoptosis when it is over expressed in cells (1). Survivin is known to be over expressed in many cancer cell lines (2) and is associated with decreased survival, increased radiation resistance, and increased recurrence (3). Hence, we were interested in survivin as a therapeutic target for radiosensitization of H460 lung cancer cells. Survivin provides an attractive target for cancer therapy because it is not expressed in most terminally differentiated tissue but highly expressed in several cancers (3, 4). In recent studies it has been found that not only is it important to determine the presence of a protein in tumors, but also the extent to which it has been over or under expressed (5). We therefore sought to determine survivin levels and found that survivin decreased in Human Umbilical Vein Endothelial Cells (HUVEC) in response to radiation but there was no such decrease in survivin in several cancer cell lines treated with radiation. Results showed that decreased survivin in HUVEC was caused by a non-p53 dependent suppression of survivin mRNA transcription. Increased survivin caused radioresistance, but this resistance could be attenuated with inhibition of survivin.
To determine the mechanism of action for this radioresistance in tumor cells we employ several methods, but we will focus on the use of luciferase reporter genes, flow cytometry quantification of cell cycle phases and antisense oligonucleotide (ASO) inhibition of survivin. Antisense Oligonucleotides (ASO) are a valuable research tool. ASO has been used to inhibit several kinases (1, 6, 7), defective gene products (8), and specific receptors (9). ASO has also shown potential for use as a therapeutic in antiangiogenesis and as a radiosensitizer in cancer (1). ASO are a useful inhibitor of mRNA translation, resulting in the attenuation of the related protein product. There are several ways this can occur, such as inhibition of ribosomal binding, and activation of RNase H. Since ASO are designed to hybridize to specific mRNA sequences, the specificity can be high. Furthermore, rather than just inhibiting a protein the ASO prevents protein formation. After transfection, protein expression can be conveniently quantified using western blot.
Materials and Methods
Cell culture, adenoviral vectors, and chemicals
HUVECs were obtained from Clonetics and were maintained in endothelial basal medium-2 (EBM-2) medium supplemented with endothelial growth medium (EGM-2) MV single aliquots (BioWhittaker). Various cancer cell lines were obtained from American Type Culture Collection and cultured in their required media. Val138 cell (a gift from Dr. Maureen Murphy, Fox Chase Cancer Center, Philadelphia, PA) originates from human lung adenocarcinoma cell line H1299 stably transfected with temperature-sensitive p53 mutant. Val138 cells were cultured in DMEM (DMEM, Invitrogen) plus 10% fetal bovine serum, 100 units/ml penicillin and streptomycin, and 0.8 mg/ml Geneticin. HEK 293 cells (American Type Culture Collection) transfected with pCDNAhis-survivin or pCDNAhis vector were selected in DMEM with 10% FCS and 0.5 mg/ml G418 (Invitrogen). Single cell clones overexpressing survivin or neomycin control were confirmed by immunoblotting. Actinomycin D (Sigma) was used at a final concentration of 5 μg/ml. Irradiation (3 Gy) was given 1 h after the drug was added, by use of a Colbalt-60 radioactive source. Adenoviral vectors overexpressing LacZ and p53 were gifts from Dr. Shuang Huang, The Scripps Research Institute (San Diego, CA).
Western immunoblots
Cells were treated with 3 Gy and various drugs and collected at various time points. The cells were counted and then were washed with ice-cold PBS twice before the addition of lysis buffer (20 nM Tris, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM phenylmethylsulfonyl fluoride, and leupeptin). Protein concentration was quantified by the Bio-Rad method. Equal amounts of protein were loaded into each well and separated by 14% SDS-PAGE gel, followed by transfer onto nitrocellulose membranes. Membranes were blocked by use of 10% nonfat dry milk in PBS for 2 h at room temperature. The blots were then incubated with the rabbit-antihuman [survivin (R&D systems), p53 and phospho-p53-serine 15 (Cell Signal), cleaved caspase 3 (Cell Signal)] antibodies overnight at 4°C. Donkey antirabbit IgG secondary antibody (1:1000; Amersham) was incubated for 1 h at room temperature. Immunoblots were developed by using the enhanced chemiluminescence (ECL) detection system (Amersham) according to the manufacturer’s protocol and autoradiography.
Flow cytometry
Cells were trypsinized, rinsed once, resuspended in PBS, and fixed with ice-cold 70% ethanol for at least 20 min. Fixed cells were then rinsed again with PBS and resuspended in 50 μg/ml propidium iodide (Sigma) with 40 kilounits (KU)/ml of DNase-free RNase (Stratagene, La Jolla, CA). Cells were then run on a Becton Dickinson FACScan flow cytometer, and the percentage of cells in each phase was calculated using ModFit software.
Real-time quantitative reverse transcription-PCR
Total RNA was isolated from cell culture, using a Qiagen RNA extraction kit. After RNA isolation, cDNA was prepared from each sample as described previously (10). Quantification of cDNA and an internal reference gene (β-actin) was conducted using a fluorescence-based real-time detection method [ABI PRISM 7700 Sequence Detection system (TaqMan); Perkin-Elmer Applied Biosystems, Foster City, CA], as described previously (11, 12). All of the quantitative reverse transcription-PCR experiments were performed as triplicates. The PCR mixture consisted of 600 nmol/liter of each primer, 200 nmol/liter probe (sequences used are given below), 5 units of AmpliTaq Gold polymerase, 200 μM each dATP, dCTP, and dGTP, 400 μM dUTP, 5.5 mM MgCl2, and 1X TaqMan buffer A containing a reference dye, to a final volume of 25 μl (all of the reagents were supplied by Perkin-Elmer Applied Biosystems). Cycling conditions were 50°C for 10 s and 95°C for 10 min, followed by 46 cycles at 95°C for 15 s and 60°C for 1 min. Colon, liver, and lung RNAs (all from Stratagene, La Jolla, CA) were used as control calibrators on each plate. Primer and probe sequences of the analyzed genes are as follows:
Survivin
Forward primer: 5'-TGC CCC GAC GTT GCC-3’
Reverse primer: 5’-CAG TTC TTG AAT GTA GAG ATG CGG T-3’
Probe: 6FAM-5’-CCT GGC AGC CCT TTC TCA AGG ACC-3’-TAMRA
β-actin
Forward primer: 5’-TGAGCGCGGCTACAGCTT-3’
Reverse primer: 5’-TCCTTAATGTCACGCACGATTT-3’
Probe: 6FAM-5’-ACCACCACGGCCGAGCGG-3’-TAMRA
COX-2
Forward primer: 5’-GCTCAAACATGATGTTTGCATTC-3’
Reverse primer: 5’-GCTGGCCCTCGCTTATGA-3’
Probe: 6FAM-5’-TGCCCAGCACTTCACGCATCAGTT-3’-TAMRA
Plasmid, AS oligonucleotides, transfection and luciferase assays
Sp1 plasmid (a gift of Dr. M. Murphy from Fox Chase Cancer Center) contains 1.1 kb of the survivin promoter in a Luciferase reporter construct, pCR2.1 vector (Invitrogen). The expression plasmid of mutant p53 was a gift of Dr. Jennifer Pietenpol, Vanderbilt University, Nashville, TN). Sp1 plasmid (2.5 μg) or 2.5 μg of SP1 plus 2.5 μg of p53 mutant plasmid was transiently transfected into 60-70% confluent HUVECs grown in 3.5-cm plates, by a F1 Targetfect (Targeting systems). After 24 h, cells were irradiated with 3 Gy. They were collected at 0, 4, 8, and 16 h after irradiation. Cells were lysed and luciferase assays were performed as the protocol from the manufacturer (Promega) on a Monolight 3010 Luminometer (PharMingen). Luciferase activity was normalized to total protein levels. The entire experiment was carried out in triplets. The luciferase activity was measured from 1.6 μg of protein lysates. Antisense (AS) oligonucleotides were synthesized (Qiagen, Inc) using the following sequences: the survivin AS oligonucleotides: 5’-TGTGC-TATTCTGTGAATT-3’; the mismatch control oligonucleotide: 5’-TAAGC-TGTTCTATGTGTT-3’. The underlined nucleotides are 2’-O-methoxyethyl modified. Subconfluent H460 cells were transfected with either of the oligonucleotides, using Lipofectin with a mixture of Lipofectin (Life Technologies, Inc., Baltimore, MD) and oligonucleotides in Opti-MEM medium (Life Technologies, Inc.) at a ratio of 3 μl Lipofectin/ml medium per 100 nM oligonucleotide. After 4 h of incubation, cells were replaced by the regular complete medium.
3-(4,5-methylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay
Briefly, various types of cells were seeded at a density of 2000-5000 cells/well in 96-well plates, were grown overnight, and were exposed to 3 Gy alone or were transfected by AS oligonucleotides before irradiation. After 48 or 72 h of incubation, 3-(4,5-methylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) was added (50 μg/well) for 4 h. Solubilization of the converted purple formazan dye was accomplished by placing cells in 100 μl of 0.01 N HCl/10% SDS and incubating overnight at 37°C. The reaction product was quantified by absorbance at 570 nm. All of the samples were assayed in triplicate, and data were analyzed by Student’s t test.
Results and Discussion
Survivin expression in HUVEC versus cancer tissue
Initially we found that human cells, such as HUVEC, showed a decrease in suvivin levels in response to radiation, however survivin levels did not decrease in cancer cells treated with radiation as shown by western blot analysis. Reduction of survivin levels in normal HUVEC was found to be due to transcriptional suppression. This was determined by isolating the total cellular RNA from HUVEC treated with 3 Gy of radiation. Real time PCR was then used to quantify survivin mRNA as shown in Figure 1.
Fig. 1. HUVEC levels of survivin after 3 Gy.
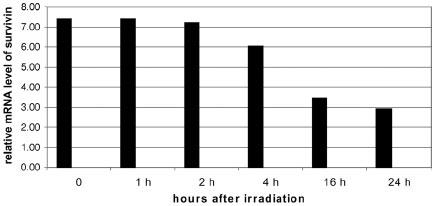
HUVEC were irradiated with 3 Gy and RNA was collected at the indicated time points. RNA was then quantified using Taqman real time PCR.
β-actin mRNA was used as an internal control. However, when colorectal cancer cell lines were tested in a similar fashion, there was no reduction in the transcription of survivin. To determine what caused the reduced transcription of survivin mRNA in HUVEC cells we first examined mitosis rates to see whether the reduced survivin level was secondary to a reduction in the number of mitotic cells. To do this cell-cycle distribution was examined via flow cytometry at various time points in HUVEC after radiation. Results showed no significant variation in the number of cells in G2-M phase post irradiation. Flow cytometry was selected due to rapid quantification of cells in each stage and its effectiveness in previous studies (13) as shown in Figure 2.
Fig. 2. Percentage of HUVEC cells in G-M phase at various time points after radiation.
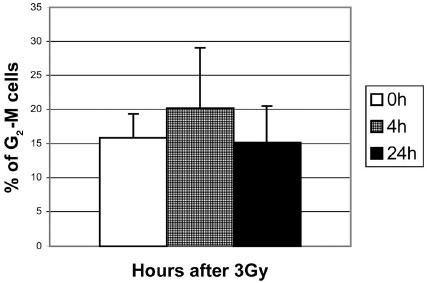
Shown are the averages with the standard deviation based on the results of 3 experiments.
Determining the mechanism of decreased survivin expression
We next examined whether reduced survivin was due to reduced stability of the mRNA secondary to radiation. We treated HUVEC with actinomycin D alone or actinomycin D with 3 Gy given 1 hour later. Neither treatment group showed decreased stability of survivin mRNA. Cox-2 mRNA was used as a control and showed a two-thirds reduction 2 hours after either treatment. These findings suggested that the reduction in survivin expression was not due to degradation of mRNA and likely due to a reduction in transcription. To test this we transfected HUVEC with a luciferase reporter under the control of a 1.1 kb survivin promoter fragment. Hence, transfected cells would only express luciferase if survivin transcription was present after irradiation. Transfected HUVEC showed a significantly reduced luciferase activity after radiation.
Lipofectin and F1 were selected as transfection agents in our experiment due to our previous experience using them. When starting to use a new transfection agent the toxicity and transfection efficiency must be balanced for each cell type. Toxicity is associated with high levels of DNA/RNA to be transfected, large amounts of transfection agent used, and long duration of transfection. Unfortunately, these factors also increase transfection efficiency; hence pilot experiments are needed to balance these two effects. Increasing the number of cells plated may also help with transfection efficiency. We have found that HUVEC and human cell lines in general are more difficult to transfect at an acceptable level without toxicity. Through pilot experiments we found that lipofectin has adequate results in cancer cells but that F1 reagent (Targeting Systems) was most effective for use in HUVEC. We have also found that greatest transfection rates are achieved with the use of Opti-MEM media (Life Technologies Inc.), a serum free media with the minimal requirements for cell survival.
Determining p53’s role in survivin expression
P53 is known to be activated in response to irradiation (14). P53 has also been shown to have activity on survivin levels in response to DNA damage (15). We wanted to examine whether reduced survivin levels in HUVEC occurred in a p53 dependent fashion. P53 levels were examined in irradiated HUVEC via western blot and showed that p53 phosphorylation increased in response to radiation. Total p53 did not change in response to radiation. Survivin level response to increased p53 was tested using an adenovirus to overexpress wild type p53. In HUVEC overexpressing p53 there was a decrease in survivin levels.
To determine how p53 over expression effects survivin levels in cancer cells, we used a lung cancer cell line, Val138, which has temperature dependent p53 mutant activity. Survivin levels were higher when defective mutant p53 was expressed at 39°C, and radiation did not reduce these levels. Furthermore, even when normal p53 was expressed at 32°C there was no decrease in survivin in response to radiation. To confirm this we cotransfected an expression plasmid mutant of p53 with a luciferase reporter activated by the 1.1 kb survivin promoter fragment into HUVEC cells. Survivin promoter was shown to have an increased expression in general with mutant p53 as shown in Figure 3. However, though the survivin promoter had greater activity in p53 mutants this did not prevent radiation induced attenuation of survivin promoter expression.
Fig. 3. Survivin mRNA levels relative to β-actin levels in Val38.
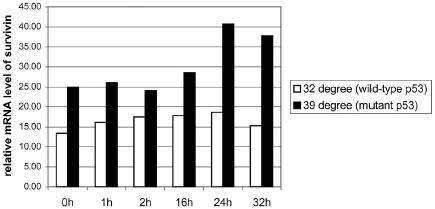
Shown are the relative levels of survivin in reference to β-actin in irradiated Val38 cells at either 32 or 39 degrees.
Survivin’s role in radiosensitization
To determine if survivin over expression affects radiosensitization, cells with normal levels of survivin and cells overexpressing survivin were treated with radiation. Western blot probing for cleaved caspase 3 showed that cells with increased survivin levels had markedly less apoptosis in response to radiation compared with cells which expressed normal levels of survivin.
To determine whether inhibition of survivin over expression could induce a radiosensitive phenotype in cancer cells, an ASO for survivin was used. To control for transfection toxicity, a missense control oligo was used. Again, optimal amounts of DNA, and transfection agent must be determined to minimize toxicity while maintaining adequate levels of transfection for each cell line used. Reduced survivin levels were confirmed with western blot. H460 cells treated with survivin ASO alone or 3 Gy alone showed a 20% decrease in viability but when 3 Gy was given concomitantly with survivin ASO there was a 2-3 fold decrease in cell viability as shown in Figure 4.
Fig. 4. MTT assay of H460 cells treated with Anti-survivin ASO.
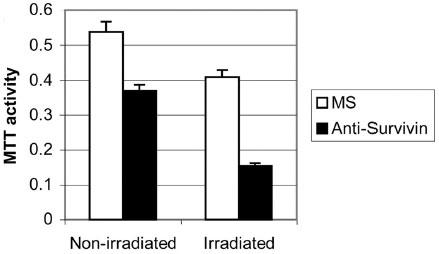
H460 cells were treated with either missense (MS) or anti-survivin ASO and then given 0 or 3 Gy. Shown are the means and standard deviations based on three separate experiments.
Discussion of antisense oligonucleotides
We have used ASO in several experiments to inhibit the expression of proteins (1). An ASO for survivin was chosen because an effective sequence had already been developed. This was done due to the variability in effectiveness of ASO. This occurs because the ASO must interact with an accessible segment of mRNA and because mRNA structure is affected by internal base composition and a number of proteins, this makes it difficult to predict in vivo mRNA structure (16). Hence, we chose a sequence that has been well established previously. Secondly, it can be difficult to get ASO to their target sequence. We used Lipofectin as a transfection agent to overcome problems with poor uptake of ASO into cells and rapid intracellular degradation.
There is great potential for the use of ASO in therapeutics. There have been several studies that have shown that ASO used in research may be useful as therapeutic agents in humans (17) and the FDA has already approved an ASO for therapeutic use (18). ASO are short oligonucleotides (13-25 nucleotides) which are thought to hybridize to a specific mRNA sequence. Pharmacologic inhibitors for survivin have been developed such as the cyclin-dependent kinase inhibitor flavopiridol which inhibits survivin phosphorylation (3). A dominant negative (DN) for survivin can also be used to inhibit its activity (19). ASO was chosen over a pharmacological agent because of the lack of availability for a specific inhibitor of survivin. Compounds such as flavopiridol inhibit survivin activity, but also have effects on numerous other proteins. ASO and DN are often both performed to confirm results of a given study. We choose ASO due to its effectiveness is previous studies.
Acknowledgments
Supported in part by Vanderbilt Discovery Grant and Vanderbilt Physician Scientist Grant. NIH grants R01-CA58508, R01-CA88076, R01-CA89674 and NCI P50-CA90949. Also sponsored in part by training grant T-32 CA93240.
Abbreviations
- HUVEC
human umbilical vein endothelial cells
- ASO
antisense oligonucleotides
- MDM2
Murine Double Minute 2
- PBS
Phosphate Buffer Saline
- IAP
inhibitors of apoptosis family
- 7-AAD
7-Aminoactinomycin D
- DN
Dominant Negative
- MTT
3-(4,5-methylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide
- EBM-2
endothelial basal medium-2
- EGM-2
endothelial growth media
Appendix
Protocols
Flow cytometry
Day 1:
Plate approximately 4 x 105 HUVEC cells into 25-cm2 flasks.
Incubate cells at 37°C overnight.
Day2:
Treat HUVEC with 3 Gy and collect cells at desired time points (0, 4 and 24 hours in this experiment).
Collect cells for analysis by washing cells with PBS 2 times and trypsinize with 0.5 ml of trypsin per flask.
After 5 minutes add 3 ml of PBS to cells and pipette mix into 5 ml Falcon tubes.
Spin cells for 5 min at 2000 rpm. Gently suction out supernatant. This step removes trypsin.
Resuspend cells in 5 ml of ice cold Ethanol (70%) for 20 minutes to fix cells.
Centrifuged cells at 2000 rpm for 5 minutes and carefully suction out supernatant.
Resuspend cells in PBS containing 50 μg/ml of propridium iodide (Sigma) with 40 kilounits (KU/ml) of DNase-free RNase (Strategene, La Jolle, CA).
Run cells on a Becton Dickinson FACScan flowcytometer and use ModFit software to calculate the percentage of cells in each phase of the cell cycle.
*Note: Do not process large number of fixed cells (more than 106) as one may get incomplete staining of cells.
Transfection of cells with luciferase reporter
To optimize transfections ideal concentrations of transfection agent and DNA must be determined.
One day prior to transfection, plate HUVEC cells (approximately 2 X 105 cells) onto 3.5 cm plates. Ideal confluence is 50-70% on the day of transfection.
Pipette 660 μl of Opti-MEM media (Life Technologies Inc.) into 8 eppendorf tubes labeled 1-8.
*Note: Do not add antibacterials or serum to transfection media.
Pipette 2.5 μg of Luciferase plasmid into tubes 1-4 and pipette 5 μg of plasmid into tubes 5-8. Flick tubes gently 15 times to vortex.
Using vortex, mix F-1 reagent (Targeting Systems) at full speed for 20 seconds.
Pipette 2.5, 5.0,10, and 15 μl of F-1 reagent into tubes 1-4 and 5-8.
Incubate tubes at 37°C for 20 minutes to allow the formation of transfection complexes.
Plated cells were washed with 2.0 ml opti-MEM and then 2.0 ml of opti-MEM media was added. The entire eppendorf of transfection complex is then added to each plate.
Incubate cells for two hours and then remove opti-MEM media. Wash cells gently with opti-MEM media and replaced with HUVEC media.
Incubate cells overnight at 37°C and then treat with 3 Gy of radiation.
Lyse cells with 200 μl of lysis buffer (20 nM Tris, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM phenylmethylsulfonyl fluoride, and leupeptin). Perform Luciferase assay according to Promega protocol on a Monolight 3010 Luminometer (Pharmingen).
Set up the same experiment with the two best F-1 concentration and vary DNA concentrations from 1, 2, 4, 8 µg to optimize DNA concentrations. Further optimization may be necessary using different concentrations. Time of transfection and number of cells plated may also need to be optimized.
Increased incubation time will generally increase transfection but toxicity as well. To attenuate toxicity serum may need to be added during transfections particularly transfections longer than 12 hours. However, serum may affect amount of transfection agent needed.
Note: β-galactosidase or GFP are effective ways to test for efficacy of transfection without a luminometer.
Ideal protein concentration was found to be 5 μg DNA and 15 μl of F-1 reagent for HUVEC cell transfection.
Adjust amounts of DNA and reagent to optimize transfection rates with minimal toxicity based on preliminary study.
Peak levels of transfection achieved in HUVEC was 30% using the F-1 reagent. If greater transfection rates are needed, Enhancer (Targeting Systems) can be used during the formation of transfection agent step.
For experimental transfection follow above protocol replacing your ideal DNA and F-1 concentrations accordingly.
Transfection of ASO with lipofectin
Note optimization of transfection will need to be performed as in the protocol for luciferase transfection.
Day 1:
Trypsinize H460 cells and replated onto 96 well plates (approximately 2000-5000 cells per well) with the goal of 50-70% confluence on day 2 of transfection protocol.
Day 2:
After optimization, it was found that 100nM of oligonucleotide and 3 μl of Lipofectin/ml was ideal for transfection of H460 cells. There is a transfection enhancer available for lipofectin (Invitrogen) if higher transfection rates are desired.
Create a stock solution sufficient for all samples. Two separate solutions need to be made, a plasmid solution and a Lipofectin solution of equal volumes. They will be combined to form the final total volume of transfection complex. Each well holds 100 μl of solution so for 30 wells, 3 ml of transfection complex would be needed and hence, 1.5 ml of plasmid and lipofectin solutions.
*Note: Opti-MEM media was used for transfection, transfection media should not contain antibiotics or serum as this will effect transfection.
Plasmid Solution: Add 300 μl of opti-MEM to an eppendorf tube and add 150 nM of oligonucleotide. Vortexed tube gently by flicking 20 times.
Lipofectin Solution: Add 4.5 μl of lipofectin to 300 μl of opti-MEM media in a separate tube and gentle vortex by flicking.
*Note: It is important to keep tubes separate. Some plasmids may interact with plastic in eppendorf tubes be sure to check plasmid specifications.
Combine the two tubes and mix gently, incubate at room temperature for 20-45 minutes to form transfection complex.
Dilute transfection complex with 2.4 ml of opti-MEM media to each tube.
*Note: If there is a long transfection and cells do not tolerate serum free media well, the dilution can be done in media with serum. However, lipofectin concentration will need to be reoptimized.
Wash cells with 100 μl of Opti-MEM media to remove serum and add 100 μl of transfection complex to each well.
Incubate for 4 hours and remove Opti-MEM media. Replace with normal cell media for cells (with serum and antibiotic).
MTT assay of transfected cells
Expose transfected and non-transfected cells to 3 Gy of radiation.
After the desired time of incubation, add 50 μg of MTT per well. Let plates incubate for 4 h at 37°C until purple precipitate forms.
Solubilize the converted purple formazan dye with 100 μl of detergent reagent (0.01 M HCL/10% SDS) per well. Incubated plates overnight at 37°C.
Quantify reaction product with spectrophotometer set at 570nm absorbance.
References
- Lu B, Mu Y, Cao C, Zeng F, Schneider S, Tan J, Price J, Chen J, Freeman M, Hallahan DE. Survivin as a therapeutic target for radiation sensitization in lung cancer. Cancer Res. 2004;64(8):2840–2845. doi: 10.1158/0008-5472.can-03-3547. [DOI] [PubMed] [Google Scholar]
- Bao R, Connolly DC, Murphy M, Green J, Weinstein JK, Pisarcik DA, Hamilton TC. Activation of cancer-specific gene expression by the survivin promoter. J Natl Cancer Inst. 2002;94(7):522–528. doi: 10.1093/jnci/94.7.522. [DOI] [PubMed] [Google Scholar]
- Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3(1):46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- Altieri DC, Marchisio PC, Marchisio C. Survivin apoptosis: an interloper between cell death and cell proliferation in cancer. Lab Invest. 1999;79(11):1327–1333. [PubMed] [Google Scholar]
- Shih AH, Dai C, Hu X, Rosenblum MK, Koutcher JA, Holland EC. Dose-dependent effects of platelet-derived growth factor-B on glial tumorigenesis. Cancer Res. 2004;64(14):4783–4789. doi: 10.1158/0008-5472.CAN-03-3831. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li M, Wang H, Agrawal S, Zhang R. Antisense therapy targeting MDM2 oncogene in prostate cancer: Effects on proliferation, apoptosis, multiple gene expression, and chemotherapy. Proc Natl Acad Sci USA. 2003;100(20):11636–11641. doi: 10.1073/pnas.1934692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RH, Randour CJ. Attenuation of matrix protein secretion by antisense oligodeoxynucleotides to the cyclin kinase inhibitor p21(Waf1/Cip1). Atherosclerosis. 2002;161(1):105–112. doi: 10.1016/s0021-9150(01)00628-1. [DOI] [PubMed] [Google Scholar]
- Hasholt L, Abell K, Norremolle A, Nellemann C, Fenger K, Sorensen SA. Antisense downregulation of mutant huntingtin in a cell model. J Gene Med. 2003;5(6):528–538. doi: 10.1002/jgm.378. [DOI] [PubMed] [Google Scholar]
- Kamiyama M, Ichikawa Y, Ishikawa T, Chishima T, Hasegawa S, Hamaguchi Y, Nagashima Y, Miyagi Y, Mitsuhashi M, Hyndman D, Hoffman RM, Ohki S, Shimada H. VEGF receptor antisense therapy inhibits angiogenesis and peritoneal dissemination of human gastric cancer in nude mice. Cancer Gene Ther. 2002;9(2):197–201. doi: 10.1038/sj.cgt.7700428. [DOI] [PubMed] [Google Scholar]
- Lord RV, Salonga D, Danenberg KD, Peters JH, DeMeester TR, Park JM, Johansson J, Skinner KA, Chandrasoma P, DeMeester SR, Bremner CG, Tsai PI, Danenberg PV. Telomerase reverse transcriptase expression is increased early in the Barrett's metaplasia, dysplasia, adenocarcinoma sequence. J Gastrointest Surg. 2000;4(2):135–142. doi: 10.1016/s1091-255x(00)80049-9. [DOI] [PubMed] [Google Scholar]
- Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6(10):995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 994;6:10–986. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Ormerod MG. Investigating the relationship between the cell cycle and apoptosis using flow cytometry. J Immunol Methods. 2002;265(1-2):73–80. doi: 10.1016/s0022-1759(02)00071-6. [DOI] [PubMed] [Google Scholar]
- Otaki M, Hatano M, Kobayashi K, Ogasawara T, Kuriyama T, Tokuhisa T. Cell cycle-dependent regulation of TIAP/m-survivin expression. Biochim Biophys Acta. 2000;1493(1-2):188–194. doi: 10.1016/s0167-4781(00)00142-1. [DOI] [PubMed] [Google Scholar]
- Zhou M, Gu L, Li F, Zhu Y, Woods WG, Findley HW. DNA damage induces a novel p53-survivin signaling pathway regulating cell cycle and apoptosis in acute lymphoblastic leukemia cells. J Pharmacol Exp Ther. 2002;303(1):124–131. doi: 10.1124/jpet.102.037192. [DOI] [PubMed] [Google Scholar]
- Gewirtz AM, Stein CA, Glazer PM. Facilitating oligonucleotide delivery: helping antisense deliver on its promise. Proc Natl Acad Sci USA. 1996;93(8):3161–3163. doi: 10.1073/pnas.93.8.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen AR, Halsey J, Fisher GA, Holmlund JT, Geary RS, Kwoh TJ, Dorr A, Sikic BI. Phase I study of an antisense oligonucleotide to protein kinase C-alpha (ISIS 3521/CGP 64128A) in patients with cancer. Clin Cancer Res. 1999;5(11):3357–3363. [PubMed] [Google Scholar]
- Orr RM. Technology evaluation: fomivirsen, Isis Pharmaceuticals Inc/CIBA vision. Curr Opin Mol Ther. 2001;3(3):288–294. [PubMed] [Google Scholar]
- Pisarev V, Yu B, Salup R, Sherman S, Altieri DC, Gabrilovich DI. Full-length dominant-negative survivin for cancer immunotherapy. Clin Cancer Res. 2003;9(17):6523–6533. [PubMed] [Google Scholar]


