Summary
We previously identified mutations in Nardilysin (dNrd1) in a forward genetic screen designed to isolate genes whose loss causes neurodegeneration in Drosophila photoreceptor neurons. Here we show that NRD1 is localized to mitochondria where it recruits mitochondrial chaperones and assists in the folding of α-ketoglutarate dehydrogenase (OGDH), a rate-limiting enzyme in the Krebs cycle. Loss of Nrd1 or Ogdh leads to an increase in α-ketoglutarate, a substrate for OGDH, which in turn leads to mTORC1 activation and a subsequent reduction in autophagy. Inhibition of mTOR activity by rapamycin or partially restoring autophagy delays neurodegeneration in dNrd1 mutant flies. In summary, this study reveals a novel role for NRD1 as a mitochondrial co-chaperone for OGDH, and provides a mechanistic link between mitochondrial metabolic dysfunction, mTORC1 signaling, and impaired autophagy in neurodegeneration.
Keywords: metabolism, alpha-ketoglutarate, mitochondrial chaperones, TCA cycle, patients with OGDHL and NRD1 mutations
Introduction
Mitochondria are essential organelles for all metazoans and defects of mitochondrial function are implicated in metabolic and neurodegenerative diseases (Pickrell and Youle, 2015; Rustin et al., 1997). The mechanisms leading to degeneration are often associated with elevated reactive oxygen species (ROS) or decreased ATP production due to defects in the electron transport chain (ETC) (Johri and Beal, 2012). However, mitochondria also play a critical role in metabolism via the Krebs or TCA cycle. Among the TCA cycle metabolites, α-ketoglutarate (α-KG or 2-oxoglutarate) lies at the intersection between carbon and nitrogen metabolism where it plays an important role in nutrition sensing and metabolic homeostasis (Huergo, 2015). α-KG, together with glutamine, have been shown to regulate mechanistic target of rapamycin (mTOR) activity and autophagy (Chin et al., 2014; Durán et al., 2012).
α-KG is produced from isocitrate by isocitrate dehydrogenase 2 and 3 (IDH2/3) and is oxidized by the α-ketoglutarate dehydrogenase complex (α-KGDHc) to succinyl-CoA in mitochondria. α-KGDHc consists of three different enzymes, including 2-oxoglutarate dehydrogenase (OGDH). Enzyme complexes consisting of multiple units in mitochondria are thought to require molecular chaperons or assembly factors since each unit imported from the cytosol into the mitochondria needs to be properly folded and protected from aggregation until assembled into a complex. For example, succinate dehydrogenase assembly factors (SDHAFs) are required for succinate dehydrogenase (SDH) complex function (Na et al., 2014; Van Vranken et al., 2014). No assembly factor, however, has yet been identified for chaperoning enzymes in α-KGDHc.
We identified mutations in Drosophila Nardilysin (N-arginine dibasic convertase; dNrd1) in an unbiased forward genetic screen designed to identify genes whose loss of function cause neurodegeneration (Yamamoto et al., 2014). NRD1 belongs to the family of zinc-metalloendopeptidases that cleave doublets of basic amino acids at the N-terminal side of arginine residue of neuropeptides in vitro (Chesneau et al., 1994; Pierotti et al., 1994). In vertebrates, NRD1 has been shown to localize to different cellular compartments including the cell surface, nucleus, and cytosol in different cells or tissues (Hiraoka et al., 2014; Hospital et al., 2000; Ma et al., 2004) NRD1 has multiple functions, including ectodomain shedding of cell surface proteins and transcriptional coregulation in the nucleus (Hiraoka et al., 2014; Nishi et al., 2006; Ohno et al., 2009).
In mammals, NRD1 is expressed in the brain (Fumagalli et al., 1998) and loss of mNrd1 in mice causes prenatal growth defects and neonatal lethality (Ohno et al., 2009). Some mutant mice escape neonatal death and live for about two years. However, these escapers show a slow progressive neurodegeneration, enhanced limb-clasping reflexes, impaired motor activity, cognitive deficits, and hypomyelination (Ohno et al., 2009). Yet, the mechanisms underlying these phenotypes are ill-defined and how the loss of NRD1 causes the phenotypes remains unclear.
In this study, we show that NRD1 localizes to mitochondria where it recruits mitochondrial chaperones to properly fold OGDH. We document that absence of NRD1 leads to a severe reduction of OGDH enzyme activity and elevated levels of its substrate, α-KG. Our findings also led to the identification of patients with neurodegenerative phenotypes that carry homozygous deleterious variants in NRD1 and OGDH-like (OGDHL). Using recently developed genetic tools in Drosophila, we show that the proteins with these variants are not functional, suggesting that these genes may be linked to neurological disorders. Hence, we uncovered a novel factor required for chaperoning OGDH, an essential enzyme in the TCA cycle. Furthermore, we show that loss of Nrd1 or Drosophila Ogdh (dOgdh) causes a progressive neurodegeneration and provide genetic data that link these genes to neurological conditions in human.
Results
Mutations in dNrd1 cause a slow demise of neuronal function and structure
To identify novel genes required for neuronal function, we performed a forward genetic screen for essential genes on the Drosophila X chromosome (Haelterman et al., 2014; Yamamoto et al., 2014) . To assess whether these mutations cause defects in neuronal function, we performed electroretinogram (ERG) recordings in homozygous mutant clones in the eye. We isolated four non-sense mutations in CG2025 that cause pupal lethality, fail to complement each other (Figure S1A), and cause similar phenotypes in ERG assays (Figure 1A-C). CG2025 is a Drosophila homologue of NRD1 (dNrd1) (Figure S1B; Figure 1E). All alleles are rescued by wild-type genomic transgenes (Venken et al., 2009, 2010) or ubiquitous expression of dNrd1 cDNA (Figure S1C).
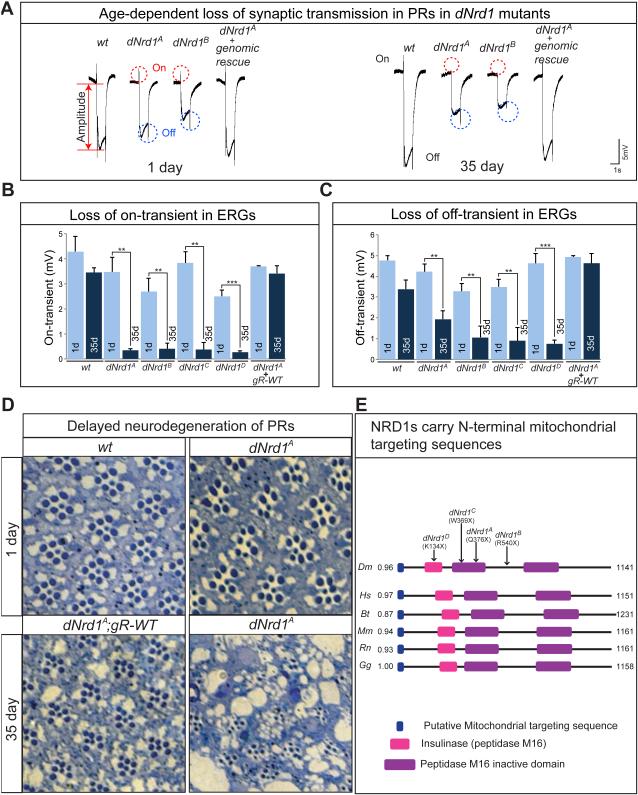
Figure 1. dNrd1 Mutants Exhibit Progressive Neurodegenerative Features in PRs.
(A) ERGs of 1 day-old and 35 day-old mutant clones in PRs of wild-type (wt), four dNrd1 alleles (dNrd1A, B, C, and D), and dNrd1A mutants carrying an 80kb P[acman] genomic rescue transgene (gR-WT). An ERG trace consists of an on-transient (red dotted circles), an amplitude (red arrow), and an off-transient (blue dotted circles). Quantification of the on-transients (B) and off-transients (C) of ERG traces in (A). Error bars indicate s.e.m. P values were calculated using Student’s t-test. **P <0.01, ***P<0.001. (D) Light micrographs of cross sections of retina of 1-day- and 35-day-old flies in wt, dNrd1A and dNrd1A; gR-WT. (E) Schematic of protein domains of fly and vertebrate NRD1. Arrows indicate molecular lesions and alleles of dNrd1. Blue boxes indicate mitochondria targeting sequences (MTSs) predicted by MitoProt and numbers beside the blue boxes indicate the probability that each NRD1 localizes to the mitochondria. Abbreviations: Dm = Drosophila melanogaster, Hs = Homo sapiens, Bt = Bos taurus, Mm = Mus musculs, Rn = Rattus norvegicus, Gg = Gallus gallus. See also Figure S1.
dNrd1 mutants exhibit a slow progressive loss of ERG on- and off-transients and a decrease in amplitude when compared to controls (Figure 1A-C). Since on- and off-transients are a measure of synaptic transmission between the photoreceptor cells (PRs) and the postsynaptic neurons (Verstreken et al., 2003), these data suggest a progressive loss of synaptic transmission in dNrd1 mutants.
To assess if dNrd1 loss is associated with morphological defects, we sectioned the retinae of one-day-old adult flies with dNrd1 mutant clones. As shown in Figure 1D, dNrd1A exhibits subtle defects in young flies and 35-day-old mutant clones exhibit gross morphological abnormalities with loss of numerous PRs (Figure 1D). Therefore, mutations in dNrd1 cause subtle developmental defects and display a striking progressive age-dependent demise of PRs.
NRD1 is a mitochondrial protein
To unravel the mechanism by which loss of NRD1 causes defects in neuronal maintenance, we examined the subcellular localization of NRD1. NRD1 has been reported to localize to the cell surface, cytoplasm (Hospital et al., 2000) and nucleus (Ma et al., 2004). As our X-chromosome screen led to the isolation of more than 30 nuclear encoded genes causing neurodegeneration and encode proteins targeted to mitochondria (Yamamoto et al., 2014), we analyzed Drosophila NRD1 (DNRD1) sequence using MitoProt (Claros and Vincens, 1996) and noted that it contains a putative mitochondrial targeting sequence (MTS) in the N-terminal 42 amino acids (Figure 1E). Similarly, vertebrate NRD1s also contain MTSs (Figure 1E).
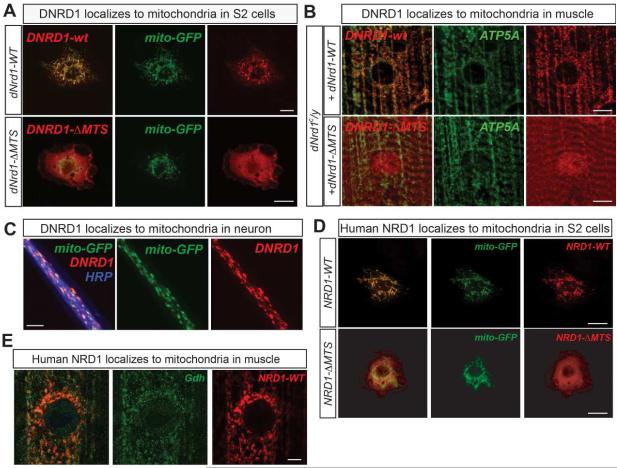
To test if DNRD1 is localized to mitochondria we expressed a C-terminally V5-tagged DNRD1 (DNRD1-V5) in Schneider 2 (S2) cells expressing mito-GFP (Rizzuto et al., 1995) and found that DNRD1-V5 colocalized with mito-GFP (Figure 2A). Similarly, DNRD1-V5 expressed in fly muscles (Figure 2B) or neurons (Figure 2C) also colocalizes with ATP5A, the α subunit of mitochondrial complex V, or with mito-GFP. C-terminally V5 tagged human NRD1 (NRD1-V5) also colocalizes with mito-GFP in S2 cells and fly muscles (Figure 2D and 2E). However, fly or human NRD1s lacking MTS (NRD1-ΔMTS) fail to localize to mitochondria in S2 cells (Figure 2A; Figure 2D). Interestingly, a small fraction of DNRD1-ΔMTS colocalizes with mitochondrial markers in muscles of dNrd1 mutants (Figure 2B), suggesting that other sequences within DNRD1 may also participate in mitochondria targeting of DNRD1 in vivo.
Figure 2. NRD1 Localizes to the Mitochondria.
(A) Confocal micrographs of S2 cells co-expressing mito-GFP (green) and full-length V5-tagged dNrd1 (DNRD1-wt), or V5-tagged dNrd1 cDNA lacking N-terminal MTS (DNRD1-ΔMTS) (red). Scale bars, 10μm. (B) Confocal micrographs of dNrd1C mutant larvae muscle expressing dNrd1-V5 (DNRD1-wt), or dNrd1-ΔMTS-V5 (DNRD1-ΔMTS) (red), using a muscle driver (C57-Gal4). ATP5A (green) labels mitochondria. Scale bars, 10μm. (C) Confocal micrographs of motor neuron axons co-expressing mito-GFP (green) and dNrd1-V5 (red) (D42-GAL4>UAS-dNrd1-V5, UAS-mito-GFP). Scale bar, 5μm. (D) Confocal micrographs of S2 cells co-expressing mito-GFP (green) and V5-tagged full-length human NRD1 (NRD1-WT), or NRD1 cDNA lacking N-terminal MTS (NRD1-ΔMTS) (red). Scale bars, 10μm. (E) Confocal micrographs of dNrd1A mutant larvae muscle expressing human NRD1 (red) (da-Gal4>UAS-human NRD1-V5). Gdh (green) labels mitochondria. Scale bars, 5μm. See also Figure S2.
Finally, we tested whether substitution of an alanine residue for the glutamate residue required for the catalytic Zn2+-binding (E161A) affects protein localization. As shown in Figure S2, this point mutation alters the protein localization in S2 cells and muscles resulting in cytoplasmic localization. In summary, our data show that both DNRD1 and human NRD1 localize to mitochondria (Figure 2A-E).
Loss of NRD1 does not affect ATP and ROS production
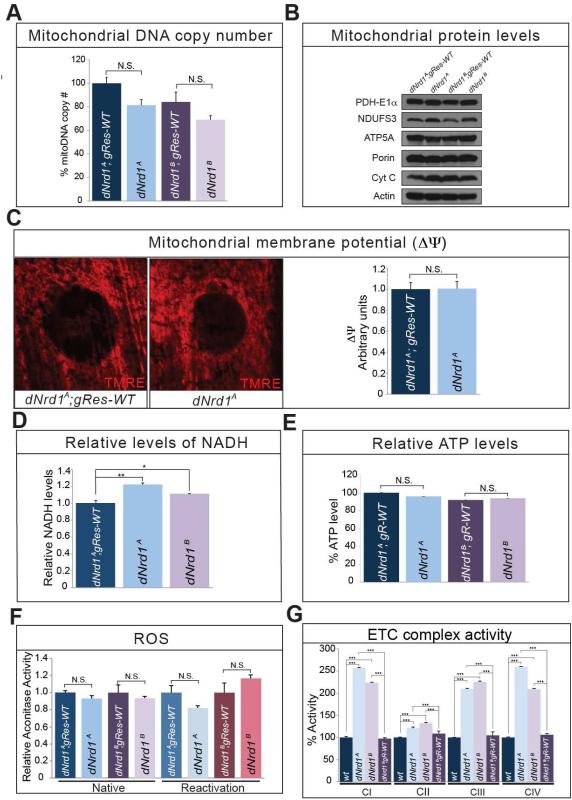
Failure to control ATP production, biogenesis or dynamics, as well as many other lesions of mitochondrial proteins are associated with neurodegenerative diseases (Burté et al., 2014; Johri and Beal, 2012; Pickrell and Youle, 2015). We therefore assessed basic mitochondrial properties in dNrd1 mutant tissues. We did not observe obvious differences in mitochondrial DNA (mtDNA) copy number (Figure 3A) or abundance of numerous mitochondrial proteins between dNrd1 mutants and controls (Figure 3B). In addition, dNrd1 mutants exhibit a normal mitochondrial membrane potential (Figure 3C). Moreover, NADH levels in dNrd1 mutants are very similar to those observed in controls (Figure 3D). Furthermore, ATP levels in dNrd1 mutants are not different from controls (Figure 3E). In addition, the activity of Aconitase, an enzyme that is sensitive to ROS, is not altered in dNrd1 mutants (Figure 3F). However, the activities of the ETC complexes I, III, and IV are increased about two-fold compared to controls (Figure 3G), but complex II activity is not significantly altered (Figure 3G). These data indicate that total energy production is not altered in dNrd1 mutants but suggest that a compensatory upregulation of complexes I, III, and IV activities may mask potential defects in energy production.
Figure 3. Loss of dNrd1 does not Interfere with ATP Levels.
(A) Relative amount of mitochondrial DNA copy number measured in dNrd1 mutant larvae and controls. (B) Western blots for mitochondrial proteins – PDH-E1α, NDUFS3, ATP5A, Porin, Cyt C and Actin in dNrd1 mutants and controls. (C) Mitochondrial membrane potential is measured by TMRE dye in larva muscle (left). Quantification of membrane potential in dNrd1A mutant and control (right). (D) Relative levels of NADH in dNrd1 mutants and control. (E) Relative levels of ATP in dNrd1 mutants and controls. (F) Aconitase activity in both native and reactivation conditions in dNrd1 mutant larvae and controls. (G) The ETC complex activity (I-IV) in dNrd1 mutant larvae and controls. Errors bars in (A), (C), (D), (E), (F), and (G) indicate s.e.m. P values were calculated using Student’s t-test. *P <0.05, **P <0.01, ***P <0.001. N.S. indicates not statistically significant.
Loss of NRD1 affects metabolites of the TCA cycle
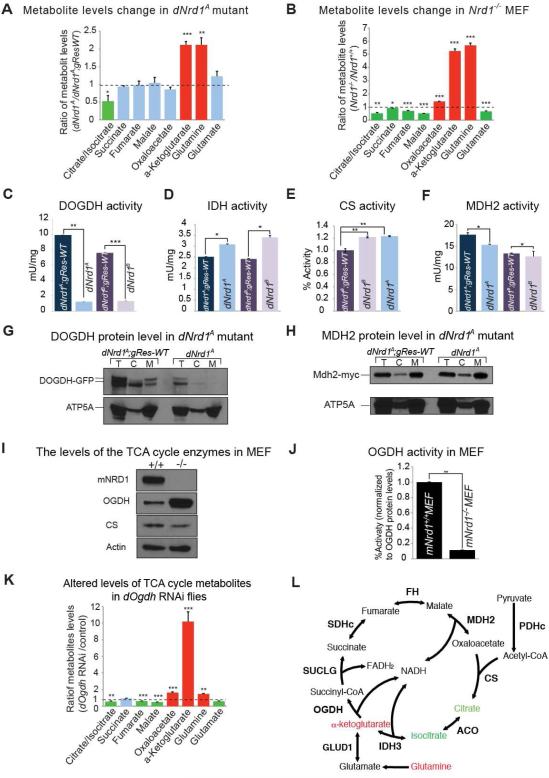
The lack of obvious mitochondrial phenotypes associated with the loss of dNrd1 prompted us to explore metabolites in dNrd1 mutant animals. The most obvious defect that we observed was a dramatic increase in α-KG and glutamine in dNrd1 mutants (Figure 4A). Similarly, mouse embryonic fibroblasts (MEFs) that lack NRD1(Ohno et al., 2009) also exhibit a significant accumulation of α-KG and glutamine (Figure 4B). Our data suggest that when NRD1 is lost, the activity or levels of enzymes of the α-KGDHc are reduced, leading to the accumulation of its substrate, α-KG (Figure 4L). An accumulation of α-KG may lead to an increase in glutamine in dNrd1 mutants and mutant MEFs through cataplerosis, a process that attempts to deplete the pool of α-KG by converting it into glutamine (Owen et al., 2002).
Figure 4. NRD1 is Required for the Activity of OGDH.
(A) Quantification of ratio of metabolites in the TCA cycle of dNrd1A mutants to those in control larvae. (B) Quantification of ratio of metabolites in the TCA cycle of Nrd1KO mutant MEF (mNrd1−/−) to those in wild-type MEF (mNrd1+/+). (C-F) The activity of enzymes in the TCA cycle is measured from purified mitochondria (C, D, and F) or total larva extract (E) in dNrd1 mutants and controls. (G-H) Western blots for protein level of DOGDH-GFP (G) or protein level of Mdh2-myc expressed from genomic rescue transgene (H) and ATP5A in total (T), cytosolic (C) and mitochondrial (M) fraction in dNrd1A mutant and control larvae. (I) Western blots for protein level of OGDH, CS and Actin in Nrd1 mutant MEF (mNrd1−/−) and wild-type MEF (mNrd1+/+). (J) The activity of OGDH in Nrd1 mutant MEF (mNrd1−/−) and wild-type MEF (mNrd1+/+). (K) Quantification of the ratio of metabolites in the TCA cycle of dOgdh RNAi larvae (Act-Gal4/UAS-dOgdh RNAi) compared to those in control larvae (Act-Gal4/+). (A-F J, and K) Error bars indicate s.e.m. P values were calculated using Student’s t-test. *P <0.05, **P <0.01, ***P <0.001. (L) Schematic representation of the TCA cycle. Metabolites that increase in both dNrd1 mutant and mNrd1−/− MEF are shown in red color. Note that in dNrd1 mutant flies (Figure 4A), dOgdh knockdown flies (Figure 4K), and in mNrd1 knockout MEFs (Figure 4B), there is a loss of some other metabolites (green) produced in the TCA cycle. Abbreviations: Citrate synthase (CS); Aconitase (ACO); Isocitrate dehydrogenase 3 (IDH3); Oxoglutarate dehydrogenase (OGDH); Glutamate dehydrogenase (GDH); Succinate-CoA ligase (SUCLG); Succinate dehydrogenase complex (SDHc); Fumarate hydratase (FH); Malate dehydrogenase 2 (MDH2); Pyruvate dehydrogenase complex (PDHc). See also Figure S3 and S4.
NRD1 is required for the activity of OGDH
To determine the cause of α-KG accumulation, we examined the enzymatic activity of Drosophila OGDH, along with other TCA cycle enzymes in dNrd1 mutants. As shown in Figure 4C, loss of dNrd1 leads to a severe loss of the OGDH activity, whereas the activity of other enzymes in the TCA cycle are not or mildly affected in dNrd1 mutants when compared to controls (Figure 4D-F; Figure 3F). These results indicate that loss of dNrd1 affects the stability, abundance, or enzymatic activity of OGDH.
To measure the protein level of DOGDH in dNrd1 mutants in vivo, we took advantage of the presence of a MiMIC (Minos-mediated integration cassette) transposon inserted in a coding intron of dOgdh (Nagarkar-jaiswal et al., 2015; Venken et al., 2011) (Figure S3A). We generated a fly strain in which the MiMIC transposon was replaced with an artificial exon encoding GFP. As shown in Figure S3B, the internally tagged DOGDH-GFP protein localizes to the mitochondria and co-localizes with ATP5A. To test whether the loss of dNrd1 affects DOGDH protein levels, we generated dNrd1 mutants carrying dOgdh-GFP. dNrd1 mutants exhibit a dramatic decrease of the protein levels of DOGDH-GFP, both in total lysate and in the mitochondrial fraction when compared to controls, whereas the level of ATP5A in the mutants is comparable to controls (Figure 4G). To test whether DNRD1 regulates the level of other enzymes in the TCA cycle, we compared the level of malate dehydrogenase 2 (MDH2) in dNrd1 mutants with that of controls, and found that they are similar (Figure 4H). Therefore, DNRD1 is required for the activity and proper protein levels of DOGDH in Drosophila.
To assess the levels and activity of OGDH in the absence of NRD1 in mammalian cells, we compared the expression of OGDH in mNrd1−/− MEFs to that in control mNrd1+/+ MEFs. Since the levels of α-KG are elevated in mNrd1−/− MEFs, we expected a loss of OGDH. Surprisingly, the level of OGDH protein in mNrd1−/− MEFs is elevated compared to wild-type MEFs, whereas the levels of citrate synthase (CS) are similar in mutant and wild-type MEFs (Figure 4I). However, the enzymatic activity of OGDH in mNrd1−/− MEFs is severely reduced compared to wild-type MEFs (Figure 4J), consistent with an elevation in α-KG (Figure 4B). These data suggest that in mammalian cells NRD1 is required for the activity of OGDH, similar to flies, but not for abundance of OGDH. Nevertheless, our data demonstrate that NRD1 is required for the proper function of OGDH in flies and mice.
Loss of dOgdh phenocopies dNrd1 mutants
If OGDH is the major target of NRD1 in vivo, we anticipated that reduction of dOgdh would phenocopy the loss of dNrd1. Knockdown of dOgdh via RNAi in larvae causes a dramatic increase in α-KG and glutamine (Figure 4K) similar to and more severe than the dNrd1 mutant (Figure 4A), suggesting that DOGDH activity is incompletely lost in dNrd1 mutants, consistent with the enzymatic assays (Figure 4C). Additionally, the profile of amino acid changes including elevated lysine level in mNrd1−/− MEFs are similarly observed in dOgdh knockdown flies (Figure S4A, S4B). Moreover, knockdown of dOgdh in PRs causes a slow progressive loss of synaptic transmission (Figure S4C), similar to the ERG phenotypes seen in dNrd1 mutant clones (Figure 1A-C). Altogether, these data reveal that OGDH is a major target of NRD1 in vivo and that loss/reduction-of-function of the two proteins have similar metabolic and neurologic phenotypes.
NRD1 interacts with OGDH as well as numerous mitochondrial chaperones
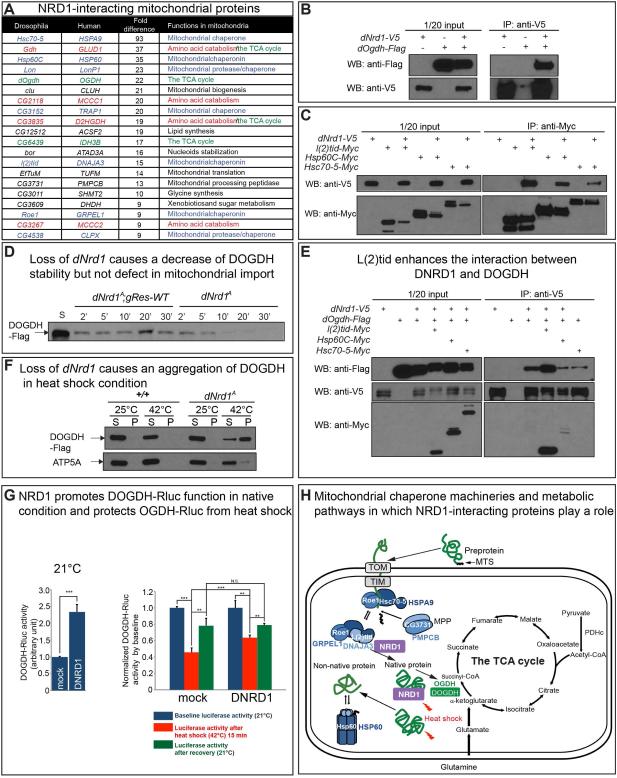
To further determine the role of NRD1, we sought to identify proteins that physically interact with NRD1. We performed IP-mass-spectrometry of V5 tagged DNRD1 and identified numerous mitochondrial proteins including Drosophila OGDH (DOGDH) (Figure 5A; Figure S5A; Supplemental Table 1). The interaction between DNRD1 and DOGDH was confirmed via co-IP in S2 cells (Figure 5B). Interestingly, 20 out of 60 proteins with spectral counts of 9-fold or more in DNRD1-V5-expressing samples when compared to controls were associated with mitochondria (Figure 5A). Bioinformatics analyses based on STRING (http://string-db.org/) (Franceschini et al., 2013) and COMPLEAT (http://www.flyrnai.org/compleat/) (Vinayagam et al., 2013) revealed that ten of the twenty mitochondrial proteins can be assigned to three different functional categories (Figure S5B). These include mitochondrial chaperones (L(2)tid/DNAJA3; Roe1/GRPEL1; Hsc70-5/HSPA9; Hsp60C/HSP60; CG3731/PMPCB), enzymes of the TCA cycle (DOGDH/OGDH; CG6439/IDH3B; Gdh/GLUD1), and proteins involved in amino acid catabolism (CG2118/MCCC1; CG3267/MCCC2) (Figure S5B). In addition, two other proteases that also function as chaperones (Lon/LonP1; CG4538/CLPX) interact with DNRD1 (Figure 5A). We verified the interaction between DNRD1 and Hsc70-5, L(2)tid and Hsp60 in S2 cells (Figure 5C). Given that DNRD1 can interact with DOGDH and a number of mitochondrial chaperone proteins, NRD1 may help chaperone OGDH.
Figure 5. NRD1 Interacts with Mitochondrial Chaperones and OGDH.
(A) The table shows 20 mitochondrial proteins that are identified by IP/MS using V5 antibody from DNRD1-expressing larvae (da-Gal4>UAS-dNrd1-V5). These mitochondrial proteins are selected from proteins whose spectral counts are at least 9-fold higher in DNRD1-expressing larvae compared to control (da-Gal4). The proteins in the TCA cycle are marked with green color, amino acid catabolism with red color and mitochondrial chaperone proteins with blue color. (B) Co-immunoprecipitation using anti-V5 antibody from S2 cell lysates overexpressing C-terminally V5-tagged DNRD1 and C-terminally Flag-tagged DOGDH. Western bots were performed using either anti-Flag or anti-V5 antibody. (C) Co-immunoprecipitation using anti-Myc antibody from S2 cell lysates overexpressing C-terminally V5-tagged DNRD1 and C-terminally Myc-tagged mitochondrial chaperone proteins. Western bots were performed using either anti-V5 or anti-Myc antibody. (D) Import of C-terminal Flag-tagged DOGDH into isolated mitochondria from dNrd1A and control larvae. An arrow indicates DOGDH-Flag proteins detected by anti-Flag antibody. S indicates 10% of starting protein for the assay. The reactions were stopped at different time points (2, 5, 10, 20, and 30 minutes) after mixing mitochondria and DOGDH-Flag protein. The isolated mitochondria were subjected to western blots with anti-Flag antibody. (E) Co-immunoprecipitation using anti-V5 antibody from S2 cell lysates overexpressing C-terminally V5-tagged DNRD1 and C-terminally Flag-tagged DOGDH, together with one of C-terminally Myc-tagged mitochondrial chaperone proteins. Western bots were performed using anti-Flag, anti-V5 or anti-Myc antibody. (F) Mitochondria isolated from wild-type or dNrd1 mutants expressing DOGDH-Flag (+/Y; UAS-dOgdh-Flag /+;da-Gal4/+ or dNrd1A/Y; UAS-dOgdh-Flag /+;da-Gal4/+) were incubated at the indicated temperature and aggregated proteins were precipitated by ultracentrifugation. Supernatants (S) and pellets (P) were analyzed by western blot with an antibody against Flag or ATP5A. (G) Mitochondrial chaperone assay of DNRD1 for DOGDH-Renilla luciferase (DOGDH-Rluc). In the presence of extra DNRD1, DOGDH-Rluc is ~2.2 fold more active (left) and upon heat-shock the loss of OGDH-Rluc activity is milder (right). Error bars indicate s.e.m. P values were calculated using Student’s t-test. **P <0.01, ***P <0.001. N.S. indicates not statistically significant. (H) Schematic representation of mitochondrial chaperones that play a role in import and protein folding, and metabolic pathways centered in the TCA cycle. See also Figure S5.
NRD1 recruits mitochondrial chaperones for OGDH
The interaction between DNRD1 with DOGDH and mitochondrial chaperones when combined with a dramatic decrease in activity of DOGDH in the absence of DNRD1 suggests that DNRD1 is required for proper mitochondrial import, folding, and/or stabilization of DOGDH (Figure 5H). Similarly, an increase in OGDH protein levels in mNrd1−/− MEFs combined with a severe decrease in OGDH activity, also suggests that the folding of the protein may be impaired. However, unlike in flies, the protein is not degraded and leads to an accumulation of OGDH (Figure 4I).
As DNRD1 interacts with Hsc70-5 and Roe1, two subunits of the presequence translocase-associated motor (PAM) complex in the inner mitochondria (Pfanner et al., 1997) (Figure 5A, 5H and Figure S5B), we performed a mitochondrial import assay in which in vitro translated DOGDH protein was mixed with purified mitochondria (Horwich et al., 1985). C-terminally Flag tagged DOGDH protein (DOGDH-Flag) produced by in vitro transcription and translation was incubated with mitochondria purified from either dNrd1 mutant or control animals. Upon incubation for different time periods, the mitochondria were isolated and washed with buffer containing protease inhibitors. Western blots allowed assessment of the abundance of DOGDH in mitochondria. The data indicate that the protein is imported into mitochondria and that the levels are maximal after 20 min in controls (Figure 5D). At early time points (Figure 5D; 2 and 5 min), the amount of imported DOGDH protein in dNrd1 mutant and control mitochondria is similar, suggesting that mitochondrial import is not defective in the absence of DNRD1 (Figure 5D). However, imported DOGDH protein is gradually lost in dNrd1 mutant mitochondria (Figure 5D). These data indicate that DNRD1 is not required for the import of DOGDH, but is essential for the stability of newly imported DOGDH in mitochondria.
The loss of DOGDH in mitochondria in the absence of DNRD1 suggests that DOGDH is misfolded and degraded. MDJ1, a yeast homologue of L(2)tid, is required for the folding of newly imported proteins in the mitochondrial matrix (Rowley et al., 1994). We find that L(2)tid protein enhances the physical interaction between DNRD1 and DOGDH proteins (Figure 5E), but other chaperones did not promote this interaction. This suggests that DNRD1, DOGDH and L(2)tid form a complex and may assist in the proper folding of DOGDH.
Misfolded mitochondrial proteins typically precipitate more easily than native proteins (Bender et al., 2011). We therefore sought to examine whether the solubility of DOGDH is affected in dNrd1 mutants in response to heat shock. Since DOGDH is significantly reduced in dNrd1 mutants (Figure 4G), we overexpressed DOGDH in wild-type and mutant larvae. Purified mitochondria expressing DOGDH-Flag from control or dNrd1 mutant animals were incubated at 25°C or 42°C. Upo n 10 min incubation, the mitochondria were lysed with detergent-containing buffer, and soluble and insoluble fractions were separated by ultracentrifugation. In control mitochondria, DOGDH-Flag was not detected in the pellet fraction, whereas in dNrd1 mutant mitochondria, a significant fraction of DOGDH-Flag was observed in the pellet at 42°C (Figure 5F). In contrast, the solubility of ATP5A that served as a negative control is much less affected than DOGDH. These data indicate that NRD1 protects OGDH from denaturation upon heat shock.
To confirm the above results, we developed a mitochondrial chaperone assay that allows us to monitor a folding state of OGDH by measuring luciferase activity of a DOGDH-Renilla luciferase fusion protein (DOGDH-Rluc). Indeed, overexpression of dNrd1 in S2 cells significantly increases DOGDH-Rluc activity (Figure 5G), suggesting that NRD1 may help fold or stabilize OGDH. To test whether NRD1 is capable of protecting OGDH from stress (holdase activity) or refolding denatured OGDH (foldase activity) (Ali et al., 2016). we incubated S2 cells in which DOGDH-Rluc is expressed with or without DRND1 at 42°C for 15 min, and retur ned the cells to 21°C for 3 hrs. Immediately after the 42°C for 15 min shift, S2 cel ls expressing DNRD1 exhibit a higher DOGDH-Rluc activity compared to control (Figure 5G), indicating that NRD1 can protect OGDH from the heat shock and has an holdase activity. However, after a 3 hrs recovery, S2 cells expressing DNRD1 do not show higher DOGDH-Rluc activity when compared to controls, indicating that NRD1 lacks foldase activity. All together, these data suggest that NRD1 form a complex with mitochondrial chaperones to fold imported OGDH in native condition, and protect OGDH from denaturation under stress condition (Figure 5H).
Patients with homozygous variants in NRD1 and OGDHL develop severe neurological features
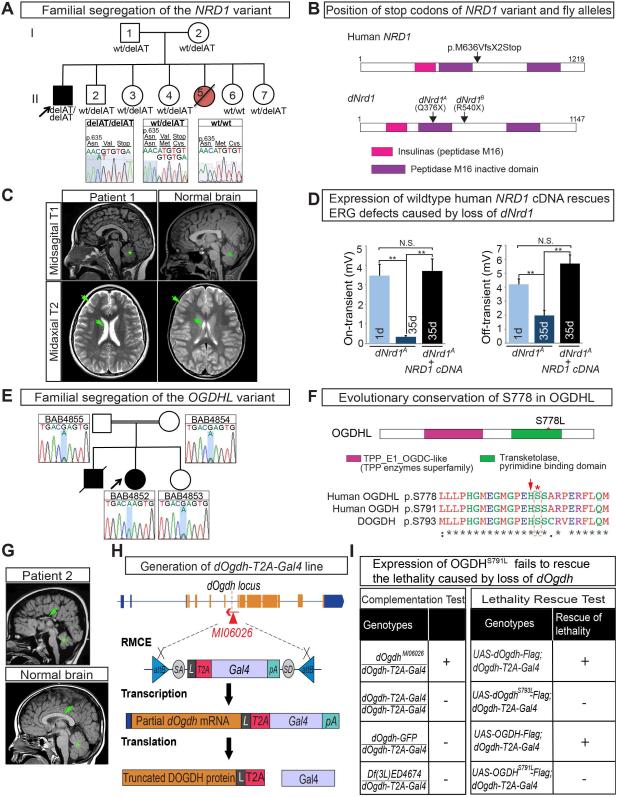
Since loss of dNrd1 or dOgdh causes a neurodegenerative phenotype in Drosophila, and mNrd1 knockout mice exhibit severe developmental and neuronal defects (Ohno et al., 2009), we hypothesized that variants in orthologous human genes may be linked to rare neurological conditions. We examined whole exome sequencing (WES) data and performed genome analyses of nearly 6,000 patients of the Baylor-Hopkins Center for Mendelian Genomics (BHCMG) database (Chong et al., 2015). We did not identify a deleterious allele. However, a posting on GeneMatcher (https://genematcher.org) (Sobreira et al., 2015) led to the identification of a patient (CDMD1122) with severe global developmental delay and ataxia referred for clinical exome sequencing to the UCLA Clinical Genomics Center (Lee et al., 2014). The patient is homozygous for a truncating variant in NRD1 (NM_002525.2:exon 17:c.1906_1907delAT: p.M636VfsX2) (Figure 6A-B). The variant causes a frameshift leading to a M636V followed by an early termination codon. Both parents and four unaffected siblings were found to be heterozygous for this variant (Figure 6A).
Figure 6. Identification of Patients with Neurodegenerative Phenotypes with Homozygous Deleterious Variants in NRD1 and OGDHL.
(A) Familial segregation of the NRD1 variant (NM_002525.2:exon 17:c.1906_1907delAT: p.M636VfsX2). The NRD1 variant is homozygous in the proband and heterozygous or wild type in the parents and unaffected siblings. A sister (Red circle) was born with early onset developmental delay and failure to thrive. She died of unknown cause at 16 months of age. No additional clinical information and DNA was available. (B) Schematic representation of protein domains of human and fly NRD1 and position of stop codons of human NRD1 variant and fly alleles. (C) A T1 sagittal MRI image of the patient at age 8 years showing mild cerebellar volume loss (green asterisk). A T2 midaxial image shows enlarged extracerebral spaces (green arrows) compared to those in a control MRI. (D) Quantification of the on- and off-transients of ERG traces in 1 day-old and 35 day-old dNrd1A clones in PRs and 35 day-old dNrd1A clones expressing human NRD1 cDNA. Error bars indicate s.e.m. P values were calculated using Student’s t-test. **P <0.01. N.S. indicates not statistically significant. (E) Familial segregation of the OGDHL variant (Chr10:50946295_G>A; OGDHL: NM_018245:exon18:c.C2333T:p.S778L). The OGDHL variant is homozygous in the proband and heterozygous in the parents and an unaffected sibling. (F) Schematic of protein domains of OGDHL and amino acid sequences around S778L. Dotted box (red) indicates a serine residue (red asterisk) that is mutated in the patient 2. Arrow (red) indicates a catalytic histidine. (G) A T1 sagittal MRI image from the patient 2 done at age 6 years showing a severely hypoplastic corpus callosum (green arrow) as well as an abnormal cerebellum (green asterisk). A control MRI image T1 sagittal sequence from a 5 year old child who was evaluated by neurology for migraine headaches showing a normal cerebellum and corpus callosum. (H) A schematic of the generation of dOgdh-T2A-Gal4 by Recombinase-Mediated Cassette Exchange (RMCE), and the translation of a Gal4 protein by a ribosomal skipping mechanism. The location of the Mi{MIC} insertion MI06026 is indicated by the red triangle. The T2A-Gal4 cassette consists of a splice acceptor (SA, light gray) followed by a linker (L, dark gray), a ribosomal skipping T2A peptide sequence (red), a Gal4 coding sequence (pale purple), a polyadenylation signal (pA, turquoise), and a splice donor (SD, light gray). Two inverted attB sites (blue) are positioned at the 5’- and 3’- end of the cassette (I) (Left table) Complementation test results of dOgdh-T2A-Gal4 alleles. +, complement; -, failure to complement. dOgdh-T2A-Gal4 complements the original MiMIC insertion (MI06026) that is not mutagenic (as it is inserted in the reverse orientation to the gene). dOgdh-T2A-Gal4 fails to complement a deficiency (Df(3L)ED4674) that lacks the dOgdh locus. These data indicate that dOgdh-T2A-Gal4 is a loss-of-function mutant. (Right table) dOgdh lethality can be rescued by expression of wild-type DOGDH or its human homologue but not by the mutant forms. Gal4 expression from dOgdh-T2A-Gal4 allowed dOgdh, dOgdhS793L, OGDH, or OGDHS791L to be expressed in dOgdh mutant background. See also Figure S6.
We performed WES on the proband, both parents and five unaffected siblings (Figure 6A) and did not discover any coding de novo variant in the proband. We found seven candidate genes with homozygous variants in the proband that were not present in the siblings. None of those genes have been previously associated with Mendelian disorders and six of the seven genes carry homozygous missense alleles that are predicted to be benign by multiple algorithms (Adzhubei et al., 2010; Kumar et al., 2009; Lek et al., 2016; Ng and Henikoff, 2003) (Supplemental Table 2). Finally, the patient is a compound heterozygote for OBSCN and PUS1, but both variants for OBSCN and one of the variants of PUS1 are classified as benign and the clinical phenotype associated with PUS1 does not fit the clinical phenotype of our patient (Supplemental Table 2). Hence, NRD1 is the only gene with a predicted homozygous loss-of-function allele. Truncating alleles in NRD1 are very rare and the gene is predicted to be highly intolerant to loss-of-function mutations (pLI score of 0.88) and there are no instances of homozygous nonsense or frameshift alleles in the Exome Aggregation Consortium (ExAC) (Lek et al., 2016).
The patient had an unremarkable prenatal and birth history, but initially presented with developmental delay and episodes of ocular deviation in the first year of life (see Supplemental Information, “Clinical Case Histories”) and developed a microcephaly (Figure 6C; Figure S6B-S6D). Altogether, the patient exhibits a progressive neurodegenerative phenotype and carries a rare, loss-of-function homozygous truncating variant in NRD1.
To determine whether molecular functions of NRD1 proteins are evolutionarily conserved, we assessed if ubiquitous expression of human NRD1 cDNA can rescue the lethality and ERG defects associated with dNrd1 fly mutants. The lethality and ERG defects were rescued (Figure S1C; Figure 6D), indicating that the function of NRD1 is conserved between fly and human.
Since NRD1 chaperones OGDH, we hypothesized that rare variants in homologous genes in human may also be associated with neurological features. Humans have two homologues of dOgdh, OGDH and OGDH-like (OGDHL). OGDH is widely expressed and present in most tissues while OGDHL is much more restricted in its expression pattern and mostly expressed in brain and liver (Bunik et al., 2008), (GeneCards: http://www.genecards.org/). No deleterious variants in OGDH or OGDHL have been previously reported. Within the BHCMG database we prioritized individuals with rare homozygous or compound heterozygous variants in these genes (Chong et al., 2015). Eight individuals were selected for segregation analysis from ~6,000 individuals. Of these, seven were found to have variants with a high frequency in public databases or variants in other genes that explained their phenotype.
Among these, one undiagnosed patient (BAB4852) carries a homozygous variant in OGDHL (Figure 6E). This patient exhibited a severe developmental delay with cerebral and cerebellar atrophy and corpus callosum abnormality (Figure 6G). A detailed phenotypic description can be found in Supplemental Information (Clinical case histories). The homozygous variant in OGDHL was considered a top candidate from a cohort of Turkish patients with brain malformations (Karaca et al., 2015). We performed WES on the proband, and both parents yielding a set of candidate variants in 13 genes, 3 of which are in known disease genes inconsistent with the phenotypes (Supplemental Table 2). The homozygous variant in OGDHL (NM_018245:exon18:c.C2333T:p.S778L) (Figure 6F) was predicted to be deleterious based on SIFT (Kumar et al., 2009; Ng and Henikoff, 2003) and polyphen2 (Adzhubei et al., 2010) and the gene is known to be expressed in the brain (Bunik et al., 2008) (GeneCards: http://www.genecards.org/). This variant segregates with the phenotype in the BAB4852 family as both parents and the unaffected sibling are heterozygous carriers, consistent with Mendelian expectations (Figure 6E). OGDHL in this patient maps within a block of absence of heterozygosity (AOH) consistent with the known consanguinity (Figure S6E). Hence, the variant in OGDHL was our top candidate for this patient and lack of functional data prompted us to experimentally test the functionality of the variant.
The p.S778 resides in a transketolase domain that is essential for activity of OGDHL (Figure 6F). Moreover, S778 (S793 in Drosophila) is evolutionarily conserved from bacteria to vertebrates, and is adjacent to the catalytic histidine (Fig. 6F, red arrow) (Bunik and Degtyarev, 2008). To assess the function of the S778L variant in flies, we created a severe loss of function or null allele of dOgdh that expresses the GAL4 gene under the control of the endogenous regulatory elements of dOgdh using the T2A-GAL4 system (dOgdh-T2A-GAL4) (Figure 6H; see Extended Results) (Bellen and Yamamoto, 2015; Diao et al., 2015; Nagarkar-jaiswal et al., 2015). We also generated transgenes that allow expression of OGDH or OGDHL under the control of UAS. Expression of human OGDHL did not rescue the lethality caused by loss of dOgdh. However, expression of human OGDH was able to rescue the lethality associated with the loss of dOgdh, whereas expression of OGDHS791L was not able to rescue (Figure 6I). In addition, expression of the fly DOGDH rescued the loss of dOgdh, but DOGDHS793L failed to rescue (Figure 6I). Moreover, the ERG defects associated with the loss of dOgdh were rescued by wild type fly DOGDH, human OGDH and OGDHL but not with the mutant isoforms (Figure S6H). Hence, the Serine to Leucine mutation in both human and fly OGDH results in a severe loss of function.
Loss of NRD1 or OGDH leads to activation of TORC1 and inhibition of autophagy
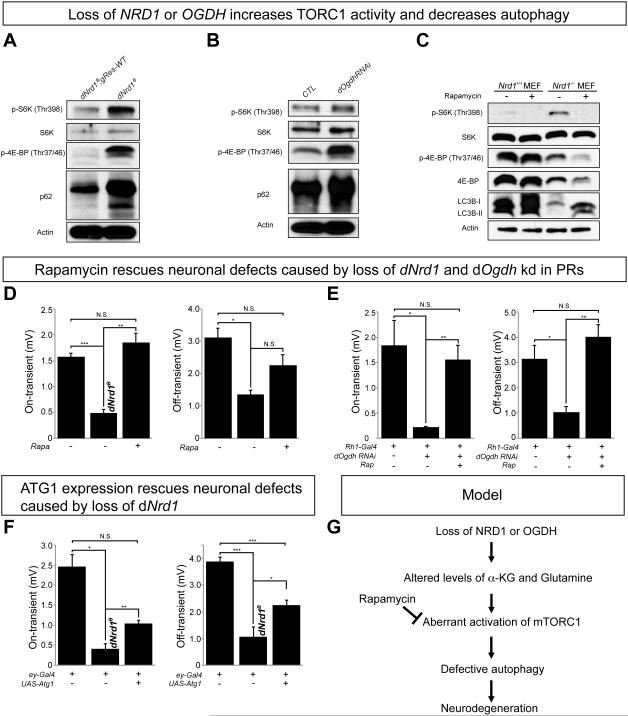
Loss of NRD1 does not affect ATP, ROS, or mitochondrial biogenesis (Figure 3) unlike several other neurodegenerative mutants (Johri and Beal, 2012). Rather, the loss of NRD1 or its target OGDH causes a specific increase of α-KG and glutamine (Figure 4A-B). Interestingly, increased levels of α-KG have been documented to promote (Durán et al., 2012) or decrease TOR activity (Chin et al., 2014). We therefore tested whether loss of dNrd1 or dOgdh affect TOR activity in fly mutants. As shown in Figure 7A and 7B, dNrd1 mutants or dOgdh RNAi animals exhibit increased levels of phospho-S6 kinase (pS6K) and phospho-eukaryotic translation initiation factor 4E binding protein (p4E-BP). Nrd1KO (mNrd1−/−) MEFs also exhibit an increased level in pS6K compared to control MEFs (Figure 7C), suggesting that loss of NRD1 or OGDH activates mTORC1 in vivo. Unlike fly mutants, however, Nrd1KO (mNrd1−/−) MEFs did not show an increase in phosphor-4E-BP levels (Figure 7C) as we observed a significant decrease of the total 4E-BP1 levels. Importantly, dNrd1 mutants or dOgdh RNAi animals exhibit an increased level of p62, an autophagy substrate (Figure 7A and 7B), and Nrd1KO (mNrd1−/−) MEFs show a decreased level in LC3B-II, an autophagy marker (Figure 7C). These data indicate that loss of NRD1 and OGDH leads to mTORC1 activation, which in turn inhibits autophagy. The decrease in autophagy in Nrd1KO (mNrd1−/−) MEFs is partially restored by treating the cells with rapamycin, an mTOR inhibitor (Figure 7C). Moreover, treating flies with eyes that carry dNrd1 mutant clones or express dOgdh RNAi, with rapamycin, rescued the ERG defects (Figure 7D and 7E). These data indicate that hyperactivation of TORC1 underlies the slow progressive neurodegenerative phenotypes observed in flies.
Figure 7. Loss of NRD1 or OGDH leads to an increase in TORC1 activity and decreases autophagy.
(A-B) Western blots for the levels of phospho-S6K, S6K, phospho-4E-BP, p62, and Actin in dNrd1 mutants (dNrd1B) and its control (A), and in dOgdh RNAi (Act-Gal4/UAS-dOgdh RNAi) and control animals (UAS-dOgdh RNAi /+) (B). (C) Western blots for the levels of phospho-S6K, S6K, phospho-4E-BP, 4E-BP, LC3B and Actin in Nrd1ko MEFs (mNrd1−/−) and wild-type MEFs (mNrd1+/+) with or without rapamycin (100 nM). (D) Quantification of the on- and off-transients of ERGs in 35-day-old dNrd1B mutant clones in PRs with or without treatment of 3μM rapamycin and control PRs (dNrd1B; gRes-WT) (E) Quantification of the on- and off-transients of ERGs in 28-day-old PRs where dOgdh is knocked-down by RNAi (Rh1-Gal4/UAS-dOgdh RNAi) with or without treatment of 3μM rapamycin and control PRs (Rh-1-Gal4/UAS-empty vector). (F) Quantification of the on- and off-transients of ERGs in 35-day-old dNrd1B mutant clones in PRs with or without expressing Atg1 (ey-Gal4>UAS-Atg1) and control PRs (dNrd1B; gRes-WT) (D-F) Error bars indicate s.e.m. P values were calculated using Student’s t-test. *P <0.05, **P <0.01, ***P <0.001. N.S. indicates not statistically significant. (G) Model of neurodegeneration in NRD1 or OGDH mutants: Loss of NRD1 or OGDH leads to an elevation of α-KG and glutamine, which induces aberrant activation of mTORC1. Hyperactive mTORC1 signaling causes a decrease in autophagy, which leads to neurodegeneration. Restoring mTORC1 activity by treatment of rapamycin rescues neuronal defects in NRD1 and OGDH mutants. See also Figure S8.
The decrease in autophagy in dNrd1 or dOgdh mutants may cause neurodegenerative phenotypes as perturbations in autophagy are associated with many neurodegenerative diseases (Menzies et al., 2015). mTORC1 activation has been shown to lead to a reduction in autophagy as well as an increase in global translation (Laplante and Sabatini, 2012). To assess if autophagy defects cause the neurodegenerative phenotypes in vivo, we expressed low levels of Atg1 in PRs. Atg1 expression, a target that TOR phosphorylates, partially rescues the ERG defects caused by loss of dNrd1 (Figure 7F). To test weather an increase in translation affects the neurodegenerative phenotypes, we expressed a dominant negative S6K (Barcelo and Stewart, 2002) that suppresses translation. This dominant negative S6K does not rescue the ERG defects (Figure S8), indicating that the reduction in autophagy and not the increase in translation, contributes to the phenotype in dNrd1. Hence, these data suggest that aberrant mTORC1 activation and subsequent autophagy defects promote the neurodegeneration associated with OGDH(L) and NRD1 mutations (Figure 7G).
Discussion
NRD1 recruits mitochondrial chaperones to promote OGDH function
NRD1 is a member of the metallopeptidases conserved from prokaryotes to eukaryotes (Pierotti et al., 1994). Several mitochondrial peptidases that belong to this family play a role in removal of the MTS of nuclear-encoded mitochondrial proteins upon import into mitochondria (Hawlitschek et al., 1988). Loss of these genes causes an accumulation of unprocessed forms of mitochondrial proteins (Mossmann et al., 2014). We, therefore, initially hypothesized that NRD1 plays a role in processing the MTS of TCA cycle enzymes. However, western blot data of OGDH and CS in mNrd1−/− MEFs did not show an alteration in molecular weight and did not support this hypothesis (Figure 4I). Instead, our IP/MS experiment in Drosophila showed that DNRD1 interacts with DOGDH and several mitochondrial chaperones (Figure 5A). These findings led us to hypothesize that NRD1 is involved in folding and/or stability of OGDH. Indeed, the following observations support this hypothesis. First, NRD1 is required for the activity of OGDH in both fly and MEFs (Figure 4C and 4J). Second, loss of mNrd1 increases OGDH protein levels in MEFs, while its loss results in a decrease in DOGDH protein levels in Drosophila (Figure 4G, 4I). Third, DNRD1 interacts directly or indirectly with numerous chaperones based on IP/MS (Figure 5A and Figure S5B) and Co-IP (Figure 5C, 5E). Fourth, an in vitro import assay shows that imported DOGDH is much less stable in the mitochondria that lack DNRD1 (Figure 5D). Fifth, DOGDH is more sensitive to heat-shock and becomes more aggregated in the absence of DNRD1 (Figure 5F). Lastly, DNRD1 partially protects DOGDH-Rluc from denaturation under heat shock (Figure 5G). Altogether, our data indicate that NRD1 acts as a mitochondrial chaperone and/or co-chaperone for OGDH.
Loss of NRD1 and OGDH leads to an activation of TORC1 and a decrease in autophagy
Loss of NRD1 or OGDH alters the profiles of metabolites including a significant elevation of α-KG. α-KG has been shown to differentially affect mTOR activity in different contexts. Durán et al. (2012) showed that glutamine or its glutaminolysis product, α-KG, activates mTORC1 in a Rag-dependent manner. They also documented that a decrease in α-KG induces autophagy in human cells. However, α-KG was documented to inhibit TOR and enhance autophagy through inhibition of ATP synthase (Chin et al., 2014). We find that loss of dNrd1 and mNrd1 causes an elevation of α-KG and glutamine, an increase in TORC1/mTORC1 activity, and a decrease in autophagy (Figure 7A, 7C). Similarly, we observe that loss of dOgdh enhances TORC1 signaling, and leads to a decrease in autophagy (Figure 7B). In addition, loss of dNrd1 does not interfere with ATP production (Figure 3E), which may be achieved by TORC1-mediated translational activation of a subset of nuclear-encoded mitochondrial proteins that include subunits of CI and CV (Morita et al., 2013). Hence, our data are consistent with the observations described in Durán et al. (2012).
Loss of NRD1 and OGDH leads to an increase in lysine levels
In addition to an elevation of α-KG, loss of NRD1 or OGDH also exhibits a very similar metabolic profile for amino acids including an elevation of lysine levels (Figure S4A, S4B), which suggests that loss of NRD1 or OGDH may impair one or more enzymes in the lysine catabolic pathway. α-KGDHc has been shown to decarboxylate α-ketoadipic acid (α-KA), an intermediate in the lysine degradation pathway (Hirashima et al., 1967; Kanzaki et al., 1969), consistent with the elevated levels of lysine. Interestingly, patients with mutations in dehydrogenase E1 and transketolase domain containing 1 (DHTKD1), another OGDH-like enzyme (Bunik and Degtyarev, 2008), fail to break down lysine and have elevated levels of α-KA (Danhauser et al., 2012). However, they present with milder intellectual disability and milder symptoms compared to the NRD1 and OGDHL patients (Danhauser et al., 2012). Together, these observations indicate that defects in lysine metabolism are unlikely to be associated with the observed phenotypes but they may exacerbate phenotypes associated with elevated levels of α-KG in NRD1 and OGDHL patients.
Rare variants in NRD1 or OGDHL are associated with progressive loss of neurological function in human
Our data show that loss of NRD1 or OGDH causes an α-KGDHc enzymatic deficiency. In flies, the phenotypes associated with the loss of either protein are very similar as they lead to a very slow, progressive neurodegeneration that is not observed in most mitochondrial mutants (Jaiswal et al., 2015; Liu et al., 2015). In mice, loss of mNrd1 also causes a slow progression of neurological phenotypes, including impairment of motor coordination and balance, impaired memory, a thin cerebral cortex, and myelination defects in the corpus callosum (Ohno et al., 2009). A number of parallels can be drawn between the phenotypes of patients with homozygous deleterious variants in NRD1 or OGDHL. They both displayed developmental defects at ~1 year of age and progressive neurodegenerative features including acquired microcephaly, cerebellar volume loss, ataxia, motor impairment, severe intellectual disability, and inability to speak (Supplementary Table 3). In these families, NRD1 and OGDHL were the best candidate genes from the WES and using flies we experimentally demonstrated that the variants in both cases were functionally deleterious, suggesting that these two variant may be linked to the pathogenesis (Figure 6D, 6I; S1C). Identification of other patients with deleterious variants in NRD1 or OGDHL will be helpful to further link these genes to the observed phenotypes.
Hyper-activation of mTOR and other human diseases
Our data show that loss of dNrd1 or dOgdh cause neurodegeneration through an hyper-activation of TORC1 and a subsequent decrease in autophagy in flies (Figure 7). Interestingly, hyper-activation of mTOR signaling has also been associated with other human diseases, and suppression of mTOR signaling using mTOR-inhibiting compounds has been shown to be effective in several trials (Crino, 2016; Mirzaa et al., 2016; Pascual, 2016). For example, everolimus, an inhibitor of mTORC1, is approved for treatment of tuberous sclerosis complex -associated subependymal giant cell astrocytomas (Franz 2013). Given that feeding rapamycin to flies that lack dNrd1 or dOgdh, partially suppresses the neurodegenerative phenotype, treatment with mTORC1 inhibitors may be a therapeutic avenue for patients with mutations in NRD1 or OGDHL.
In summary, we identify a previously undocumented function of NRD1 in mitochondria in flies and mice. We show that NRD1 binds to and chaperones OGDH, a key enzyme in the TCA cycle. In the absence of NRD1, OGDH is misfolded, leading to an upregulation of a key metabolite, α-KG. Loss of dNrd1 or dOgdh in flies, or loss of NRD1 or OGDHL in human, is associated with progressive neurodegenerative phenotypes with striking similarities. We further show that elevated levels of α-KG activate TORC1/mTORC1, and decrease autophagy. Consistent with these findings, some aspects of the neurodegenerative phenotypes caused by a decrease in DNRD1 or DOGDH activity can be rescued by providing rapamycin to the flies. Our data provide compelling evidence that intertwining cross species genomic approaches in species as diverse as Drosophila and human provide a solid foundation to unravel potential molecular pathogenic mechanisms.
Experimental Procedures
Fly strains and maintenance
dNrd1mutants (mut=A,B,C,D) – y1 w* dNrd1mut P{neoFRT}19A/FM7c, Kr-Gal4, UAS-GFP and X-chromosome isogenized control flies – y1 w* P{neoFRT}19A (wt in figures) were generated and mapped as previously described (Haelterman et al., 2014; Yamamoto et al., 2014) For eye mosaic experiments, y1 w* dNrd1A or B, P{neoFRT}19A/FM7c, Kr-Gal4, UAS-GFP were crossed with y w cl(1) P{neoFRT}19A/Dp(1;Y)y+; ey-FLP. dOgdh-GFP was generated as previously described (Nagarkar-jaiswal et al., 2015). da-Gal4 (BDSC_5460), C57-Gal4 (a gift from Vivian Budnik), D42-Gal4 (RRID:BDSC_8816), Actin-Gal4, w1118; Dp(1;3)DC245, PBac{DC245}VK00033, Df(3L) ED4674 (RRID:BDSC_30363), and VALIUM20-EGFP-shRNA (RRID:BDSC_41552) were obtained from the Bloomington Drosophila Stock Center. dOgdh RNAi lines (v12778 (RRID:FlyBase_FBst0450668) for ERG assay; v50393 (RRID:FlyBase_FBst0469014) for metabolic profiling) were obtained from the Vienna Drosophila Resource Center (Dietzl et al., 2007). Mdh2-myc genomic rescue flies were generously provided by Carl Thummel (Wang et al., 2010). All flies were maintained at room temperature (21°C). Crosses were kept at 25°C.
Aggregation assay
An aggregation assay was performed as previously descried (Bender et al., 2011). Briefly, a total of 200 μg isolated mitochondria was resuspended in 400 μl of an aggregation assay buffer (250mM sucrose, 10 mM MOPS/KOH, pH7.2, 3 mM MgCl2, 80 mM KCl, 2 mM NADH, 1 mM ATP). Mitochondria in buffer was divided into two tubes: One tube was incubated at 25°C min and the other tu be was incubated at 42°C. After incubation, mitochondria were isolated in mitochondria isolation buffer by centrifugation at 8,000g for 10 min at 4°C. Isolated mitochondria were lysed in 100 μl of lysis buffer (0.5% Triton X-100, 30 mM Tris, pH 7.4; 200 mM KCl, 5 mM EDTA) containing cOmplete protease inhibitor (Roche) and subjected to ultracentrifugation at 125K g for 30 min at 4°C. Supernatants were removed and the pe llets were re-extracted by vigorous shaking in the lysis buffer. Supernatants and pellets were analyzed by western blot using anti-Flag antibody (Sigma-Aldrich Cat# F1804 RRID:AB_262044) and anti-ATP5A antibody (Abcam Cat# ab14748 RRID:AB_301447).
Ethics Statement
Informed consent was obtained prior to participation from all subjects or parents of recruited subjects under an Institutional Review Board approved protocol at Baylor College of Medicine (H-29697) through the Center for Mendelian Genomics73 or at the University of California in Los Angeles (11-001087).
Supplementary Material
Acknowledgments
We thank L. Duraine, P-T Li, Y. He for technical support; A. Leibfried, A. Ephrussi, C. Thummel for reagents; Y. Kwon for an assistance for bioinformatics; E. Seto for MRI images; K. Venkatachalam, H-T Chao for critical reading of this manuscript; K. Schulze for proofreading. This study was supported by the NIH 5R01GM067858, NIH T32 NS043124-11 and the Research Education and Career Horizon Institutional Research and Academic Career Development Award Fellowship 5K12GM084897 (H.S.), the Jan and Dan Duncan Neurological Research Institute at Texas Children’s Hospital (S.Y.), the CPRIT Metabolomics Core Facility Support Award RP120092 (N.P. and A.S.K.), NCI/2P30CA125123-09 Shared Resources Metabolomics core, Dan L. Duncan Cancer Center (DLDCC), Alkek Center for Molecular Discovery (AS, NP), Mass Spectrometry COE by Agilent (AS, NP) and the NIH R01GM098387 (B.H.G.), the research grants (26293068, 26670139 and 26116715) and a research program of the P-Direct from the MEXT of Japan (E.N.), and the NIH K08NS076547 funded by NINDS (M.F.W.). We acknowledge support of the NIH (1RC4GM096355), the Robert A. and Renee E. Belfer Family Foundation, the Huffington Foundation and Target ALS to H.J.B. H.J.B. is an Investigator of the Howard Hughes Medical Institute. This work was also supported in part by the National institute of Neurological Disease and Stroke (NINDS) R01NS058529 (J.R.L.) and the Baylor-Hopkins Center for Mendelian Genomics, US National Human Genome Research Institute (NHGRI) National Heart Lung and Blood Institute (NHBLI) grant U54HG006542.
J.R.L. has stock ownership in 23andMe and Lasergen, is a paid consultant for Regeneron Pharmaceuticals and is a coinventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Baylor College of Medicine derives revenue from the chromosomal microarray analysis and clinical exome sequencing offered in the Baylor Miraca Genetics Laboratory http://www.bmgl.com/BMGL/Default.aspx) Confocal microscopy was supported by the Baylor College of Medicine IDDRC grant number 1U54 HD083092 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental Information includes Supplemental Experimental Procedures and seven figures.
Author Contributions
W.H.Y. and H.J.B. conceived and designed the project. W. H. Y. designed and conducted most experiments and analyzed the data. H.S., M.J., and S.Y performed the genetic screen and mapping. N.A.H mapped several alleles. S.N.J. generated dOgdh-GFP reagent. N.P, V.P, and A.S. performed metabolomic profiling and analysis. T.D. and B.H.G. performed mitochondrial copy number and ETC assays. E.N., and M.O. generated Nrd1 knockout MEF reagent. D.M.M. performed WES: J. H. interpreted MRI data. T.T., A.A., J.C., J.S. V. A. A., and E.K. collected clinical data. M.F.W. and E.K. analyzed clinical and genomic data. V.A.A. S.F.N., E.K. and J.R.L. collected human subjects, performed segregation analysis and identified the patients. W.H.Y., M.F.W., S.Y, and H.J.B. wrote the manuscript.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali YO, Allen HM, Yu L, Li-Kroeger D, Bakhshizadehmahmoudi D, Hatcher A, McCabe C, Xu J, Bjorklund N, Taglialatela G, et al. NMNAT2:HSP90 Complex Mediates Proteostasis in Proteinopathies. PLoS Biol. 2016;14:e1002472. doi: 10.1371/journal.pbio.1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcelo H, Stewart MJ. Altering Drosophila S6 kinase activity is consistent with a role for S6 kinase in growth. Genesis. 2002;34:83–85. doi: 10.1002/gene.10132. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Yamamoto S. BenchMarks Morgan ’ s Legacy : Fruit Flies and the Functional Annotation of Conserved Genes. Cell. 2015;163:12–14. doi: 10.1016/j.cell.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender T, Lewrenz I, Franken S, Baitzel C, Voos W. Mitochondrial enzymes are protected from stress-induced aggregation by mitochondrial chaperones and the Pim1/LON protease. Mol Biol Cell. 2011;22:541–554. doi: 10.1091/mbc.E10-08-0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunik VI, Degtyarev D. Structure-function relationships in the 2-oxo acid dehydrogenase family: Substrate-specific signatures and functional predictions for the 2-oxoglutarate dehydrogenase-like proteins. Proteins Struct. Funct. Genet. 2008;71:874–890. doi: 10.1002/prot.21766. [DOI] [PubMed] [Google Scholar]
- Bunik V, Kaehne T, Degtyarev D, Shcherbakova T, Reiser G. Novel isoenzyme of 2-oxoglutarate dehydrogenase is identified in brain, but not in heart. FEBS J. 2008;275:4990–5006. doi: 10.1111/j.1742-4658.2008.06632.x. [DOI] [PubMed] [Google Scholar]
- Burté F, Carelli V, Chinnery PF, Yu-Wai-Man P. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat. Rev. Neurol. 2014;11:11–24. doi: 10.1038/nrneurol.2014.228. [DOI] [PubMed] [Google Scholar]
- Chesneau V, Pierotti a R., Prat a, Gaudoux F, Foulon T, Cohen P. N-arginine dibasic convertase (NRD convertase): a newcomer to the family of processing endopeptidases. An overview. Biochimie. 1994;76:234–240. doi: 10.1016/0300-9084(94)90151-1. [DOI] [PubMed] [Google Scholar]
- Chin RM, Fu X, Pai MY, Vergnes L, Hwang H, Deng G, Diep S, Lomenick B, Meli VS, Monsalve GC, et al. The metabolite α-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR. Nature. 2014;510:397–401. doi: 10.1038/nature13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JX, Buckingham KJ, Jhangiani SN, Boehm C, Sobreira N, Smith JD, Harrell TM, McMillin MJ, Wiszniewski W, Gambin T, et al. The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Crino PB. The mTOR signalling cascade: paving new roads to cure neurological disease. Nat. Rev. Neurol. 2016;12:379–392. doi: 10.1038/nrneurol.2016.81. [DOI] [PubMed] [Google Scholar]
- Danhauser K, Sauer SW, Haack TB, Wieland T, Staufner C, Graf E, Zschocke J, Strom TM, Traub T, Okun JG, et al. DHTKD1 mutations cause 2-aminoadipic and 2-oxoadipic aciduria. Am. J. Hum. Genet. 2012;91:1082–1087. doi: 10.1016/j.ajhg.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao F, Ironfield H, Luan H, Diao F, Shropshire WC, Ewer J, Marr E, Potter CJ, Landgraf M, White BH. Plug-and-Play Genetic Access to Drosophila Cell Types using Exchangeable Exon Cassettes. Cell Rep. 2015;10:1410–1421. doi: 10.1016/j.celrep.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su K-C, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Durán RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, Hall MN. Glutaminolysis activates Rag-mTORC1 signaling. Mol. Cell. 2012;47:349–358. doi: 10.1016/j.molcel.2012.05.043. [DOI] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, Von Mering C, et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli P, Accarino M, Egeo A, Scartezzini P, Rappazzo G, Pizzuti A, Avvantaggiato V, Simeone A, Arrigo G, Zuffardi O, et al. Human NRD Convertase : A Highly Conserved Metalloendopeptidase Expressed at Specific Sites during Development and in Adult Tissues. 1998;245:238–245. doi: 10.1006/geno.1997.5078. [DOI] [PubMed] [Google Scholar]
- Haelterman NA, Jiang L, Li Y, Bayat V, Sandoval H, Ugur B, Tan KL, Zhang K, Bei D, Xiong B, et al. Large-scale identification of chemically induced mutations in Drosophila melanogaster. Genome Res. 2014;24:1707–1718. doi: 10.1101/gr.174615.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawlitschek G, Schneider H, Schmidt B, Tropschug M, Hartl FU, Neupert W. Mitochondrial protein import: identification of processing peptidase and of PEP, a processing enhancing protein. Cell. 1988;53:795–806. doi: 10.1016/0092-8674(88)90096-7. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Matsuoka T, Ohno M, Nakamura K, Saijo S, Matsumura S, Nishi K, Sakamoto J, Chen PM, Inoue K, et al. Critical roles of nardilysin in the maintenance of body temperature homoeostasis. Nat Commun. 2014;5:3224. doi: 10.1038/ncomms4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirashima M, Hayakawa T, Koike M. Mammalian alpha-keto acid dehydrogenase complexes. II. An improved procedure for the preparation of 2-oxoglutarate dehydrogenase complex from pig heart muscle. J. Biol. Chem. 1967;242:902–907. [PubMed] [Google Scholar]
- Horwich a L., Kalousek F, Mellman I, Rosenberg LE. A leader peptide is sufficient to direct mitochondrial import of a chimeric protein. EMBO J. 1985;4:1129–1135. doi: 10.1002/j.1460-2075.1985.tb03750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital V, Chesneau V, Balogh a, Joulie C, Seidah NG, Cohen P, Prat a. N-arginine dibasic convertase (nardilysin) isoforms are soluble dibasic-specific metalloendopeptidases that localize in the cytoplasm and at the cell surface. Biochem. J. 2000;349:587–597. doi: 10.1042/0264-6021:3490587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huergo LF. The Emergence of 2-Oxoglutarate as a Master Regulator Metabolite. 2015;79:419–435. doi: 10.1128/MMBR.00038-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal M, Haelterman N. a., Sandoval H, Xiong B, Donti T, Kalsotra A, Yamamoto S, Cooper T. a., Graham BH, Bellen HJ. Impaired Mitochondrial Energy Production Causes Light-Induced Photoreceptor Degeneration Independent of Oxidative Stress. PLOS Biol. 2015;13:e1002197. doi: 10.1371/journal.pbio.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johri A, Beal MF. Mitochondrial Dysfunction in Neurodegenerative Diseases. 2012;342:619–630. doi: 10.1124/jpet.112.192138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki T, Hayakawa T, Hamada M, Fukuyoshi Y, Koike M. Mammalian alpha-keto acid dehydrogenase complexes. IV. Substrate specificities and kinetic properties of the pig heart pyruvate and 2-oxyoglutarate dehydrogenase complexes. J. Biol. Chem. 1969;244:1183–1187. [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR Signaling. Cold Spring Harb. Perspect. Biol. 2012;4:a011593. doi: 10.1101/cshperspect.a011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-rivera F, Das K, Toy T, Harry B, Yourshaw M, et al. Clinical Exome Sequencing for Genetic Identification of Rare Mendelian Disorders. Jama. 2014;90095:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang K, Sandoval H, Yamamoto S, Jaiswal M, Sanz E, Li Z, Hui J, Graham BH, Quintana A, et al. Article Glial Lipid Droplets and ROS Induced by Mitochondrial Defects Promote Neurodegeneration. Cell. 2015;160:177–190. doi: 10.1016/j.cell.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Chow KM, Yao J, Hersh LB. Nuclear shuttling of the peptidase nardilysin. Arch. Biochem. Biophys. 2004;422:153–160. doi: 10.1016/j.abb.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Menzies FM, Fleming A, Rubinsztein DC. Compromised autophagy and neurodegenerative diseases. Nat. Publ. Gr. 2015;16:345–357. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- Millecamps S, Julien J-P. Axonal transport deficits and neurodegenerative diseases. Nat. Rev. Neurosci. 2013;14:161–176. doi: 10.1038/nrn3380. [DOI] [PubMed] [Google Scholar]
- Mirzaa GM, Campbell CD, Solovieff N, Goold CP, Jansen LA, Menon S, Timms AE, Conti V, Biag JD, Olds C, et al. Association of MTOR Mutations With Developmental Brain Disorders, Including Megalencephaly, Focal Cortical Dysplasia, and Pigmentary Mosaicism. JAMA Neurol. 2016;73:836–845. doi: 10.1001/jamaneurol.2016.0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Gravel S-P, Cheénard V, Sikström K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, et al. MTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013;18:698–711. doi: 10.1016/j.cmet.2013.10.001. rie. [DOI] [PubMed] [Google Scholar]
- Mossmann D, Vögtle F-N, Taskin AA, Teixeira PF, Ring J, Burkhart JM, Burger N, Pinho CM, Tadic J, Loreth D, et al. Amyloid-β Peptide Induces Mitochondrial Dysfunction by Inhibition of Preprotein Maturation. Cell Metab. 2014;20:662–669. doi: 10.1016/j.cmet.2014.07.024. [DOI] [PubMed] [Google Scholar]
- Na U, Yu W, Cox J, Bricker DK, Brockmann K, Rutter J, Thummel CS, Winge DR. The LYR factors sdhaf1 and SDHAF3 mediate maturation of the iron-sulfur subunit of succinate dehydrogenase. Cell Metab. 2014;20:253–266. doi: 10.1016/j.cmet.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkar-jaiswal S, Lee P, Campbell ME, Chen K, Anguiano-zarate S, Gutierrez MC, Busby T, Lin W, He Y, Schulze KL, et al. A library of MiMICs allows tagging of genes and reversible , spatial and temporal knockdown of proteins in Drosophila. Elife. 2015;4:e05338. doi: 10.7554/eLife.05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi E, Hiraoka Y, Yoshida K, Okawa K, Kita T. Nardilysin enhances ectodomain shedding of heparin-binding epidermal growth factor-like growth factor through activation of tumor necrosis factor-alpha-converting enzyme. J. Biol. Chem. 2006;281:31164–31172. doi: 10.1074/jbc.M601316200. [DOI] [PubMed] [Google Scholar]
- Ohno M, Hiraoka Y, Matsuoka T, Tomimoto H, Takao K, Miyakawa T, Oshima N, Kiyonari H, Kimura T, Kita T, et al. Nardilysin regulates axonal maturation and myelination in the central and peripheral nervous system. Nat. Neurosci. 2009;12:1506–1513. doi: 10.1038/nn.2438. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- Pascual JM. Genetic Gradients in Epileptic Brain Malformations. JAMA Neurol. 2016;73:787. doi: 10.1001/jamaneurol.2016.1039. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Craig EA, Angelika H. Mitochondrial Preprotein Translocase. Annu Rev Cell Dev Biol. 1997;13:25–51. doi: 10.1146/annurev.cellbio.13.1.25. [DOI] [PubMed] [Google Scholar]
- Pickrell AM, Youle RJ. The Roles of PINK1, Parkin, and Mitochondrial Fidelity in Parkinson’s Disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierotti AR, Prat A, Chesneau V, Gaudoux F, Leseney AM, Foulon T, Cohen P. N-arginine dibasic convertase, a metalloendopeptidase as a prototype of a class of processing enzymes. Proc. Natl. Acad. Sci. U. S. A. 1994;91:6078–6082. doi: 10.1073/pnas.91.13.6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr. Biol. 1995;5:635–642. doi: 10.1016/s0960-9822(95)00128-x. [DOI] [PubMed] [Google Scholar]
- Rowley N, Prip-Buus C, Westermann B, Brown C, Schwarz E, Barrell B, Neupert W. Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell. 1994;77:249–259. doi: 10.1016/0092-8674(94)90317-4. [DOI] [PubMed] [Google Scholar]
- Rustin P, Bourgeron T, Parfait B, Chretien D, Munnich A, Rötig A. Inborn errors of the Krebs cycle: A group of unusual mitochondrial diseases in human. Biochim. Biophys. Acta - Mol. Basis Dis. 1997;1361:185–197. doi: 10.1016/s0925-4439(97)00035-5. [DOI] [PubMed] [Google Scholar]
- Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, Popodi E, Holtzman SL, Schulze KL, Park S, Carlson JW, Hoskins RA, Bellen HJ, Kaufman TC. A molecularly defined duplication set for the X chromosome of Drosophila melanogaster. Genetics. 2010;186:1111–1125. doi: 10.1534/genetics.110.121285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstreken P, Koh T, Schulze KL, Zhai RG, Hiesinger PR, Zhou Y, Mehta SQ, Cao Y, Roos J, Bellen HJ. Synaptojanin Is Recruited by Endophilin to Promote Synaptic Vesicle Uncoating. 2003;40:733–748. doi: 10.1016/s0896-6273(03)00644-5. [DOI] [PubMed] [Google Scholar]
- Vinayagam A, Hu Y, Kulkarni M, Roesel C, Sopko R, Mohr SE, Perrimon N. Protein complex-based analysis framework for high-throughput data sets. Sci. Signal. 2013;6:rs5. doi: 10.1126/scisignal.2003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vranken JG, Bricker DK, Dephoure N, Gygi SP, Cox JE, Thummel CS, Rutter J. SDHAF4 promotes mitochondrial Succinate dehydrogenase activity and prevents neurodegeneration. Cell Metab. 2014;20:241–252. doi: 10.1016/j.cmet.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lam G, Thummel CS. Med24 and Mdh2 are required for Drosophila larval salivary gland cell death. Dev. Dyn. 2010;239:954–964. doi: 10.1002/dvdy.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Jaiswal M, Charng W, Gambin T, Karaca E, Mirzaa G, Wiszniewski W, Sandoval H, Haelterman N. a, Xiong B, et al. Resource A Drosophila Genetic Resource of Mutants to Study Mechanisms Underlying Human Genetic Diseases. Cell. 2014;159:200–214. doi: 10.1016/j.cell.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.