Abstract
Protein kinase inhibitors can be effective in treating selected cancers, but most suppress several kinases. Imatinib mesylate has been useful in the treatment of Philadelphia chromosome-positive chronic myelogenous leukemia and B cell acute lymphoblastic leukemia through the inhibition of BCR-ABL tyrosine kinase activity. Imatinib mesylate has also been shown to inhibit KIT, ARG, and platelet-derived growth factor receptors α and β, and potentially other tyrosine kinases. We have produced a mutant allele of BCR-ABL (T315A) that is uniquely inhibitable by the small molecule 4-amino-1-tert-butyl-3-(1-naphthyl)pyrazolo[3,4-d]pyrimidine and used it to demonstrate that sole suppression of BCR-ABL activity was insufficient to eliminate BCR-ABL+ KIT+-expressing immature murine myeloid leukemic cells. In contrast, imatinib mesylate effectively eliminated BCR-ABL+ KIT+-expressing leukemic cells. In the cellular context of mature myeloid cells and Pro/Pre B cells that do not express KIT, monospecific BCR-ABL inhibition was quantitatively as effective as imatinib mesylate in suppressing cell growth and inducing apoptosis. These results suggest that the therapeutic effectiveness of small molecule drugs such as imatinib mesylate could be due to the inhibitor's ability to suppress protein kinases in addition to the dominant target.
Deregulation of protein kinase activity has been shown to induce cell proliferation, suppress apoptosis, and block differentiation leading to carcinogenesis (1). Small molecule inhibitors with specificity toward suppression of particular oncoprotein kinases such as epidermal growth factor receptor, Flt-3, and BCR-ABL have helped to demonstrate that deregulated kinase activity of these proteins is critical for the development of specific cancers (2–4).
BCR-ABL is the fusion gene product of the Philadelphia (Ph) chromosome generated from a reciprocal translocation between chromosome 9 containing the tyrosine kinase ABL and chromosome 22 with the BCR gene (5, 6). BCR-ABL tyrosine kinase activity has been shown to be essential for the induction of in vitro cellular transformation (7) and in vivo leukemogenesis (8, 9). Deregulated BCR-ABL tyrosine kinase activity accounts for >90% of chronic myelogenous leukemia (CML) cases and 5–15% of acute B cell lymphoblastic leukemias (B-ALL) (10).
Suppression of BCR-ABL tyrosine kinase activity by imatinib mesylate (Gleevec, ST1571, or CP57148B) has been shown to correlate with hematological remission in Ph positive CML and B-ALL patients (3). However, imatinib mesylate can suppress the tyrosine kinase activity of KIT, platelet-derived growth factor receptors, c-ABL, and c-ARG, in addition to BCR-ABL (3). The effectiveness of imatinib mesylate could be due to inhibition of other kinases, in addition to BCR-ABL, dependent on patterns of expression.
KIT has been shown to be up-regulated and activated by BCR-ABL (11, 12). A recent study of Ph+ CML patients revealed that 6 of 80 patient cell samples contained activating mutations in KIT (13).
BCR-ABL tyrosine kinase activity is essential for in vitro cellular transformation, in vitro myeloid progenitor cell expansion, and B-ALL development (3, 14). New small molecule inhibitors that suppress BCR-ABL tyrosine kinase activity, such as pyrido[2,3-d]pyrimidine inhibitor analogues, have been developed (15–17), which block at least BCR-ABL and KIT.
Inhibitors with monospecificity toward engineered protein kinase alleles have been developed. These protein kinases generally contain a glycine or alanine silent mutation within a conserved residue in the ATP-binding pocket that does not alter phosphoacceptor specificity or biological function of the kinase (18). These mutant kinases possess an expanded ATP-binding site that sensitizes the kinase domain to inhibition by bulky inhibitors that cannot bind WT kinase domains (18). Oligonucleotide DNA arrays used to measure genome-wide transcription demonstrated that these mutant kinases regulated a very similar set of genes as the WT kinase (19).
Specificity of these inhibitors has been well documented. v-SRC, CamKII, and CDK2 containing an amino acid substitution in the conserved ATP binding site have been shown to be selectively inhibited by C-3 pyrazolo[3,4-d]pyrimidine-derivatized inhibitors (18–20). These inhibitors have been demonstrated to selectively suppress kinase activity of protein kinase A (PKA) and v-erbB inhibitor-sensitized forms (21, 22). Inhibitor administration suppressed the growth of NIH 3T3 cells expressing a v-erbB inhibitor-sensitive form and not v-erbB WT form in mice injected s.c. with these cells (22). Primary T cells expressing large amounts of an inhibitor sensitive form of LCK were shown to developmentally progress upon drug addition coincident with suppression of kinase activity (23).
We applied this technology to examine cellular responses caused by BCR-ABL inhibition versus those caused by inhibition of BCR-ABL, KIT, and potentially other kinases by the drug imatinib mesylate. Monospecific inhibition of BCR-ABL tyrosine kinase activity was sufficient to abrogate expansion of cells directly dependent on BCR-ABL activity for growth and survival. However, BCR-ABL-induced in vivo myeloproliferative disorder (MPD) progenitors that also used KIT signaling pathways for cellular expansion continued to grow in the presence of monospecific BCR-ABL inhibition. These cells were eliminated upon imatinib mesylate addition. These results demonstrate that sole BCR-ABL inhibition is insufficient to eliminate all BCR-ABL-expressing progenitors and suggests that part of the efficacy of imatinib mesylate in treating CML may be due to the inhibitor's ability to suppress multiple kinases.
Materials and Methods
Generation of BCR-ABL T315A Form. The 5′ primer GAAGACTGCAGTCATGAAAGAGATCAAAC and 3′ primer CCCGTAGGTCATGAACTCTGCGATG, containing a mutation of 315 (ACT) threonine to 315 (GCA) alanine, were used to convert p210BCR-ABL WT to p210BCR-ABL T315A (numbering of ABL is based on isoform 1a throughout). The 85-bp PCR fragment contained BspHI sites at both ends and was cloned into a 3′ BCR-ABL fragment cut with BspHI. This mutant 3′ABL fragment was subsequently cut with EcoRI and HindIII and directionally cloned into a 5′ BCR-ABL fragment cut with EcoRI and HindIII in the pMSCV vector. Sequencing verified the presence of the T315A mutation in the BCR-ABL clone.
Generation of Virus Stocks, BCR-ABL-Expressing BaF3 Cells, MPD, and B-ALL. The retroviral vector pMSCV (24) containing a 5′ LTR-driven BCR-ABL internal ribosome entry site enhanced GFP (EGFP) was used to generate high-titer helper-free retrovirus stocks prepared by transient cotransfection of 293T cells (25). Generation of BCR-ABL-expressing BaF3 cells (26) and BCR-ABL-induced MPD (27) were as described. MPD was transferred into secondary recipients with 106 frozen splenic cells from primary recipients being injected intravenously into Balb/C sublethally irradiated at 450 rad. Generation of BCR-ABL-induced B-ALL was as described (28) except that primary BCR-ABL-expressing Pro/Pre B cells were cultured for 2 weeks before transplantation into mice. Flow cytometry, pathological evaluation, and cell morphology analyses were conducted as described (26–28).
Inhibitor Studies. In BCR-ABL-expressing MPD cells, cultures were set up at 106 cells per well of a six-well plate from frozen samples and cultured in Iscove's modified Dulbecco's medium/10% FBS/50 ng/ml stem cell factor (R & D Systems catalog no. 455MC). BCR-ABL-expressing BaF3 cells and Pro/Pre B cells were maintained as described (28). Imatinib mesylate was dissolved to 10 μM in water and stored at 4°C. NaPP1 (4-amino-1-tert-butyl-3-(1-naphthyl)pyrazolo[3,4-d]pyrimidine) was made as described (18) and dissolved to 10 μM in DMSO and stored at 25°C. ACK45 (BD Pharmingen catalog no. 553867) was used at 10 μg/ml. Inhibitors were added daily at described concentrations, and control cells were given an equivalent volume of DMSO.
Cell Cycle, Apoptosis, and Protein Analyses. Cells were incubated with Hoechst dye 33342 in Hanks' balanced salt solution (HBSS) at 10 μg/ml at 37°C for 30 min followed by one wash with ice-cold HBSS. Cells were subsequently stained for surface markers as described (26–28). Apoptosis analysis was performed as described (26). Immunoblotting was performed as described (26) with the addition of antibodies: BAX (catalog no. 2772), BAD (catalog no. 9292), cleaved caspase 3 and 7 (catalog no. 9915 kit), and cleaved poly(ADP ribose) polymerase (PARP) (catalog no. 9544) from Cell Signaling Technology (Beverly, MA); cyclin D2 (sc 593), cyclin D3 (sc 182), CDK4 (sc 260), and p27KIP (sc 528) from Santa Cruz Biotechnology; cyclin E (catalog no 06-459) and A (catalog no. 06-138) from Upstate Biotechnology (Lake Placid, NY); and CDK2 (catalog no. 610145) from Transduction Laboratories (Lexington, KY). Antibodies were used according to manufacturers' protocol.
Results
The Small Molecule Inhibitor NaPP1 Selectively Suppresses a BCR-ABL T315A Inhibitor-Sensitive Form Without Affecting WT BCR-ABL or KIT Activity. We engineered a BCR-ABL T315A allele (ABL isoform 1a numbering) containing a silent mutation in the ATP-binding site corresponding to v-SRC I338G, c-ABL T315A, and CDK2 F80A mutations shown to be sensitive to inhibition by C-3 derivatized pyrazolo[3,4-d]pyrimidine-based inhibitors (18, 19, 29). This BCR-ABL T315A form was constructed into the murine stem cell virus internal ribosome entry site EGFP retroviral vector (27).
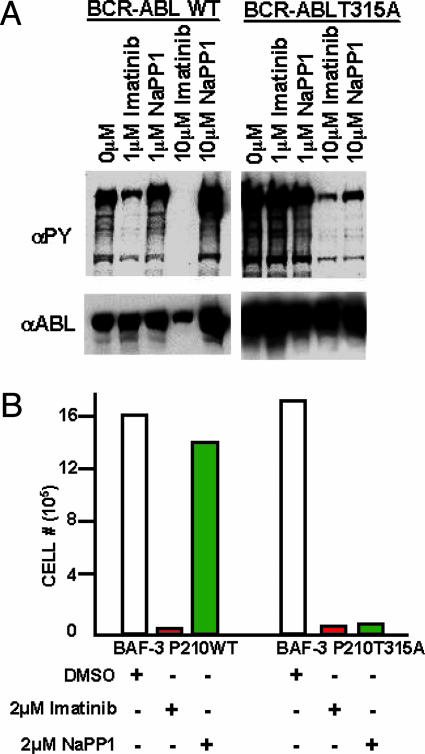
BCR-ABL T315A expression in the IL-3-dependent Pro-B BaF3 cell line led to elevated BCR-ABL tyrosine phosphorylation and IL-3-independent cell growth, similar to BCR-ABL WT (Fig. 1). NaPP1 (10 μM) inhibitor was sufficient to suppress BCR-ABL T315A tyrosine phosphorylation in levels similar to 10 μM imatinib mesylate (Fig. 1 A). NaPP1 was unable to suppress BCR-ABL WT tyrosine phosphorylation unlike imatinib mesylate, demonstrating NaPP1 selectivity for inhibiting the BCR-ABL T315A form (Fig. 1 A).
Fig. 1.
NaPP1 selectively suppresses BCR-ABL T315A-induced tyrosine phosphorylation and cytokine-independent growth without affecting BCR-ABL WT. (A) Concentrations (1 and 10 μM) of NaPP1 and imatinib mesylate were added to BCR-ABL T315A and BCR-ABL WT expressing BaF3 cells in the presence of 1% WeHI IL-3 containing medium, and total levels of tyrosine phosphorylation and BCR-ABL expression were measured 24 h after inhibitor addition as described (26). Equivalent volumes of DMSO were added to control cells. (B) Concentrations (2 μM) of NaPP1 and imatinib mesylate were added to BCR-ABL T315A and BCR-ABL WT expressing BaF3 cells in the absence of WeHI IL-3 containing medium every 24 h, and cell counts as measured by trypan blue exclusion were obtained 48 h after inhibitor addition. One set of data from a representative experiment repeated three times is shown.
These BCR-ABL expressing BaF3 cells expressed extremely high levels of BCR-ABL, and 1 μM concentrations of NaPP1 or imatinib mesylate was inefficient at inhibiting all BCR-ABL tyrosine phosphorylation (Fig. 1 A). NaPP1 concentrations >2 μM can be generally toxic to cells (C. Zhang and K.M.S., unpublished observations). To determine whether the maximum dose of 2 μM NaPP1 or imatinib mesylate was sufficient for inhibiting BCR-ABL T315A and WT activity, we generated BaF3 cells that express lower levels of BCR-ABL by fluorescence-activated cell sorting and used them as described below.
Concentrations of 2 μM NaPP1 or imatinib mesylate suppressed BCR-ABL T315A-induced IL-3-independent cell growth by >10-fold (Fig. 1B). Concentrations of 2 μM NaPP1 did not suppress BCR-ABL WT-induced IL-3-independent cell growth, unlike 2 μM imatinib mesylate, demonstrating specificity of NaPP1 at selectively inhibiting the BCR-ABL T315A form (Fig. 1B).
KIT can be up-regulated and activated by BCR-ABL (11, 12), and some CML patients have activating mutations in KIT (13). These results bring into question whether the effectiveness of imatinib mesylate in treating Ph+ patients is due to the inhibitor's ability to suppress both BCR-ABL and KIT or solely BCR-ABL tyrosine kinase activity. To determine whether NaPP1 could be used to selectively inhibit BCR-ABL T315A activity without affecting KIT signaling, we examined whether NaPP1 was capable of suppressing KIT activity, similar to the KIT monospecific inhibitor ACK45, a monoclonal antibody directed to the ectodomain of the receptor (ACK45 product 553867, BD Pharmingen), and the BCR-ABL and KIT inhibitor imatinib mesylate.
WT KIT was introduced to BaF3 cells, which do not express KIT. ACK45 and imatinib mesylate suppressed stem cell factor (SCF)-induced KIT tyrosine phosphorylation and IL-3-independent growth (Supporting Text and Fig. 6, which are published as supporting information on the PNAS web site). NaPP1 did not demonstratively inhibit SCF-induced KIT activation (Supporting Text and Fig. 6).
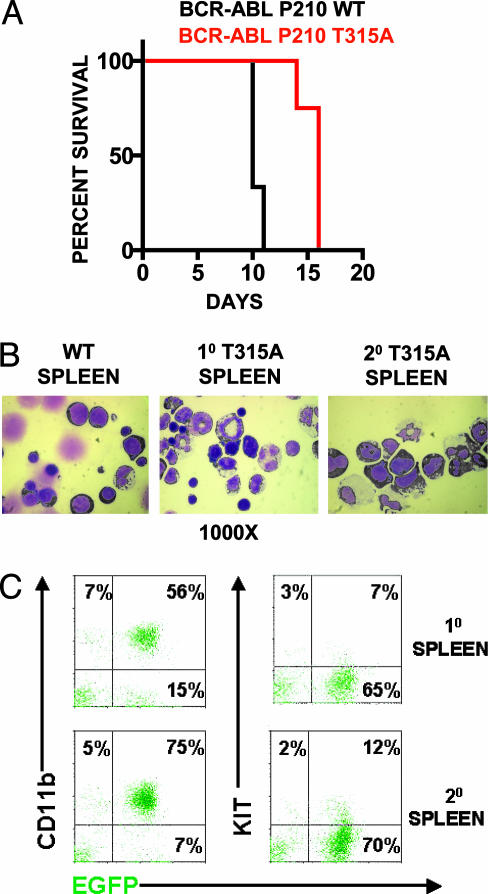
BCR-ABL T315A Generates a Transplantable MPD similar to BCR-ABL WT. To critically determine whether BCR-ABL T315A had similar in vivo oncogenic effects as BCR-ABL WT, we compared BCR-ABL T315A and BCR-ABL WT expression for the development of a murine MPD. All mice transplanted with either BCR-ABL T315A (4) or BCR-ABL WT (3) transduced hematopoietic stem cell and progenitor-enriched populations developed indistinguishable fatal MPDs (Fig. 2A). Both groups of mice succumbed to disease 10–16 days after transplantation with enlarged spleens weighing up to 1,000 mg, leukemic cell infiltration in the liver and spleen (data not shown), and pulmonary hemorrhages. The small significant difference in time to death, as analyzed by log rank test, is likely due to differences in virus titers used in the experiment. Both groups had a dominance of leukemic cells (50–90%) in the peripheral blood, spleen, and bone marrow (Fig. 2B and data not shown). A total of 55–75% of the cells were composed of mature leukemic myeloid cells characterized by EGFP expression and CD11b expression, with undetectable expression of KIT, B220, Ter-119, CD4, or CD8 (Fig. 2C and data not shown). A total of 7–15% of cells had blast-like morphology and expressed EGFP and KIT in the absence of other mature cell markers, which is suggestive of immature leukemic progenitors (Fig. 2 B and C and data not shown).
Fig. 2.
BCR-ABL T315A generates a transplantable MPD similar to BCR-ABL WT. (A) Survival curves of mice transplanted with BCR-ABL T315A or BCR-ABL WT cells. Generation of MPD was as described (27). Cell morphology (B) and cell surface expression (C) of CD11b and KIT in leukemic cells from mice transplanted BCR-ABL T315A expressing cells were conducted as described (26–28). Similar patterns of expression were observed for BCR-ABL WT cell populations. Slides were photographed at ×1,000. Cytospins and flourescence-activated cell sorter plots represent an animal from each group, and similar results were obtained in all leukemic mice from each group.
BCR-ABL T315A-expressing primary leukemic spleen cells were transplanted into secondary recipients and generated phenotypically indistinguishable MPDs in 10 of 10 mice 3–4 weeks after transplantation (Fig. 2B). Leukemic cells from secondary recipients were largely composed of BCR-ABL-expressing mature myeloid cells with a small population of BCR-ABL-expressing hematopoietic progenitors similar to the population of primary leukemic cells (Fig. 2C). These results demonstrate that BCR-ABL T315A generates a MPD phenotype indistinguishable from that of BCR-ABL WT.
Imatinib Mesylate Eliminates a Wider Range of BCR-ABL T315A-Expressing MPD Cells Than NaPP1. To examine whether BCR-ABL inhibition alone was as effective as both BCR-ABL and KIT inhibition in suppressing BCR-ABL induced MPDs, the expansion of BCR-ABL T315A-expressing primary splenic MPD cells 48 h after NaPP1, ACK45, NaPP1 plus ACK45, or imatinib mesylate addition was analyzed. Imatinib mesylate suppressed cell expansion 5-fold, a level similar to both NaPP1 and ACK45 addition (Fig. 3A). However, sole inhibition of BCR-ABL T315A by NaPP1 consistently suppressed cell expansion to a lesser degree of 3- to 4-fold (Fig. 3A). Specific KIT inhibition by ACK45 led to a slight reduction in cell expansion (Fig. 3A). These results suggest that there may be a BCR-ABL- and KIT-expressing MPD population that is resistant to elimination by sole BCR-ABL inhibition and may require both BCR-ABL and KIT inhibition for elimination.
Fig. 3.
Imatinib mesylate eliminates a wider range of BCR-ABL T315A-expressing MPD cells than NaPP1. (A) Inhibitor concentrations as shown were added to BCR-ABL T315A-expressing MPD cells as described in Materials and Methods, and cell counts as measured by trypan blue exclusion were obtained 48 h after inhibitor addition. (B) Inhibitor concentrations as shown in A were added to BCR-ABL T315A-expressing MPD cells, and EGFP, KIT and CD11b levels were measured 48 h after inhibitor addition as described (26–28). Two separate experiments were conducted in duplicate with similar results.
To investigate whether NaPP1, ACK45, NaPP1 plus ACK45, and imatinib mesylate inhibitors differentially eliminated specific populations of primary MPD cells, the distribution of BCR-ABL T315A-expressing KIT- and CD11b-positive live cells 48 h after inhibitor addition was examined. Imatinib mesylate or NaPP1 plus ACK45 addition to cell cultures similarly lead to a dramatic reduction in the percentage of BCR-ABL- and KITexpressing progenitors from 15–17% to between 0.7% and 2% (Fig. 3B). Surprisingly, NaPP1 did not lead to a demonstrable decrease in the percentage of BCR-ABL T315A- and KIT-expressing progenitors (Fig. 3B). ACK45 addition reduced the percentage of BCR-ABL T315A- and KIT-expressing progenitors to between 6% and 7% (Fig. 3B). These results demonstrate that elimination of BCR-ABL- and KIT-expressing progenitors requires both BCR-ABL and KIT inhibition.
These results are in contrast to BCR-ABL T315A expression in the cellular context of more mature myeloid cells, where NaPP1 was quantitatively as effective as imatinib mesylate or NaPP1 plus ACK45 at reducing the percentage of live CD11b-expressing cells from 69–65% to 55% 48 h after inhibitor(s) addition and increasing the fraction of sub-G1 apoptotic cells to 28–29% (Fig. 3B). In this cell population, which does not express KIT (data not shown), ACK45 did not lead to a demonstrable decrease in the percentage of mature myeloid cells or increase apoptotic cells (Fig. 3B).
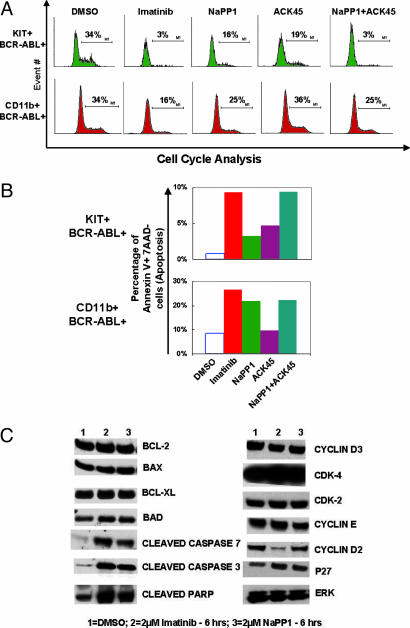
Simultaneous KIT and BCR-ABL Inhibition Has a Synergistic Effect in Cell Cycle Suppression and an Additive Effect in Apoptotic Induction of KIT-Positive MPD Cells. CML inhibitors have been shown to suppress leukemic cell expansion by both suppressing cell proliferation and inducing apoptosis (3). We examined the contribution of selective BCR-ABL, KIT, and BCR-ABL plus KIT inhibition in reducing the percentage of cells in the S/G2/M phase of the cell cycle and in inducing cell death in specific KIT- or CD11b-positive MPD populations.
NaPP1 or ACK45 addition reduced the percentage of KIT-positive cells in the S/G2/M phase of the cell cycle by 2-fold (Fig. 4A). In contrast, imatinib mesylate or NaPP1 plus ACK45 reduced the percentage of KIT-positive cells in the S/G2/M phase of the cell cycle by 10-fold (Fig. 4A). NaPP1 or ACK45 addition induced apoptosis in 3–4% of KIT-positive MPD cells (Fig. 4B). Imatinib mesylate or NaPP1 plus ACK45 addition induced apoptosis in 9% of KIT-positive MPD cells (Fig. 4B). These results demonstrate that BCR-ABL and KIT inhibition have a synergistic effect at blocking S/G2/M-phase cell cycle progression and an additive effect in apoptotic induction of KIT-expressing MPD cells.
Fig. 4.
Simultaneous KIT and BCR-ABL inhibition has a synergistic effect in cell cycle suppression and an additive effect in apoptotic induction of KIT-positive MPD cells. (A) NaPP1 (2 μM), imatinib mesylate (2 μM), and ACK45 (10 μg/ml) inhibitor concentrations were added in the combinations shown to BCR-ABL T315A-expressing MPD cells every 24 h, and cell cycle analyses were conducted 48 h after inhibitor addition as described in Materials and Methods. (B) Inhibitor concentrations as shown in A were added in combinations shown to BCR-ABL T315A-expressing MPD cells every 24 h, and levels of annexin V staining were measured 48 h after inhibitor addition as described (26). (C) Concentrations of NaPP1 (2 μM) and imatinib mesylate (2 μM) were added to BCR-ABL T315A-expressing MPD cells, and levels of examined proteins were measured 6 h after inhibitor addition as described in Materials and Methods. Two separate experiments were conducted in duplicate with similar results.
In the cellular context of mature myeloid MPD cells, ACK45 inhibition of KIT does not additively or synergistically reduce the percentage of cells in the S/G2/M phase of the cell cycle beyond that of sole BCR-ABL inhibition (Fig. 4A). ACK45 addition did not induce apoptosis above control cells, and NaPP1 addition induced apoptosis to similar levels as NaPP1 plus ACK45 addition (Fig. 4B).
We next examined levels of cell cycle and apoptotic regulatory protein expression upon NaPP1 or imatinib mesylate addition in total MPD cells. NaPP1 or imatinib mesylate addition induced the generation of cleaved proapoptotic molecules Caspase 3, Caspase 7, and PARP to similar levels (Fig. 4C). NaPP1 or imatinib mesylate addition did not lead to significant changes in the levels of proapoptotic molecules BAX and BAD, and antiapoptotic molecules BCL-2 and BCL-XL (Fig. 4C).
In contrast, imatinib mesylate, but not NaPP1, dramatically reduced levels of the G1/S phase cell cycle regulator cyclin D2 (Fig. 4C). Expression of other cell cycle regulators examined were not significantly altered upon imatinib mesylate or NaPP1 addition (Fig. 4C). These results suggest that cyclin D2 may be a critical molecule suppressed upon simultaneous BCR-ABL and KIT inhibition to reduce cell cycle progression.
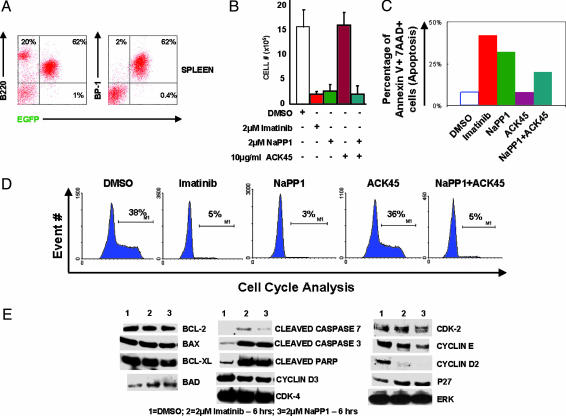
In the Cellular Context of Pro/Pre B cells, NaPP1 Is Quantitatively and Qualitatively as Effective as Imatinib Mesylate in Suppressing BCR-ABL-Induced Cell Expansion. In addition to generating CML, BCR-ABL expression accounts for 5–15% of human B-ALL (10). We first demonstrated that BCR-ABL T315A-expressing primary Pro/Pre B cells generated an in vivo B-ALL-like disease indistinguishable from BCR-ABL WT (with enlarged spleens and lymph nodes). Cells in the peripheral blood, spleen, and bone marrow of moribund mice were dominated by 50–80% of BCR-ABL-expressing Pro/Pre B cells characterized by EGFP, BP-1, and B220 expression and lymphoid cell morphology (Fig. 5A and data not shown).
Fig. 5.
In the cellular context of Pro/Pre B cells, NaPP1 is quantitatively and qualitatively as effective as imatinib mesylate in suppressing BCR-ABL-induced cell expansion. Generation of B-ALL was as described in Materials and Methods. (A) Flow cytometry analysis of EGFP, BP-1, and B220 expression was as described (26–28). (B) NaPP1 (2 μM), imatinib mesylate (2 μM), and ACK45 (10 μg/ml) inhibitor concentrations were added every 24 h in combinations shown to BCR-ABL T315A-expressing Pro/Pre B cells, and viable cell counts as measured by trypan blue exclusion were obtained 48 h after inhibitor addition. BCR-ABL T315A-expressing Pro/Pre B cells were maintained as described (28). (C) Inhibitor concentrations as shown in B were added, and levels of annexin V staining were measured 48 h after inhibitor addition as described in Materials and Methods and ref. 26. (D) Inhibitor concentrations as described in B and DNA content for cell cycle analysis measured as described in Materials and Methods. (E) Concentrations of NaPP1 (2 μM) and imatinib mesylate (2 μM) were added to BCR-ABL T315A-expressing Pro/Pre B cells, and levels of noted proteins were measured 6 h after inhibitor addition as described in Materials and Methods. Two separate experiments were conducted in duplicate with similar results.
Inhibition of BCR-ABL T315A activity by NaPP1 or imatinib mesylate suppressed cell expansion at quantitatively similar levels of 6- to 7-fold 48 h after inhibitor addition (Fig. 5B). Addition of ACK45 and NaPP1 did not quantitatively suppress cell expansion beyond that of cell cultures solely inhibited by NaPP1 (Fig. 5B). Monospecific inhibition of KIT by ACK45 did not suppress cell growth of BCR-ABL T315A-expressing Pro/Pre B cells (Fig. 5B). Inhibition of additional protein kinases by imatinib mesylate or ACK45 did not increase the level of apoptosis beyond that induced by NaPP1 monospecific inhibition of BCR-ABL T315A in Pro/Pre B cells (Fig. 5C).
NaPP1 or imatinib mesylate reduced the percentage of S/G2/M-phase cells from 37–39% to 3–5% 48 h after inhibitor addition (Fig. 5D). Addition of ACK45 and NaPP1 did not quantitatively reduce the percentage of cells in S/G2/M phase of the cell cycle beyond that of cell cultures solely inhibited by NaPP1, and monospecific inhibition of KIT by ACK45 did not reduce the percentage of cells in S/G2/M phase of the cell cycle (Fig. 5D).
In the cellular context of BCR-ABL T315A-expressing Pro/Pre B cells, imatinib mesylate, or NaPP1 up-regulated the cleaved proapoptotic protein products Caspase 3 and PARP at quantitatively similar levels without affecting BAX, BAD, BCL-2, and BCL-XL expression (Fig. 5E). Imatinib mesylate or NaPP1 suppressed cyclin D2 at quantitatively similar levels without significantly altering the expression of cyclin D3, cyclin E, p27KIP, CDK-2, and CDK-4 (Fig. 5E).
Discussion
Strong evidence for the essential role of BCR-ABL in the Ph+ CML and ALL comes from the spectrum of mutations selected in CML patients who become resistant to imatinib mesylate (30, 31). We examined whether monospecific BCR-ABL inhibition by the drug NaPP1 was as effective as imatinib mesylate in eliminating leukemic cells derived from a BCR-ABL-induced MPD. We demonstrated that NaPP1 and imatinib mesylate were quantitatively equivalent in eliminating BCR-ABL expressing mature myeloid cells. However, NaPP1 was unable to eliminate KIT-positive progenitors, unlike imatinib mesylate. These results suggest that imatinib mesylate's effectiveness in eliminating Ph+ CML populations may be due to the drug's ability to suppress multiple signaling pathways required for the survival of distinct leukemic cell populations, in addition to BCR-ABL. Imatinib mesylate treatment is not sufficient to eliminate primitive Ph+ hematopoietic stem cells (32, 33). These findings suggest that inhibition of additional pathways not suppressed by imatinib mesylate will be required to eliminate all Ph+ CML clones.
KIT signaling has been shown to be essential in normal Pro B and Pro T cell development (34). BCR-ABL expression accounts for 5–15% of human B-ALL cases (3). In our murine model, we could not detect KIT expression on the Pro B/Pre B populations transformed by BCR-ABL. We examined whether the effectiveness of imatinib mesylate in suppressing Ph+ B-ALL could be due to the drug's ability in blocking low levels of KIT or other unknown kinases, in addition to BCR-ABL activity. We demonstrated that monospecific BCR-ABL inhibition by NaPP1 was quantitatively as effective as imatinib mesylate in eliminating BCR-ABL-expressing Pro/Pre B populations. Other oncogenic changes outside the spectrum of imatinib mesylate may contribute to the refractory nature of this disease.
In BCR-ABL-expressing Pro/Pre B cells, cyclin D2 down-regulation correlated with suppression of cell growth and apoptosis. Cyclin D2-deficient bone marrow cells have been shown to be resistant to BCR-ABL-induced Pro/Pre B cell transformation (35). Our laboratory has previously demonstrated that overexpression of cyclin D1 complements BCR-ABL transformation (36). In the cellular context of BCR-ABL MPD-expressing KIT-positive cells, suppression of KIT activity correlated with cell death and cyclin D2 down-regulation. These results suggest that cyclin D2 up-regulation could be a common mechanism used by BCR-ABL-expressing KIT-positive and Pro/Pre cells for cell growth and survival. A significant percentage of Ph+ acute phase CML and B-ALL patients relapse from imatinib mesylate because of mutations in BCR-ABL rendering the protein resistant to drug inhibition. Cyclin D2 induces cell cycle progression by activation of its associated serine kinases CDK4 and -6 (37, 38). Inactivation of this signaling mechanism may be an effective approach in suppressing disease development in imatinib mesylate-resistant Ph+ CML and B-ALL patients.
Recently, C. Kung, D. M. Kenski, S. H. Dickerson, R. W. Howson, L. F. Kuyper, H. D. Madhani, and K.M.S. (unpublished work) demonstrated that the effectiveness of the drug GW400426 in blocking cell growth was due to the inhibitor's ability to simultaneously suppress both CDK1 and PHO85 activity. These results provide further support that the effectiveness of some small molecule kinase inhibitors can be due to multiple targets for inhibition. The use of allele-specific protein kinase drug inhibitory systems can help to identify distinct therapeutic targets for cancer and other diseases.
Supplementary Material
Acknowledgments
We are grateful to James Johnson for excellent technical assistance with animal work and Barbara Anderson for excellent preparation of the manuscript. O.N.W. is an Investigator of the Howard Hughes Medical Institute. This work was partially supported by National Institutes of Health Grants CA76204 (to O.N.W.) and AI044009 (to K.M.S.).
Author contributions: S.W. wrote the paper.
Abbreviations: Ph, Philadelphia; CML, chronic myelogenous leukemia; ALL, acute lymphoblastic leukemia; B-ALL, B cell ALL; MPD, myeloproliferative disorder; EGFP, enhanced GFP; NaPP1, 4-amino-1-tert-butyl-3-(1-naphthyl)pyrazolo[3,4-d]pyrimidine; PARP, poly(ADP ribose) polymerase.
References
- 1.Blume-Jensen, P. & Hunter, T. (2001) Nature 411, 355–365. [DOI] [PubMed] [Google Scholar]
- 2.Gilliland, D. G. & Griffin, J. D. (2002) Blood 100, 1532–1542. [DOI] [PubMed] [Google Scholar]
- 3.Wong, S. & Witte, O. N. (2004) Annu. Rev. Immunol. 22, 247–306. [DOI] [PubMed] [Google Scholar]
- 4.Blencke, S., Ullrich, A. & Daub, H. (2003) J. Biol. Chem. 278, 15435–15440. [DOI] [PubMed] [Google Scholar]
- 5.Nowell, P. C. & Hungerford, D. A. (1960) Science 132, 1497–1501. [Google Scholar]
- 6.Rowley, J. D. (1973) Nature 243, 290–293. [DOI] [PubMed] [Google Scholar]
- 7.Pendergast, A. M., Gishizky, M. L., Havlik, M. H. & Witte, O. N. (1993) Mol. Cell. Biol. 13, 1728–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pear, W. S., Miller, J. P., Xu, L., Pui, J. C., Soffer, B., Quackenbush, R. C., Pendergast, A. M., Bronson, R., Aster, J. C., Scott, M. L. & Baltimore, D. (1998) Blood 92, 3780–3792. [PubMed] [Google Scholar]
- 9.Zhang, X. & Ren, R. (1998) Blood 92, 3829–3840. [PubMed] [Google Scholar]
- 10.Deininger, M. W., Goldman, J. M. & Melo, J. V. (2000) Blood 96, 3343–3356. [PubMed] [Google Scholar]
- 11.Hallek, M., Danhauser-Riedl, S., Herbst, R., Warmuth, M., Winkler, A., Kolb, H. J., Druker, B., Griffin, J. D., Emmerich, B. & Ullrich, A. (1996) Br. J. Haematol. 94, 5–16. [DOI] [PubMed] [Google Scholar]
- 12.Pierce, A., Spooncer, E., Ainsworth, S. & Whetton, A. D. (2002) Oncogene 21, 3068–3075. [DOI] [PubMed] [Google Scholar]
- 13.Inokuchi, K., Yamaguchi, H., Tarusawa, M., Futaki, M., Hanawa, H., Tanosaki, S. & Dan, K. (2002) Leukemia 16, 170–177. [DOI] [PubMed] [Google Scholar]
- 14.Shah, N. P., Tran, C., Lee, F. Y., Chen, P., Norris, D. & Sawyers, C. L. (2004) Science 305, 399–401. [DOI] [PubMed] [Google Scholar]
- 15.Wisniewski, D., Lambek, C. L., Liu, C., Strife, A., Veach, D. R., Nagar, B., Young, M. A., Schindler, T., Bornmann, W. G., Bertino, J. R., et al. (2002) Cancer Res. 62, 4244–4255. [PubMed] [Google Scholar]
- 16.Huang, M., Dorsey, J. F., Epling-Burnette, P. K., Nimmanapalli, R., Landowski, T. H., Mora, L. B., Niu, G., Sinibaldi, D., Bai, F., Kraker, A., et al. (2002) Oncogene 21, 8804–8816. [DOI] [PubMed] [Google Scholar]
- 17.Huron, D. R., Gorre, M. E., Kraker, A. J., Sawyers, C. L., Rosen, N. & Moasser, M. M. (2003) Clin. Cancer Res. 9, 1267–1273. [PubMed] [Google Scholar]
- 18.Bishop, A. C., Shah, K., Liu, Y., Witucki, L., Kung, C. & Shokat, K. M. (1998) Curr. Biol. 8, 257–266. [DOI] [PubMed] [Google Scholar]
- 19.Bishop, A. C., Ubersax, J. A., Petsch, D. T., Matheos, D. P., Gray, N. S., Blethrow, J., Shimizu, E., Tsien, J. Z., Schultz, P. G., Rose, M. D., et al. (2000) Nature 407, 395–401. [DOI] [PubMed] [Google Scholar]
- 20.Weiss, E. L., Bishop, A. C., Shokat, K. M. & Drubin, D. G. (2000) Nat. Cell Biol. 2, 677–685. [DOI] [PubMed] [Google Scholar]
- 21.Niswender, C. M., Ishihara, R. W., Judge, L. M., Zhang, C., Shokat, K. M. & McKnight, G. S. (2002) J. Biol. Chem. 277, 28916–28922. [DOI] [PubMed] [Google Scholar]
- 22.Fan, Q. W., Zhang, C., Shokat, K. M. & Weiss, W. A. (2002) Curr. Biol. 12, 1386–1394. [DOI] [PubMed] [Google Scholar]
- 23.Denzel, A., Hare, K. J., Zhang, C., Shokat, K., Jenkinson, E. J., Anderson, G. & Hayday, A. (2003) J. Immunol. 171, 519–523. [DOI] [PubMed] [Google Scholar]
- 24.Hawley, R. G., Lieu, F. H. L., Fong, Z. C. & Hawley, T. S. (1994) Gene Therapy 1, 136–138. [PubMed] [Google Scholar]
- 25.Pear, W. (1996) in Current Protocols in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (Wiley, Boston), Vol. 2, pp. 9.11.1–9.11.18. [Google Scholar]
- 26.Wong, S., McLaughlin, J., Cheng, D. & Witte, O. (2003) Blood 101, 4088–4097. [DOI] [PubMed] [Google Scholar]
- 27.Wong, S., McLaughlin, J., Cheng, D., Shannon, K., Robb, L. & Witte, O. N. (2003) Proc. Natl. Acad. Sci. USA 100, 11630–11635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kharas, M. G., Deane, J. A., Wong, S., O'Bosky, K. R., Rosenberg, N., Witte, O. N. & Fruman, D. A. (2004) Blood 103, 4268–4275. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Y., Witucki, L. A., Shah, K., Bishop, A. C. & Shokat, K. M. (2000) Biochemistry 39, 14400–14408. [DOI] [PubMed] [Google Scholar]
- 30.Shah, N. P., Nicoll, J. M., Nagar, B., Gorre, M. E., Paquette, R. L., Kuriyan, J. & Sawyers, C. L. (2002) Cancer Cell 2, 117–125. [DOI] [PubMed] [Google Scholar]
- 31.Shah, N. P. & Sawyers, C. L. (2003) Oncogene 22, 7389–7395. [DOI] [PubMed] [Google Scholar]
- 32.Graham, S. M., Jorgensen, H. G., Allan, E., Pearson, C., Alcorn, M. J., Richmond, L. & Holyoake, T. L. (2002) Blood 99, 319–325. [DOI] [PubMed] [Google Scholar]
- 33.Bhatia, R., Holtz, M., Niu, N., Gray, R., Snyder, D. S., Sawyers, C. L., Arber, D. A., Slovak, M. L. & Forman, S. J. (2003) Blood 101, 4701–4707. [DOI] [PubMed] [Google Scholar]
- 34.Agosti, V., Corbacioglu, S., Ehlers, I., Waskow, C., Sommer, G., Berrozpe, G., Kissel, H., Tucker, C. M., Manova, K., Moore, M. A., et al. (2004) J. Exp. Med. 199, 867–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jena, N., Deng, M., Sicinska, E., Sicinski, P. & Daley, G. Q. (2002) Cancer Res. 62, 535–541. [PubMed] [Google Scholar]
- 36.Afar, D. E. H., McLaughlin, J., Sherr, C. J., Witte, O. N. & Roussel, M. F. (1995) Proc. Natl. Acad. Sci. USA 92, 9540–9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherr, C. J. (1996) Science 274, 1672–1677. [DOI] [PubMed] [Google Scholar]
- 38.Sherr, C. J. (2004) Cell 116, 235–246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.