One of the most important chemical concepts is defining how one molecule recognizes and controls the properties of another molecule or ion. Nowhere is this issue more significant than in the field of biomolecular recognition. Metalloproteins efficiently control the geometry and coordination number of metal ions as well as the types of ligands bound to them. Incorporation of metals in the correct binding site is essential for proper biological activity of the protein, regardless of whether the function is catalytic, structural, or regulatory. Despite the importance of metalloproteins, the major factors determining metal-ion selectivity and specificity are not yet well understood. Recent studies on metalloregulatory proteins and carbonic anhydrase suggest that coordination number and geometry of the metal ion are key elements.[1,2] This control becomes even more significant when proteins can bind two metal ions with different coordination environments. This is the case for the enzyme 5′-aminolevulinic acid dehydratase, which contains two ZnII centers, one bound in a five-coordinate geometry and the other bound in a pseudotetrahedral geometry. Interestingly, lead toxicity is mainly due to the replacement by a PbII ion of the ZnII ion bound to the tetrahedral site.[3,4] Thus, a deep knowledge of the factors determining this delicate ion recognition is crucial to understanding the principles that govern metalloprotein structure and function.
Using the de novo designed TRI peptide family (TRI = Ac-G-(LKALEEK)4-G-NH2), we have been investigating the influence of protein environment on metal-ion specificity and how subtle changes in the amino acid sequence can fully control the coordination number of the metal ion. By exploiting our previous observations that we could influence the metal binding affinity in the TRI coiled coils by placing cysteine in either position a or d of the heptad repeat unit, we designed a TRI derivative with two cysteine binding sites (TRI L9CL19C, Table 1) that shows sequential and selective binding of CdII ions, first to the a site and then to the d site.[5] This result highlights how site-specific binding of CdII ions can be achieved in a de novo designed peptide containing two binding sites that are defined by the same first-coordination-sphere ligands, three cysteine residues in both cases, solely by changing the conformation and environment of the metal-binding cavity. Later, using the a binding sites, we designed two peptides, TRI L16Pen and TRI L12AL16C (Table 1), that are able to bind CdII ions exclusively in a trigonal-planar structure and a pseudotetrahedral geometry, respectively. This precise control of the coordination number and geometry of the CdII centers was achieved by controlling the steric constraints in the metal-binding pocket.[6] We know that these complexes, [Cd(TRI L16Pen)3] and [Cd(TRI L12AL16C)3], have different physical properties, as shown by their UV and CD spectra at pH 8.5 ([Cd(TRI L16Pen)3] UV band: λ227 (28 900 m−1 cm−1), CD band: λ227 (5000 deg dmol−1 cm2) and λ257 (−3500 deg dmol−1 cm2); [Cd(TRI L12AL16C)3], UV band: λ230 (21 800 m−1 cm−1), CD band: λ237 (5500 deg dmol−1 cm2) and λ260 (−590 deg dmol−1 cm2)) and also by their different pH profiles for binding CdII centers, as explained below.
Table 1.
Derivatives of the TRI peptide.
| Peptide | Sequence[a] |
|---|---|
| TRI L9CL19C | Ac-G LKALEEK CKALEEK LKACEEK LKALEEK G-NH2 |
| TRI L12AL16C | Ac-G LKALEEK LKAAEEK CKALEEK LKALEEK G-NH2 |
| TRI L16Pen | Ac-G LKALEEK LKALEEK XKALEEK LKALEEK G-NH2 |
| TRI L12AL16CL23Pen | Ac-G LKALEEK LKAAEEK CKALEEK XKALEEK G-NH2 |
| TRI L16PenL23CL26A | Ac-G LKALEEK LKALEEK XKALEEK CKAAEEK G-NH2 |
| Grand L16PenL26AL30C | Ac-G LKALEEK LKALEEK XKALEEK LKAAEEK CKALEEK G-NH2 |
| Grand L26AL30C | Ac-G LKALEEK LKALEEK LKALEEK LKAAEEK CKALEEK G-NH2 |
| Grand L16Pen | Ac-G LKALEEK LKALEEK XKALEEK LKALEEK LKALEEK G-NH2 |
X = Penicillamine. Residues in bold indicate modifications.
While being able to impose a specific coordination number on a metal was a signal achievement, a more challenging construct would be a single polypeptide that utilizes subtle sequence perturbations to bind selectively the same metal into distinct coordination structures. Specifically, our next goal was to design a single short peptide capable of simultaneously binding two CdII ions, one with a trigonal-planar geometry and the second one with a pseudotetrahedral geometry, within the same three-stranded coiled coil. While other investigations have shown that de novo peptides can bind two metals in separate mononuclear sites[5, 7] or in binuclear centers,[8–10] they have not addressed the question of selective binding of the same type of metal ion in different centers having distinct geometries. Two important issues to be addressed with our studies are whether the presence of one metal center influences the properties of a second center and whether different properties can be observed for the same metal in distinct sites. We designate heterochromic peptides as those capable of binding two equivalents of the same metal ion in a similar way while enforcing different physical properties in those metals. This is equivalent to an individual having two eyes, but of different color. These studies are of fundamental importance to understand the factors that control metal-ion specificity, an important objective of de novo designed metalloproteins.
Our initial strategy was to introduce the amino acid modifications that led to the trigonal planar {CdS3} unit and to the pseudotetrahedral {CdS3O} unit into the TRI peptide sequence. The first modification involves the replacement of the cysteine residue with penicillamine, and the second one calls for the substitution of the leucine residue above the cysteine layer for alanine.[6] The 113Cd NMR spectrum of a solution containing our first construct, TRI L12AL16CL23Pen (Table 1), and two equivalents of CdII at pH 9.6 shows two resonances with 113Cd chemical shifts of δ = 593 ppm and 574 ppm. Based on our previous results,[6, 11] these 113Cd chemical shifts are consistent with a peptide that binds two CdII ions, both as primarily the {CdS3O} species (Table 2). Thus, while we were successful obtaining the four-coordinate site, we did not succeed in designing the three-coordinate center. We reasoned that if position 23 was too near the edge of the coiled coil and the coordination of water to the CdII center were promoted by the fraying of the α-helices, then moving the three-coordinate site towards the center of the coiled coil might avoid this situation and yield a binding site with a higher percentage of the three-coordinate species. Hence, we switched the positions of cysteine and penicillamine to obtain the new construct TRI L16PenL23CL26A (Table 1). At pH 9.6, the 113Cd NMR spectrum of a solution containing this peptide and two equivalents of CdII shows two resonances with δ = 676 ppm and 609 ppm. These chemical shifts are consistent with a peptide that binds two CdII centers, one present to 90–95%as the {CdS3} species (δ = 676 ppm) and the other to approximately 80%as the {CdS3O} species (δ = 609 ppm; Table 2). These results indicate that to define the three-coordinate site, it is important to control not only the first coordination sphere around the CdII center but also its location inside the coiled coil.
Table 2.
Percentages of the four-coordinate CdII species {CdS3O} for different peptides of the TRI family.
| Peptide | Four-coordinate CdII {CdS3O}[a][%] | |
|---|---|---|
| L#AL#C or L#CL#A site | L#Pen site | |
| TRI L12AL16C | 100 | – |
| TRI L16Pen | – | 0 |
| TRI L12AL16CL23Pen | 100 | 90 |
| TRI L16PenL23CL26A | 80 | 5–10 |
| Grand L16PenL26AL30C | 100 | 0 |
Values determined based on correlation between the percentages of three- and four-coordinate CdII species from 111mCd PAC spectroscopy (PAC = perturbed angular correlation of γ rays) and the corresponding 113Cd NMR chemical shifts obtained for different CdII complexes of the TRI peptide family.[6, 11]
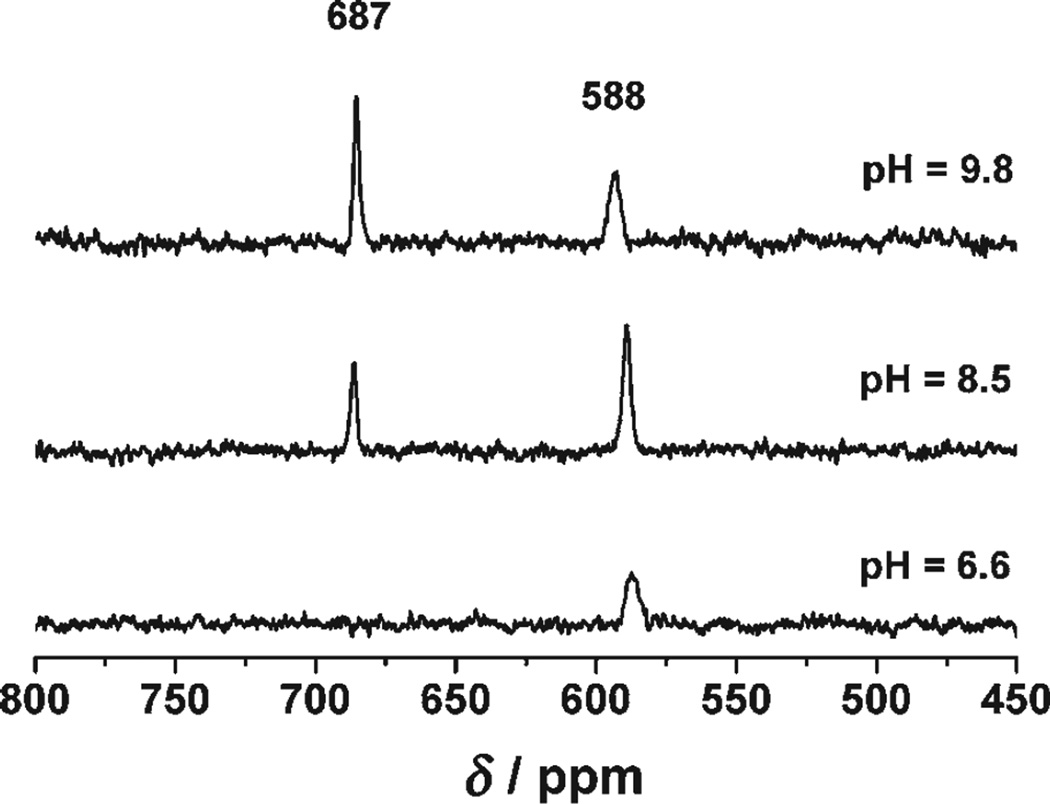
Our next goal was to improve the design of the four-coordinate site, keeping the two binding sites strictly independent. To achieve this aim, we moved to the Grand peptides, which contain an extra heptad repeat unit, thus allowing the introduction of the cysteine residue seven amino acids further down the sequence (L30C). It was felt that this new construct, Grand L16PenL26AL30C (Table 1), should tolerate the introduction of an alanine residue above the cysteine without disrupting the three-coordinate site. The corresponding 113Cd NMR spectrum at pH 9.8 shows two resonances, one at δ = 687 ppm and the other at δ = 588 ppm (Figure 1). These results are consistent with a peptide that binds two CdII ions, one with a pure trigonal planar {CdS3} coordination geometry (δ = 687 ppm) and the other with a pure pseudotetrahedral {CdS3O} geometry (δ = 588 ppm; Table 2).
Figure 1.
113Cd NMR spectra of a solution containing 2.6 mm trimer Grand L16PenL26AL30C and 5.2 mm 113Cd(NO3)2 at different pH values.
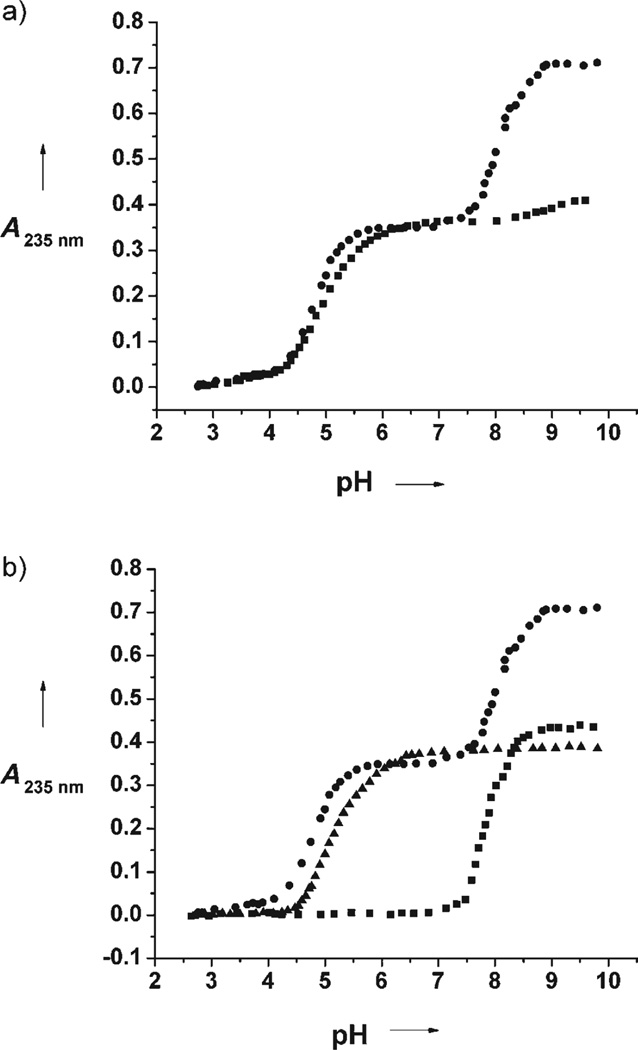
Having succeeded in making a single, short peptide capable of binding CdII ions in a trigonal S3 environment in one position and in a pseudotetrahedral S3O environment in a second position, we next investigated the control of metal-ion binding and the impact on the physical properties of the centers. The pH dependence of the formation of the complex [Cd2(Grand L16PenL26AL30C)3]2− was monitored at 235 nm, and the pH titration curve is shown in Figure 2a. Interestingly, this UV/Vis pH titration reveals a very different pH profile for CdII ions binding to each of the sites in Grand L16PenL26AL30C. Two well-defined regions are observed, and their shapes are consistent with the simultaneous release of two protons, as was observed previously for the TRI peptides and related monosubstituted peptides.[11, 12] The best fit to the experimental data was obtained by analyzing each region independently and using the reported model, which is shown in Equation (1).[12]
| (1) |
Figure 2.
UV/Vis pH titrations: a) pH dependence of 2 equiv (●) and 1 equiv (■) of CdII binding to Grand L16PenL26AL30C (20 µm trimer); b) pH dependence of 2 equiv of CdII binding to Grand L16PenL26AL30C (20 µm trimer; ●), of 1 equiv of CdII binding to Grand L26AL30C (20 µm trimer; ▲), and of 1 equiv of CdII binding to Grand L16Pen (20 µm trimer; ■).
The apparent pKa2 values determined for each region are reported in Table 3. Repetition of the experiment with only one equivalent of CdII in solution indicates selective binding of CdII ions to the lower pKa2 binding site (Figure 2a); however, UV spectral data alone can not establish which site corresponds to the {CdS3} or {CdS3O} geometries.
Table 3.
Apparent pKa2values for TRI and Grand peptides.
| Peptide | Apparent pKa2[a] |
|---|---|
| TRI L12AL16C | 12.2±0.2 |
| TRI L16Pen | 15.3±0.2 |
| Grand L26AL30C | 9.9±0.2 |
| Grand L16Pen | 15.7±0.2 |
| Grand L16PenL26AL30C | 9.6±0.2 (L12AL30C site) |
| 16.1±0.2 (L16Pen site) |
The best fit to the experimental data is consistent with the model given in Equation (1), which corresponds to the simultaneous release of two protons. A = (cMε)/(10(pKa2−2pH)+1), where A is the observed absorbance, cM is the total metal concentration, and ε is the extinction coefficient of the complex.[11, 12]
To assign binding sites to these different pKa2 values, we synthesized two Grand peptides, each containing only one CdII binding site: Grand L26AL30C (four-coordinate site) and Grand L16Pen (three-coordinate site). The pH-dependent curves of CdII binding to these Grand peptides are shown in Figure 2b. These pH profiles are also consistent with the simultaneous release of two protons, and the fit of the experimental data using the model mentioned above [Eq. (1)] give the apparent pKa2 values reported in Table 3. These titrations provide two useful pieces of information. First, the pKa2 values suggest that the low-pKa2 species in Grand L16PenL26AL30C corresponds to the four-coordinate site (pKa2 = 9.6 vs. pKa2 = 9.9 for Grand L26AL30C) while the high pKa2 (pKa2 = 16.1 vs. pKa2 = 15.7 for Grand L16Pen) is associated with the three-coordinate CdII center. Second, the close similarities between the pKa2 Is of CdII ions binding to the monosubstituted peptides with those determined for the disubstituted peptide indicate that the two binding sites in Grand L16PenL26AL30C behave independently of each other.
To confirm our assignments, we carried out 113Cd NMR spectroscopic pH titrations (Figure 1). At pH 6.6 only a single resonance is observed with a 113Cd chemical shift of δ = 588 ppm, indicating binding of CdII ions to the four-coordinate site. When the pH value is raised to 9.8, a second resonance appears at δ = 687 ppm, the hallmark signal for {CdS3}. These results corroborate the UV/Vis pH titrations and clearly show that the binding site capable of sequestering CdII ions at low pH values corresponds to the site with {CdS3O} coordination. The apparent pKa2 value of 9.6 obtained for the CdII ion binding to this site is the lowest observed to date for our TRI peptide family.
Because we know from the pH titrations that the two metal sites in Grand L16PenL26AL30C behave independently of one another, there must be another explanation for the lowering of the pKa2 value of the {CdS3O site}. The most reasonable candidate is the difference in affinity of the Grand versus TRI peptide frameworks. We determined the pH profile of CdII ions binding to TRI L12AL26C and TRI L16Pen. Consistent with the previous data, two protons are released in each system; the apparent pKa2 values are reported in Table 3. All these data indicate that while the pH dependence of CdII ions binding to the three-coordinate site {CdS3} is not affected to a large extent in moving from Grand to TRI peptides, the four-coordinate site is perturbed. This effect is most likely a reflection of different self-association affinities and pH-dependent aggregation state preferences of the peptides themselves. The Grand peptides show a higher self-association affinity than TRI peptides,[13] which could result in the formation of three-stranded coiled coils at pH values lower than 6.0. This observation suggests that the formation and stabilization of the four-coordinate {CdS3O} unit may be achieved at more acidic pH values than those reported herein by increasing the self-association energy of the three-stranded coiled coil relative to the two-stranded coiled coil.
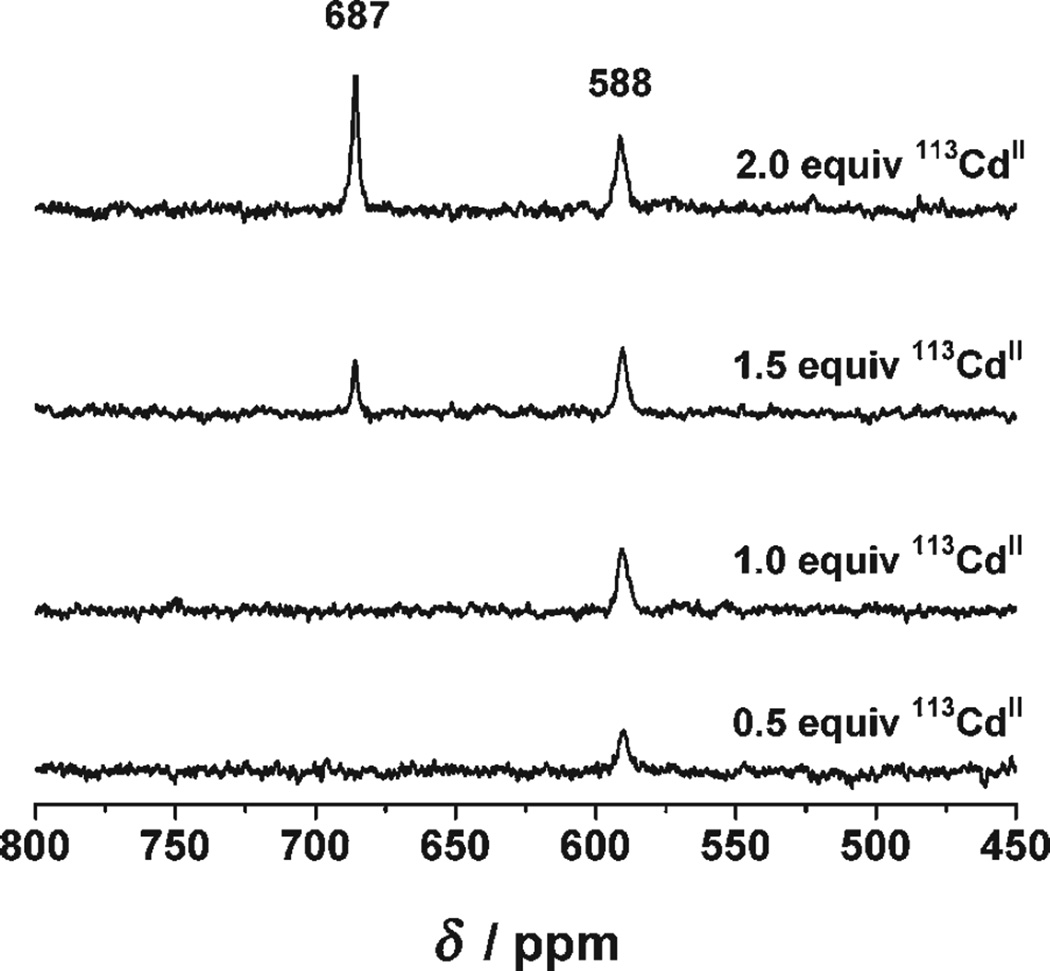
Figure 3 shows the 113Cd NMR spectra corresponding to the titration of successive aliquots of 0.5 equivalents of 113Cd(NO3)2 into a solution containing 2.6 mm trimer Grand L16PenL26AL30C at pH 9.6. A single peak at δ = 588 ppm is observed up to one equivalent of CdII. A second resonance at δ = 687 ppm appears on addition of the second equivalent of CdII. We conclude that under conditions in which CdII ions will bind exclusively as either {CdS3O} (pseudotetrahedral) or {CdS3} (trigonal planar; i.e. pH > 9.0), these data show unambiguously that this metal ion binds selectively to the four-coordinate site. Thus, regardless of the pH value, the four-coordinate site is the first filled by CdII. This result most likely reflects the preference of CdII for this coordination environment. These data, together with the pH titration experiments explained above, point out how CdII ions bound to these two binding sites possess different physical properties and how they behave independently of one another.
Figure 3.
113Cd NMR spectra of solutions containing 2.6 mm trimer Grand L16PenL26AL30C and different amounts of 113Cd(NO3)2 at pH 9.6.
In conclusion, we are able to design a single, short peptide (Grand L16PenL26AL30C) capable of binding two CdII ions within the same three-stranded coiled coil, one with a pseudotetrahedral and the other with a trigonal planar coordination geometry. More interestingly, we have shown how CdII ions bound to the two different sites possess different physical properties. Binding of CdII ions to the four-coordinate site occurs at much lower pH value (pKa2 = 9.6) than binding to the three-coordinate site (pKa2 = 16.1). Moreover, CdII binds selectively to the four-coordinate site under all pH conditions. Thus, the heterochromic peptide Grand L16PenL26AL30C shows site-selective metal-ion recognition and controls the physical properties of CdII centers by controlling their coordination geometry. This advance portends a new generation of heterochromic metallo-biomolecules that may be used as selective bifunctional switches.
Experimental Section
Peptide synthesis and purification: The different peptides of the TRI family (see Table 1 for sequences) were synthesized on an Applied Biosystems 433 A peptide synthesizer using standard protocols[14] and purified and characterized as described previously.[15]
UV/Vis spectroscopy: Fresh solutions of the purified peptides were prepared for each experiment using doubly distilled water. The peptide concentration of the solutions was determined by quantization of the cysteine thiol groups using 4,4′-dipyridyl disulphide.[16] The pH titrations were performed as previously described.[12] A concentration of 60 µm monomer peptide and the corresponding amount of CdCl2 were used in all experiments. In all cases, reverse titrations were carried out to verify the reversibility of the process. The pH-dependent absorption spectra were fit using the same models and procedure as in our previous studies.[11, 12]
113Cd NMR spectroscopy : All spectra were collected at room temperature on a Varian Inova 500 spectrometer (110.92 MHz for 113Cd) equipped with a 5-mm broadband probe. 113Cd NMR spectra were externally referenced to a 0.1m Cd(ClO4)2 solution in D2O. A spectral width of 847 ppm (93897 Hz) was sampled using a 5.0-µs 90° pulse and 0.05-s acquisition time with no delay between scans. Samples were prepared under a flow of argon by dissolving 30–35 mg of the lyophilized and degassed peptides in 450–500 µL 15% D2O solution. The peptide concentrations were determined using 4,4′-dipyridyl disulphide.[16] The final samples were prepared by the addition of the appropriate amount of 250 mm 113Cd(NO3)2 solution (prepared from 95%isotopically enriched 113CdO obtained from Oak Ridge National Laboratory), and the pH value was adjusted with KOH or HCl solution. An argon atmosphere was maintained when possible, but the samples came in contact with O2 while the pH value was adjusted. The actual final concentrations for each experiment are indicated in the text and the figure captions. The data were analyzed using the software MestRe-C.[17] All free induction decays (FIDs) were zero-filled to double the original points and were processed by application of 50-Hz line broadening prior to Fourier transformation.
Acknowledgments
V.L.P. thanks the National Institute of Health for support of this research (R01 ES0 12236) and O.I. thanks the Margaret and Herman Sokol Foundation for a Postdoctoral Award.
Contributor Information
Dr. Olga Iranzo, Department of Chemistry, University of Michigan, Ann Arbor, Michigan 48109-1055 (USA), Fax: (+1) 734-936-7628
Chris Cabello, Department of Chemistry, University of Michigan, Ann Arbor, Michigan 48109-1055 (USA), Fax: (+1) 734-936-7628.
Prof. Vincent L. Pecoraro, Department of Chemistry, University of Michigan, Ann Arbor, Michigan 48109-1055 (USA), Fax: (+1) 734-936-7628 Biophysics Research Division, University of Michigan, Ann Arbor, Michigan 48109-1055 (USA).
References
- 1.McCall KA, Fierke CA. Biochemistry. 2004;43:3979. doi: 10.1021/bi0498914. and references therein. [DOI] [PubMed] [Google Scholar]
- 2.Pennella MA, Arunkumar AI, Giedroc DP. J. Mol. Biol. 2006;356:1124. doi: 10.1016/j.jmb.2005.12.019. and references therein. [DOI] [PubMed] [Google Scholar]
- 3.Erskine PT, Senior N, Awan S, Lambert R, Lewis G, Tickle IJ, Sarwar M, Spencer P, Thomas P, Warren MJ, Shoolingin-Jordan PM, Wood SP, Cooper JB. Nat. Struct. Biol. 1997;4:1025. doi: 10.1038/nsb1297-1025. [DOI] [PubMed] [Google Scholar]
- 4.Jaffe EK, Martins J, Li J, Kervinen J, Dunbrack RLJ. J. Biol. Chem. 2001;276:1531. doi: 10.1074/jbc.M007663200. [DOI] [PubMed] [Google Scholar]
- 5.Matzapetakis M, Pecoraro VL. J. Am. Chem. Soc. 2005;127:18229. doi: 10.1021/ja055433m. [DOI] [PubMed] [Google Scholar]
- 6.Lee K-H, Cabello C, Hemmingsen L, Marsh ENG, Pecoraro VL. Angew. Chem. 2006;118:2930. doi: 10.1002/anie.200504548. Angew. Chem. Int. Ed.2006, 45, 2864. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka T, Mizuno T, Fukui S, Hiroaki H, Oku J-I, Kanaori K, Tajima K, Shirakawa M. J. Am. Chem. Soc. 2004;126:14023. doi: 10.1021/ja047945r. [DOI] [PubMed] [Google Scholar]
- 8.Lombardi A, Summa CM, Geremia S, Randaccio L, Pavone V, DeGrado WF. Proc. Natl. Acad. Sci. USA. 2000;97:6298. doi: 10.1073/pnas.97.12.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Summa CM, Rosenblatt MM, Hong J-K, Lear JD, DeGrado WF. J. Mol. Biol. 2002;321:923. doi: 10.1016/s0022-2836(02)00589-2. [DOI] [PubMed] [Google Scholar]
- 10.Spiegel K, De Grado WF, Klein ML. Proteins Struct. Funct. Bioinf. 2006;65:317. doi: 10.1002/prot.21113. [DOI] [PubMed] [Google Scholar]
- 11.Iranzo O, Ghosh D, Pecoraro VL. Inorg. Chem. 2006;45:9959. doi: 10.1021/ic061183e. [DOI] [PubMed] [Google Scholar]
- 12.Matzapetakis M, Ghosh D, Weng T-C, Penner-Hahn JE, Pecoraro VL. J. Biol. Inorg. Chem. 2006;11:876. doi: 10.1007/s00775-006-0140-7. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh D, Lee K-H, Demeler B, Pecoraro VL. Biochemistry. 2005;44:10732. doi: 10.1021/bi0506674. [DOI] [PubMed] [Google Scholar]
- 14.Chan WC, White PD. Fmoc Solid Phase Peptide Synthesis: A Practical Approach. New York: Oxford University Press; 2000. [Google Scholar]
- 15.Farrer BT, Harris NP, Balchus KE, Pecoraro VL. Biochemistry. 2001;40:14 696. doi: 10.1021/bi015649a. [DOI] [PubMed] [Google Scholar]
- 16.Mantle M, Stewart G, Zayas G, King M. Biochem. J. 1990;266:597. [PMC free article] [PubMed] [Google Scholar]
- 17.Cobas C, Cruces J, Sardina FJ. MestRe-C version 2–3. Spain: Universidad de Santiago de Compostela; 2000. [Google Scholar]