Abstract
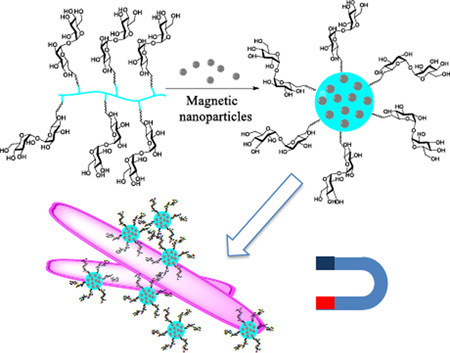
A multivalent trehalose-grafted biodegradable poly(lactic acid) was successfully synthesized and encapsulated with iron oxide magnetic nanoparticles to give stable micelles (87.8 ± 7.4 nm). The magnetic micelles, when treated with concanavalin A, caused a rapid precipitation, indicating the multivalent presentation of trehalose ligands on the micelles. When the magnetic micelles were treated with Mycobacterium smegmatis mc2 155, orange clusters were observed resulting from the interactions between the magnetic micelles and the mycobacteria. Very little particle interaction was found on Staphylococcus epidermidis 35984 or E. coli ORN 208. With the aid of a household magnet, the orange precipitates could be easily seen by the naked eyes at 106 CFU/mL, and 104 CFU/mL under an optical microscope. The method developed represents a new approach to the detection of mycobacteria, which does not require the use of molecular biology reagents or sophisticated instruments.
Keywords: Trehalose, glycopolymer, poly(lactic acid), magnetic micelles, mycobacterium
Graphical abstract
Tuberculosis (TB), caused by Mycobacterium (M.) tuberculosis, has surpassed HIV to become the top infectious disease, killing 1.5 million people and creating 10 million new cases each year.[1] The resurgence of TB, especially the rapid increase in multidrug-resistance TB cases, calls for new diagnostic and therapeutic strategies. A timely and rapid diagnosis is the first critical step to ensure effective treatment. However, this remains a significant challenge, especially in resource-limited regions. Globally, one third of all TB cases are unidentified.[2] This results in delays in providing patients with the appropriate treatment, and contributes to the ongoing global TB epidemic. A number of methods are currently in use for TB diagnosis. Culturing has been the gold standard, which involves growing the bacteria from the patient’s sample in a growth medium or on an agar plate.[3] It has high sensitivity, but results can take several weeks to obtain due to the slow-growing nature of TB.[4] The nucleic acid test employs the PCR (polymerase chain reaction) technique to identify the nucleic acid sequence that is specific for M. tuberculosis.[5] The recently developed GeneXpert, a fully automated PRC-based TB detection instrument, can give the results quickly and with reasonably high accuracy.[6] Other novel approaches are also reported, however, most relying on immunoassays and/or special instruments.[7,8] Therefore, the infrastructure and material costs are still relatively high, and the operation requires trained staff and uninterrupted power, air conditioning and security provisions for external computers. In low-income countries where 95% of the TB cases and 98% of the death occur, the sputum smear microscopy has been the primary method for the diagnosis of pulmonary tuberculosis, owing to its low material and infrastructure cost.[9] The method, however, has a number of drawbacks. The process is labor intensive and time consuming. The lab technician must meticulously examine each slide and then record the number of organisms observed. Moreover, the method has poor sensitivity of 20–80%, and especially in HIV-co-infected patients.[10]
In this work, we report the development of a new assay based on the selective targeting of trehalose for mycobacteria. Trehalose is a disaccharide of d-glucose connected through an α-1,1-glycosidic bond. It is a key component of the mycobacterial cell wall as well as the cytosol.[11] The mycobacterium uses trehalose as an osmoprotectant and produces various trehalose-containing glycolipids in its cell wall, which are essential for its growth and proliferation.[12] In addition, trehalose dimycolate is proposed to be possibly involved in cell infection by specifically interacting with macrophage inducible C-type lectin (Mincle).[13] In previous studies, we found that trehalose-functionalized nanoparticles interacted strongly with M. smegmatis.[14] Furthermore, the trehalose-functionalized nanoparticles selectively targeted M. smegmatis in the presence of mammalian cells. This opens up opportunities to develop trehalose-functionalized nanomaterials for diagnosis and therapeutics. For example, we have shown that trehalose-functionalized mesoporous nanoparticles were able to increase the antimicrobial efficacy of antibiotics.[15] Here, we designed a trehalose-grafted multivalent glycopolymer based on poly(lactic acid) (PLA). Micelles were prepared from the glycopolymer and were encapsulated with magnetic nanoparticles for the capture and detection of mycobacteria.
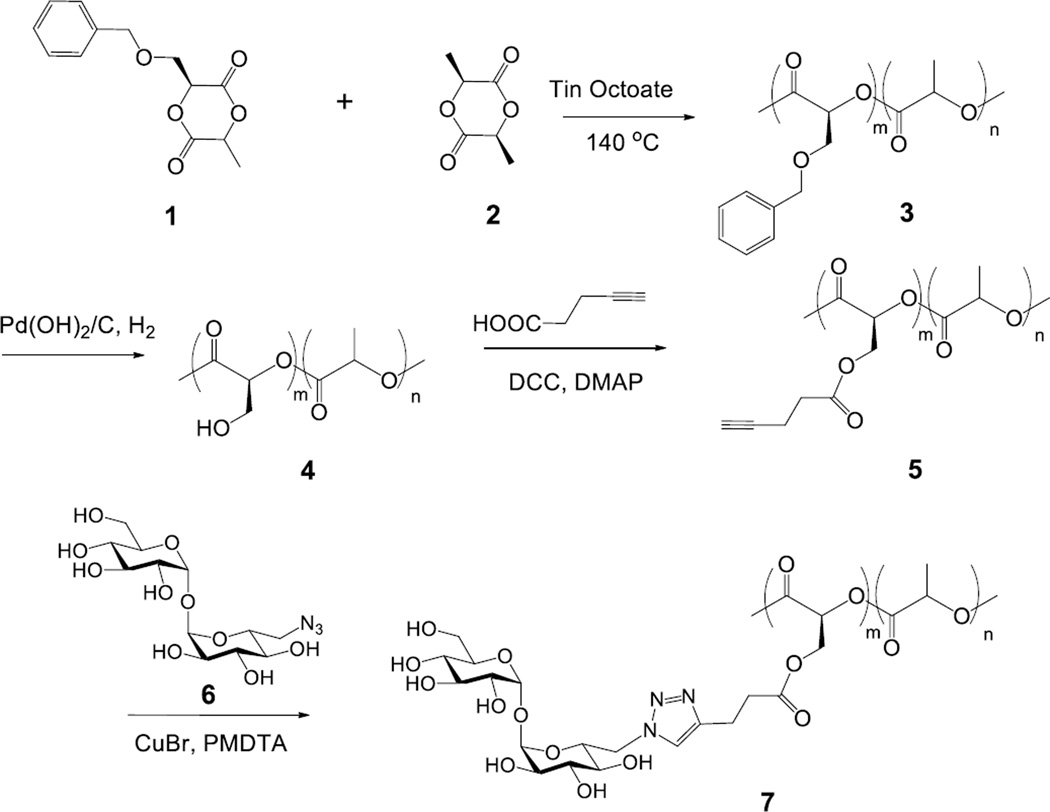
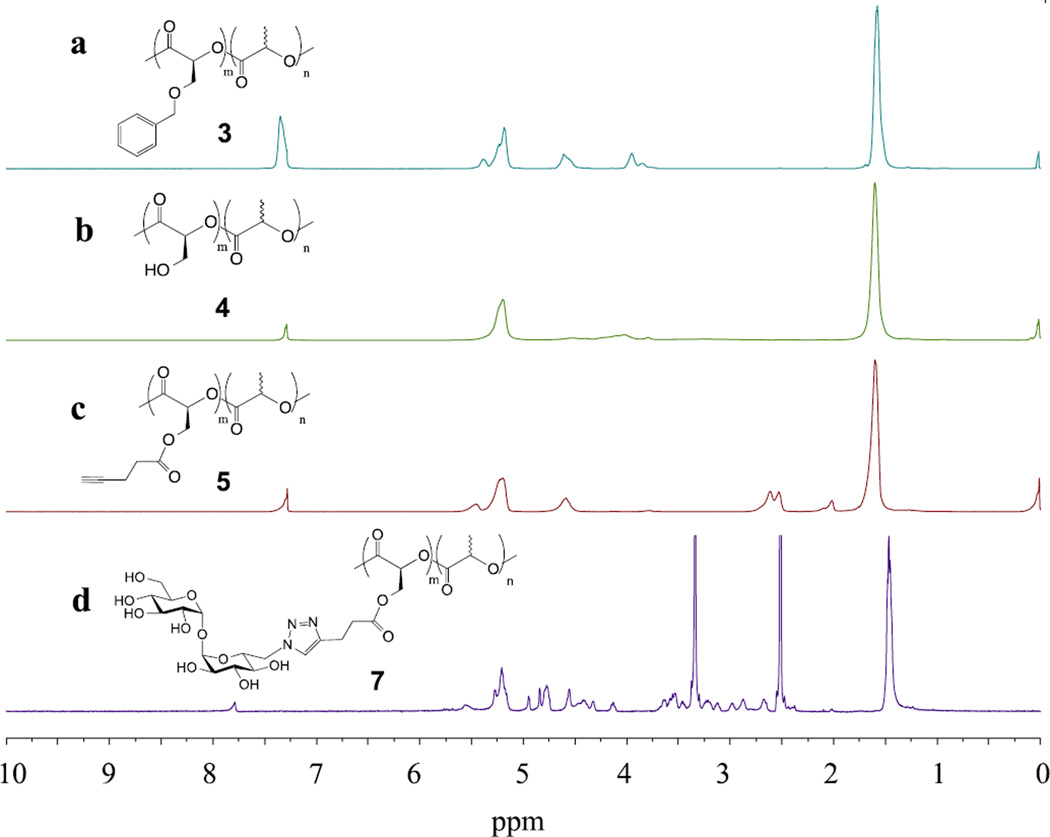
The trehalose glycopolymer was synthesized following the reaction sequence in Scheme 1. The benzyloxy-functionalized lactide monomer 1 was prepared following a literature procedure (Scheme S1).[16] The monomer 1 and L-lactide 2 were copolymerized in bulk with a catalytic amount of tin octoate at 140 °C to give a random copolymer 3. Since the monomer 1 had a bulky benzyloxy group, a higher reaction temperature and longer reaction time were needed to achieve high conversion. The composition of copolymer 3 (m:n) was calculated from 1H NMR to be 1:7 from the integrals at 4.6 ppm (PhCH2O-) and 1.6 ppm (-CH3) (Figure 1a). Deprotection of the benzyl group by catalytic hydrogenation gave copolymer 4, which was accompanied by the disappearance of the peak at 4.6 ppm (Figure 1b). A decrease in the molecular weight was observed from the GPC (gel permeation chromatography) after 3 was converted to 4 (Table 1 and Figure S1). 4-Pentynoic acid was then coupled onto copolymer 4, yielding the alkyne-derivatized copolymer 5. The 1H NMR spectrum showed the shifts of -O(-CH2)CH(CO)- and -O(-CH2)CH(CO)- from 4.0/5.2 ppm (Figure 1b) to 4.6/5.4 ppm, together with a new peak at 2.0 ppm, which corresponds to the alkyne proton (Figure 1c). Trehalose was then grafted to the copolymer by Cu(I)-catalyzed click reaction of alkyne-derivatized copolymer 5 and 6-azido-6-deoxy-trehalose (6) synthesized following a literature procedure (Scheme S2).[17] The 1H NMR spectrum indicated 90% conversion after overnight reaction. The product was purified by dialyzing in water followed by freeze-drying to give trehalose-grafted glycopolymer 7 (Figure 1d). The glycopolymer was soluble only in a polar solvent such as DMSO or DMF due to the high trehalose content (29 wt%, see calculation in SI). Without trehalose, the copolymers 3, 4 and 5 were soluble in most common organic solvents. GPC showed an increase of M̅n from 19,800 to 34,100, and a slight increase of PDI from 1.38 to 1.76 after trehalose conjugation (Table 1).
Scheme 1.
Synthesis of trehalose glycopolymer.
Figure 1.
1H NMR spectra of copolymer a) 3, b) 4, c) 5 in CDCl3, and d) 7 in DMSO-d6.
Table 1.
Molecular weights of copolymers measured by GPC using PMMA in DMF as the calibration standard.
| Copolymer | M̅n | M̅w | PDI |
|---|---|---|---|
| 3 | 16500 | 26200 | 1.60 |
| 4 | 12100 | 19400 | 1.60 |
| 5 | 19800 | 27300 | 1.38 |
| 7 | 34100 | 60000 | 1.76 |
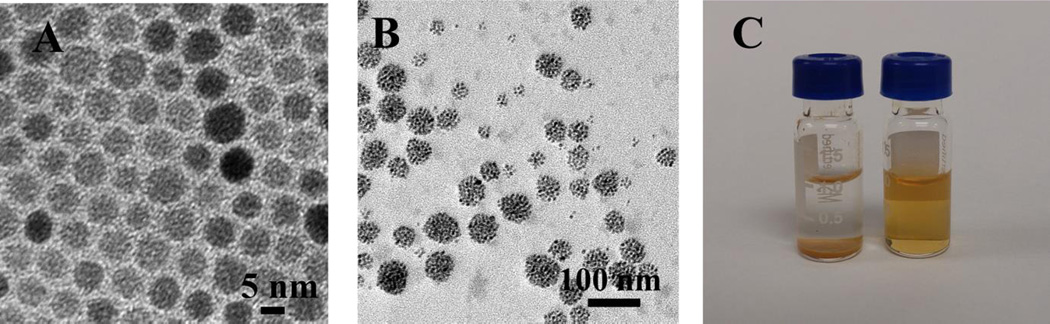
The glycopolymer 7 carrying multiple trehalose ligands could form stable micelles in water in the absence of any surfactant. To prepare the micelles, the glycopolymer was dissolved in DMSO, and the solution was slowly added into water under vigorous stirring.[18] After dialysis, micelles with a hydrodynamic diameter of 34.1 ± 3.3 nm were obtained (Table 2). The magnetic micelles were next prepared. Oleic acid-coated superparamagnetic iron oxide nanoparticles of 6.4 ± 0.7 nm (measured by TEM, Figure 2A) were synthesized following a previous protocol.[19] Without the addition of the glycopolymer, the nanoparticles immediately precipitated out of water and turned into large agglomerates (Figure S2).
Table 2.
Characterization of micelles.
| Hydrodynamic diameter [nm][a] |
PDI[a] | Zeta potential [mV][a] |
Diameter by TEM [nm] |
|
|---|---|---|---|---|
| Glycopolymer micelles |
34.1 ± 3.3 | 0.13 ± 0.03 | −11.9 ± 1.2 | / |
| Magnetic micelles | 87.8 ± 7.4 | 0.15 ± 0.02 | −12.9 ± 0.9 | 35.4 ± 5.6 |
| Magnetic micelles (after 4 weeks)[b] |
91.6 ± 5.1 | 0.14 ± 0.03 | −14.0 ± 1.3 | / |
Measured by DLS
In D.I. water at 2–8 °C
Figure 2.
TEM images of A) oleic acid-coated iron oxide nanoparticles, B) magnetic micelles, and C) oleic acid-coated iron oxide nanoparticles (I) and magnetic micelles (II) in water.
To prepare the magnetic micelles, glycopolymer 7 was dissolved in 1:1 v/v DMSO/H2O (5 mL, 1 mg/mL), and iron oxide nanoparticles in THF (2 mg/mL, ~ 1 mL) was slowly added while stirring until the solution became turbid. After dialysis and centrifugation, micelles containing the magnetic nanoparticles, i.e., magnetic micelles, were obtained (Figure 2B). The magnetic micelles could be freely dispersed in water, whereas the oleic acid-coated magnetic nanoparticles precipitated quickly (Figure 2C). After nanoparticle encapsulation, the hydrodynamic diameter of the micelles increased to 87.8 ± 7.4 nm (Table 2). The negative zeta potential value of −12.9 ± 0.9 mV can be attributed to the terminal –COOH on the glycopolymer.[20] The stability of the micelles was investigated by storing the sample in D.I. water at 2–8 °C for 4 weeks. No precipitates were observed, and no statistically significant increase in the hydrodynamic diameter (91.6 ± 5.1 nm) was detected by DLS (Table 2). Thus, the glycopolymer acted as a protective layer preventing the hydrophobic nanoparticles from agglomerating. Since polylactide could undergo hydrolysis to yield–COOH, if the glycopolymer degraded, the decomposed micelles would cause the zeta potential to become more negative (< −20 mV).[21] However, the zeta potential of the magnetic micelles after storage remained largely unchanged (Table 2), further supporting the high storage stability of the magnetic micelles.
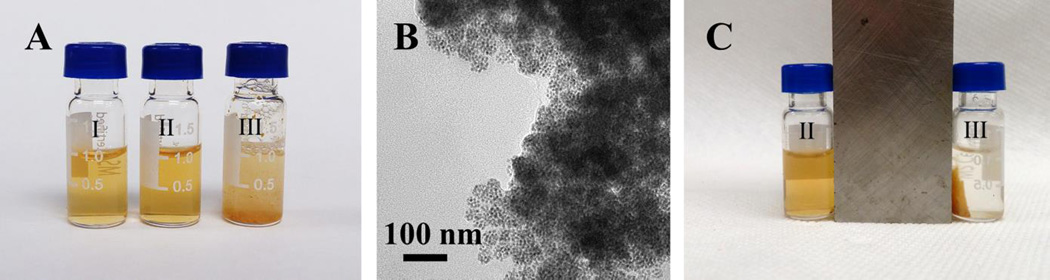
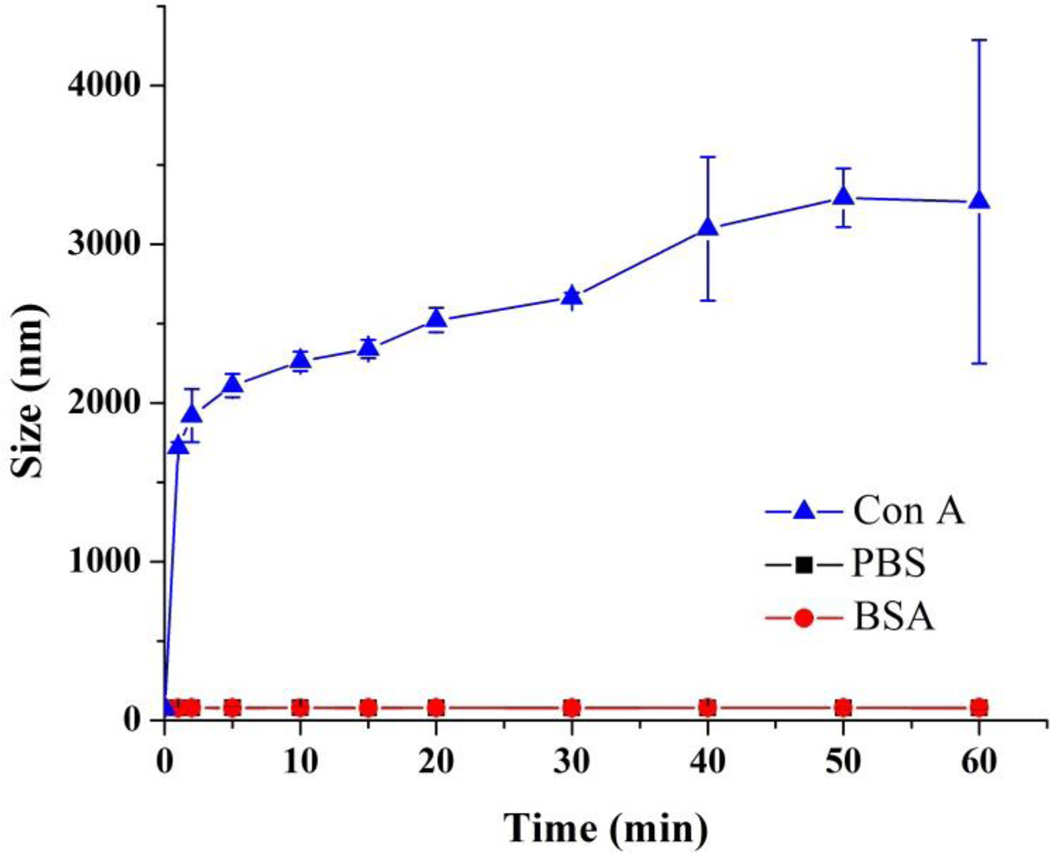
We hypothesize that the non-polar segments of the glycopolymer interacted with the hydrophobic iron oxide nanoparticles to form the core, whereas the polar segments, i.e., the grafted trehalose would be mostly exposed at the exterior of the micelle. To test this, a lectin binding study was conducted using Con A (concanavalin A). Con A binds to α-glucosides including trehalose with a Kd of 0.22 mM.[22] The magnetic micelles were incubated with Con A at 25 °C for 1 h. PBS (phosphate buffered saline, pH 7.4) and BSA (bovine serum albumin), which is a highly adhesive protein that tends to adhere to many materials and surfaces, were used as the negative controls. When the magnetic micelles were treated with PBS or BSA, the suspension remained clear and no agglomeration was observed (Figure 3A, I and II). After the magnetic micelles were treated with Con A, the suspension turned turbid instantly (Figure 3A, III). TEM images revealed the formation of large clusters, consisting of densely packed micelles (Figure 3B). In the clusters, individual micelles were visible, suggesting that the cluster formation was the result of surface trehalose ligands interacting with Con A. When a magnet was placed next to the sample, the orange micelles moved towards the magnet. In less than 2 minutes, all micelles clustered next to the magnet and the supernatant solution was clear and colorless (Figure 3C). Micelles without encapsulated iron oxide nanoparticles also formed aggregates upon treating with Con A. In this case, however, the aggregates remained dispersed in the solution and did not move in the presence of a magnet (Figure S3). To monitor the lectin binding, the magnetic micelles were treated with Con A at different incubation time, and the particle size was measured. In PBS or BSA, the particle size remained at around 80 nm even after incubation for 1 hour. On the other hand, after mixing with Con A for 1 minute, the particle size increased drastically from 80 nm to 1.7 µm and continued to grow to 3.3 µm in 1 hour. Con A is a homotetramer with four binding units.[20] The micelles have multiple trehalose ligands on the surface, and when treated with the tetrameric Con A, could be crosslinked by Con A, leading to the formation of the large agglomerates observed in the DLS and TEM. This is similar to the observation from other glyconanomaterial systems where the multivalent α-glucoside ligands on nanomaterials cause particle agglomeration when treated with Con A.[23]
Figure 3.
A) Magnetic micelles in PBS (I), BSA (II) and Con A (III). B) TEM image of sample III. C) Samples II and III after applying a magnet.
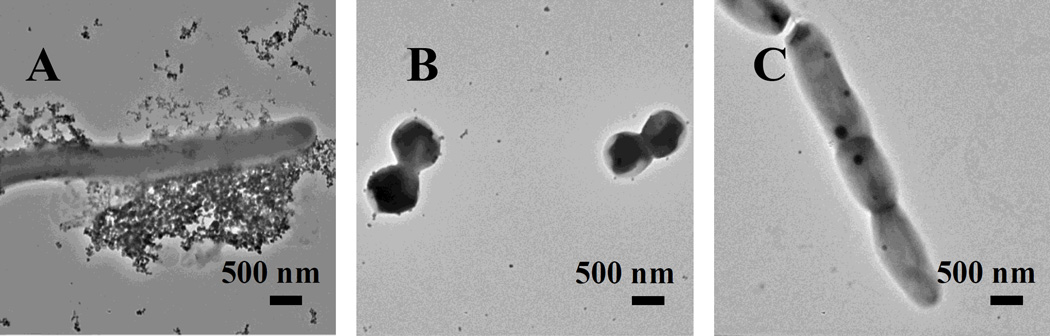
To test the ability of magnetic micelles to detect mycobacteria, the micelles were treated with M. smegmatis mc2 155. M. smegmatis is a widely used non-pathogenic model for M. tuberculosis studies.[24] Gram-positive S. epidermidis 35984 and Gram-negative E. coli ORN 208 were used as the negative controls. For M. smegmatis mc2 155, many magnetic micelles were seen around the bacteria (Figure 5A), and the membrane deformation was also observed (Figure S4). On the contrary, very few particles were found on S. epidermidis (Figure 5B) or E. coli ORN 208 (Figure 5C), indicating that the magnetic micelles could indeed selectively target the mycobacterium. Furthermore, the number of magnetic micelles increased with the incubation time (Figure S5). Both the magnetic micelles and the bacteria had a negative zeta potential (-12.9 ± 0.9 and -27.6 ±1.2 mV,[25] respectively), and therefore, the surface charge should not be the reason for the observation. The strong interactions of the magnetic micelles with mycobacteria could only be attributed to the trehalose ligands on the micelle surface.
Figure 5.
TEM images of (A) M. smegmatis mc2 155, (B) S. epidermidis and (C) E. coli ORN 208 after incubating with magnetic micelles overnight.
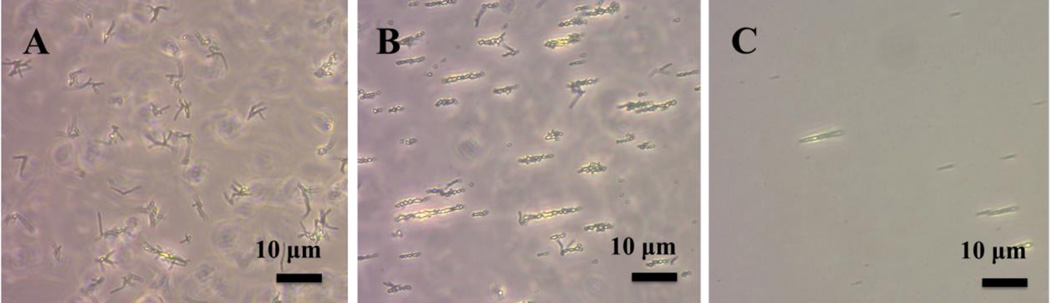
To investigate the utility of the magnetic micelles in detecting mycobacterium, a magnet was placed near the magnetic micelle-treated bacteria solution. The bacteria clusters, which were randomly oriented initially (Figure 6A), became aligned with the magnetic field (Figure 6B and 6C) and moved towards the magnet. In contrast, no movement was detected in the case of S. epidermidis 35984 or E. coli ORN 208 (Figure S6). At the bacteria concentrations of 106 CFU/mL or above, when a magnet was placed at the bottom of the vials for 15 min, orange precipitates were clearly visible (Figure 7, III and IV). At the bacteria concentration of 104 CFU/mL or below, no obvious precipitates were visible by the naked eye (Figure 7, I and II). However, at 104 CFU/mL, the magnet-induced alignment/movement of the bacteria clusters could still be seen under a microscope (Figure 6C). This is also supported by the TEM image where a large number of magnetic micelles were observed around the mycobacteria (Figure S7).
Figure 6.
Optical images of magnetic micelles treated with M. smegmatis mc2 155 (108 CFU/mL) A) before, and B) after placing a magnet to the left of the sample. C) M. smegmatis mc2 155 (104 CFU/mL) after applying a magnet.
Figure 7.
Magnet-induced agglomeration after treating magnetic micelles with M. smegmatis mc2 155 at (I) 103, (II) 104, (III) 106, and (IV) 108 CFU/mL.
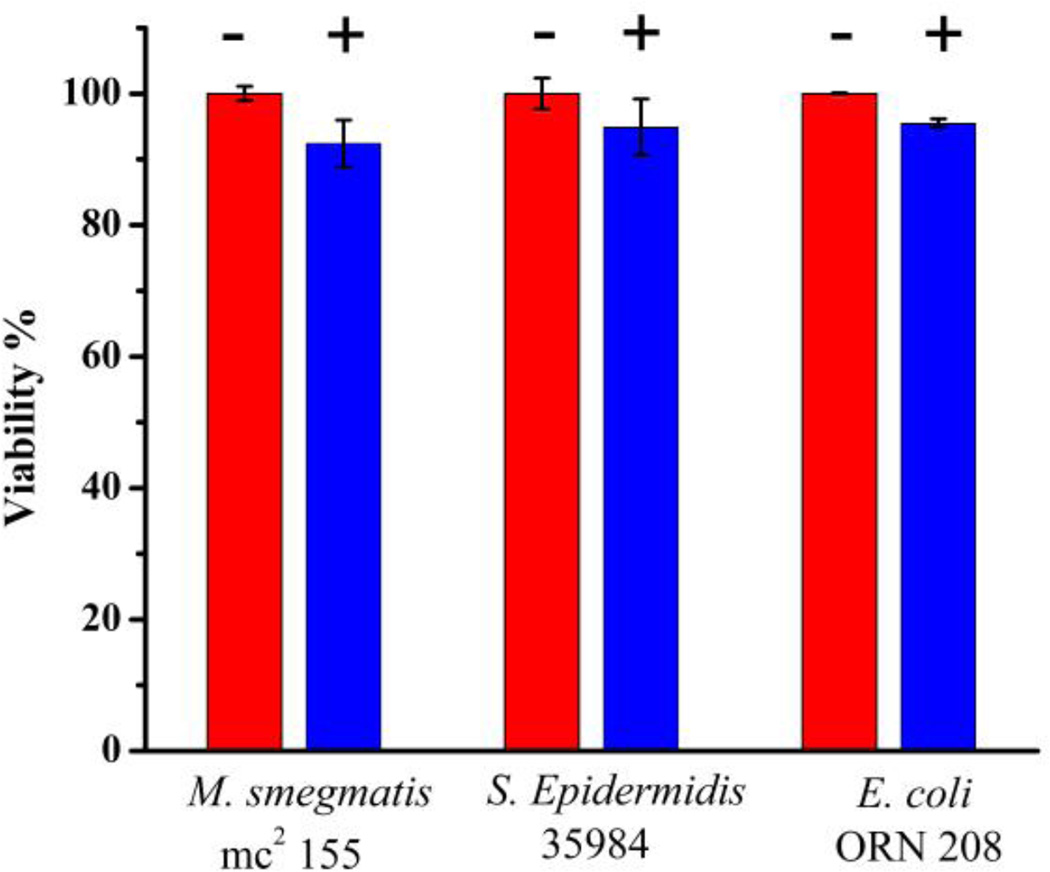
The viability of the bacteria after treating with the magnetic micelles for 24 hours was determined to test whether the magnetic micelles are toxic to the bacteria. High viabilities of 92.4 ± 3.3% 94.9 ± 4.2% and 95.5 ± 0.7% were found for M. smegmatis mc2 155, S. epidermidis 35984 and E. coli ORN 208, respectively (Figure 8). The results are consistent with the non-toxic nature of PLA reported by others.[26]
Figure 8.
Cell viabilities of M. smegmatis mc2 155, S. epidermidis 35984 and E. coli ORN 208 before (−) and after (+) treating with magnetic micelles (0.25 mg/mL) for 24 hours.
In summary, we have synthesized a multivalent trehalose-grafted PLA polymer, and encapsulated iron oxide magnetic nanoparticles. The glycopolymer protected the hydrophobic magnetic nanoparticles from precipitating from aqueous solutions and formed stable magnetic micelles. Lectin binding experiment with Con A supported both the surface presentation of trehalose ligands and the magnetic property of the micelles. The micelles bound to M. smegmatis, but minimally to S. epidermidis or E. coli. Using the magnetic micelles, M. smegmatis was successfully captured by applying a magnet. The detection can be directly visualized by the naked eyes at 106 CFU/mL and above, and at 104 CFU/mL with the aid of a microscope. These magnetic multivalent micelles prepared from biocompatible and biodegradable PLA could serve as a new platform having potentials for TB detection and therapy.
Supplementary Material
Figure 4.
The hydrodynamic diameter (measured by DLS) vs. incubation time of magnetic micelles in PBS, BSA or Con A.
Acknowledgments
The authors thank the financial support of this work from the National Institutes of Health (R01GM080295, R21AI109896), and a startup grant from University of Massachusetts Lowell. We thank Professor William R. Jacobs of Albert Einstein College of Medicine for the kind gift of M. smegmatis mc2 155.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.a) Zumla A, Raviglione M, Hafner R, Fordham von Reyn C. N Engl. J. Med. 2013;368:745. doi: 10.1056/NEJMra1200894. [DOI] [PubMed] [Google Scholar]; b) Kwan CK, Ernst JD. Clin. Microbiol. Rev. 2011;24:351. doi: 10.1128/CMR.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cattamanchi A, Davis JL, Pai M, Huang L, Hopewell PC, Steingart KR. Clin. Microbiol. Rev. 2010;48:2433. doi: 10.1128/JCM.00208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caviedes L, Lee T-S, Gilman RH, Sheen P, Spellman E, Lee EH, Berg DE, Montenegro-James S. Clin. Microbiol. Rev. 2000;38:1203. doi: 10.1128/jcm.38.3.1203-1208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keeler E, Perkins MD, Small P, Hanson C, Reed S, Cunningham J, Aledort JE, Hillborne L, Rafael ME, Girosi F, Dye C. Nat. Rev. Microbiol. 2007;49 doi: 10.1038/nature05446. [DOI] [PubMed] [Google Scholar]
- 5.Niemz A, Boyle DS. Expert Rev. Mol. Diagn. 2012;12:687. doi: 10.1586/erm.12.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kambashi B, Mbulo G, McNerney R, Tembwe R, Kambashi A, Tihon V, Godfrey-Faussett P. Int. J. Tuberc. Lung Dis. 2001;5:364. [PubMed] [Google Scholar]
- 7.Chen Y, Xianyu Y, Wang Y, Zhang X, Cha R, Sun J, Jiang X. ACS Nano. 2015;9:3184–3191. doi: 10.1021/acsnano.5b00240. [DOI] [PubMed] [Google Scholar]
- 8.Hamasur B, Bruchfeld J, van Helden P, Källenius G, Svenson S. PloS ONE. 2015;10(4):e0123457. doi: 10.1371/journal.pone.0123457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steingart KR, Ng V, Henry M, Hopewell PC, Ramsay A, Cunningham J, Urbanczik R, Perkins MD, Aziz MA, Pai M. Lancet Infect. Dis. 2006;6:664. doi: 10.1016/S1473-3099(06)70602-8. [DOI] [PubMed] [Google Scholar]
- 10.Lawn SD, Kerkhoff AD, Vogt M, Wood R. Lancet Infect. Dis. 2012;12:201. doi: 10.1016/S1473-3099(11)70251-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodruff PJ, Carlson BL, Siridechadilok B, Pratt MR, Senaratne RH, Mougous JD, Riley LW, Williams SJ, Bertozzi CR. J Biol. Chem. 2004;279:28835. doi: 10.1074/jbc.M313103200. [DOI] [PubMed] [Google Scholar]
- 12.De Smet KAL, Weston A, Brown IN, Young DB, Robertson BD. Microbiology. 2000;146:199. doi: 10.1099/00221287-146-1-199. [DOI] [PubMed] [Google Scholar]
- 13.Ishikawa E, Ishikawa T, Morita YS, Toyonaga K, Yamada H, Takeuchi O, Kinoshita T, Akira S, Yoshikai Y, Yamasaki S. J Exp. Med. 2009;206:2879. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayawardana KW, Jayawardena HSN, Wijesundera SA, De Zoysa T, Sundhoro M, Yan M. Chem. Comm. 2015;51:12028. doi: 10.1039/c5cc04251h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Zhou J, Jayawardana KW, Kong N, Ren Y, Hao N, Yan M, Ramstrom O. ACS Biomater. Sci. Eng. 2015;1:1250. doi: 10.1021/acsbiomaterials.5b00274. [DOI] [PubMed] [Google Scholar]; b) Hao N, Chen X, Jeon S, Yan M. Adv. Healthcare Mater. 2015;4:2797. doi: 10.1002/adhm.201500491. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hao N, Jayawardana KW, Chen X, Yan M. ACS Appl. Mater. Interfaces. 2015;7:1040. doi: 10.1021/am508219g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerhardt WW, Noga DE, Hardcastle KI, García AJ, Collard DM, Weck M. Biomacromolecules. 2006;7:1735. doi: 10.1021/bm060024j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Richardson AC, Tarelli E. J Chem. Soc. C. 1971:3733. [Google Scholar]; b) Backus KM, Boshoff HI, Barry CS, Boutureira O, Patel MK, D'Hooge F, Lee SS, Via LE, Tahlan K, Barry CE, Davis BG. Nat. Chem. Biol. 2011;7:228. doi: 10.1038/nchembio.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Letchford K, Burt H. Eur. J. Pharm. Biopharm. 2007;65:259. doi: 10.1016/j.ejpb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Jayawardena HSN, Jayawardana KW, Chen X, Yan M. Chem. Comm. 2013;49:3034. doi: 10.1039/c3cc40491a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruan G, Feng S. Biomaterials. 2003;24:5037. doi: 10.1016/s0142-9612(03)00419-8. [DOI] [PubMed] [Google Scholar]
- 21.Hu Y, Zhang L, Cao Y, Ge H, Jiang X, Yang C. Biomacromolecules. 2004;5:1756. doi: 10.1021/bm049845j. [DOI] [PubMed] [Google Scholar]
- 22.Oda Y, Kasai K, Ishii S. J Biochem. 1981;89:285. doi: 10.1093/oxfordjournals.jbchem.a133192. [DOI] [PubMed] [Google Scholar]
- 23.a) Wang X, Ramström O, Yan M. Anal. Chem. 2010;82:9082. doi: 10.1021/ac102114z. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang X, Ramstrom O, Yan M. Analyst. 2011;136:4174. doi: 10.1039/c1an15469a. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang X, Matei E, Gronenborn AM, Ramström O, Yan M. Anal. Chem. 2012;84:4248. doi: 10.1021/ac3006632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyagi JS, Sharma D. Trends Microbiol. 2002;10:68. doi: 10.1016/s0966-842x(01)02296-x. [DOI] [PubMed] [Google Scholar]
- 25.a) Etienne G, Villeneuve C, Billman-Jacobe H, Astarie-Dequeker C, Dupont M, Daffé M. Microbiology. 2002;148:3089. doi: 10.1099/00221287-148-10-3089. [DOI] [PubMed] [Google Scholar]; b) Hayden SC, Zhao G, Saha K, Phillips RL, Li X, Miranda OR, Rotello VM, El-Sayed MA, Schmidt-Krey I, Bunz UHF. J Am. Chem. Soc. 2012;134:6920. doi: 10.1021/ja301167y. [DOI] [PubMed] [Google Scholar]; c) Wilson WW, Wade MM, Holman SC, Champlin FR. J.Microbiol. Methods. 2001;43:153. doi: 10.1016/s0167-7012(00)00224-4. [DOI] [PubMed] [Google Scholar]
- 26.a) Athanasiou KA, Niederauer GG, Agrawal CM. Biomaterials. 1996;17:93. doi: 10.1016/0142-9612(96)85754-1. [DOI] [PubMed] [Google Scholar]; b) Anderson JM, Shive MS. Adv. Drug. Deliv. 2012;64(Supplement):72. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.