Abstract
Diseases are often a result of multiple malfunctions in complex, nonlinear biological/biochemical networks. As such, these processes are far more complicated to understand because they tend to give rise to functions that are emergent in nature, i.e., higher-level (mal)functions that are more than the sum of their parts. Systems biology provides a new approach to understanding biological systems and diseases from a holistic perspective.
DYNAMICAL, WHOLE-CELL SYSTEMS BIOLOGY
A system of intracellular biochemical networks has been shown to have emergent functions such as nontrivial cellular decision making.1 A function such as this is not detectable by isolated observation of individual network elements; there is no single protein or gene, biochemical reaction, area, or substructure that is responsible for or correlated with the ultimate function or malfunction (i.e., disease) of the system. Therefore, understanding how diseases occur in these systems and coming up with a reasonable therapy require an integrated, systems analysis strategy. A key component of the systems approach lies in understanding not only the topology of the various networks but also the associated dynamics as the system evolves in time. This “dynamical” systems biology has the potential to shed new light on the dynamics of healthy cells and also to open doors to the development of more efficient/effective drug therapies.
To exploit the full potential of systems biology, significantly larger and eventually whole-cell models will be needed. Recently, the first complete whole-cell model of Mycoplasma genitalium was created.2 This model incorporates all molecular constituents and their interactions and demonstrates how comprehensive whole-cell models can be used to further biomedical research via computer simulations and predictions of previously unobserved biological phenomena. This model was constructed by a single group of researchers; however, modeling the substantially larger and more complex human cells will likely require a large-scale, collaborative approach.
A CROWD-SOURCING PLATFORM FOR SYSTEMS BIOLOGY
The dual problem of understanding emergent cellular function and rational drug discovery is a perfect candidate for the research application of the modern phenomenon of “crowd-sourcing.” Scientific crowd-sourcing (e.g., EyeWire, http://www.eyewire.org; Galaxy Zoo, http://www.galaxyzoo.org; or the Polymath Project, http://www.polymathprojects.org) is a distributed problem-solving and production model in which the understanding of large-scale systems is created via a community of scientists contributing their detailed understanding of local areas of the system to create the large-scale system. Thus, knowledge of the individual system constituents is integrated into a global system that then has the potential to display emergent properties (including disease states as system malfunction) and that can be used to discover new drug targets. With such a system the promise of the systems biology approach can become a reality. It is with this idea in mind that we have developed the online community modeling system called the Cell Collective.3
Dynamical modeling frameworks range from differential equations, able to capture high levels of detail, to more coarse-grained approaches such as Boolean networks in which species are described digitally as either active or inactive. The rich information of kinetics contained in differential equations comes with the price of limited scalability and dependency on many difficult-to-obtain biological (kinetic) parameters. On the other hand, Boolean networks are discrete and qualitative (based on the logic of individual interactions), free of kinetic parameters, and relatively easy to construct, making them more suitable for large-scale system modeling. One of the drawbacks of traditional Boolean networks is that they are deterministic whereas biological systems are stochastic. The Cell Collective combines the easy-to-construct and parameter-free nature of qualitative modeling methods with the continuous data input/output and stochastic features of more detailed modeling approaches.
The software is designed to allow modeling of complex biological networks based on understanding of the logic of the interactions of the individual components. For instance, Akt, an important regulator of apoptosis, can be activated via phosphorylation by phosphoinositide-dependent protein kinase-1 (PDK1) or integrin-linked kinase (ILK). This activation step also requires the recruitment of Akt to the plasma membrane via phosphatidylinositol 4,5 bisphosphate (PIP2) or phosphatidylinositol 3,4,5-trisphosphate (PIP3). This regulatory mechanism of Akt can be easily and manually abstracted into a logic expression as (PDK1 OR ILK) AND (PIP2 OR PIP3). However, as models become more complex, larger, and interconnected, the number of regulatory components of a single species and hence the representing logic expression can become complex. For example, Rac, a regulator of cell motility, was included in our recent large-scale models in which its regulatory mechanism is governed by 16 interacting partners, resulting in a logic expression that is cumbersome, errorprone, and inefficient to create manually:
(RasGRF AND ¬(RhoGDI AND ¬PAK) AND ¬(p190RhoGAP AND Rac) AND ¬(RalBP1 AND Rac) AND ECM AND Integrins) OR (Tiam AND ¬(RhoGDI AND ¬PAK) AND ¬(p190RhoGAP AND Rac) AND ¬(RalBP1 AND Rac) AND (ECM AND Integrins)) OR (PixCool AND ¬(RhoGDI AND ¬PAK) AND ((PAK AND Gβγ AND ((¬Cdc42 AND ¬Rac) AND (Integrins AND ECM))) OR (¬Gβγ AND (Cdc42 AND (Integrins AND ECM) AND ¬Rac)) OR (¬PAK AND (¬RhoGDI AND (¬DOCK180 AND ¬(RhoGDI AND ¬PAK) AND ¬(p190RhoGAP AND Rac) AND ¬(RalBP1 AND Rac) AND ¬RasGRF AND ¬(RhoGDI AND ¬PAK) AND ¬(p190RhoGAP AND Rac) AND ¬(RalBP1 AND Rac) AND ¬Tiam AND ¬(RhoGDI AND ¬PAK) AND ¬(p190RhoGAP AND Rac) AND ¬(RalBP1 AND Rac)) AND (Integrins AND ECM) AND Cdc42 AND ¬Rac)))) OR (DOCK180 AND ¬(RhoGDI AND ¬PAK) AND ¬(p190RhoGAP AND Rac) AND ¬(RalBP1 AND Rac) AND (ECM AND Integrins)).
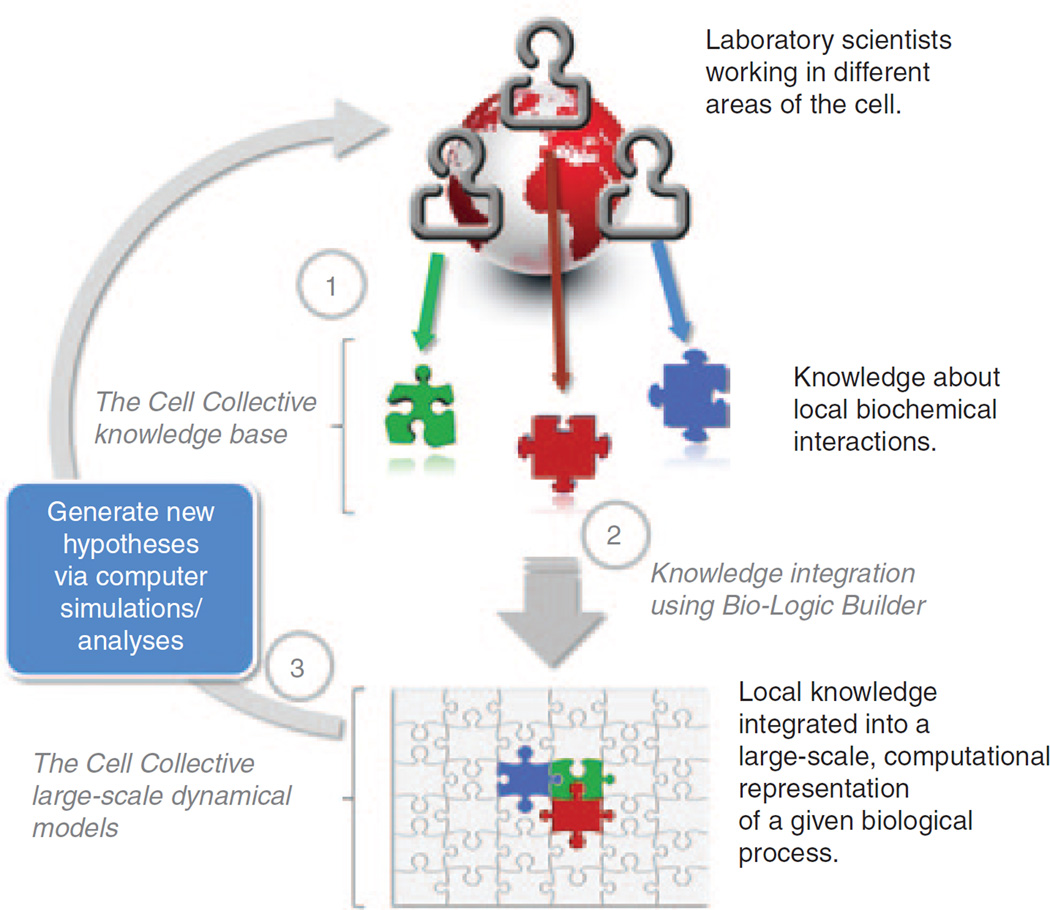
Using the Cell Collective’s nontechnical, plain-English, and logic-based user interface, the creation of large-scale dynamical computational models eliminates the need to enter complex mathematics or computer code, making it a particularly useful tool for mainstream laboratory scientists (a brief tutorial on how to construct a simple model is available on the Cell Collective website; http://www.thecellcollective.org). The Web-based nature of the software provides a unique environment to support a crowd-sourcing approach to the construction, simulation, and analysis of complex computational models of biological systems (Figure 1). New laboratory results and/or results generated/predicted with the models (and subsequently verified experimentally) can be presented to the community as a candidate for incorporation into models published in the platform. Multiple rounds of modification in this manner result in continuous improvement and growth of these models, with the goal of reaching the scale of whole cells (Figure 1). Note that in order to account for interactions that may differ across different cell types, users can create new or “fork” existing models to build models representing different cell types. Although the platform has been designed as a collaborative modeling approach, users can also create private models or share their models with a defined group of collaborators.
Figure 1.
Overview of the flow of knowledge about biological processes and the role of the Cell Collective in integrating and understanding this knowledge in the context of the biological processes as a whole. (1) Laboratory scientists with expert understanding of various parts of the cell or cell types collectively integrate their local knowledge into the platform. (2) Using the Bio-Logic Builder (ref. 5), this local knowledge is integrated into central models using qualitative information (and without the need to enter mathematical expressions or computer code). (3) Dynamical models created in the Cell Collective can be simulated and interrogated to pretest existing hypotheses as well as generate new ones, including the predictions and ranking of potential drug targets and their effects. Adapted from ref. 3.
DRUG TARGET IDENTIFICATION VIA COMPUTER SIMULATIONS
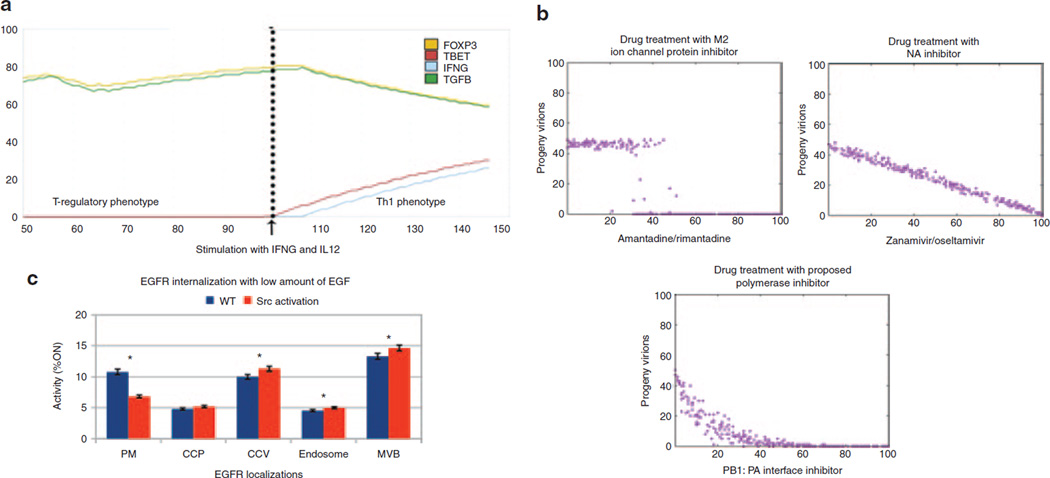
Because models created in the Cell Collective are dynamical (with a time dimension), one can ask questions critical to drug therapy design, including “what parts of the system are most/least sensitive to perturbation?” and “what parts of the system are most/least influential on the rest of the system when perturbed?” The real-time, interactive simulation features enables scientists to test many what-if scenarios while instantly observing the effects of changing environmental, pathogenic, and/or drug-induced conditions (Figure 2a).
Figure 2.
Examples of the Cell Collective applications to large-scale models of biological systems. (a) Real-time simulation of a model of a biochemical network governing CD4+ T-cell differentiation. The model was simulated to study the reprogrammability of T-regulatory cells to T-helper 1 (Th1) cells on stimulation of the biochemical network with interferon-γ (IFNG) and interleukin 12–Th1 cell–inducing stimuli. This model is currently being used to better understand the plasticity and reprogrammability of T cells (data not shown). (b) Simulations of a large-scale dynamical model of the influenza A replication cycle and comparison of the effects of existing drugs with a proposed drug target, the PB1:PA interface, on the generation of progeny virions suggest that drugging the proposed target would be more efficient. (c) Predicted increase of epidermal growth factor receptor (EGFR) internalization induced by constitutive activation of the Src nonreceptor protein kinase. The y-axis in a–c and the x-axis in b correspond to the activity level of the noted model components; note that this measure does not correspond to biochemical concentrations but is rather a semiquantitative way to capture the effects of varying biological conditions. The x-axis in a is an arbitrary time scale and does not correspond to real time. CCP, clathrin-coated pits; CCV, clathrin-coated vesicles; EGF, epidermal growth factor; FOXP3, forkhead box P3; MVB, multivesicular bodies; NA, neuraminidase; PB1:PA, polymerase basic 1 protein polymerase acidic protein complex; PM, plasma membrane; TBET, T-box expressed in T cells; WT, wild type.
Since its release, hundreds of models have been created in the Cell Collective, and a number of these models are now available to the scientific community for additional studies and contributions. For example, simulations of a new model of the molecular dynamics in influenza A replication reproduced a number of dynamical properties of the viral replication cycle under normal conditions as well as during treatment with various inhibitors.4 The model was also simulated to compare the effectiveness of the antiviral drugs amantadine/rimantadine and zanamivir/oseltamivir with a drug targeting the polymerase acidic protein-polymerase basic 1 protein (PA:PB1) complex (Figure 2b). In another study, to better understand the role of ErbB signaling in the progression of breast cancer, a multiscale model of signal transduction in human mammary epithelial cells consisting of 240+ molecular species and 1,100+ biochemical interactions was developed and used to predict inducing effects of Src overexpression on the internalization of epidermal growth factor receptor; this prediction was subsequently verified experimentally (Figure 2c). (See Supplementary Data online for additional references and application use cases.)
Because the computational framework behind the Cell Collective is based on a widely used modeling and simulation schema, the platform is compatible with a plethora of previously published modeling efforts (some of which are now available in the Cell Collective and are summarized and referenced in the Supplementary Data online), including studies on migration of keratinocytes, Fanconi anemia/breast cancer pathway, yeast cell cycle, T-cell differentiation (Figure 2a), and drug-target identification in the G1/S cell-cycle transition, as well as a 300+ node model of signal transduction in macrophages.
SUMMARY
Rather than applying relatively simplistic strategies for direct intervention at intuitively obvious sites, the ability to interrogate biological networks in a systemic context in order to search for nonintuitive approaches cannot be underestimated. Current and future discoveries in quantitative analysis will be difficult or even impossible to apply in the absence of large-scale, accurate models of biochemical/biological systems. We hope that as models in the Cell Collective become more comprehensive, they will provide new opportunities not only to identify and rank individual potential drug targets but also to explore the possibilities of drug repurposing and combinatorial therapies.
Rational drug design/discovery is a major problem that has yet to succumb to the power of modern laboratory science. It is likely that more tools will be required to attack this problem, including new ones such as systems and computational approaches. The merging of the laboratory and quantitative analysis has the potential for exciting synergies, and the Cell Collective is designed as a platform on which these synergies can become a reality.
Supplementary Material
Acknowledgments
The authors acknowledge the College of Arts and Sciences at the University of Nebraska at Omaha, the University of Nebraska Foundation, the Patrick J. Kerrigan and Donald F. Dillon foundations, the National Aeronautics and Space Administration Nebraska Space Grant Consortium, and National Institutes of Health grant no. 5R01DA030962.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/cpt
CONFLICT OF INTEREST
The authors declared no conflict of interest.
References
- 1.Helikar T, Konvalina J, Heidel J, Rogers JA. Emergent decision-making in biological signal transduction networks. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1913–1918. doi: 10.1073/pnas.0705088105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karr JR, et al. A whole-cell computational model predicts phenotype from genotype. Cell. 2012;150:389–401. doi: 10.1016/j.cell.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helikar T, et al. The Cell Collective: toward an open and collaborative approach to systems biology. BMC Syst. Biol. 2012;6:96. doi: 10.1186/1752-0509-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madrahimov A, Helikar T, Kowal B, Lu G, Rogers J. Dynamics of influenza virus and human host interactions during infection and replication cycle. Bull. Math. Biol. 2012 doi: 10.1007/s11538-012-9777-2. e-pub ahead of print 19 October 2012. [DOI] [PubMed] [Google Scholar]
- 5.Helikar T, et al. Bio-Logic Builder: a non-technical tool for building dynamical, qualitative models. PLoS ONE. 2012;7:e46417. doi: 10.1371/journal.pone.0046417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.