Abstract
Although inflammation and protease/antiprotease imbalance have been postulated to be critical in cigarette smoke–induced (CS-induced) emphysema, oxidative stress has been suspected to play an important role in chronic obstructive pulmonary diseases. Susceptibility of the lung to oxidative injury, such as that originating from inhalation of CS, depends largely on its upregulation of antioxidant systems. Nuclear factor, erythroid-derived 2, like 2 (Nrf2) is a redox-sensitive basic leucine zipper protein transcription factor that is involved in the regulation of many detoxification and antioxidant genes. Disruption of the Nrf2 gene in mice led to earlier-onset and more extensive CS-induced emphysema than was found in wild-type littermates. Emphysema in Nrf2-deficient mice exposed to CS for 6 months was associated with more pronounced bronchoalveolar inflammation; with enhanced alveolar expression of 8-oxo-7,8-dihydro-2′-deoxyguanosine, a marker of oxidative stress; and with an increased number of apoptotic alveolar septal cells — predominantly endothelial and type II epithelial cells — as compared with wild-type mice. Microarray analysis identified the expression of nearly 50 Nrf2-dependent antioxidant and cytoprotective genes in the lung that may work in concert to counteract CS-induced oxidative stress and inflammation. The responsiveness of the Nrf2 pathway may act as a major determinant of susceptibility to tobacco smoke–induced emphysema by upregulating antioxidant defenses and decreasing lung inflammation and alveolar cell apoptosis.
Introduction
Pulmonary emphysema is a major manifestation of chronic obstructive pulmonary disease (COPD), which affects more than 16 million Americans and is the fourth highest cause of death in United States (1). COPD is the only disease among the top 10 causes of death with rising incidence in the United States, and it is predicted to reach worldwide epidemic proportions (2). Cigarette smoking accounts for most of this debilitating disease, but other environmental risk factors include air pollution and chronic occupational exposure to various dusts (3).
Permanent destruction of peripheral air spaces distal to terminal bronchioles is the hallmark of emphysema (4). Emphysema is also characterized by accumulation of inflammatory cells such as macrophages and neutrophils (1) in bronchioles and alveolar structures. In humans, a deficiency in antiproteinase inhibitors produced by inflammatory cells, such as α1-antitrypsin, has been shown to contribute to a protease/antiprotease imbalance, thereby favoring destruction of alveolar extracellular matrix in cigarette smoke–induced (CS-induced) emphysema (5, 6). MMPs play a central role in experimental emphysema, as documented by the resistance of macrophage metalloelastase knockout mice against emphysema caused by chronic inhalation of CS (7). Moreover, pulmonary overexpression of interleukin-13 in transgenic mice results in MMP- and cathepsin-dependent emphysema (8). There is recent evidence that apoptosis of alveolar septal cells also contributes to human emphysema and is required for experimental emphysema caused by inhibition of the vascular endothelial growth factor receptor (4).
Markers of oxidative stress (e.g., hydrogen peroxide and the end products of lipid peroxidation such as ethane, pentane, and 8-isoprostane) are elevated in the breath and serum of patients with COPD (9). Oxidative stress enhances inflammation, inactivates critical antiproteinase inhibitors such as α1-antitrypsin (10), and enhances apoptosis of alveolar cells (4). Inflammatory mediators such as interleukin-8 and tumor necrosis factor–α, which are increased in bronchoalveolar samples obtained from patients with COPD (10, 11), are regulated by proinflammatory redox-sensitive transcription factors, including nuclear factor–κB and activator protein-1. Numerous studies have demonstrated that the susceptibility of the lung to oxidative injury depends largely on the upregulation of protective antioxidant systems (10). Although oxidative stress, which originates from CS and infiltrating inflammatory cells, is suspected to be involved in the etiopathogenesis of the disease (10), there is no conclusive experimental evidence that supports a central role for oxidative stress in pathogenesis of CS-induced emphysema. Critical host factors that protect the lungs against oxidative stress may either directly determine susceptibility to alveolar tissue destruction in emphysema or act as modifiers of risk by affecting the intensity of inflammation associated with chronic CS inhalation.
Linkage analysis of hyperoxia-resistant and -sensitive mouse strains have identified nuclear factor, erythroid-derived 2, like 2 (Nrf2) as a candidate gene for resistance to hyperoxic injury (12). Nrf2 encodes a basic leucine zipper protein (bZIP) transcription factor which, upon activation in response to oxidative or electrophilic stress, detaches from its cytosolic inhibitor, Keap1, translocates to the nucleus, and binds to the antioxidant response element (ARE) in the promoter of target genes, leading to their transcriptional induction (13). Though little is known about Nrf2-regulated genes in the lungs, the recognized members of this group include several critical antioxidant genes, such as heme oxygenase-1 (HO-1), γ–glutamyl cysteine synthase (γ-GCS), and several members of the glutathione S-transferase (GST) family (13).
We have postulated that Nrf2 is a critical transcription factor that determines susceptibility to lung inflammation, oxidative stress, and alveolar cell apoptosis caused by chronic exposure to CS. In the present study, we demonstrate that disruption of the Nrf2 gene led to earlier-onset and more extensive CS-induced emphysema in mice. Thus, responsiveness of the Nrf2 pathway in lung cells plays a critical role in attenuating the development of CS-induced emphysema.
Results
Histological and lung morphometric studies.
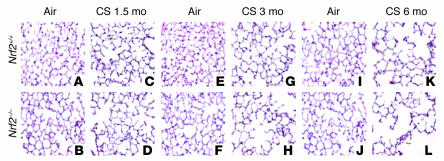
Lungs from air-exposed Nrf2-disrupted (Nrf2–/–) and wild-type (Nrf2+/+) mice showed normal alveolar structure (Figure 1). Since the alveolar diameter of air-exposed Nrf2–/– mice was slightly smaller than that of air-exposed wild-type mice (Table 1), we undertook detailed lung morphometric measurements, as well as light microscopic and ultrastructural studies, to ensure that Nrf2–/– lung does not have delayed development or compromised structural integrity when maintained in normal room air. There were no significant differences in alveolar diameter and mean linear intercept between Nrf2+/+ and Nrf2–/– lungs at 3 days, 10 days, 2 months, or 6 months of age (Supplemental Figure 1, A–C; supplemental material available at http://www.jci.org/cgi/content/full/114/1248/9/DC1). Histochemical staining for reticulin and elastin showed similar alveolar architecture in the wild-type and knockout lungs, with progressive attenuation of alveolar septa occurring between day 10 and 2 months of age in both genetic backgrounds (Supplemental Figure 1A). At 2 months of age, there was no significant difference in the total lung capacity (Supplemental Methods) between the air-exposed Nrf2+/+ (1.19 ± 0.16 ml; average weight of mice, 23 ± 1.4 g) and Nrf2–/– mice (1.12 ± 0.19 ml; average weight of mice, 23 ± 1.2 g), and the proliferation rate was similar in Nrf2+/+ and Nrf2–/– lungs (Supplemental Figure 1D). Finally, Nrf2+/+ and Nrf2–/– lungs had similar ultrastructural alveolar organization, with alveolar-capillary membranes lined by type I epithelial cells, and both had normal alveolar type II cell populations (Supplemental Figure 2, A and B). Histological examination of the lung sections did not reveal any tumors in air- or CS-exposed mice. Furthermore, H&E-stained lung sections did not show any significant inflammation in the lungs of air-exposed Nrf2+/+ or Nrf2–/– mice (Figure 1 and Supplemental Figure 1A).
Figure 1.
Increased susceptibility of Nrf2–/– mice to CS-induced emphysema. Shown are H&E-stained lung sections from Nrf2+/+ and Nrf2–/– mice exposed to air alone (A and B, E and F, and I and J) and to CS (C and D, G and H, and K and L) at the indicated times. Sections from the air-exposed Nrf2+/+ and Nrf2–/– mice show normal alveolar structure (n = 5 per group). Lung sections from the CS-treated (6 months) Nrf2–/– mice show increased air space enlargement when compared with the lung sections from the CS-treated Nrf2+/+ mice. Original magnification, ×20.
Table 1.
Effect of chronic exposure to CS on lung morphometry
To determine the role of Nrf2 in susceptibility to CS-induced emphysema, Nrf2-disrupted and wild-type Nrf2 (ICR strain) mice were exposed to CS for 1.5 to 6 months, and CS-induced lung damage was assessed by computer-assisted morphometry. There was a dramatic increase in alveolar destruction in the lungs of Nrf2-disrupted mice when compared to wild-type ICR mice after 6 months of exposure to CS. Both the alveolar diameter (increased by 33.1% in Nrf2 –/– vs. 8.5% in Nrf2+/+ mice) and mean linear intercept (increased by 26.1% in Nrf2 –/– vs. 8.3% in Nrf2+/+ mice) were significantly higher in CS-exposed Nrf2-disrupted mice (Table 1 and Figure 1). Alveolar enlargement was detected in the lungs of Nrf2–/– mice as early as 3 months after exposure to CS began (Table 1 and Figure 1), suggesting an earlier onset of emphysema in Nrf2-disrupted mice. Long-term (6 months) exposure of Nrf2+/+ mice to CS resulted in an increase of less than 10% in the mean linear intercept and alveolar diameter (Table 1), highlighting the intrinsic resistance of Nrf2+/+ ICR mice to CS-induced pulmonary emphysema.
Apoptosis assays.
To determine whether chronic exposure to CS (6 months) induced apoptosis of alveolar septal cells in vivo, we conducted TUNEL on lung sections from air- and CS-exposed mice. Labeling of DNA strand breaks in situ by fluorescent TUNEL demonstrated a higher number of TUNEL-positive cells in the alveolar septa of CS-exposed Nrf2–/– mice (154.27 TUNEL-positive cells per 1,000 DAPI-positive cells) than in CS-exposed Nrf2+/+ mice (26.42 TUNEL-positive cells per 1,000 DAPI-positive cells) or in air-exposed Nrf2–/– or Nrf2+/+ mice (Figure 2, A and B). Double staining of the TUNEL-labeled lung sections (Figure 2C) with antibody to surfactant protein C (SpC) to label type II epithelial cells, anti-CD34 to label endothelial cells, and anti–Mac-3 to label macrophages revealed the occurrence of apoptosis, predominantly in endothelial (Nrf2–/– = 52 ± 3.6 vs. Nrf2+/+ = 8 ± 1.8 TUNEL-positive and CD34-positive cells per 1,000 DAPI-positive alveolar cells) and type II epithelial cells (Nrf2–/– = 43 ± 4.3 vs. Nrf2+/+ = 6 ± 0.96 TUNEL-positive and SpC-positive cells per 1,000 DAPI-positive alveolar cells) in the lungs of CS-exposed Nrf2–/– mice when compared with Nrf2+/+ mice. Most alveolar macrophages in CS-exposed lungs did not show evidence of apoptosis (Nrf2–/– = 5 ± 0.42 Mac-3–positive cells per 1,000 DAPI-positive cells vs. Nrf2+/+ = 3 ± 0.96 Mac-3–positive cells per 1,000 DAPI-positive cells).
Figure 2.
Cigarette smoke exposure causes lung cell apoptosis as assessed by TUNEL in Nrf2–/– lungs. (A) Lung sections (n = 5 per group) of room air– or CS-exposed (6 months) Nrf2+/+ or Nrf2–/– mice were subjected to TUNEL (left column) and DAPI stain (middle column). Merged images are shown in the right column. CS-exposed Nrf2–/– mice show abundant TUNEL-positive cells (arrows) in the alveolar septa. Magnification, ×20. (B) Quantification of TUNEL-positive cells (per 1,000 DAPI-stained cells). The number of TUNEL-positive cells was significantly higher in the CS-exposed Nrf2–/– mice as compared with their wild-type counterparts (*P – 0.05). Values represent mean ± SEM. (C) Identification of apoptotic (TUNEL-positive) type II epithelial cells (left column), endothelial cells (middle column), and alveolar macrophages (right column) in the lungs of CS-exposed (6 months) Nrf2+/+ and Nrf2–/– mice. Type II epithelial cells, endothelial cells, and alveolar macrophages were detected with anti-SpC, anti-CD34, and anti–Mac-3 antibodies, respectively, as outlined in Methods. Nuclei were detected with DAPI (blue). Shown are the merged images, with colocalization (yellow arrows) of cell-specific markers (cytoplasmic red signal) and apoptosis (nuclear green + blue DAPI signal, resulting in a lavender-like signal); non-apoptotic (TUNEL-negative) cells with positive cell specific marker (red signal) are highlighted with a red arrow. TUNEL-positive apoptotic cells lacking a cell-specific marker are highlighted by white arrowheads. The majority of TUNEL-positive cells consisted of endothelial and type II epithelial cells, whereas most alveolar macrophages were TUNEL negative. Scale bars: 5 μm.
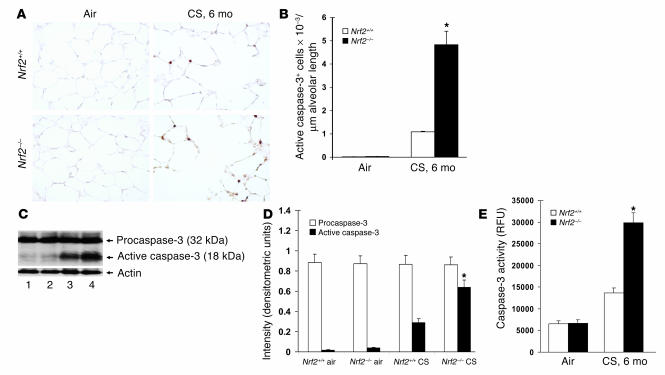
Immunohistochemical analysis showed a higher number of caspase-3–positive cells in the alveolar septa of CS-exposed Nrf2–/– mice (4.83 active caspase-3–positive cells/mm alveolar length) than in CS-exposed Nrf2+/+ mice (1.09 active caspase-3–positive cells/mm alveolar length). Lung sections from the air-exposed control Nrf2–/– and wild-type mice showed few or no caspase-3–positive cells (Figure 3, A and B). Enhanced activation of caspase-3 in Nrf2–/– lungs exposed to CS for 6 months was further documented by the increased detection of the 18-kDa active caspase-3 peptide in whole-lung lysates (increase in Nrf2–/– mice was 2.3-fold that of CS-exposed Nrf2+/+ mice; Figure 3, C and D), as well as by increased caspase-3 enzymatic activity (increase in activity in Nrf2–/– mice was 2.1-fold that of CS-exposed Nrf2+/+ mice; Figure 3E).
Figure 3.
CS treatment leads to activation of caspase-3 in Nrf2–/– lungs. (A) Active caspase-3 expression in lung sections from CS-exposed (6 months) Nrf2+/+ and Nrf2–/– mice. CS-exposed Nrf2 –/– mice show increased numbers of caspase-3–positive cells in the alveolar septa (n = 5 per group). Magnification, ×40. (B) Number of caspase-3–positive cells in the lungs of air- and CS-exposed mice. Caspase-3–positive cells were significantly higher in the lungs of CS-exposed Nrf2–/– mice. (C) Increased expression of the 18-kDa active form of caspase-3 in lungs of CS-exposed (6 months) Nrf2–/– mice (Western blot; lanes 1 and 3: air- and CS-exposed Nrf2+/+ mice, respectively; lanes 2 and 4: air- and CS-exposed Nrf2–/– mice, respectively). (D) Quantification of procaspase-3 and active caspase-3 obtained in Western blots of air- or CS-exposed Nrf2+/+ and Nrf2–/– lungs. Values are represented as mean ± SEM. (E) Caspase-3 activity in the lungs of air- or CS-exposed (6 months) Nrf2+/+ and Nrf2–/– mice. Caspase-3 activity was significantly higher in the lungs of CS-exposed Nrf2–/– mice than in the lungs of their wild-type counterparts (n = 3 per group). Values (relative fluorescence units [RFU]) are represented as mean ± SEM. *P – 0.05 vs. CS-exposed Nrf2+/+ mice.
Marker of oxidative stress in the lungs.
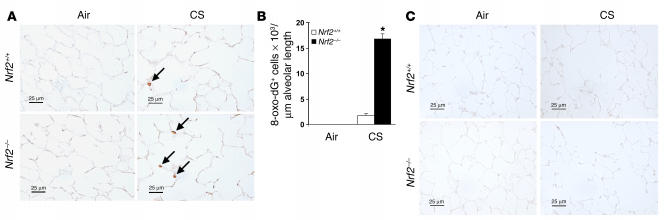
Immunohistochemical staining with anti–8-oxo-7,8-dihydro-2′-deoxyguanosine (anti–8-oxo-dG) antibody was used to assess oxidative stress in both Nrf2–/– and Nrf2+/+ lungs after inhalation of CS. A small number of alveolar septal cells (1.78 cells/mm alveolar length) exhibited staining for 8-oxo-dG in lung sections from the Nrf2+/+ mice, whereas significantly more (16.8 cells/mm alveolar length) were stained in the Nrf2–/– mice (Figure 4, A and B). Lung sections from air-exposed Nrf2+/+ and Nrf2–/– mice showed few or no 8-oxo-dG–positive cells. Immunostaining with normal mouse IgG antibody did not show any IgG-reactive cells in the lungs of air- or CS-exposed mice (Figure 4C). These results indicate that exposure to CS for 6 months enhanced oxidative damage to the lungs of the Nrf2-disrupted mice.
Figure 4.
Increased sensitivity of Nrf2–/– mice to oxidative stress after CS exposure. (A) Immunohistochemical staining for 8-oxo-dG in lung sections from the mice exposed to CS (6 months) (n = 5 per group). Lung sections from the CS-exposed Nrf2–/– mice show increased staining for 8-oxo-dG (indicated by arrows) when compared with lung sections from CS-exposed Nrf2+/+ mice and the respective air-exposed control mice. Magnification, ×40. (B) Quantification of 8-oxo-dG–positive alveolar septal cells in lungs after 6 months of CS exposure. The number of cells that reacted with anti–8-oxo-dG antibody was significantly higher in the lung tissues of the CS-exposed Nrf2–/– mice than in the lung tissues of the CS-exposed Nrf2+/+ mice and air-exposed control mice. Values (positive cells/mm alveolar length) represent mean ± SEM. *P – 0.05 vs. CS-exposed Nrf2+/+ mice. (C) Immunohistochemical staining with normal mouse-IgG1 antibody in lung sections from air- or CS-exposed Nrf2+/+ and Nrf2–/– mice. Magnification, ×40. Scale bars: 25 μm.
Inflammatory cells in the lungs.
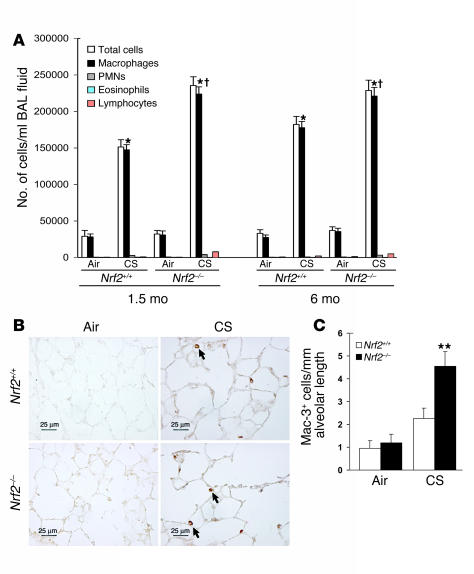
Analysis of differential cell counts in the bronchoalveolar lavage (BAL) fluid revealed a significant increase in the number of total inflammatory cells in the lungs of CS-exposed (1.5 or 6 months) Nrf2+/+ and Nrf2–/– mice when compared to their respective air-exposed control littermates (Figure 5A). However, the total number of inflammatory cells in BAL fluid from the CS-exposed Nrf2–/– mice was significantly higher than in CS-exposed wild-type mice. Among the inflammatory cell population, macrophages were the predominant cell type, constituting as much as 87–90% of the total inflammatory cell population in the BAL fluid of both genotypes exposed to CS. Other inflammatory cells such as polymorphonuclear leukocytes, eosinophils, and lymphocytes constituted 10–13% of the total inflammatory cells in the BAL fluid of both genotypes. Immunohistochemical staining of the lung sections with Mac-3 antibody revealed the presence of an increased number of macrophages (Figure 5, B and C) in the lungs of CS-exposed Nrf2–/– mice at 6 months (4.54 Mac-3–positive cells/mm alveolar length) when compared with lungs of their wild-type counterparts (2.27 Mac-3–positive cells/mm alveolar length). However, the immunohistochemical staining did not show any significant difference in the number of alveolar macrophages in the lungs of air-exposed Nrf2+/+ (0.96 Mac-3–positive cells/mm alveolar length) and Nrf2–/– mice (1.18 Mac-3–positive cells/mm alveolar length). There were significantly fewer neutrophils and lymphocytes than there were macrophages. There were 0.92 versus 0.49 neutrophils and 0.78 versus 0.43 lymphocytes per millimeter alveolar length in CS-exposed Nrf2–/– and wild-type mice, respectively (Supplemental Figure 3, A–D).
Figure 5.
Increased inflammation in the lungs of CS-exposed Nrf2–/– mice. (A) Lavaged inflammatory cells from control and CS-exposed mice. The number of macrophages in BAL fluid collected from the CS-exposed (both 1.5 months and 6 months) Nrf2–/– mice was significantly higher than in the BAL fluid from CS-exposed Nrf2+/+ mice and the respective age-matched control mice. Values represent mean ± SEM (n = 8). PMNs, polymorphonuclear leukocytes. *P – 0.05 vs. control of the same genotype; P – 0.05 across the genotypes in CS-exposed group. (B) Immunohistochemical detection of macrophages (arrows) in lungs of Nrf2+/+ and Nrf2–/– mice exposed to CS for 6 months. Magnification, ×40. Scale bars: 25 μm. (C) Quantification of macrophages in lungs after 6 months of CS exposure. The lung sections from the CS-exposed Nrf2–/– mice showed significantly more macrophages than did those from wild-type counterparts exposed to CS (**P – 0.025). However, there was no significant difference in the number of alveolar macrophages between the air-exposed Nrf2+/+ and Nrf2–/– mice (P > 0.9).
Activation of Nrf2 in the lungs of Nrf2+/+ mice.
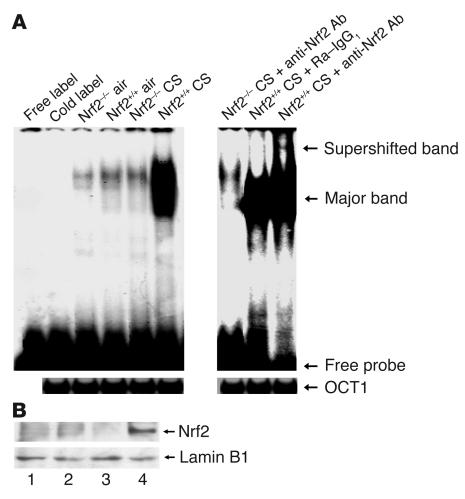
An electrophoretic mobility shift assay (EMSA) was used to determine the activation and DNA binding activity of Nrf2 in the lungs in response to acute exposure of the mice to CS (5 hours). In response to CS, there was an increased binding of nuclear proteins isolated from the lungs of CS-exposed Nrf2+/+ mice to an oligonucleotide probe containing the ARE consensus sequence, as compared to the binding of nuclear proteins isolated from CS-exposed Nrf2–/– mice or air-exposed control mice. Supershift analysis with anti-Nrf2 antibody also showed the binding of Nrf2 to the ARE consensus sequence, suggesting the activation of Nrf2 in the lungs of Nrf2+/+ mice in response to CS exposure (Figure 6A). However, supershift analysis of the nuclear proteins from the lungs of CS-exposed Nrf2–/– mice with anti-Nrf2 antibody did not show any super-shifted band, consistent with the absence of Nrf2 in the ARE-nuclear protein complex.
Figure 6.
Activation of Nrf2 in CS-exposed Nrf2+/+ lungs. (A) EMSA to determine the DNA binding activity of Nrf2. For gel shift analysis, 10 μg of nuclear protein from the lungs of air-and CS-exposed mice was incubated with the labeled human NQO1 ARE sequence and analyzed on a 5% non-denaturing polyacrylamide gel. For supershift assays, the labeled NQO1 ARE was first incubated with 10 μg of nuclear extract and then with 4 μg of anti-Nrf2 antibody for 2 hours. Nuclear protein of Nrf2+/+ lungs showed increased binding to the ARE-containing sequence (lower arrow) after CS exposure, with a supershifted band caused by preincubation with anti-Nrf2 antibody, thus confirming the binding of Nrf2 to the ARE sequence (upper arrow). Ra–IgG1, rabbit IgG1. (B) Nuclear accumulation of Nrf2. Western blot analysis with anti-Nrf2 antibody showed the nuclear accumulation of the transcription factor Nrf2 in the lungs of Nrf2+/+ mice in response to CS exposure (lanes 1 and 3: air-exposed Nrf2–/– and Nrf2+/+ mice, respectively; lanes 2 and 4: CS-exposed Nrf2–/– and Nrf2+/+ mice, respectively; lamin B1 was used as the loading control). Western blot analysis was carried out 3 times with the nuclear proteins isolated from the lungs of 3 different air- or CS-exposed Nrf2+/+ and Nrf2–/– mice.
Western blot analysis was performed to determine the nuclear accumulation of Nrf2 in the lungs in response to CS exposure. Immunoblot analysis (Figure 6B) showed increased levels of Nrf2 in the nuclei isolated from the lungs of CS-exposed Nrf2+/+ mice, suggesting the nuclear accumulation of Nrf2 in the lungs of wild-type mice in response to CS exposure. Increase of nuclear Nrf2 is needed for the activation of ARE and the transcriptional induction of various antioxidant genes.
Transcriptional induction of Nrf2-dependent genes.
To uncover the Nrf2-dependent genes that may account for the emphysema-sensitive phenotype of the Nrf2–/– background, we examined the pulmonary expression profile of air- and CS-exposed (5 hours) mice by oligonucleotide microarray analysis using the Affymetrix mouse gene chip U74A. The complete gene expression data set is available at http://faculty.jhsph.edu/biswal2.xls. Table 2 lists the genes that were significantly upregulated in the lungs of Nrf2+/+ mice, but not in those of Nrf2–/– mice, in response to CS. The regions upstream of the transcription start site of these Nrf2-dependent genes were analyzed for the presence of putative AREs using the Genamics Expression 1.1 Pattern Finder Tool software. The location of the AREs in these Nrf2-dependent genes are also presented in Table 2. Nrf2 regulates about 50 antioxidant and cytoprotective genes. The majority of these Nrf2-regulated genes contain possible functional AREs in the genomic sequences upstream of their transcription start sites.
Table 2.
Nrf2-dependent protective genes induced by CS in the lungs of Nrf2 wild-type mice
Validation of microarray data by Northern blot and enzyme assay.
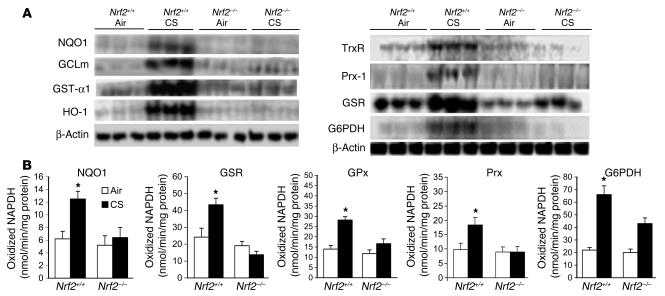
Validation of the microarray data was performed using the samples used in the arrays. Northern hybridization confirmed the transcriptional induction of genes involved in glutathione synthesis (glutamate cysteine ligase modifier subunit [GCLm]), NADPH regeneration (glucose-6-phosphate dehydrogenase [G6PDH]), detoxification of oxidative stress–inducing components of CS (by NADPH: quinone oxidoreductase-1 [NQO1], GST-α1, HO-1, thioredoxin reductase [TrxR], and peroxiredoxin-1 [Prx-1]) in the lungs of CS-exposed Nrf2+/+ but not Nrf2–/– mice (Figure 7A). Glutathione reductase (GSR) was also induced in CS-exposed Nrf2–/– mice; however, the magnitude of the induction was significantly higher in Nrf2 wild-type mice than in Nrf2-disrupted mice. The increases in these induced genes (NQO1, 7.2-fold; GST-α1, 2-fold; heavy subunit of γ-GCS [γ-GCS(h)], 4.8-fold; TrxR, 4.8-fold; G6PDH, 2.2-fold; HO-1, 3.4-fold; GSR, 1.8 fold; Prx-1, 1.6-fold), as measured by Northern analysis, were comparable to those determined by microarray.
Figure 7.
Validation of microarray data by Northern blot and enzyme assays. (A) Analysis of mRNA levels of NQO1, GCLm, GST-α1, HO-1, TrxR, Prx-1, GSR, and G6PDH in the lungs of Nrf2+/+ and Nrf2–/– mice exposed to either air or CS (n = 3 per group). (B) Effect of CS on the specific activities of selected enzymes in the lungs of Nrf2+/+ and Nrf2–/– mice. Values represent mean ± SE (n = 3 per group). *P – 0.05 vs. control of the same genotype.
Enzyme assays of selected gene products (NQO1, GSR, Prx, glutathione peroxidase [GPx] and G6PDH) were carried out to determine the extent to which their transcriptional induction in the lung paralleled changes in their activities (Figure 7B). There were significant increases in the activities of all enzymes in the lungs of CS-exposed Nrf2+/+ mice when compared to those of CS-exposed Nrf2–/– mice as well as those of the air-exposed mice of both genotypes. Moreover, the basal activities of these enzymes were significantly lower in the air-exposed Nrf2-disrupted mice than in the air-exposed wild-type mice.
Discussion
Our findings indicate that Nrf2, as previously shown for MMP-12, is a critical determinant of susceptibility to emphysema caused by CS. Because oxidative stress has overarching effects on several important mechanisms involved in CS-induced emphysema, particularly inflammation and apoptosis, Nrf2-induced gene expression may afford wide protection against the injurious effects of CS in the lung. The enhanced susceptibility of Nrf2-disrupted mice to CS-induced emphysema is remarkable, since the ICR parental strain (14) has been shown to be intrinsically resistant to CS-induced alveolar destruction and air space enlargement, as compared to susceptible strains such as C57BL/6J and DBA/2 (15). The higher levels of antioxidants and α1-antitrypsin in the lungs of ICR strain mice, as compared to those of susceptible strains, may contribute to this resistance (15).
We found that deletion of Nrf2 resulted in increased alveolar inflammation, alveolar septal cell apoptosis involving predominantly endothelial and type II epithelial cells, enhanced alveolar oxidative stress, and ultimately more pronounced emphysema following exposure to CS, when compared with wild-type mice. Such broad lung pathogenic effects resulting from elimination of Nrf2 indicate that, in CS-induced emphysema, oxidative stress regulates the intensity of alveolar inflammation, the extent of alveolar cell appoptosis, and ultimately the rate of onset and severity of the emphysema. In fact, the increased severity of CS-induced emphysema caused by deletion of Nrf2 in ICR mice is equivalent to that seen in MMP-12–competent C57Bl/6J mice (when compared to MMP12–/– mice) (7). Despite abundant evidence of the elevation of markers of pulmonary and systemic oxidative stress in chronic smokers, there is a paucity of mechanistic support for the centrality of oxidative stress in the pathogenesis of CS-induced emphysema. An oxidative burden in the smoker’s lungs is generated by the 4,700 chemical components present in CS and by the inflammatory cells that accumulate in the smoker’s airways. This oxidative burden elicits a protective response that is dependent upon the ability of the lung cells to upregulate antioxidant defenses. The upregulation and activation of transcription factors such as Nrf2 might be one of these protective mechanisms. Prior and concomitant to airspace enlargement caused by CS exposure, there was an increased infiltration of inflammatory cells, predominantly macrophages, which may have contributed to the alveolar injury through the activity of their elastolytic enzymes, particularly MMP-12 (7). A decrease in the activity and levels of antiproteases might have followed the enhanced oxidative stress in our model and thus contributed to protease/antiprotease imbalance. However, future studies will determine the role of specific proteases in the development of emphysema in Nrf2–/– mice in response to CS.
In addition to inflammation, the lungs of Nrf2-disrupted mice show increased alveolar cell apoptosis when compared to wild-type lungs. Alveolar cell apoptosis has been progressively recognized as a critically important mechanism of alveolar septal destruction in emphysema. Apoptosis is required for emphysema caused by VEGF receptor inhibition and is sufficient to cause emphysema, as demonstrated in mice instilled intrabronchially with active caspase-3 (16). However, the contribution of alveolar cell apoptosis has not been addressed in animal models of CS-induced emphysema. Long-term exposure to CS resulted in increased apoptosis of endothelial and type II epithelial cells at the 6-month time point in the lungs of CS-exposed Nrf2–/– mice when compared with CS-exposed Nrf2+/+ mice. Staining of the TUNEL-labeled lung sections with Mac-3 antibody showed the presence of few or no apoptotic macrophages in the lungs of CS-exposed Nrf2+/+ or Nrf2–/– mice. Immunohistochemical staining, enzyme assay, and Western blot analysis have also revealed the increased number or activity of caspase-3 in the lungs of CS-exposed Nrf2–/– mice, suggesting the occurrence of excessive apoptosis. The presence of enhanced lung apoptosis in these Nrf2–/– mouse lungs might be related to enhanced oxidative stress, inflammation, or excessive lung proteolysis. Oxidative stress and apoptosis are part of a mutually interactive feedback loop in VEGF receptor blockade–induced emphysema (4). Furthermore, reactive oxygen and nitrogen species can modify and inactivate survival cell signaling molecules and cause apoptosis. Inflammation and protease/antiprotease imbalance may also promote apoptosis by means of activated T lymphocytes, which are increased in COPD and seem to correlate with the degree of emphysema (17) and the amount of unopposed leukocyte elastase (18), gelatinase, or collagenase (19) activity.
Consistent with a central role for Nrf2 in the upregulation of antioxidant defenses during CS-related stress, in Nrf2–/– mice exposed to CS, there was an enhanced formation of 8-oxo-dG, one of the most abundant DNA adducts in response to oxidative stress (20). The importance of Nrf2 in affording protection during oxidative stress has already been highlighted by the enhanced susceptibility of Nrf2–/– mice to hyperoxia (12, 21) and chemically induced lung injury (22). The majority of the genes that were significantly upregulated in the lungs of CS-exposed Nrf2 wild-type mice, but not Nrf2-disrupted mice, have functions likely to confer protection against oxidative stress and inflammation.
Nrf2 — in association with several other proteins such as small maf proteins, c-jun, ARE binding protein-1, CBP/p300, and p160 family coactivators — binds to ARE, leading to transcriptional induction of target genes (23, 24). In response to CS exposure, there was an increased binding of nuclear proteins from the lungs of Nrf2+/+ mice to the ARE sequence. This binding is presumably due to the interaction of nuclear proteins with the ARE, particularly of Nrf2 in the lungs of CS-exposed Nrf2+/+ mice, as confirmed by the supershift analysis with anti-Nrf2 antibody. The slightly increased binding of nuclear proteins to ARE in knockout lung extracts is probably due to the involvement of proteins other than Nrf2, since the supershift analysis failed to detect a complex of Nrf2 in Nrf2–/– lung extracts. Western blot analysis confirmed the nuclear accumulation of Nrf2 in the lungs of CS-exposed Nrf2+/+ mice. Increased nuclear Nrf2 is critical for the activation of ARE and the transcriptional induction of various antioxidant genes.
Nrf2, in response to CS, regulates genes involved in two major redox systems, the glutathione and thioredoxin systems (25, 26). Enzymes involved in glutathione synthesis (γ-GCS catalytic and regulatory subunits), members of the GST family, GSR, GPx2, GPx3, and genes that constitute the thioredoxin system (TrxR and Prx-1) were all induced in the lungs of Nrf2+/+ mice in response to CS. The members of these redox systems interact with various transducers and effector molecules to bring about antioxidant-specific responses. The regeneration of reduced Trx and glutathione by TrxR and GSR, respectively, utilizes NADPH as a reducing equivalent generated by G6PDH and phosphogluconate dehydrogenase, both of which are also induced in Nrf2+/+ lungs. Prx-1 and GPx reduce hydroperoxides by utilizing two electrons provided by Trx and glutathione, respectively. In addition, GPx and peroxiredoxins have been shown to play a potential role in protection against peroxynitrite (27), a potent oxidant generated from the reaction of superoxide and nitrous oxide present in CS. Furthermore, the oxidized forms of GPx and peroxiredoxins are reduced back to their functional forms by Trx (28). These results suggest a cross talk between the thioredoxin and glutathione redox systems and the NADPH regenerating system.
Several GSTs, as well as UDP-glucuronosyl transferase (UGT) and NQO1, were selectively induced only in Nrf2+/+ mice in response to CS. Various isoforms of GSTs and UGTs play important roles in the detoxification of tobacco smoke carcinogens such as 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, benzo(a)pyrene and other polycyclic aromatic hydrocarbons that act as electrophiles and cause DNA damage and cytotoxicity (29, 30). NQO1 blocks redox cycling of polyaromatic hydrocarbons and benzoquinones present in CS (31), thereby reducing the levels of ROS and presumably 8-oxo-dG. Various enzymes, including aldehyde dehydrogenase and aldo-keto reductase, which are involved in the detoxification of reactive aldehydes such as acetaldehyde and acrolein, were selectively induced in the lungs of CS-exposed Nrf2+/+ mice. HO-1, a critical enzyme involved in protection against oxidant-mediated cellular injury, as well as the iron-sequestering protein ferritin light chain 1, which prevents uncontrolled surges in the intracellular free concentration of the highly reactive yet poorly soluble ferric iron (32, 33), were induced only in the lungs of CS-exposed Nrf2+/+ mice. Reduction of ferric iron by superoxide can generate reactive hydroxyl radicals via the Fenton reaction. Superoxide dismutase 3, the major extracellular antioxidant enzyme in the lung that attenuates ROS-mediated lung cell injury and inflammation, is also selectively upregulated in Nrf2+/+ mice in response to CS (34). CS also induced heat shock proteins such as HSP40 and mitochondrial stress-70 protein, as well as ubiquitin C, a protein involved in the degradation of oxidized proteins. Other Nrf2-regulated genes included the DNA damage repair protein GADD45G, lung structural proteins such as tropoelastin and procollagen type IV, α2, and endomucin-1, sequestosome 1, MafF, HIF-1α–related factor and α1-antitrypsin proteinase inhibitor. Mutations in the α1-antitrypsin proteinase inhibitor have been associated with an increased risk of COPD (35). Furthermore, we have located one or more AREs in the upstream regions of most of these differentially expressed genes, indicating the possibility of a direct role for Nrf2 in their transcriptional induction.
Taken together, these results provide a clear link between a defective response of the transcription factor Nrf2 and several lung problems: excessive oxidative stress, increased apoptosis, inflammation, and worsened emphysema. Nrf2 is activated in response to CS in the lungs of wild-type mice, leading to transcriptional induction of target genes that might provide resistance against the development of emphysema. Conversely, a lack of responsiveness of the Nrf2 pathway confers susceptibility to severe emphysema due to CS exposure in this model. The identification of Nrf2 as a determinant of susceptibility can have wide implications in the area of tobacco smoke–related lung diseases, where oxidative stress and inflammation play important roles.
Methods
Antibodies and reagents.
We used the following antibodies and reagents: anti–caspase-3 polyclonal antibody for immunohistochemistry (Idun Pharmaceuticals); biotinylated anti-mouse IgG, peroxidase–conjugated streptavidin, Vectashield HardSet mounting medium, and RTU HRP-avidin complex (Vector Laboratories); rabbit anti-SpC antibody (Chemicon International Inc.); rat anti–mouse Mac-3 antibody (BD Biosciences); anti–rabbit Texas red antibody, streptavidin–Texas red conjugated complex, and DAPI (Molecular Probes Inc.); biotinylated rabbit anti-mouse secondary antibody (DakoCytomation); octamer transcription factor 1 (OCT1) and CaspACE Assay kit (Promega Corp.); leupeptin, pepstatin A, and normal mouse IgG1 (Sigma-Aldrich); rat anti–mouse neutrophil antibody (Serotec); actin and anti–mouse CD45R primary antibody (Santa Cruz Biotechnology Inc.); rabbit anti–caspase-3 antibody for Western blot (Cell Signaling Technology Inc.); anti-CD34 and anti–lamin B1 antibody (Zymed Laboratories Inc.); and CH11 monoclonal antibody (Beckman Coulter Inc.).
Animals and care.
Nrf2-deficient ICR mice were generated as described (14). Mice were genotyped for Nrf2 status by PCR amplification of genomic DNA extracted from blood (36). PCR amplification was carried out using 3 different primers: 5′-TGGACGGGACTATTGAAGGCTG-3′ (sense for both genotypes), 5′-CGCCTTTTCAGTAGATGGAGG-3′ (antisense for Nrf2+/+ mice), and 5′-GCGGATTGACCGTAATGGGATAGG-3′ (antisense for LacZ) (36). Mice were fed AIN-76A diet (Harlan Teklad) and had access to water ad libitum; they were housed under controlled conditions (23 ± 2°C; 12-hour light/dark cycles). All experimental protocols conducted on the mice were performed in accordance with the standards established by the US Animal Welfare Acts, as set forth in NIH guidelines and in the Policy and Procedures Manual of the Johns Hopkins University Animal Care and Use Committee.
Exposure to CS.
The CS machine for smoke exposure was similar to the one used by Witschi et al. (37). However, the exposure regimen in terms of chamber atmosphere and duration of CS exposure was considerably more intense. At 8 weeks of age, the mice were divided into four groups (n = 40 per group): control Nrf2 wild-type mice, experimental Nrf2 wild-type mice, control Nrf2-disrupted mice, and experimental Nrf2-disrupted mice. The control groups were kept in a filtered air environment, and the experimental groups were subjected to CS for various time periods. CS exposure was carried out (7 hours/day, 7 days/week for up to 6 months) by burning 2R4F reference cigarettes (2.45 mg nicotine per cigarette; purchased from the Tobacco Research Institute, University of Kentucky) using a smoking machine (Model TE-10, Teague Enterprises). Each smoldering cigarette was puffed for 2 seconds, once every minute for a total of 8 puffs, at a flow rate of 1.05 l/min, to provide a standard puff of 35 cm3. The smoke machine was adjusted to produce a mixture of sidestream smoke (89%) and mainstream smoke (11%) by burning five cigarettes at one time. Chamber atmosphere was monitored for total suspended particulates and carbon monoxide, with concentrations of 90 mg/m3 and 350 ppm, respectively.
Morphologic and morphometric analyses.
After exposure to CS for various time periods (1.5, 3, and 6 months), the mice (n = 5 per group) were anesthetized with halothane (Halocarbon Laboratories) and the lungs were inflated with 0.5% low-melting agarose at a constant pressure of 25 cm as previously described (38). The inflated lungs were fixed in 10% buffered formalin and embedded in paraffin. Sections (5 μm) were stained with H&E. Mean alveolar diameter, alveolar length, and mean linear intercepts were determined by computer-assisted morphometry with Image Pro Plus software (Media Cybernetics). The lung sections in each group were coded, and representative images (15 per lung section) were acquired with a Nikon E800 microscope (lens magnification, ×20) by an investigator who was blind to the identity of the slides (4).
TUNEL assay.
Apoptotic cells in the tissue sections from the agarose-inflated lungs were detected by the Fluorescein-FragEL DNA Fragmentation Detection Kit (Oncogene Research Products) according to the recommendations of the manufacturer. The lung sections (n = 5 per group) were stained with the TdT labeling reaction mixture and mounted with Fluorescein-FragEL mounting medium (Oncogene Research Products). DAPI and fluorescein were visualized at 330–380 nm and 465–495 nm, respectively. Overlapping DAPI in red and FITC in green create a yellow, apoptotic-positive signal. Images (15 per lung section) of the lung sections were acquired with a Nikon E800 microscope (lens magnification, ×20). In each image, the number of DAPI-positive (red) and apoptotic cells (yellow) were counted manually. Apoptotic cells were normalized by the total number of DAPI-positive cells.
Identification of alveolar apoptotic cell populations in the lungs.
To identify the different alveolar cell types undergoing apoptosis in the lungs, we performed fluorescent TUNEL labeling in the lung sections from the air- and CS-exposed (6 months) Nrf2+/+ and Nrf2–/– mice, using the Fluorescein-FragEL DNA Fragmentation Detection Kit (Oncogene Research Products). To identify the apoptotic type II epithelial cells in the lungs after TUNEL labeling, we incubated the lung sections first with an anti–mouse SpC antibody and then with an anti–rabbit Texas red antibody. Apoptotic endothelial cells were identified by incubating the fluorescent TUNEL-labeled sections first with the anti–mouse CD34 antibody and then with the biotinylated rabbit anti–mouse secondary antibody. The lung sections were rinsed in PBS and then incubated with the streptavidin–Texas red conjugated complex. The apoptotic macrophages in the lungs were identified by incubating the TUNEL-labeled lung sections first with the rat anti–mouse Mac-3 antibody and then with the anti–rat Texas red antibody. Finally, DAPI was applied to all lung sections, incubated for 5 minutes, washed, and mounted with Vectashield HardSet mounting medium (Vector Laboratories). DAPI and fluorescein were visualized at 330–380 nm and 465–495 nm, respectively. Images of the lung sections were acquired with the Nikon E800 microscope, lens magnification ×40.
Immunohistochemical localization of active caspase-3.
Immunohistochemical staining of active caspase-3 was performed using anti–active caspase-3 antibody (39), and the active caspase-3–positive cells were counted with a macro using the Image Pro Plus program (Media Cybernetics) (4). The counts were normalized by the sum of the alveolar profiles, herein named as alveolar length, and expressed in μm or mm. Alveolar length correlates inversely with the mean linear intercept, that is, as the alveolar septa are destroyed, the mean linear intercept increases as total alveolar length decreases.
Caspase-3 activity assay.
Caspase-3 activity was assessed using a fluorometric CaspACE Assay commercial kit (Promega Corp.) according to the manufacturer’s instructions. Briefly, the frozen lung tissues were immediately homogenized with hypotonic lysis buffer (25 mM HEPES [pH 7.5], 5 mM MgCl2, 5 mM EDTA, 5 mM DTT, 2 mM PMSF, 10 μg/ml pepstatin A, and 10 μg/ml leupeptin) using a mechanical homogenizer on ice and centrifuged at 12,000 g for 15 minutes at 4°C. The clear supernatant was collected and frozen in liquid nitrogen. The protein was quantified using Bradford’s reagent (Bio-Rad). Lung supernatant containing 30 μg of protein was added to a reaction buffer (98 μl) containing 2 μl DMSO, 10 μl of 100 mM DTT, and 32 μl of caspase assay buffer in a 96-well flat-bottom microtiter plate (Corning-Costar Corp.). The reaction mixture was incubated at 30°C for 30 minutes. Then, 2 μl of 2.5 mM caspase-3 substrate (Ac-DEVD-AMC) was added to the wells and incubated for 60 minutes at 30°C. The fluorescence of the reaction was measured at an excitation wavelength of 360 nm and an emission wavelength of 460 nm. We used 30 μg of protein from anti-Fas antibody–treated Jurkat cells (treated with 1 μg CH11 monoclonal antibody per milliliter of RPMI medium containing 5 × 105 cells for 16 hours at 37°C) as a positive control. A caspase-3–specific inhibitor (2 μl of 2.5 mM DEVD-CHO) was used to show specificity of caspase-3 activity. The activity was below background levels after the addition of caspase-3 inhibitor. These experiments were performed in triplicate and repeated 3 times.
Immunohistochemical localization of 8-oxo-dG.
For the immunohistochemical localization and quantification of 8-oxo-dG, lung sections (n = 5 per group) from the mice exposed to CS for 6 months were incubated with anti–8-oxo-dG antibody and stained using the Iso-IHC DAB kit (InnoGenex) using mouse antibodies. Normal mouse-IgG1 antibody was used as a negative control. The 8-oxo-dG–positive cells were counted with a macro (using Image Pro Plus), and the counts were normalized by alveolar length as described (4).
BAL and phenotyping.
Immediately following exposure to CS for 1.5 months or 6 months, mice (n = 8 per group) were anesthetized with sodium pentobarbital. The BAL fluid collected from the lungs of the mice was centrifuged (500 g at 4°C), and the cell pellet was resuspended in PBS. The total number of cells in the lavage fluid was determined, and 2 × 104 cells were cytocentrifuged (Shandon Southern Products) onto glass slides and stained with Wright-Giemsa stain (Diff-Quik; Baxter Scientific Products). Differential cell counts were performed on 300 cells, according to standard cytologic techniques (40).
Immunohistochemical localization of inflammatory cells in the lungs.
Macrophages were identified by the rat anti–mouse Mac-3 and secondary biotinylated anti-rat antibody immunostaining using the Vector RTU HRP-avidin complex with 3,3′-diaminobenzidine as the chromogenic substrate. The number of Mac-3–positive cells in the lung sections (n = 3 per group and 10 fields per lung section) were counted manually and normalized by alveolar length.
EMSA.
EMSA was carried out according to a procedure described previously (41). For gel shift analysis, 10 μg of nuclear protein that had been prepared from the lungs of mice exposed to air or to CS for 5 hours was incubated with the labeled human NQO1 ARE, and the mixtures were analyzed on a 5% nondenaturing polyacrylamide gel. To determine the specificity of protein(s) binding to the ARE sequence, 50-fold excess of unlabeled competitor oligo (ARE consensus sequence) was incubated with the nuclear extract for 10 minutes prior to the addition of radiolabeled probe. For supershift analysis, the labeled NQO1 ARE was first incubated for 30 min with 10 μg of nuclear proteins and then with 4 μg of anti-Nrf2 antibody for 2 hours. Normal rabbit IgG1 (4 μg) was used as a control for the supershift assay. The mixtures were separated on native polyacrylamide gel and developed by autoradiography. The P32-labeled consensus sequence for OCT1 was used as a control for gel loading. The EMSA was performed 3 times with the nuclear proteins isolated from 3 different air- or CS-exposed Nrf2+/+ and Nrf2–/– mice.
Western blot analysis.
Western blot analysis was performed according to previously published procedures (41). To determine the nuclear accumulation of Nrf2, we used 10% SDS-PAGE to separate 50 μg of the nuclear proteins isolated from the lungs of air- or CS-exposed (5 hours) Nrf2+/+ and Nrf2–/– mice. Then, we electrophoretically transferred them onto a PVDF membrane (Millipore). The membranes were blocked with 5% (w/v) BSA in Tris-buffered saline (20 mM Tris/HCl [pH 7.6] and 150 mM NaCl) with 0.1% [v/v] Tween-20 for 2 hours at room temperature, and then incubated overnight at 4°C with polyclonal rabbit anti-Nrf2 antibody, followed by incubation with HRP-conjugated secondary antibody. The blots were developed using an enhanced chemiluminescence Western blotting detection kit (Amersham Biosciences). Then, the blots were stripped and reprobed with anti–lamin B1 antibody.
To identify the active caspase-3, the lung tissues (n = 3) were homogenized with the lysis buffer (containing 50 mM Tris/HCl [pH 8.0], 150 mM NaCl, 0.5% (v/v) Nonidet P40, 2 mM EDTA, and a protease inhibitor cocktail) on ice using a mechanical homogenizer. Following centrifugation at 12,000 g for 15 minutes, the protein concentration of the supernatant was determined using Bradford’s reagent. Equal amounts of protein (30 μg) were resolved on 15% SDS-PAGE and transferred onto a PVDF membrane (Millipore). The membranes were incubated with rabbit anti–caspase-3 antibody and then with secondary anti-rabbit antibody linked to HRP conjugate. The blots were developed using the enhanced chemiluminescence Western blotting detection kit (Amersham Biosciences). Thereafter, blots were stripped and reprobed with antibodies to actin. Western blot was performed thrice with protein extracts from 3 different air- or CS- exposed (6 months) Nrf2+/+ and Nrf2–/– mice. Band intensities of procaspase-3 and active caspase-3 of the 3 blots were determined using NIH Image-Pro Plus software. Values are represented as mean ± SEM.
Transcriptional profiling using oligonucleotide microarrays.
Lungs were excised from control (air-exposed) and CS-exposed (5 hours) mice (n = 3 per group) and processed for total RNA extraction using the TRIzol reagent (Invitrogen). The isolated RNA was used for gene expression profiling with Murine Genome U74A version 2 arrays (Affymetrix) using procedures described earlier (42). To identify the differentially expressed transcripts, pairwise comparison analyses were carried out with the Data Mining Tool 3.0 program (Affymetrix). Only those differentially expressed genes that appeared in at least 6 of the 9 comparisons and showed a change of more than 1.4-fold were selected. In addition, the Mann-Whitney pairwise comparison test was performed to rank the results by concordance as an indication of the significance (P ≤ 0.05) of each identified change in gene expression. Genes which were upregulated only in the lungs of wild-type mice in response to CS were selected and used for the identification of AREs in their upstream sequence.
Identification of AREs in Nrf2-regulated genes.
To identify the presence and location of AREs in Nrf2-dependent genes, the murine homologs of human genes were employed (Human Genome build 34, version 1, the National Center for Biotechnology Information [NCBI] database). For every gene, a 10-kb sequence upstream from the transcription start site (TSS) was used to search for AREs with the help of Genamics Expression 1.1 Pattern Finder Tool software (Genamics) using the primary core sequence of ARE (RTGAYNNNGCR) (43) as the probe. TSS for all the genes was determined by following the Human Genome build 34, version 1, of the NCBI database.
Northern blotting.
Northern blotting was performed according to the procedure described earlier (42). In brief, 10 μg of total RNA isolated from the lungs of air- and CS-exposed (5 hours) mice (n = 3) was separated on 1.2% agarose gel, transferred to nylon membranes (NYTRAN Super Charge; Schleicher & Schuell), and ultraviolet-crosslinked. Full-length probes for NQO1, γ-GCS (regulatory subunit), GST-α1, HO-1, TrxR, Prx-1, GSR, G6PDH, and β-actin were generated by PCR from the cDNA of murine liver. These PCR products were radiolabeled with [α-32P] cytidine triphosphate (CTP) and hybridized using QuickHyb solution (Stratagene) according to the manufacturer’s protocol. After the films were exposed to the phosphoimager screen for 24 hours, hybridization signals were detected using a Bioimaging system (BAS1000, Fuji Photo Film). Quantification of mRNA was performed using Scion image analysis software (Scion Corporation). Levels of RNA were quantified and normalized for RNA loading by stripping and reprobing the blots with a probe for β-actin.
Enzyme activity assays.
For measuring enzyme activity of selected genes, mice were exposed to CS for 5 hours and sacrificed after 24 hours. The lungs were excised (n = 3 per group) and processed as described (21) to measure the activities of NQO1, G6PDH, GPx, Prx, and GSR. GPx activity was measured according to the procedure of Flohe and Gunzler (44). NQO1 activity was determined using menadione as a substrate (45). The peroxidase activity of Prx was measured by monitoring the oxidation of NADPH as described (46). G6PDH activity was determined from the rate of glucose 6-phosphate–dependent reduction of NADP+ (47). GSR activity was determined from the rate of oxidation of NADPH by using oxidized glutathione as substrate (48). Protein concentration was determined by using the Biorad reagent, with bovine serum albumin as the standard. The values for enzyme-specific activities are given as means ± SE. Student’s t test was used to determine statistical significance.
Statistical analysis.
Statistical analysis was performed by ANOVA, with the selection of the most conservative pairwise multiple comparison method using the program SigmaPlot 2000 (SPSS Inc.), and differences between groups were determined by Student’s t test using the InStat program (GraphPad Software Inc.).
Supplementary Material
Acknowledgments
This work was supported by National Institute of Environmental Health Sciences Center grant P30 ES 03819; the Maryland Cigarette Restitution Fund (to S. Biswal); the Flight Attendant Research Institute (to S. Biswal); Alpha — 1 Foundation (to R.M. Tuder and I. Petrache); the American Thoracic Society and the American Lung Association (to I. Petrache); and NIH grants P50 CA058184, R01 66554 (to R.M. Tuder), and R01 CA94076 (to T.W. Kensler). We thank Wayne Mitzner for the total lung capacity measurements.
Footnotes
Chung Y. Cho and Rajesh K. Thimmulappa contributed equally to this work.
Nonstandard abbreviations used: ARE, antioxidant response element; BAL, bronchoalveolar lavage; COPD, chronic obstructive pulmonary disease; CS, cigarette smoke; EMSA, electrophoretic mobility shift assay; G6PDH, glucose-6-phosphate dehydrogenase; γ–GCS, γ-glutamyl cysteine synthase; GPx, glutathione peroxidase; GSR, glutathione reductase; GST, glutathione S-transferase; HO-1, heme oxygenase-1; NQO1, NADPH: quinone oxidoreductase-1; Nrf2, nuclear factor, erythroid-derived 2, like 2; OCT1, octamer transcription factor 1; 8-oxo-dG, 8-oxo-7,8-dihydro-2′-deoxyguanosine; Prx-1, peroxiredoxin-1; SpC, surfactant protein C; TrxR, thioredoxin reductase; TSS, transcription start site; UGT, UDP-glucuronosyl transferase.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Mahadeva R, Shapiro SD. Chronic obstructive pulmonary disease * 3: Experimental animal models of pulmonary emphysema. Thorax. 2002;57:908–914. doi: 10.1136/thorax.57.10.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petty TL. Definition, epidemiology, course, and prognosis of COPD. Clin. Cornerstone. 2003;5:1–10. doi: 10.1016/s1098-3597(03)90003-2. [DOI] [PubMed] [Google Scholar]
- 3.Viegi G, Scognamiglio A, Baldacci S, Pistelli F, Carrozzi L. Epidemiology of chronic obstructive pulmonary disease (COPD) Respiration. 2001;68:4–19. doi: 10.1159/000050456. [DOI] [PubMed] [Google Scholar]
- 4.Tuder RM, et al. Oxidative stress and apoptosis interact and cause emphysema due to vascular endothelial growth factor receptor blockade. Am. J. Respir. Cell Mol. Biol. 2003;29:88–97. doi: 10.1165/rcmb.2002-0228OC. [DOI] [PubMed] [Google Scholar]
- 5.Eriksson S. Pulmonary emphysema and α1-antitrypsin deficiency. Acta Med. Scand. 1964;175:197–205. doi: 10.1111/j.0954-6820.1964.tb00567.x. [DOI] [PubMed] [Google Scholar]
- 6.Joos L, Pare PD, Sandford AJ. Genetic risk factors of chronic obstructive pulmonary disease. Swiss Med. Wkly. 2002;132:27–37. doi: 10.4414/smw.2002.09752. [DOI] [PubMed] [Google Scholar]
- 7.Hautamaki RD, Kobayashi DK, Senior RM, Shapiro SD. Requirement for macrophage elastase for cigarette smoke-induced emphysema in mice. Science. 1997;277:2002–2004. doi: 10.1126/science.277.5334.2002. [DOI] [PubMed] [Google Scholar]
- 8.Zheng T, et al. Inducible targeting of IL-13 to the adult lung causes matrix metalloproteinase- and cathepsin-dependent emphysema. J. Clin. Invest. 2000;106:1081–1093. doi: 10.1172/JCI10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horvath I, et al. “Haemoxygenase-1 induction and exhaled markers of oxidative stress in lung diseases”, summary of the ERS Research Seminar in Budapest, Hungary, September, 1999. Eur. Respir. J. 2001;18:420–430. doi: 10.1183/09031936.01.00231201. [DOI] [PubMed] [Google Scholar]
- 10.MacNee W, Rahman I. Is oxidative stress central to the pathogenesis of chronic obstructive pulmonary disease? Trends Mol. Med. 2001;7:55–62. doi: 10.1016/s1471-4914(01)01912-8. [DOI] [PubMed] [Google Scholar]
- 11.Li J, et al. Signaling intermediates required for NF-κB activation and IL-8 expression in CF bronchial epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2003;284:L307–L315. doi: 10.1152/ajplung.00086.2002. [DOI] [PubMed] [Google Scholar]
- 12.Cho HY, et al. Linkage analysis of susceptibility to hyperoxia. Nrf2 is a candidate gene. Am. J. Respir. Cell Mol. Biol. 2002;26:42–51. doi: 10.1165/ajrcmb.26.1.4536. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 2002;17:17. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 14.Itoh K, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 15.Cavarra E, et al. Effects of cigarette smoke in mice with different levels of alpha(1)-proteinase inhibitor and sensitivity to oxidants. Am. J. Respir. Crit. Care Med. 2001;164:886–890. doi: 10.1164/ajrccm.164.5.2010032. [DOI] [PubMed] [Google Scholar]
- 16.Aoshiba K, Yokohori N, Nagai A. Alveolar wall apoptosis causes lung destruction and emphysematous changes. Am. J. Respir. Cell Mol. Biol. 2003;28:555–562. doi: 10.1165/rcmb.2002-0090OC. [DOI] [PubMed] [Google Scholar]
- 17.Finkelstein R, Fraser RS, Ghezzo H, Cosio MG. Alveolar inflammation and its relation to emphysema in smokers. Am. J. Respir. Crit. Care Med. 1995;152:1666–1672. doi: 10.1164/ajrccm.152.5.7582312. [DOI] [PubMed] [Google Scholar]
- 18.Trevani AS, et al. Neutrophil apoptosis induced by proteolytic enzymes. Lab. Invest. 1996;74:711–721. [PubMed] [Google Scholar]
- 19.Segura-Valdez L, et al. Upregulation of gelatinases A and B, collagenases 1 and 2, and increased parenchymal cell death in COPD. Chest. 2000;117:684–694. doi: 10.1378/chest.117.3.684. [DOI] [PubMed] [Google Scholar]
- 20.Kasai H. Analysis of a form of oxidative DNA damage, 8-hydroxy-2’- deoxyguanosine, as a marker of cellular oxidative stress during carcinogenesis. Mutat. Res. 1997;387:147–163. doi: 10.1016/s1383-5742(97)00035-5. [DOI] [PubMed] [Google Scholar]
- 21.Cho HY, et al. Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell Mol. Biol. 2002;26:175–182. doi: 10.1165/ajrcmb.26.2.4501. [DOI] [PubMed] [Google Scholar]
- 22.Chan K, Kan YW. Nrf2 is essential for protection against acute pulmonary injury in mice. Proc. Natl. Acad. Sci. U. S. A. 1999;96:12731–12736. doi: 10.1073/pnas.96.22.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhakshinamoorthy S, Jaiswal AK. Small maf (MafG and MafK) proteins negatively regulate antioxidant response element-mediated expression and antioxidant induction of the NAD(P)H:Quinone oxidoreductase1 gene. J. Biol. Chem. 2000;275:40134–40141. doi: 10.1074/jbc.M003531200. [DOI] [PubMed] [Google Scholar]
- 24.He CH, et al. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J. Biol. Chem. 2001;276:20858–20865. doi: 10.1074/jbc.M101198200. [DOI] [PubMed] [Google Scholar]
- 25.Das KC, White CW. Redox systems of the cell: possible links and implications. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9617–9618. doi: 10.1073/pnas.162369199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casagrande S, et al. Glutathionylation of human thioredoxin: a possible crosstalk between the glutathione and thioredoxin systems. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9745–9749. doi: 10.1073/pnas.152168599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sies H, Arteel GE. Interaction of peroxynitrite with selenoproteins and glutathione peroxidase mimics. Free Radic. Biol. Med. 2000;28:1451–1455. doi: 10.1016/s0891-5849(00)00253-7. [DOI] [PubMed] [Google Scholar]
- 28.Nordberg J, Arner ES. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic. Biol. Med. 2001;31:1287–1312. doi: 10.1016/s0891-5849(01)00724-9. [DOI] [PubMed] [Google Scholar]
- 29.Kim PM, Wells PG. Genoprotection by UDP-glucuronosyltransferases in peroxidase-dependent, reactive oxygen species-mediated micronucleus initiation by the carcinogens 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and benzo[a]pyrene. Cancer Res. 1996;56:1526–1532. [PubMed] [Google Scholar]
- 30.Engel LS, et al. Pooled analysis and meta-analysis of glutathione S-transferase M1 and bladder cancer: a HuGE review. Am. J. Epidemiol. 2002;156:95–109. doi: 10.1093/aje/kwf018. [DOI] [PubMed] [Google Scholar]
- 31.Dinkova-Kostova AT, Talalay P. Persuasive evidence that quinone reductase type 1 (DT diaphorase) protects cells against the toxicity of electrophiles and reactive forms of oxygen. Free Radic. Biol. Med. 2000;29:231–240. doi: 10.1016/s0891-5849(00)00300-2. [DOI] [PubMed] [Google Scholar]
- 32.Vogt BA, Alam J, Croatt AJ, Vercellotti GM, Nath KA. Acquired resistance to acute oxidative stress. Possible role of heme oxygenase and ferritin. Lab. Invest. 1995;72:474–483. [PubMed] [Google Scholar]
- 33.Brittenham, G.M., et al. 2000. Clinical consequences of new insights in the pathophysiology of disorders of iron and heme metabolism. Hematology (Am. Soc. Hematol. Educ. Program). 39–50. [DOI] [PubMed]
- 34.Folz RJ, Abushamaa AM, Suliman HB. Extracellular superoxide dismutase in the airways of transgenic mice reduces inflammation and attenuates lung toxicity following hyperoxia. J. Clin. Invest. 1999;103:1055–1066. doi: 10.1172/JCI3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poller W, et al. Mis-sense mutation of alpha 1-antichymotrypsin gene associated with chronic lung disease. Lancet. 1992;339:1538. doi: 10.1016/0140-6736(92)91301-n. [DOI] [PubMed] [Google Scholar]
- 36.Ramos-Gomez M, et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witschi H, Espiritu I, Maronpot RR, Pinkerton KE, Jones AD. The carcinogenic potential of the gas phase of environmental tobacco smoke. Carcinogenesis. 1997;18:2035–2042. doi: 10.1093/carcin/18.11.2035. [DOI] [PubMed] [Google Scholar]
- 38.Kasahara Y, et al. Inhibition of VEGF receptors causes lung cell apoptosis and emphysema. J. Clin. Invest. 2000;106:1311–1319. doi: 10.1172/JCI10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasahara Y, et al. Endothelial cell death and decreased expression of vascular endothelial growth factor and vascular endothelial growth factor receptor 2 in emphysema. Am. J. Respir. Crit. Care Med. 2001;163:737–744. doi: 10.1164/ajrccm.163.3.2002117. [DOI] [PubMed] [Google Scholar]
- 40.Saltini C, et al. Accurate quantification of cells recovered by bronchoalveolar lavage. Am. Rev. Respir. Dis. 1984;130:650–658. doi: 10.1164/arrd.1984.130.4.650. [DOI] [PubMed] [Google Scholar]
- 41.Tirumalai R, Rajesh Kumar T, Mai KH, Biswal S. Acrolein causes transcriptional induction of phase II genes by activation of Nrf2 in human lung type II epithelial (A549) cells. Toxicol. Lett. 2002;132:27–36. doi: 10.1016/s0378-4274(02)00055-3. [DOI] [PubMed] [Google Scholar]
- 42.Thimmulappa RK, et al. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- 43.Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proc. Natl. Acad. Sci. U. S. A. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flohe L, Gunzler WA. Assays of glutathione peroxidase. Meth. Enzymol. 1984;105:114–121. doi: 10.1016/s0076-6879(84)05015-1. [DOI] [PubMed] [Google Scholar]
- 45.Prochaska HJ, Santamaria AB. Direct measurement of NAD(P)H:quinone reductase from cells cultured in microtiter wells: a screening assay for anticarcinogenic enzyme inducers. Anal. Biochem. 1988;169:328–336. doi: 10.1016/0003-2697(88)90292-8. [DOI] [PubMed] [Google Scholar]
- 46.Chae HZ, Kang SW, Rhee SG. Isoforms of mammalian peroxiredoxin that reduce peroxides in presence of thioredoxin. Meth. Enzymol. 1999;300:219–226. doi: 10.1016/s0076-6879(99)00128-7. [DOI] [PubMed] [Google Scholar]
- 47.Lee CY. Glucose-6-phosphate dehydrogenase from mouse. Meth. Enzymol. 1982;89:252–257. doi: 10.1016/s0076-6879(82)89045-9. [DOI] [PubMed] [Google Scholar]
- 48.Carlberg I, Mannervik B. Glutathione reductase. Meth. Enzymol. 1985;113:484–490. doi: 10.1016/s0076-6879(85)13062-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.