Abstract
Fli-1 is a member of the Ets transcription factor family and is expressed during T-cell development; however, the role Fli-1 plays in early T-cell differentiation has not been elucidated. In this report, we demonstrate that in mouse, Fli-1 overexpression retards the CD4−CD8− double negative (DN) to CD4+CD8+ double positive (DP) transition by deregulating normal DN thymocyte development. Specifically, Fli-1 expression moderates the DN2 and DN3 developmental transitions. We further show that Fli-1 overexpression partially mimics strong TCR signals in developing DN thymocytes and thereby enhances γδ T-cell development. Conversely, Fli-1 knockdown by shRNA reverses the lineage bias from γδ T cells and directs DN cells to the αβ lineage by attenuating TCR signalling. Therefore, Fli-1 plays a critical role in both the DN2 to DN3 transition and αβ/γδ lineage commitment.
Keywords: Fli-1, T-cell development, β-selection, pre-TCR, γδ T-cell lineage, T cells, thymic selection, γδ T cells, transcription factors, thymopoiesis
Introduction
T-cell development begins when DN precursors enter the thymus [1]. Here they undergo an ordered development and can be subdivided into consecutive stages based on c-Kit, CD44 and CD25 expression. DN1 cells (CD25−CD44+) are heterogeneous and can become T cells, B cells, dendritic cells (DCs) and natural killer (NK) cells [2, 3]. True DN1 T-cell progenitors express high levels of c-Kit [4]. DN2 cells (CD25+CD44+) are mainly T-lineage restricted with no B-cell potential, but some DC and NK-cell potential in the c-KitHigh DN2a cells and only limited NK-cell potential in the c-KitInt DN2b cells [3, 5–8]. DN3 cells (CD25+CD44−) are committed T cells with fully rearranged TCRγ, δ or β loci. Expression of a productively rearranged γδTCR allows development along the (mostly DN) γδ T-cell lineage whereas a functional TCRβ promotes adoption of the αβ T-cell lineage [9]. Further development of αβ T cells and initiation of the DN to DP transition requires signalling through the pre-TCR complex, which comprises a rearranged β chain associated with pre-Tα [10–12]. Based on CD28 expression levels, DN3 cells can be subdivided into those immediately before β or γδ-selection (DN3a=CD28lo) and those after selection (DN3b=CD28hi) [13]. Finally, DN4 cells (CD25−CD44−) are primed to become DP cells [14], which then through positive selection further develop into either mature CD4+CD8− or CD4−CD8+ single positive (SP) T cells [1]. Differentiation into the αβ or γδ T-cell lineage has been shown to be influenced by the strength of the pre-TCR/TCR signal [15, 16]. This model proposes that strong TCR signals result in development of γδ T cells whilst weaker TCR signals lead to αβ T-cell development.
Fli-1 is an Ets transcription factor, which is primarily expressed in haematopoietic cells, including most myeloid, B and T cells [17–19]. We have recently published that Fli-1 overexpression induces pre-T-cell lymphoblastic leukaemia/lymphoma [20] and previous reports have suggested a role for Fli-1 in autoimmune disease [21, 22]. However, the precise role of Fli-1 in normal early T-cell development remains unclear.
Here we show that Fli-1 overexpression inhibits the DN to DP transition following a β-selection checkpoint arrest at the DN3a stage. Gain-of-function analysis using the Scid.adh cell line revealed that enforced expression Fli-1 could mimic a strong TCR signal, thereby leading to enhanced γδ T-cell development in normal T-cell precursors. Conversely, decreased Fli-1 expression by shRNA knockdown in a transgenic γδTCR background resulted in biased development to αβ CD4+CD8+ cells confirming an essential role for Fli-1 in early T-cell development and αβ/γδ T-cell lineage commitment.
Results and Discussion
Fli-1 functions during commitment and at the β-selection checkpoint of DN T-cell development
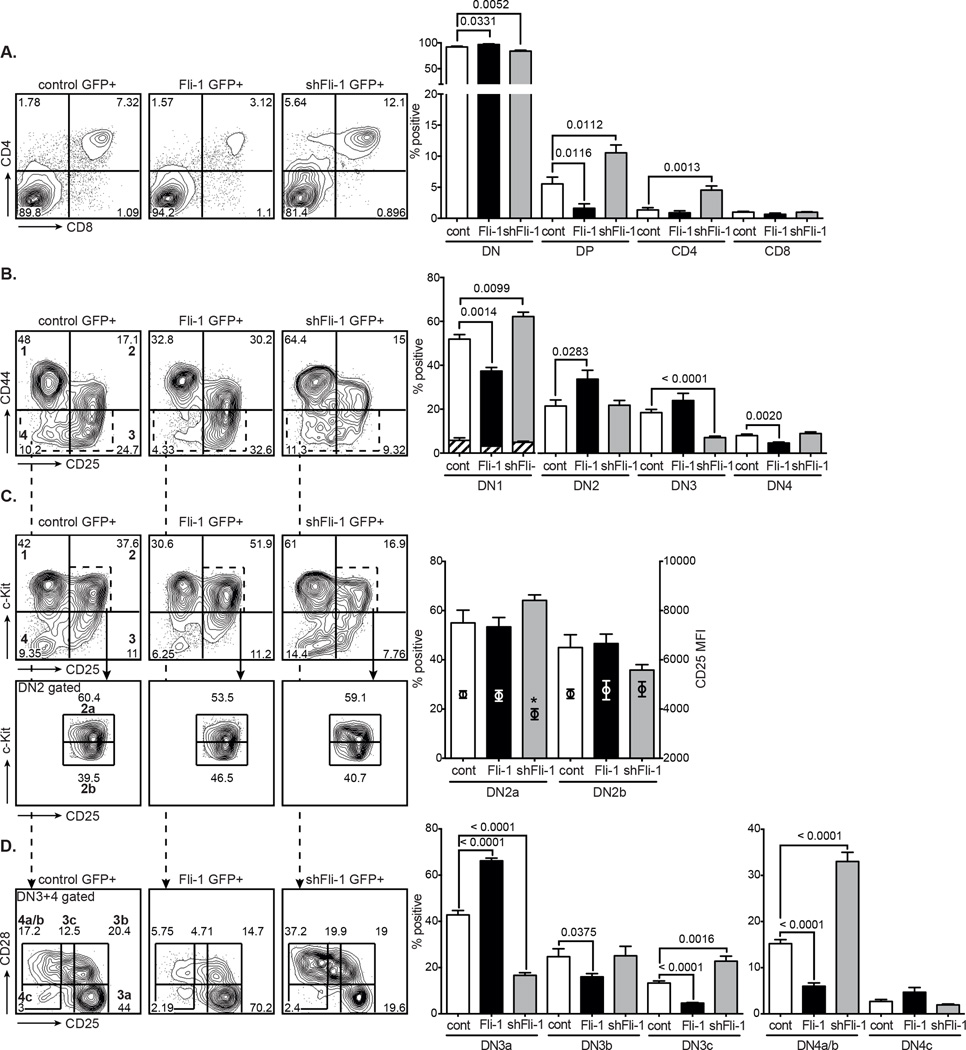
To study the function of Fli-1 in T-cell development we retrovirally overexpressed or knocked down Fli-1 in developing T-cell progenitors in vitro using the OP9-DL1 system [23]. Whole E15 foetal liver (FL) cells were expanded on OP9-DL1 stroma for 6 days, transduced with control, Fli-1 overexpression or shRNA Fli-1 knockdown constructs and cultured on OP9-DL1 stroma for an additional 6 days. The resultant differentiated T cells were analysed for developmental differences by flow cytometry. Fli-1 overexpression inhibited the DN to DP transition and caused a decrease in the percentage of DP T cells, whereas knockdown of Fli-1 resulted in an increase in DP cells (Fig. 1A). Further examination of the DN subpopulations revealed that Fli-1 overexpression significantly decreased the percentage of c-Kit positive DN1 cells, enhanced the percentage of DN2 and DN3 cells and decreased DN4 cells (Fig. 1B). Downregulation of Fli-1 showed the opposite effect with more DN1 cells and an accelerated transition through the DN3 and DN4 stages. A kinetic analysis, examining the cells every 6 days up to 24 days after transduction, indicated that the DN2 and DN3 stages were most critically affected. After 12 days of cell culture, little change was observed and Fli-1 overexpressing as well as Fli-1 knockdown cells displayed decreased DP percentages due to partial arrests at the DN3 and DN2 stages respectively (Supporting Information Fig. 1) [20].
Figure 1. Effects of Fli-1 overexpression and Fli-1 knockdown on in vitro T-cell development.
GFP+ MigR1 control, Fli-1 and shFli-1-transduced FL cells were grown on OP9-DL1 cells for 6 days and analysed for expression of T-cell markers by flow cytometry. (A) GFP+-gated analysis of CD4 and CD8 expression. Flow cytometry plots are representative of 5 independent experiments. Data in graphs are represented as mean + SEM of 5 independent experiments, each comprised of a single sample per group. p values as indicated. (B) Lineage negative-gated GFP+ DN1–4 populations (bold numbers in left panel) as determined by CD44 and CD25 expression. Flow cytometry plots are representative of 7 independent experiments. Hatched portions of the bar graph indicate the fraction of c-Kit-negative DN1 cells and data are shown as mean + SEM of 7 independent experiments, each comprised of a single sample per group. (C) Lineage negative-gated GFP+ DN1–4 populations (bold numbers in top left panel) as determined by c-Kit and CD25 expression and DN2-gated c-Kithigh and c-KitInt DN2a and b subpopulations (bottom panels). Flow cytometry plots are representative of 5 independent experiments. Circles in bar graphs indicate CD25 median fluorescence intensity (MFI) (right y-axis) (mean + SEM of 5 independent experiments, each comprised of a single sample per group). *p < 0.05, unpaired two-tailed student’s t-test. (D) DN3+4-gated (see B) GFP+ DN3 and DN4 subpopulations (indicated in bold in left panel) based on CD28 and CD25 expression. Flow cytometry plots are representative of 7 independent experiments. Data in bar graphs shown as mean + SEM of 7 independent experiments, each comprised of a single sample per group. p values as indicated, unpaired t-test.
Fli-1 overexpression and shRNA downregulation were shown to be within physiological range in T-cell progenitors, namely 2-fold increased or 3 to 2-fold decreased in DN2 and DN3 cells respectively (Supporting Information Fig. 2A,B). The specificity of 3 different shRNAs for Fli-1 was first confirmed in 3T3 cells by real-time PCR and Western blotting (Supporting Information Fig. 2A,B). Construct #2 targets the Fli-1 cDNA and inhibited expression of Fli-1 from the overexpression construct (Supporting Information Fig. 2C). Functional specificity in T cells was therefore tested using Fli-1 shRNA constructs #1 and 3, which only target the Fli-1 3’UTR. Six days after a first transduction with MLS control and shFli-1, GFP+ DN2 cells were sorted, transduced with MSCV control dsRed or Fli-1 dsRed and grown on OP9-DL1 for another 12 days. Overexpression of the Fli-1 cDNA allowed the shFli-1 DN2 cells to progress to DN3 cells and rescued the knockdown phenotype, corroborating the specificity of the constructs and the role of Fli-1 in DN development (Supporting Information Fig. 2D).
Endogenous Fli-1 mRNA expression levels throughout DN thymocyte development are also consistent with both the ectopic Fli-1-induced DN3 accumulation, as cells may need to downregulate Fli-1 in order to transit from the DN3 to the DN4 stage, as well as the Fli-1 knockdown-induced delay at the DN1 and DN2 stages, where higher levels of Fli-1 are required for proper DN development (Supporting Information Fig. 2E) [17, 20, 24]. Similar expression patterns are found in a group of genes associated with stem cell or progenitor cell functions such as Lmo2, SCL/Tal1 and Lyl1 [25]. Overexpression of these genes has been linked to increased stem cell-like features and decreased differentiation and as a result disturbed DN-DP development, more specifically at the DN2 and DN3 stages [26–30].
Consequently we analysed the DN2 and DN3 stages in more detail using c-Kit and CD28 respectively [8, 13]. Fli-1 overexpression resulted in a significant increase in the percentage of DN3a cells and a concomitant decrease in the post-β-selection DN3b, DN3c and DN4a/b stages (Fig. 1C,D). As the DN4a are the most efficient DP precursors this explains why fewer DP thymocytes are generated in Fli-1 transduced cells [13]. shRNA Fli-1 knockdown, in contrast, resulted in slightly more immature DN2a (with significantly lower CD25) and fewer DN3a cells but considerably more DN3c and DN4a/b cells (the most immediate DP precursors) leading to more downstream DP thymocytes (Fig. 1C,D). These results indicate Fli-1 plays a role at the commitment and β-selection stages of early T-cell development. Detailed analysis of in vivo Fli-1 expression in T cells also agrees with a checkpoint role for Fli-1 in pre-TCR/TCR signalling [31, 32].
Fli-1 overexpression enhances pre-TCR signalling in Scid.adh cells
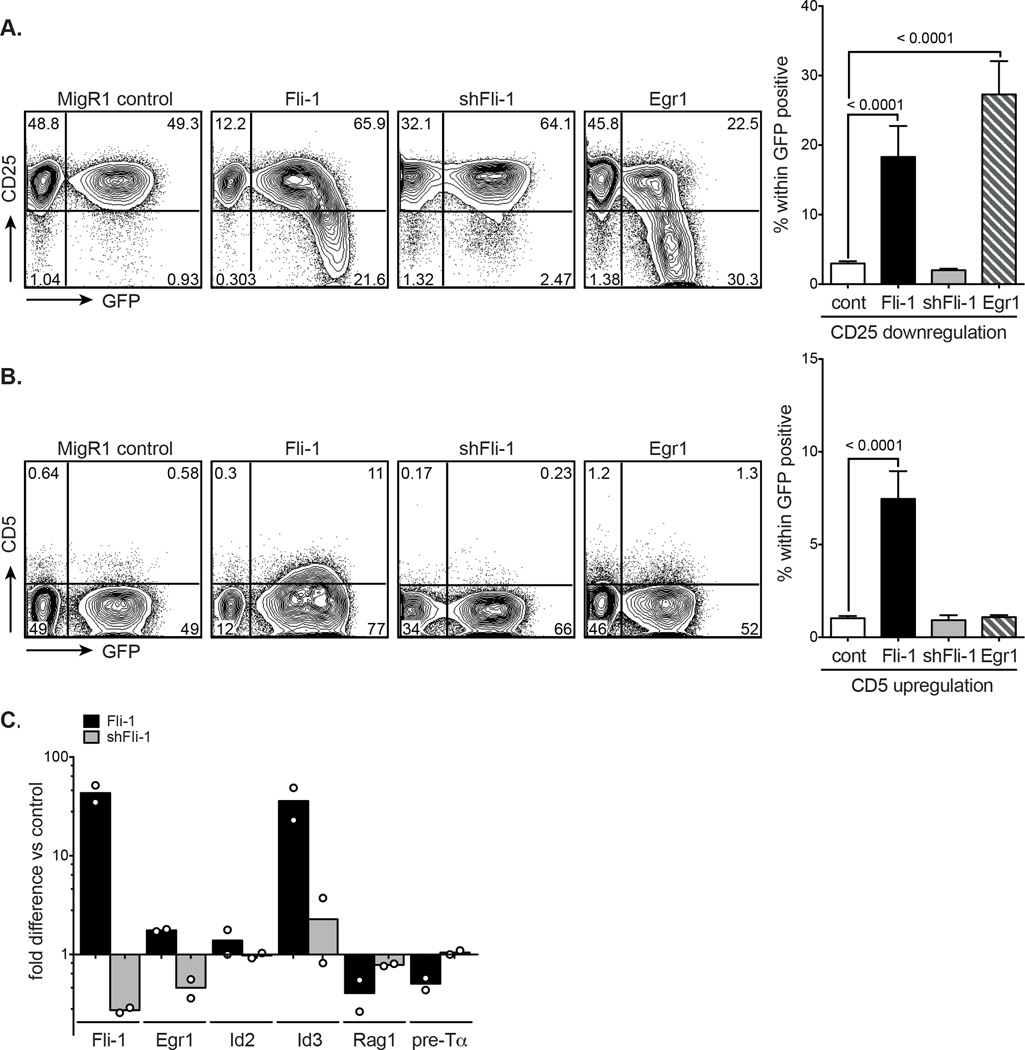
As a tool to study pre-TCR signalling, we utilised the Scid.adh cell line which resembles DN3a thymocytes and downregulates CD25 and upregulates CD5 (an activation and pro-survival antigen [33, 34]), when its pre-TCR is signalled [35, 36]. The Scid.adh cell line was retrovirally transduced with MigR1, Fli-1, shRNA Fli-1 and Egr1 as a positive control. Forty-two hours post-transduction, CD25 and CD5 expression of the Scid.adh cell line were assessed by flow cytometry. It could be clearly seen that Fli-1 overexpression significantly induced CD25 downregulation similar to Egr1 (Fig. 2A) as well as CD5 upregulation, which was not observed in Egr1 expressing cells (Fig. 2B). The highest levels of GFP, corresponding with the highest levels of Fli-1 expression had the strongest effect on CD25 downregulation (Fig. 2A Fli-1 panel) and high levels of CD5 in particular are indicative of stronger TCR complex signals [33, 37–39]. Fli-1 downregulation in contrast had no effect. Fli-1 overexpression, like Egr1 [36], thus replicates many of the molecular events induced by pre-TCR signalling.
Figure 2. Effects of Fli-1 overexpression on pre-TCR signalling in Scid.adh cells.
MigR1 control, Fli-1, shRNA Fli-1 and Egr1-transduced Scid.adh cells were analysed for CD25 and CD5 expression by flow cytometry, 42 hours after transduction. Graphs show percentage (A) CD25 downregulation or (B) CD5 upregulation in GFP+ Scid.adh cells and data are shown as mean + SEM of ≥ 7 independent experiments, each comprised of a single sample per group. p values in A and B were determined by unpaired two-tailed student’s t-tests. (C) mRNA expression of Fli-1, Egr1, Id2, Id3, Rag1 and pre-Tα was determined by quantitative real-time PCR in sorted GFP+ Fli-1 and shRNA Fli-1-transduced Scid.adh cells at 42 hours post-transduction. Expression levels are shown as the mean fold difference versus sorted GFP+ control. Circles represent independent experiments; n = 2, each comprised of a single sample per group.
To analyse the expression levels of genes known to be involved in or modulated by pre-TCR signalling [36, 40], we subsequently sorted GFP+ MigR1 control, Fli-1 and shRNA Fli-1 expressing Scid.adh cells two days after transduction. Levels of Egr1, Id2, and more significantly, Id3 were increased when Fli-1 was overexpressed and Rag1 and pre-Tα levels were decreased, reflective of pre-TCR signalling. Fli-1 knockdown resulted in either no effect or the opposite effect (Fig. 2C). This strongly suggests that enforced Fli-1 overexpression can mimic pre-TCR signalling and enhance the downstream consequences thereby producing a strong signal. The observation of a DN to αβ DP block induced by Fli-1 overexpression (Fig. 1A) could then perhaps be better explained by a diversion to γδ rather than a block of αβ DP T-cell development. This notion would be further supported by the increase in Id3 expression level, which has been shown to promote γδ T-cell fate (Fig. 2C) [41].
Ectopic Fli-1 biases TCRβ+ T cells to the γδ lineage whilst knockdown redirects γδ to αβ DP T cells
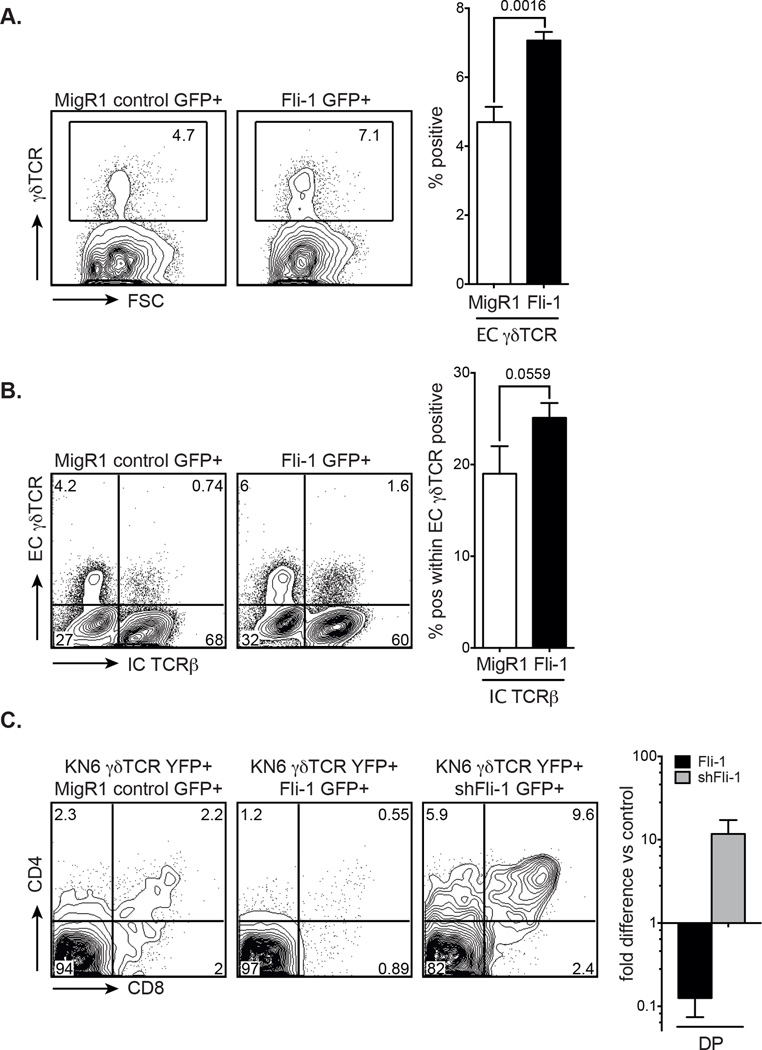
Strong TCR signalling has been clearly associated with γδ development. Indeed, it is thought that strong signals preferentially lead to adoption of the γδ fate and oppose αβ T-cell commitment [15, 16, 42]. Even stronger signalling leads to apoptosis. Weak signalling through the pre-TCR, on the contrary, favours αβ over γδ T-cell commitment [43]. As Fli-1 RNA levels were found to be high in wild type γδ T cells (Supporting Information Fig. 2E) and Fli-1 overexpression in Scid.adh cells resulted in strong CD25 downregulation and CD5 upregulation it was hypothesised that Fli-1 may be redirecting uncommitted DN cells to the γδ lineage by enhancing TCR signalling. As predicted, analysis of Fli-1 transduced FL cells grown in the OP9-DL1 system (which accurately reproduces γδ T-cell development in vitro [38]), revealed that there was a statistically significant increase in γδ T cells compared with the MigR1 control (Fig. 3A). Sorted GFP+ Fli-1 DN3 cells (normally already committed to either the αβ or γδ lineage [38]) additionally showed an increase in intracellular TCRβ (i.e. pre-TCR) within the γδTCR positive population, reinforcing the theory that a strong signal induced by Fli-1 overexpression can divert cells towards the γδ T-cell lineage (Fig. 3B) [44]. A recent report has shown that DN2 cells can develop into both IL-17 and IFN-γ producing γδ T cells, but only IFN-γ producing γδ T cells develop from DN3 cells [45]. Given that the vast majority of Fli-1 overexpressing T cells are DN3 cells, Fli-1 signals (like strong TCR signals [46]) may be preferentially involved in the development of IFN-γ producing T cells. This hypothesis should be tested in the future.
Figure 3. Effects of Fli-1 overexpression and Fli-1 knockdown on in vitro γδ versus αβ T-cell development.
(A) GFP+-gated γδTCR expression in MigR1 control and Fli-1-transduced FL cells determined by flow cytometry 12 days after transduction. Flow cytometry plots are representative of 5 independent experiments. Graphed data are shown as mean + SEM of 5 independent experiments, each comprised of a single sample per group. p value as indicated. Unpaired two-tailed student’s t-test. (B) Extracellular γδTCR and intracellular TCRβ expression in MigR1 and Fli-1-transduced FL cells determined by flow cytometry, 6 days after GFP+ DN3 cells were sorted by flow cytometry. Flow cytometry plots are representative of 5 independent experiments. Graphed data are shown as mean + SEM of 5 independent experiments, each comprised of a single sample per group. p value as indicated. Unpaired one-tailed student’s t-test. (C) Representative flow cytometry plots (5 independent experiments) of DP development in Rag-1−/− cells (gated on YFP+ and GFP+ cells), 8 days after co-transduction of sorted DN3 cells with KN6-γδTCR and MigR1 control, Fli-1 or shRNA Fli-1 retrovirus. Graphed data show mean + SEM of fold difference in DP development in Rag-1−/− KN6-γδTCR/Fli-1 or shRNA Fli-1 co-transduced cells versus control of 5 independent experiments, each comprised of a single sample per group.
If a strong signal by the γδTCR commits developing DN cells to the γδ T-cell lineage, then attenuation of this signal should bias cells towards the αβ lineage and more CD4+CD8+ DP cells should be generated. To test this, we chose to knockdown Fli-1 expression in Rag-1−/− cells transduced with a KN6-γδTCR construct [41]. For this experiment, Rag-1−/− DN3 cells were sorted and co-transduced with MigR1 control, Fli-1 or shRNA Fli-1-GFP and KN6-γδTCR-YFP constructs and cultured on OP9-DL1 cells for 8 days. The KN6-γδTCR-YFP construct alone produced some DP thymocytes [38] but the shRNA Fli-1 construct co-transduced with the KN6-γδTCR generated significantly more DP cells, whereas Fli-1 overexpression seemed to further reduce DP development (Fig. 3C). Therefore, knockdown of Fli-1 reduced the capacity of the γδTCR signal to promote adoption of the γδ fate and as a result, enhanced the diversion of progenitors towards the αβ lineage, as evidenced by the increased generation of DP cells. This result is consistent with a model of T-cell development whereby Fli-1 expression converts the normally weak pre-TCR signal to a strong signal, thereby favouring γδ over αβ lineage choice.
Concluding remarks
Our data have demonstrated that Fli-1 overexpression can replicate many aspects of TCR signalling at the molecular and functional level. Decreasing levels of Fli-1 expression in DN3/DN4 αβ lineage precursors and much higher levels in γδ T cells support a role for Fli-1 throughout early T-cell development. Enforced Fli-1 expression indeed promotes adoption of the γδ fate by turning a normally weak pre-TCR signal into a strong signal. Conversely, knockdown of Fli-1 results in diversion to the αβ fate. Taken together, we report here the first evidence that Fli-1 modulates the intensity of TCR signalling and is an important effector of αβ/γδ lineage choice. Further studies into the in vivo function of Fli-1 should be of particular relevance in light of the recent interest in γδ T cells and their role in disease and therapy [47, 48].
Materials and Methods
Antibodies
Fluorochromes used for surface staining included PE, APC, Alexa Fluor 647, PeCy7, PerCP, and eFluor450. Conjugated anti-mouse CD4 (GK1.5), CD8 (53-6.7), CD25 (PC61, eBio 3C7), CD44 ((IM7), c-Kit (2B8), CD28 ((E18), CD5 (53-7.3), γδTCR (GL3) and TCRβ (H57-597) antibodies were from Biolegend (San Diego, CA, USA) eBioscience (San Diego, CA, USA) and BD-Pharmingen (San Diego, CA, USA). Lineage depletion for CD25/44 staining was accomplished by staining cells with a cocktail of biotinylated mAb (anti-mouse CD3, CD4, CD8, B220, Mac-1, Gr-1, Ter119 and NK1.1; (Biolegend, eBioscience and BD-Pharmingen) and gating out Streptavidin (SA-) PeCy7 or SA-Alexa Fluor 594 positive cells (Invitrogen, Carlsbad, CA, USA).
Flow Cytometry
Filtered cell suspensions from C57BL/6 E15 FL-stromal cell co-cultures or Scid.adh suspension cultures, were washed in phosphate-buffered saline (PBS) with 1% foetal calf serum (FCS) and 0.01% NaN3 (FACS buffer). Fc receptors were blocked with 2.4G2 (anti-FcRII clone). Cells were then stained with the appropriately diluted antibodies, washed with FACS buffer and where necessary stained with SA. Labelled cells were resuspended in FACS buffer plus Propidium Iodide (PI), acquired on a Becton Dickinson FACScalibur or LSRII, and analysed using FlowJo software (Treestar, Inc., San Carlos, CA). Routinely, 1×105 to 2×105 events were collected. Fluorescent-activated cell sorting of DN subpopulations was performed on a FACSAria (Becton-Dickinson).
Plasmids and Retrovirus
MigR1-GFP was a gift from Warren Pear and was used as described previously [49]. MSCV-dsRed was constructed by replacing the GFP by a monomeric dsRed fragment. MSCV-YFP and KN6-γδTCR have been described previously [41]. Wild type murine Fli-1 cDNA was cloned from 129Sv/J embryoid bodies, ligated into MigR1 and the insert confirmed by sequencing. For Fli-1 knockdown, two shRNAs targeting the 3’UTR (#1 and 3) and one targeting the ORF (#2) of the murine Fli-1 cDNA were generated by PCR, cloned into the LMS and LMP vectors and sequence verified [50]. #1 and 3 showed maximum efficiency and were chosen for further experiments. Retrovirus was produced using the Phoenix packaging cell line (www.stanford.edu/group/nolan).
Retroviral Transduction and Cell Culture
E15 foetal liver (FL) cells were cultured on OP9-DL1 cells [23] for 6 days in 5ng/ml FLT3L and 0.25ng/ml IL-7. On day 6, 5×105 FL cells were retrovirally transduced by centrifugation with 50% (v/v) retroviral supernatant at 1100g for 90 min in 50ng/ml SCF, 5ng/ml FLT3L, 0.25ng/ml IL-7 and 8µg/ml polybrene. One day later, cells were reseeded onto OP9-DL1 stroma in 5ng/ml FLT3L and 0.25ng/ml IL-7 and analysed by flow cytometry and reseeded every 6 days. Scid.adh cells were maintained as described previously [35] and 2×105 exponentially growing cells were transduced by centrifugation with 50% (v/v) retroviral supernatant at 700g for 90 min in 8µg/ml polybrene. The spinofection medium was replaced by fresh medium on the next day and cells were analysed by flow cytometry on day 2, 42 hours after transduction.
Quantitative real-time PCR
Total RNA was extracted using the NucleoSpin RNA II kit (Macherey Nagel, Düren, Germany). First-strand cDNA synthesis was carried out with 1µg of total RNA using random hexamers and MuMLV RT (New England Biolabs, Ipswich, MA USA). Real-time PCR was performed with a Rotor-Gene 3000 (Corbett Robotics, Brisbane, QLD, Australia) and SYBR Green for GAPDH, Fli-1, Egr1, Id2, Id3 Rag1 and pre-Tα (primer sequences available upon request). Gene expression levels were calculated relative to GAPDH and data were normalized by dividing each expression value by the median of gene expression of MigR1 control.
Statistics
Unpaired two-tailed student’s t-tests were performed on all data (except figure 3B, where a one-tailed student’s t-test was applied), using Prism (GraphPad Software, LaJolla, CA, USA). Significance was determined as p<0.05 or less.
Supplementary Material
Acknowledgments
This work was supported by an NHMRC Project Grant #559004 and in part by the Victorian Government’s OIS Program. DI receives support from the Cancer Council of Victoria and the Leukaemia Foundation of Australia.
Abbreviations
- Fli-1
Friend Leukaemia Integration 1
- DN
CD4−CD8− double negative thymocyte
- DP
CD4+CD8+ double positive thymocyte
- shRNA
small hairpin RNA
- UTR
untranslated region
Footnotes
Conflict of Interest
The authors declare no financial or commercial conflict of interest.
References
- 1.Anderson G, Harman BC, Hare KJ, Jenkinson EJ. Microenvironmental regulation of T cell development in the thymus. Semin Immunol. 2000;12:457–464. doi: 10.1006/smim.2000.0260. [DOI] [PubMed] [Google Scholar]
- 2.Carlyle JR, Michie AM, Furlonger C, Nakano T, Lenardo MJ, Paige CJ, Zuniga-Pflucker JC. Identification of a novel developmental stage marking lineage commitment of progenitor thymocytes. J Exp Med. 1997;186:173–182. doi: 10.1084/jem.186.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu L, Li CL, Shortman K. Thymic dendritic cell precursors: relationship to the T lymphocyte lineage and phenotype of the dendritic cell progeny. J Exp Med. 1996;184:903–911. doi: 10.1084/jem.184.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, Petrie HT. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2004;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Balciunaite G, Ceredig R, Rolink AG. The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood. 2005;105:1930–1936. doi: 10.1182/blood-2004-08-3087. [DOI] [PubMed] [Google Scholar]
- 6.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 7.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 8.Yui MA, Feng N, Rothenberg EV. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol. 2010;185:284–293. doi: 10.4049/jimmunol.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godfrey DI, Kennedy J, Mombaerts P, Tonegawa S, Zlotnik A. Onset of TCR-beta gene rearrangement and role of TCR-beta expression during CD3−CD4−CD8− thymocyte differentiation. J Immunol. 1994;152:4783–4792. [PubMed] [Google Scholar]
- 10.Groettrup M, Ungewiss K, Azogui O, Palacios R, Owen MJ, Hayday AC, von Boehmer H. A novel disulfide-linked heterodimer on pre-T cells consists of the T cell receptor beta chain and a 33 kd glycoprotein. Cell. 1993;75:283–294. doi: 10.1016/0092-8674(93)80070-u. [DOI] [PubMed] [Google Scholar]
- 11.Saint-Ruf C, Ungewiss K, Groettrup M, Bruno L, Fehling HJ, von Boehmer H. Analysis and expression of a cloned pre-T cell receptor gene. Science. 1994;266:1208–1212. [PubMed] [Google Scholar]
- 12.Kruisbeek AM, Haks MC, Carleton M, Michie AM, Zuniga-Pflucker JC, Wiest DL. Branching out to gain control: how the pre-TCR is linked to multiple functions. Immunol Today. 2000;21:637–644. doi: 10.1016/s0167-5699(00)01744-8. [DOI] [PubMed] [Google Scholar]
- 13.Teague TK, Tan C, Marino JH, Davis BK, Taylor AA, Huey RW, Van De Wiele CJ. CD28 expression redefines thymocyte development during the pre-T to DP transition. Int Immunol. 2010;22:387–397. doi: 10.1093/intimm/dxq020. [DOI] [PubMed] [Google Scholar]
- 14.Wilson A, Petrie HT, Scollay R, Shortman K. The acquisition of CD4 and CD8 during the differentiation of early thymocytes in short-term culture. Int Immunol. 1989;1:605–612. doi: 10.1093/intimm/1.6.605. [DOI] [PubMed] [Google Scholar]
- 15.Haks MC, Lefebvre JM, Lauritsen JP, Carleton M, Rhodes M, Miyazaki T, Kappes DJ, et al. Attenuation of gammadeltaTCR signaling efficiently diverts thymocytes to the alphabeta lineage. Immunity. 2005;22:595–606. doi: 10.1016/j.immuni.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Hayes SM, Li L, Love PE. TCR signal strength influences alphabeta/gammadelta lineage fate. Immunity. 2005;22:583–593. doi: 10.1016/j.immuni.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126:3131–3148. doi: 10.1242/dev.126.14.3131. [DOI] [PubMed] [Google Scholar]
- 18.Ben-David Y, Giddens EB, Letwin K, Bernstein A. Erythroleukemia induction by Friend murine leukemia virus: insertional activation of a new member of the ets gene family, Fli-1, closely linked to c-ets-1. Genes Dev. 1991;5:908–918. doi: 10.1101/gad.5.6.908. [DOI] [PubMed] [Google Scholar]
- 19.Watson DK, Smyth FE, Thompson DM, Cheng JQ, Testa JR, Papas TS, Seth A. The ERGB/Fli-1 gene: isolation and characterization of a new member of the family of human ETS transcription factors. Cell Growth Differ. 1992;3:705–713. [PubMed] [Google Scholar]
- 20.Smeets MF, Chan AC, Dagger S, Bradley CK, Wei A, Izon DJ. Fli-1 overexpression in hematopoietic progenitors deregulates T cell development and induces pre-T cell lymphoblastic leukaemia/lymphoma. PLoS One. 2013;8:e62346. doi: 10.1371/journal.pone.0062346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathenia J, Reyes-Cortes E, Williams S, Molano I, Ruiz P, Watson DK, Gilkeson GS, et al. Impact of Fli-1 transcription factor on autoantibody and lupus nephritis in NZM2410 mice. Clin Exp Immunol. 2010;162:362–371. doi: 10.1111/j.1365-2249.2010.04245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richard EM, Thiyagarajan T, Bunni MA, Basher F, Roddy PO, Siskind LJ, Nietert PJ, et al. Reducing FLI1 levels in the MRL/lpr lupus mouse model impacts T cell function by modulating glycosphingolipid metabolism. PLoS One. 2013;8:e75175. doi: 10.1371/journal.pone.0075175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 24.David-Fung ES, Yui MA, Morales M, Wang H, Taghon T, Diamond RA, Rothenberg EV. Progression of regulatory gene expression states in fetal and adult pro-T-cell development. Immunol Rev. 2006;209:212–236. doi: 10.1111/j.0105-2896.2006.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothenberg EV, Zhang J, Li L. Multilayered specification of the T-cell lineage fate. Immunol Rev. 2010;238:150–168. doi: 10.1111/j.1600-065X.2010.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cleveland SM, Smith S, Tripathi R, Mathias EM, Goodings C, Elliott N, Peng D, et al. Lmo2 induces hematopoietic stem cell-like features in T-cell progenitor cells prior to leukemia. Stem Cells. 2013;31:882–894. doi: 10.1002/stem.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tatarek J, Cullion K, Ashworth T, Gerstein R, Aster JC, Kelliher MA. Notch1 inhibition targets the leukemia-initiating cells in a Tal1/Lmo2 mouse model of T-ALL. Blood. 2011;118:1579–1590. doi: 10.1182/blood-2010-08-300343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat Immunol. 2000;1:138–144. doi: 10.1038/77819. [DOI] [PubMed] [Google Scholar]
- 29.Gothert JR, Brake RL, Smeets M, Duhrsen U, Begley CG, Izon DJ. NOTCH1 pathway activation is an early hallmark of SCL T leukemogenesis. Blood. 2007;110:3753–3762. doi: 10.1182/blood-2006-12-063644. [DOI] [PubMed] [Google Scholar]
- 30.Zohren F, Souroullas GP, Luo M, Gerdemann U, Imperato MR, Wilson NK, Gottgens B, et al. The transcription factor Lyl-1 regulates lymphoid specification and the maintenance of early T lineage progenitors. Nat Immunol. 2012;13:761–769. doi: 10.1038/ni.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 32.Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, Bendall S, et al. The transcriptional landscape of alphabeta T cell differentiation. Nat Immunol. 2013;14:619–632. doi: 10.1038/ni.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azzam HS, Grinberg A, Lui K, Shen H, Shores EW, Love PE. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J Exp Med. 1998;188:2301–2311. doi: 10.1084/jem.188.12.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Axtell RC, Xu L, Barnum SR, Raman C. CD5-CK2 binding/activation-deficient mice are resistant to experimental autoimmune encephalomyelitis: protection is associated with diminished populations of IL-17-expressing T cells in the central nervous system. J Immunol. 2006;177:8542–8549. doi: 10.4049/jimmunol.177.12.8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carleton M, Ruetsch NR, Berger MA, Rhodes M, Kaptik S, Wiest DL. Signals transduced by CD3epsilon, but not by surface pre-TCR complexes, are able to induce maturation of an early thymic lymphoma in vitro. J Immunol. 1999;163:2576–2585. [PubMed] [Google Scholar]
- 36.Carleton M, Haks MC, Smeele SA, Jones A, Belkowski SM, Berger MA, Linsley P, et al. Early growth response transcription factors are required for development of CD4(−)CD8(−) thymocytes to the CD4(+)CD8(+) stage. J Immunol. 2002;168:1649–1658. doi: 10.4049/jimmunol.168.4.1649. [DOI] [PubMed] [Google Scholar]
- 37.Azzam HS, DeJarnette JB, Huang K, Emmons R, Park CS, Sommers CL, El-Khoury D, et al. Fine tuning of TCR signaling by CD5. J Immunol. 2001;166:5464–5472. doi: 10.4049/jimmunol.166.9.5464. [DOI] [PubMed] [Google Scholar]
- 38.Ciofani M, Knowles GC, Wiest DL, von Boehmer H, Zuniga-Pflucker JC. Stage-specific and differential notch dependency at the alphabeta and gammadelta T lineage bifurcation. Immunity. 2006;25:105–116. doi: 10.1016/j.immuni.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Tarakhovsky A, Kanner SB, Hombach J, Ledbetter JA, Muller W, Killeen N, Rajewsky K. A role for CD5 in TCR-mediated signal transduction and thymocyte selection. Science. 1995;269:535–537. doi: 10.1126/science.7542801. [DOI] [PubMed] [Google Scholar]
- 40.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 41.Lauritsen JP, Wong GW, Lee SY, Lefebvre JM, Ciofani M, Rhodes M, Kappes DJ, et al. Marked induction of the helix-loop-helix protein Id3 promotes the gammadelta T cell fate and renders their functional maturation Notch independent. Immunity. 2009;31:565–575. doi: 10.1016/j.immuni.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong GW, Zuniga-Pflucker JC. gammadelta and alphabeta T cell lineage choice: resolution by a stronger sense of being. Semin Immunol. 2010;22:228–236. doi: 10.1016/j.smim.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Hayes SM, Love PE. Strength of signal: a fundamental mechanism for cell fate specification. Immunol Rev. 2006;209:170–175. doi: 10.1111/j.0105-2896.2006.00356.x. [DOI] [PubMed] [Google Scholar]
- 44.Kreslavsky T, Garbe AI, Krueger A, von Boehmer H. T cell receptor-instructed alphabeta versus gammadelta lineage commitment revealed by single-cell analysis. J Exp Med. 2008;205:1173–1186. doi: 10.1084/jem.20072425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibata K, Yamada H, Nakamura M, Hatano S, Katsuragi Y, Kominami R, Yoshikai Y. IFN-gamma-Producing and IL-17-Producing gammadelta T Cells Differentiate at Distinct Developmental Stages in Murine Fetal Thymus. J Immunol. 2014;192:2210–2218. doi: 10.4049/jimmunol.1302145. [DOI] [PubMed] [Google Scholar]
- 46.Turchinovich G, Hayday AC. Skint-1 identifies a common molecular mechanism for the development of interferon-gamma-secreting versus interleukin-17-secreting gammadelta T cells. Immunity. 2011;35:59–68. doi: 10.1016/j.immuni.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 47.Prinz I, Silva-Santos B, Pennington DJ. Functional development of gammadelta T cells. Eur J Immunol. 2013;43:1988–1994. doi: 10.1002/eji.201343759. [DOI] [PubMed] [Google Scholar]
- 48.Vantourout P, Hayday A. Six-of-the-best: unique contributions of gammadelta T cells to immunology. Nat Rev Immunol. 2013;13:88–100. doi: 10.1038/nri3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Izon DJ, Punt JA, Xu L, Karnell FG, Allman D, Myung PS, Boerth NJ, et al. Notch1 regulates maturation of CD4+ and CD8+ thymocytes by modulating TCR signal strength. Immunity. 2001;14:253–264. doi: 10.1016/s1074-7613(01)00107-8. [DOI] [PubMed] [Google Scholar]
- 50.Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.