Abstract
Purpose
Chemokine receptor CXCR4 plays an important role in tumor aggressiveness, invasiveness, and metastasis formation. Quantification of CXCR4 expression by tumors may have an impact on prediction and evaluation of tumor response to therapies. In this study, we developed a robust and straightforward F-18 labeling route of T140, a CXCR4 peptide-based antagonist.
Procedures
T140 derivative was conjugated to 1,4,7-triazacyclononane-triacetic acid (NOTA) and labeled with Al[18F]. Al[18F]NOTA-T140 was evaluated in vitro in cell-based assay and stability in mouse serum and in vivo using CXCR4 positive and negative tumor xenograft models.
Results
Labeling of Al[18F]NOTA-T140 was completed within 30 min with a radiochemical yield of 58±5.3 % at the end of synthesis, based on fluoride-18 activity. Al[18F]NOTA-T140 accumulated in CHO-CXCR4 positive but not negative tumors. Al[18F]NOTA-T140 uptake in the tumors correlated with CXCR4 protein expression. Moreover, Al[18F]NOTA-T140 had high accumulation in CXCR4-positive metastatic tumors.
Conclusions
The simplicity of Al[18F]NOTA-T140 labeling along with its properties to specifically image CXCR4 expression by tumors warrant further clinical application for the diagnosis of CXCR4 clinically.
Keywords: T140, CXCR4, Fluoride-aluminum chelate, F-18, PET
Introduction
Tumor metastasis is the major cause of cancer lethality. More than 90 % of all cancer suffering and death are associated with metastasis [1]. Chemokine (C-X-C motif) receptor 4 (CXCR4), the prominent G protein-coupled receptor (GPCR) for chemokine stromal cell-derived factor 1 (SDF-1/CXCL12), is upregulated in more than 20 different human cancers [2–7]. The CXCL12/CXCR4 signaling pathway plays important roles in tumor migration, invasion, and proliferation [8–13].
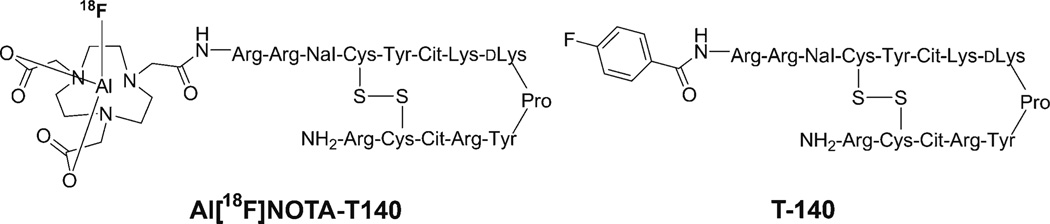
Positron emission tomography (PET) allows noninvasive visualization of cellular or molecular processes with excellent sensitivity and quantification. To date, a variety of radiolabeled CXCR4 antagonists, such as peptides [14–21] and small molecules [22–25], have been specifically developed for visualizing and quantifying CXCR4 expression noninvasively in tumors with PET. T140 is a 14-residue peptide containing a single disulfide bridge, an amidated C terminus, and a 4-fluorobenzoyl group at the N terminus (Fig. 1). T140 was found to possess high CXCR4 antagonistic activity and high binding affinity (nM range); therefore, it has been subjected to various modifications for the purposes of imaging CXCR4 [26–28].
Fig. 1.
Chemical structure of Al[18F]NOTA-T140.
We have previously labeled T140 with F-18 indirect labeling which included three steps for the synthesis of N-succinimidyl-4-[18F]fluorobenzoate prosthetic group and then an additional step for the peptide conjugation. This labeling methodology is time-consuming as it requires two HPLC purifications and is obtained in low radiochemical yield [14].
Fluorine-18 has preferable properties for clinical purposes mainly because of its suitable half-life (109.8 min) and its high positron efficiency (97 %) [29]; therefore, in this study, we report on a simple and rapid F-18 labeling of T140 derivative, Al[18F]NOTA-T140 (Fig. 1). This tracer was evaluated in vitro in cells and in vivo in subcutaneous and metastatic xenograft tumor models in mice. Moreover, the tumor uptake of Al[18F]NOTA-T140 was correlated with CXCR4 expression level.
Materials and Methods
General
2,2′-(7-(2-((2,5-Dioxopyrrolidin-1-yl)oxy)-2-oxoethyl)-1,4,7-triazonane-1,4-diyl)diacetic acid (NOTA-NHS ester) was purchased from CheMatech (Dijon, France). T140 peptide with free amino terminus and N-[1-(4,4-dimethyl-2,6-dioxocyclohex-1-ylidene) ethyl] (Dde) protected lysine residue was purchased from C.S. Bio (Menlo Park, CA). All other solvents and chemicals were purchased from Sigma-Aldrich (St. Louis, MO). C18 Sep-Pak plus (Waters Corporation, Milford, MA) was activated with 5 ml of EtOH and 10 ml of water. Na-acetate pH 4.0 buffer (0.4 M) was prepared using 18.2 Ω Milli-Q water (Millipore Corporation, Massachusetts, USA) and glacial acetic acid to adjust the pH.
Chemistry
Synthesis of NOTA-T140
T140 conjugation with NOTA-NHS ester chelator was done similarly to the published procedures [20]. Briefly, 1 eq of Dde protected peptide in dimethylformamide (200 µl) was reacted with 1.2 eq of NOTA-NHS ester and 5 eq of diisopropylethylamine at room temperature for 4–5 h. Subsequently, 2 % (v/v) of hydrazine was added in order to remove the Dde protecting groups.
The conjugated peptide was purified on an HPLC system using preparative C18 column (Higgins, 5 µm, 20×250 mm), flow rate of 12 ml/min, and a linear gradient system, starting from 95 % of solvent A [0.1 % trifluoroacetic acid (TFA) in water] and 5 % of solvent B [0.1 % TFA in CH3CN] and ending with 65 % solvent B at 35 min. The effluent was monitored by UV absorbance (210 and 280 nm). The purity of the NOTA-T140 conjugate as well as the labeled Al[18F]NOTA-T140 was determined using the same linear gradient and UV absorbance on an analytical C18 column (Phenomenex, Luna 150×4.6 mm, 3 µm) with a flow of 1 ml/ min. The purity was found to be greater than 95 % with a retention time of 15.3 min. The mass spectrum of NOTA-T140 was determined using a Waters LC–MS system (Waters, Milford, MA) to give molecular mass of 1161.98 [(M+H+)/2].
Radiolabeling of NOTA-T140 with Al[18F]
To 120 µl of CH3CN in a 1 ml plastic Eppendorf tube were added 50 µg of NOTA-T140 in 12 µl 0.4 M Na-acetate (pH 4.0) followed by 4 µl of 2 mM AlCl3 solution in 0.4 M Na-acetate (pH 4.0) and 40 µl of [18F]/H2[18O] (23–28 mCi, 851–1036 MBq). The tube was placed in a heating block at 100 °C for 15 min. Then, the reaction was cooled and diluted with 10 ml of water. Then, the crude reaction mixture was loaded onto C18 cartridge. The cartridge was further washed with 6 ml of water, and the labeled peptide was eluted using 0.5–0.7 ml of 10 mM HCl/ethanol.
Biology
Cell Culture
Wild-type Chinese hamster ovarian (CHO) cells and CXCR4-transfected CHO-CXCR4 cells were kindly given to us by Dr. David McDermott from the National Institute of Allergy and Infectious Diseases (NIAID), NIH. Both cell lines were grown in F-12K medium (American Type Culture Collection, Manassas, VA) containing 10 % fetal bovine serum and 1 % penicillin/ streptomycin at 37 °C under 5 % CO2 atmosphere.
Competition Binding Assay
CHO-CXCR4 cells were harvested and resuspended in chemokine binding buffer consisting of phosphate-buffered saline (PBS), 50 mM N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid), 1 mM CaCl2, 5 mM MgCl2, 0.5 % (w/v) bovine serum albumin, and 0.3 mM NaN3. They were placed in a 96-well membrane plate (Corning, Tewksbury, MA) to have 2×104 cells per well. 125I-CXCL12 (0.6 kBq/well, PerkinElmer, Inc.) and different concentrations of T140 and NOTA-T140, ranging from 1 pM to 1 mM were added to the wells. After 1 h of gentle shaking at room temperature, the plate was washed three times with PBS and the radioactivity that remained on the membrane was measured by γ-counter (Wallac Wizard 1480, PerkinElmer Inc.). IC50 values were calculated by GraphPad Prism software (GraphPad Software Inc., CA).
Cell Uptake, Internalization, and Efflux
Cell uptake, internalization, and efflux were done similarly to the published procedures [20]. Briefly, the uptake studies were performed in 24-well plates. Seeded were 1×105 cells per well and incubated with 11.1 kBq (0.3 µCi) of Al[18F]NOTA-T140 per well at 37 °C for 5, 15, 30, 60, and 120 min. After each time point, the cells were washed with cold PBS twice (1 ml for each wash) and cells were removed from the plates by addition of 0.5 ml of 0.1 M NaOH. For blocking studies, 10 µg/well (22 µM) of unlabeled peptide was incubated for 2 h with Al[18F]NOTA-T140.
The internalization studies were done similarly except for an additional wash with acid buffer (50 mM glycine, 0.1 M NaCl, pH 2.8, which adjusted by conc. HCl) for 1 min, which was conducted after the two PBS washes in order to remove surface-bound Al[18F]NOTA-T140. After the 1 min incubation with the acid, the cells were washed again with cold PBS and removed from the plate using NaOH. For the cell efflux studies, the cells were incubated with 11.1 kBq (0.3 µCi) per well of Al[18F]NOTA-T140 for 2 h at 37 °C. Then, the radioactive medium was removed and replaced by F-12K medium without any radioactivity and the cells incubated for different time points. After each time point, the cells were washed and harvested as described above. The cells were collected into Eppendorf tubes and were counted in a γ-counter (Wallac Wizard 1480, PerkinElmer Inc). Results are shown as % of total amount of radioactivity added to the cells at time zero.
Animal Model
Mice were purchased from Harlan Laboratories (Frederick, MD). Female athymic nude mice were housed in an animal facility under pathogen-free conditions. In vivo studies were conducted under a protocol approved by the NIH Clinical Center Animal Care and Use Committee (animal protocol NIBIB 13– 01) and in accordance with the NIH Guide for the Care and Use of Animals.
For tumor xenograft model, mice were injected subcutaneously (s.c.) at two sites: the left and right shoulders, with 107 CXCR4-positive or CXCR4-negative CHO tumor cells per site (n=5). In order to study the correlation between CXCR4 expression and tumor uptake of Al[18F]-NOTA-T140, mice were injected with 20, 40, 60, and 80 % CHO-CXCR4 cells mixed with CHO. Each of these four mixtures was injected s.c. into a single mouse at four sites on the shoulders and flanks (n=4). To set up the metastatic tumor model, mice were injected intravenously via the tail vein with 106 CHO-CXCR4 cells (n=4). All the tumors were allowed to develop for 2 weeks prior to imaging or biodistribution studies.
PET Imaging Animals were anesthetized using a mixture of isoflurane and oxygen. PET imaging studies were performed using an Inveon small animal PET scanner (Siemens Medical Solutions). Tumor-bearing mice were each injected intravenously with 100 µCi (approximately 0.2 nmol) of Al[18F]NOTA-T140. A 10-min scan was acquired at 60 and 120 min after tracer injection.
For receptor-blocking experiments, tumor mice were co-injected with 10, 50, and 100 µg of unlabeled NOTA-T140 and 100 µCi of Al[18F]NOTA-T140. Ten-minute static PET scans were then acquired at 60 and 120 min post-injection. Reconstruction of PET images was done without correction for attenuation or scatter using a three-dimensional ordered subset expectation maximization algorithm. ASI Pro VM™ software was used for image analysis. Regions of interest were drawn for each organ on the coronal images to calculate %ID/g, assuming a density of 1 for all tissues.
Ex Vivo Biodistribution
Tumor-bearing mice (n=5) were injected with Al[18F]NOTA-T140 via tail vein (100 µCi/mouse). At 2 h after injection of tracer, animals were sacrificed, then tumors and other tissues (heart, lung, spleen, stomach, intestine, pancreas, kidney, muscle, and bone) and blood were harvested and wet-weighed. The radioactivity of samples was measured using a γ-counter, and the results were expressed as percentages of the injected dose per gram of tissue (%ID/g).
Western Blotting
Tumors (n=24, four samples from each group) were excised, homogenized, and extracted with T-PER Tissue Protein Extraction Reagent (Pierce Biotechnology Inc, Rockford, IL). Protein concentration was determined according to BCA Protein Assay Kit (Pierce Biotechnology Inc.) and adjusted to equivalent values using T-PER buffer. After 4– 12 % precasted Bis-Tris, SDS-PAGE Protein Gel (Life Technology, Frederick, MD) was used for protein separation, and 0.45 µm pore size PVDF membrane (Life Technology) was used for protein transferring. Immunoblotting was carried out using a rabbit polyclonal anti-CXCR4 antibody (1:1000; Abcam, Cambridge, MA) and Donkey anti-rabbit secondary antibody (IgG) (1:3000, Jackson Immunoresearch Laboratories Inc, West Grove, PA). Detection was done using SuperSignal West Pico Chemiluminescent Substrate (Pierce Biotechnology Inc.). The image signal was collected from Amersham Imager 600 (GE Healthcare Life Science). Data were processed and analyzed by ImageJ software (NIH).
Statistical Analysis
Results were presented as mean±standard deviation (SD). Group comparisons were made using Student’s t test for unpaired data. P values<0.05 were considered statistically significant.
Results
Chemistry and Radiochemistry
Conjugation of NOTA-NHS ester to the α-amine of T140 peptide resulted in a chemical conjugation yield of 60–62 % after HPLC purification. Al[18F] complexation with NOTA was rather straightforward (Supplemental Fig. S1). The total radiosynthesis time was 30 min with an overall radiochemical yield of 58±5.3 % (not decay-corrected, n=7), calculated from the start of synthesis to the elution of the labeled peptide from the C18 cartridge. Al[18F]NOTA-T140 was obtained with a radiochemical purity 995 % as determined by an analytical HPLC with a retention time of 15.8 min and specific activity of 511±30 mCi/µmol (18.9 ±1.1 GBq/µmol).
In Vitro Assays
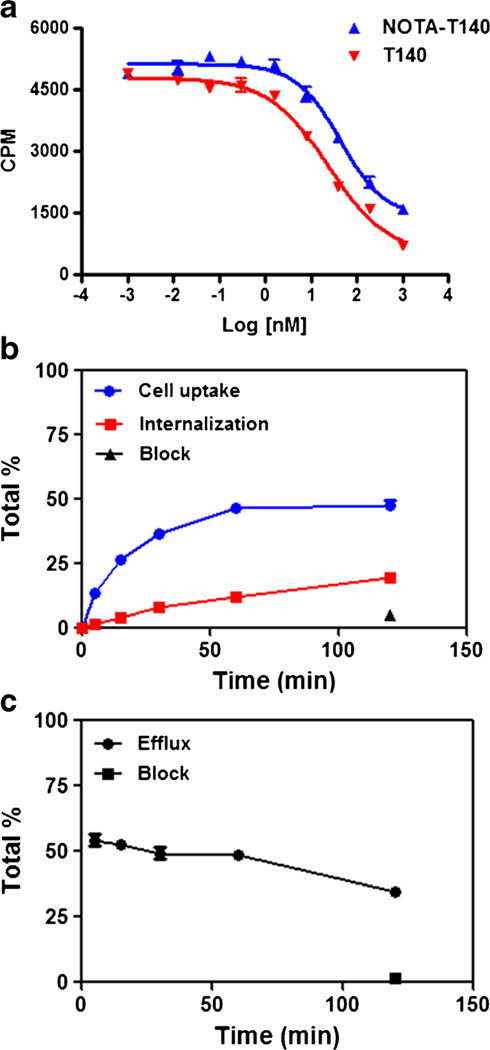
All in vitro assays were conducted in CHO-CXCR4 cells which express high levels of CXCR4 [25]. The CXCR4 expression by these cells was tested using flow cytometry (Supplemental Fig. S2). Cell binding assays using CHO-CXCR4 showed that NOTA-T140 had similar binding affinity to CXCR4 as T140 (43.32 vs. 24.25 nM, Fig. 2a).
Fig. 2.
a [125I]CXCL12 competitive cell binding assays of NOTA-T140 vs. T140 in CHO-CXCR4 cells. b Cell uptake, internalization, and c efflux studies of Al[18F]NOTA-T140 in CHO-CXCR4 cells (results are shown as average of three experiments ± standard deviations).
Cellular uptake, internalization, and efflux of Al[18F]-NOTA-T140 were further evaluated (Fig. 2b, c). Al[18F]-NOTA-T140 had rapid uptake with values of 26.4±0.57 % at 15 min (Fig. 2b). This uptake was increased at 1 h (46.4± 1.33 %) and then remained steady for 2 h (44.5±2.41 %, Fig. 2b). Almost 50 % of the uptake at 2 h was due to internalization (Fig. 2b). Blocking with unlabeled peptide significantly decreased the cell uptake to 5.11±0.35 % (Fig. 2b). The efflux of Al[18F]NOTA-T140 from the cells was slow (Fig. 2c).
Animal Studies
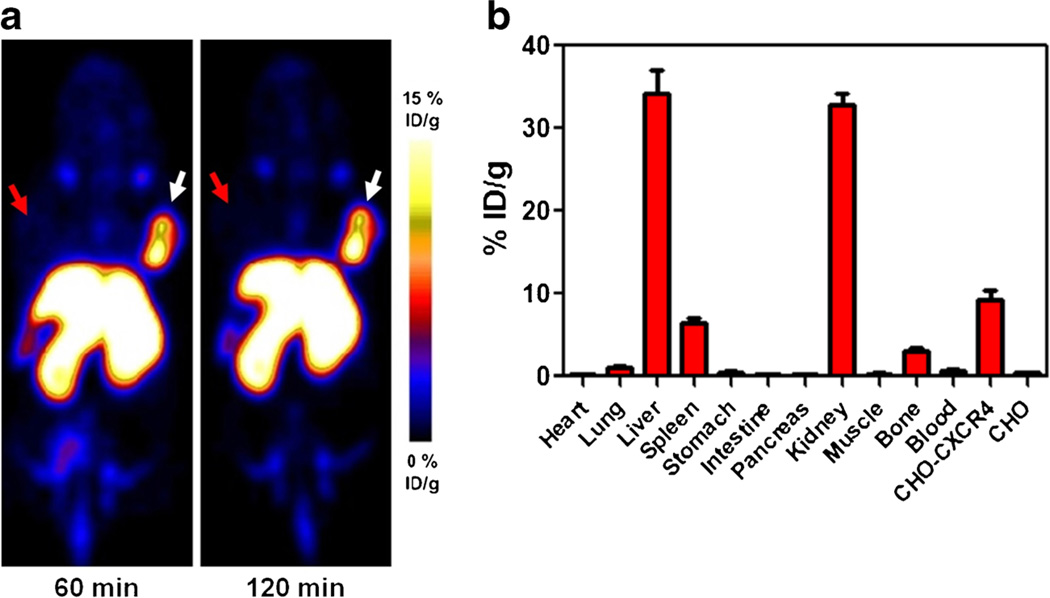
Al[18F]NOTA-T140 PET imaging clearly visualized CHO-CXCR4-positive tumors but not CHO-negative tumors (Fig. 3a). Al[18F]NOTA-T140 had high uptake in the positive tumors (8.34±0.93 %ID/g) at 1 h post-injection which remained steady up to 2 h post-injection (8.63±1.08 %ID/g, Fig. 3a). This uptake was 26-fold greater than the uptake in the negative tumors (0.33± 0.03 %ID/g at 1 h and 0.34±0.01 %ID/g at 2 h post-injection, Fig. 3a). Al[18F]NOTA-T140 also had prominent uptake in the liver and kidneys that were not related to CXCR4 expression [22]. Biodistribution studies of Al[18F]NOTA-T140 at 2 h post-injection showed accumulation in CXCR4-expressing organs as the CHO-CXCR4 tumors (9.20±2.08 %ID/g) and the spleen (6.37±1.18 %ID/g) and the bone (2.95±0.74 %ID/g, Fig. 3b), which relates to CXCR4 expression in the bone marrow. Al[18F]NOTA-T140 uptake in the blood and muscle was very low, which resulted in high image contrast (Fig. 3a, b). Nevertheless, its uptake in the metabolic organs (liver and kidneys) was still high at 2 h post-injection (34.24±4.98 %ID/g and 32.82±2.62 %ID/g, respectively).
Fig. 3.
a Representative PET images of a mouse bearing CHO-CXCR4-positive tumor (right) and CHO-negative tumor (left) at 1 and 2 h post-injection of Al[18F]NOTA-T140. White arrow indicates CHO-CXCR4 tumor, and red arrow indicates CHO tumor locations. b Biodistribution of Al[18F]NOTA-T140 at 2 h post-injection. Results are an average of five mice ± standard deviations.
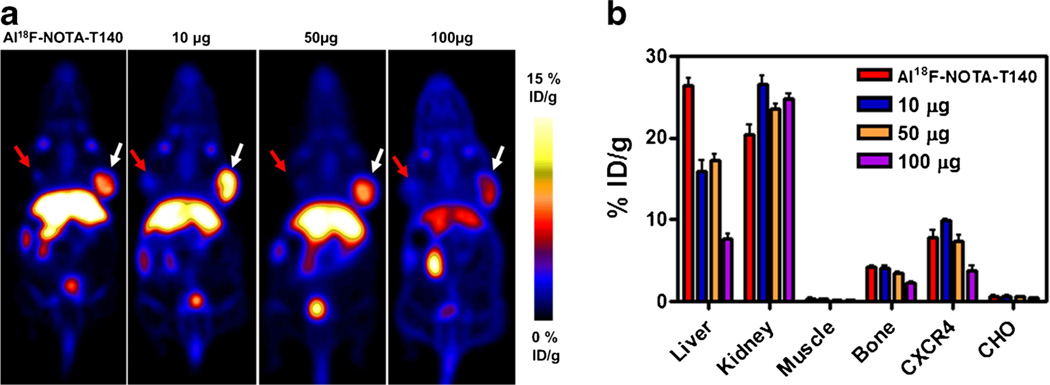
In order to test the specificity of Al[18F]NOTA-T140 uptake, several blocking doses were co-administered to the mice (Fig. 4a, b). Co-injection of the labeled peptide with 10 and 50 µg of unlabeled peptide resulted in almost no blocking in the positive tumor. Upon co-administration of 100 µg of unlabeled peptide, a reduction of 50–60 % in CHO-CXCR4 tumor uptake (3.79±1.05 %ID/g) was observed. This co-administration of 100 µg also increased the uptake in the kidneys and decreased the uptake in the liver (Fig. 4b).
Fig. 4.
a Representative PET images and b Al[18F]NOTA-T140 uptake calculation of mice bearing CHO-CXCR4 (white arrow) and CHO (red arrow) tumors after co-injection of Al[18F]NOTA-T140 with different doses of unlabeled peptide at 1 h post-injection. Results are shown as the average of five mice ± standard deviations.
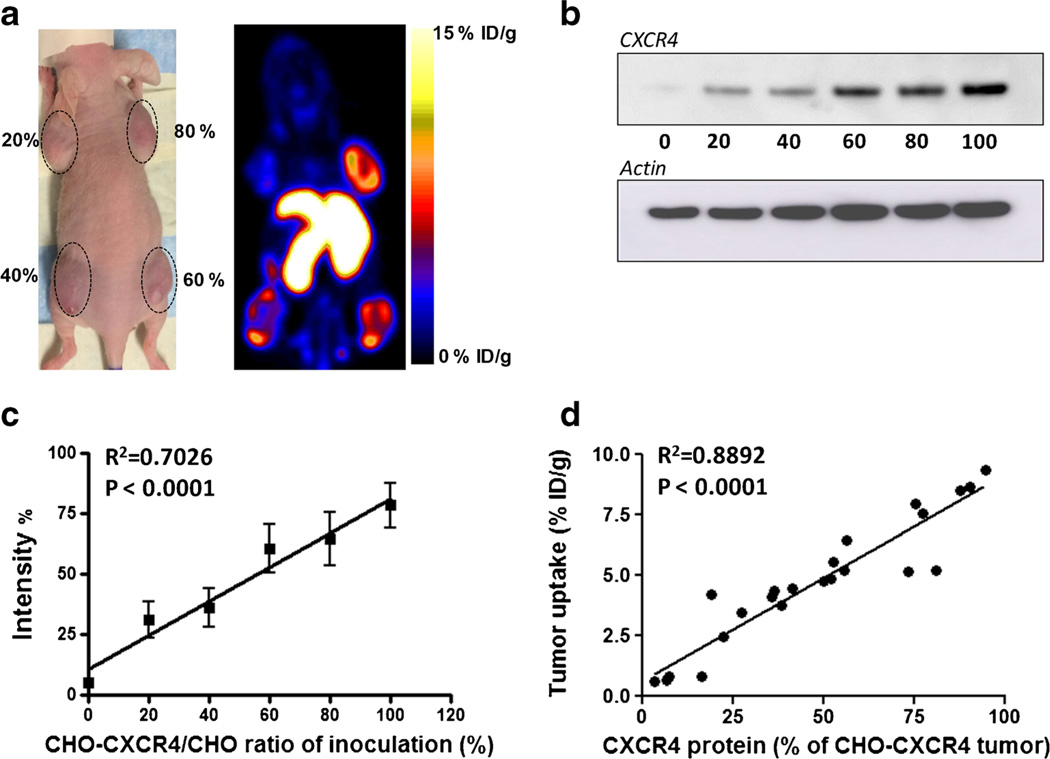
We also tested the correlation between Al[18F]NOTA-T140 uptake in CHO-CXCR4 tumors and CXCR4 protein expression (Fig. 5). Each mouse was injected with CHO-CXCR4 cells which were premixed with negative CHO cells to have different levels of CXCR4 expression (20, 40, 60, and 80 %). PET images showed increasing tracer uptake in the tumor corresponding to the percentage of receptor-positive cells (Fig. 5a). Western blot analysis confirmed an excellent correlation between Al[18F]NOTA-T140 uptake and CXCR4 concentration (Fig. 5b – d).
Fig. 5.
a Representative photograph (left) and PET image (right) of a mouse bearing tumors with different amounts of CHO-CXCR4 cells (20, 40, 60, and 80 %) at 1 h post-injection of Al[18F]NOTA-T140. b Western blot of CXCR4 expression by tumors generated using increasing proportion of CHO-CXCR4 cells. c Quantification of Western blots shown in B from six groups. Tumors with only CHO-CXCR4 cells were defined as 100 %. d Correlation of CXCR4 receptor levels (determined by Western blot) and Al[18F]NOTA-T140 uptake (determined by PET scan) in the tumor.
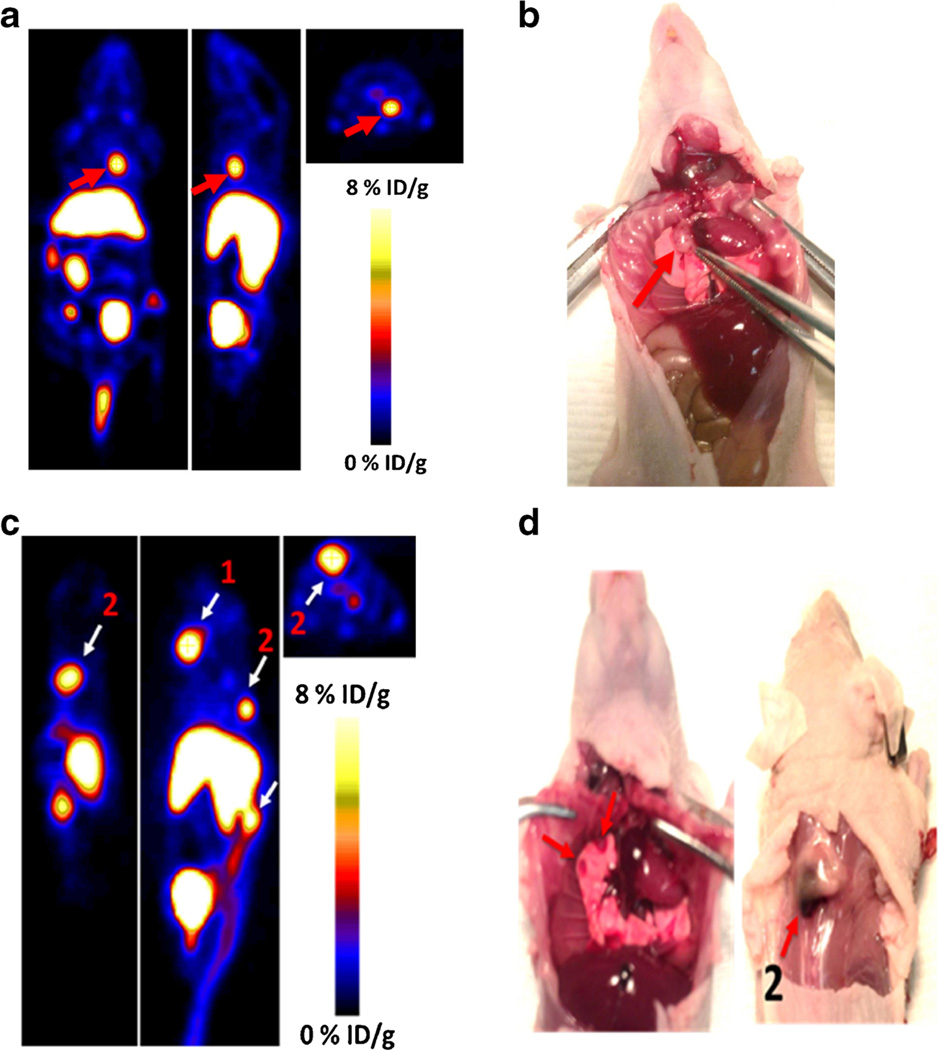
We also evaluated the ability of Al[18F]NOTA-T140 to accumulate and distinguish CXCR4 tumors which are not subcutaneous (Fig. 6). At 2 weeks after intravenous injection of CHO-CXCR4 cells, several foci in the neck, chest, and spine were detected by injection of Al[18F]-NOTA-T140. These tumors had moderate to high uptake of the peptide (Fig. 6).
Fig. 6.
a Coronal (left), sagittal (middle), and axial (left) PET images of metastatic model of CHO-CXCR4 tumors at 1 h post-injection of Al[18F]NOTA-T140 with a %ID/g of 9.33, calculated from the PET images. Red arrow represents lesion location. b Photograph of a, tumor location shown with red arrow. c Mouse bearing two CXCR4-positive metastatic tumors on the spine, with %ID/g of 12.2 (for 1) and 9.8 (for 2), at 1 h post-injection of Al[18F]NOTA-T140. d Ventral (left) and dorsal (right) photograph of c. Tumor locations are shown with white arrows.
Discussion
CXCR4 plays a pivotal role in tumor proliferation and metastasis [30]. Widely expressed in various tumors, CXCR4 has been studied as a prognostic marker and target for therapy. Detection of CXCR4 expression by noninvasive methods, such as molecular imaging, could be a valuable tool for evaluation of disease.
T140 peptide has very high binding affinity to CXCR4 and favorable pharmacokinetics. We have previously labeled T140 without changing its chemical structure. Interestingly, the peptide showed red blood cell (RBC) binding that was not CXCR4-specific, and we overcame that limitation by injection of the tracer with low specific activity (SA). Imaging studies with the low SA [18F]T140 were promising, clearly visualizing CXCR4-positive tumors and displaying low accumulation in metabolic organs (G3–4 %ID/g in the kidneys and liver); however, the labeling was laborious and had very low radiochemical yield [14, 18].
By replacing the 4-fluorobenzoyl group at the N terminus with NOTA or DOTA chelators, we were able to eliminate the binding to RBCs, yet still maintain the specificity toward CXCR4 [20]. These two new T140 derivatives were easily labeled with PET metal isotopes [64Cu] and [68Ga]. However, they possessed prominent uptakes in the liver and kidneys (about 30 %ID/g) that were not CXCR4 dependent [20].
In 2009, McBride et al. reported on a robust and straightforward [18F]-labeling route by chelation of Al[18F] with NOTA [31]. Since then, many publications reported the usage of this methodology in labeling peptides, small molecules, and proteins [32–35]. Moreover, this methodology has been applied in the clinic in lung cancer patients, by labeling RGD-based peptide, Alfatide [35]. Taking into consideration the significance and importance of CXCR4 in cancer diagnosis, we decided to apply this Al[18F] methodology on CXCR4 peptide antagonist, T140, which indeed resulted in reasonable radiochemical yield with short reaction time and high specific activity.
In this study, we have chosen to use CHO-CXCR4 as our positive cell line. These engineered cells express high level of CXCR4 (6.8×105 per cell), which is comparable to that on Jurkat cells (105 per cell) [25, 36]. In tumor xenografts, Al[18F]NOTA-T140 had rapid clearance from the blood and high-specific accumulation in CHO-CXCR4 tumors but not CXCR4-negative CHO tumors (Fig. 3).
To evaluate the ability of the tracer to discern a range of CXCR4 receptor concentrations, we made tumor xenografts with different ratios of CXCR4-negative and CXCR4-positive cells (Fig. 5). Consequently, tumors that had about 20 % CXCR4-expressing cells, shown to express ~1.5×105 CXCR4 molecules per cell by Western blotting, had uptake of 2–3 %ID/g. Moreover, a significant correlation was found between tumor uptake and CXCR4 protein level (Fig. 5d), suggesting that the tracer would allow detection and quantification of CXCR4 in cancer lesions that have low expression of the receptor, as well as in cancer lesions where only a small amount of cells express CXCR4.
Moreover, Al[18F]NOTA-T140 also accumulated in metastatic tumors that were CXCR4-positve (Fig. 6). This result is important because the metastatic model is more physiologically relevant than the subcutaneous tumor model, simulating metastatic tumor cell migration and seeding in various organs. The ability to image such lesions will allow evaluating the role of CXCR4 in metastasis to specific organs that express CXCR4 and its ligand CXCL12 such as the bone marrow. Unfortunately, Al[18F]NOTA-T140 also had high and undesired uptake in the liver and kidneys, which is probably not due to CXCR4 expression, because the receptor expression in the liver is limited to cells of the hematopoietic lineage and few other cells such as stellate cells [22]. As mentioned above, T140 was previously labeled by us similarly to the method described in this paper using [64Cu] (on the lysine groups and the N terminus) and on the fluorobenzoyl group using F-18 [14]. Comparison of the uptake by the liver using different T140 derivatives suggests that the uptake is probably due to the presence of metal chelator, which may undergo transchelation under physiological condition.
Al[18F]NOTA-T140 showed significant uptake in the bone (Fig. 3b), which is due to CXCR4 expression by multiple cell lineages of the hematopoietic system [37]. To eliminate the possibility that some of the accumulation in the bone is due to defluorination, we performed stability studies (Supplemental Fig. S3). No significant defluorination was observed with up to 2-h incubation of Al[18F]NOTA-T140 in mouse serum. Moreover, we carried out blocking experiment (Fig. 4b). Co-injection of unlabeled NOTA-T140 with the labeled tracer significantly reduced tracer uptake in the bone.
A number of other CXCR4 PET tracers were reported previously, including [64Cu]AMD3100 and its derivatives and [68Ga]CPCR4.2 [17, 22, 25, 38, 39]. Similar to AMD3100, its derivatives labeled with either [64Cu] or [11C] displayed very high accumulation in the liver that is probably CXCR4-independent. [64Cu]AMD3100 was first described on 2009 [22], and it is currently being evaluated in human cancer patients [40]. Under clinical settings, Al[18F]-NOTA-T140 will have an important advantage over [64Cu]AMD3100 and its [64Cu]-labeled derivatives because it has less accumulation in the liver and should have a more favorable dosimetry because of the usage of F-18 rather than [64Cu].
Other than [18F]T140, the only CXCR4 tracer that was reported in the literature and does not have high uptake in the liver is [68Ga]CPCR4.2 [17, 41]. [68Ga]CPCR4.2 showed promising results in mice with high tumor uptake and almost no accumulation in the liver. Interestingly, despite the high conservation between human CXCR4 and mouse CXCR4, [68Ga]CPCR4.2 does not bind to mouse CXCR4. This is not a limitation for future clinical evaluation; however, it limits the evaluation of the tracer in mice and therefore also limits our ability to compare [68Ga]CPCR4.2 with Al[18F]NOTA-T140. Importantly, Al[18F]NOTA-T140 is based on a drug that was previously evaluated in cancer patients as a stem cell/leukemic cell-mobilizing agent, with negligible side effect [42]. [68Ga]CPCR4.2 on the other hand was never evaluated as a drug targeting CXCR4, and it is unknown whether it has any functional activity as an agonist or antagonist of the receptor.
In conclusion, Al[18F]NOTA-T140 was shown to specifically accumulate in CXCR4-expressing tumors and provide high imaging contrast. Along with the ease of the radiosynthesis, high radiochemical yield, and specific activity, it may be suitable for clinical translation. Nevertheless, its high uptake in the liver and kidneys may hinder its usefulness to distinguish tumors in these organs.
Supplementary Material
Acknowledgments
This work was supported, in part, by the National Basic Research Program of China (2015CB931800 and 2015CB931803), National Natural Science Foundation of China (81130028 and 31210103913), the Key Grant Project of Heilongjiang Province (GA12C302), the Ph.D. Programs Foundation of Ministry of Education of China (201123071100203), the Key Laboratory of Molecular Imaging Foundation (College of Heilongjiang Province), and the Intramural Research Program (IRP) of the National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11307-015-0872-2) contains supplementary material, which is available to authorized users.
Conflict of Interest. The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material. ESM 1(PDF 676 kb)
References
- 1.Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer. 2006;6:449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- 2.Muller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 3.Taichman RS, Cooper C, Keller ET, et al. Use of the stromal cell-derived factor-1/CXCR4 pathway in prostate cancer metastasis to bone. Cancer Res. 2002;62:1832–1837. [PubMed] [Google Scholar]
- 4.Vicari AP, Caux C. Chemokines in cancer. Cytokine Growth Factor Rev. 2002;13:143–154. doi: 10.1016/s1359-6101(01)00033-8. [DOI] [PubMed] [Google Scholar]
- 5.Rubin JB, Kung AL, Klein RS, et al. A small-molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors. Proc Natl Acad Sci U S A. 2003;100:13513–13518. doi: 10.1073/pnas.2235846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka T, Bai Z, Srinoulprasert Y, et al. Chemokines in tumor progression and metastasis. Cancer Sci. 2005;96:317–322. doi: 10.1111/j.1349-7006.2005.00059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Redjal N, Chan JA, Segal RA, Kung AL. CXCR4 inhibition synergizes with cytotoxic chemotherapy in gliomas. Clin Cancer Res. 2006;12:6765–6771. doi: 10.1158/1078-0432.CCR-06-1372. [DOI] [PubMed] [Google Scholar]
- 8.Murphy PM. Chemokines and the molecular basis of cancer metastasis. N Engl J Med. 2001;345:833–835. doi: 10.1056/NEJM200109133451113. [DOI] [PubMed] [Google Scholar]
- 9.Barbero S, Bonavia R, Bajetto A, et al. Stromal cell-derived factor 1alpha stimulates human glioblastoma cell growth through the activation of both extracellular signal-regulated kinases 1/2 and Akt. Cancer Res. 2003;63:1969–1974. [PubMed] [Google Scholar]
- 10.Tamamura H, Hori A, Kanzaki N, et al. T140 analogs as CXCR4 antagonists identified as anti-metastatic agents in the treatment of breast cancer. FEBS Lett. 2003;550:79–83. doi: 10.1016/s0014-5793(03)00824-x. [DOI] [PubMed] [Google Scholar]
- 11.Schols D. HIV co-receptor inhibitors as novel class of anti-HIV drugs. Antivir Res. 2006;71:216–226. doi: 10.1016/j.antiviral.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Burger JA, Burkle A. The CXCR4 chemokine receptor in acute and chronic leukaemia: a marrow homing receptor and potential therapeutic target. Br J Haematol. 2007;137:288–296. doi: 10.1111/j.1365-2141.2007.06590.x. [DOI] [PubMed] [Google Scholar]
- 13.Jacobson O, Weiss ID. CXCR4 chemokine receptor overview: biology, pathology and applications in imaging and therapy. Theranostics. 2013;3:1–2. doi: 10.7150/thno.5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobson O, Weiss ID, Kiesewetter DO, et al. PET of tumor CXCR4 expression with 4-18F–T140. J Nucl Med. 2010;51:1796–1804. doi: 10.2967/jnumed.110.079418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demmer O, Dijkgraaf I, Schumacher U, et al. Design, synthesis, and functionalization of dimeric peptides targeting chemokine receptor CXCR4. J Med Chem. 2011;54:7648–7662. doi: 10.1021/jm2009716. [DOI] [PubMed] [Google Scholar]
- 16.Demmer O, Gourni E, Schumacher U, et al. PET imaging of CXCR4 receptors in cancer by a new optimized ligand. ChemMed-Chem. 2011;6:1789–1791. doi: 10.1002/cmdc.201100320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gourni E, Demmer O, Schottelius M, et al. PET of CXCR4 expression by a 68Ga-labeled highly specific targeted contrast agent. J Nucl Med. 2011;52:1803–1810. doi: 10.2967/jnumed.111.098798. [DOI] [PubMed] [Google Scholar]
- 18.Jacobson O, Weiss ID, Szajek LP, et al. PET imaging of CXCR4 using copper-64 labeled peptide antagonist. Theranostics. 2011;1:251–262. doi: 10.7150/thno/v01p0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hennrich U, Seyler L, Schafer M, et al. Synthesis and in vitro evaluation of 68Ga-DOTA-4-FBn-TN14003, a novel tracer for the imaging of CXCR4 expression. Bioorg Med Chem. 2012;20:1502–1510. doi: 10.1016/j.bmc.2011.12.052. [DOI] [PubMed] [Google Scholar]
- 20.Jacobson O, Weiss ID, Szajek LP, et al. Improvement of CXCR4 tracer specificity for PET imaging. J Control Release. 2012;157:216–223. doi: 10.1016/j.jconrel.2011.09.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang XX, Sun Z, Guo J, et al. Comparison of 18F-labeled CXCR4 antagonist peptides for PET imaging of CXCR4 expression. Mol Imaging Biol. 2013;15:758–767. doi: 10.1007/s11307-013-0640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobson O, Weiss ID, Szajek L, et al. 64Cu-AMD3100—a novel imaging agent for targeting chemokine receptor CXCR4. Bioorg Med Chem. 2009;17:1486–1493. doi: 10.1016/j.bmc.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nimmagadda S, Pullambhatla M, Stone K, et al. Molecular imaging of CXCR4 receptor expression in human cancer xenografts with [64Cu]AMD3100 positron emission tomography. Cancer Res. 2010;70:3935–3944. doi: 10.1158/0008-5472.CAN-09-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Silva RA, Peyre K, Pullambhatla M, et al. Imaging CXCR4 expression in human cancer xenografts: evaluation of monocyclam 64Cu-AMD3465. J Nucl Med. 2011;52:986–993. doi: 10.2967/jnumed.110.085613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss ID, Jacobson O, Kiesewetter DO, et al. Positron emission tomography imaging of tumors expressing the human chemokine receptor CXCR4 in mice with the use of 64Cu-AMD3100. Mol Imaging Biol. 2012;14:106–114. doi: 10.1007/s11307-010-0466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuil J, Buckle T, van Leeuwen FW. Imaging agents for the chemokine receptor 4 (CXCR4) Chem Soc Rev. 2012;41:5239–5261. doi: 10.1039/c2cs35085h. [DOI] [PubMed] [Google Scholar]
- 27.Weiss ID, Jacobson O. Molecular imaging of chemokine receptor CXCR4. Theranostics. 2013;3:76–84. doi: 10.7150/thno.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nayak TR, Hong H, Zhang Y, Cai W. Multimodality imaging of CXCR4 in cancer: current status towards clinical translation. Curr Mol Med. 2013;13:1538–1548. doi: 10.2174/1566524013666131111121325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobson O, Chen X. PET designated flouride-18 production and chemistry. Curr Top Med Chem. 2010;10:1048–1059. doi: 10.2174/156802610791384298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demmer O, Frank AO, Hagn F, et al. A conformationally frozen peptoid boosts CXCR4 affinity and anti-HIV activity. Angew Chem Int Ed Engl. 2012;51:8110–8113. doi: 10.1002/anie.201202090. [DOI] [PubMed] [Google Scholar]
- 31.McBride WJ, Sharkey RM, Karacay H, et al. A novel method of 18F radiolabeling for PET. J Nucl Med. 2009;50:991–998. doi: 10.2967/jnumed.108.060418. [DOI] [PubMed] [Google Scholar]
- 32.Lipowska M, Klenc J, Shetty D, et al. Al18F-NODA-butyric acid: biological evaluation of a new PET renal radiotracer. Nucl Med Biol. 2014;41:248–253. doi: 10.1016/j.nucmedbio.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lang L, Li W, Guo N, et al. Comparison study of [18F]FAl-NOTA-PRGD2, [18F]FPPRGD2, and [68Ga]Ga-NOTA-PRGD2 for PET imaging of U87MG tumors in mice. Bioconjug Chem. 2011;22:2415–2422. doi: 10.1021/bc200197h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laverman P, D’Souza CA, Eek A, et al. Optimized labeling of NOTA-conjugated octreotide with F-18. Tumour Biol. 2012;33:427–434. doi: 10.1007/s13277-011-0250-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan W, Guo N, Pan D, et al. First experience of 18F-alfatide in lung cancer patients using a new lyophilized kit for rapid radio-fluorination. J Nucl Med. 2013;54:691–698. doi: 10.2967/jnumed.112.113563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee B, Sharron M, Montaner LJ, et al. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci USA. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aiuti A, Tavian M, Cipponi A, et al. Expression of CXCR4, the receptor for stromal cell-derived factor-1 on fetal and adult human lympho-hematopoietic progenitors. Eur J Immunol. 1999;29:1823–1831. doi: 10.1002/(SICI)1521-4141(199906)29:06<1823::AID-IMMU1823>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 38.Woodard LE, De Silva RA, Behnam Azad B, et al. Bridged cyclams as imaging agents for chemokine receptor 4 (CXCR4) Nucl Med Biol. 2014;41:552–561. doi: 10.1016/j.nucmedbio.2014.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartimath SV, van Waarde A, Dierckx RA, de Vries EF. Evaluation of N-[11C]Methyl-AMD3465 as a PET tracer for imaging of CXCR4 receptor expression in a C6 glioma tumor model. Mol Pharm. 2014;11:3810–3817. doi: 10.1021/mp500398r. [DOI] [PubMed] [Google Scholar]
- 40.Clinical, trials. Imaging CXCR4 expression in subjects with cancer using 64Cu-Plerixafor. [Updated December 3, 2014. Accessed February 20, 2014];Clinical trials. gov website. 2014 http://clinicaltrials.gov/ct2/show/NCT02069080.
- 41.Wester HJ, Keller U, Schottelius M, et al. Disclosing the CXCR4 expression in lymphoproliferative diseases by targeted molecular imaging. Theranostics. 2015;5:618–630. doi: 10.7150/thno.11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peled A, Abraham M, Avivi I, et al. The high-affinity CXCR4 antagonist BKT140 is safe and induces a robust mobilization of human CD34+ cells in patients with multiple myeloma. Clin Cancer Res. 2014;20:469–479. doi: 10.1158/1078-0432.CCR-13-1302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.