Abstract
Bone morphogenetic proteins (BMPs) function in most tissues but have cell type-specific effects. Given the relatively small number of BMP receptors, this exquisite signaling specificity requires additional molecules to regulate this pathway’s output. The receptor tyrosine kinase MuSK (muscle-specific kinase) is critical for neuromuscular junction formation and maintenance. Here, we show that MuSK also promotes BMP signaling in muscle cells. MuSK bound to BMP4 and related BMPs with low nanomolar affinity in vitro and to the type I BMP receptors ALK3 and ALK6 in a ligand-independent manner both in vitro and in cultured myotubes. High-affinity binding to BMPs required the third, alternatively spliced MuSK immunoglobulin-like domain. In myoblasts, endogenous MuSK promoted BMP4-dependent phosphorylation of SMADs and transcription of Id1, which encodes a transcription factor involved in muscle differentiation. Gene expression profiling showed that MuSK was required for the BMP4-induced expression of a subset of genes in myoblasts, including regulator of G protein signaling 4 (Rgs4). In myotubes, MuSK enhanced the BMP4-induced expression of a distinct set of genes, including transcripts characteristic of slow muscle. MuSK-mediated stimulation of BMP signaling required type I BMP receptor activity but was independent of MuSK tyrosine kinase activity. MuSK-dependent expression of Rgs4 resulted in the inhibition of Ca2+ signaling induced by the muscarinic acetylcholine receptor in myoblasts. These findings establish that MuSK has dual roles in muscle cells, acting both as a tyrosine kinase-dependent synaptic organizing molecule and as a BMP co-receptor that shapes BMP transcriptional output and cholinergic signaling.
INTRODUCTION
Bone morphogenetic proteins (BMPs) function in virtually all tissues in developing and mature organisms, and cell type-specific output of BMP signaling is essential for proper tissue function and differentiation. However, the large BMP family is served by only a handful of BMP receptors. Thus, signaling specificity in distinct cellular and developmental contexts requires additional molecules to modulate pathway output. Here, we identify a muscle-specific kinase (MuSK) as a co-receptor that potentiates BMP signaling in myogenic cells.
BMPs are a large subfamily of conserved signaling molecules within the transforming growth factor-β (TGFβ) superfamily Two different classes of receptors, type I (ALK2, ALK3, or ALK6) and type II (BMPRII, ActRIIB, and ActRII), bind BMPs on the cell surface (1, 2). After ligand binding, the type I receptor is phosphorylated by the constitutively active type II receptor (2). The activated type I receptor phosphorylates small mothers against decapentaplegic 1 (SMAD1), SMAD5, or SMAD8, which then associates with SMAD4 (3). This complex translocates to the nucleus where it can function as part of the transcriptional activator or repressor complexes in a cell type-specific fashion (4, 5). For example, BMP4 induces osteoblast differentiation but inhibits myoblast differentiation (6, 7).
The canonical BMP signaling pathway is modulated at several levels. The best understood mechanism is the control of ligand availability by either secreted or cell surface–associated BMP-binding molecules. For example, the secreted proteins noggin, chordin, and members of the DAN (differential screening-selected gene in neuroblastoma) family sequester BMPs and prevent their association with signaling receptors (8). The glycophosphatidylinositol-anchored RGM/Dragon family co-receptors bind BMPs and positively regulate signaling, whereas the transmembrane protein BAMBI (BMP and activin membrane-bound inhibitor) acts as a type I pseudoreceptor to inhibit BMP signaling (9). BMP pathway modulators can also act in SMAD-dependent and SMAD-independent manners (10, 11). Finally, receptor tyrosine kinases (RTKs) such as c-KIT and ROR-2 can modulate signaling by binding TGFβ family ligands and receptors (12).
MuSK is an RTK that is highly abundant at the postsynaptic membrane of the neuromuscular junction (NMJ) and is well established as the master regulator of the formation and maintenance of this synapse (13–15). Proper signaling in this context requires neuronal agrin, low-density lipoprotein receptor-related protein 4 (LRP4), docking protein 7 (DOK7), and MuSK tyrosine kinase activity (13). MuSK is also present outside the NMJ, notably in intact slow-twitch muscle, denervated fast-twitch muscle, and brain (16–18). However, MuSK function in these nonsynaptic contexts is poorly understood.
Here, we report that MuSK binds BMPs and influences the BMP4-mediated gene expression signature in muscle cells. MuSK promoted the BMP4-induced phosphorylation of SMAD1/5/8 and expression of Id1, which encodes a transcriptional coactivator. In myoblasts, MuSK was required for the BMP4-induced expression of a subset of genes, including the guanosine triphosphatase (GTP)–activating protein (GAP) Rgs4. This MuSK-dependent, BMP4-responsive GAP regulated muscarinic acetylcholine receptor (mAChR)–mediated Ca2+ signaling in these cells. In myotubes, MuSK stimulated the BMP4-induced expression of a large set of genes, including the slow muscle-enriched genes myosin heavy chain 15 (Myh15) and carbonic anhydrase 3 (Car3). MuSK bound to the type I BMP receptors ALK3 and ALK6 and the type I activin receptor ALK4 in a ligand-independent manner. Type I BMP receptor activity was necessary for the regulation of MuSK-dependent transcripts, but MuSK tyrosine kinase activity was dispensable. We propose that MuSK acts as a BMP co-receptor to confer cell type–specific signaling in muscle. These findings also establish that MuSK has dual roles in muscle cells, acting as both a tyrosine kinase-dependent synaptic organizing molecule and a BMP co-receptor that shapes transcriptional output and Ca2+ signaling in myogenic cells.
RESULTS
MuSK binds to BMPs
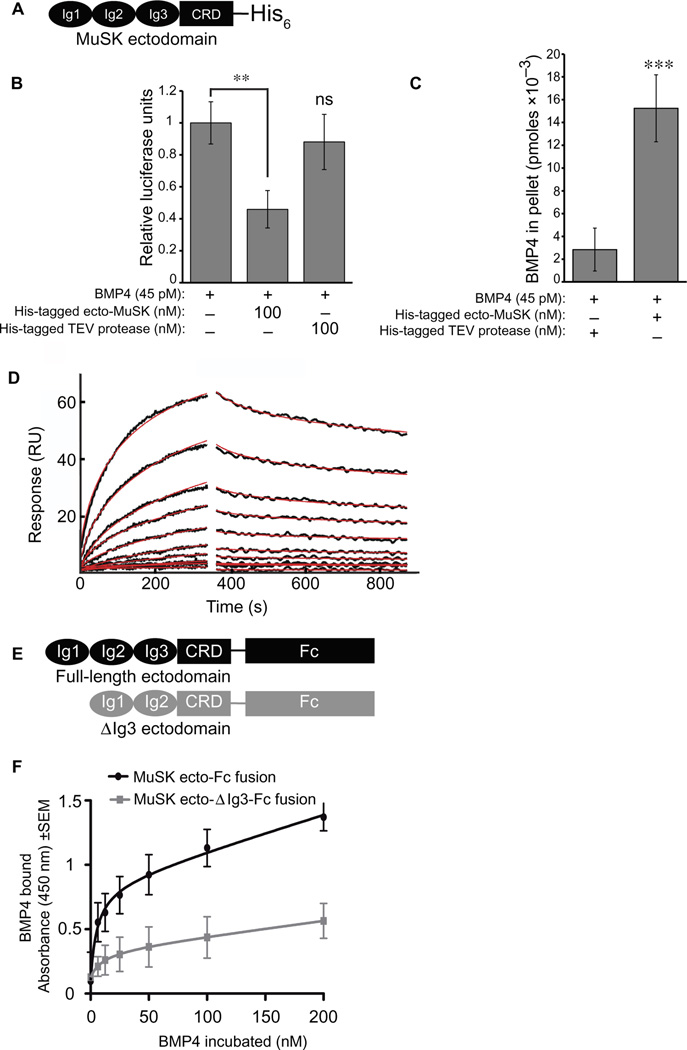
Because both BMP4 and MuSK are important for muscle differentiation (19), we tested whether MuSK and BMP interact in vitro. We preincubated purified BMP4 with the His-tagged MuSK ectodomain (Fig. 1A) or a control protein (His-tagged tobacco etch virus protease). After depletion of the tagged MuSK or control protein with nickel beads, we assessed BMP4 activity in the supernatants using a cell line harboring a luciferase reporter gene under the control of the BMP-responsive elements of the Id1 promoter [C2C12BRA (20)]. Luciferase activity in the supernatants was reduced about 50%, indicating that BMP4 had coprecipitated with MuSK on the nickel beads (Fig. 1B). We observed no significant inhibition of BMP4 activity with the control protein. Enzyme-linked immunosorbent assay (ELISA) analysis of the bead-containing pellets confirmed the specific pull-down of BMP4 by MuSK (Fig. 1C). Thus, BMP4 and MuSK bind in solution.
Fig. 1. MuSK ectodomain binds to BMP4.
(A) Schematic representation of the MuSK ectodomain fusion protein used in the reporter and the solution-binding assays. The fusion protein contains the three immunoglobulin (Ig)-like domains (Ig1, Ig2, and Ig3) and the cysteine-rich domain (CRD) of the extracellular portion of MuSK fused to a His-tag at the C terminus. (B) BMP4 depletion. Soluble BMP4 was co-incubated with His-tagged MuSK ectodomain or a control protein [His-tagged tobacco etch virus (TEV) protease] followed by precipitation with nickel beads. Residual soluble BMP4 activity was measured using the C2C12BRA reporter cell line. The average value of untreated cells was set as 100% activity. Data are means ± SD from three independent experiments with eight replicates in each [n = 3; **p < 0.01; ns, not significant versus BMP4-only treatment, Bonferroni-adjusted one-way analysis of variance (ANOVA)]. (C) BMP4 coprecipitates with the MuSK ectodomain. The amount of BMP4 in eluates from the bead pellets was analyzed by ELISA. Data are representative of three independent experiments. Values indicate the average from eight replicates and are means ± SD (***P < 0.0001, unpaired, two-sided Student’s t test). RU, response units. (D) SPR binding analysis of MuSK with BMP4. Representative SPR profiles are shown for various concentrations of MuSK binding to BMP4. Sensorgrams were normalized for MuSK binding to a mock-coupled flow cell. The black lines show the experimental measurements of a twofold serial dilution over the concentration range 2 mM to 1.96 nM of each sen-sorgram, and the red lines correspond to global fits of the data to a 1:1 model using a heterogeneous surface model with the program EVILFIT. (E) Schematics of Fc-fusion of full length (FL) and Ig3-lacking (Δlg3) MuSK ectodomain proteins used in (F). (F) The MuSK Ig3 domain is required for BMP4 binding. Immobilized FL or Δlg3 MuSK ectodomain Fc-fusions were incubated with BMP4 (0 to 200 nM). Data are means ± SD from three independent experiments with four technical replicates in each (n = 3).
To determine the kinetics and binding affinity of the MuSK-BMP4 interaction, we used surface plasmon resonance (SPR). BMP4 was immobilized as the ligand, and the MuSK ectodomain was used as the analyte. A kinetic analysis using a heterogeneous ligand model revealed high-affinity binding between these molecules, with a dissociation constant (Kd) of 6.1 nM, similar in magnitude to the high-affinity interaction of BMP2 with its type I receptors (Fig. 1D and table S1) (21). We also used SPR to determine whether the MuSK ectodomain binds to the closely related family members BMP2 and BMP7 (22, 23). These proteins bound MuSK with comparable affinity to BMP4 (5.6 and 11.8 nM for BMP2 and BMP7, respectively) (fig. S1 and table S1). Thus, the MuSK ectodomain binds with high affinity to a closely related set of the BMP family members that includes BMP2, BMP4, and BMP7.
As a further test of the interaction between MuSK and BMP4, we treated C2C12BRA cells either with BMP4 alone or with purified recombinant MuSK ectodomain. Coincubation with 50 or 100 nM MuSK ectodomain inhibited BMP4-induced luciferase activity by 50% (fig. S2). In contrast, no inhibition was observed with the His-tagged control protein (200 nM; fig. S2). Thus, the MuSK ectodomain inhibits the BMP4-induced activation of the Id1 reporter in this cell line, possibly by sequestering BMP4.
The Ig3 domain of MuSK is required for high-affinity BMP4 binding
We next determined which regions of the MuSK extracellular domain were required for binding to BMP4. One candidate domain was suggested by MuSK alternative splicing. A major splice isoform of MuSK lacks the Ig3 domain in the extracellular portion of the molecule. The biological importance of this ΔIg3 form of MuSK has been unclear because the naturally occurring splice form with an intact cytoplasmic domain is sufficient for forming postsynaptic specializations in muscle cells in culture and in vivo (24). To test the potential role of this domain in MuSK-BMP4 binding, we generated constructs encoding Fc-fusion ectodomain proteins either containing or lacking the Ig3 domain (FL and ΔIg3, respectively; Fig. 1E) and tested their binding to soluble BMP4 over a range of concentrations. BMP4 displayed saturable, high-affinity binding to FL-ecto-MuSK with a half maximum of ~30 nM (Fig. 1F). In contrast, only low, nonsaturable BMP4 binding was observed with ΔIg3-ecto-MuSK.
We considered the possibility that the failure to observe specific binding of BMP4 to Fc-ΔIg3-ecto-MuSK might be due to artifactual misfolding of the ectodomain. To control for such potential misfolding, we assessed the binding of both proteins to biglycan, which associates with MuSK in a manner that is independent of the Ig3 domains (25). Biglycan bound equivalent amounts of both the FL-ecto-MuSK and ΔIg3-ecto-MuSK proteins (fig. S3). Together, these data indicate that the alternatively spliced Ig3 domain is necessary for MuSK binding to BMP4.
MuSK affects the expression of distinct sets of BMP4-induced genes in myoblasts
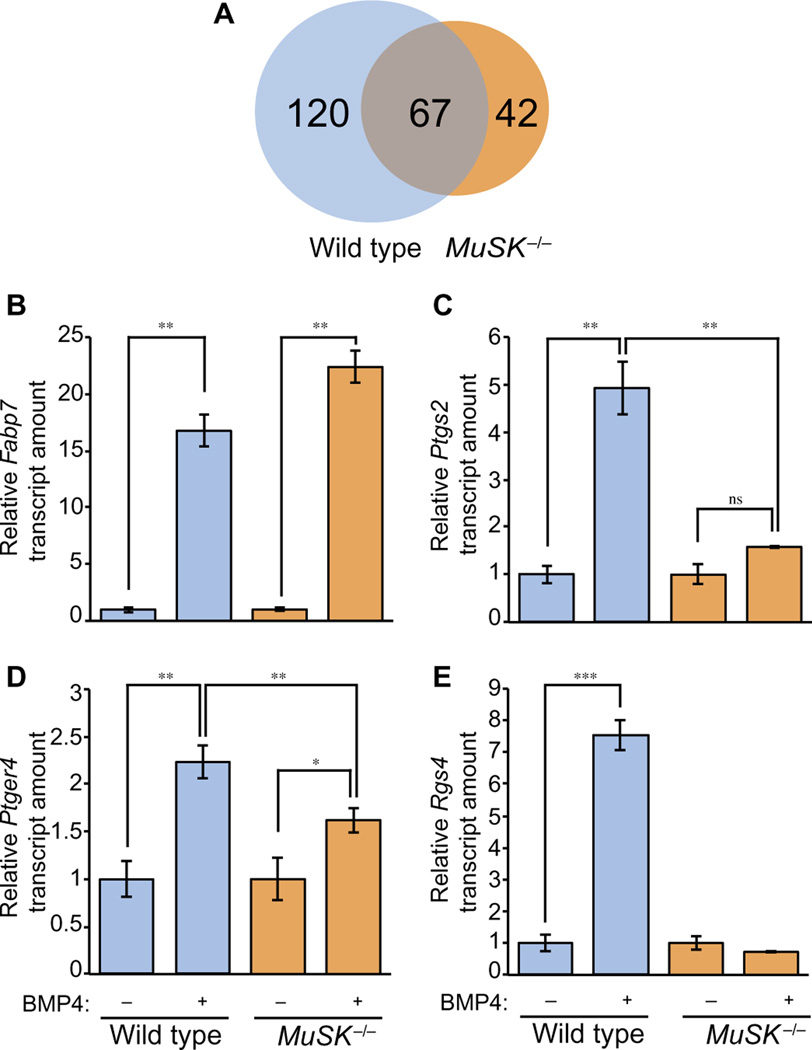
BMPs are potent and selective regulators of gene transcription. We therefore determined whether MuSK modulates BMP-induced gene transcription in myogenic cells. As a first test of this possibility, we treated either wild-type or MuSK−/− myoblasts with BMP4 and analyzed their gene expression profiles using microarrays. We identified 120 transcripts that increased in abundance in wild-type but not in MuSK−/− cells after BMP4 treatment [≥1.5-fold; false discovery rate (FDR)–corrected P < 0.05]. Thus, these BMP-regulated transcripts are MuSK-dependent (Fig. 2A and table S2). In addition, 67 transcripts increased in abundance both in wild-type and MuSK−/− myoblasts and therefore, were not qualitatively regulated by MuSK (Fig. 2A and table S3). Notably, many of the genes induced in both genotypes encoded proteins active in the canonical BMP pathway including Id1, Id2, and Id3 and Smad6, Smad7, and Smad9. Finally, 42 transcripts increased abundance only in MuSK−/− myoblasts (Fig. 2A and table S4).
Fig. 2. MuSK influences BMP4-induced expression of a subset of genes in myoblasts.
(A) Wild-type H-2Kb-tsA58 and MuSK−/− myoblasts were serum-deprived for 4 hours and then treated with BMP4. Microarray analysis identified differentially expressed genes upon BMP4 treatment in both genotypes. The number of genes up-regulated in response to BMP4 in wild-type and MuSK−/− myoblasts are grouped into a Venn diagram as wild type only, shared, and MuSK−/− only. Data represent the averages of three independent biological replicates. FDR-corrected P < 0.05 and ±1.5-fold change were used as selection criteria to identify genes differentially expressed between samples. (B to E) Validation of microarray results for four genes. Transcript abundances for MuSK-independent expression of Fabp7 (B) and MuSK-regulated expression of Ptgs2 (C), Ptger4 (D), and Rgs4 (E) were measured by qRT-PCR. Data are means ± SD from three biological replicate experiments (***P < 0.0001; **P < 0.01; *P < 0.05; ns, not significant versus untreated conditions, Bonferroni-adjusted one-way ANOVA).
Because the arbitrary fold change and statistical significance cutoffs might lead to false-positive hits in gene array studies, we validated the responses revealed by the microarrays for a subset of transcripts. Using quantitative reverse transcription polymerase chain reaction (qRT-PCR), the relative amount of transcripts of these genes was measured and normalized to 18S ribosomal RNA (rRNA). Fabp7, a previously unidentified BMP4 target, was prominent among the 67 MuSK-independent transcripts. qRT-PCR analysis showed that Fabp7 transcript abundance increased more than 15-fold between untreated and treated wild-type myoblasts, as well as between untreated and treated MuSK−/− myoblasts, after an 8-hour BMP4 treatment (Fig. 2B). Next, we validated a group of MuSK-dependent genes. BMP4 selectively increased the expression of Ptgs2 and Ptger4 in wild-type as compared to MuSK−/− myoblasts (Fig. 2, C and D). Ptger2 encodes cyclooxygenase-2 (COX2), the key enzyme in the prostaglandin pathway, and Ptger4 encodes a prostaglandin receptor. BMP4 also induced an eightfold increase in the expression of Rgs4, a regulator of heterotrimeric guanine nucleotide-binding protein (G protein) signaling, in wild-type cells, but this transcript did not change in abundance in MuSK−/− cells (Fig. 2E). Finally, Rgs4 also showed a strict MuSK dependence when cells were stimulated with BMP4 for a shorter period (2 hours; fig. S4). Together, these results confirm the microarray results and demonstrate that MuSK-dependent transcripts show robust and selective responses to BMP stimulation in myoblasts.
MuSK affects the expression of distinct sets of genes in myoblasts and myotubes
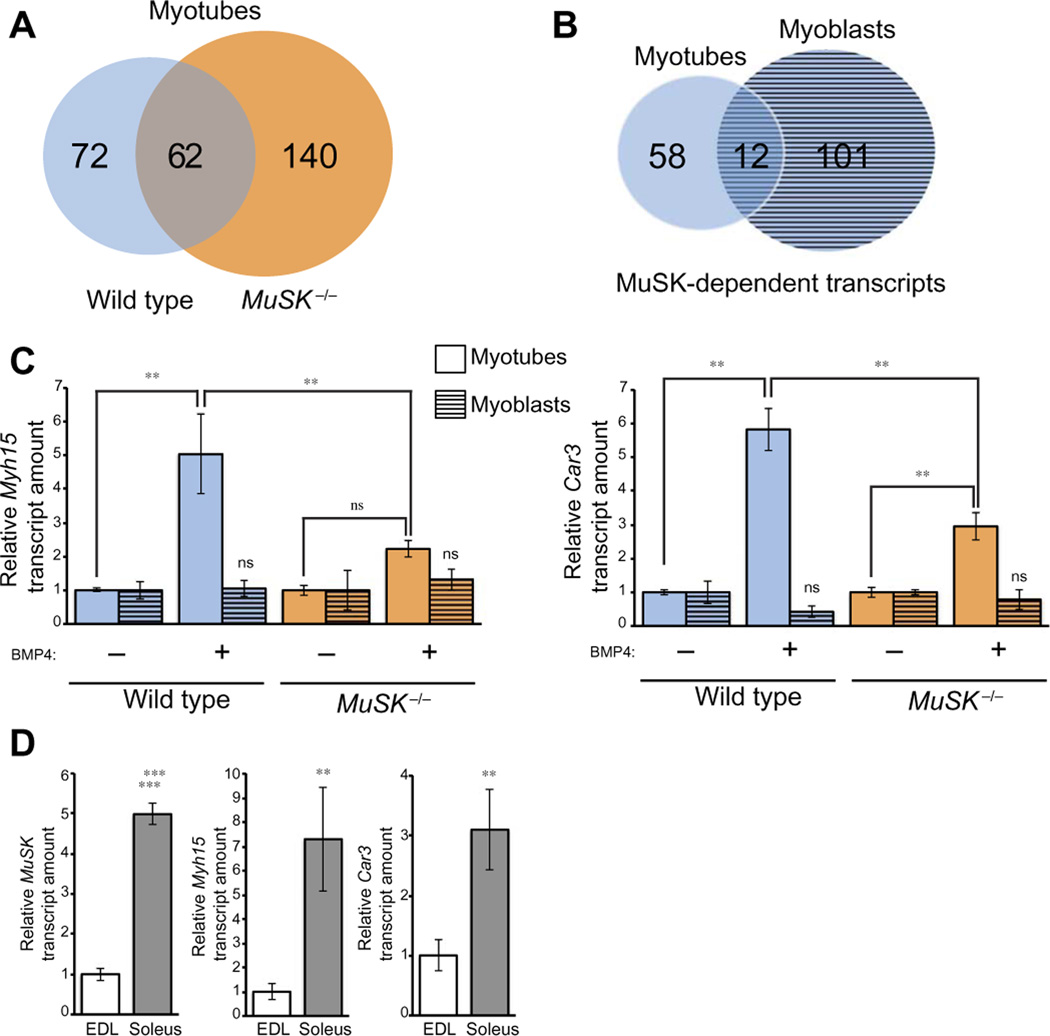
MuSK is highly abundant at the NMJ in differentiated muscle, where its role as a synapse-organizing molecule is well established. We next determined whether MuSK also modulates BMP-induced gene transcription in differentiated muscle cells. We stimulated wild-type or MuSK−/− myotubes with BMP4 and the profiled gene expression in these cells by microarray. BMP4 induced the up-regularion of 134 and 202 transcripts in wild-type and MuSK−/− myotubes, respectively (≥1.5-fold increase; FDR, <0.05). There were 72 transcripts that increased in abundance only in wild-type myotubes, whereas 62 transcripts increased in abundance in both genotypes (Fig. 3A and tables S5 and S6). Further 140 transcripts increased abundance only in MuSK−/− myotubes (Fig. 3A and table S7). Finally, a comparison of transcripts that increased in wild-type myoblasts and myotubes in a manner that depended on both BMP4 and MuSK revealed 113 and 70 transcripts in these cell types, respectively, with only 12 common to both (Fig. 3B and tables S8 to 10). Together, these results indicate that MuSK regulates transcriptional output of the BMP pathway in myoblasts and myotubes in a cell type-specific manner.
Fig. 3. MuSK influences BMP4-induced expression of a subset of genes in myotubes.
(A) Wild-type H-2Kb-tsA58 and MuSK−/− myotube cultures were treated with BMP4. RNA was isolated and subjected to transcriptomic analysis. The number of genes up-regulated in response to BMP4 responses for up-regulated genes in wild-type and MuSK−/− myotubes are grouped in a Venn diagram as wild type only, shared, and MuSK−/− only. Data represent the averages of three independent biological replicates. FDR-corrected P < 0.05 and ±1.5-fold change were used as selection criteria to identify genes differentially expressed between samples. (B) Venn diagram showing the number of transcripts that increased in abundance in response to BMP4 in a MuSK-dependent manner in myoblasts and myotubes. (C) Validation of microarray results. Wild-type H-2Kb-tsA58 and MuSK−/− myotubes were treated with BMP4, and the abundance of Myh 15 and Car3 transcripts was analyzed in myoblasts and myotubes by qRT-PCR. Data are means ± SD from five biological replicate experiments (**P < 0.01; *P < 0.05; ns, not significant versus untreated condition, one-way ANOVA with Bonferroni correction). (D) MuSK, Myh15, and Car3 expression in soleus muscles compared to extensor digitorum longus (EDL) muscles. The abundance of MuSK, Myh15, and Car3 transcripts was analyzed by qRT-PCR. Data are means ± SD from five different animals (**P < 0.0001; **P< 0.01, unpaired, two-sided Student’s t test).
We then analyzed the MuSK-dependent BMP4 responses in myoblasts and myotubes according to gene functions and localization patterns in the cell. These transcripts included a large number of signaling molecules; thus, we first manually attributed each MuSK-dependent transcriptional response to a functional or localization-based category related to signaling. This analysis demonstrated that half of these specific responses in myoblasts and more than half of those in myotubes affected signaling-related molecules such as growth factors, transcription factors, cell surface receptors, extracellular matrix proteins, and intracellular signaling proteins (fig. S5, A and B). In addition, a Gene Ontology (GO) analysis showed that several limb morphogenesis and transcription-related terms are enriched for MuSK-dependent BMP4 responses in myoblasts (fig. S5C). Similarly, transcription-related terms were among the most significantly enriched GO terms within the MuSK-dependent BMP4 responses in myotubes, which agrees with the high percentage of transcription factors that are up-regulated in the presence of MuSK (fig. S5, B and D).
MuSK affects the expression of genes expressed in slow-twitch muscle fibers
Although MuSK is abundant at the NMJ in all muscles, it is also localized extrasynaptically in slow-twitch, but not fast-twitch, muscle fibers (17). These observations suggest that MuSK may have unique, nonsynaptic functions in slow muscle. One possibility is that MuSK could play a role in regulating the expression of slow muscle-specific genes. Examination of the microarray data revealed the MuSK-dependent and BMP4-induced up-regulation of Myh15 and Car3, both of which have been previously suggested to be slow-type fiber markers (26, 27). We validated the MuSK-dependent regulation of these transcripts by qRT-PCR in cultured myotubes (Fig. 3C). Finally, we confirmed that MuSK, Myh15, and Car3 were enriched in slow muscles as compared to fast muscles isolated from mice (soleus and extensor digitorum longus, respectively; Fig. 3D). Notably, neither of these genes was up-regulated by BMP4 in myoblasts (Fig. 3C), further reinforcing the cell type selectivity of MuSK-dependent transcription in the myogenic lineage. Together, these results suggest that MuSK selectively regulates the expression of transcripts expressed in slow muscle.
MuSK kinase activity is not required for modulating BMP4 signaling
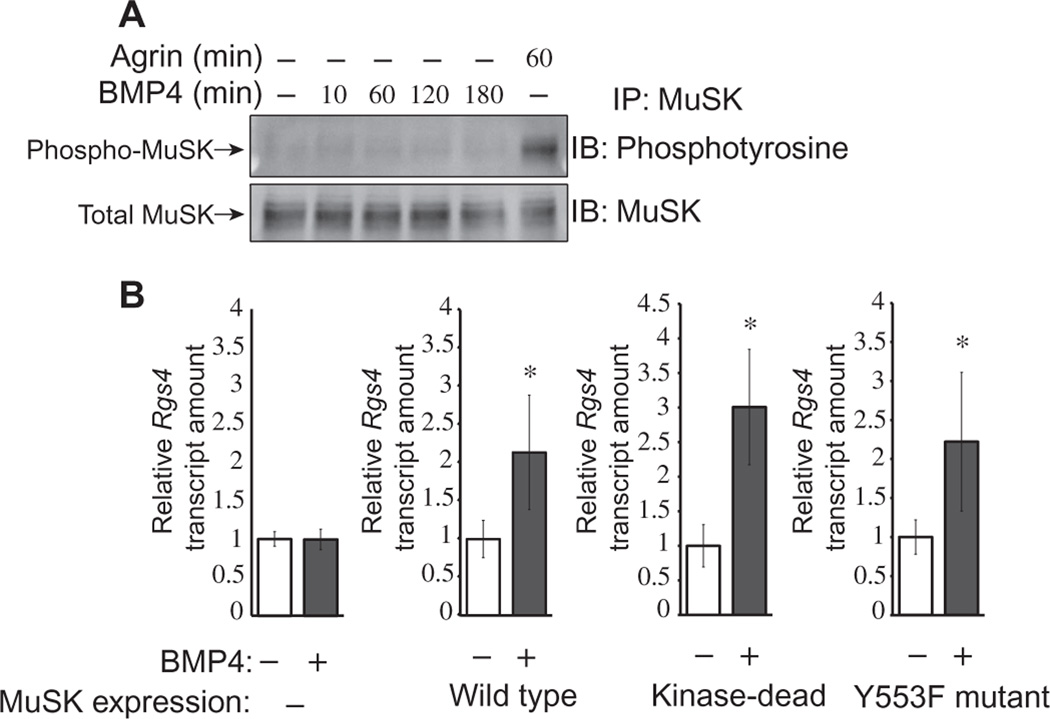
NMJ differentiation requires MuSK activation, as well as both an active MuSK tyrosine kinase domain and the juxtamembrane tyrosine in an NPXY motif at position 553 (28, 29). We therefore investigated the role of MuSK tyrosine kinase activity in the regulation of BMP-induced transcription. As a first test, we determined whether BMP4 stimulated MuSK kinase activity. We treated wild-type myotubes with BMP4 for intervals ranging from 10 to 180 min and then assessed MuSK activation (phosphorylation). We detected no increase in MuSK tyrosine phosphorylation at any of these time points (Fig. 4A). As a positive control, we observed robust MuSK kinase activation in parallel cultures treated with agrin for 60 min (Fig. 4A). The time intervals and ligand concentrations in these experiments were the same as those used to demonstrate MuSK-dependent BMP-mediated transcription (Figs. 2 and 3). Thus, BMP4 does not induce MuSK phosphorylation under conditions where it stimulates MuSK-dependent transcription.
Fig. 4. MuSK regulation of BMP signaling is independent of MuSK kinase activity.
(A) Wild-type H-2Kb-tsA58 myotubes were treated with BMP4 (25 ng/ml) or with agrin for the indicated times. MuSK was then immuno-precipitated, and the tyrosine kinase activation was assessed by Western blotting with a phosphotyrosine-specific antibody (upper panel). The blots were stripped and reprobed with a MuSK-specific antibody to assess total MuSK (bottom panel). Blots are representative of three independent experiments. IP, immunoprecipitation; IB, immunoblot. (B) MuSK−/− myoblasts or MuSK−/− myoblasts transgenically expressing wild-type, kinase-dead (K608A), or Y553F MuSK were treated with BMP4 (3.25 ng/ml for 2.5 hours), and Rgs4 transcript abundance was measured by qRT-PCR. Data are means ± SD from three biological replicate experiments (*P < 0.05, unpaired, two-sided Student’s t test).
We next used a genetic approach to test the potential role of MuSK activation in BMP4-mediated transcription. We used MuSK−/− cell lines that had been stably transfected with constructs encoding wild-type, kinase-dead (K608A), or Y553F variants of MuSK. Previous work has shown that the wild-type mutants, but neither the kinase-dead nor Y553F mutants, are activated in response to agrin in these rescue lines (28, 29). In agreement with the results presented previously (Fig. 2), BMP4 did not increase Rgs4 expression in MuSK−/− myoblasts. However, all the MuSK transgenic cell lines—including those lacking kinase activity— showed robust BMP4-stimulated Rgs4 expression (Fig. 4B). Together, these results demonstrate that MuSK is not activated by BMP4 and that its tyrosine kinase activity is dispensable for MuSK-dependent, BMP4-induced transcription.
MuSK-dependent gene regulation requires the activity of canonical BMP pathway members
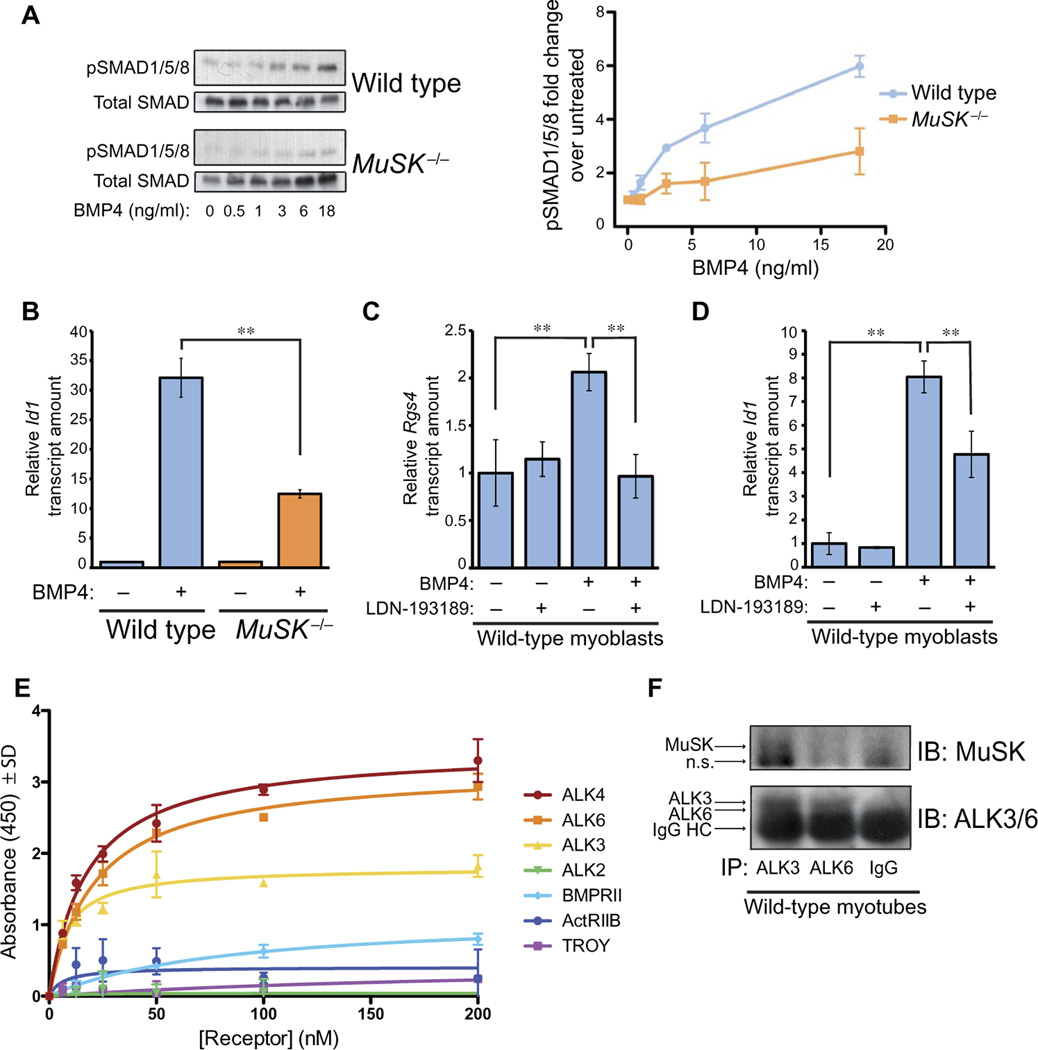
We next explored the relationship between MuSK and the BMP signaling mediators. The core BMP pathway involves ligand activation of the type I and type II receptor complex followed by phosphorylation and translocation of SMAD1/5/8 to the nucleus, where it regulates transcription. We first examined the role of MuSK in SMAD phosphorylation. We treated either wild-type or MuSK−/− myoblasts with BMP4 and assessed SMAD phosphorylation by Western blotting (Fig. 5A, left panel). SMAD1/5/8 phosphorylation was reduced in MuSK−/− cells at all ligand concentrations tested (Fig. 5A, right panel). These results indicate that MuSK enhances BMP signaling, at least in part, by promoting SMAD1/5/8 phosphorylation.
Fig. 5. MuSK is a BMP co-receptor and regulates the canonical BMP4 pathway.
(A) Wild-type H-2Kb-tsA58 and MuSK−/− myoblasts were treated with BMP4 at the indicated concentrations, and total and phosphorylated SMAD1/5/8 (pSMAD1/5/8) were then assessed by Western blotting (left panel). The abundance of pSMAD1/5/8 in Western blots was normalized to total SMAD1/5/8 for each condition. Fold change in pSMAD1/5/8 was calculated and plotted as the ratio of BMP4-treated conditions to the untreated controls (right panel). Data are means ± SEM from three biological replicate experiments and their independent Western blots. (B) Wild-type H-2Kb-tsA58 and MuSK−/− myoblasts were treated with BMP4 (3.25 ng/ml) for 2 hours. Id1 transcript abundance was quantified by qRT-PCR. Data are means ± SD from three biological replicates (**P < 0.01, one-way ANOVA with Bonferroni correction). (C) Serum-deprived wild-type H-2Kb-tsA58 myoblasts were treated with BMP4 in the presence or absence of the BMP type I receptor inhibitor LDN-193189, and Rgs4 transcript abundance was quantified by qRT-PCR. Data are means ± SD from three biological replicate experiments (**P < 0.01, one-way ANOVA with Bonferroni correction). (D) Serum-deprived wild-type H-2Kb-tsA58 myoblasts were treated with BMP4 in the presence or absence of LDN-193189, and Id1 transcript abundance was measured by qRT-PCR. Data are means ± SD from three biological replicate experiments (**P < 0.01, one-way ANOVA with Bonferroni correction). (E) Immobilized His-tagged MuSK ectodomain was incubated with the indicated concentrations of recombinant purified Fc-fusion versions of ALK2, ALK3, ALK4, ALK6, BMPRII, ActRIIB, and TROY. Bound receptors were detected with horseradish peroxidase (HRP)-conjugated antibodies recognizing human and mouse IgG. Data are representative of three independent experiments. Values indicate the average from four replicates and are means ± SD. (F) Pooled detergent extracts of cultured H-2Kb-tsA58 myotubes were divided into equal volumes and incubated with antibodies recognizing ALK3 or ALK6 or with normal IgG and immunoprecipitated. Immunoprecipitates were immunoblotted to show MuSK or ALK3 and ALK6 as indicated. A nonspecific band (n.s.) in the MuSK immunoblots is indicated. Data are representative of three biological replicate experiments. HC, heavy chain.
The attenuated phosphorylated SMAD1/5/8 response in MuSK−/− myoblasts suggested that canonical BMP-induced gene transcription might also be reduced. To test this possibility, we compared the abundance of Id1 transcripts after BMP stimulation in both wild-type and MuSK−/− myoblasts. Although Id1 transcripts were increased in both genotypes, there was a threefold greater response in the wild-type cells as compared to the MuSK−/− cells (Fig. 5B). Thus, both BMP-induced changes in SMAD1/5/8 phosphorylation and Id1 transcript abundance are reduced in the absence of MuSK. We conclude that MuSK promotes canonical BMP signaling.
We next determined whether type I BMP receptor activity plays a role in regulating MuSK-dependent transcripts. We used a selective inhibitor of type I BMP receptors, LDN-193189 (30). Treatment of myoblasts with this compound inhibited BMP4-induced up-regulation of Rgs4 expression (Fig. 5C). As a control, we also examined the expression of the canonical BMP target Id1 and showed that BMP4-induced up-regulation of this transcript was also inhibited by LDN-193189 (Fig. 5D). Thus, BMP4-induced up-regulation of MuSK-dependent transcripts requires type I BMP receptor activity.
MuSK binds to the type I BMP receptors ALK3 and ALK6 and the type I activin receptor ALK4
The ability of MuSK to quantitatively and qualitatively modulate the BMP signaling pathway in a tyrosine kinase-independent fashion raised the possibility that MuSK acts as a co-receptor in these myogenic cells. To test this idea, we assessed the binding of the MuSK ectodomain to the type I BMP receptors ALK2, ALK3, and ALK6 and the type II BMP receptors ActRIIB and BMPRII in a solid-phase binding assay. ALK3 and ALK6 bound to the ectodomain of MuSK in a saturable and high-affinity manner (Fig. 5E). In contrast, no specific binding was observed with ALK2, the type II receptors ActRIIB and BMPRII, or the tumor necrosis factor family receptor TROY. We also tested MuSK binding to ALK4, a type I activin receptor that is closely related to the type I BMP receptors ALK3 and ALK6. ALK4 bound MuSK in a high-affinity manner, similar to ALK6 (Fig. 5E). Finally, to determine whether endogenous MuSK and BMP receptors associate in muscle cells, we prepared detergent extracts from cultured myotubes and immunoprecipitated ALK3 and ALK6. MuSK coimmunoprecipitated with ALK3, but not ALK6, under these conditions (Fig. 5F). Therefore, we conclude that in myotubes, MuSK selectively associates with ALK3, one of the preferred type I receptors for BMP4 (1). Together, these data demonstrate that MuSK is a co-receptor for BMP in muscle cells.
Finally, we asked whether MuSK regulates compartmentalization of the BMP signaling mediators SMAD1, SMAD5, and SMAD8. Immunostaining for phosphorylated SMAD1/5/8 revealed that the subcellular localization of these key BMP signaling components differed in wild-type and MuSK−/− myoblasts. In wild-type myoblasts, phosphorylated SMAD1/5/8 was distributed in abundant, distinctive puncta in over 70% of the cells (fig. S6, A and B). In contrast, such phosphorylated SMAD1/5/8 puncta were sparse in MuSK−/− cells and could only be detected in ~20% of the mutant cells. Together, these observations suggest that MuSK might favor cytoplasmic retention of phosphorylated SMAD1/5/8.
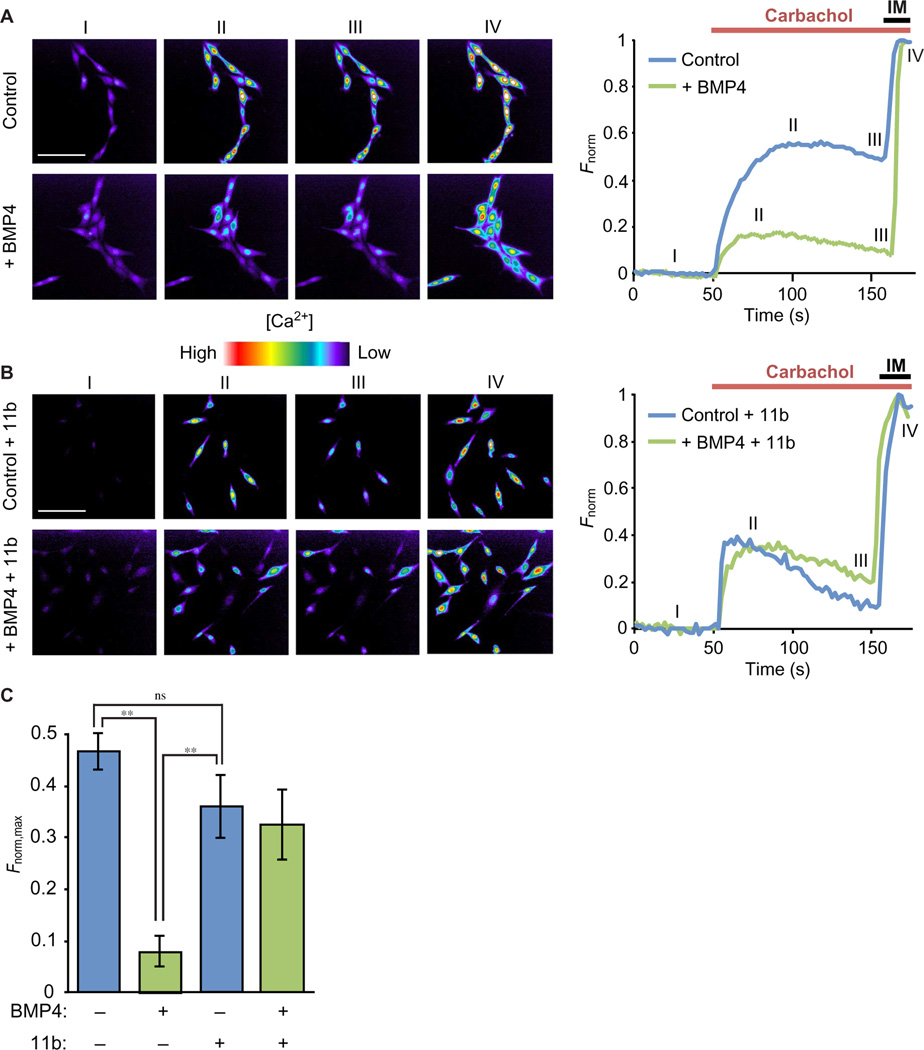
MuSK- and BMP4-dependent expression of Rgs4 inhibits carbachol-induced Ca2+ responses in myoblasts
We next sought to determine the physiological role of MuSK-dependent BMP4 gene regulation in myogenic cells. Regulators of G protein signaling (RGS) proteins are GAPs that directly bind to and stimulate the GTPase activity of the Gα subunits of G proteins, thus reducing G protein activity. GAPs regulate many intracellular signaling events, including Ca2+ oscillations in the cytosol (31). Therefore, we hypothesized that MuSK-dependent, BMP4-induced expression of Rgs4 may modulate Ca2+ signaling (32). To test this idea, we first treated wild-type myoblasts with the cholinergic agonist carbachol, which increases intracellular Ca2+ concentration by activating Gαq-coupled muscarinic receptors (33). As expected, carbachol treatment increased intracellular Ca2+ in resting myoblasts (Fig. 6A, upper panel images; blue trace in the graph on the right). In contrast, the carbachol-mediated Ca2+ signal was attenuated in BMP4-treated myoblasts (Fig. 6A, lower panel images; green trace in the graph on the right). To test whether this reduction was due to the action of RGS4, we treated the myoblasts with the selective RGS4 inhibitor 11b (34) and analyzed their Ca2+ responses with or without BMP4 pretreatment. BMP4 pretreatment did not alter the Ca + response evoked by carbachol in myoblasts treated with the RGS4 inhibitor, suggesting that RGS4 mediates the reduction in carbachol-induced Ca2+ response by BMP4 (Fig. 6, B and C). Together, these results indicate that MuSK-dependent transcription of the BMP4-responsive gene Rgs4 mediates the inhibition of carbachol-induced Ca2+ response in myoblasts.
Fig. 6. BMP4 treatment inhibits carbachol-induced Ca2+ responses in wild-type H-2Kb-tsA58 myoblasts in an RGS4-dependent manner.
(A) Pseudochrome Fluo-4 fluorescence images of H-2Kb-tsA58 myoblasts stimulated with carbachol in the absence (top) or presence (bottom) of BMP4. The images were acquired during baseline (I), peak carbachol response (II), before ionomycin (IM) addition (III), and at peak IM response (IV). Scale bar, 100 µm. The representative average of normalized Ca2+ imaging responses (Fnorm) after a 4-hour BMP4 treatment (right). (B) Pseudochrome Fluo-4 fluorescence images of wild-type myoblasts treated with the RGS4 inhibitor 11b and stimulated with carbachol in the absence (top) or presence (bottom) of BMP4 treatment. The images were acquired during baseline (I), peak carbachol response (II), before IM addition (III), and at peak IM response (IV) (left). Scale bar, 100 µm. The representative average of normalized Ca2+ imaging responses (Fnorm) after BMP4 treatment in the presence of the RGS4 inhibitor 11b (right). (C) The normalized peak fluorescence of carbachol-induced Ca2+ responses (Fnorm,max) was significantly decreased by BMP4 treatment only in the absence of the RGS4 inhibitor 11 b. Data are means ± SD from seven independent experiments for each condition (**P < 0.01, one-way ANOVA with Bonferroni correction).
DISCUSSION
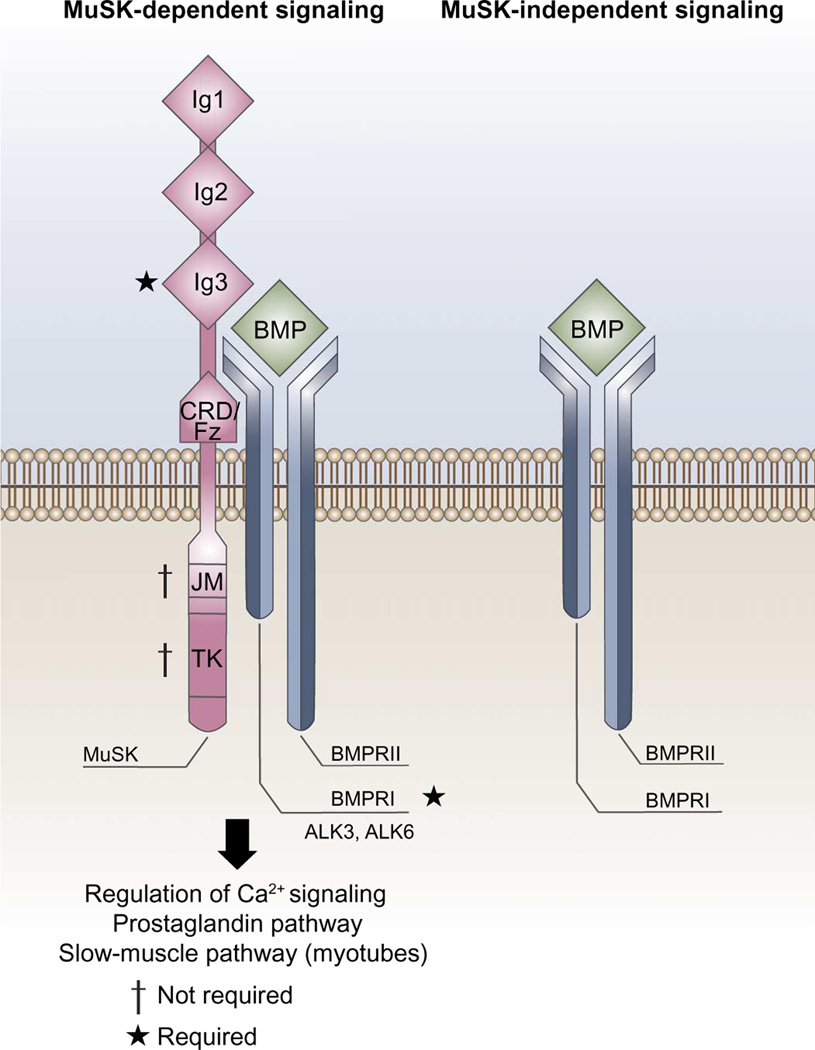
Here, we show that the RTK MuSK, previously established as the central organizer of the NMJ, binds BMPs and BMP receptors and shapes the transcriptional response to these factors in myogenic cells. We propose that MuSK acts as a tissue-specific BMP co-receptor that is required for the BMP-responsive transcription of a subset of genes in a cell context-dependent manner (Fig. 7). Several observations indicate that MuSK is a BMP co-receptor (Figs. 1, 4, and 5). (i) MuSK binds BMPs with high affinity and associates with the type I receptors ALK3 and ALK6 in a saturable, high-affinity, and BMP-independent fashion. (ii) Endogenous MuSK and ALK3 are associated in muscle cells. (iii) BMP-induced SMAD phosphorylation and Id1 transcription are reduced in the absence of MuSK. (iv) BMP does not stimulate MuSK tyrosine kinase activity, and the stimulation of BMP-induced MuSK-dependent transcripts is sensitive to ALK inhibitors but independent of MuSK tyrosine kinase activity.
Fig. 7. Model for MuSK regulation of BMP4 signaling.
MuSK binds to BMPs and the BMP receptors ALK3 and ALK6 and regulates the transcriptional output of BMP signaling in muscle cells. MuSK regulation of BMP signaling is cell type-specific because BMP induced the expression of different sets of genes in myoblasts and myotubes in a manner that depended on MuSK. The kinase activity of MuSK, which is required for proper NMJ formation, is dispensable for MuSK-mediated regulation of BMP signaling. JM, juxta-membrane; TK, tyrosine kinase; CRD/Fz, CRD/frizzled-like domain.
MuSK binds to BMP2, BMP4, and BMP7 with affinities ranging from 5.6 to 11.8 nM, which are comparable to the affinities with which BMPs bind to other physiologically relevant BMP-interacting molecules (11, 35). Moreover, the presence of MuSK increases SMAD1/5/8 activation in response to a wide range of BMP4 concentrations, and the expression of transcripts encoding canonical BMP pathway components is greater in the presence of MuSK. These results indicate that MuSK is a positive regulator of canonical BMP signaling.
The role of MuSK in shaping the BMP-mediated transcriptional profile is marked. We observed 101 and 58 MuSK-dependent transcripts unique to myoblasts and myotubes, respectively (Figs. 2 and 3 and tables S3 and S6). The selectivity and robustness of this response are underscored by the comparison of MuSK-dependent and MuSK-independent transcripts (Fig. 2). For example, BMP increased the abundance of Fabp7 >15-fold in either the presence or absence of MuSK. In contrast, BMP induced the expression of Rgs4 and Ptgs2 eight- and fourfold, respectively, in a manner that was strictly dependent on MuSK.
The BMP-MuSK pathway is likely to shape the transcriptional output through multiple mechanisms. Increased expression of Rgs4 was observed within 2 hours of BMP4 treatment, consistent with this gene being a direct target of MuSK-dependent BMP signaling. However, many transcription factors were also regulated in a MuSK- and BMP-dependent manner (fig. S5), suggesting that other MuSK-dependent, BMP4-induced transcripts are the product of a BMP-induced transcription factor cascade.
There are at least three, nonmutually exclusive mechanisms by which a MuSK co-receptor could regulate BMP signaling. First, MuSK could augment signaling by presenting a ligand to the BMP receptor complex. Second, MuSK may create subcellular signaling compartments that generate unique outputs, a mechanism supported by the distinct repertoire of MuSK-dependent transcripts in myoblasts and myotubes (Figs. 2 and 3) and the presence of MuSK-dependent intracellular granules containing phosphorylated SMAD1/5/8 (fig. S6). Finally, as an ALK binding protein, MuSK could shape the composition and thus the output of BMP signaling complexes in myogenic cells.
MuSK-dependent BMP signaling modulates the muscle cell response to mAChR-mediated Ca2+ signaling through the regulation of RGS4 production. RGS proteins stimulate Gα GTPase activity, thus diminishing G protein signaling (31). Activation of Gαq-coupled mAChRs by carbachol results in a transient increase in Ca2+ that was inhibited by BMP4 treatment (Fig. 6). Direct pharmacological blockade of RGS4 reversed this inhibition, suggesting a direct link between the MuSK-BMP pathway and the cellular response to mAChR activation. However, it should be noted that the complete mechanism by which MuSK-dependent BMP signaling induces Rgs4 expression remains to be elucidated.
MuSK may regulate multiple subclasses of the BMP superfamily. For example, MuSK binds ALK4 (Fig. 4), a type I activin receptor and one of the preferred type I receptors for myostatin, a potent negative regulator of skeletal muscle growth (36). Other RTKs may play a similar role in BMP signaling. For example, the RTK ROR2 has been reported to bind ALK6 in heterologous cells (10, 37, 38).
The role of MuSK as a BMP co-receptor is mechanistically distinct from its function in synapse formation. Agrin, LRP4, and the tyrosine kinase activity of MuSK are absolutely required for NMJ formation and stability. Moreover, this pathway is restricted to differentiated muscle cells, and agrin-LRP4-MuSK-dependent AChR clustering does not require de novo transcription (39). In contrast, the role of MuSK as a BMP co-receptor is characterized by a robust transcriptional response in both myoblasts and myotubes that requires neither its tyrosine kinase activity nor a juxtamembrane tyrosine that is critical for NMJ differentiation (Fig. 4). Moreover, the MuSK Ig3 domain is dispensable for its ability to mediate AChR clustering (40) but is required for BMP binding (Fig. 1). Although these two MuSK pathways are distinct, they could collaborate in fully differentiated synapses. For example, the expression of Dok7, which encodes an essential binding partner of MuSK in postsynaptic differentiation, is regulated in a MuSK- and BMP-dependent fashion (table S6). Finally, the BMP pathway plays a critical role in the differentiation of nerve muscle synapses in Drosophila melanogaster (41). However, in this case, the BMP ortholog Gbb acts as a muscle-derived retrograde signal to promote presynaptic differentiation (41, 42).
The MuSK-BMP pathway could play a role in the regulation of myofiber size and composition. BMP signaling promotes muscle hypertrophy and counters denervation-induced atrophy (43–45). After denervation or immobilization, MuSK abundance increases along the entire length of the muscle (46), suggesting that MuSK plays a role in BMP-mediated signaling in these pathological contexts. MuSK is also present extrasynaptically in slow-oxidative muscle fibers (17), and two slow-muscle genes, Myh15 and Car3, are stimulated by BMP in a MuSK-dependent manner. Finally, the BMP-MuSK pathway regulates the production of both COX2, which is important for myoblast proliferation, fusion, and growth (47–50), and RGS4, which can inhibit cell growth and myofilament organization in neonatal cardiac myocytes (51).
The data presented here show that MuSK is a BMP co-receptor that modulates mAChR-mediated Ca2+ signaling in myogenic cells as well as pathways important for muscle differentiation and size. Understanding such regulation could be beneficial for treating pathological conditions, such as Duchenne muscular dystrophy, insulin-resistant type 2 diabetes, and disuse atrophy (52), in which muscle mass is lost or muscle function is compromised. Finally, future studies may also reveal whether MuSK modulates BMP signaling in nonmuscle tissues such as brain (18).
MATERIALS AND METHODS
Antibodies and recombinant proteins
Purified recombinant human BMP4, ALK6-Fc, ALK3-Fc, ALK4-Fc, BMPRII-Fc, and TROY-Fc chimeras, mouse ALK2-Fc and ActRIIB-Fc chimeras, and rat agrin were obtained from R&D systems. Antibodies specific for MuSK, ALK3, ALK6, and BMP4 (both unlabeled and bioti-nylated) were obtained from R&D Systems. Streptavidin-HRP was obtained from Thermo and used at 1:15,000 dilution. Mouse HRP and human IgG(Fc) secondary antibodies were obtained from KPL and used at 1:400 dilution. SMAD5 and phosphorylated SMAD1/5/8 antibodies were obtained from Epitomics, and phosphotyrosine (4G10) antibody was obtained from EMD Millipore Corporation and used at 1:1000, 1:2000, and 1:1000 dilutions, respectively. Alexa Fluor 555–conjugated goat antibody against rabbit IgG was obtained from Invitrogen and used at 1:1000. Constructs encoding full-length and Ig3-lacking Fc-Fusion MuSK ectodomains were obtained from GenScript and were used at the indicated concentrations. A construct encoding the His-tagged MuSK ectodomain was expressed in human embryonic kidney (HEK) 293 PEAKrapid cells from the American Type Culture Collection, and secreted recombinant His-MuSK ectodomain was purified from the cell culture supernatant by immobilized nickel ion affinity chromatography. LDN-193189 was obtained from Stemgent and was used at 50 nM.
Mammalian cell culture and mice
Mouse C2C12BRA cells (18) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 2% l-glutamine, and 1% penicillin-streptomycin and cultured at 37°C in 8% CO2. Immortalized myoblast cultures of wild-type mouse H-2Kb-tsA58 (53) and MuSK−/− and MuSK rescue lines [wild-type MuSK (B1), kinase-dead MuSK (K608A), and MuSK Y553A] were cultured on gelatin-coated dishes in DMEM supplemented with 20% fetal bovine serum, 2% l-glutamine, 1% penicillin-streptomycin, 1% chicken embryo extract, and 1 U of interferon-γ under permissive temperature at 33°C in 8% CO2. Myotubes were obtained by switching the confluent myoblast cultures to a medium with DMEM supplemented with 5% horse serum, 2% l-glutamine, and antibiotics at 37°C in 8% CO2. For the LDN-193189 treatments, myoblasts were treated with the drug (50 nM) for 30 min before BMP4 treatment, and the drug was kept in cultures during the course of the treatment. Soleus and extensor digitorum longus muscles were harvested from 5.5-week-old adult C57BL/6 mice. All protocols were conducted under accordance and with the formal approval of Brown University’s Institutional Animal Care and Use Committee.
Luciferase reporter assays
C2C12BRA cells were plated in 96-well culture dishes at 4 × 103 to 5 × 103 cells per well and allowed to adhere overnight. The indicated recombinant proteins were premixed in DMEM containing 0.1% bovine serum albumin (BSA) for 20 min at 4°C, and the original culture medium was replaced with this solution. The cells were incubated for 8 hours and washed twice with phosphate-buffered saline (PBS) before the extracts were prepared with 1× cell lysis buffer (50 µl per well; Roche). The lysate (40 µl) was transferred to an opaque white 96-well microplate and mixed with 100 µl of luciferase substrate (Roche). The luciferase activity was read in a luminometer and reported as relative luciferase unit (RLU), which is the value for each condition after subtraction of mock treatment (no-BMP4) value and normalization to BMP4-only condition. All assays were performed in eight biological replicates and repeated at least two times with similar results.
For testing of supernatants in the solution-binding experiments, the indicated recombinant proteins were premixed in DMEM containing 0.1% BSA for 2 hours at 4°C, followed by addition of magnetic nickel-bound beads (Promega) and a further 2-hour incubation at 4°C The same protocol detailed above was followed after this step to test BMP4 activity. The average value of untreated cells was subtracted from each condition, and everything was normalized to BMP4-only condition (100% activity).
Immunoprecipitation and coimmunoprecipitation
For MuSK immunoprecipitation, treated H-2Kb-tsA58 myotubes were lysed in extraction buffer [10 mM tris-HCl (pH 7.4), 1% Triton X-100, 0.5% NP-40, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1 mM sodium orthovanadate, 10 mM sodium fluoride and 1× EDTA-free protease inhibitor cocktail (Roche cOmplete)]. For MuSK-ALK coimmunoprecipitation, H-2Kb-tsA58 myotubes were lysed in extraction buffer [10 mM tris-HCl (pH 7.4), 1% Triton X-100, 150 mM NaCl, 1 mM sodium orthovanadate, 10 mM sodium fluoride, and 1× EDTA-free protease inhibitor cocktail (Roche cOmplete)]. Lysates for immunoprecipitation and coimmunoprecipitation experiments were precleared with protein G-bound magnetic beads (Invitrogen); bicinchoninic acid assay (BCA) was used to quantify total protein amounts in the lysates (Pierce). For immunoprecipitation experiments, lysates with equal amounts of total protein and the MuSK-specific antibody were mixed and tumbled overnight at 4°C. For coimmunoprecipitation, pooled lysates from three to four T75 flasks were divided into equal volumes for the pull-downs with ALK3-and ALK6-specific antibodies or the control (normal IgG) antibody overnight at 4°C After the addition of protein G–bound magnetic beads, lysates were tumbled for 4 to 6 hours at 4°C The beads were washed with extraction buffer, and 2× sample buffer was added for elution of the immunoprecipitated proteins from the beads. Proteins were eluted by boiling the samples at 98°C for 5 min. Western blots were run for the samples as indicated below.
Western blots
For phosphorylated SMAD1/5/8 Western blotting, lysates from cells that were serum-deprived in DMEM containing 0.1% BSA for 5 to 6 hours and treated with BMP4 for 15 min were prepared in extraction buffer containing 10 mM tris-HCl (pH 7.4), 1% Triton X-100, 0.5% NP-40, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1 mM sodium orthovanadate, 10 mM sodium fluoride, and 1× EDTA-free protease inhibitor cocktail (Roche cOfmplete). Lysates were cleared by centrifugation at 13,000 rpm for 10 min at 4°C. Total protein was measured by BCA (Pierce). Equal amounts of protein were separated on 5 to 15% gradient SDS–polyacrylamide gel electrophoresis gels and immunoblotted with phosphorylated SMAD1/5/8 antibody (Cell Signaling Technology). Antiphosphotyrosine antibody (4G10) was used for immunoprecipitations of phosphorylated MuSK (Millipore). Membranes were then stripped and reprobed with SMAD1/5/8 antibody (Cell Signaling Technology) or MuSK antibody for phospho-MuSK immunoprecipitation (R&D Systems).
ELISA assays
For MuSK binding to BMP4, recombinant MuSK proteins (FL-ecto-MuSK and ΔIg3-ecto-MuSK) were immobilized on 96-well plates at 2 µg/ml overnight. Plates were blocked with 1% BSA in PBS and incubated with BMP4 (0 to 200 nM). Bound BMP4 was detected with biotinylated anti-BMP4 antibody (R&D Systems) followed by streptavidin-conjugated HRP (Thermo). Graphs were generated with absorbance values, each data point representing the average of three independent ELISAs with four replicate wells for all data points in each independent assay. For MuSK binding to biglycan, His-tagged nonglycanated biglycan (54) was immobilized, and MuSK ectodomain Fc-fusion proteins (full-length MuSK and ΔIg3 MuSK) were incubated with immobilized biglycan. Bound MuSK was detected with HRP-conjugated human Fc antibody. For MuSK binding to ALK6, His-tagged MuSK was immobilized and Fc-fusion ALK3, ALK6, and TROY proteins were incubated with immobilized MuSK. Bound receptors were detected with HRP-conjugated and Fc-specific anti-human IgG antibody (KPL).
RNA extraction, reverse transcription, and qRT-PCR
Total RNA was isolated from cells with TRIzol (Invitrogen), cleaned up and deoxyribonuclease-treated in Qiagen RNeasy columns, and reverse-transcribed into first-strand complementary DNA (cDNA) (Invitrogen). The qRT-PCR reaction consisted of initial incubation at 50°C for 2 min and denaturation at 95°C for 5 min. The cycling parameters were as follows: 95°C for 15 s and 60°C for 30 s. After 40 cycles, the reactions underwent a final dissociation cycle as follows: 95°C for 15 s, 60°C for 1 min, 95°C for 15 s, and 60°C for 15 s. Expression of each gene was normalized to 18.S rRNA expression. The primer sequences used in qRT-PCR reactions were as follows: 5′-AGGAGTGGGCCTGCGGCTTA-3′ and 5′-AACGGCCATGCACCACCACC-3′ for mouse 18S rRNA; 5′-GGG-ATCTCTGGGAAAGACAC-3′ and 5′-TCTCTGGAGGCTGAAAGGTG-3′ for mouse Id1; 5′-GTATTTCCATCGCTCCTTGG-3′ and 5′-TGA-GGCCTATAAAGCACATGG-3′ for mouse Rgs4; 5′-TCTTCGGGCAA-GAAACTCTG-3′ and 5′-TTGCATGTGACTGCTTCTCC-3′ for mouse Car3; 5′-CAGGCACACTTCTCCTTTCC-3′ and 5′-CCTTCCTCATCATG-GACCAG-3′ for mouse Myh15; 5′-CATCAGCTGGATTGAAAACG-3′ and 5′-CAGCCTTTGCGGTACTGAAC-3′ for mouse MuSK; 5′-TCC-TCTCTGTTGCGTGTGTC-3′ and 5′-CGTTAAGCAACAGGACATGC-3′ for mouse Ptger4; 5′-CGCTGATTGGGTTTTCGTAG-3′ and 5′-CCTGA-GCTGAGGTTTTCCTG-3′ for mouse Ptgs2; 5′-CTTTGGGGATAT-CGTTGCTG-3′ and 5′-GCTGGCTAACTCTGGGACT-3′ for mouse Fabp7.
Microarrays and bioinformatic analysis
Total RNA from cultures of wild-type and MuSK−/− myoblasts and myotubes treated with BMP4 (25 ng/ml) for 8 hours was prepared by the TRIzol extraction method (Invitrogen). The quality of the input RNA (150 to 200 ng) for the microarrays was checked by a bioanalyzer [RNA integrity number (RIN) scores of >9]. Total RNA was converted to double-stranded cDNA and then in vitro-transcribed overnight using the Whole Transcript expression kit from Invitrogen (catalog #4411981). After cleanup, 10 µg of in vitro-transcribed cRNA was converted to deoxyuridine triphosphate-labeled cDNA, and 5.5 µg of the generated single-stranded cDNA was enzymatically fragmented followed by TdT-mediated biotin end-labeling using the Whole Transcript terminal labeling kit (Affymetrix, catalog #900670). Successful fragmentation (~75 nucleotides) was demonstrated on the bioanalyzer with RIN scores of 2.6. About 2.5 µg of cDNA was hybridized at 45°C to Affymetrix Mouse 1.0 Gene ST chip (catalog #901168). The arrays were washed and stained following the Affymetrix standard protocol and scanned on an Affymetrix 3000 7G scanner.
The Affymetrix Expression console was used to analyze the overall performance and quality of the arrays, and Partek Genomics Suite was used to detect differentially expressed genes. To identify genes differentially expressed between samples, we used a combination of two selection criteria: ±1.5-fold change and FDR-adjusted P < 0.05, such that the FDR was controlled at 5%. FDR amounts to a statistical adjustment to allow for multiple testing, whereas the fold change cutoff serves as an additional filter to identify biologically meaningful hits. GO analysis was performed using the DAVID (Database for Annotation, Visualization, and Integrated Discovery) database. (55)
Surface plasmon resonance
The binding affinities and kinetic parameters between BMPs and MuSK were determined by SPR spectroscopy using the BIAcore3000 optical biosensor instrument (GE Healthcare Life Sciences). The carboxymethylated surface of the sensor chip (CM5) was activated with N-hydroxysuccinimide and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride. The CM5 chip contains four flow cells, and among these four cells, three were used for the assay. Flow cell 1 was used as a control surface, whereas flow cells 2, 3, and 4 were used as test surfaces. Recombinant human BMP2 (2466 RU), BMP4 (2108 RU), and BMP7 (2050 RU) were covalently coupled in flow cells 2, 3, and 4, respectively. Unreacted active ester groups were blocked with 1 M ethanolamine hydrochloride (pH 8.5). The control flow cell 1 was treated in an identical manner but without coupling protein. The binding assays were carried out at 25°C in 20 mM Hepes buffer (pH 7.5), 500 mM NaCl, 3.4 mM EDTA, and 0.005% surfactant P-20. Various concentrations of MuSK were applied over the biosensor chip at a flow rate of 20 µl/min for 360 s to measure the association phase followed by buffer only for 600 s to measure the dissociation phase. The sensor chip was regenerated with four short pulses of guanidine hydrochloride (2 M) at 100 µl/min. Data were evaluated using the software BIAevaluation 4.1.1 (BIAcore AB). Because primary amine coupling can result in a heterogeneous population of BMP ligand with certain molecules being affected by the coupling process, SPR sensorgrams were globally analyzed using a distribution model for continuous affinity and rate constant analysis (kon and koff) with the program EVILFIT (56). This model helps account for BMP molecules that have reduced affinity due to the coupling procedure.
Calcium imaging
H-2Kb-tsA58 myoblasts plated on glass coverslips and pretreated with BMP4 (25 ng/ml) or vehicle for 4 hours were incubated in the dark for 20 min in extracellular solution with 2 µM Fluo-4 AM (Molecular Probes/ Life Technologies) and 250 µM sulfinpyrazone (uridine 5′-diphospho-glucuronosyltransferase inhibitor, Sigma-Aldrich) to prevent the loss of Fluo-4 from cells. Coverslips were then transferred to the imaging chamber, and time-lapse fluorescence images were acquired every 2 s using MetaMorph software (Molecular Devices). The M1 receptor agonist carbachol (50 µM) was added after acquiring 25 baseline images (50 s), followed by addition of ionomycin (2 µM) to elicit maximal Fluo-4 fluorescence, used for normalization. The RGS4 inhibitor 11b (CCG-203769, 3 µM) was added together with Fluo-4 AM and sulfinpyrazone for 20 min.
For each Ca 2+ imaging experiment, the fluorescence intensity of more than seven individual cells from each coverslip was measured as a function of time and was averaged. Fluorescence intensities were quantified and normalized as Fnorm(t) = (Fcell(t) - Fmin)/Fiono- Fmin), where Fcell is the fluorescence of an intracellular region of interest (>25% of total cell area), Fiono is the maximal fluorescence with ionomycin, and Fmin is the baseline fluorescence before stimulation. Carbachol-induced changes in fluorescence intensity were quantified using MetaMorph (Molecular Devices), MATLAB (MathWorks), and Excel software (Microsoft).
Statistical analysis
The average values of the replicate experiments are given as means ± SD/ SEM. Statistical differences among the experimental groups were analyzed by one-way ANOVA for multiple comparisons and by unpaired, two-sided Student’s t test when comparing two experimental groups. To prevent the identification of false-positives due to increased number of comparisons, ANOVA results were corrected by the conservative Bonferroni post hoc test. Significance was defined as P < 0.05 (***P < 0.0001; **P < 0.01; *P < 0.05).
Supplementary Material
Acknowledgments
We are grateful to D. B. Rifkin at New York University for providing the C2C12BRA reporter line, M. Ruegg at the University of Basel for the His-tagged MuSK construct, R. Herbst at the Medical University of Vienna for providing mutant MuSK rescue lines, and R. Neubig at Michigan State University for the 11b compound. We thank M. Dechene for the purification of the His-tagged MuSK construct. We are also grateful to B. McKechnie for technical assistance and Z. Wu at Brown University for advice on statistical analyses.
Funding: The research was supported by NIH grants HD23924 and NS064295 (J.R.F.), GM084186 and GM114640 (T.B.T.), and a fellowship from The Suna and Inan Kirac Foundation to R.N.O. The Genomics Core Facility at Brown University has received partial support from the NIH (National Institute of General Medical Sciences grant number P30GM103410 and National Center for Research Resources grant numbers P30RR031153, P20RR018728, and S10RR02763), NSF (Experimental Program to Stimulate Competitive Research grant number 0554548), Lifespan Rhode Island Hospital, and the Division of Biology and Medicine, Brown University.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/9/444/ra87/DC1
Fig. S1. SPR binding analysis of MuSK with BMP2 and BMP7.
Fig. S2. Soluble MuSK ectodomain inhibits BMP4 activity.
Fig. S3. Equivalent binding of biglycan to MuSK ectodomain either containing (FL) or lacking (Δlg3) the third Ig domain.
Fig. S4. BMP4 treatment stimulates Rgs4 expression in wild-type but not MuSK−/− myoblasts.
Fig. S5. MuSK favors the transcription of signaling- and transcription-related genes.
Fig. S6. Abundant phosphorylated SMAD1/5/8 in cytosolic granules in wild-type but not MuSK−/− myoblasts.
Table S1. Association and dissociation rate constants and overall Kd values for the interaction of MuSK with BMP2, BMP4, and BMP7, as determined by SPR.
Table S2. Transcripts induced by BMP4 only in wild-type myoblasts.
Table S3. Transcripts induced by BMP4 in both wild-type and MuSK−/− myoblasts.
Table S4. Transcripts induced by BMP4 only in MuSK−/− myoblasts.
Table S5. Transcripts induced by BMP4 only in wild-type myotubes.
Table S6. Transcripts induced by BMP4 in both wild-type and MuSK−/− myotubes.
Table S7. Transcripts induced by BMP4 only in MuSK−/− myotubes.
Table S8. Transcripts induced by BMP4 in a MuSK-dependent manner in both myoblasts and myotubes.
Table S9. Transcripts induced by BMP4 in a MuSK-dependent manner only in myoblasts.
Table S10. Transcripts induced by BMP4 in a MuSK-dependent manner only in myotubes.
Author contributions: A.Y. and J.R.F. conceived the study. A.Y., T.B.T., E.O., and J.R.F. designed the experiments. A.Y., C.K., C. Schmiedel, R.N.O., S.M., and C. Schorl, conducted the experiments. A.Y., C.K., C. Schmiedel, R.N.O., C. Schorl, E.O., T.B.T., and J.R.F. analyzed the data. A.Y. and J.R.F. wrote the paper with input from all the authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Microarray expression data files can be obtained from the NIH Gene Expression Omnibus with accession number GSE59567.
REFERENCES AND NOTES
- 1.Wakefield LM, Hill CS. Beyond TGFβ: Roles of other TGFβ superfamily members in cancer. Nat Rev. Cancer. 2013;13:328–341. doi: 10.1038/nrc3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bragdon B, Moseychuk O, Saldanha S, King D, Julian J, Nohe A. Bone morpho-genetic proteins: A critical review. Cell. Signal. 2011;23:609–620. doi: 10.1016/j.cellsig.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Conidi A, Cazzola S, Beets K, Coddens K, Collart C, Cornells F, L Cox, Joke D, Dobreva MP, Dries R, Esguerra C, Francis A, Ibrahimi A, Kroes R, Lesage F, Maas E, Moya I, Pereira PNG, Stappers E, Stryjewska A, van den Berghe V, Vermeire L, Verstappen G, Seuntjens E, Umans L, Zwijsen A, Huylebroeck D. Few Smad proteins and many Smad-interacting proteins yield multiple functions and action modes in TGFβ/BMP signaling in vivo. Cytokine Growth Factor Rev. 2011;22:287–300. doi: 10.1016/j.cytogfr.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Korchynskyi O, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 5.Heldin C-H, Moustakas A. Role of Smads in TGFβ signaling. Cell Tissue Res. 2012;347:21–36. doi: 10.1007/s00441-011-1190-x. [DOI] [PubMed] [Google Scholar]
- 6.Miyama K, Yamada G, Yamamoto TS, Takagi C, Miyado K, Sakai M, Ueno N, Shibuya H. A BMP-inducible gene, dlx5, regulates osteoblast differentiation and mesoderm induction. Dev. Biol. 1999;208:123–133. doi: 10.1006/dbio.1998.9197. [DOI] [PubMed] [Google Scholar]
- 7.Ono Y, Calhabeu F, Morgan JE, Katagiri T, Amthor H, Zammit PS. BMP signalling permits population expansion by preventing premature myogenic differentiation in muscle satellite cells. Cell Death Differ. 2011;18:222–234. doi: 10.1038/cdd.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nolan K, Thompson TB. The DAN family: Modulators of TGF-β signaling and beyond. Protein Sci. 2014;23:999–1012. doi: 10.1002/pro.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corradini E, Babitt JL, Lin HY. The RGM/DRAGON family of BMP co-receptors. Cytokine Growth Factor Rev. 2009;20:389–398. doi: 10.1016/j.cytogfr.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sammar M, Stricker S, Schwabe GC, Sieber C, Hartung A, Hanke M, Oishi I, Pohl J, Minami Y, Sebald W, Mundlos S, Knaus P. Modulation of GDF5/BRI-b signalling through interaction with the tyrosine kinase receptor Ror2. Genes Cells. 2004;9:1227–1238. doi: 10.1111/j.1365-2443.2004.00799.x. [DOI] [PubMed] [Google Scholar]
- 11.Hagihara M, Endo M, Hata K, Higuchi C, Takaoka K, Yoshikawa H, Yamashita T. Neogenin, a receptor for bone morphogenetic proteins. J. Biol. Chem. 2011;286:5157–5165. doi: 10.1074/jbc.M110.180919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassel S, Yakymovych M, Hellman U, Rönnstrand L, Knaus P, Souchelnytskyi S. Interaction and functional cooperation between the serine/threonine kinase bone morphogenetic protein type II receptor with the tyrosine kinase stem cell factor receptor. J. Cell. Physiol. 2006;206:457–467. doi: 10.1002/jcp.20480. [DOI] [PubMed] [Google Scholar]
- 13.Hubbard SR, Gnanasambandan K. Structure and activation of MuSK, a receptor tyrosine kinase central to neuromuscular junction formation. Biochim. Biophys. Acta. 2013;1834:2166–2169. doi: 10.1016/j.bbapap.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghazanfari N, Fernandez KJ, Murata Y, Morsch M, Ngo ST, Reddel SW, Noakes PG, Phillips WD. Muscle specific kinase: Organiser of synaptic membrane domains. Int. J. Biochem. Cell Biol. 2011;43:295–298. doi: 10.1016/j.biocel.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Singhal N, Martin PT. Role of extracellular matrix proteins and their receptors in the development of the vertebrate neuromuscular junction. Dev. Neurobiol. 2011;71:982–1005. doi: 10.1002/dneu.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowen DC, Park JS, Bodine S, Stark JL, Valenzuela DM, Stitt TN, Yancopoulos GD, Lindsay RM, Glass DJ, DiStefano PS. Localization and regulation of MuSK at the neuromuscular junction. Dev. Biol. 1998;199:309–319. doi: 10.1006/dbio.1998.8936. [DOI] [PubMed] [Google Scholar]
- 17.Punga AR, Maj M, Lin S, Meinen S, Rüegg MA. MuSK levels differ between adult skeletal muscles and influence postsynaptic plasticity. Eur. J. Neurosci. 2011;33:890–898. doi: 10.1111/j.1460-9568.2010.07569.x. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Osta A, Tsokas P, Pollonini G, Landau EM, Blitzer R, Alberini CM. MuSK expressed in the brain mediates cholinergic responses, synaptic plasticity, and memory formation. J. Neurosci. 2006;26:7919–7932. doi: 10.1523/JNEUROSCI.1674-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukada S-i, Uezumi A, Ikemoto M, Masuda S, Segawa M, Tanimura N, Yamamoto H, Miyagoe-Suzuki Y, Takeda S. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- 20.Zilberberg L, ten Dijke P, Sakai LY, Rifkin DB. A rapid and sensitive bioassay to measure bone morphogenetic protein activity. BMC Cell Biol. 2007;8:41. doi: 10.1186/1471-2121-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinecke K, Seher A, Schmitz W, Mueller TD, Sebald W, Nickel J. Receptor oligomerization and beyond: A case study in bone morphogenetic proteins. BMC Biol. 2009;7:59. doi: 10.1186/1741-7007-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki A, Kaneko E, Maeda J, Ueno N. Mesoderm induction by BMP-4 and −7 heterodimers. Biochem. Biophys. ResCommun. 1997;232:153–156. doi: 10.1006/bbrc.1997.6219. [DOI] [PubMed] [Google Scholar]
- 23.von Bubnoff A, Cho KW. Intracellular BMP signaling regulation in vertebrates: Pathway or network? Dev. Biol. 2001;239:1–14. doi: 10.1006/dbio.2001.0388. [DOI] [PubMed] [Google Scholar]
- 24.Hesser BA, Sander A, Witzemann V. Identification and characterization of a novel splice variant of MuSK. FEBS Lett. 1999;442:133–137. doi: 10.1016/s0014-5793(98)01641-x. [DOI] [PubMed] [Google Scholar]
- 25.Amenta AR, Creely HE, Mercado MLT, Hagiwara H, McKechnie BA, Lechner BE, Rossi SG, Wang Q, Owens RT, Marrero E, Mei L, Hoch W, Young MF, McQuillan DJ, L Rotundo R, Fallon JR. Biglycan is an extracellular MuSK binding protein important for synapse stability. J. Neurosci. 2012;32:2324–2334. doi: 10.1523/JNEUROSCI.4610-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desjardins PR, Burkman JM, Shrager JB, Allmond LA, Stedman HH. Evolutionary implications of three novel members of the human sarcomeric myosin heavy chain gene family. Mol. Biol. Evol. 2002;19:375–393. doi: 10.1093/oxfordjournals.molbev.a004093. [DOI] [PubMed] [Google Scholar]
- 27.Lyons GE, Buckingham ME, Tweedie S, Edwards YH. Carbonic anhydrase III, an early mesodermal marker, is expressed in embryonic mouse skeletal muscle and notochord. Development. 1991;11:233–244. doi: 10.1242/dev.111.1.233. [DOI] [PubMed] [Google Scholar]
- 28.Mazhar S, Herbst R. The formation of complex acetylcholine receptor clusters requires MuSK kinase activity and structural information from the MuSK extracellular domain. Mot. Celt. Neurosci. 2012;49:475–486. doi: 10.1016/j.mcn.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herbst R, Burden SJ. The juxtamembrane region of MuSK has a critical role in agrin-mediated signaling. EMBO J. 2000;19:67–77. doi: 10.1093/emboj/19.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogt J, Traynor R, Sapkota GP. The specificities of small molecule inhibitors of the TGFβ and BMP pathways. Cell. Signal. 2011;23:1831–1842. doi: 10.1016/j.cellsig.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 31.Hollinger S, Hepler JR. Cellular regulation of RGS proteins: Modulators and integrators of G protein signaling. Pharmacol. Rev. 2002;54:527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- 32.Berchtold MW, Brinkmeier H, Müntener M. Calcium ion in skeletal muscle: Its crucial role for muscle function, plasticity and disease. Physiol. Rev. 2000;80:1215–1265. doi: 10.1152/physrev.2000.80.3.1215. [DOI] [PubMed] [Google Scholar]
- 33.Mayerhofer A, Föhr KJ, Sterzik K, Gratzl M. Carbachol increases intracellular free calcium concentrations in human granulosa-lutein cells. J. Endocrinol. 1992;135:153–159. doi: 10.1677/joe.0.1350153. [DOI] [PubMed] [Google Scholar]
- 34.Blazer LL, Storaska AJ, Jutkiewicz EM, Turner EM, Calcagno M, Wade SM, Wang Q, Huang X-P, Traynor JR, Husbands SM, Morari M, Neubig RR. Selectivity and anti-Parkinson’s potential of thiadiazolidinone RGS4 inhibitors. ACS Chem. Neurosci. 2015;6:911–919. doi: 10.1021/acschemneuro.5b00063. [DOI] [PubMed] [Google Scholar]
- 35.Samad TA, Rebbapragada A, Bell E, Zhang Y, Sidis Y, Jeong S-J, Campagna JA, Perusini S, Fabrizio DA, Schneyer AL, Lin HY, Brivanlou AH, Attisano L, Woolf CJ. DRAGON, a bone morphogenetic protein co-receptor. J. Biol. Chem. 2005;280:14122–14129. doi: 10.1074/jbc.M410034200. [DOI] [PubMed] [Google Scholar]
- 36.Rebbapragada A, Benchabane H, Wrana JL, Celeste AJ, Attisano L. Myostatin signals through a transforming growth factor β-like signaling pathway to block adipo-genesis. Mol. Cell. Biol. 2003;23:7230–7242. doi: 10.1128/MCB.23.20.7230-7242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raz R, Strieker S, Gazzerro E, L Clor J, Witte F, Nistala H, Zabski S, Pereira RC, Stadmeyer L, Wang X, Gowen L, Sleeman MW, Yancopoulos GD, Canalis E, Mundlos S, Valenzuela DM, Economides AN. The mutation R0R2W749X, linked to human BDB, is a recessive mutation in the mouse, causing brachydactyly, mediating patterning of joints and modeling recessive Robinow syndrome. Development. 2008;135:1713–1723. doi: 10.1242/dev.015149. [DOI] [PubMed] [Google Scholar]
- 38.Sammar M, Sieber C, Knaus P. Biochemical and functional characterization of the Ror2/BRIb receptor complex.. Biochem. Biophys. Res. Commun. 2009;381:1–6. doi: 10.1016/j.bbrc.2008.12.162. [DOI] [PubMed] [Google Scholar]
- 39.Wallace BG. Regulation of agrin-induced acetylcholine receptor aggregation by Ca++ and phorbol ester. J. Cell Biol. 1988;107:267–278. doi: 10.1083/jcb.107.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou H, Glass DJ, Yancopoulos GD, Sanes JR. Distinct domains of MuSK mediate its abilities to induce and to associate with postsynaptic specializations. J. Cell Biol. 1999;146:1133–1146. doi: 10.1083/jcb.146.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu H, Xiong WC, L Mei. To build a synapse: Signaling pathways in neuromuscular junction assembly. Development. 2010;137:1017–1033. doi: 10.1242/dev.038711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCabe BD, Marqués G, Haghighi AP, Fetter RD, Crotty ML, Haerry TE, Goodman CS, O’Connor MB. The BMP homolog Gbb provides a retrograde signal that regulates synaptic growth at the Drosophila neuromuscular junction. Neuron. 2003;39:241–254. doi: 10.1016/s0896-6273(03)00426-4. [DOI] [PubMed] [Google Scholar]
- 43.Sartori R, Schirwis E, Blaauw B, Bortolanza S, Zhao J, Enzo E, Stantzou A, Mouisel E, Toniolo L, Ferry A, Strieker S, Goldberg LA, Dupont S, Piccolo S, Amthor H, Sandri M. BMP signaling controls muscle mass. Nat. Genet. 2013;45:1309–1318. doi: 10.1038/ng.2772. [DOI] [PubMed] [Google Scholar]
- 44.Winbanks CE, Chen JL, Qian H, Liu Y, Bernardo BC, Beyer C, Watt KI, Thompson RE, Connor T, Turner BJ, McMullen JR, Larsson L, McGee SL, Harrison CA, Gregorevic P. The bone morphogenetic protein axis is a positive regulator of skeletal muscle mass. J. Cell Biol. 2013;203:345–357. doi: 10.1083/jcb.201211134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sartori R, Gregorevic P, Sandri M. TGFβ and BMP signaling in skeletal muscle: Potential significance for muscle-related disease. Trends Endocrinol. Metab. 2014;25:464–471. doi: 10.1016/j.tem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Valenzuela DM, Stitt TN, DiStefano PS, Rojas E, Mattsson K, Compton DL, Nuñez L, Park JS, L Stark J, Gies DR, Thomas S, Le Beau MM, Fernald AA, Copeland NG, Jenkins NA, Burden SJ, Glass DJ, Yancopoulos GD. Receptor tyrosine kinase specific for the skeletal muscle lineage: Expression in embryonic muscle, at the neuromuscular junction, and after injury. Neuron. 1995;15:573–584. doi: 10.1016/0896-6273(95)90146-9. [DOI] [PubMed] [Google Scholar]
- 47.Otis JS, Burkholder TJ, Pavlath GK. Stretch-induced myoblast proliferation is dependent on the COX2 pathway. Exp. Cell Res. 2005;310:417–425. doi: 10.1016/j.yexcr.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 48.Bondesen BA, Mills ST, Pavlath GK. The COX-2 pathway regulates growth of atrophied muscle via multiple mechanisms. Am. J. Physiol. Cell Physiol. 2006;290:C1651–C1659. doi: 10.1152/ajpcell.00518.2005. [DOI] [PubMed] [Google Scholar]
- 49.Horsley V, Pavlath GK. Prostaglandin F2α stimulates growth of skeletal muscle cells via an NFATC2-dependent pathway. J. Cell Biol. 2003;161:111–118. doi: 10.1083/jcb.200208085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yilmaz A, Engeler R, Constantinescu S, Kokkaliaris KD, Dimitrakopoulos C, Schroeder T, Beerenwinkel N, Paro R. Ectopic expression of Msx2 in mammalian myo-tubes recapitulates aspects of amphibian muscle dedifferentiation. Stem Cell Res. 2015;15:542–553. doi: 10.1016/j.scr.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 51.Tamirisa P, Blumer KJ, Muslin AJ. RGS4 inhibits G-protein signaling in cardio-myocytes. Circulation. 1999;99:441–447. doi: 10.1161/01.cir.99.3.441. [DOI] [PubMed] [Google Scholar]
- 52.Schiaffino S, Sandri M, Murgia M. In: Skeletal Muscle Plasticity in Health and Disease: From Genes to Whole Muscle. Bottinelli R, Reggiani C, editors. Dordrecht, The Netherlands: Springer; 2007. pp. 91–119. [Google Scholar]
- 53.Morgan JE, Beauchamp JR, Pagel CN, Peckham M, Ataliotis P, Jat PS, Noble MD, Farmer K, Partridge TA. Myogenic cell lines derived from transgenic mice carrying a thermolabile T antigen: A model system for the derivation of tissue-specific and mutation-specific cell lines. Dev. Biol. 1994;162:486–498. doi: 10.1006/dbio.1994.1103. [DOI] [PubMed] [Google Scholar]
- 54.Amenta AR, Yilmaz A, Bogdanovich S, McKechnie BA, Abedi M, Khurana TS, Fallon JR. Biglycan recruits utrophin to the sarcolemma and counters dystrophic pathology in mdx mice. Proc. Natl. Acad. Sci. U.S.A. 2011;108:762–767. doi: 10.1073/pnas.1013067108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 56.Svitel J, Balbo A, Mariuzza RA, Gonzales NR, Schuck P. Combined affinity and rate constant distributions of ligand populations from experimental surface binding kinetics and equilibria. Biophys. J. 2003;84:4062–4077. doi: 10.1016/S0006-3495(03)75132-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.