Abstract
Decades of anatomical studies on the primate primary visual cortex (V1) have led to a detailed diagram of V1 intrinsic circuitry, but this diagram lacks information about the output targets of V1 cells. Understanding how V1 local processing relates to downstream processing requires identification of neuronal populations defined by their output targets. In primates, V1 layers (L)2/3 and 4B send segregated projections to distinct cytochrome oxidase (CO) stripes in area V2: neurons in CO blob columns project to thin stripes while neurons outside blob columns project to thick and pale stripes, suggesting functional specialization of V1-to-V2 CO streams. However, the conventional diagram of V1 shows all L4B neurons, regardless of their soma location in blob or interblob columns, as projecting selectively to CO blobs in L2/3, suggesting convergence of blob/interblob information in L2/3 blobs and, possibly, some V2 stripes. However, it is unclear whether all L4B projection neurons show similar local circuitries. Using viral-mediated circuit tracing, we have identified the local circuits of L4B neurons projecting to V2 thick stripes in macaque. Consistent with previous studies, we found the somata of this L4B subpopulation to reside predominantly outside blob columns; however, unlike previous descriptions of local L4B circuits, these cells consistently projected outside CO blob columns in all layers. Thus, the local circuits of these L4B output neurons, just like their extrinsic projections to V2, preserve CO streams. Moreover, the intra-V1 laminar patterns of axonal projections identify two distinct neuron classes within this L4B subpopulation, including a rare novel neuron type, suggestive of two functionally specialized output channels.
SIGNIFICANCE STATEMENT Conventional diagrams of primate primary visual cortex (V1) depict neuronal connections within and between different V1 layers, but lack information about the cells' downstream targets. This information is critical to understanding how local processing in V1 relates to downstream processing. We have identified the local circuits of a population of cells in V1 layer (L)4B that project to area V2. These cells' local circuits differ from classical descriptions of L4B circuits in both the laminar and functional compartments targeted by their axons, and identify two neuron classes. Our results demonstrate that both local intra-V1 and extrinsic V1-to-V2 connections of L4B neurons preserve CO-stream segregation, suggesting that across-stream integration occurs downstream of V1, and that output targets dictate local V1 circuitry.
Keywords: cell types, connections, cytochrome oxidase, G-deleted rabies virus, primary visual cortex, primate
Introduction
Cortical computations emerge from the activity of specific neural circuits. Therefore, knowledge of the anatomical diversity of cortical neurons and their specific wiring is critical for a mechanistic understanding of cortical computations. Decades of research on the anatomy of the primate primary visual cortex (V1) have led to a detailed diagram of V1 local circuits (Lund et al., 1994, 2003; Callaway, 2014). However, these diagrams were derived from labeling and reconstruction of V1 neurons without any knowledge of their long-distance targets. This is because labeling of V1 cells in previous studies was achieved by bulk injections of traditional neuroanatomical tracers into V1 in vivo (Blasdel et al., 1985; Lachica et al., 1992; Yoshioka et al., 1994), random intracellular fills in slices (Callaway and Wiser, 1996; Wiser and Callaway, 1996), or Golgi staining (Lund, 1973; Lund and Boothe, 1975). However, V1 sends projections to multiple cortical and subcortical targets, and most excitatory V1 neurons project outside V1. Therefore, a comprehensive understanding of how the local V1 circuitry relates to downstream processing requires identification of neuronal populations defined by their projection targets. Indeed, studies of the mouse visual system have revealed that, even within the same V1 layer, neurons that project to different targets can be morphologically and functionally distinct, and belong to unique local and long-range cortical microcircuits (Glickfeld et al., 2013; Vélez-Fort et al., 2014; Kim et al., 2015).
In primate V1, excitatory neurons in layers (L) 2/3 and L4B send segregated projections to distinct cytochrome oxidase (CO) stripes in area V2, with thin stripes receiving projections from neurons whose somata reside primarily inside CO blob columns, and thick and pale stripes from neurons residing primarily outside blob columns (Sincich et al., 2007, 2010; Federer et al., 2009, 2013). This anatomical segregation suggests parallel processing of specific stimulus attributes by different V1-to-V2 CO streams. Classical diagrams of V1 for all L4B excitatory neurons depict a single axonal branching motif consisting of projections to both supragranular (L2/3 and L4B) and infragranular (mainly L5) layers (Callaway and Wiser, 1996), with projections to L2/3 selectively targeting CO blobs, regardless of the L4B cells' soma location inside or outside blob columns (Lachica et al., 1992; Yoshioka et al., 1994; Callaway and Wiser, 1996). Projections from L4B interblobs to L2/3 blobs suggest convergence of CO streams in L2/3 blobs and, possibly, in the V2 stripes receiving inputs from V1 blobs. However, it is unclear whether a stereotyped axonal branching motif is seen for all L4B cells regardless of their downstream targets. Answering this question requires labeling and reconstructing the local V1 circuitry of L4B cells identified by their V2 stripe target.
The recent emergence of viral-mediated fluorescent labeling methods has enabled researchers to label projection neurons fully and at high resolution, revealing the neurons' morphology and local axon projections (Luo et al., 2008). In this study, to understand how local V1 circuits of L4B cells relate to their target V2 stripe type, we have used a green fluorescent protein (GFP)-expressing G-deleted rabies virus (GdRV; Nassi and Callaway, 2007; Wickersham et al., 2007; Briggs et al., 2016) to obtain Golgi-like fills of L4B neurons that specifically project to the thick CO stripes of V2, and have fully reconstructed these cells' dendritic and axonal processes. We find morphological evidence for at least two distinct classes of neurons, including a novel neuron type not previously described. Both cell classes differ from previous descriptions of L4B neurons in two important ways. First, while only Class 1 cells project to supragranular and infragranular layers, as previously described, the rare Class 2 cells mainly project to L5 and do not project to L2/3. This suggests two specialized L4B output channels related to the V2 thick stripes. Second, the somata of both cell classes reside predominantly outside blob columns, and their axons exclusively project outside blob columns. This demonstrates that thick-stripe projecting L4B neurons largely preserve CO-stream segregation in both their intra-V1 and interareal V1-to-V2 connectivity.
Materials and Methods
To fully label the axon and dendrites of L4B neurons projecting to the V2 thick stripes, we targeted injections of GdRV expressing the gene for enhanced green fluorescent protein (EGFP), GdRV-EGFP (based on the SAD-B19 strain), to the thick V2 stripes, identified in vivo using intrinsic signal optical imaging. The modified virus lacks the glycoprotein necessary for trans-synaptic infection. Therefore, it effectively behaves as a retrograde tracer, infecting cells at their synapses and producing vast amounts of GFP, yielding Golgi-like labeling of the cell soma, dendritic, and axonal processes (Wickersham et al., 2007). Labeled L4B cells and their processes in V1 were reconstructed through serial sections, and their distribution within V1 layers and CO compartments in V1 were analyzed quantitatively.
Animals
We made a total of eight injections of GdRV-EGFP into six animals, and selected for neuronal reconstruction and analysis only those cases for which the viral injection was confined to a V2 thick stripe and produced high-quality labeling of L4B neurons, namely three injections from three adult male macaque monkeys (Macaca fascicularis). Two of these animals received a single injection of GdRV-EGFP in dorsal V2 of one hemisphere, while the third animal received two such injections separated by 7 mm (one of these two injections was in a pale stripe; therefore, it was not included in this study). Injections were targeted at the exposed portion of dorsal V2, posterior to the lunate sulcus, guided by optical imaging. Additional injections of different tracers were made in the same or opposite hemisphere of the same animals for unrelated experiments.
Surgical and tracer injection procedures
All experimental procedures were in accordance with protocols approved by the University of Utah Institutional Animal Care and Use Committee. Surgical procedures were performed as described previously (Federer et al., 2013). Briefly, animals were preanesthetized with ketamine (25 mg/kg, i.m.), intubated, and artificially ventilated with a 70:30 mixture of N2O and O2. The head of each animal was fixed by positioning the animal in a stereotaxic apparatus. Isoflurane (1.0–2.5%) was used to maintain anesthesia, and depth of anesthesia was assessed continuously by monitoring end-tidal CO2, saturation of peripheral oxygen, electrocardiogram, blood pressure, withdrawal, and corneal reflexes. Temperature was also monitored and the animal was wrapped in a water blanket. To expose area V2 and the anterior half of V1 on the opercular surface, a large (∼15 × 10 mm) craniotomy and a durotomy were performed posterior to the lunate sulcus. On completion of the surgery, the craniotomy was filled with 3% agar and sealed using a glass coverslip glued to the skull. At this point isofluorane was turned off, anesthesia maintained with sufentanil citrate (5–10 μg/kg/h), and paralysis induced by continuous infusion of vecuronium bromide (0.3 mg/kg/h) to prevent eye movements. The pupils were dilated with topical atropine, the eyes were protected with gas-permeable contact lenses and refracted, and optical imaging was started. Once functional regions corresponding to the V2 stripes were identified (see below, Optical imaging), the glass coverslip was removed, and GdRV-EGFP (donated by Prof. E. Callaway or purchased from the Salk Institute Viral Vector Core) was pressure injected into V2 using a picospritzer and glass micropipettes of 35–45 μm tip diameter. Since our goal was to fully reconstruct axonal processes of labeled neurons, we needed to obtain sparse labeling of cells in V1. To accomplish this, we injected small volumes (375–500 nl) of the viral solution at 0.6 or 1 mm depth, over a 30–40 min period. The pipette was left in place for 5–10 min on completion of the injection before being retracted. On completion of the viral injections, the animals were kept under anesthesia for an additional 3–5 d, during which additional optical imaging was performed, and finally killed with sodium pentobarbital (150 mg/kg, i.v.), and perfused transcardially with saline for 3 min, followed by fixative.
Optical imaging
Intrinsic signal optical imaging was used to identify the thick V2 stripes. Specifically, imaging for ocular dominance enabled us to identify the V1–V2 border, imaging for orientation identified the thick and pale stripes (which have well defined orientation maps, unlike thin stripes; Fig. 1A; Federer et al., 2009), and imaging for color delineated the thin stripes (Lu and Roe, 2008). Acquisition of intrinsic signals was performed using the Imager 3001 and VDAQ software (Optical Imaging) under red-light illumination (630 nm). Orientation maps were obtained by presenting full-field, high-contrast (100%), achromatic drifting square-wave gratings of eight different orientations and 1.0–2.0 cycles/° spatial frequency, moving back and forth at 1.5–2.0 Hz in directions perpendicular to the grating orientation. Responses to the same oriented stimuli were averaged across trials, first-frame subtracted, and orientation difference images (Fig. 1A) were generated by subtracting responses to two orthogonal orientations. Imaging for color responses was performed by flashing red–green, blue–yellow, and black–white pairs of full field squares every 500 ms. Averaged color responses were then subtracted from the averaged response to the blank and the achromatic stimuli. Thick stripes were identified as the middle of the regions containing strong orientation signals and no color responses. In each case, reference images of the surface vasculature were taken under 546 nm illumination (green light); these images were later used to align the top histological tissue section to the optical maps obtained in vivo for postmortem identification of injection site location on CO-stained tissue.
Figure 1.
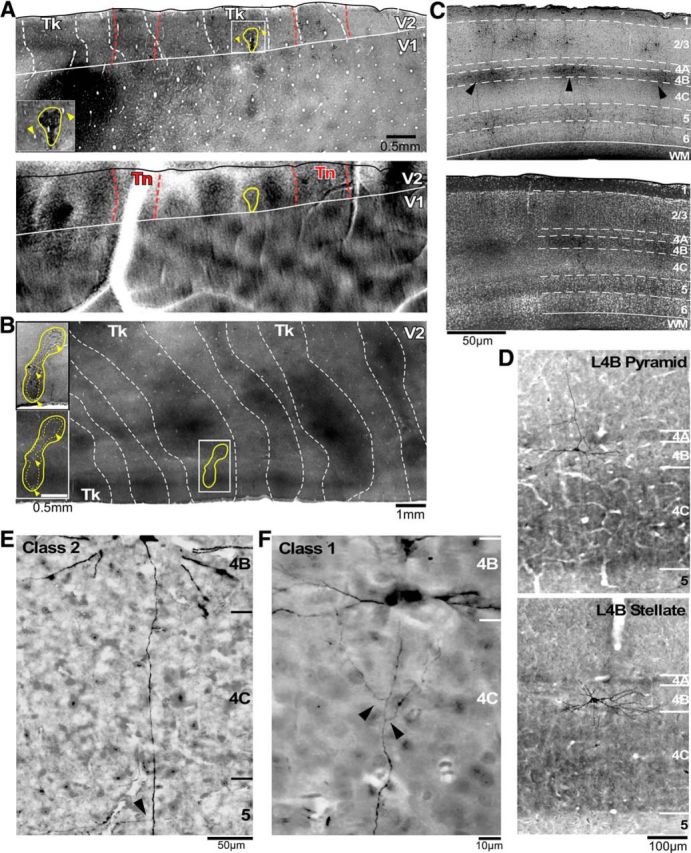
Injection sites and expression of GdRV-EGFP in V1 L4B neurons. A, top, Photograph of an example viral injection site (outlined in yellow) on a tangential section through V1 and V2 immunoreacted with DAB, to enhance GFP signals, and stained for CO, to reveal CO compartments and layers. The injection site is also shown at higher magnification in the inset. Arrowheads in the inset and top panel in A point at the same blood vessels, for referencing. The white solid contour outlines the V1–V2 border, the white dashed contours are the dark CO stripes, and the red dashed contours are the outlines of the thin stripes obtained from the optical images (bottom) and superimposed onto the CO-stained section. Registration of the optical images with the CO-stained sections was obtained by aligning in vivo images of the surface vasculature (taken under green light) with the surface vasculature in the most superficial tangential tissue section. Note the good correspondence of thin CO stripes with functional thin stripes defined by optical imaging. Tk, Thick stripes; Tn, thin stripes. Bottom, Differential orientation map generated by subtracting single condition maps in response to gratings of 45 and 135° orientation, with superimposed outline of the viral injection site (yellow) and the position of the V1–V2 border (white solid contour), both taken from the CO map in the top panel. Orientation responsive domains are present in both thick and pale stripes, but not thin stripes (outlined in red), and smaller orientation domains are also visible within V1. B, A viral injection site (dashed yellow lines, core of the injection site; solid yellow lines, halo of the injection site) in a different case. Top inset, Viral injection site shown on a section immunoreacted for DAB to enhance GFP signals. The yellow contour outlines the full extent of the injection site through all cortical layers. Bottom inset, Same field of view as in the top inset, but from an adjacent section stained for CO with the injection outline superimposed from the top inset. Arrowheads in the two insets point at same blood vessels in the two sections. The larger image shows a wider field of view at lower magnification of the same CO section as in the bottom inset, to reveal the CO stripe pattern. Note that the injection is confined to a thick stripe. C, top, Low-magnification photograph of pia-to-white matter section of V1 immunoreacted using DAB to reveal the laminar distribution of GFP-labeled V1 neurons resulting from a viral injection in a thick stripe. Arrowheads point at labeled L4B neurons. White dashed contours delineate borders between cortical layers (labeled on the right). Cortical layer boundaries were obtained from staining the same section with fluorescent Nissl (bottom). D, Photographs of example GFP-labeled pyramidal and spiny stellate L4B neurons projecting to thick stripes, shown on a pia-to-white matter section of V1 stained for CO (to reveal layers) and immunoreacted with DAB (to enhance GFP signals). E, F, High-power photographs of the two main morphological classes of thick-stripe-projecting L4B cells. The Class 2 cell shows only a single branch coming off the main axon in L5 (arrowhead), but no axonal branches in the layers above. In contrast, the Class 1 cell shows two branches (arrowheads) coming off each side of the main axon in L4C and rising upward toward the supragranular layers. This cell's axon also sent off branches in the infragranular layers (data not shown).
Histology
Brain fixation was obtained by transcardial perfusion with 0.5% paraformaldehyde (PFA) in 0.1 m phosphate buffer for 5 min in two animals, and 4% PFA for 15 min in the third animal. In the first two animals, V1 was dissected away from V2 by cutting along the V1–V2 border under a dissecting microscope. The V1 block was postfixed in 4% PFA for 1–2 h, immersed in 30% sucrose for cryoprotection, embedded in agar, and frozen-sectioned at 40 μm in a pia-to-white matter plane parallel to the V1–V2 border. This plane was chosen to best reveal V1 laminae. To best reveal the CO stripe pattern, the V2 blocks for these two cases were unfolded, flattened, postfixed between glass slides for 1–2 h, and sectioned tangentially to the pial surface at 40 μm. In the third animal, a block containing both V1 and V2 was gently flattened in a plane parallel to the imaging camera, postfixed between glass slides, and sectioned tangentially at 40 μm. In two cases, alternate tissue sections from the V1 block (one case) or the V1/V2 block (the other case) were stained for CO (Sigma-Aldrich) to reveal layers and CO compartments, and then immunoreacted to enhance GFP signals using DAB (Vector Laboratories, RRID:AB_2336382). All remaining sections were immunoreacted for GFP. In the third case, all V1 sections were immunoreacted for GFP and costained with fluorescent Nissl stain (Neuro Trace diluted 1:100; Invitrogen) to reveal layers. The sections from the separated V2 blocks were stained for CO and immunoreacted for GFP to reveal the stripe location of the injection sites. Digitized images of the CO stripes were taken on wet mounted sections before immunoreacting the same sections, and, in most cases, again after GFP immunohistochemistry. Immunohistochemistry to enhance and permanently stain GFP signals was performed by incubating sections for 48 h in primary antibody (1:2000 chicken anti-GFP; Aves Labs, RRID:AB_10000240), then for 1 h in 1:200 biotinylated donkey anti-chicken secondary IgG (Jackson Laboratories, RRID:AB_2313596), followed by a standard nickel-enhanced ABC-DAB reaction (yielding a black reaction product), using an ABC-avidin-peroxidase kit (Vector Laboratories, RRID:AB_2336382).
Data analysis
Stripe location of injection sites.
Viral injection sites were identified as a region of black glial cell label visible around the pipette track (Fig. 1A, inset, B, top inset), and their diameter in each section was estimated to extend to the nearest labeled neuronal somata (which would be labeled by retrograde transport, as the virus does not infect somata). A composite injection site was obtained by aligning, using radial blood vessels, the outlines of the injection site in each section throughout the depth of the cortex. To determine the location of these injection sites with respect to V2 stripes, CO staining was first digitized at low magnification; then, using IRtweak warping software (NCRtoolset, Scientific Computing and Imaging Institute, University of Utah, RRID:SCR_014639; Anderson et al., 2009), 3–6 images of CO sections were aligned using the radial blood vessels as reference, and then merged in Adobe Photoshop (using the multiply function). The V2 stripe pattern is much clearer in these composite images than in any single CO-stained section where parts of stripes may be missing (Federer et al., 2009, 2013). Finally, the composite outline of each injection site was aligned to the CO image stack. From these images, we selected the injection sites clearly confined to a thick stripe for further analysis.
Selection and reconstruction of L4B neurons.
To avoid any systematic sampling bias for our neuronal reconstructions, in the case in which both V1 and V2 were sectioned in the tangential plane we fully reconstructed all L4B neurons (n = 11) with somata contained in one section through the middle of L4B. The location of L4B was assessed by aligning this section to adjacent CO-stained sections. Identification of L4B for the two cases with V1 sectioned in the pia-to-white matter plane was based either on CO or fluorescent Nissl stains: in both stains, L4B is identifiable as a pale layer sandwiched between two darkly stained layers (L4A and L4C; Fig. 1C, bottom, D). For one of these cases, we reconstructed seven of 10 labeled L4B cells (one fully and six partially, due to inability to follow the axon through darkly stained CO sections). The amount of axon arbors we were able to reconstruct for these six cells was sufficient to classify them as Class 1 cells, but inadequate for subclassification into 1a or 1b (see Results). In the third case, we fully reconstructed all L4B cells (n = 5) with somata at the posterior edge of the labeled field, where axonal label was sparser, allowing full neuronal reconstruction and unambiguous assignment of processes to the correct neuron.
Selected cells were reconstructed through serial sections on a Zeiss Axioskop light microscope connected to a Qimaging color CCD camera, using 40× and 63× objectives, and Neurolucida software (MicroBrightField, RRID:SCR_001775). The axon of each neuron was initially identified as a small-caliber, spine-devoid process exiting the bottom of the soma (Fig. 1E,F). The extensive interlaminar branching and subsequent projection into white matter confirmed the identity of the process as an axon. Most axons were reconstructed well into white matter, and a few were followed all the way back to the V2 injection site. To reconstruct cells across multiple serial sections, the section outline and large radial blood vessels were initially used for coarse alignment across sections. Fine-scale alignment was then achieved by matching local blood vessels and axon exit points to the adjacent section's local blood vessels and axon entry points. This process of using global and very local fiducial points allowed for highly accurate and complete reconstruction of axons.
In addition to the axons, for the neurons reconstructed in the tangentially sectioned case we also plotted boutons. Boutons were identified as punctations rising from, yet attached to, the axon (bouton-termineaux), or large swellings along the axon that were greater in diameter than the thickest proximal portion of the axon (bouton-en-passant). The overwhelming majority of boutons were of the termineaux variety, and due to the possible ambiguity associated with marked en-passant boutons, all boutons were analyzed together.
Measurements of axon length and cluster analysis.
For each fully reconstructed neuron, we measured the percentage of total axon length dedicated to each V1 layer, by extracting from Neurolucida the axon length in each layer for each section, and subsequently summing these measurements across sections. Axonal length measurements only included branches off the primary descending axon, and were not corrected for tissue shrinkage.
Hierarchical cluster analysis (Cauli et al., 2000; Briggs et al., 2016) was performed to classify neurons based on the percentage of total axon length in L2/3, L4A, L4B, L4C, L5, and L6. Although L4A and L4C generally contained axonal branches passing through to other layers, and L6 was generally not a primary target, all layers were used for this analysis to avoid any subjective biasing of the data. The hierarchical cluster analysis plots the position of each cell in an N-dimensional space based on the number of metrics used to describe the cell, which in this study was axon length across six different layers. The distance between cells in this space was calculated using squared Euclidian distance, and linkages between cells were created using the Wards method (Briggs et al., 2016). Statistical comparisons of axon lengths in different layers, for different cell classes identified by the cluster analysis, were performed using a Kruskal–Wallis test corrected for multiple comparisons using Bonferroni's correction, unless otherwise specified in the Results. For each cell, we also measured the largest extent of axonal projections in each layer along two lines, one parallel to the V1–V2 border, one orthogonal to the V1–V2 border, and both passing through the cell's soma, (thus, one line was orthogonal to the ocular dominance bands, while the other was parallel to the ocular dominance bands). The reported measurements (in micrometers) encompass the full extent of the axon, i.e., on both sides of the soma. Statistical comparisons of axonal extents across different cell classes were also performed using a Kruskal–Wallis test with Bonferroni's correction.
Quantitative analysis of soma and bouton location relative to CO compartments.
To analyze soma and bouton position relative to CO compartments, we used the radial blood vessels nearest the label to align the nearest CO section containing well stained blobs to each section containing the reconstructed neuron. In practice, we primarily used one section of tissue in which L3 showed clear blobs over the entire region of interest. Images were scaled in Neurolucida and warped when necessary, using IRtweak software. For boutons in the upper layers, we generally performed minimal scaling or warping, but for boutons in the lower layers warping was typically needed, due to differential shrinkage across tissue sections.
Outlines of CO blobs and the middle of the interblobs were obtained using methods previously described (Federer et al., 2009). Briefly, digitized images of CO-stained tissue were processed using low-pass, equalization, and exponential filters (Image-Pro Plus, Media Cybernetics). The blobs were outlined by segmenting out the darkest 33% of pixels in the gray scale CO image (Boyd and Casagrande, 1999; Sincich and Horton, 2005a). Blob centers were defined as the darkest spot within each blob and the lightest pixels represented interblob centers. All automated CO compartment segmentation was visually inspected for accuracy and manually corrected, if necessary.
Tracings of blob outlines, blob centers, and interblob centers were exported and analyzed in Matlab (MathWorks, RRID:SCR_001622) using custom scripts. For all somata and boutons, we measured the distance to the nearest blob outline. For somata and boutons located inside blobs, this distance was divided by the distance of the blob outline to the blob center (measured along a line intersecting the position of the measured bouton or soma). For somata and boutons located outside blobs, the distance to the nearest blob outline was divided by the distance of the blob outline to the interblob center. This yielded a border distance index (BDI) with values ranging from −1 to +1, with −1 indicating a soma at a blob center, 0 a soma at a blob border, and +1 a soma at an interblob center. For statistical analysis, we defined a “blob-border region” as the region encompassing a BDI from −0.4 to 0.4, which, for an average blob radius of ∼ 143 μm in our material, corresponds to a region ∼57 μm on each side of the blob border. This definition of the blob-border region was primarily driven by the data. Specifically, we consistently found distribution peaks in the 0.0–0.4 range (border), or the 0.6–1.0 range (interblob); while there were rarely peaks in the 0.0 to −0.4 range, we chose this range to keep the size of the blob-border region symmetrical on either side of the blob border. All statistical comparisons of bouton distributions as a function of soma location of the parent cells or laminar location of the boutons were performed using a Kruskal–Wallis test with Bonferroni's correction.
Results
Golgi-like fills of thick-stripe-projecting V1 L4B neurons were obtained by injecting GdRV-EGFP into the thick stripes of V2 identified in vivo by intrinsic signal optical imaging (Fig. 1). We report data from three injections confined to thick stripes in three different animals. Two such injection sites are shown in Figure 1A,B. The top panel of Figure 1A shows the location of a GdRV-EGFP injection site on the V2 CO map, and the bottom panel shows the outline of the same injection site overlaid onto a differential orientation map obtained using optical imaging in vivo from the same animal. The injection site was in a V2 region having an orientation map, which coincided with a dark thick stripe in CO staining. A second injection case is shown in Figure 1B. All injection sites extended into the infragranular layers. Labeled neurons resulting from these injections were in V1 layers known to provide inputs to the V2 thick stripes (Sincich et al., 2010), mainly L2/3 and L4B, with sparser labels in L4A and L5/6 (preferentially at the L5–L6 border; Fig. 1C). The Golgi-like fills of V1 neurons resulting from these injections (Fig. 1D–F) enabled morphological reconstruction of the dendritic and axonal processes of many neurons. We reconstructed 23 neurons, 17 completely and 6 partially, but sufficiently to allow for neuronal classification (see below). For the fully reconstructed neurons, we quantitatively analyzed the intra-V1 axonal arborization patterns with respect to V1 layers and CO compartments.
V1 laminar patterns of axonal arborization reveal distinct classes of thick-stripe-projecting L4B neurons
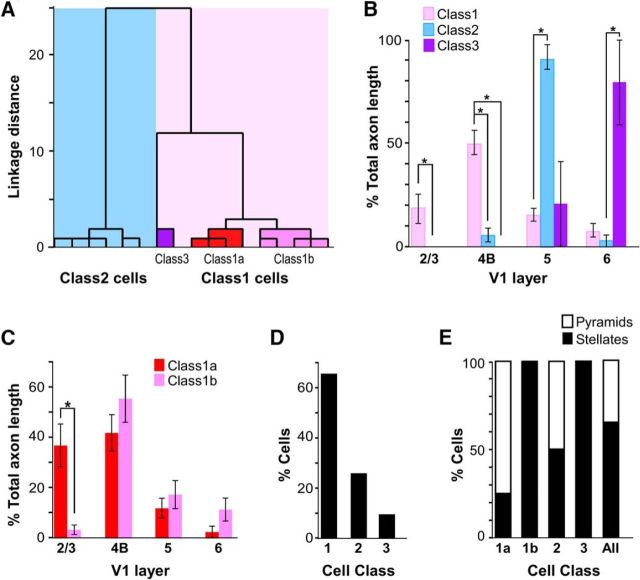
Previous reconstructions of the intra-V1 axons of L4B neurons, labeled by random intracellular fills in slices, showed all L4B cells as projecting primarily to L2/3, within L4B, and to L5, with some cells additionally sending sparser projections to L6 (Callaway and Wiser, 1996). The output targets of these reconstructed cells were, however, unknown. To determine whether thick-stripe-projecting L4B neurons show this same stereotyped axonal arborization motif, we reconstructed dendrites and axons of 23 cells (17 completely, 6 partially) selected in unbiased fashion (see Materials and Methods). For each of the 17 completely reconstructed cells, we quantified the percentage of total axonal length dedicated to each layer. Subjective analysis of the quantitative laminar distribution of axon arbors revealed two broad categories of cells, distinguished primarily by the presence (Class 1) or absence (Class 2) of projections to L2/3. Class 1 cells corresponded to classical descriptions of L4B neurons, with primary projections to L2/3, L4B, and L5 (Figs. 1F, 2A,B). By comparison, Class 2 cells predominantly targeted L5, with little or no branching in L4B and no projections to L2/3 (Figs. 1E, 2C). For both cell classes, the projections to L5 appeared to target mainly L5B.
Figure 2.
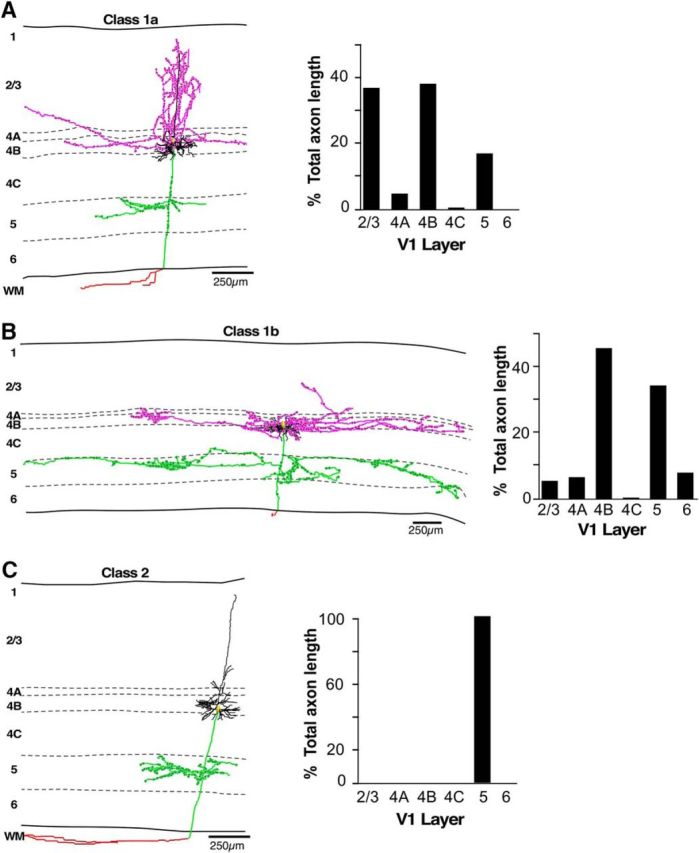
Example of each L4B neuron class. A–C, Example reconstructions of labeled V1 L4B neurons for each class as determined by cluster analysis, and including a Class 1a pyramidal cell (A), a Class 1b stellate cell (B), and a Class 2 pyramidal cell (C). Bar charts to the right of each reconstructed cell indicate the percentage of total axon length in each layer for that cell. Dendrites are illustrated in black, supragranular axon branches in pink, infragranular axon branches in green, the axon in the white matter in red, and the soma in yellow. Solid black lines indicate the top and bottom of V1, and the dashed lines the laminar boundaries, with layers labeled for each cell. Note different scale bar for the more extensive Class 1b cell.
To obtain a quantitative and unbiased assessment of whether the axonal branching patterns of our cell sample varied systematically, and indeed defined distinct classes of cells, we performed independent hierarchical cluster analysis on our sample of completely reconstructed cells (n = 17), using as metrics the percentage of total axonal length in each layer. This analysis separated our population of neurons into two major clusters based on the presence or absence of projections to L2/3 (Fig. 3A), supporting our initial qualitative assessment of the data. Class 1 cells (n = 11) had on average 18% of their axon in L2/3, 50% in L4B, and 15% in L5. Class 2 neurons (n = 6) had on average 5% of their axon in L4B (with many cells having no projections in L4B), 91% in L5, and 0% in L2/3 (Fig. 3B). Additionally, the cluster analysis separated Class 1 cells into two subgroups, which we have named Class 1a and 1b (Fig. 3A). Class 1a cells (n = 4) had on average 37% of their axon in L2/3 reaching the top part of these layers, 42% in L4B, 12% in L5, and 3% in L6 (Fig. 3C; for an example of a Class 1a cell, see Fig. 2A). Class 1b cells (n = 5) differed from Class 1a primarily for the paucity of projections to L2/3 (which additionally were confined to the bottom half of these layers) and the dominant axon projections within L4B itself (Fig. 2B); the population averages of percentage of total axon length for this subclass were 4% in L2/3, 56% in L4B, 17% in L5, and 12% in L6 (Fig. 3C). The cluster analysis also identified an additional subgroup of cells (Class 3; Fig. 3A), as having predominant axon branching in L6, but also some branching in L5, and no branching in L2/3 or L4B. Our initial qualitative assessment placed these neurons in Class 2, as they lacked projections to L2/3. However, the cluster analysis identified these cells as a subgroup of Class 1, perhaps due to similarity with Class 1 in the percentage of axon in L5. However, as the axonal distribution of these cells differed fundamentally from Class 1 cells in all other layers (Fig. 3B), we consider these cells as a separate class and, thus, we refer to them as Class 3. Note that we only found two such Class 3 cells. Figure 3B shows the percentage distribution of axon length in each layer for the three different cell classes. Compared with Class 1 cells, Class 2 cells had no projections to L2/3 (p = 0.016), significantly less projections within L4B (p = 0.005), and significantly more axon dedicated to L5 (p = 0.003). Unlike Class 1 cells, Class 3 cells had no projections to L2/3 and L4B (p = 0.042), however, compared with Class 2 cells, Class 3 cells had significantly more axon in L6 (p = 0.024). Figure 3C shows the percentage distribution of axon length in each layer for the two different subclasses of Class 1. Compared with Class 1b cells, Class 1a cells had significantly more axon branching in L2/3 (p = 0.016, Wilcoxon rank-sum test). All cell classes had passing axons in L4A and L4C (the latter two were included in the computation of total axon length, but not in the bar chart of Fig. 3B,C).
Figure 3.
Classification of thick-stripe-projecting L4B neurons. A, Dendrogram of our population of fully reconstructed L4B neurons (n = 17), illustrating the linkage distances, a measurement of clustering, of Class 1 (light pink shading) and Class 2 (blue shading) cells. Distinct subclusters are also clearly separated within the Class 1 cluster, and included Class 1a cells (red shading), Class 1b cells (dark pink shading), and a distinct cluster of neurons (Class 3; purple shading). B, Histogram of the percentage of axonal branching in each layer for each of the three cell classes (n = 17). Error bars represent SEMs. Asterisks indicate statistically significant differences. C, Histogram of the percentage of axonal branching in each layer for each of the two subclasses of Class 1 cells. D, Percentage of cells in each class across the population (n = 23). E, Percentage of pyramids and stellates in each cell class and subclass, and across the entire population of fully reconstructed neurons (n = 17).
Figure 3D shows the frequency of occurrence of each cell class within our population of thick-stripe-projecting L4B neurons (n = 23). We included in this analysis the six partially reconstructed neurons, which could be reliably classified as Class 1 cells based on the presence of axon branching in L2/3. Class 1 cells represented 65% of our sample, whereas Class 2 and Class 3 cells were 26 and 9%, respectively.
L4B includes two morphological types of excitatory neurons: pyramidal cells, which make up the majority cell type within L4B (Callaway and Wiser, 1996), and which receive functional inputs from both the magnocellular (M) and parvocellular (P) divisions of L4C (Yabuta et al., 2001), and spiny stellate cells, which receive only M inputs from L4Cα. Therefore, we were interested in assessing the proportion of each morphological cell type within the different classes of L4B cells projecting to thick stripes. Cells were defined as pyramids or stellates based on full reconstructions of their dendrites, with pyramidal cells defined as those having an apical dendrite extending into L3 and above (Figs. 1D, 2A,C), and cells without such an apical dendrite classified as stellates (Figs. 1D, 2B). Across our entire population of cells (n = 23), 61% of cells had stellate morphology; that is 60% of Class 1 cells (n = 9 of 15), 50% of Class 2 cells (n = 3 of 6), and 100% of Class 3 cells (n = 2 of 2) were stellates. Across the population of fully reconstructed cells (n = 17; Fig. 3E), stellates represented 25% of Class 1a, 100% of Class 1b, and 50% of Class 2.
We found no significant differences across cell classes in soma area (p = 0.67; one-way ANOVA), soma perimeter (p = 0.97), or total dendritic length (p = 0.78).
Lateral extent of axonal projections in different layers
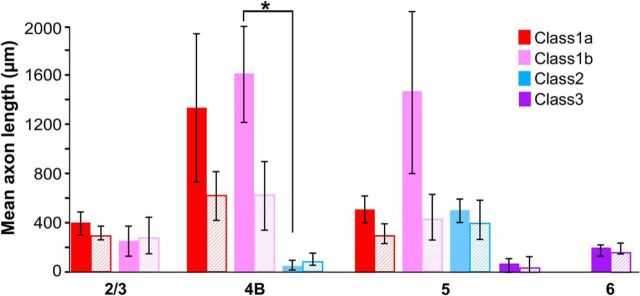
We next examined the lateral extent of axonal projections made by each cell class to understand whether the different cell classes take part in local or long-range processing in V1. Because V1 has a well known retinotopic organization, long-range connections can integrate information across distant visual field regions. For each fully reconstructed cell (n = 17), we measured the end-to-end extent of axonal projections (i.e., on each side of the soma) within each layer along two axes, one parallel to the V1–V2 border (and therefore orthogonal to the ocular dominance bands) and the other orthogonal to it (and therefore parallel to the ocular dominance bands). Figure 4 summarizes these measurements across our cell sample. All Class 1 cells sent narrowly focused projections to L2/3 (extent across vs along ocular dominance columns: Class 1a, mean ± SEM, 401 ± 92 × 309 ± 42 μm; Class 1b, 262 ± 123 × 285 ± 146 μm), but typically long-range projections within L4B (Class 1a, 1341 ± 602 × 617 ± 194 μm; Class 1b, 1615 ± 397 × 611 ± 276 μm). Class 1a cells sent short-range projections within L5 (mean, 518 ± 109 × 309 ± 87 μm), unlike Class 1b cells (mean, 1479 ± 673 × 447 ± 188 μm), which could send either short-range or long-range projections to this layer. In contrast, both Class 2 and Class 3 cells had very narrowly focused projections in all layers to which they projected (Class 2: L4B mean, 60 ± 41 × 92 ± 62 μm; L5 mean, 507 ± 95 × 420 ± 159 μm; Class 3: L5 mean, 70 ± 70 × 54 ± 62 μm; L6 mean, 199 ± 50 × 189 ± 44 μm). One can estimate the corresponding visuotopic extent of these connections by dividing their cortical extent by the magnification factor at the retinal eccentricity of our injection sites (∼5–6°), and adding the mean receptive field size at the same eccentricity. We performed these estimates for extent of connections across ocular dominance columns; specifically, at 5.5° eccentricity, the magnification factor across ocular dominance columns is ∼2.09 mm/° and the mean receptive field size is 0.27° (Van Essen et al., 1984). Accordingly, Class 1a cells have axonal projections in L4B extending up to 1° from the soma (2° diameter), and Class 1b cells send projections to L5 extending up to 1.2° from the soma, indicating that these long-range projections contact neurons with nonoverlapping receptive fields. The narrowly focused projections of Class 2 and 3 cells and the projections to L2/3 of Class 1 cells instead extend only 0.3–0.4° on each side of the soma, indicating that these cells send only local connections that do not extend beyond the receptive field of the parent cell.
Figure 4.
Extent of axons in each layer for different cell classes. Histogram of the mean lateral extent of axonal projections in each V1 layer for each cell class and subclass. Solid and hatched bars indicate extent of connections measured parallel and orthogonal, respectively, to the V1/V2 border. Asterisk indicates statistically significant differences (p = 0.029). Error bars indicate SEMs.
Intra-V1 axonal arborizations of thick-stripe-projecting L4B neurons avoid CO blob columns
For each reconstructed neuron, we next determined the position of its soma and boutons over the CO map of V1.
Soma location relative to CO compartments
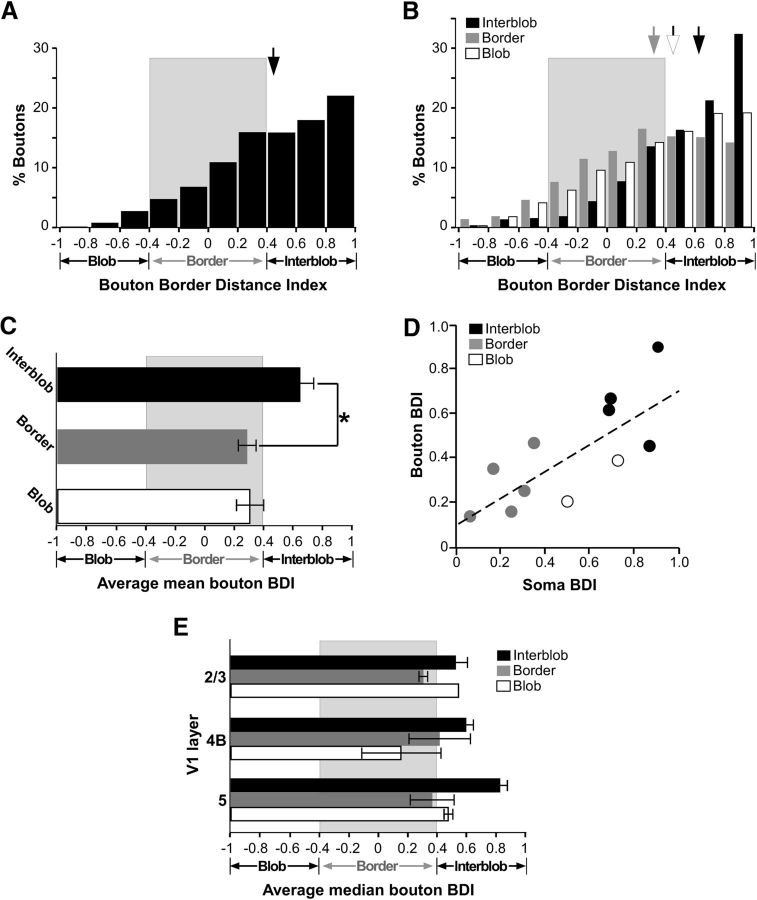
For each reconstructed cell, we measured the distance of its soma from the border of the nearest CO blob, and computed a BDI (see Materials and Methods). An index of 0 indicates a soma located at a blob border, an index of +1 indicates a soma located in the center of an interblob, and an index of −1 indicates a soma in the center of a blob. We used a data-driven definition of the blob border region as the region of BDI values between −0.4 and +0.4 (see Materials and Methods). Figure 5A shows the distribution of BDI values for the somata across the population of cells, demonstrating a bias for somata to be located at blob borders. We found that 18% of somata were located inside blobs, 47% at blob borders, and 35% inside interblobs (n = 17 including 6 somata classified qualitatively in pia-to-white matter sections; Fig. 5B).
Figure 5.
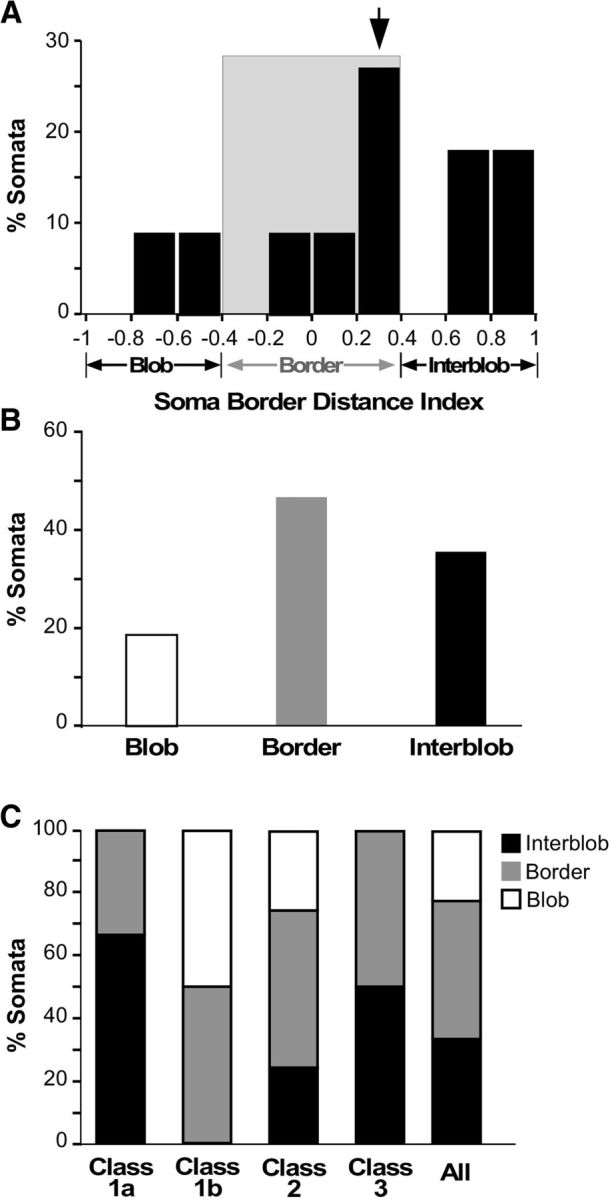
Soma location relative to CO compartments. A, Distribution of L4B neurons according to the BDI of their somata. B, Percentage of somata in each CO compartment. C, Percentage of somata in each CO compartment for each L4B neuron class and subclass.
We found no unique relationship between cell class and soma position over the CO map. However, 50% of Class 1b and Class 2 neurons had somata located at borders, while 67% of Class 1a neurons were inside interblobs; only Class 1b and 2, but not Class 1a, included neurons with somata inside blobs. The two Class 3 cells in our sample resided in an interblob and at a blob border, respectively (Fig. 5C). However, given our small sample size for each subclass, we do not think this distribution necessarily represents a thorough sampling of the CO location of cells in each class and subclass.
Bouton location relative to CO compartments
For each fully reconstructed cell, we measured the distance of each bouton from the border of the nearest CO blob, and computed a BDI, as above. This analysis was based on a smaller population of cells (n = 11) for which we had both complete reconstructions as well as CO staining in tangential sections. Figures 6–8 show examples of axons and boutons aligned to CO-stained tissue sections for each neuron class, in each main target layer (L3, L4B, L5), and corresponding quantitative analyses (distribution of bouton BDI values in each layer). For all three example cells, the boutons in each layer were predominantly located outside CO blobs. The soma of the Class 1a cell (Fig. 6) was located at a blob border (BDI, 0.19; Fig. 6C); the number of boutons for this cell was largest at blob borders in L2/3 and L5 (Fig. 6B,D), and in interblobs within L4B (Fig. 6C). Interestingly, the Class 1b cell (Fig. 7), despite its soma being in the center of a blob (BDI, −0.75; Fig. 7C), distributed the large majority of its boutons outside blobs, both at borders and in interblobs, in all layers. The Class 2 cell (Fig. 8), whose soma was located at a border (BDI, 0.29; Fig. 8B), also distributed its boutons in L5 predominantly at borders and interblobs.
Figure 6.
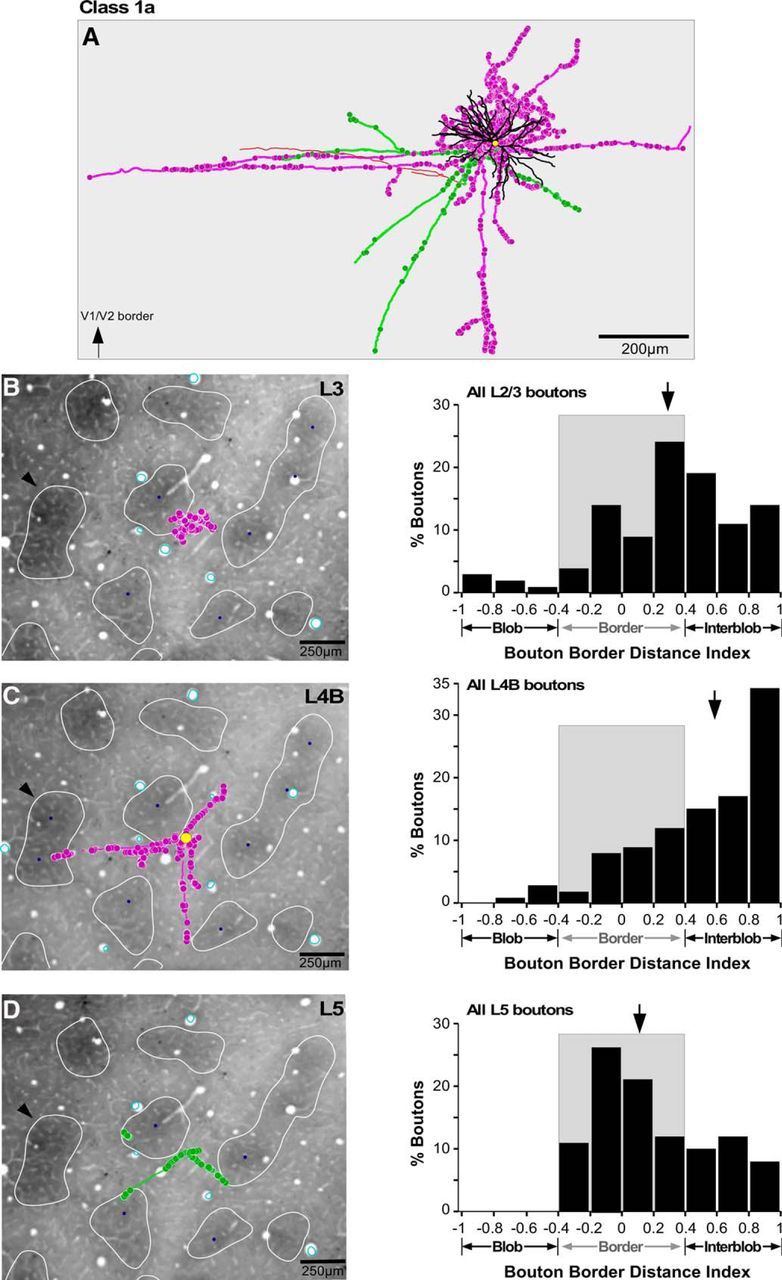
Location of axons and boutons relative to CO compartments for an example Class 1a cell. A, Example reconstruction of a Class 1a pyramidal neuron whose soma was located at a blob border (same cell as in Fig. 2A, but viewed from the top). The arrow at the bottom left corner points to the V1–V2 border, which for all cells shown in Figures 6–8, is located toward the top of A. Conventions are as in Figure 2. B, C, The cell's axon and boutons (pink) from a single section in L3 (B) and a single section in L4B (C) are shown over a CO-stained tissue section of V1 to illustrate their location relative to CO compartments. The yellow dot in C marks the location of the soma. The CO blobs are outlined in white, and the blob centers (blue dots) are indicated only for the blobs used in each layer for the quantitative analysis of the BDI. Black arrows in B–D point at the same CO blob for referencing. Cyan ovals are the outlines of the radial blood vessels used for aligning the cell's drawings to the CO-stained tissue section. The histogram to the right of each panel indicates for that cell the distribution of BDI values for all boutons in the respective layer. Gray shading marks the CO blob border region. Arrows, Medians. D, Same as in B and C but for axon and boutons (green) in L5.
Figure 7.
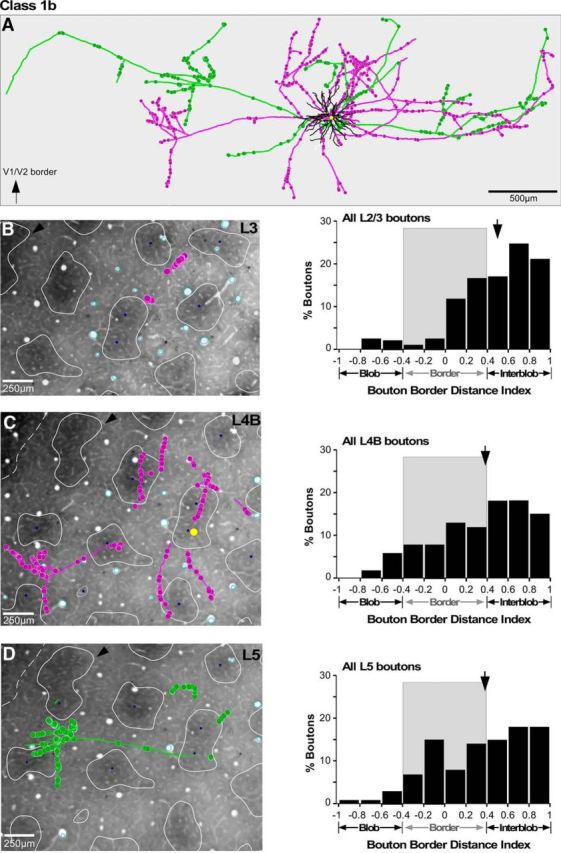
Location of axons and boutons relative to CO compartments for an example Class 1b cell. A, Example reconstruction of a Class 1b stellate neuron whose soma was located inside a blob (same cell as in Fig. 2B, but viewed from the top). B–D, The cell's axon and boutons from single sections through L3 (B), L4B (C), and L5 (D) shown over a CO-stained tissue section of V1 to illustrate their location relative to CO compartments. Other conventions are as in Figure 6.
Figure 8.
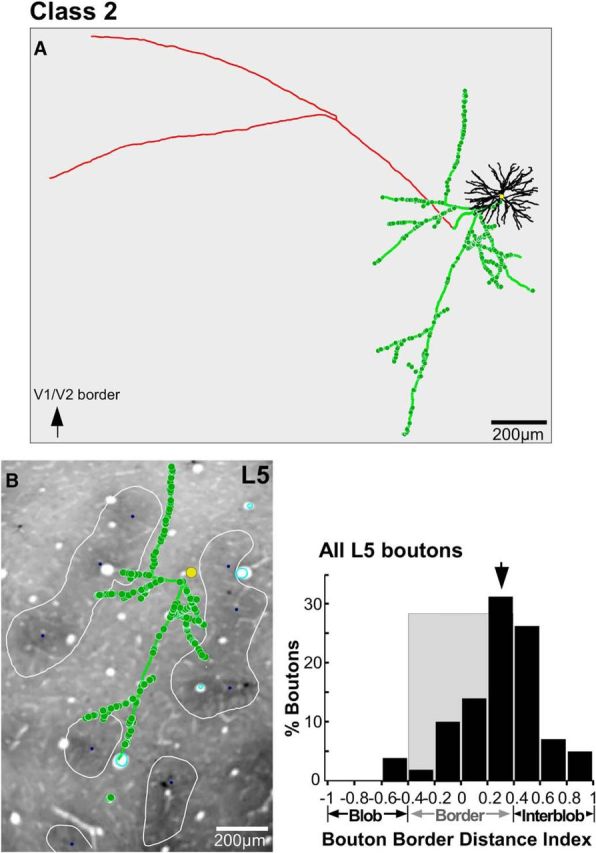
Location of axons and boutons relative to CO compartments for an example Class 2 cell. A, Example reconstruction of a Class 2 pyramidal neuron whose soma was located at a blob border (same cell as in Fig. 2C, but viewed from the top). B, The cell's axon and boutons in a single section through L5 are shown over a CO-stained tissue section of V1. Other conventions are as in Figure 6.
Across the population, when all boutons were pooled together (n = 8479) regardless of cell class, CO location of the parent soma, and layers, we found that most boutons were located outside blobs (Fig. 9A). In Figure 9B, we show the distribution of boutons according to their BDI values for L4B cells with somata located in blobs, borders, and interblobs. The distribution for cells with somata in the interblobs peaked at larger BDI values (median, 0.63; Fig. 9B, black arrow) than the peak of the distribution for cells with somata located at borders (median, 0.32; Fig. 9B, gray arrow), and the difference between the means of these two distributions was statistically significant (p < 0.001), indicating a bias toward like-to-like connectivity.
Figure 9.
Location of boutons relative to CO compartments: quantitative analysis. A, Distribution of BDI values for all boutons from all cells and layers pooled together. B, Distribution of BDI values for all boutons grouped by the soma location of their parent cell in blobs, borders, or interblobs, regardless of layers. Arrows, Medians. C, Average of mean BDI values for boutons computed separately for each cell (see Results), with cells grouped by CO location of their somata regardless of layers. Asterisk indicates a statistically significant difference. Error bars here and in E are SEM. D, Scatterplot of BDI values for soma versus boutons (across all layers) for each cell. White dots mark cells with somata in blobs whose BDI values are plotted on the positive, instead of the negative, x-axis. E, Average of median BDI values for boutons in each layer computed across cells, and grouped by CO location of the parent somata.
Because the number of boutons varied from cell to cell, and because each layer contained a different proportion of boutons, these distributions may have been biased by cells and layers with unusually high numbers of boutons. Therefore, for each cell we also calculated a median BDI value for the boutons in each layer, and averaged these median values across layers to obtain a mean median BDI value for the cell; the mean values for cells residing in the same CO compartments were, then, averaged to obtain mean population BDI values (Fig. 9C). This analysis supported and strengthened the results shown in Figure 9B, as the population bouton BDI values for cells in interblobs was significantly greater than for cells located at blob borders (p = 0.05). Figure 9D further plots for each cell the absolute values of the soma BDI versus the bouton BDI, demonstrating a positive and significant correlation between the two indices (r = 0.762; p < 0.006; Pearson's correlation). Note that the soma BDI values for the cells residing inside blobs are shown on the positive, instead of the negative, x-axis to demonstrate that a correlation between the two absolute BDI values is maintained even for these cells. A positive correlation between the two indices, however, was still observed after exclusion of the blob cells (r = 0.817; p = 0.013). These results demonstrate a bias for a like-to-like local connectivity pattern, as neurons located at border regions distributed their boutons preferentially at border regions, while neurons located in interblobs distributed their boutons preferentially in interblobs. However, this like-to-like connectivity did not strictly apply to the few cells in our sample whose somata resided inside blobs. The axons and boutons of these cells also clearly avoided blobs (Figs. 7B–D, 9B,C); however, their boutons still terminated closer to a blob border if their soma resided closer to a blob border and, conversely, farther from a blob border if their soma resided farther from a blob border. It is noteworthy that the branching pattern of these two cells is consistent with the border/interblob targeting pattern of all other cells in our sample, suggesting that this axonal branching motif is a feature of L4B neurons projecting to V2 thick stripes. In summary, all thick-stripe-projecting L4B neurons projected largely outside blobs, regardless of their soma location in blobs, at borders or interblobs. This was also the case when the analysis was performed separately for boutons in each layer. Figure 9E shows the average of the median bouton BDI in each layer for the population of cells, calculated by estimating for each cell the median BDI value in each layer, and averaging the specific layer median BDI values across cells with somata in the same CO compartment. In all layers, including L2/3, boutons dominated outside blobs, and were biased toward interblobs if their parent somata were in interblobs, or toward borders if their parent somata were located at borders.
Discussion
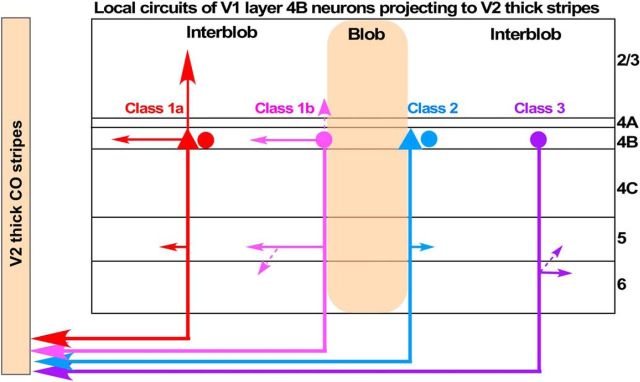
We found that V1 L4B neurons that project to V2 thick stripes consist of two major morphological types defining at least two distinct cell classes (Fig. 10). Class 1 cells send projections to supragranular and infragranular layers, with narrowly focused projections to L2/3, and projections within L4B and to L5 often extending for long distances. Class 1 cells, in turn, comprise two subclasses distinguished by the relative amount of projections they send to L2/3, with Class 1b cells sending no or few projections mainly to L3 and Class 1a cells sending denser projections up to L2/3A. Moreover, Class 1b cells have exclusively spiny stellate dendritic morphology, while Class 1a cells are predominantly, but not exclusively, pyramidal. Class 2 cells can be spiny stellates or pyramids, send no projections to L2/3, and project mainly to L5. A rare type, the Class 3, resembles cells in Class 2, but send projections mainly to L6, and are exclusively spiny stellates; however, as we only found two such cells, we are unsure whether this constitutes a distinct class or whether this is a variant of Class 2. Finally, we found that all cell classes have somata located predominantly at blob borders and in interblob columns, and in all layers send axonal projections outside blob columns, with a bias toward targeting predominantly blob borders if the soma is at a border, and interblobs if the soma is in an interblob.
Figure 10.
Summary diagram of the local circuits made by V1 L4B cells projecting to V2 thick stripes. Different colors indicate the different L4B cell classes. Triangles, Pyramidal cells; ovals, spiny stellate cells. A triangle next to an oval indicates that both pyramids and stellates contribute to that specific cell class. Dashed arrows indicate weaker projections, and solid arrows stronger projections. Class 1 cells (red, Class 1a; pink, Class 1b) make narrowly focused projections to L2/3 (mainly Class 1a), long-range horizontal connections in L4B, and short or long projections primarily in L5, with Class 1b cells sending sparse projections also to L6. Class 2 and Class 3 cells make narrowly focused projections primarily to L5 or L6, respectively. For each cell class and subclass the diagram indicates only the dominant soma position relative to blobs (but see Fig. 5C for actual distribution of soma position). All cell classes project predominantly outside CO blob columns.
Our findings differ from classical descriptions of L4B local circuits. Previous random intracellular fills of L4B neurons in V1 slices demonstrated a single axonal branching motif: projections to supragranular and infragranular layers, with those to L2/3 targeting selectively blobs, regardless of the cell's soma position under a blob or interblob (Callaway and Wiser, 1996). This axon motif resembles that of our Class 1 cells, except for the projections to CO blobs, which we failed to observe. Previous single L4B neuron reconstructions, based on Golgi impregnation (Lund, 1973; Lund and Boothe, 1975), identified two branching motifs: pyramids with laminar projection patterns similar to our Class 1a cells, and spiny stellate cells similar to our Class 1b cells. However, Lund and colleagues identified the infragranular projections as targeting predominantly L6, rather than L5, which is inconsistent with this study, the findings of Callaway and Wiser (1996), and later studies from Lund and colleagues themselves (Blasdel et al., 1985) using bulk injections of tracers in V1 laminae. Therefore, we interpret this discrepant result as an incorrect layer assignment, likely due to the difficulty of accurately identifying layers in Golgi-impregnated tissue, which is incompatible with CO or Nissl counterstains. Moreover, whereas Lund and colleagues identified the Class 1a axon branching motif as unique to pyramids, we found that Class 1a comprises both morphological types, albeit pyramids predominated in this class. None of the previous studies described a branching motif corresponding to our Class 2 or Class 3 cells, which, thus, represent new cell types.
What distinguishes our studies from these previous studies is that we only reconstructed a specific subpopulation of L4B neurons identified by their projection target (V2 thick stripes), whereas the extrinsic targets of the L4B cells in previous studies were unknown. Our approach enabled us to identify rare cell types, such as Class 2 cells, which represent only 26% of our thick-stripe-projecting L4B population. Indeed, using antidromic electrical microstimulation in V2, El-Shamayleh et al. (2013) estimated that only ∼8% of V1 cells in L2–L4 project to V2. Assuming that each V2 stripe type receives similar amounts of projections from V1 (but see Sincich and Horton, 2002b for a demonstration that pale stripes receive the densest V1 projections), only 2% of V1 supragranular cells project to thick stripes. It is known that only 23–24% of the V1 supragranular projections to thick stripes arise from L4B (Federer et al., 2009; Sincich et al., 2010), i.e., <0.5% of V1 supragranular cells. These estimates are similar to the anatomical estimates of Rockland (1997). Class 2 cells further represent only 26% of this L4B population, i.e., only 0.1% of V1 supragranular cells. These estimates suggest that previous studies, in which L4B neurons were randomly filled intracellularly, or randomly reconstructed in Golgi-impregnated tissue, could have easily missed the rare Class 2 and 3 L4B cells that we found in this study.
It is unlikely that our Class 2 cells simply represent Class 1 cells whose projections to L2/3 were missed due to incomplete GFP labeling. We found clear examples within a single tissue section of cells whose main axon trunk ran from L4B to the white matter sending collaterals to L5 but not to other layers (Fig. 1, compare E, F). In addition, the axon of some of these cells could be followed all the way to V2, indicating complete filling of the neurons. These cells clearly represent a different neuron class, as their projections avoided blob columns, unlike previous descriptions of L4B neurons.
Two classes of thick-stripe-projecting L4B cells suggest specialized functional circuits (Fig. 10). We speculate that Class 2 cells, with narrow projections predominantly to L5 interblobs, may send fast, retinotopically precise information to the L5 neurons that project to the superior colliculus (SC; Lund et al., 1975), known to reside predominantly in L5B interblobs (Lia and Olavarria, 1996), and/or to the pulvinar, possibly via previously described branching axons to both subcortical targets (Rockland, 1998). Class 2 cells clearly avoid the supragranular layers, where most of the corticocortical-projecting V1 neurons reside. This pathway, thus, via its contacts with L5 cells, may be concerned with directing eye movements toward incoming visual stimuli, and/or sending “efferent copies” to the pulvinar and SC (Sherman, 2016). Instead, Class 1 cells, while also projecting to L5, send most of their projections to the supragranular layers, and therefore are in a position to more directly affect the responses of corticocortical-projecting V1 neurons. Moreover, the long-range projections of Class 1 cells to L4B and/or L5 may serve to integrate visual signals across distant regions of space.
Another important result of this study is that our population of L4B neurons sent intra-V1 projections outside CO blob columns, in all V1 layers. This contrasts with previous studies demonstrating projections from L4B to blobs regardless of the CO location of the parent soma. For example, Callaway and Wiser (1996) found all 15 L4B cells in their sample to project to CO blobs; Lachica et al. (1992) found retrograde label in L4B after bulk injections of biocytin in L3 blobs, but not after injections in interblobs. Yoshioka et al. (1994) similarly found dominant retrograde label in L4B after biocytin injections into blobs, but also sparser label at the top of L4B after injections in interblobs. It is likely that in these previous studies, biocytin, which is not an effective retrograde tracer, only labeled the densest L4B-to-L3 blob projections, leaving unlabeled the sparser L4B-to-interblob projections. The estimates above that thick-stripe-projecting L4B neurons represent only ∼0.5% of all V1 supragranular cells suggest this is possible.
The finding of Lachica et al. (1992) that M-dominated L4B projects specifically to L3 blobs represented the first demonstration that the CO blobs, then thought to be P-dominated, instead also receive M inputs. The interblobs were still thought to be P-dominated, via inputs from L4Cβ and L4A. However, later studies demonstrated convergence of M and P inputs into both L3 blobs and interblobs, via direct relays from L4Cα and L4Cβ (Yabuta and Callaway, 1998a). Our finding of L4B projections to interblobs from both spiny stellate cells (which receive only M inputs) and pyramidal cells [which can receive both M and P inputs (Yabuta et al., 2001)] provides additional evidence for convergence of M and P streams in interblobs.
While it is now well recognized that M and P channels mix in blobs and interblobs (Sincich and Horton, 2005b), there exists anatomical segregation in the projections from the CO blobs and interblobs of V1 to the V2 CO stripes (Sincich and Horton, 2002a; Federer et al., 2009, 2013); this suggests that other functional properties, perhaps color (Tootell et al., 1988a, 2004; Xiao et al., 2007; Lu and Roe, 2008), spatial frequency (Tootell et al., 1988b; Silverman et al., 1989; Edwards et al., 1995; Nauhaus et al., 2016), and/or orientation (Economides et al., 2011), are segregated in CO compartments. An important question is where in the visual system these parallel streams converge to generate unitary percepts. Previous observations that CO blobs receive inputs from L4B neurons located in either blob or interblob columns suggested that local V1 circuits may integrate blob and interblob information. However, these studies lacked knowledge of the extrastriate targets of the identified L4B cells. In contrast, here we have shown that the local circuits of L4B output neurons that project to V2 thick stripes maintain CO-stream specificity, in that they arise from somata located largely outside blob columns and send axonal projections largely outside blob columns. Although it remains to be demonstrated whether the intralaminar and interlaminar circuits of specific V2-projecting L2/3 cells are also CO-stream specific, previous studies have typically shown L2/3 neurons to preferentially connect similar CO compartments, at least within L2/3 (Yoshioka et al., 1996; Yabuta and Callaway, 1998b). If the local circuits of L4B neurons projecting to thin and pale stripes also maintain CO-stream specificity, then interactions between functional CO streams likely occur downstream of V1, perhaps within V2, where long-range horizontal connections link different CO stripes (Levitt et al., 1994).
Our population of L4B neurons also showed a bias for like-to-like local connectivity: cells in interblobs projected preferentially to interblobs, and cells at blob borders projected preferentially to border regions. These results support the notion, previously suggested by studies of V1-to-V2 projections in marmosets (Federer et al., 2009), and horizontal projections in L2/3 of macaque (Yabuta and Callaway, 1998b), that the blob border region represents a distinct compartment.
Footnotes
This work was supported by the National Institutes of Health (National Eye Institute Grants R01 EY019743 and R01 EY026812, and National Institute of Neurological Disorders and Stroke Grant U01 NS099702), the National Science Foundation (Grants IOS 1355075 and EAGER 1649923), the University of Utah Research Foundation (Seed Grant 10040877), the University of Utah Neuroscience Initiative, and by a grant from Research to Prevent Blindness, Inc., to the Department of Ophthalmology, University of Utah. We thank Dr. Sam Merlin for help with experiments, Maryam Bijanzadeh for help with Matlab scripts, and Kesi Sainsbury for expert technical assistance. No PubMed ID found.
The authors declare no competing financial interests.
References
- Anderson JR, Jones BW, Yang J, Shaw MV, Watt CB, Koshevoy P, Spltenstein J, Jurrus E, UV K, Whitaker RT, Mastronarde D, Tasdizen T, Marc RE. A computational framework for ultrastructural mapping of neural circuitry. PLoS Biol. 2009;7:e1000074. doi: 10.1371/journal.pbio.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasdel GG, Lund JS, Fitzpatrick D. Intrinsic connections of macaque striate cortex: axonal projections of cells outside lamina 4C. J Neurosci. 1985;5:3350–3369. doi: 10.1523/JNEUROSCI.05-12-03350.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JD, Casagrande VA. Relationships between cytochrome oxidase (CO) blobs in primate primary visual cortex (V1) and the distribution of neurons projecting to the middle temporal area (MT) J Comp Neurol. 1999;409:573–591. doi: 10.1002/(SICI)1096-9861(19990712)409:4<573::AID-CNE5>3.0.CO%3B2-R. [DOI] [PubMed] [Google Scholar]
- Briggs F, Kiley CW, Callaway EM, Usrey WM. Morphological substrates for parallel streams of corticogeniculate feedback originating in both V1 and V2 of the macaque monkey. Neuron. 2016;90:388–399. doi: 10.1016/j.neuron.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway EM. Cell types and local circuits in the primary visual cortex of the macaque monkey. In: Chalupa LM, Werner JS, editors. The New Visual Neurosciences. Cambridge, MA: MIT; 2014. pp. 353–366. [Google Scholar]
- Callaway EM, Wiser AK. Contributions of individual layer 2–5 spiny neurons to local circuits in macaque primary visual cortex. Vis Neurosci. 1996;13:907–922. doi: 10.1017/S0952523800009159. [DOI] [PubMed] [Google Scholar]
- Cauli B, Porter JT, Tsuzuki K, Lambolez B, Rossier J, Quenet B, Audinat E. Classification of fusiform neocortical interneurons based on unsupervised clustering. Proc Natl Acad Sci USA. 2000;97:6144–6149. doi: 10.1073/pnas.97.11.6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economides JR, Sincich LC, Adams DL, Horton JC. Orientation tuning of cytochrome oxidase patches in macaque primary visual cortex. Nat Neurosci. 2011;14:1574–1580. doi: 10.1038/nn.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DP, Purpura KP, Kaplan E. Contrast sensitivity and spatial frequency response of primate cortical neurons in and around the cytochrome oxidase blobs. Vision Res. 1995;35:1501–1523. doi: 10.1016/0042-6989(94)00253-I. [DOI] [PubMed] [Google Scholar]
- El-Shamayleh Y, Kumbhani RD, Dhruv NT, Movshon JA. Visual response properties of V1 neurons projecting to V2 in macaque. J Neurosci. 2013;33:16594–16605. doi: 10.1523/JNEUROSCI.2753-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federer F, Ichida JM, Jeffs J, Schiessl I, McLoughlin N, Angelucci A. Four projections streams from primate V1 to the cytochrome oxidase stripes of V2. J Neurosci. 2009;29:15455–15471. doi: 10.1523/JNEUROSCI.1648-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federer F, Williams D, Ichida JM, Merlin S, Angelucci A. Two projection streams from macaque V1 to the pale cytochrome oxidase stripes of V2. J Neurosci. 2013;33:11530–11539. doi: 10.1523/JNEUROSCI.5053-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickfeld LL, Andermann ML, Bonin V, Reid RC. Cortico-cortical projections in mouse visual cortex are functionally target specific. Nat Neurosci. 2013;16:219–226. doi: 10.1038/nn.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Juavinett AL, Kyubwa EM, Jacobs MW, Callaway EM. Three types of cortical layer 5 neurons that differ in brain-wide connectivity and function. Neuron. 2015;88:1253–1267. doi: 10.1016/j.neuron.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachica EA, Beck PD, Casagrande VA. Parallel pathways in macaque monkey striate cortex: anatomically defined columns in layer III. Proc Natl Acad Sci USA. 1992;89:3566–3570. doi: 10.1073/pnas.89.8.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt JB, Yoshioka T, Lund JS. Intrinsic cortical connections in macaque area V2: evidence for interaction between different functional streams. J Comp Neurol. 1994;342:551–570. doi: 10.1002/cne.903420405. [DOI] [PubMed] [Google Scholar]
- Lia B, Olavarria JF. The distribution of corticotectal projection neurons correlates with the interblob compartment in macaque striate cortex. Vis Neurosci. 1996;13:461–466. doi: 10.1017/S0952523800008130. [DOI] [PubMed] [Google Scholar]
- Lu HD, Roe AW. Functional organization of color domains in V1 and V2 of macaque monkey revealed by optical imaging. Cereb Cortex. 2008;18:516–533. doi: 10.1093/cercor/bhm081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund JS. Organization of neurons in the visual cortex, area 17, of the monkey (Macaca mulatta) J Comp Neurol. 1973;147:455–496. doi: 10.1002/cne.901470404. [DOI] [PubMed] [Google Scholar]
- Lund JS, Boothe RG. Interlaminar connections and pyramidal neuron organisation in the visual cortex, area 17, of the macaque monkey. J Comp Neurol. 1975;159:305–334. doi: 10.1002/cne.901590303. [DOI] [Google Scholar]
- Lund JS, Lund RD, Hendrickson AE, Bunt AH, Fuchs AF. The origin of efferent pathways from the primary visual cortex, area 17, of the macaque monkey as shown by retrograde transport of horseradish peroxidase. J Comp Neurol. 1975;164:287–303. doi: 10.1002/cne.901640303. [DOI] [PubMed] [Google Scholar]
- Lund JS, Yoshioka T, Levitt JB. Substrates for interlaminar connections in area V1 of macaque monkey cerebral cortex. In: Peters A, Rockland KS, editors. Primary visual cortex in primates. New York: Plenum; 1994. pp. 37–60. [Google Scholar]
- Lund JS, Angelucci A, Bressloff PC. Anatomical substrates for functional columns in macaque monkey primary visual cortex. Cereb Cortex. 2003;13:15–24. doi: 10.1093/cercor/13.1.15. [DOI] [PubMed] [Google Scholar]
- Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassi JJ, Callaway EM. Specialized circuits from primary visual cortex to V2 and area MT. Neuron. 2007;55:799–808. doi: 10.1016/j.neuron.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauhaus I, Nielsen KJ, Callaway EM. Efficient receptive field tiling in primate V1. Neuron. 2016;91:893–904. doi: 10.1016/j.neuron.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockland KS. Elements of cortical architecture: hierarchy revisited. In: Jones EG, Peters A, editors. Extrastriate cortex in primates. New York: Plenum; 1997. [Google Scholar]
- Rockland KS. Convergence and branching patterns of round, type 2 corticopulvinar axons. J Comp Neurol. 1998;390:515–536. doi: 10.1002/(SICI)1096-9861(19980126)390:4<515::AID-CNE5>3.0.CO%3B2-3. [DOI] [PubMed] [Google Scholar]
- Sherman SM. Thalamus plays a central role in ongoing cortical functioning. Nat Neurosci. 2016;19:533–541. doi: 10.1038/nn.4269. [DOI] [PubMed] [Google Scholar]
- Silverman MS, Grosof DH, De Valois RL, Elfar SD. Spatial frequency organization in the primate striate cortex. Proc Natl Acad Sci USA. 1989;86:711–715. doi: 10.1073/pnas.86.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sincich LC, Horton JC. Divided by cytochrome oxidase: a map of the projections from V1 to V2 in macaques. Science. 2002a;295:1734–1737. doi: 10.1126/science.1067902. [DOI] [PubMed] [Google Scholar]
- Sincich LC, Horton JC. Pale cytochrome oxidase stripes in V2 receive the richest projection from macaque striate cortex. J Comp Neurol. 2002b;447:18–33. doi: 10.1002/cne.10174. [DOI] [PubMed] [Google Scholar]
- Sincich LC, Horton JC. Input to V2 thin stripes arises from V1 cytochrome oxidase patches. J Neurosci. 2005a;25:10087–10093. doi: 10.1523/JNEUROSCI.3313-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sincich LC, Horton JC. The circuitry of V1 and V2: integration of color, form and motion. Annu Rev Neurosci. 2005b;28:303–326. doi: 10.1146/annurev.neuro.28.061604.135731. [DOI] [PubMed] [Google Scholar]
- Sincich LC, Jocson CM, Horton JC. Neurons in V1 patch columns project to V2 thin stripes. Cereb Cortex. 2007;17:935–941. doi: 10.1093/cercor/bhl004. [DOI] [PubMed] [Google Scholar]
- Sincich LC, Jocson CM, Horton JC. V1 interpatch projections to V2 thick stripes and pale stripes. J Neurosci. 2010;30:6963–6974. doi: 10.1523/JNEUROSCI.5506-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootell RB, Silverman MS, Hamilton SL, De Valois RL, Switkes E. Functional anatomy of macaque striate cortex. III. Color. J Neurosci. 1988a;8:1569–1593. doi: 10.1523/JNEUROSCI.08-05-01569.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootell RB, Silverman MS, Hamilton SL, Switkes E, De Valois RL. Functional anatomy of macaque striate cortex. V. Spatial frequency. J Neurosci. 1988b;8:1610–1624. doi: 10.1523/JNEUROSCI.08-05-01610.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootell RB, Nelissen K, Vanduffel W, Orban GA. Search for color “center(s)” in macaque visual cortex. Cereb Cortex. 2004;14:353–363. doi: 10.1093/cercor/bhh001. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Newsome WT, Maunsell JH. The visual field representation in striate cortex of the macaque monkey: asymmetries, anisotropies, and individual variability. Vision Res. 1984;24:429–448. doi: 10.1016/0042-6989(84)90041-5. [DOI] [PubMed] [Google Scholar]
- Vélez-Fort M, Rousseau CV, Niedworok CJ, Wickersham IR, Rancz EA, Brown AP, Strom M, Margrie TW. The stimulus selectivity and connectivity of layer six principal cells reveals cortical microcircuits underlying visual processing. Neuron. 2014;83:1431–1443. doi: 10.1016/j.neuron.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007;4:47–49. doi: 10.1038/nmeth999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiser AK, Callaway EM. Contributions of individual layer 6 pyramidal neurons to local circuitry in macaque primary visual cortex. J Neurosci. 1996;16:2724–2739. doi: 10.1523/JNEUROSCI.16-08-02724.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Casti A, Xiao J, Kaplan E. Hue maps in primate striate cortex. Neuroimage. 2007;35:771–786. doi: 10.1016/j.neuroimage.2006.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta NH, Callaway EM. Functional streams and local connections of layer 4C neurons in primary visual cortex of the macaque monkey. J Neurosci. 1998a;18:9489–9499. doi: 10.1523/JNEUROSCI.18-22-09489.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta NH, Callaway EM. Cytochrome-oxidase blobs and intrinsic horizontal connections of layer 2/3 pyramidal neurons in primate V1. Vis Neurosci. 1998b;15:1007–1027. doi: 10.1017/s0952523898156018. [DOI] [PubMed] [Google Scholar]
- Yabuta NH, Sawatari A, Callaway EM. Two functional channels from primary visual cortex to dorsal visual cortical areas. Science. 2001;292:297–300. doi: 10.1126/science.1057916. [DOI] [PubMed] [Google Scholar]
- Yoshioka T, Levitt JB, Lund JS. Independence and merger of thalamocortical channels within macaque monkey primary visual cortex: anatomy of interlaminar projections. Vis Neurosci. 1994;11:467–489. doi: 10.1017/S0952523800002406. [DOI] [PubMed] [Google Scholar]
- Yoshioka T, Blasdel GG, Levitt JB, Lund JS. Relation between patterns of intrinsic lateral connectivity, ocular dominance and cytochrome oxidase reactive regions in macaque monkey striate cortex. Cereb Cortex. 1996;6:297–310. doi: 10.1093/cercor/6.2.297. [DOI] [PubMed] [Google Scholar]