Abstract
The SrrAB two-component regulatory system (TCRS) positively influences the transcription of genes involved in aerobic respiration in response to changes in respiratory flux. Hydrogen peroxide (H2O2) can arise as a byproduct of spontaneous interactions between dioxygen and components of respiratory pathways. H2O2 damages cellular factors including protein associated iron-sulfur cluster prosthetic groups. We found that a Staphylococcus aureus strain lacking the SrrAB two-component regulatory system (TCRS) is sensitive to H2O2 intoxication. We tested the hypothesis that SrrAB manages the mutually inclusive expression of genes required for aerobic respiration and H2O2 resistance. Consistent with our hypothesis, a ΔsrrAB strain had decreased transcription of genes encoding for H2O2 resistance factors (kat, ahpC, dps). SrrAB was not required for the inducing the transcription of these genes in cells challenged with H2O2. Purified SrrA bound to the promoter region for dps suggesting that SrrA directly influences dps transcription. The H2O2 sensitivity of the ΔsrrAB strain was alleviated by iron chelation or deletion of the gene encoding for the peroxide regulon repressor (PerR). The positive influence of SrrAB upon H2O2 metabolism bestowed protection upon the solvent accessible iron-sulfur (FeS) cluster of aconitase from H2O2 poisoning. SrrAB also positively influenced transcription of scdA (ytfE), which encodes for a FeS cluster repair protein. Finally, we found that SrrAB positively influences H2O2 resistance only during periods of high dioxygen-dependent respiratory activity. SrrAB did not influence H2O2 resistance when cellular respiration was diminished as a result of decreased dioxygen availability, and negatively influenced it in the absence of respiration (fermentative growth). We propose a model whereby SrrAB-dependent regulatory patterns facilitate the adaptation of cells to changes in dioxygen concentrations, and thereby aids in the prevention of H2O2 intoxication during respiratory growth upon dixoygen.
Introduction
Staphylococcus aureus is a human pathogen that has the ability infect nearly every tissue of the body [1]. The ability of S. aureus to sense various environmental stimuli, and rapidly calibrate its cellular physiology in response, is a cornerstone of its success as a pathogen. S. aureus is a facultative anaerobe and can respire dioxygen. Hydrogen peroxide (H2O2) is a deleterious by-product of aerobic respiration, and can arise as a result of interactions between dioxygen and components of respiratory pathways [2–5]. H2O2 can cause damage to cellular membranes and biological polymers, as well as oxidize protein bound iron-sulfur (FeS) cluster prosthetic groups [6, 7]. Oxidation of FeS clusters by H2O2 results in cluster disintegration and enzyme inactivation [7–9]. Ultimately, H2O2 exposure can result in metabolic standstill and eventually cell death [10, 11]. The importance of reactive oxygen species (ROS) in preventing S. aureus infections is evidenced by the fact that individuals carrying metabolic or genetic defects affecting ROS formation by polymorphonuclear neutrophils, the first line of defense in human innate immunity, often have chronic and reoccurring S. aureus infections [12].
The S. aureus genome encodes for a variety of mechanisms to detoxify ROS and to repair the damage caused by ROS to cellular molecules. Staphylococcal cells maintain high titers of the H2O2 scavenging enzymes catalase (Kat), alkylhydroperoxidase (Ahp) and the superoxide scavengers SodA and SodM [13–15]. In contrast to Escherichia coli, Kat is proposed to be the primary intracellular H2O2 scavenger in S. aureus [16]. SodA is proposed to be the dominant aerobic superoxide dismutase [15, 17, 18]. The findings that strains lacking ROS-scavenging enzymes display attenuated virulence in models of infection underscores the value of these ROS scavenging mechanisms [16, 19]. H2O2 can oxidize ferrous iron resulting in the production of hydroxyl radicals (Fenton reaction), which readily damage DNA [20]. The effects of Fenton chemistry are suppressed by producing a bi-functional protein (Dps) that binds and sequesters iron, as well as binds and protects DNA from oxidative damage [21–23]. S. aureus also encodes for a di-iron RIC protein ScdA (YtfE in Escherichia coli) that has a role in repairing FeS proteins damaged by H2O2 [24, 25].
The genome of S. aureus encodes for sixteen two-component regulatory systems (TCRS). TCRS permit bacteria to integrate multiple signals into cellular signaling circuits allowing for a rapid and robust response to stimuli [26]. TCRS typically consist of two proteins: a histidine kinase (HK) and a response regulator (RR). The HK can be either cytosolic or membrane-associated and the RR is typically a cytosolic DNA binding protein. A phosphorylation cascade between the HK and RR results in conformational changes in the RR protein, altered affinity towards target DNA sequences, modified gene expression, and a tailored physiological response [27]. TCRS have been implicated in the oxidative stress response in multiple bacterial species [28–30].
The staphylococcal respiratory regulator (SrrAB) is a TCRS that is thought to be the dominant transcriptional regulator mediating the aerobic to anaerobic switch in S. aureus [31–34]. SrrAB is a pleiotropic regulator of aerobic and anaerobic respiration and energy metabolism and is capable of activating as well as repressing gene transcription [31, 34–37]. SrrAB also modulates the response to nitric oxide stress [36, 38]. Alterations in cellular respiratory flux serve as one stimulus for SrrAB [36]. S. aureus strains lacking SrrAB display attenuated survival in models of infection [34, 38, 39].
In this study we focused upon defining a role for the SrrAB TCRS in mediating resistance to H2O2. Previous studies found that SrrAB positively influences expression of multiple genes required for aerobic respiration and changes in cellular respiratory flux act as a stimulus for SrrAB [34, 36]. We tested a model wherein S. aureus use SrrAB to co-regulate genes required for aerobic respiration and H2O2 resistance. Such a regulatory tuning mechanism would allow respiring cells to achieve physiological homeostasis by protecting and repairing cellular macromolecules from H2O2 damage. In support of this model, SrrAB positively influenced the transcription of genes involved in H2O2 resistance and aerobic respiration (ahpC, kat, dps, scdA, cydB) during respiratory growth. However, SrrAB was not required for inducing the transcription of H2O2 resistance genes upon challenge with H2O2. Purified SrrA bound to the promoter region for dps suggesting that SrrAB directly modulates transcription of at least one H2O2 resistance factor. Intriguingly, SrrAB negatively modulated H2O2 resistance during conditions of low respiration or in its absence. During respiratory growth the positive influence of SrrAB upon H2O2 resistance bestowed protection upon the solvent accessible FeS cluster of aconitase from ROS poisoning.
Material and Methods
Materials
Restriction enzymes, quick DNA ligase kit, deoxynucleoside triphosphates, and Phusion DNA polymerase were purchased from New England Biolabs. The plasmid mini-prep kit, gel extraction kit, and RNAprotect were purchased from Qiagen. DNase I was purchased from Ambion. Lysostaphin was purchased from Ambi products. Oligonucleotides were purchased from Integrated DNA Technologies and sequences are listed in S1 Table. Trizol and High-Capacity cDNA Reverse Transcription Kits were purchased from Life Technologies. Tryptic Soy broth (TSB) was purchased from MP biomedical. Unless specified, all chemicals were purchased from Sigma-Aldrich and were of the highest purity available.
Bacterial growth conditions
Unless otherwise stated, the S. aureus strains used in this study (Table 1) were constructed in the S. aureus community-associated USA300_LAC strain that was cured of the native plasmid pUSA03, which confers erythromycin resistance (USA300_LAC or WT) [40]. S. aureus were cultured overnight aerobically in 10 mL culture tubes with a culture volume of 1 mL. For subcultures in the presence of dioxygen, overnight cultures were used to inoculate either 10 or 30 mL culture tubes covered with a loosely fitting stainless-steel cap (Kap-Uts) that allowed for free diffusion of gases. Unless otherwise indicated, the 10 mL tubes contained 1 mL of media and the 30 mL tubes contained 5 mL of media. Liquid shake-tube cultures were grown at 37°C with shaking at 250 rpm unless otherwise indicated. Aerobic phenotypic analyses in 96-well plates were assessed in 200 μL cultures using a BioTek 808E visible absorption spectrophotometer with medium shake speed and the culture optical density was monitored at 600 nm. Difco agar was added (15 g L-1) for solid medium. The staphylococcal defined media contained per 100 mL: 1 g (NH4)2SO4, 4.5 g KH2PO4, 10.5 g K2HPO4, 110 mM NaCl, 30 mM KCl, 50 μg nicotinic acid, 50 μg thiamine, 0.3 μg biotin and 0.025 mg of each of the 20 canonical amino acids. Glucose (14 mM) was added as carbon sources. When required antibiotics were added at the following concentrations: 150 μg mL-1 ampicillin; 30 μg mL-1 chloramphenicol (Cm); 10 μg mL-1 erythromycin (Erm); 3 μg mL-1 tetracycline (Tet); 125 μg mL-1 kanamycin (Kan); 150 ng mL-1 anhydrotetracycline (Atet). To maintain plasmids media was supplemented with either 15 μg mL-1 or 5 μg mL-1 of chloramphenicol or erythromycin, respectively.
Table 1. Strains and plasmids used in this study.
| S. aureus Strains | Genotype/Description | Genetic Background | Source/Reference |
|---|---|---|---|
| JMB1100 | USA300_LAC (erm sensitive) | LAC | [40] |
| RN4220 | Restriction minus | NCTC8325 | [86] |
| JMB 1467 | ΔsrrAB (SAUSA300_1441–42) | LAC | [87] |
| JMB 2047 | ΔsrrAB::tet | LAC | This work |
| JMB 1209 | Δdps (SAUSA300_2092) | LAC | This work |
| JMB6326 | sodA::Tn (ermB) (SAUSA300_1513) | LAC | NARSA [88] |
| JMB 2968 | sodM::Tn (ermB) (SAUSA300_0135) | LAC | NARSA [88] |
| JMB2080 | ΔahpC::erm (SAUSA300_0379) | LAC | V. Torres |
| JMB 2078 | Δkat::erm (SAUSA300_1232) | LAC | V. Torres |
| JMB2151 | ΔperR::kan (SAUSA300_1842) | LAC | V. Torres |
| JMB2615 | ΔsrrAB ΔperR::kan | LAC | This work |
| JMB7103 | ΔsrrAB ΔahpC::erm | LAC | This work |
| JMB 5562 | ΔsrrAB Δkat::erm | LAC | This work |
| JMB6069 | cidA::Tn (ermB) (SAUSA300_2479) | LAC | [88] |
| JMB6070 | ΔsrrAB cidA::Tn (ermB) | LAC | This work |
| JMB 6023 | cidB::Tn (ermB) | LAC | NARSA [88] |
| JMB 6024 | ΔsrrAB cidB::Tn (ermB) (SAUSA300_2479) | LAC | This work |
| SH1000 | parent | SH1000 | [77] |
| JMB4556 | ΔsrrAB::tet | SH1000 | This work |
| Newman | parent | Newman | [89] |
| JMB 4751 | ΔsrrAB::tet | Newman | This work |
| JMB 2030 | ΔsrrAB::tet | RN4220 | This work |
| JMB1432 | fur::tet | LAC | [90] |
| JMB 1163 | acn::tet | LAC | [91] |
| JMB 7105 | ΔahpC::erm acn::tet | LAC | This work |
| JMB 3537 | acn::Tn erm | LAC | NARSA [88] |
| JMB 3538 | Δnfu acn::erm | LAC | This work |
| JMB 7107 | Δkat::erm acn::tet | LAC | This work |
| JMB 4367 | ΔsrrAB acn::erm | LAC | This work |
| JMB1254 | ΔscdA (SAUSA300_0253) | LAC | This work |
| Other Strains | |||
| Escherichia coli PX5 | Protein Express | ||
| Escherichia coli BL21-AI | Life Technologies | ||
| Plasmids used in this study | |||
| Plasmid name | Insert Locus/function | Source/Reference | |
| pJB38 | construction of chromosomal gene deletions | [92] | |
| pJB38_ΔscdA | Construction of ΔscdA | This work | |
| pJB38_Δdps | Construction of Δdps | This work | |
| pJB38_ΔsrrAB::tet | Construction of srrAB::tet allele | This work | |
| pCM28 | Cloning vector for genetic complementation | A. Horswill | |
| pCM11 | Cloning vector for trascriptional reporters | [45] | |
| pCM11_PsufC | Reporter construct transcriptional activity | [46] | |
| pCM11_Pdps | Reporter construct transcriptional activity | This work | |
| pCM28_srrAB | Complementing vector | This work | |
| pEPSA5 | Xylose inducible over-expression | [93] | |
| pEPSA5_acnA (pacnA) | Aconitase over-production | [47, 94] | |
| pET20b | Cloning vector for protein production | EMD Millipore | |
| pET20b(+)_srrA | Protein production | This work | |
Growth analyses for hydrogen peroxide resistance
Strains were cultured overnight in TSB at the indicated culture vessel headspace to the medium volume ratio (HVR). Optical density (OD) on all strains was standardized to OD 2.5 (A600) in a final volume of 1 mL of 1X PBS. Two μL of the resuspended culture was used to inoculate 198 μL media in a 96-well microtiter plate. A multichannel pipettor was used to amend the media with hydrogen peroxide or vehicle control and the cultures were rapidly mixed. Bacterial strains were then cultured with constant shaking in a heated microplate reader.
Challenge during early exponential growth
Strains cultured overnight in TSB were diluted into fresh TSB medium to a final OD of 0.1 (A600) at a HVR of 10 and allowed to grow with shaking for two doublings (OD of 0.4 (A600)). Subsequently, the strains were challenged with H2O2 or vehicle control as outlined above.
Challenge during fermentative (anaerobic) growth
Strains were cultured overnight in TSB at a HVR of 0, as described earlier [41–43]. At point of harvest the cultures tubes were placed inside a COY anaerobic chamber (<1 ppm of O2). Optical density on all strains was standardized in 1X PBS. The strains were then removed from the COY chamber and immediately sub-cultured into fresh TSB medium and challenged with H2O2 or vehicle control as outlined above.
H2O2 killing assays
Bacterial strains were cultured with shaking to OD 10 (A600) in TSB. Cells were pelleted by centrifugation and resuspended in an equal volume of PBS. The optical density for all the strains was adjusted to an OD of 0.7 (A600) in a total volume of 1 mL of 1X PBS. Cells were subsequently challenged with a bolus of H2O2 and incubated for two hours at room temperature. Fifty μL of the reaction mixture was diluted 1:20 into PBS buffer containing catalase (1300 units mL-1). Colony forming units (CFU) were determined by serially diluting cells and spot plating upon TSB agar plates.
Recombinant DNA and genetic techniques
All clones were passaged through RN4220 and subsequently transduced into the appropriate strains using bacteriophage 80α [44]. All S. aureus mutant strains and plasmids were verified using PCR or by sequencing PCR products or plasmids. DNA sequencing was performed at Genewiz, (South Plainfield, NJ).
Creation of plasmids and mutant strains
Chromosomal DNA from JMB 1100 was used as the template for PCR reactions for the construction of plasmids. To create the scdA chromosomal deletion, approximately 500 base pairs upstream and downstream of the scdA gene (SAUSA300_0253) was amplified using PCR the following primer pairs; 0253up5EcoRI and up3NheI; down5MluI and down3KpnI. Amplicons were gel purified and joined by PCR using the 0253up5EcoRI and down3KpnI primer pair. The amplicon was digested with EcoRI and KpnI, and ligated into similarly digested pJB38 [10]. The ligation was transformed into E. coli DH5α and colonies were screened by PCR for the correct plasmid. Plasmid pJB38_ΔscdA was isolated and transformed into RN4220 by selection upon TSA-Cm at 30°C. Plasmid pJB38_ΔscdA was transduced into JMB1100, and single colonies were inoculated into 5 mL of TSB-Cm. Cultures were grown at 42°C overnight before plating upon TSA-Cm to select for single recombinants. Single colonies were inoculated into 5 mL of TSB medium, grown overnight, and cultures were diluted 1:25,000 before plating 100 μL on TSA-anhydrotetracycline to select for the loss of plasmid. Cm sensitive colonies were screened for the double recombination event using PCR with primers 0253up5EcoRI and down3KpnI.
To create the dps chromosomal deletion, approximately 500 base pairs upstream and downstream of the dps gene (SAUSA300_2092) was amplified using PCR the following primer pairs; 2092up5EcoRI and 2092up3NheI; 2092down5MluI and 2092down3SalI. Amplicons were gel purified and joined by PCR using the 2092up5EcoRI and 2092down3SalI primer pair. The amplicon was digested with EcoRI and SalI, and ligated into similarly digested pJB38. Thereafter the Δdps strain was created as outlined above.
The pJB38_ΔsrrAB::tet plasmid was created by using PCR to amplify the tetM allele from strain JMB1432 using primers G+tetMluI and G+tetNheI. The PCR product was digested with MluI and NheI and ligated into similarly digested pJB38_ΔsrrAB (pJB38_ΔsrrAB::tetM). The ΔsrrAB::tetM strain was created as outlined above.
Transcriptional reporter plasmids for sufC and dps were constructed using the pCM11 vector backbone [45, 46]. Primers were designed to amplify the upstream regions (200–800 base pairs) upstream of the annotated translational start site for the genes. PCR amplicons were digested with HindIII and KpnI and subsequently ligated into similarly digested plasmid.
Quantitative real-time PCR assays
Overnight cultures were diluted to an OD of 0.1 (A600) in a final volume of 5 mL of fresh TSB (HVR 6) and cDNA libraries were constructed as previously described [47].
ROS challenge
Culture optical density was standardized to an OD of 0.1 (A600) in a final volume of 5 mL of fresh TSB (HVR 6) and strains were subsequently grown with shaking to the desired optical densities and treated with either 10 mM H2O2 or the vehicle control and grown for an additional 25 minutes. Aliquots of cells were then harvested by centrifugation and treated with RNAprotect (Qiagen) reagent prior to RNA extraction and cDNA library preparation.
Anaerobic work
Unless otherwise mentioned anaerobic work was performed using a Coy anaerobic glove-box (Grass Lake, MI). Solutions, plastic-ware, and liquid and solid growth medium was allowed to equilibrate for >6 h inside the glove-box before use.
Cell-free extract enzyme assays
Aconitase (AcnA) assays
Overnight cultures were diluted to a final OD of 0.1 (A600) in fresh TSB growth at HVR of 2.5, 5, 10 or 20 (ratios were altered as per experimental requirement) in the presence or absence of 1% xylose. Strains were cultured to an OD of 8 (A600) and cell pellets were harvested by centrifugation. Cell-free lysates/extracts were prepared inside a COY anaerobic chamber using anaerobic lysis buffer (25 mM Tris, 150 mM NaCl, pH 7.4), as described previously [47]. The assay was initiated by addition of 25 μL of lysate to 975 μL of lysis buffer containing 20 mM DL-isocitrate. Aconitase activity was determined by monitoring the conversion of isocitrate to cis-aconitate spectrophotometrically using a Beckman Coulter DU530 UV-Vis absorption spectrophotometer (cis-aconitate ε240 nm = 3.6 mM-1cm-1 [48]).
For AcnA assays under anaerobic conditions, strains were cultured in 2 mL microcentrifuge tubes containing TSB and 1% xylose at a headspace to volume ratio of zero, as described earlier [47].
The effect of dioxygen on AcnA activity was assessed as described earlier [42]. Briefly, strains were cultured anaerobically as described above. Protein synthesis was inhibited by the addition of anaerobic 100 μg mL-1 rifampicin inside a COY chamber. Dioxygen was introduced to one set of cultures by decanting into tubes at a HVR of 15 and vigorous shaking for 35 minutes. The control set of cultures experienced continued anaerobic incubation.
Catalase assays
Overnight cultures were diluted to a final OD of 0.1 (A600) in fresh TSB and cultured to an OD of 8 (A600), at the desired HVR. Cell lysis and clarification of the supernatant were as described earlier [47]. The cell-free lysate was further diluted 50-fold and catalase activity was assayed by the addition of 5 μL of the diluted extract to 975 μL of assay buffer A (50 mM Tris, pH 8.0, 10 mM MgCl2, and 18 mM H2O2). The decomposition of H2O2 was monitored spectrophotometrically, as described elsewhere [49].
Superoxide dismutase assays
Cells were cultured, harvested, and cell pellets obtained as described above for aconitase assays or as specified in the figure legend. SOD activity in the cell lysates was determined using the xanthine oxidase-cytochrome c method as described elsewhere [50].
Succinate dehydrogenase assays
Cells were cultured, harvested, and cell pellets obtained as described above for aconitase assays or as specified in the figure legend. Sdh activity was determined as described previously [46]. The reaction mixture contained PBS (pH 7.5), 2 mM KCN, 742 nM XTT, 19.5 nM phenazine methosulfate and 130 mM sodium succinate (pH 7.1). The reaction was initiated by the addition of cell-free extract and reduction of XTT was monitored at 490 nm.
For all data examining enzymatic activity that is presented in this study, activity was standardized with respect to the protein concentration of the cell-free lysate and subsequent normalizations were as mentioned in the individual panels.
Protein purification
SrrA containing a C-terminal poly-histidine affinity tag was overproduced in and purified from E. coli. E.coli BL21-AI* cells containing the pET20b-srrA plasmid were grown to an optical density of 0.6 (A600) before induction by the addition of 0.3% arabinose. Cells were harvested by centrifugation and flash frozen in liquid nitrogen. Cells were lysed by resuspension in 50 mL of Buffer A (50 mM Tris, pH 7.5, 150 mM NaCl and 5% glycerol) and passage through a chilled French pressure cell. Lysates were subsequently clarified by centrifugation and loaded onto a column of NI-NTA resin (Qiagen), equilibrated with Buffer A. The column was washed with 25 column volumes of wash buffer (50 mM Tris, pH 7.5, 2 M NaCl and 5% glycerol) before equilibration with 5 column volumes of Buffer A. Protein was eluted using a linear gradient with Buffer A containing 200 mM Imidazole. Fractions were collected and analyzed by SDS-PAGE. Fractions containing SrrA protein were pooled and dialyzed overnight at 4°C in Buffer B (50 mM Tris, pH 7.5, 110 mM KCl, 5% glycerol and 1 mM DTT). Dialyzed protein was loaded onto a column containing Q-Sepharose resin pre-equilibrated with Buffer B. The column was washed with Buffer B and SrrA was eluted using a linear gradient of Buffer B containing 2 M KCl. Fractions were collected and analyzed using SDS-PAGE. Fractions containing SrrA protein at >95% purity were pooled and dialyzed overnight against Buffer B containing 0.01 M EDTA. SrrA was concentrated over a 3,000 Dalton molecular-mass-cutoff membrane (Amicon YM-3). Purified protein was further verified using western blots as previously described [51]. Concentrated protein was flash frozen in single use aliquots in liquid nitrogen. Concentration for proteins was routinely determined using micro-Biuret assay [52].
DNA mobility shift assays
Primers were used amplify 150 base pairs upstream of the annotated translational start sites of the rpsC, srrA, and dps genes using PCR. A biotin labeled primer was used to create all amplicons except those used as cold competitor controls. All amplicons were purified using a 3.5% NATIVE-PAGE gel, as described elsewhere (National Diagnostics). Prior to binding assays, phosphorylated SrrA (Srr~P) was obtained by incubating SrrA with 90 mM acetylphosphate at room temperature for 100 minutes. Binding assays were carried out in a reaction volume of 27 μL containing 15–146 ng of SrrA protein, 8 fmol of labeled DNA, 30 ng non-specific poly(dI-dC) DNA, 20 mM EDTA and 5 mM MgSO4. After 20 minutes of incubation at room temperature the binding reactions were subjected to electrophoresis on a 3.5% TBE gel. The DNAs were subsequently transferred to a Hybond N membrane (GE Healthcare) and the blots were developed using chemiluminescent detection (Lightshift Kit, Pierce). Specific DNA::protein interactions were verified using 200-fold molar excess of a cold competitor control.
Results
A strain lacking the staphylococcal respiratory regulatory system (SrrAB) has increased susceptibility to H2O2
We screened a library of S. aureus strains lacking individual two-component regulatory systems (TCRS) for H2O2 sensitivity. A strain lacking the SrrAB TCRS had an extended lag-time before outgrowth when compared to the parental USA300_LAC strain (wild-type; WT) when challenged with H2O2 (Fig 1A and 1B). Returning srrAB in trans negated the H2O2 sensitivity of the ΔsrrAB strain verifying that the absence of SrrAB was resulting in the sensitivity phenotype (Fig 1A and 1B). The time necessary for the WT to initiate outgrowth increased in synchrony with increasing concentrations of H2O2 (S1A Fig). The lag-times for the ΔsrrAB strain also increased in synchrony with increasing H2O2, but the lag-times were significantly greater than those of the WT (S1B Fig). However, challenge with H2O2 did not significantly alter the generation times of either strain (data not shown).
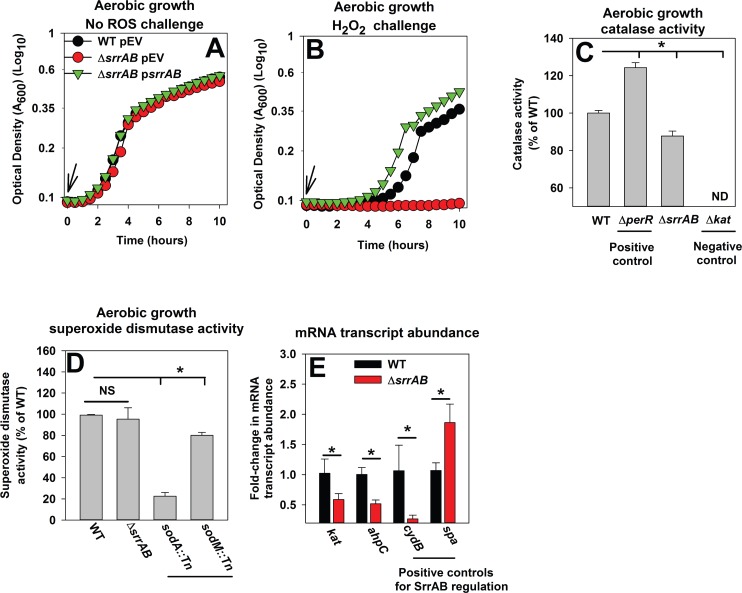
Fig 1. A ΔsrrAB strain is deficient in growth upon H2O2 challenge and has decreased transcription of ahpC and kat during aerobic growth.
Panels A-B; A ΔsrrAB strain is deficient in growth upon H2O2 challenge. The WT (JMB1100) with pCM28 empty vector (pEV) and the ΔsrrAB strain (JMB1467) with pCM28 (pEV) or psrrAB were diluted into defined medium to a final optical density of 0.025 (A600) and challenged with vehicle control (Panel A) or 1.32 mM hydrogen peroxide (H2O2) (Panel B) at the point of inoculation (indicated by arrow). Panel C; Catalase (Kat) activity is decreased in a ΔsrrAB strain. Kat activity was assessed in cell-free lysates from the WT, ΔsrrAB, ΔperR (JMB2151), and Δkat (JMB2078) strains. ND represents no detectable activity. Panel D; Superoxide dismutase (Sod) activity is not decreased in a ΔsrrAB strain. Sod activity was assessed in cell-free lysates from the WT, ΔsrrAB, sodA::Tn (JMB 6326), and sodM::Tn (JMB 2968) strains. Panel E; The abundances of mRNA transcripts corresponding to genes that encode for H2O2 detoxification proteins are lower in the ΔsrrAB strain. The abundances of the ahpC, kat, cydB and spa transcripts were determined in the WT and ΔsrrAB strains. The data were normalized to 16s rRNA transcript levels and are presented as fold-change relative to the WT. Representative data are shown in Panels A-B and experiments were repeated on three independent occasions. Data in Panel C-E represent the average of biological triplicates with standard deviations shown. Where indicated, two-tail student t-tests were performed on the data and * represents statistically significant data with P< 0.05.
To further examine this phenomenon, cells were resuspended in buffer and challenged with a bolus of H2O2. The stress was terminated by the addition of catalase and the viable cells were quantified. The ΔsrrAB strain had a ~10-fold decrease in survival post H2O2 challenge (S1C Fig) and this phenotype could be genetically complemented. Moreover, increasing the initial inoculum of the ΔsrrAB strain 10-fold resulted in a lag-time before outgrowth that was similar to that of the WT upon H2O2 challenge (data not shown). We concluded that the increased lag-time of the ΔsrrAB strain upon H2O2 challenge was likely an outcome of decreased cell viability.
SrrAB positively influences basal expression of H2O2 resistance factors
Ahp and Kat detoxify H2O2 and SodA has been proposed to facilitate resistance to H2O2 in alternate bacteria [16, 53–56]. PerR represses expression of peroxide resistance factors including Kat [57]. Catalase (Kat) activity was measured in cell-free lysates harvested from the WT, ΔsrrAB, Δkat (negative control) and ΔperR (positive control) strains, cultured aerobically. The ΔsrrAB strain displayed ~15% lower catalase activity than the WT (P<0.05) (Fig 1C). Kat activity was increased by ~25% in the ΔperR mutant while no activity was detected in the Δkat strain. Superoxide dismutase activity was measured in cell-free lysates harvested from the WT, ΔsrrAB, sodA::Tn, and sodM::Tn strains cultured aerobically. Sod activity was not decreased in the ΔsrrAB strain, but it was decreased by ~75% in the sodA::Tn strain and by ~20% in the sodM::Tn strain (Fig 1D).
We next examined whether SrrAB modulated the transcript levels of ahpC and kat. SrrAB is known to alter transcription of spa and cydB and SrrA directly modulates transcription of spa. The spa and cydB genes are repressed and activated by SrrAB, respectively [31, 34, 35]. The abundances of the transcripts corresponding to ahpC, kat, and cydB were reduced in the ΔsrrAB strain (~2–3 fold for each gene) (P<0.05) (Fig 1E). The abundance of the spa transcript was increased in the ΔsrrAB strain (~2 fold) (P<0.05) (Fig 1E).
We assessed whether SrrAB was required for the induction of ahpC and kat in cells challenged with H2O2. The WT and ΔsrrAB strains were cultured to post-exponential growth phase, challenged with a bolus of H2O2, and the transcript abundances were assessed. The changes in transcript levels for ahpC and kat, upon H2O2 challenge, were indistinguishable between the WT and ΔsrrAB strains. (S2 Fig).
From Fig 1, S1 Fig and S2 Fig, we concluded that SrrAB a) positively influences the basal transcript levels for ahpC and kat, and b) manages the mutually inclusive regulation of genes required for H2O2 resistance, aerobic respiration (cydB) and virulence (spa) during aerobic growth.
SrrAB positively influences Dps expression and iron chelation or introduction of a null perR allele alleviates the deficient survival of the ΔsrrAB strain upon H2O2 challenge
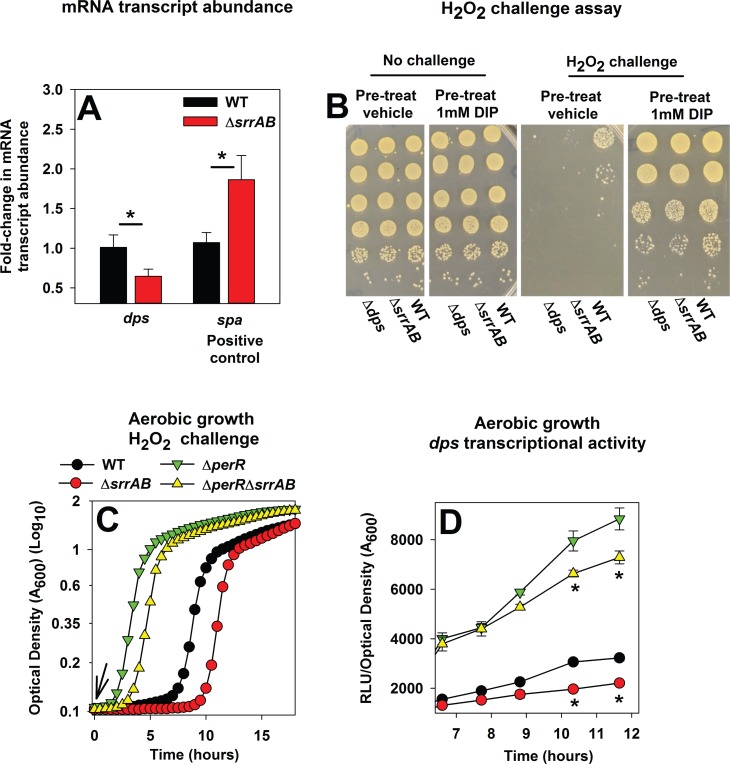
Dps protects cells from H2O2 toxicity [21–23]. SrrAB could positively influence dps transcription, and thereby influence H2O2 resistance. Consistent with this theory, the abundance of the dps transcript was decreased in the ΔsrrAB strain (Fig 2A). In part, Dps imparts cellular protection by binding and sequestering free Fe, and thereby suppressing Fenton chemistry. We examined whether pre-incubation of the ΔsrrAB and Δdps strains with an iron chelator would mitigate the survival of these strains upon H2O2 challenge. The ΔsrrAB and Δdps strains that had been pre-incubated with vehicle-control, followed by H2O2 challenge, displayed decreased survival when compared to the WT. However, pre-incubation of strains with the cell permeable metal chelator 2,2-dipyrydyl, followed by H2O2 challenge, rescued the phenotypes of the ΔsrrAB and Δdps strains (Fig 2B).
Fig 2. SrrAB positively influences Dps expression and iron chelation or introduction of a ΔperR allele alleviates the deficient survival of a ΔsrrAB mutant upon H2O2 challenge.
Panel A; The abundance of the dps transcript is lower in the ΔsrrAB strain. The abundances of the dps and spa transcripts were determined in the WT (JMB1100) and ΔsrrAB (JMB1467) strains from the cDNA libraries described in Fig 1E. The data were normalized to 16s rRNA transcript levels and are presented as fold-change relative to the WT. Panel B; Pre-incubation of the ΔsrrAB and Δdps strains with a metal chelator alleviates their sensitivity towards H2O2 challenge. The WT, ΔsrrAB, and Δdps (JMB2092) strains were cultured aerobically and subsequently incubated in buffer with vehicle control or 1 mM 2,2 dipyrydyl. Cells were then challenged with H2O2 and survival was determining colony-forming units (CFU). Panel C; Introduction of a ΔperR allele mitigates the H2O2 sensitivity phenotype of the ΔsrrAB strain. The WT, ΔsrrAB, ΔperR (JMB2151), and ΔperR ΔsrrAB (JMB2615) strains were diluted into TSB and challenged with 1.57 mM H2O2 at the point of inoculation (indicated by arrow). Panel D; SrrAB and PerR influence dps transcriptional activity independent of one another. The transcriptional activity of dps was assessed in the WT, ΔsrrAB, ΔperR and ΔperR ΔsrrAB strains containing gfp under the transcriptional control of the dps promoter (pCM11_dps). Representative data are displayed in Panels B and C and experiments were performed on least three independent occasions. Data in Panels A and D represent the average of biological triplicates with standard deviations shown. Two-tail student t-tests were performed on the data in Panel A and * represents statistically significant data with P< 0.05.
Next, we examined whether increasing the expression of H2O2 resistance factors in the ΔsrrAB strain would mitigate the H2O2 sensitivity phenotype of this strain. PerR represses transcription of ahpC, kat, and dps (52). As expected, the ΔperR strain displayed increased resistance to H2O2 challenge (Fig 2C). Introduction of the ΔperR mutation mitigated the H2O2 sensitivity phenotype of the ΔsrrAB strain. The ΔperR ΔsrrAB strain displayed increased resistance towards H2O2 relative to the WT; however, this resistance was lower than that of the ΔperR mutant. These data suggested that PerR and SrrAB influence the expression of peroxide resistance factors independent of one another. Consistent with this premise, Kat activity was lower in the ΔperR ΔsrrAB strain than in the ΔperR strain (S3 Fig). Moreover, the transcriptional activity of dps was lower in the ΔperR ΔsrrAB strain than the ΔperR strain (Fig 2D).
From the data in Fig 2 and S3 Fig we concluded that a) the H2O2 sensitivity of the ΔsrrAB strain was the result of decreased expression of H2O2 resistance factors, and b) SrrAB influences H2O2 resistance factors independently of PerR.
SrrA binds to the promoter regions of dps and srrA
SrrA is an OmpR family response regulator that binds to the srrA promoter [31]. OmpR type regulators may bind inverted repeat sequences and RR binding regions can have an internal spacer of a varying length and sequence [58–62]. Visual inspection of the srrA promoter revealed the presence of an inverted repeat sequence separated by a four base-pair spacer region (AAATAN4TTTAT). The potential binding site was also found in the promoter regions of icaA (SAUSA300_2600), tsst (SA1819), and agrB (P2 promoter, SAUSA300_1989) (S4 Fig); each of which is a direct binding target for SrrA~P [31, 35]. The inverted repeat was not found to occur in the promoter regions for four randomly selected genes in LAC (SAUSA300- 2112, 1112, 0124 and 2198). Allowing for a maximum of one mismatch, the inverted repeat was found to be present in the promoter region for dps (S4 Fig). A putative SrrA binding sequence was not observed in the promoter regions for kat or ahpC.
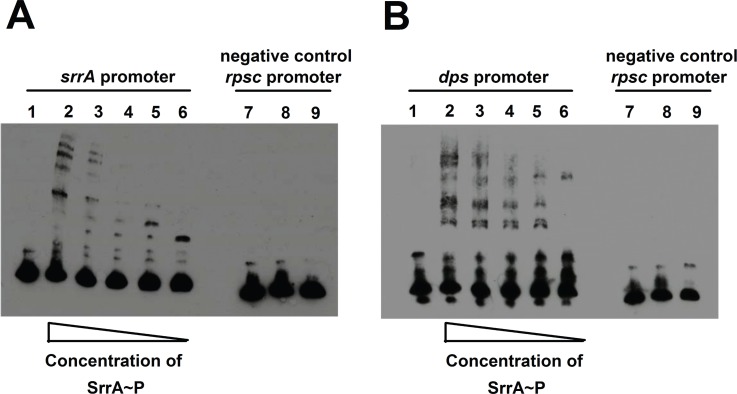
Electromobility gel shift assays (EMSAs) were used to assess whether SrrA is capable of binding to the promoter region of dps. Binding to the srrA promoter was assessed as a positive control. The promoter of rpsC lacks the putative SrrA binding region, and was included as a negative control for non-specific DNA binding. SrrA bound to 150 base pair DNA fragments corresponding to the sequences located upstream of the annotated transcriptional start sites for srrA and dps in a concentration dependent manner (Fig 3A and 3B). SrrA did not bind to the rpsC promoter at the highest protein concentrations examined (Fig 3A and 3B). Consistent with the SrrA::DNA interactions being specific, the addition of excess unlabeled promoter DNA decreased the amount of biotin labeled DNA bound by SrrA. The data presented in Fig 2 and Fig 3 suggested that SrrAB directly modulates the transcription of dps.
Fig 3. SrrA binds to DNA fragments immediately preceding the annotated transcriptional start sites for srrA and dps.
Panels A and B; Electromobility gel shift assays (EMSAs) demonstrating binding of SrrA to DNAs that correspond to the 150 base pair segments immediately preceding the annotated transcriptional start sites for the srrA (A) and dps (B). EMSAs were performed with SrrA (15–146 ng) and 8 fM of biotin labeled DNA. For each gel, the samples in lane 1 contain 146 ng SrrA with labeled sample DNA and a 125-fold excess of non-labeled (cold) competitor DNA. The samples in lanes 2–5 contain labeled DNA with varying amounts of SrrA protein (15–146 ng). The samples in lane 6 contain labeled DNA, but no SrrA. The samples in lanes 7–9 show that the interaction of SrrA with DNA is specific. The samples in lane 7 contain 146 ng SrrA with rpsC promoter DNA and a 125-fold excess of non-labeled (cold) competitor DNA. The samples in lanes 8 contain rpsC promoter DNA with 146 ng of SrrA. The samples in lane 9 contain rpsC promoter DNA, but no SrrA.
SrrAB influences H2O2 resistance independent of CidA and CidB in S. aureus LAC
While this manuscript was under preparation, a study was published investigating the influence of SrrAB upon programmed cell death mediated by cidABC [63]. The study used a special medium (TSB medium amended with 35 mM glucose) that induces cell death phenotypes [63]. The authors found that SrrAB represses the transcription of cidABC when cultured in the cell death media. They also found that a ΔsrrAB mutant was sensitive to H2O2 intoxication and this phenotype was mitigated by the introduction of a null cidA or a null cidB allele. These data led to the hypothesis that the H2O2 sensitivity of the ΔsrrAB strain was the result of increased CidA and CidB activity. However, we found that the introduction of a cidA::Tn or a cidB::Tn into the LAC ΔsrrAB strain did not alleviate H2O2 sensitivity when cultured in TSB medium (S5 Fig). Strains lacking Kat or PerR displayed increased sensitivity and resistance phenotypes towards H2O2, as expected (S5 Fig). We concluded that the H2O2 sensitivity of the S. aureus LAC ΔsrrAB strain cultured under our growth conditions and in TSB medium is independent of CidA or CidB.
SrrAB negatively influences ahpC and kat transcription during early exponential growth in TSB and during fermentative growth
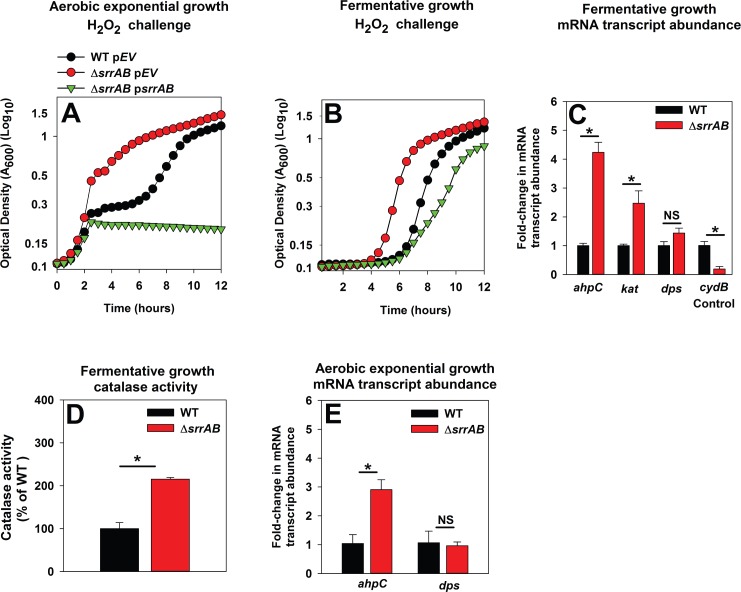
While we were revising this manuscript, Oogai et al. published a report finding that during aerobic exponential-growth in TSB medium a S. aureus ΔsrrAB mutant in the MW2 background had increased transcription of ahpC and kat (but not dps), as well as increased resistance towards H2O2 [64]. In contrast to Oogai et al., the H2O2 sensitivity phenotypes reported herein and by Windham et al. were conducted upon cells cultured to post-exponential growth. We tested the hypothesis that SrrAB negatively modulates H2O2 resistance during exponential growth. Consistent with our reasoning, a LAC ΔsrrAB strain, cultured to exponential phase, followed by challenge with H2O2, displayed increased resistance and this phenotype could be genetically complemented (Fig 4A).
Fig 4. SrrAB negatively influences ahpC and kat during the early exponential growth and fermentative growth phases upon culture in TSB medium.
Panels A-B; The ΔsrrAB strain is resistant towards H2O2 challenge when cultured aerobically to exponential growth (Panel A) or fermentatively (Panel B). The WT (JMB1100) with pCM28 empty vector (pEV) and the ΔsrrAB strain (JMB1467) with pCM28 (pEV) or psrrAB were cultured in TSB aerobically to exponential growth phase (2 doublings) (Panel A) or fermentatively (Panel B). The cells were subsequently challenged with 2.6 mM (Panel A) or 0.22 mM H2O2 (Panel B) and growth was monitored aerobically. Panels C; The mRNA transcript abundances corresponding to ahpC and kat are increased in the ΔsrrAB strain cultured fermentatively. The abundances of the ahpC, kat, dps, and cydB mRNA transcripts were determined in the WT and ΔsrrAB strains cultured fermentatively. Panel D; Catalase (Kat) activity is increased in a ΔsrrAB strain cultured fermentatively. Kat activity was assessed in cell-free lysates from the WT and ΔsrrAB strains after fermentative culture. Panel E; The abundance of the ahpC transcript is increased in the ΔsrrAB strain cultured aerobically to exponential growth. Transcript abundances corresponding to ahpC and dps were quantified in the WT and ΔsrrAB strains cultured aerobically to exponential growth phase. The data in Panels C and E were normalized to 16s rRNA transcript levels and are presented as fold-change relative to the WT. Data shown in panels C-E represent the average of biological triplicates with standard deviations shown. Representative growth profiles are presented in Panels A and B and experiments were performed on least three independent occasions. Where indicated, two-tail student t-tests were performed on data and * denotes p< 0.05 and NS denotes not significant.
When S. aureus is sub-cultured into TSB medium the TCA cycle is largely repressed during the initial period of growth (exponential growth) and fermentative by-products accumulate in the extracellular milleu [65, 66]. Subsequently, TCA cycle function increases as the fermentative byproducts produced during initial growth are utilized to support respiratory growth (post-exponential growth) [42, 65, 66]. Decreased TCA cycle activity during exponential growth would result in reduced flux through respiratory pathways. We examined whether SrrAB modulates H2O2 resistance negatively during fermentative growth. Strains were cultured anaerobically in the absence of a terminal electron acceptor (fermentative growth), followed by challenge with H2O2. Subsequent growth was monitored aerobically. The ΔsrrAB strain displayed greater resistance to H2O2 challenge and this phenotype could be genetically complemented (Fig 4B). The growth of the non-challenged ΔsrrAB and WT strains was indistinguible (data not shown).
Transcriptional analyses supported the H2O2 resistance phenotypes observed during fermentative and aerobic exponential growth. The transcript levels for kat and ahpC were increased ~5- and ~3-fold in the ΔsrrAB strain during fermentative growth (P<0.05), while those for dps were not significantly altered (Fig 4C). Moreover, Kat activity in cell-free extracts of the fermentatively cultured ΔsrrAB strain was ~200% higher (Fig 4D). Likewise, transcript levels for ahpC were increased ~3 fold in the ΔsrrAB strain during aerobic exponential growth, while those for dps were not significantly altered (Fig 4E). These findings led us to conclude that SrrAB negatively influences expression of AhpC and Kat during growth conditions when respiratory activity is low or absent.
A ΔsrrAB mutant incurs increased damage to aconitase during post-exponential aerobic growth
Aconitase (AcnA) requires a solvent exposed [Fe4-S4] cluster for function [48]. Enzymes with solvent exposed FeS cofactors are poisoned by H2O2 [2–5]. We tested the hypothesis that SrrAB manages the expression of genes required for H2O2 resistance during respiratory growth, and thereby imparts protection to AcnA from ROS-induced damage.
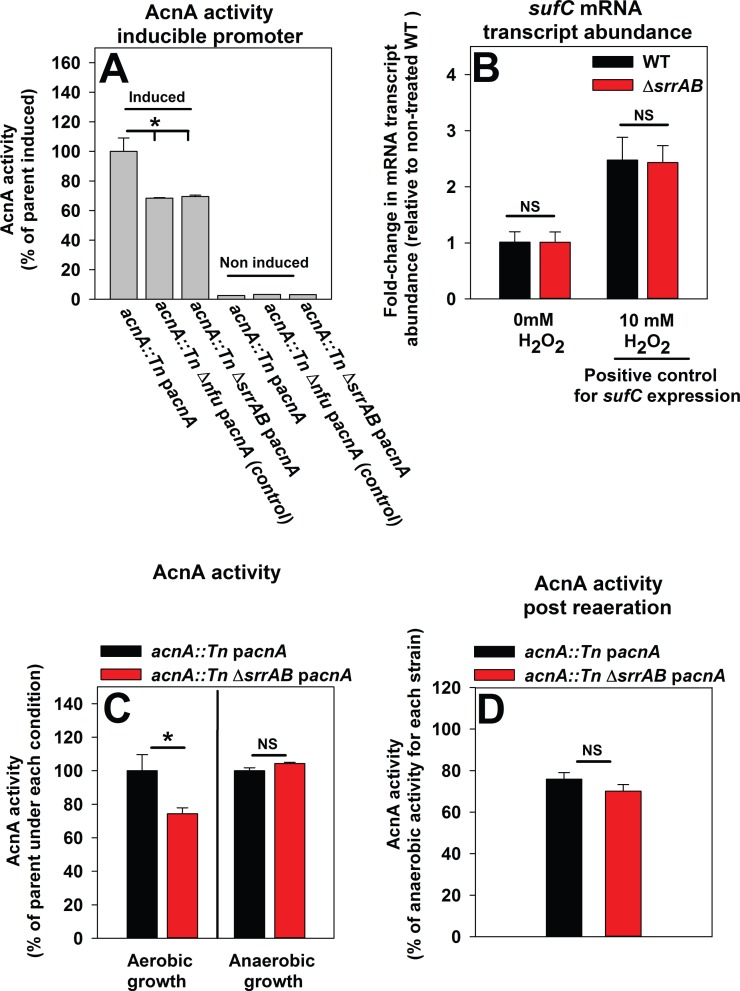
We constructed acnA::Tn and acnA::Tn ΔsrrAB strains and returned acnA into the strains under the transcriptional control of a xylose inducible promoter (pacnA). Introduction of pacnA allows for the control of acnA transcription, thereby negating potential changes in acnA transcription between strains. Nfu is necessary for the maturation of holo-AcnA; therefore, an acnA::Tn Δnfu strain carrying pacnA was included as a positive control [47]. AcnA activity was assessed during post-exponential growth and found to be ~25% lower in the Δnfu and ΔsrrAB strains (Fig 5A).
Fig 5. A ΔsrrAB strain incurs increased damage to aconitase when cultured aerobically.
Panel A; The activity of aconitase (AcnA) is decreased in a ΔsrrAB mutant and this phenotype is independent of acnA transcription levels. AcnA activity was assessed in the acnA::Tn (JMB 3537; parent), acnA::Tn Δnfu (JMB 3538), and acnA::Tn ΔsrrAB (JMB 4367) strains carrying pacnA, which contains acnA under the transcriptional control of a xylose inducible promoter. Strains were cultured aerobically in the presence (induced) or absence (not induced) of 1% xylose. Panel B; The sufC mRNA transcript accumulates to similar levels in the WT and ΔsrrAB strains. The WT (JMB1100) and ΔsrrAB (JMB1467) strains were cultured aerobically and challenged with either 10 mM H2O2 or vehicle control and the abundance of the sufC mRNA transcript was quantified. Data were normalized to the 16s rRNA transcript levels and are presented as fold-change relative to the WT strain. Panel C; The activity of AcnA is similar in the WT and ΔsrrAB strains when cultured anaerobically. AcnA activity was assessed in the acnA::Tn (JMB 3537; parent) and acnA::Tn ΔsrrAB (JMB 4367) strains carrying pacnA that were cultured either aerobically or anaerobically. Panel D; A ΔsrrAB mutant does not display increased dioxygen damage to AcnA. The acnA::Tn (JMB 3537; parent) and acnA::Tn ΔsrrAB (JMB 4367) strains carrying pacnA were cultured anaerobically for 4.5 hours, treated with a protein synthesis inhibitor (100 μg mL-1 rifampicin) and either exposed to dioxygen or incubated anaerobically for 35 minutes subsequent to determining the activity of AcnA. Data in Panels A-D represent the average of biological triplicates. Where indicated, two-tail student t-tests were performed on data and * denotes p< 0.05 and NS denotes not significant.
Altered transcription of genes required for the synthesis of FeS clusters could result in decreased AcnA activity in the ΔsrrAB strain. The sufCDSUB gene products are responsible for the synthesis of FeS clusters and decreased transcription of sufCDSUB results in decreased FeS enzyme activity (Roberts C., et al., in revision). Transcription of sufC was not altered in the ΔsrrAB strain (Fig 3B). Transcription of sufC increases upon H2O2 challenge [47]. However, transcription of sufC was induced to a similar extent (~2 fold) in both the WT and ΔsrrAB strains upon H2O2 challenge (Fig 5B) [47]. Likewise, suf transcriptional activity was similar in the WT and ΔsrrAB strains over time (S6 Fig).
Two experiments were conducted to determine whether increased damage by dioxygen underlies the low AcnA activity observed in the ΔsrrAB mutant. First, AcnA activity was assessed in the acnA::Tn and acnA::Tn ΔsrrAB strains carrying pacnA and cultured in the presence and absence of dioxygen. Culturing the ΔsrrAB mutant anaerobically restored AcnA activity (Fig 5C). Second, the same strains were cultured anaerobically, treated with a protein synthesis inhibitor (rifampicin), and one-half of the cultures were exposed to dioxygen [8], while the remainder were incubated anaerobically. Exposure of cells to dioxygen resulted in a decrease in AcnA activity, but importantly, the activity was indistinguishable between the two strains (Fig 5D). From Fig 5 we concluded that the ΔsrrAB mutant had decreased AcnA activity when cultured aerobically and this phenotype was a) not an outcome of decreased transcription of sufC, or b) increased damage by dioxygen.
SrrAB increases the ability of S. aureus to resist H2O2 intoxication during periods of high respiratory flux protecting AcnA
Respiratory flux is subject to the concentration of the terminal electron acceptor in the culture medium and SrrAB is responsive to changes in respiratory status [36, 66, 67]. SrrAB did not positively influence H2O2 resistance factors during fermentative growth and AcnA did not incur damage in a ΔsrrAB mutant following fermentative growth. Thus, we hypothesized that the influence of SrrAB upon the expression of H2O2 resistance factors, and the subsequent protection imparted to AcnA, would be altered with respect to dioxygen concentrations in the medium and the ensuing changes in respiratory flux.
Dioxygen diffusion into growth media is a function of the aeration experienced by cultures. Aeration of batch cultures can be modified by altering the culture vessel headspace to the medium volume ratio (HVR). Decreased HVR leads to decreased aeration and less dissolved dioxygen in the medium [66–69]. Culturing S. aureus under low aeration (HVR 1.25) results in a metabolic block at the TCA cycle [66], which would result in reduced flux through respiratory pathways.
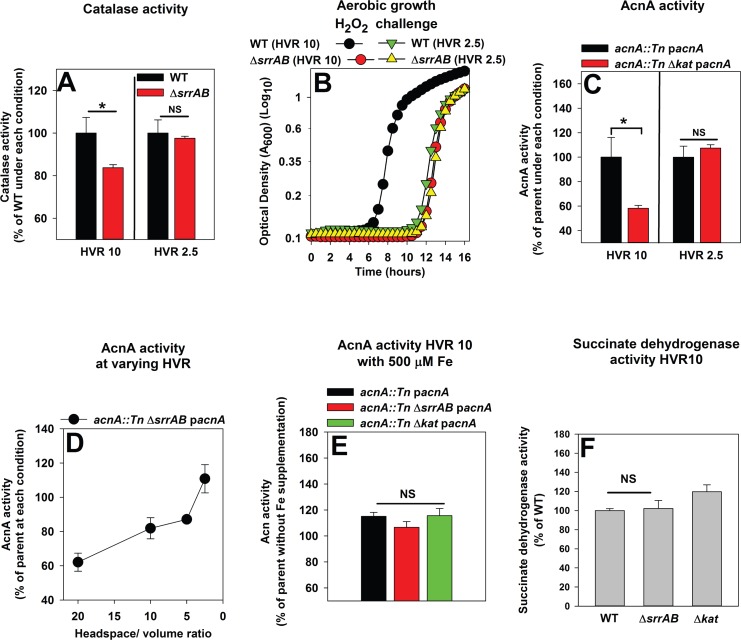
We examined whether SrrAB alters the expression of H2O2 resistance factors in response to culture aeration. Kat activity was reduced by ~20% in the ΔsrrAB strain when cultured under high aeration (HVR 10), but was statistically indistinguishable from that of the WT when cultured under low aeration (HVR 2.5) (Fig 6A). Increased expression of H2O2 resistance factors would be predicted to result in greater resistance towards H2O2 in cells cultured with high aeration. Consistent with this idea, WT cells cultured under high aeration (HVR 10) were more resistant to H2O2 challenge than cells cultured under low aeration (HVR 2.5). In contrast, the sensitivity of the ΔsrrAB strain towards H2O2 was not altered as a variable of HV ratio (Fig 6B).
Fig 6. SrrAB upregulates H2O2 resistance during periods of high aeration and imparts protection to AcnA.
Panel A; SrrAB positively influences catalase (Kat) expression in cells cultured under high aeration, but not under low aeration. Kat activity was assessed in cell-free lysates generated from the WT (JMB1100) and ΔsrrAB (JMB1467) strains cultured at a culture vessel headspace to liquid volume ratios (HVR) of 2.5 or 10. Panel B; SrrAB increases the ability of S. aureus to withstand H2O2 intoxication when cells are cultured under high aeration. The WT and ΔsrrAB strains were cultured at a HVR of 10 or 2.5, diluted into fresh TSB medium and challenged with 1.57 mM H2O2 at the point of inoculation. Panel C; AcnA activity is decreased in a strain lacking catalase when cultured under high aeration, but not under low aeration. AcnA activity was assessed in cell-free lysates harvested from the acnA::Tn (JMB3537; parent) and the acnA::Tn Δkat (JMB7107) strains carrying pacnA cultured at a HVR of 2.5 or 10. Panel D; AcnA activity is decreased in a ΔsrrAB mutant cultured under high aeration, but not when cultured under low aeration. AcnA activity was assessed in cell-free lysates from the acnA::Tn (JMB3537; parent) and acnA::Tn ΔsrrAB (JMB4367) strains carrying pacnA cultured at various HVRs. Panel E; The decreased AcnA activity under high aeration in strains defective in H2O2 resistance is restored by supplementation of growth medium with Fe. AcnA activity was assessed in cell-free lysates harvested from the acnA::Tn (JMB3537; parent), acnA::Tn ΔsrrAB (JMB4367), and acnA::Tn Δkat (JMB7107) strains carrying pacnA. Cells were cultured at HVR of 10 and in the presence or absence of 500 μM Fe. Panel F; Succinate dehydrogenase (Sdh) activity is not decreased in strains deficient in H2O2 resistance. Sdh activity was assessed in cell-free lysates from the WT, ΔsrrAB, and Δkat (JMB2078) strains cultured at a HVR of 10. Data shown in Panels A, C and D-F represent the average of biological triplicates with standard deviations shown. Representative growth profiles are presented in Panel B and experiments were performed on least three independent occasions. Where indicated, two-tail student t-tests were performed on data and * denotes p< 0.05 and NS denotes not significant.
We reasoned that a strain deficient in scavenging H2O2 would have increased AcnA damage resulting in decreased AcnA activity when cultured under high aeration (HVR 10). The acnA::Tn Δkat strain carrying pacnA displayed lower AcnA activity than the acnA::Tn strain carrying pacnA when cultured under high aeration (HVR 10), but not when cultured under low aeration (HVR 2.5) (Fig 6C). AcnA activity was also assessed in the acnA::Tn and acnA::Tn ΔsrrAB strains carrying pacnA following culture under varying HVRs. AcnA activity in the ΔsrrAB acnA::Tn strain with pacnA was significantly lower than that of the acnA::Tn strain with pacnA (~60%) when cultured under high aeration (HVR 20). However, as the HVR was decreased, the difference in AcnA activity between the two strains diminished. AcnA activity was fully restored in ΔsrrAB mutant after culture at a HVR of 2.5 (Fig 6D).
Supplementing the growth medium with excess iron (Fe) restores the activities of FeS cluster requiring dehydratase enzymes damaged by oxidation [70, 71]. The acnA::Tn, acnA::Tn ΔsrrAB, and acnA:Tn Δkat strains carrying pacnA were cultured under high aeration (HVR 10) with and without Fe supplementation (500 μM). Supplementation of media with Fe recovered AcnA activity in the ΔsrrAB and Δkat strains (Fig 6E).
Succinate dehydrogenase (Sdh) also requires Fe-S clusters for function. However, the Fe-S clusters of Sdh are protected by the polypeptide providing increased stability [72, 73]. Consequently, exposure of up to 1mM H2O2 does not inactivate Sdh [72, 73]. Sdh activity was not decreased in the ΔsrrAB or Δkat strains cultured under high aeration (HVR 10) (Fig 6F). These findings confirmed that a) the ΔsrrAB and Δkat strains do not have a broad deficiency in the function of FeS proteins, and b) H2O2 accumulation in S. aureus predominantly damages solvent exposed FeS clusters.
The data presented in Fig 6 demonstrated that S. aureus increased the ability to resist H2O2 when cultured under high aeration and these alterations arose due to regulatory changes mediated by SrrAB. Further, strains deficient in detoxifying endogenously produced H2O2 (ΔsrrAB and Δkat) displayed decreased AcnA activity when cultured under high aeration, but not low aeration.
SrrAB positively influences transcription of scdA, which encodes for a FeS cluster repair protein
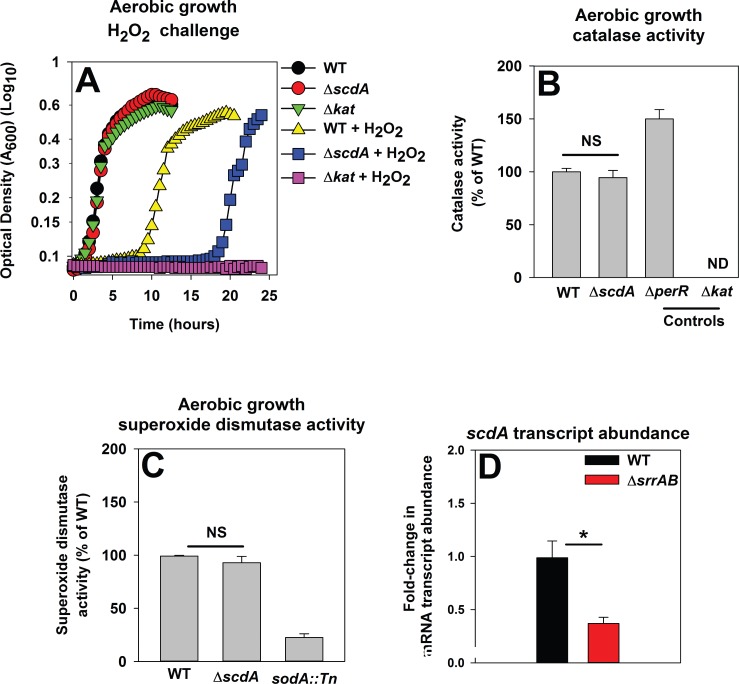
The S. aureus di-iron RIC protein ScdA (YtfE in Escherichia coli) has a role in the repair of FeS proteins damaged by H2O2 [24, 25]. Previous studies on ScdA were conducted in the S. aureus strain RN4220, which lacks Sigma B and Agr funtion, which control expression of ROS resistance genes [74, 75]. We conducted experiments to determine whether a) ScdA is required for H2O2 resistance in S. aureus LAC, b) the H2O2 sensitivity of a ΔscdA strain is independent of altered expression of detoxification factors, and c) SrrAB modulates scdA transcription. S. aureus LAC ΔscdA and Δkat strains displayed increased sensitivity towards H2O2 (Fig 7A). However, the ΔscdA strain did not display decreased Kat (Fig 7B) or Sod activity (Fig 7C). The transcript level corresponding to scdA was decreased (~3 fold) in the ΔsrrAB strain during post-exponential growth (Fig 7D).
Fig 7. SrrAB positively influences the transcription of scdA.
Panel A; A S. aureus USA300_LAC ΔscdA mutant is sensitive to H2O2 intoxication. The WT (JMB1100), ΔscdA (JMB1254), and Δkat (JMB2078) strains were cultured aerobically, diluted into fresh medium, and challenged with 1.57 mM H2O2 at the point of inoculation. Panel B; The ΔscdA strain is not defective in catalase (Kat) activity. Kat activity was assessed in cell-free lysates from the WT, ΔscdA, ΔperR (JMB2151), and Δkat strains cultured at a HVR of 10. Panel C; The ΔscdA strain does not have decreased superoxide dismutase (Sod) activity. Sod activity was assessed in cell-free lysates from the WT, ΔscdA, and ΔsodA::Tn (JMB6326) strains cultured at a HVR of 10. Panel D; The abundance of the scdA mRNA transcript is lower in the ΔsrrAB strain during post-exponential growth. The mRNA abundance corresponding to scdA was determined using the same cDNA libraries as described in Fig 2A. The data were normalized to 16s rRNA transcript levels and are presented as fold-change relative to the WT strain. Representative growth profiles are presented in Panel A and experiments were performed on least three independent occasions. The data in Panels B-D represent the average of biological triplicates with standard deviations shown.
The positive influence of SrrAB upon H2O2 metabolism is conserved in diverse S. aureus isolates
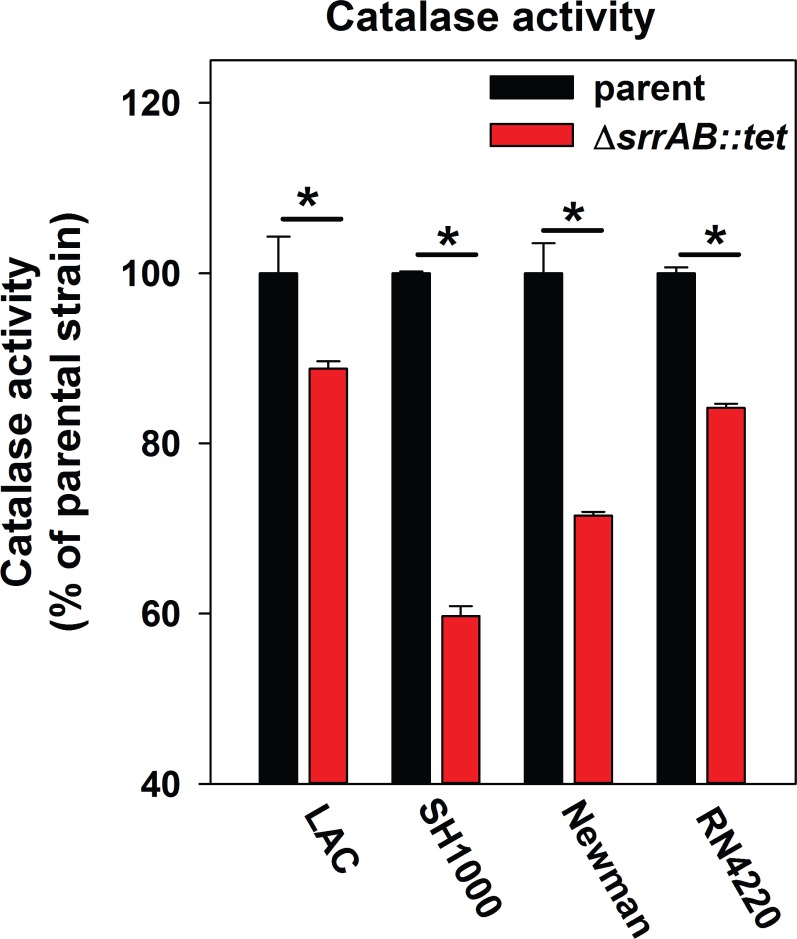
Regulatory networks that are integral to S. aureus physiology differ between isolates [76, 77]. We examined the influence of SrrAB upon ROS metabolism by assessing catalase activity in the SH1000, Newman, and RN4220 genetic backgrounds and their isogenic ΔsrrAB mutants. Strains lacking SrrAB displayed lower catalase activity relative to their parent strains (~10–60% lower catalase activity). When compared to a LAC background, the catalase activity was substantially lower in the Newman and SH1000 ΔsrrAB mutants (~30% and ~60% lower, respectively) (Fig 8). The ΔsrrAB mutants were also more sensitive than their isogenic parent strains to H2O2 challenge (data not shown).
Fig 8. The influence of SrrAB upon aerobic respiration and H2O2 metabolism is conserved in diverse S. aureus isolates.
Catalase (Kat) activity is decreased in diverse S. aureus isolates lacking SrrAB. The USA_300 LAC (JMB1100), USA300_LAC ΔsrrAB (JMB2047), Newman (JMB1422), Newman ΔsrrAB (JMB4751), SH1000 (JMB1323), SH1000 ΔsrrAB (JMB4556), RN4220 (JMB1103), RN4220 ΔsrrAB (JMB2030) strains were cultured aerobically to post-exponential growth phase and Kat activities were determined in the cell-free lysates. The data represent the average of biological triplicates with standard deviations shown.
Discussion
The goal of this study was to further our understanding of the role of SrrAB in H2O2 resistance. During post-exponential growth a ΔsrrAB strain was sensitive to H2O2 challenge. In S. aureus, AhpC and Kat are required for scavenging H2O2 [16]. The iron-sequestering and DNA-binding protein Dps is utilized to suppress Fenton chemistry [21–23]. ScdA (YtfE) also provides resistance to H2O2 by aiding in the repair of H2O2 damaged FeS proteins [24, 25]. The ΔsrrAB strain has decreased transcription of ahpC, kat, dps and scdA; therefore, the H2O2 sensitivity phenotype of the ΔsrrAB strain is likely to arise, in part, as a result of the combined effect of decreased expression of these genes. In support of this idea we found that a) increasing the expression of H2O2 resistance factors by the introduction of a null perR allele into the ΔsrrAB strain increased H2O2 tolerance, and b) pre-incubation of ΔsrrAB strain with a cell permeable iron chelator prior to H2O2 challenge, alleviated the H2O2 sensitivity phenotype. Further, purified SrrA was capable of binding to the promoter region of dps in EMSA assays, suggesting that SrrAB directly modulates the transcription of at least one H2O2 resistance factor.
While our manuscript was under revision, Oogai et al. published a study that both confirmed our findings, and yet, seemingly contradicted them [64]. Oogai et al. found that SrrAB negatively influences AhpC and Kat expression in the S. aureus isolate MW2 [64]. We further investigated this phenomenon in order to reconcile our findings with those of Oogai et al. We found that the negative influence of SrrAB upon H2O2 detoxification occurs during periods of decreased TCA cycle activity (aerobic exponential growth in TSB), which would ultimately lead to decreased flux through respiratory pathways or during fermentative growth. Moreover, SrrAB increased the ability of cells to withstand H2O2 stress when respiratory activity was high. The results presented herein confirm the findings of Oogai et al., as well as significantly expand upon them. The finding that SrrAB altered the H2O2 resistance of S. aureus in response to respiration are in agreement with the prevailing hypothesis that alterations in the cellular respiratory status serve as a stimulus for SrrAB ([36] and Mashruwala et al. in review). Our findings extend the findings of others in emphasizing the importance of careful reporting of the culture media, growth parameters, and growth phases in the study of bacterial physiology [66, 67].
It is currently unclear how SrrAB negatively influences ahpC, kat and scdA transcription. We did not identify putative SrrA binding sites in the promoter regions for ahpC, kat, or scdA suggesting SrrAB indirectly modulates their expression. Oogai et al. reached a similar conclusion for AhpC and Kat [64]. SrrAB, as well as it's ortholog ResDE in Bacillus subtilis, modulate the expression of the small non-coding regulatory RNA called RsaE/RoxS [78]. B. subtilis strains lacking RsaE/RoxS display altered expression of peroxide metabolism genes [78]. Thus, one explanation would be that SrrAB influences the expression of kat, ahpC and ytfe via RsaE/RoxS. We are currently testing this hypothesis. A strain lacking SrrAB also has altered respiration, redirected carbon flux, and by inference an altered redox status [36, 37]. Thus, an alternate explanation is that the physiological changes that are the result of the absence of SrrAB indirectly affect the expression of AhpC, Kat, and Ytfe.
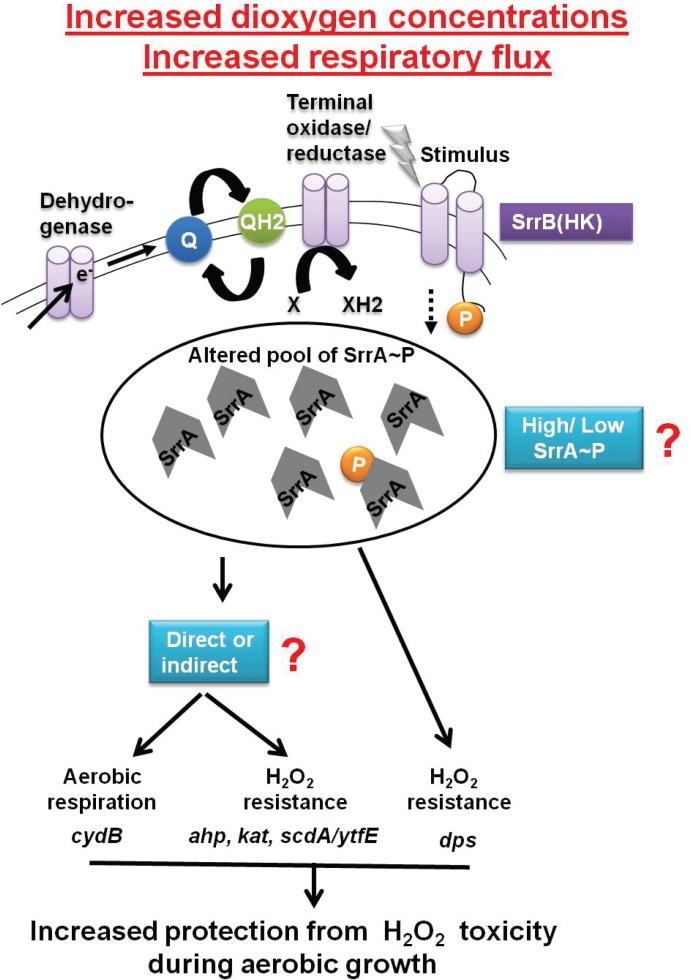
Why would SrrAB regulate H2O2 resistance factors in S. aureus? SrrAB influences the transcription of multiple genes necessary for aerobic respiration [34] in response to respiratory flux [36]. H2O2 is a by-product of spontaneous interactions between dioxygen and components of respiratory pathways [2–5]. Therefore, H2O2 arises spontaneously during aerobic growth [2, 3]. Accumulation of as little as two μM H2O2 has been proposed to be sufficient to inhibit the growth of E. coli [79]. S. aureus cells lacking the ability to synthesize both Ahp and Kat accumulate ~25 μM H2O2. The accumulation of H2O2 is maximal during post-exponential growth, which coincides with maximal respiratory processes [16]. Consequently, during aerobic growth, when S. aureus cells are respiring, it is imperative for the cell to detoxify H2O2 and repair macromolecules damaged by H2O2. This led us to propose a working model displayed in Fig 9 wherein SrrAB manages the mutually inclusive expression of genes required for aerobic respiration, H2O2 detoxification, and the repair of damaged cellular assets (FeS clusters), thereby facilitating proficient growth.
Fig 9. Working model for the role of SrrAB in modulating the transcription of genes utilized in H2O2 resistance and dioxygen respiration.
SrrAB modulates gene transcription in response to cellular respiratory flux [36]. We propose that increased culture aeration leads to increased respiratory flux during post-exponential growth, which results in altered kinase activity of SrrB and variation in the cellular pool of SrrA~P. An altered SrrA~P pool results in increased expression of genes under the SrrAB regulon that are utilized for H2O2 resistance and dioxygen respiration. The resultant physiological changes allow for cellular homeostasis by protecting macromolecules against H2O2 toxicity that arise during dioxygen respiration.
Consistent with our model, SrrAB modulated the transcription of H2O2 detoxification factors (kat and ahpC). Protein-associated solvent accessible FeS clusters, such as the cofactor of AcnA, are among the primary cellular targets of H2O2 [7, 8, 80, 81]. A ΔsrrAB strain had decreased AcnA activity when cultured aerobically and AcnA was restored by either a) decreasing culture aeration, which results in less dissolved dioxygen and decreased cellular respiration [66, 67], b) anaerobic growth, or c) supplementing the growth medium with Fe, which aids in the repair of oxidatively damaged FeS clusters [70, 71]. Moreover, SrrAB positively influenced the transcription of scdA, which functions in the repair of ROS damaged FeS clusters [24, 25]. The oxidation of FeS clusters can result in increased cytosolic free Fe, which can catalyze Fenton chemistry. Data presented suggest that SrrAB directly influences the transcription of dps and Dps is utilized to bind and sequester cytosolic free Fe suppressing Fenton chemistry.
Similar to SrrAB, the ArcAB system in gram-negative bacteria is responsive to the respiratory status of the cell [36, 82]. ArcAB controls the transcription of genes required for TCA cycle activity and fermentative processes. ArcAB also influences the transcription of ROS resistance genes in E. coli, Haemophilus influenzae, and Salmonella enterica [83–85]. In a direct parallel to SrrAB, ArcAB also alters the transcription of dps
SrrAB coregulated the transcription of genes encoding for virulence factors along with genes encoding for aerobic respiration and H2O2 resistance factors. The significance underlying the regulatory tethering of these diverse processes is not clear. However, SrrAB is responsive to cellular respiratory status [36]. Cellular respiration is directly connected to cellular energy homeostasis. Therefore, one explanation would be that S. aureus reprograms its virulence repertoire in accordance with respiratory status and the energetic demands or energetic potential of the cell. However, this idea remains to be tested.
In summary, the results presented in this study show that SrrAB manages the mutually inclusive transcription of genes involved in aerobic respiration and H2O2 resistance. Our data suggest that SrrAB alters expression of H2O2 resistance factors in response to cellular respiratory status. This regulatory tuning imparts protection to AcnA from H2O2 damage and we propose that it is likely to also facilitate the adaptation of S. aureus to shifts in dioxygen concentrations and changes in respiratory flux.
Supporting Information
Panel A; The lag-time necessary for S. aureus to initiate outgrowth is increased as a function of H2O2 concentration. The WT (JMB1100) strain was cultured aerobically and then diluted into fresh defined medium and challenged with varying concentrations of H2O2. Panel B; The lag-times necessary for the ΔsrrAB strain to initiate outgrowth post H2O2 challenge is greater than the lag-times required for the WT strain. The WT (JMB1100) and ΔsrrAB (JMB1467) strains were diluted into fresh defined medium and challenged with various concentrations of H2O2. The difference in lag-phase (relative to the WT) was determined by measuring the time to grow to an optical density (OD) of 0.2 (A600) for each strain and subtracting these values from the time taken for WT to grow to the same OD. Panel C; The WT (JMB1100) with pCM28 (empty vector; pEV) and the ΔsrrAB strain (JMB1467) with pEV or pCM28_srrAB (srrAB psrrAB) were cultured in TSB, standardized and challenged with H2O2 for 2 hours. The H2O2 stress was terminated with catalase addition and the surviving colony-forming units (CFU) were determined. Representative data are shown in Panels A and B and experiments were repeated on three independent occasions. Data in Panel C represent average of biological triplicates with standard deviations presented for all data, but not visible in some cases. Note that the differences in H2O2 concentrations between data in Panel A and C is due to the fact that cells were adjusted to a higher optical density for the survival assay.
(TIF)
The accumulation of mRNA transcripts corresponding to H2O2 resistance genes is similar in the WT and ΔsrrAB strains upon hydrogen peroxide challenge. The WT (JMB1100) and ΔsrrAB (JMB1467) strains were cultured to an optical density (OD) of 6.5 (A600) at a HVR of 6 and challenged with 10 mM H2O2 or vehicle control. The mRNA transcript abundances corresponding to kat and ahpC were assessed post H2O2 treatment. The data were normalized to 16s rRNA transcript and are presented as a ratio of the transcript abundance upon challenge with ROS to the transcript abundance of the non-treated control for each strain. Data represent the average of biological triplicates with standard deviations shown. Two-tail student t-tests were performed on all samples P> 0.05 and is denoted as non-significant (NS).
(TIF)
Kat activity was assessed in cell-free lysates from the WT (JMB1100), ΔsrrAB (JMB1467), ΔperR (JMB2151), and ΔperR ΔsrrAB (JMB2615) strains.
(TIF)
Depiction of the conserved inverted repeat sequence (bold and underlined) found in promoter regions bound by SrrA. The inverted repeat sequence was found to be separated by a variable spacer region of between 3–6 base-pairs. The sequence within the proposed dps promoter region had one mismatch (blue font).
(TIF)
The WT (JMB1100), ΔsrrAB (JMB1467), ΔsrrAB cidB::Tn (JMB6024), ΔsrrAB cidA::Tn (JMB6070), ΔperR (JMB2151), and Δkat (JMB2078) strains were cultured aerobically, diluted into fresh TSB medium, and challenged with H2O2 at the point of inoculation. Representative growth profiles are presented and experiments were performed on least three independent occasions.
(TIF)
The transcriptional activity of the sufC gene was assessed in the WT (JMB1100) and ΔsrrAB (JMB1467) strains containing gfp under the transcriptional control of the sufC promoter (pCM11_sufC). Data represent the average of biological triplicates with standard deviations shown.
(TIF)
(DOCX)
Acknowledgments
The Boyd lab is supported by Rutgers University, the Charles and Johanna Busch foundation and USDA MRF project NE−1028. A.A.M. is supported by the Douglas Eveleigh fellowship from the Microbial Biology Graduate Program and an Excellence Fellowship from Rutgers University. The authors would like to thank Dr. Ann M. Stock for helpful discussions and Christina Roberts for help preparing the manuscript. We also thank Dr. William Belden for use of his real-time thermocycler.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Support was provided by: Rutgers University School of Environmental and Biological Sciences; Busch Biomedical Grant from Rutgers University; United States Department of Agriculture grant NE-1028. The Boyd lab is supported by Rutgers University, the Charles and Johanna Busch foundation and USDA MRF project NE−1028. AAM is supported by the Douglas Eveleigh fellowship from the Microbial Biology Graduate Program and an Excellence Fellowship from Rutgers University.
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520–32. Epub 1998/08/26. 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- 2.Imlay JA, Fridovich I. Superoxide production by respiring membranes of Escherichia coli. Free Radic Res Commun. 1991;12–13 Pt 1:59–66. Epub 1991/01/01. [DOI] [PubMed] [Google Scholar]
- 3.Boveris A, Chance B. The mitochondrial generation of hydrogen peroxide. General properties and effect of hyperbaric oxygen. Biochem J. 1973;134(3):707–16. Epub 1973/07/01. PubMed Central PMCID: PMC1177867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Messner KR, Imlay JA. Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. J Biol Chem. 2002;277(45):42563–71. Epub 2002/08/30. 10.1074/jbc.M204958200 [DOI] [PubMed] [Google Scholar]
- 5.Messner KR, Imlay JA. The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J Biol Chem. 1999;274(15):10119–28. Epub 1999/04/03. [DOI] [PubMed] [Google Scholar]
- 6.Imlay JA. How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Advances in Microbial Physiology. 2002;46:111–53. [DOI] [PubMed] [Google Scholar]
- 7.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nature Reviews Microbiology. 2013;11(7):443–54. Epub 2013/05/29. PubMed Central PMCID: PMC4018742. 10.1038/nrmicro3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Djaman O, Outten FW, Imlay JA. Repair of oxidized iron-sulfur clusters in Escherichia coli. J Biol Chem. 2004;279(43):44590–9. Epub 2004/08/17. 10.1074/jbc.M406487200 [DOI] [PubMed] [Google Scholar]
- 9.Kennedy MC, Kent TA, Emptage M, Merkle H, Beinert H, Munck E. Evidence for the formation of a linear [3Fe-4S] cluster in partially unfolded aconitase. J Biol Chem. 1984;259(23):14463–71. Epub 1984/12/10. [PubMed] [Google Scholar]
- 10.Imlay JA. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol. 2006;59(4):1073–82. Epub 2006/01/25. MMI5028 [pii]. 10.1111/j.1365-2958.2006.05028.x [DOI] [PubMed] [Google Scholar]
- 11.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. Epub 2003/10/07. 10.1146/annurev.micro.57.030502.090938 [DOI] [PubMed] [Google Scholar]
- 12.Song E, Jaishankar GB, Saleh H, Jithpratuck W, Sahni R, Krishnaswamy G. Chronic granulomatous disease: a review of the infectious and inflammatory complications. Clin Mol Allergy. 2011;9(1):10 Epub 2011/06/01. 1476-7961-9-10 [pii].PubMed Central PMCID: PMC3128843. 10.1186/1476-7961-9-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clements MO, Watson SP, Foster SJ. Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J Bacteriol. 1999;181(13):3898–903. Epub 1999/06/29. PubMed Central PMCID: PMC93877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armstrong-Buisseret L, Cole MB, Stewart GS. A homologue to the Escherichia coli alkyl hydroperoxide reductase AhpC is induced by osmotic upshock in Staphylococcus aureus. Microbiology. 1995;141 (Pt 7):1655–61. Epub 1995/07/01. [DOI] [PubMed] [Google Scholar]
- 15.Clements MO, Foster SJ. Stress resistance in Staphylococcus aureus. Trends Microbiol. 1999;7(11):458–62. Epub 1999/11/05. S0966-842X(99)01607-8 [pii]. [DOI] [PubMed] [Google Scholar]
- 16.Cosgrove K, Coutts G, Jonsson IM, Tarkowski A, Kokai-Kun JF, Mond JJ, et al. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J Bacteriol. 2007;189(3):1025–35. Epub 2006/11/23. JB.01524-06 [pii] PubMed Central PMCID: PMC1797328. 10.1128/JB.01524-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annual Review of Biochemistry. 2008;77:755–76. Epub 2008/01/05. PubMed Central PMCID: PMC3057177. 10.1146/annurev.biochem.77.061606.161055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaupp R, Ledala N, Somerville GA. Staphylococcal response to oxidative stress. Front Cell Infect Microbiol. 2012;2:33 Epub 2012/08/25. PubMed Central PMCID: PMC3417528. 10.3389/fcimb.2012.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu GY, Essex A, Buchanan JT, Datta V, Hoffman HM, Bastian JF, et al. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202(2):209–15. Epub 2005/07/13. jem.20050846 [pii].PubMed Central PMCID: PMC2213009. 10.1084/jem.20050846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240(4852):640–2. Epub 1988/04/29. [DOI] [PubMed] [Google Scholar]
- 21.Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5(4):294–303. Epub 1998/04/18. [DOI] [PubMed] [Google Scholar]
- 22.Park S, You X, Imlay JA. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx- mutants of Escherichia coli. Proc Natl Acad Sci U S A. 2005;102(26):9317–22. Epub 2005/06/22. 0502051102 [pii]. PubMed Central PMCID: PMC1166606. 10.1073/pnas.0502051102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ilari A, Ceci P, Ferrari D, Rossi GL, Chiancone E. Iron incorporation into Escherichia coli Dps gives rise to a ferritin-like microcrystalline core. Journal of Biological Chemistry. 2002;277(40):37619–23. 10.1074/jbc.M206186200 [DOI] [PubMed] [Google Scholar]
- 24.Justino MC, Almeida CC, Teixeira M, Saraiva LM. Escherichia coli di-iron YtfE protein is necessary for the repair of stress-damaged iron-sulfur clusters. J Biol Chem. 2007;282(14):10352–9. Epub 2007/02/10. M610656200 [pii]. 10.1074/jbc.M610656200 [DOI] [PubMed] [Google Scholar]
- 25.Overton TW, Justino MC, Li Y, Baptista JM, Melo AM, Cole JA, et al. Widespread distribution in pathogenic bacteria of di-iron proteins that repair oxidative and nitrosative damage to iron-sulfur centers. J Bacteriol. 2008;190(6):2004–13. Epub 2008/01/22. JB.01733-07 [pii]. PubMed Central PMCID: PMC2258886. 10.1128/JB.01733-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annual Review of Biochemistry. 2000;69:183–215. 10.1146/annurev.biochem.69.1.183 [DOI] [PubMed] [Google Scholar]
- 27.Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–54. Epub 2009/07/07. PubMed Central PMCID: PMC3645274. 10.1146/annurev.micro.091208.073214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng DM, Liu MJ, ten Cate JM, Crielaard W. The VicRK system of Streptococcus mutans responds to oxidative stress. J Dent Res. 2007;86(7):606–10. Epub 2007/06/26. 86/7/606 [pii]. [DOI] [PubMed] [Google Scholar]
- 29.Hwang S, Kim M, Ryu S, Jeon B. Regulation of oxidative stress response by CosR, an essential response regulator in Campylobacter jejuni. PLoS One. 2011;6(7):e22300 Epub 2011/08/04. PubMed Central PMCID: PMC3139631. 10.1371/journal.pone.0022300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCluskey J, Hinds J, Husain S, Witney A, Mitchell TJ. A two-component system that controls the expression of pneumococcal surface antigen A (PsaA) and regulates virulence and resistance to oxidative stress in Streptococcus pneumoniae. Mol Microbiol. 2004;51(6):1661–75. Epub 2004/03/11. 3917 [pii]. [DOI] [PubMed] [Google Scholar]
- 31.Pragman AA, Yarwood JM, Tripp TJ, Schlievert PM. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. Journal of bacteriology. 2004;186(8):2430–8. Epub 2004/04/03. PubMed Central PMCID: PMC412142. 10.1128/JB.186.8.2430-2438.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pragman AA, Ji Y, Schlievert PM. Repression of Staphylococcus aureus SrrAB using inducible antisense srrA alters growth and virulence factor transcript levels. Biochemistry. 2007;46(1):314–21. Epub 2007/01/03. 10.1021/bi0603266 [DOI] [PubMed] [Google Scholar]
- 33.Yarwood JM, McCormick JK, Schlievert PM. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J Bacteriol. 2001;183(4):1113–23. Epub 2001/02/07. PubMed Central PMCID: PMC94983. 10.1128/JB.183.4.1113-1123.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilde AD, Snyder DJ, Putnam NE, Valentino MD, Hammer ND, Lonergan ZR, et al. Bacterial Hypoxic Responses Revealed as Critical Determinants of the Host-Pathogen Outcome by TnSeq Analysis of Staphylococcus aureus Invasive Infection. PLoS Pathog. 2015;11(12):e1005341 Epub 2015/12/20. PubMed Central PMCID: PMC4684308. 10.1371/journal.ppat.1005341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ulrich M, Bastian M, Cramton SE, Ziegler K, Pragman AA, Bragonzi A, et al. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol Microbiol. 2007;65(5):1276–87. Epub 2007/08/19. 10.1111/j.1365-2958.2007.05863.x [DOI] [PubMed] [Google Scholar]
- 36.Kinkel TL, Roux CM, Dunman PM, Fang FC. The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. MBio. 2013;4(6):e00696–13. Epub 2013/11/14. mBio.00696-13 [pii]. PubMed Central PMCID: PMC3892780. 10.1128/mBio.00696-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Throup JP, Zappacosta F, Lunsford RD, Annan RS, Carr SA, Lonsdale JT, et al. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry. 2001;40(34):10392–401. Epub 2001/08/22. [DOI] [PubMed] [Google Scholar]
- 38.Richardson AR, Dunman PM, Fang FC. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol Microbiol. 2006;61(4):927–39. Epub 2006/07/25. MMI5290 [pii]. 10.1111/j.1365-2958.2006.05290.x [DOI] [PubMed] [Google Scholar]
- 39.Richardson AR, Libby SJ, Fang FC. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science. 2008;319(5870):1672–6. Epub 2008/03/22. 319/5870/1672 [pii]. 10.1126/science.1155207 [DOI] [PubMed] [Google Scholar]
- 40.Boles BR, Thoendel M, Roth AJ, Horswill AR. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One. 2010;5(4):e10146 Epub 2010/04/27. PubMed Central PMCID: PMC2854687. 10.1371/journal.pone.0010146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choby JE, Mike LA, Mashruwala AA, Dutter BF, Dunman PM, Sulikowski GA, et al. A Small-Molecule Inhibitor of Iron-Sulfur Cluster Assembly Uncovers a Link between Virulence Regulation and Metabolism in Staphylococcus aureus. Cell Chem Biol. 23(11):1351–61. Epub 2016/10/25. S2451-9456(16)30347-6 [pii]. 10.1016/j.chembiol.2016.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mashruwala AA, Bhatt S, Poudel S, Boyd ES, Boyd JM. The DUF59 Containing Protein SufT Is Involved in the Maturation of Iron-Sulfur (FeS) Proteins during Conditions of High FeS Cofactor Demand in Staphylococcus aureus. PLoS genetics. 12(8):e1006233 Epub 2016/08/16. PubMed Central PMCID: PMC4982691. 10.1371/journal.pgen.1006233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuchs S, Pane-Farre J, Kohler C, Hecker M, Engelmann S. Anaerobic gene expression in Staphylococcus aureus. J Bacteriol. 2007;189(11):4275–89. 10.1128/JB.00081-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novick RP. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. Epub 1991/01/01. 0076-6879(91)04029-N [pii]. [DOI] [PubMed] [Google Scholar]
- 45.Malone CL, Boles BR, Lauderdale KJ, Thoendel M, Kavanaugh JS, Horswill AR. Fluorescent reporters for Staphylococcus aureus. J Microbiol Methods. 2009;77(3):251–60. Epub 2009/03/07. S0167-7012(09)00066-9 [pii]. PubMed Central PMCID: PMC2693297. 10.1016/j.mimet.2009.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mashruwala AA, Roberts CA, Bhatt S, May KL, Carroll RK, Shaw LN, et al. Staphylococcus aureus SufT: an essential iron-sulphur cluster assembly factor in cells experiencing a high-demand for lipoic acid. Molecular microbiology. Epub 2016/10/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mashruwala AA, Pang YY, Rosario-Cruz Z, Chahal HK, Benson MA, Mike LA, et al. Nfu facilitates the maturation of iron-sulfur proteins and participates in virulence in Staphylococcus aureus. Mol Microbiol. 2015;95(3):383–409. Epub 2014/11/13. 10.1111/mmi.12860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kennedy MC, Emptage MH, Dreyer JL, Beinert H. The role of iron in the activation-inactivation of aconitase. Journal of Biological Chemistry. 1983;258(18):11098–105. [PubMed] [Google Scholar]
- 49.Beers RF Jr., Sizer IW. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952;195(1):133–40. Epub 1952/03/01. [PubMed] [Google Scholar]
- 50.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J Biol Chem. 1969;244(22):6049–55. Epub 1969/11/25. [PubMed] [Google Scholar]
- 51.Joska TM, Mashruwala A, Boyd JM, Belden WJ. A universal cloning method based on yeast homologous recombination that is simple, efficient, and versatile. J Microbiol Methods. 2014;100:46–51. Epub 2014/01/15. S0167-7012(13)00364-3 [pii]. 10.1016/j.mimet.2013.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olson BJ, Markwell J. Assays for determination of protein concentration. Curr Protoc Protein Sci. 2007;Chapter 3:Unit 3 4. Epub 2008/04/23. [DOI] [PubMed] [Google Scholar]
- 53.Dukan S, Touati D. Hypochlorous acid stress in Escherichia coli: resistance, DNA damage, and comparison with hydrogen peroxide stress. J Bacteriol. 1996;178(21):6145–50. Epub 1996/11/01. PubMed Central PMCID: PMC178483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maalej S, Dammak I, Dukan S. The impairment of superoxide dismutase coordinates the derepression of the PerR regulon in the response of Staphylococcus aureus to HOCl stress. Microbiology-Sgm. 2006;152:855–61. [DOI] [PubMed] [Google Scholar]
- 55.Mahawar M, Tran V, Sharp JS, Maier RJ. Synergistic roles of Helicobacter pylori methionine sulfoxide reductase and GroEL in repairing oxidant-damaged catalase. J Biol Chem. 286(21):19159–69. Epub 2011/04/05. M111.223677 [pii]. PubMed Central PMCID: PMC3099729. 10.1074/jbc.M111.223677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ballal A, Manna AC. Regulation of superoxide dismutase (sod) genes by SarA in Staphylococcus aureus. J Bacteriol. 2009;191(10):3301–10. Epub 2009/03/17. JB.01496-08 [pii]. PubMed Central PMCID: PMC2687179. 10.1128/JB.01496-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect Immun. 2001;69(6):3744–54. Epub 2001/05/12. PubMed Central PMCID: PMC98383. 10.1128/IAI.69.6.3744-3754.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhattacharya M, Das AK. Inverted repeats in the promoter as an autoregulatory sequence for TcrX in Mycobacterium tuberculosis. Biochemical and Biophysical Research Communications. 2011;415(1):17–23. 10.1016/j.bbrc.2011.09.143 [DOI] [PubMed] [Google Scholar]
- 59.Wosten MMSM Groisman EA. Molecular characterization of the PmrA regulon. Journal of Biological Chemistry. 1999;274(38):27185–90. [DOI] [PubMed] [Google Scholar]
- 60.Glover RT, Kriakov J, Garforth SJ, Baughn AD, Jacobs WR. The two-component regulatory system senX3-regX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. Journal of bacteriology. 2007;189(15):5495–503. 10.1128/JB.00190-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eraso JM, Kaplan S. Half-Site DNA Sequence and Spacing Length Contributions to PrrA Binding to PrrA Site 2 of RSP3361 in Rhodobacter sphaeroides 2.4.1. Journal of bacteriology. 2009;191(13):4353–64. 10.1128/JB.00244-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Been M, Bart MJ, Abee T, Siezen RJ, Francke C. The identification of response regulator-specific binding sites reveals new roles of two-component systems in Bacillus cereus and closely related low-GC Gram-positives. Environmental Microbiology. 2008;10(10):2796–809. 10.1111/j.1462-2920.2008.01700.x [DOI] [PubMed] [Google Scholar]
- 63.Windham IH, Chaudhari SS, Bose JL, Thomas VC, Bayles KW. SrrAB modulates Staphylococcus aureus cell death through regulation of cidABC transcription. J Bacteriol. Epub 2016/01/27. JB.00954-15 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oogai Y, Kawada-Matsuo M, Komatsuzawa H. Staphylococcus aureus SrrAB Affects Susceptibility to Hydrogen Peroxide and Co-Existence with Streptococcus sanguinis. PloS one. 11(7):e0159768 Epub 2016/07/22. PubMed Central PMCID: PMC4956065. 10.1371/journal.pone.0159768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Somerville GA, Said-Salim B, Wickman JM, Raffel SJ, Kreiswirth BN, Musser JM. Correlation of acetate catabolism and growth yield in Staphylococcus aureus: implications for host-pathogen interactions. Infect Immun. 2003;71(8):4724–32. Epub 2003/07/23. PubMed Central PMCID: PMC166023. 10.1128/IAI.71.8.4724-4732.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ledala N, Zhang B, Seravalli J, Powers R, Somerville GA. Influence of Iron and Aeration on Staphylococcus aureus Growth, Metabolism, and Transcription. J Bacteriol. 2014;196(12):2178–89. Epub 2014/04/08. JB.01475-14 [pii] 10.1128/JB.01475-14. PubMed Central PMCID: PMC4054190. 10.1128/JB.01475-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Somerville GA, Proctor RA. Cultivation conditions and the diffusion of oxygen into culture media: the rationale for the flask-to-medium ratio in microbiology. BMC Microbiol. 13:9 Epub 2013/01/18. 1471-2180-13-9 [pii]. PubMed Central PMCID: PMC3551634. 10.1186/1471-2180-13-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McDaniel LE, Bailey EG, Zimmerli A. Effect of Oxygen-Supply Rates on Growth of Escherichia Coli. Ii. Comparison of Results in Shake Flasks and 50-Liter Fermentor. Appl Microbiol. 1965;13:115–9. Epub 1965/01/01. PubMed Central PMCID: PMC1058203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McDaniel LE, Bailey EG, Zimmerli A. Effect of Oxygen Supply Rates on Growth of Escherichia Coli. Appl Microbiol. 1965;13:109–14. Epub 1965/01/01. PubMed Central PMCID: PMC1058202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benov L, Fridovich I. Growth in iron-enriched medium partially compensates Escherichia coli for the lack of manganese and iron superoxide dismutase. J Biol Chem. 1998;273(17):10313–6. Epub 1998/05/30. [DOI] [PubMed] [Google Scholar]
- 71.Benov L, Fridovich I. Why superoxide imposes an aromatic amino acid auxotrophy on Escherichia coli. The transketolase connection. J Biol Chem. 1999;274(7):4202–6. Epub 1999/02/06. [DOI] [PubMed] [Google Scholar]
- 72.Jang S, Imlay JA. Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulphur assembly system, and OxyR induces the Suf system to compensate. Mol Microbiol. 2010;78(6):1448–67. Epub 2010/12/15. PubMed Central PMCID: PMC3051806. 10.1111/j.1365-2958.2010.07418.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, et al. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299(5607):700–4. 10.1126/science.1079605 [DOI] [PubMed] [Google Scholar]
- 74.Sun F, Liang H, Kong X, Xie S, Cho H, Deng X, et al. Quorum-sensing agr mediates bacterial oxidation response via an intramolecular disulfide redox switch in the response regulator AgrA. Proc Natl Acad Sci U S A. 2012;109(23):9095–100. Epub 2012/05/16. 1200603109 [pii]. PubMed Central PMCID: PMC3384213. 10.1073/pnas.1200603109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nair D, Memmi G, Hernandez D, Bard J, Beaume M, Gill S, et al. Whole-genome sequencing of Staphylococcus aureus strain RN4220, a key laboratory strain used in virulence research, identifies mutations that affect not only virulence factors but also the fitness of the strain. J Bacteriol. 193(9):2332–5. Epub 2011/03/08. JB.00027-11 [pii]. PubMed Central PMCID: PMC3133102. 10.1128/JB.00027-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Memmi G, Nair DR, Cheung A. Role of ArlRS in autolysis in methicillin-sensitive and methicillin-resistant Staphylococcus aureus strains. J Bacteriol. 2012;194(4):759–67. Epub 2011/12/06. JB.06261-11 [pii]. PubMed Central PMCID: PMC3272946. 10.1128/JB.06261-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Herbert S, Ziebandt AK, Ohlsen K, Schafer T, Hecker M, Albrecht D, et al. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect Immun. 2010;78(6):2877–89. Epub 2010/03/10. IAI.00088-10 [pii]. PubMed Central PMCID: PMC2876537. 10.1128/IAI.00088-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Durand S, Braun F, Lioliou E, Romilly C, Helfer AC, Kuhn L, et al. A nitric oxide regulated small RNA controls expression of genes involved in redox homeostasis in Bacillus subtilis. PLoS Genet. 11(2):e1004957 Epub 2015/02/03. 10.1371/journal.pgen.1004957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seaver LC, Imlay JA. Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J Bacteriol. 2001;183(24):7182–9. Epub 2001/11/22. PubMed Central PMCID: PMC95567. 10.1128/JB.183.24.7182-7189.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem. 2007;282(2):929–37. Epub 2006/11/15. M607646200 [pii]. 10.1074/jbc.M607646200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Flint DH, Tuminello JF, Emptage MH. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem. 1993;268(30):22369–76. Epub 1993/10/25. [PubMed] [Google Scholar]
- 82.Georgellis D, Kwon O, Lin ECC. Quinones as the redox signal for the Arc two-component system of bacteria. Science. 2001;292(5525):2314–6. 10.1126/science.1059361 [DOI] [PubMed] [Google Scholar]
- 83.Wong SM, Alugupalli KR, Ram S, Akerley BJ. The ArcA regulon and oxidative stress resistance in Haemophilus influenzae. Mol Microbiol. 2007;64(5):1375–90. Epub 2007/06/05. MMI5747 [pii]. PubMed Central PMCID: PMC1974803. 10.1111/j.1365-2958.2007.05747.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Loui C, Chang AC, Lu S. Role of the ArcAB two-component system in the resistance of Escherichia coli to reactive oxygen stress. BMC Microbiol. 2009;9:183 Epub 2009/09/01. 1471-2180-9-183 [pii]. PubMed Central PMCID: PMC2748088. 10.1186/1471-2180-9-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu S, Killoran PB, Fang FC, Riley LW. The global regulator ArcA controls resistance to reactive nitrogen and oxygen intermediates in Salmonella enterica serovar Enteritidis. Infect Immun. 2002;70(2):451–61. Epub 2002/01/18. PubMed Central PMCID: PMC127680. 10.1128/IAI.70.2.451-461.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, et al. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305(5936):709–12. Epub 1983/10/20. [DOI] [PubMed] [Google Scholar]
- 87.Pang YY, Schwartz J, Bloomberg S, Boyd JM, Horswill AR, Nauseef WM. Methionine sulfoxide reductases protect against oxidative stress in Staphylococcus aureus encountering exogenous oxidants and human neutrophils. J Innate Immun. 2014;6(3):353–64. Epub 2013/11/20. 000355915 [pii]. PubMed Central PMCID: PMC3972283. 10.1159/000355915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio. 2013;4(1):e00537–12. Epub 2013/02/14. mBio.00537-12 [pii]. PubMed Central PMCID: PMC3573662. 10.1128/mBio.00537-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Duthie ES, Lorenz LL. Staphylococcal coagulase; mode of action and antigenicity. J Gen Microbiol. 1952;6(1–2):95–107. Epub 1952/02/01. 10.1099/00221287-6-1-2-95 [DOI] [PubMed] [Google Scholar]
- 90.Horsburgh MJ, Ingham E, Foster SJ. In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and Is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J Bacteriol. 2001;183(2):468–75. Epub 2001/01/03. PubMed Central PMCID: PMC94901. 10.1128/JB.183.2.468-475.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sadykov MR, Mattes TA, Luong TT, Zhu Y, Day SR, Sifri CD, et al. Tricarboxylic acid cycle-dependent synthesis of Staphylococcus aureus Type 5 and 8 capsular polysaccharides. J Bacteriol. 2010;192(5):1459–62. Epub 2010/01/12. PubMed Central PMCID: PMC2820852. 10.1128/JB.01377-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bose JL, Fey PD, Bayles KW. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol. 2013;79(7):2218–24. Epub 2013/01/29. AEM.00136-13 [pii]. PubMed Central PMCID: PMC3623228. 10.1128/AEM.00136-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, et al. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol. 2002;43(6):1387–400. Epub 2002/04/16. 2832 [pii]. [DOI] [PubMed] [Google Scholar]
- 94.Mashruwala AA, Boyd JM. De Novo Assembly of Plasmids Using Yeast Recombinational Cloning. Methods Mol Biol. 1373:33–41. Epub 2015/07/22. 10.1007/7651_2015_275 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Panel A; The lag-time necessary for S. aureus to initiate outgrowth is increased as a function of H2O2 concentration. The WT (JMB1100) strain was cultured aerobically and then diluted into fresh defined medium and challenged with varying concentrations of H2O2. Panel B; The lag-times necessary for the ΔsrrAB strain to initiate outgrowth post H2O2 challenge is greater than the lag-times required for the WT strain. The WT (JMB1100) and ΔsrrAB (JMB1467) strains were diluted into fresh defined medium and challenged with various concentrations of H2O2. The difference in lag-phase (relative to the WT) was determined by measuring the time to grow to an optical density (OD) of 0.2 (A600) for each strain and subtracting these values from the time taken for WT to grow to the same OD. Panel C; The WT (JMB1100) with pCM28 (empty vector; pEV) and the ΔsrrAB strain (JMB1467) with pEV or pCM28_srrAB (srrAB psrrAB) were cultured in TSB, standardized and challenged with H2O2 for 2 hours. The H2O2 stress was terminated with catalase addition and the surviving colony-forming units (CFU) were determined. Representative data are shown in Panels A and B and experiments were repeated on three independent occasions. Data in Panel C represent average of biological triplicates with standard deviations presented for all data, but not visible in some cases. Note that the differences in H2O2 concentrations between data in Panel A and C is due to the fact that cells were adjusted to a higher optical density for the survival assay.
(TIF)
The accumulation of mRNA transcripts corresponding to H2O2 resistance genes is similar in the WT and ΔsrrAB strains upon hydrogen peroxide challenge. The WT (JMB1100) and ΔsrrAB (JMB1467) strains were cultured to an optical density (OD) of 6.5 (A600) at a HVR of 6 and challenged with 10 mM H2O2 or vehicle control. The mRNA transcript abundances corresponding to kat and ahpC were assessed post H2O2 treatment. The data were normalized to 16s rRNA transcript and are presented as a ratio of the transcript abundance upon challenge with ROS to the transcript abundance of the non-treated control for each strain. Data represent the average of biological triplicates with standard deviations shown. Two-tail student t-tests were performed on all samples P> 0.05 and is denoted as non-significant (NS).
(TIF)
Kat activity was assessed in cell-free lysates from the WT (JMB1100), ΔsrrAB (JMB1467), ΔperR (JMB2151), and ΔperR ΔsrrAB (JMB2615) strains.
(TIF)
Depiction of the conserved inverted repeat sequence (bold and underlined) found in promoter regions bound by SrrA. The inverted repeat sequence was found to be separated by a variable spacer region of between 3–6 base-pairs. The sequence within the proposed dps promoter region had one mismatch (blue font).
(TIF)
The WT (JMB1100), ΔsrrAB (JMB1467), ΔsrrAB cidB::Tn (JMB6024), ΔsrrAB cidA::Tn (JMB6070), ΔperR (JMB2151), and Δkat (JMB2078) strains were cultured aerobically, diluted into fresh TSB medium, and challenged with H2O2 at the point of inoculation. Representative growth profiles are presented and experiments were performed on least three independent occasions.
(TIF)
The transcriptional activity of the sufC gene was assessed in the WT (JMB1100) and ΔsrrAB (JMB1467) strains containing gfp under the transcriptional control of the sufC promoter (pCM11_sufC). Data represent the average of biological triplicates with standard deviations shown.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.